Abstract
We generated a large number of mutations in the hepatitis B virus (HBV) core gene inserted into a bacterial expression vector. The new mutagenesis procedure generated deletions and insertions (as sequence repeats) of various lengths at random positions between M1 and E145 but not substitutions. The R-rich 30-amino-acid C-terminal domain was not analyzed. A total of 50,000 colonies were tested with a polyclonal human serum for the expression of hepatitis B core or e antigen. A total of 110 mutants randomly chosen from 1,500 positive colonies were genotyped. Deletions and insertions were clustered in four regions: D2 to E14, corresponding to the N-terminal loop in a model for the core protein fold (B. Bottcher, S. A. Wynne, and R. A. Crowther, Nature 386:88–91, 1997); V27 to P50 (second loop); L60 to V86 (upper half of the alpha helix forming the N-terminal part of the spike and the tip of the spike); and V124 to L140 (C-terminal part of the C-terminal helix and downstream loop). Deletions or insertions in the remaining parts of the molecule forming the compact center of the fold seemed to destabilize the protein. Of the 110 mutations, 38 allowed capsid formation in Escherichia coli. They mapped exclusively to nonhelical regions of the proposed fold. The mutations form a basis for subsequent analysis of further functions of the HBV core protein in the viral life cycle.
Hepatitis B virus (HBV) is a human blood-borne pathogen causing acute and chronic liver inflammation, which is associated with the development of hepatocellular carcinoma (for a review, see reference 4). This DNA virus is able to persistently replicate in hepatocytes through an RNA intermediate by reverse transcription (for a review, see reference 18). The spherical virion has a diameter of 42 nm and consists of an outer envelope and an internal nucleocapsid, which is composed of a protein shell surrounding a circular, partially double-stranded DNA genome of 3.2 kb and a viral reverse transcriptase (for a review, see reference 17).
The shell of the capsid is formed by multiple copies of a single core protein of 185 amino acids (aa) for genotype A (19), used in this work (27). The core protein forms homodimers (31) which self-assemble at micromolar concentrations (24) into icosahedral capsids (7) in heterologous expression systems in the absence of other viral proteins. These recombinant particles are morphologically and immunologically indistinguishable from natural capsids. Two kinds of particles are formed; one type, composed of 90 dimers, has a T=3 symmetry and a diameter of 32 nm, and the other type, formed by 120 dimers, has a T=4 symmetry and a diameter of 36 nm (10, 32). During the assembly process in heterologous systems, nonspecific host RNA is packaged into the particles by interaction with the protamine-like R-rich C-terminal domain of 30 aa (3). Deletion of this domain still allows capsid formation but prevents RNA packaging.
Recently, models for the fold of C-terminally truncated core proteins assembled in bacteria to capsids were proposed (5, 8) (see Fig. 7). These models are based on computer-aided processing of cryoelectron microscopic pictures of particles with T=4 symmetry. A prominent part of the structure is a spike protruding from the surface of the capsids; the spike is formed by two long antiparallel alpha-helical regions connected by a short loop around A80 at the tip of the spike. The base of the spike is girdled by the loop-helix-loop structure of the N-terminal 50 aa of the protein. At the C terminus of the spike, the peptide chain sharply bends and forms another alpha helix almost perpendicular to the spike, followed by another nonhelical region. In the dimer, the two core protein subunits interact with each other mainly in the region of the spike-forming helices. The four helices are combined into a compact bundle. The two C-terminal helices extrude from the structure on opposite sites. They establish the interdimer contacts at the fivefold and local sixfold symmetries of the capsids (13).
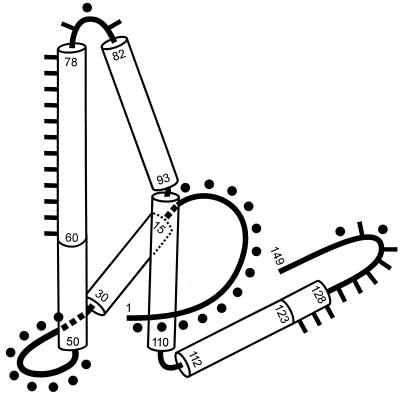
FIG. 7.
Comparison of the core protein domains defined in this work (Table 2) with the proposed fold of the C-terminally truncated protein (5), using the handedness determined in reference 7a. Alpha-helical regions are drawn as cylinders. Numbers indicate amino acid residues. Regions where insertions or deletions were compatible with capsid formation are marked by dots (compare with Fig. 4 and Table 2). They correspond to the nonhelical domains. Areas where insertions or deletions allowed stable HBcAg or HBeAg expression but no capsid formation are indicated by bars. No insertions or deletions could be identified in the center of the structure formed by the first helix (L15-L30), the N-terminal third of the second helix (P50-L60), the fourth helix (G94-F110), and the N-terminal part of the fifth helix (R112-G123). Adapted with permission of Macmillan Magazines Ltd. (reference 5, copyright 1997).
The capsids react as the highly immunogenic T-cell-independent as well as the T-cell-dependent hepatitis B core antigen (HBcAg) (16). The HBcAg determinant is conformational and is formed by amino acid residues around A80 (21). Denaturation of capsids destroys HBcAg reactivity and generates a distinct antigen specificity (hepatitis B e antigen [HBeAg]). Two linear HBeAg determinants (HBe1 and HBe2) have been defined around A80 and P138, respectively (21). Because of their high immunogenicity, HBV capsids are used as carriers for foreign epitopes in experimental recombinant vaccines (e.g., 23). Fusions to the N terminus of the core protein, internal insertions around A80, and fusions to the C terminus are compatible with particle formation and result in external exposure of the foreign domains.
The two forms of capsids having different diameters can also be found in infected liver (12). In this situation, however, a complex of the RNA pregenome and reverse transcriptase forms a nucleus for efficient capsid assembly (1) at submicromolar concentrations of core protein dimers, ensuring predominantly packaging of the pregenome. A complex of the core protein and a chaperonin has been identified as an intermediate in capsid assembly in a eukaryotic cell-free expression system (15). Other steps in the HBV life cycle besides capsid formation also depend on the core protein. For example, the transport of the nucleocapsid to the nucleus (11) and disassembly are important for establishing infection and for intracellular amplification of the viral genome during infection (25). Reverse transcription and second-strand DNA synthesis of the viral genome in the lumen of the cytoplasmic capsid are influenced by mutations in the core protein (2). During genomic DNA synthesis, a maturation signal which is necessary for envelopment is generated on the surface of the nucleocapsid (28). Finally, the mature nucleocapsid presumably interacts with internal membranes carrying viral envelope proteins, and its envelopment probably requires a direct interaction with envelope proteins (6).
We started to investigate the functions of the core protein in the HBV life cycle by a genetic approach. The aim of this work was to generate a panel of core gene mutations which would allow capsid formation and which could be used in subsequent studies to characterize later functions of the core protein, such as pregenome packaging, genome synthesis, intracellular trafficking, or nucleocapsid envelopment. To achieve this aim, a large number of quasi-random mutations were generated in the HBV core gene inserted in a bacterial expression vector. A new mutagenesis procedure which generated deletions or insertions (as sequence repeats) but no amino acid substitutions in the core gene was used. The mutants were first screened for the expression of HBcAg or HBeAg. Positive variants were subsequently tested for capsid formation. By this approach, 38 core gene mutations which were compatible with capsid formation were identified. These mutations were found exclusively in four regions of the primary sequence forming nonhelical areas, on the basis of a model for the core protein fold (5).
MATERIALS AND METHODS
Expression plasmid for bacterial core protein synthesis.
The hybrid bacterial tac promoter (9) was isolated as an 89-bp HindIII-BamHI fragment from plasmid pDR540 (PL Biochemicals, Freiburg, Germany) and inserted into the HindIII and BamHI sites of plasmid pBluescript KS(+) (Stratagene). The HBV core gene was PCR amplified from a plasmid containing a copy of a genotype A HBV genome (27) (GenBank-EMBL data bank accession no. X02763) with the oligonucleotides 5′GGTAGGATCCGGGATGGACATTG (BamHI site underlined; start codon of core gene boldfaced) and 5′GTGAGTGATTGG (nucleotide [nt] 336 to nt 325 of the HBV genome), cleaved with BamHI at the 5′ end in the primer region and at the 3′ end at nt 26 in the HBV sequence, and inserted into the BamHI site of the tac promoter plasmid. (Numbering of the plus strand of the HBV genome starts with the G of the single EcoRI site.) The XbaI site in the polylinker sequence of pBluescript KS(+) was destroyed by cleaving, filling in, and religation. The BamHI site 3′ of the core gene was destroyed by partial BamHI digestion, filling in, and religation. Six restriction enzyme recognition sites were then introduced stepwise by site-directed in vitro mutagenesis (14) without changing the amino acid sequence of the core protein (see Fig. 2), resulting in plasmid pMK8.
FIG. 2.
Single restriction enzyme cleavage sites introduced into the core gene without changing the coding. Six unique restriction enzyme cleavage sites were introduced by site-directed in vitro mutagenesis. The sites were used for the mutagenesis of different regions of the core gene (see Fig. 3) by the method shown in Fig. 1. Numbers indicate the positions of the peptides in the core protein (top line) and the positions of the WT nucleotide sequences in the HBV genome (middle line). The nucleotide sequences in the bottom line show the point mutations; the recognition sites for the enzymes are underlined.
Mutagenesis.
Five micrograms of plasmid pMK8 (3.46 pmol of DNA ends) linearized in the core gene at one of eight sites (see Fig. 2 and 3) was incubated with 0.1 U of exonuclease Bal 31 (New England Biolabs) in 0.1-ml total volume at 37°C. Thirty microliters was removed after 5, 10, and 15 min, and the reaction was immediately stopped by the addition of 170 μl of 15 mM EGTA, phenol-chloroform extraction, incubation at 65°C for 5 min, and another phenol-chloroform extraction. The DNA was recovered by ethanol precipitation. To check the extent of the exonuclease reaction, 1 μg of each sample was incubated with a restriction enzyme cleaving approximately 300 bp away from the site used for linearization and truncation, and the length distributions of the corresponding fragments were determined by electrophoresis through a 5% polyacrylamide gel. Usually, after 5 or 10 min of incubation, the number of base pairs removed from one DNA end peaked at approximately 50. Samples containing appropriate fragment lengths were combined, the single-stranded ends were filled in with Klenow DNA polymerase, and the DNA was cleaved with PflMI at nt 3207 in the HBV sequence (see site X in Fig. 1). Fragments from the upstream and downstream truncations (see Fig. 3 for fragment pairs used) were isolated, ligated, and used for the electroporation of Escherichia coli DH5α. To monitor the extent and distribution of the truncations produced by the exonuclease treatment, plasmids from 24 unselected colonies were prepared, and the ligation sites were determined by sequencing. The experiment was repeated with altered reaction conditions for the exonuclease treatment if the distribution was not satisfactory.
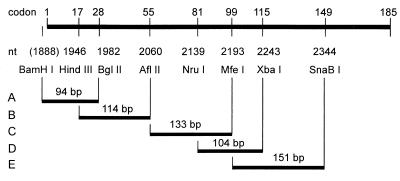
FIG. 3.
Restriction enzyme pairs used for mutagenesis. The upper bar represents the HBV core gene. Numbers above the bar indicate codons where the DNA is cleaved by the indicated restriction enzymes. Numbers below the bar indicate the first base pair of the corresponding recognition sites introduced in part by site-directed in vitro mutagenesis (Fig. 2). Restriction enzyme pairs used for mutagenesis (corresponding to sites 1 and 2 in Fig. 1) are connected by vertical and horizontal bars and define regions A to E where deletions and insertions were introduced.
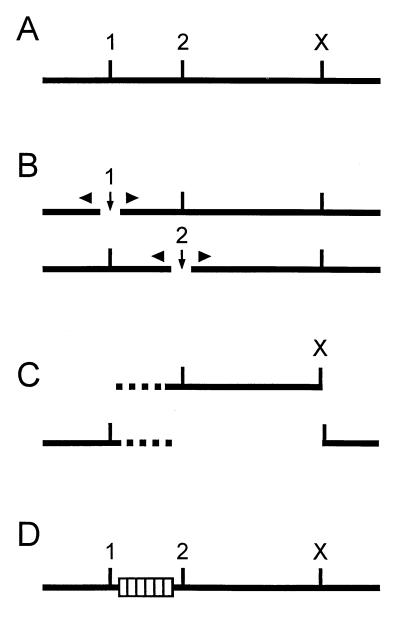
FIG. 1.
Mutagenesis procedure. (A) Mutations were introduced between two single restriction enzyme cleavage sites (sites 1 and 2). A third, remote single restriction enzyme cleavage site (X) was required. (B) The plasmid was separately linearized at sites 1 and 2 (vertical arrows), and variable numbers of base pairs were removed from the DNA ends by exonuclease treatment (triangles). (C) The DNA was cut with restriction enzyme X, and the fragments containing the regions of interest (broken lines) were isolated. (D) The fragments from both samples were recombined by sticky-end ligation at site X and blunt-end ligation randomly joining a 5′ segment of the region to a 3′ segment (hatched box). This procedure generated deletions and insertions (as sequence repeats) of variable lengths at random positions.
Antigen assay on filters.
Bacterial colonies were transferred from agar plates to nitrocellulose filter replicas (approximately 2,000 colonies per 12-cm-diameter filter) and incubated for 12 h on Luria broth plates supplemented with ampicillin and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were lysed in situ with lysozyme-chloroform vapor (22). After cells were washed in 10% (vol/vol) fetal calf serum–phosphate-buffered saline, HBcAg or HBeAg was detected on the filter with human serum F1451 (positive for antibody to HBcAg [anti-HBc] and for antibody to HBeAg [anti-HBe]; dilution, 1:1,000) and a peroxidase-labeled secondary antibody (DAKO Diagnostika, Hamburg, Germany). The colonies around a positive signal on the master plate were individually transferred to a nitrocellulose filter, and the antigen assay was repeated to finally identify the positive colony.
Detection of capsids by agarose gel electrophoresis.
Bacteria were grown in 1.5 ml of Terrific broth (22) supplemented with ampicillin and IPTG overnight and harvested by centrifugation. The weight of the cell pellet was determined, and the cells were frozen in liquid nitrogen and thawed at room temperature three times. The material was resuspended in 1 μl of 10 mM Tris-Cl (pH 8.0)–150 mM NaCl–1 mg of DNase I (Boehringer GmbH, Mannheim, Germany) per ml–10 mg of lysozyme (Sigma) per ml per 1 mg of cell pellet. After incubation at 37°C for 30 min, the sample was spun for 10 min at 4,600 × g, and the cleared lysate was recovered. RNase A (2 μl; 1 mg/ml; Boehringer) was added to 20 μl of the cleared lysate. After incubation for 15 min at room temperature, 4 μl of loading buffer (50% [vol/vol] glycerin–0.1% [wt/vol] bromphenol blue in 50 mM NaPO4 [pH 7.4]) was added, and the sample was loaded onto a 10-cm 0.8% (wt/vol) agarose gel. The gel buffer and electrophoresis buffer were 50 mM NaPO4 (pH 7.4). Electrophoresis was done at 80 V for 30 min. The gel was stained by soaking in ethidium bromide (2 μg/ml) for 30 min and destained by soaking in water for 10 min. The bacterial RNA in core particles was visualized by UV light, and a picture was taken.
Detection of capsids by sucrose gradient centrifugation.
The cleared lysate from core protein-expressing E. coli was prepared as described above. Lysate (0.5 ml) diluted 1:10 in phosphate-buffered saline was layered on top of a sucrose gradient (1.3 ml of 15% [wt/vol], 1.3 ml of 30%, 1.3 ml of 45%, and 0.5 ml of 60% sucrose in phosphate-buffered saline) in an SW60 rotor (Beckman) and spun for 2 h at 20°C and 38,000 rpm. The gradient was fractionated from the top (11 by 0.44 ml).
HBcAg was measured in the fractions by an enzyme-linked immunosorbent assay (ELISA). A microtiter plate was coated with a human anti-HBc- and anti-HBe-positive serum (F1451; 1:1,000 dilution). The samples were applied at a 1:50 dilution. The peroxidase-labeled secondary antibody was prepared from sheep serum containing a high anti-HBc titer (1:100,000) and a low anti-HBe titer (1:128). The sheep had been immunized with bacterially expressed HBcAg.
HBcAg in combination with HBeAg was detected by a dot blot. Two microliters of each sucrose gradient fraction was dotted onto a nitrocellulose filter and dried. Detection of the antigen was done as with lysed bacterial colonies on filters (see above).
RESULTS
Mutagenesis procedure.
We used the following generally applicable procedure to introduce deletions or insertions (as sequence repeats) of variable lengths at random positions between two restriction enzyme cleavage sites (Fig. 1A, sites 1 and 2) in a plasmid. The molecules were linearized separately at each site, and the DNA ends were truncated by exonuclease Bal 31 (Fig. 1B). The reaction conditions were chosen so that between a few and approximately 100 bp were removed from each end (for details, see Materials and Methods). After the exonuclease treatment was terminated, both samples were cleaved at a third remote restriction enzyme site (Fig. 1C, site X), and the DNA fragments from both samples containing the target region were isolated. The fragments were mixed and recombined by ligation (Fig. 1D). The 5′ and 3′ fragments were joined randomly; therefore, the distribution and length of deletions or repeats depended primarily on the distribution of the DNA ends produced during the exonuclease treatment. The mutants are referred to by the C-terminal amino acids encoded by the upstream fragment of the core gene and the N-terminal amino acids encoded by the fused downstream fragment; e.g., mutant A11-E8 contains peptide E8-F9-G10-A11 as a tandem repeat inserted between K7 and T12, and mutant A11-V13 has a deletion of T12. If the fusion created a single new codon at the fusion site, the corresponding amino acid is indicated (e.g., P50-L-D32). Sole amino acid substitutions cannot be generated by the method because two ligated fragments creating the wild-type (WT) length of the DNA molecule also generate the WT sequence.
If the region shortened by the exonuclease is 100 bp long at each DNA end, the maximum number of mutants attainable by this technique is (100 × 100) − 100, or 9,900. This number increases exponentially with the length of the truncated DNA segment. In order to keep this number within reasonable limits and to avoid the generation of very long insertions or deletions, six single restriction enzyme sites were introduced into the HBV core gene by in vitro mutagenesis without changing the amino acid sequence (Fig. 2). These six sites, together with the natural BglII site at nt 1983 and a BamHI site between the bacterial promoter and the core gene, were used to apply the mutagenesis procedure separately to five regions of the gene (Fig. 3, regions A to E). The regions are between 94 and 153 bp long, and adjacent regions overlap, except for regions B and C. (The BglII site at nt 1983 could not be used for downstream mutagenesis because the core gene carries two additional BglII sites, at nt 2403 and nt 2427.) The five regions cover codons 1 to 149 of the core gene. The R-rich C terminus, which can be deleted without blocking capsid formation (10), was omitted.
The approximate distribution of the DNA ends produced by the Bal 31 treatment was determined after each round of mutagenesis by sequencing the plasmids of a number of unselected colonies (data not shown). When the distribution was unsatisfactory, additional mutagenesis experiments were carried out with the same region.
Genotype of stable antigen-positive mutants.
For each region, between 6,200 and 13,600 colonies were tested for the expression of HBcAg or HBeAg (Table 1) with a polyclonal human serum containing the corresponding antibodies, anti-HBc and anti-HBe. The ratio of antigen-positive colonies varied from 6.3% (region E) to 0% (region D). It is unlikely that mutagenesis in region D destroyed all core protein epitopes recognizable by the human antiserum because the main HBcAg and HBeAg epitopes have been mapped to sequences around A80 (at the extreme 5′ boundary of region D) and P144 (approximately 30 aa downstream of region D). It is more likely that all insertions and deletions in this area destabilized the protein.
TABLE 1.
Number of colonies screened after mutagenesis of different regions of the core gene
| Regiona | No. of colonies that were
|
||
|---|---|---|---|
| Tested | HBcAg or HBeAg positive | Randomly selectedb | |
| A | 13,600 | 593 | 35 |
| B | 11,300 | 152 | 28 |
| C | 8,000 | 240 | 34 |
| D | 6,200 | 0 | 0 |
| E | 9,100 | 573 | 13 |
| Total | 48,200 | 1,558 | 110 |
Letters indicate the regions defined in Fig. 3.
Used for genotyping and phenotyping.
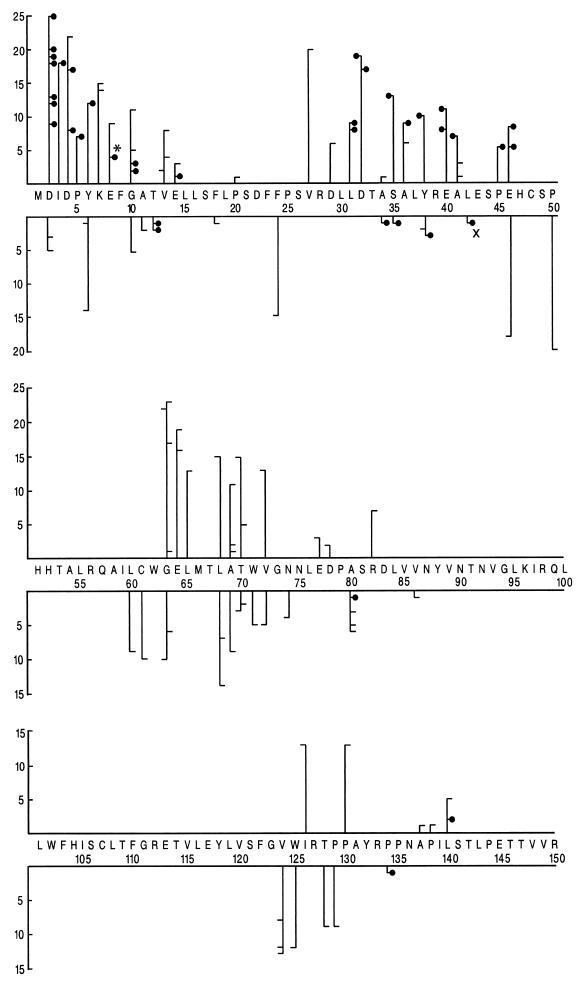
Antigen-positive colonies were randomly chosen (Table 1), and the fusion sites of the upstream and downstream core gene fragments ligated in individual mutants were determined by sequencing (Fig. 4). WT genes were found at a low frequency (approximately 5%) in each mutagenesis round. The genotyped 110 mutations (69 insertions and 41 deletions) formed four clusters which fixed the limits of four variable domains (domain I from D2 to E14, domain III from F24 to P50, domain V from L60 to R86, and domain VII from V124 to L140) and three intervening constant domains carrying few mutations (domain II from L15 to F23) or no mutations (domain IV from H51 to I59 and domain VI from N87 to G123) (Fig. 4 and Table 2).
FIG. 4.
Genotypes and phenotypes of 110 HBcAg- or HBeAg-positive core gene mutants. The abscissa shows the numbered HBV core protein sequence up to R150 in the one-letter code. The ordinate above (below) the sequence denotes the number of amino acids repeated (deleted) in individual mutants. A mutant forming capsids is represented by a dot; a capsid-defective mutant is represented by a dash. A dot (dash) points to the N-terminal amino acid of the peptide which is repeated (deleted) at that site. The position of a dot or dash on the upper (lower) ordinate indicates the length of the repeat (deletion). For example, mutant A11-E8 (marked by an asterisk) carries a 4-aa repeat and contains the sequence …Y6-K7-E8-F9-G10-A11-E8-F9-G10-A11-T12-V13…, and mutant A41-E43 (marked by multiplication sign) carries a deletion of 1 aa (L42). A dot or dash connected to the left side of a vertical bar indicates that the ligation of the upstream and downstream core gene fragments (Fig. 1) generated a single new codon at the fusion point.
TABLE 2.
Number of mutants identified in different core protein domains
| Domain | Span | No. of mutants that were
|
||
|---|---|---|---|---|
| HBcAg or HBeAg positive | Capsid forminga | Ratio (%) | ||
| I | D2–E14 | 34 | 18 | 53 |
| II | L15–F23 | 2 | 0 | 0 |
| III | F24–P50 | 27 | 17 | 63 |
| IV | H51–I59 | 0 | ||
| V | L60–R86 | 34 | 1 | 3 |
| VI | N87–G123 | 0 | ||
| VII | V124–L140 | 13 | 2 | 15 |
Number of mutants that scored positive for capsid formation in the assay shown in Fig. 5.
Assays for capsid formation.
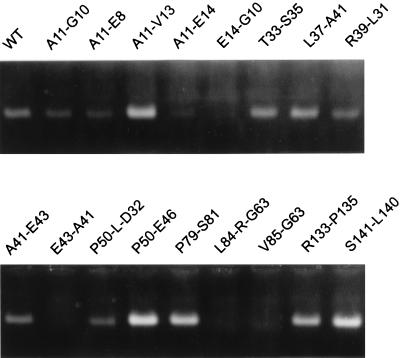
The 110 mutants were tested for their ability to form capsids in E. coli by the following indirect method, which has the advantage that a relatively large number of assays can be done in one experiment. Cleared bacterial lysates were treated with RNase and DNase. The RNA in the lumen of core particles is protected from the nuclease attack, while all RNA is destroyed if no particles are formed. During separation of the treated lysates by electrophoresis through native agarose gels, the intact capsids run as a band and can finally be visualized by ethidium bromide staining under UV light due to the packaged RNA (Fig. 5).
FIG. 5.
Detection of core particles by agarose gel electrophoresis. Cleared lysates from bacteria expressing WT or mutant core proteins were treated with DNase and RNase and separated by native agarose gel electrophoresis. Capsids ran as a band. Only the bacterial RNA encapsidated in core particles was nuclease protected and became visible by ethidium bromide staining under UV light.
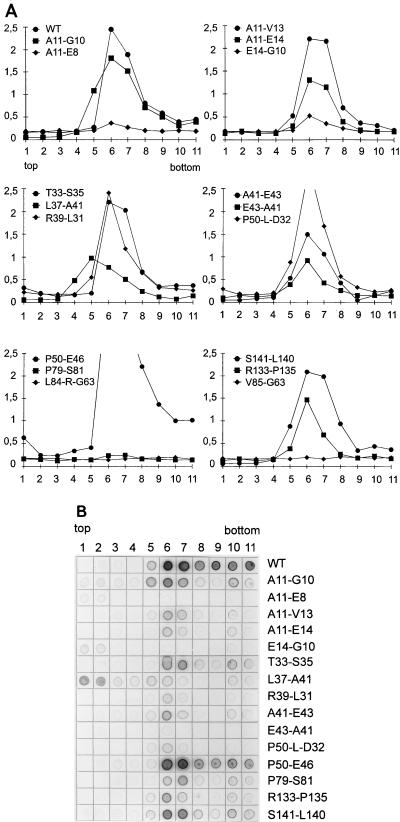
In order to evaluate this assay, 13 capsid-forming mutants and 4 non-capsid forming mutants also were tested by sucrose gradient centrifugation for capsid formation. The fractions were assayed (i) for HBcAg in an HBcAg ELISA which did not recognize HBeAg (Fig. 6A) and (ii) for HBcAg or HBeAg by blotting aliquots from the fractions on membranes and detecting the antigen with the same polyclonal human serum as that used in the initial screening of bacterial colonies (Fig. 6B).
FIG. 6.
Detection of core particles by sucrose gradient centrifugation. Cleared lysates from bacteria expressing WT and mutant core proteins were separated by sucrose gradient centrifugation. Eleven fractions were harvested from the top. The fractions were assayed with an HBcAg ELISA (A) (numbers on the ordinate indicate optical densities) or by blotting of an aliquot onto a membrane and detecting HBcAg or HBeAg in a Western blot assay (B). Core particles peaked in fractions 6 and 7. Mutant P79-S81 was negative in the HBcAg ELISA, but capsids became detectable in the HBcAg or HBeAg blot.
The results of the two assays for capsid formation (gel electrophoresis and sucrose gradient centrifugation) were in good accord and justified the use of agarose gel electrophoresis for detecting capsids (Table 3). Two of the four mutants negative in the agarose gel assay (L84-R-G63 and V85-G63) also were negative in the HBcAg ELISA of the sucrose gradients (Fig. 6A). The other two mutants negative in the agarose gel assay (E14-G10 and E43-A41) showed a very weak HBcAg signal in the central fractions of the sucrose gradients (Fig. 6A) and were scored negative in the dot blot. The amount of core particles produced by these mutants probably was very low and could be detected by the sensitive HBcAg ELISA but not by the less sensitive agarose gel assay and dot blot. For mutant E14-G10, the HBcAg or HBeAg dot blot (Fig. 6B) demonstrated that the majority of the antigen was in a nonparticulate state and appeared in the upper fractions of the gradient.
TABLE 3.
Results of core particle assays with randomly selected HBcAg- or HBeAg-positive core gene mutants
| Mutant | Detection of core particles on:
|
||
|---|---|---|---|
| Agarose gela | Sucrose gradientb
|
||
| HBcAg | HBc/eAg | ||
| None (WT) | + | + | + |
| A11-G10 | + | + | + |
| A11-E8 | + | (+) | − |
| A11-V13 | + | + | + |
| A11-E14 | (+) | + | + |
| E14-G10 | − | (+) | − |
| T33-S35 | + | + | + |
| L37-A41 | + | (+) | (+) |
| R39-L31 | + | + | + |
| A41-E43 | + | + | + |
| E43-A41 | − | (+) | − |
| P50-L-D32 | + | + | + |
| P50-E46 | + | + | + |
| P79-S81 | + | − | + |
| L84-R-G63 | − | − | ND |
| V85-G63 | − | − | ND |
| R133-P135 | + | + | + |
| S141-L140 | + | + | + |
All 13 mutants positive in the agarose gel assay also demonstrated capsid formation in the sucrose gradients (Table 3). Three mutants, however, had a unique pattern. Mutant A11-E8 produced only a very weak signal in the HBcAg ELISA and was negative in the HBcAg or HBeAg dot blot. The reason for this finding is not clear. The second mutant, L37-A41, repeatedly produced an antigen peak in fraction 5 instead of fraction 6 or 7 for unknown reasons (Fig. 6A and B). Also, this mutant showed a relatively large amount of nonparticulate HBcAg or HBeAg (top fractions in Fig. 6B). Clearly, the capsid assembly of this variant, although allowing protection of RNA (Fig. 5), was abnormal. The third mutant, P79-S81, formed particles but was negative in the HBcAg ELISA. This result, however, was not surprising because the HBcAg determinant has been mapped to the region around A80. Consequently, this mutant was detectable in the HBcAg or HBeAg dot blot (Fig. 6B).
Of the 110 core gene mutants, 38 scored positive in the nuclease treatment-agarose gel assay for capsid formation (Fig. 4). The ratio of capsid-forming mutants varied between different domains, from 3% (domain V) to 63% (domain III) (Table 2).
Distribution of mutations relative to a proposed core protein fold.
During the course of this work, models for the folding of a C-terminally truncated HBV core protein in bacterially expressed capsids were proposed by others (5, 8). In our study, domains I to VII were defined in the core protein primary amino acid sequence according to the effect of insertions and deletions on stable antigen expression and capsid formation (Table 2). Comparison of these domains from the N to the C terminus with a model for the protein fold (5) provided the following results (Table 2 and Fig. 7). Domains I and III correspond to the N-terminal and second loops of the model, respectively. Mutations in these domains were partly compatible with capsid formation. Domain II, in which only two small mutations which blocked capsid formation were identified, coincides with the first alpha helix. The nonchangeable domain IV forms a basal part of the spike. The variable domain V corresponds to the C-terminal one-third of helix 2 and the third short loop at the tip of the spike. With the exception of mutation P79-S81, which maps to the very tip of the spike, mutations in this domain blocked particle formation. The next three helices, interrupted by a kink at G94 and a turn at G111, form the constant domain VI. Finally, domain VII corresponds to the C-terminal part of the C-terminal helix and the next loop. All mutations in the helical part of this domain (V124 to T128) blocked particle formation. The two capsid-forming mutants with mutations in this domain carried a small deletion (1 aa) and a short insertion (2 aa), and their mutations mapped to the nonhelical region.
In summary, mutations allowing capsid formation were found exclusively in nonhelical regions of the proposed fold. Such mutations could be identified in all four loops. Most of the helical regions were devoid of insertions or deletions; only two of them contained mutations which blocked capsid formation.
DISCUSSION
The HBV core protein, only 185 aa long, supports an astonishing number of complex functions in the life cycle of the virus, such as capsid formation, selective packaging of the pregenome-reverse transcriptase complex, trafficking of the capsid in the cell, and envelopment. We were interested in analyzing these functions by characterizing the phenotypes of core gene mutants. One problem with this approach is that it is not a trivial matter to generate core gene mutants with an informative phenotype: mild mutations, such as single-amino-acid substitutions, often maintain the complete WT phenotype. More drastic mutations, such as deletions or insertions, are more likely to generate a mutant phenotype; however, they have the disadvantage that the resulting proteins are often unstable or blocked in early functions, such as capsid formation (29), so that later functions cannot be scored.
We tried to circumvent this problem by generating a large number of quasi-random mutants carrying insertions and deletions but no amino acid substitutions and by selecting those allowing capsid formation with two simple screening steps. With this approach, 38 capsid-forming mutants were obtained; 30 of them carried insertions of up to 25 aa, and 8 carried deletions of between 1 and 3 aa (Fig. 4). A new region which tolerated insertions with respect to capsid formation and which has not been described earlier (domain III) (Table 2) was identified. The mutants form the basis for a subsequent analysis of core protein-dependent functions in the HBV life cycle (unpublished data).
The distribution of the identified mutations reflects to some extent the general ability of the core protein to be mutated by insertions and deletions. This notion is based on two facts. (i) The size and position of insertions or deletions produced in the mutagenesis procedure were quasi-random and relatively even throughout the analyzed core gene sequence. These characteristics were achieved by applying the mutagenesis protocol to different overlapping areas (Fig. 3) and by controlling the distribution of the DNA ends generated during the exonuclease treatment by polyacrylamide gel electrophoresis of digested fragments as well as by sequencing of unselected clones. (ii) All four domains which tolerated mutations with respect to stable antigen expression in E. coli (domains I, III, V, and VII; Table 2) corresponded mainly to nonhelical structures in the proposed fold of the protein (Fig. 7).
Apparently, insertions and deletions in the less flexible helical parts of the fold destabilized the protein. An exception was the C-terminal part of the first spike-forming helix (C61 to D78), where a large number of repeats and deletions were found. Interestingly, these insertions or deletions did not preferentially represent multiples of helix turns (e.g., 3, 7, or 10 aa). These mutant proteins probably are quite stable in E. coli because the mutations found in this area did not destroy the central, compact part of the fold. All mutations in this area, however, prevented core particle morphogenesis. Also, mutations in the small C-terminal part of the proposed C-terminal helix from V124 to T128 resulted in this phenotype.
A striking finding is that no single HBcAg- or HBeAg-positive mutant was found among 6,200 colonies tested after mutagenesis of region D (Table 1). Apparently, even small insertions and deletions in this part of the molecule (S81 to V115) destabilized the protein. This observation is supported by naturally occurring core gene mutants which carried deletions corresponding to region D and which were also unstable (30).
It has been shown that insertions into the core protein are compatible with capsid formation when fused to the N or C terminus or introduced at the HBcAg determinant around A80 (reviewed in reference 26). The C-terminal R-rich region was not analyzed by us. However, the tolerance of the N terminus for elongation was found in the present study. The insertion site around A80, however, was not clearly identified by our approach. In contrast, three mutants carrying repeats with a length of 2 to 7 aa between E77 and R82 were blocked in capsid formation (Fig. 4). The simplest explanation for this discrepancy is that the target site tolerating insertions is extremely small (3 aa, from P79 to S81), greatly reducing the likelihood of finding corresponding mutants among the 34 randomly selected HBcAg- or HBeAg-positive clones from the mutagenesis of region C (Table 1).
Potentially, our approach would be limited if the mutagenesis were to destroy the antigenicity of stable and possibly capsid-forming protein variants completely. However, this situation is unlikely in the case of the core protein and the antiserum used in the initial screening on filters. The two antigen determinants exposed on the surface of capsids are apart from each other in the primary sequence and could therefore not be destroyed simultaneously in the same mutant by our approach: determinant HBc, near A80 (21), and a determinant between R127 and R133 (20). Apparently, the human serum used in the screening recognized both determinants because seven mutations (one allowing capsid formation) between E77 and R82 and four mutations (one allowing capsid formation) between R127 and R134 were identified. This notion is also supported more directly by the antigenicity of the capsid-forming mutant P79-S81. This mutant was negative in an HBcAg ELISA with sheep anti-HBc serum because the main HBcAg epitope that mapped to the sequence around A80 was destroyed. However, the mutant was recognized by the human antiserum used in the initial screening (Table 3).
The second screening round designed to find capsid-forming mutants was done with an indirect assay that identified RNase-protected RNA in the lumen of capsids (Fig. 5). Clearly, this test was not as specific with respect to WT capsid formation as, e.g., sucrose gradient centrifugation. Mutant L37-A41 was scored as WT in the agarose gel assay and produced a large amount of nonparticulate HBcAg or HBeAg (Fig. 6B), and the mutant capsids moved in the sucrose gradient differently from the WT capsids (Fig. 6A). However, the phenotype of most of the doubly-checked mutants was confirmed by sucrose gradient centrifugation (Table 3). We therefore believe that the overall picture of the distribution of capsid-forming mutants is correct even if a small fraction of mutants is, like L37-A41, not exactly WT with respect to capsid formation.
We did not investigate at which step in particle morphogenesis the capsid-negative, HBcAg- or HBeAg-positive mutants were blocked. It is likely that mutants with changes in domain VII assemble into dimers because the dimer interface is formed mainly by the spike region. They are probably defective in establishing interdimer contacts, a step which is mediated by the C-terminal loop, which corresponds to domain VII (13).
Twelve of the capsid-forming mutants were characterized by complementation of a core-negative HBV genome in eukaryotic cell cutures (unpublished data). None of them was WT for all of the functions assayed. This result demonstrates that the HBV core protein, which is involved in many steps of the viral life cycle, cannot readily mutate without losing a vital function.
ACKNOWLEDGMENT
This work was supported in part by the Fritz Thyssen Stiftung.
REFERENCES
- 1.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beames B, Lanford R E. Carboxy-terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology. 1993;194:597–607. doi: 10.1006/viro.1993.1299. [DOI] [PubMed] [Google Scholar]
- 3.Birnbaum F, Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumberg B S. Hepatitis B virus, the vaccine, and the control of primary cancer of the liver. Proc Natl Acad Sci USA. 1997;94:7121–7125. doi: 10.1073/pnas.94.14.7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottcher B, Wynne S A, Crowther R A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 6.Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350–9357. doi: 10.1128/jvi.71.12.9350-9357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen B J, Richmond J E. Electron microscopy of hepatitis B core antigen synthesized in E. coli. Nature. 1982;296:677–679. doi: 10.1038/296677a0. [DOI] [PubMed] [Google Scholar]
- 7a.Conway J F, Cheng N, Zlotnick A, Stahl S J, Wingfield P T, Belnap D M, Kanngiesser U, Noah M, Steven A C. Hepatitis B virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c- and e-antigens J. Mol Biol. 1998;279:1111–1121. doi: 10.1006/jmbi.1998.1845. [DOI] [PubMed] [Google Scholar]
- 8.Conway J F, Cheng N, Zlotnick A, Wingfield P T, Stahl S J, Steven A C. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 9.de Boer H A, Comstock L J, Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci USA. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallina A, Bonelli F, Zentilin L, Rindi G, Muttini M, Milanesi G. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J Virol. 1989;63:4645–4652. doi: 10.1128/jvi.63.11.4645-4652.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kann M, Bischof A, Gerlich W H. In vitro model for the nuclear transport of the hepadnavirus genome. J Virol. 1997;71:1310–1316. doi: 10.1128/jvi.71.2.1310-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenney J M, von Bonsdorff C H, Nassal M, Fuller S D. Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure. 1995;3:1009–1019. doi: 10.1016/s0969-2126(01)00237-4. [DOI] [PubMed] [Google Scholar]
- 13.Konig S, Beterams G, Nassal M. Mapping of homologous interaction sites in the hepatitis B virus core protein. J Virol. 1998;72:4997–5005. doi: 10.1128/jvi.72.6.4997-5005.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 15.Lingappa J R, Martin R L, Wong M L, Ganem D, Welch W J, Lingappa V R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B virus capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milich D R, McLachlan A. The nucleocapsid of hepatitis B virus is both a T-cell-independent and a T-cell-dependent antigen. Science. 1986;234:1398–1401. doi: 10.1126/science.3491425. [DOI] [PubMed] [Google Scholar]
- 17.Nassal M. Hepatitis B virus morphogenesis. Curr Top Microbiol Immunol. 1996;214:297–337. doi: 10.1007/978-3-642-80145-7_10. [DOI] [PubMed] [Google Scholar]
- 18.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepat. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 19.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo R I, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 20.Pushko P, Sallberg M, Borisova G, Ruden U, Bichko V, Wahren B, Pumpens P, Magnius L. Identification of hepatitis B virus core protein regions exposed or internalized at the surface of HBcAg particles by scanning with monoclonal antibodies. Virology. 1994;202:912–920. doi: 10.1006/viro.1994.1413. [DOI] [PubMed] [Google Scholar]
- 21.Salfeld J, Pfaff E, Noah M, Schaller H. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J Virol. 1989;63:798–808. doi: 10.1128/jvi.63.2.798-808.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Schodel F, Moriarty A M, Peterson D L, Zheng J A, Hughes J L, Will H, Leturcq D J, McGee J S, Milich D R. The position of heterologous epitopes inserted in hepatitis B virus core particles determines their immunogenicity. J Virol. 1992;66:106–114. doi: 10.1128/jvi.66.1.106-114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seifer M, Zhou S, Standring D N. A micromolar pool of antigenically distinct precursors is required to initiate cooperative assembly of hepatitis B virus capsids in Xenopus oocytes. J Virol. 1993;67:249–257. doi: 10.1128/jvi.67.1.249-257.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuttleman J S, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 26.Ulrich R, Nassal M, Meisel H, Kruger D H. Core particles of hepatitis B virus as carrier for foreign epitopes. Adv Virus Res. 1998;50:141–182. doi: 10.1016/s0065-3527(08)60808-8. [DOI] [PubMed] [Google Scholar]
- 27.Valenzuela P, Quiroga M, Zaldivar J, Gray R, Rutter W. The nucleotide sequence of the hepatitis B viral genome and the identification of the major viral genes. UCLA Symp Mol Cell Biol. 1980;18:57–70. [Google Scholar]
- 28.Wei Y, Tavis J E, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Guo J, Ying Z, Hua S, Dong W, Chen H. Capsid assembly and involved-function analysis of 12 core protein mutants of duck hepatitis B virus. J Virol. 1994;68:338–345. doi: 10.1128/jvi.68.1.338-345.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan T T, Lin M H, Qiu S M, Shih C. Functional characterization of naturally occurring variants of human hepatitis B virus containing the core internal deletion mutation. J Virol. 1998;72:2168–2176. doi: 10.1128/jvi.72.3.2168-2176.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou S, Standring D N. Hepatitis B virus capsid particles are assembled from core-protein dimer precursors. Proc Natl Acad Sci USA. 1992;89:10046–10050. doi: 10.1073/pnas.89.21.10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zlotnick A, Cheng N, Conway J F, Booy F P, Steven A C, Stahl S J, Wingfield P T. Dimorphism of hepatitis B virus capsids is strongly influenced by the C-terminus of the capsid protein. Biochemistry. 1996;35:7412–7421. doi: 10.1021/bi9604800. [DOI] [PubMed] [Google Scholar]