Abstract
We have used cryoelectron microscopy and image reconstruction to study B-capsids recovered from both the nuclear and the cytoplasmic fractions of cells infected with simian cytomegalovirus (SCMV). SCMV, a representative betaherpesvirus, could thus be compared with the previously described B-capsids of the alphaherpesviruses, herpes simplex virus type 1 (HSV-1) and equine herpesvirus 1 (EHV-1), and of channel catfish virus, an evolutionarily remote herpesvirus. Nuclear B-capsid architecture is generally conserved with SCMV, but it is 4% larger in inner radius than HSV-1, implying that its ∼30% larger genome should be packed more tightly. Isolated SCMV B-capsids retain a relatively well preserved inner shell (or “small core”) of scaffolding-assembly protein, whose radial-density profile indicates that this protein is ∼16-nm long and consists of two domains connected by a low-density linker. As with HSV-1, the hexons but not the pentons of the major capsid protein (151 kDa) bind the smallest capsid protein (∼8 kDa). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed cytoplasmic B-capsid preparations to contain proteins similar in molecular weight to the basic phosphoprotein (∼119 kDa) and the matrix proteins (65 to 70 kDa). Micrographs revealed that these particles had variable amounts of surface-adherent material not present on nuclear B-capsids that we take to be tegument proteins. Cytoplasmic B-capsids were classified accordingly as lightly, moderately, or heavily tegumented. By comparing the three corresponding density maps with each other and with the nuclear B-capsid, two interactions were identified between putative tegument proteins and the capsid surface. One is between the major capsid protein and a protein estimated by electron microscopy to be 50 to 60 kDa; the other involves an elongated molecule estimated to be 100 to 120 kDa that is anchored on the triplexes, most likely on its dimer subunits. Candidates for the proteins bound at these sites are discussed. This first visualization of such linkages makes a step towards understanding the organization and functional rationale of the herpesvirus tegument.
Herpesvirus form an extensive family of DNA viruses, whose host range encompasses much of the animal kingdom. They are classified into three subfamilies alpha-, beta-, and gammaherpesviruses, on the basis of biological properties [53]. Despite this diversity, their assembly pathway is closely conserved: the nucleocapsid is formed in the nucleus and follows a pathway that bears a marked resemblance to those of DNA bacteriophages (8, 14). First, a procapsid is assembled, which then releases its morphogenic internal scaffolding protein and becomes filled with DNA, concomitant with a major conformational transition of the capsid shell. Subsequent events, however, are not phage-like. The nucleocapsid exits the nucleus and acquires its tegument (53) and lipoprotein envelope. The latter events remain a focus of active research and some controversy (see, for example, references 13, 26, 51, and 63).
Although virion assembly is a continuous sequential process, several kinds of capsids have been identified as representing stable endpoints or long-lived states. They include A-capsids, empty shells thought to arise from abortive attempts to package DNA or its aberrant release; C-capsids, which are filled with DNA; and B-capsids, which contain internal proteins but little or no DNA (for reviews, see references 28, 49 and 62). “B-capsid” refers either to an intranuclear capsid visualized in situ that contains internal material with different staining properties from the DNA in C-capsids (which we interpret as mainly scaffolding-assembly protein) or to a capsid isolated from the nuclear fraction that contains scaffolding proteins but little or no DNA. Here, we relax the requirement for nuclear provenance and refer to nuclear and cytoplasmic B-capsids, respectively. It now appears that there are several kinds of B-capsids. “Large-cored” B-capsids correspond to the normally short-lived and infrequently observed procapsid (42, 47). “Small-cored” B-capsids (49) derive from large-cored B-capsids and have a mature surface shell. It remains unclear whether these particles represent an assembly intermediate or an abortive byproduct (56) or how many subclasses they may comprise.
As noted above, herpesvirus capsid assembly is a conservative process. The current paradigm is that the surface shell is a T=16 icosahedron containing three essential proteins. Nine hundred sixty copies of the major capsid protein (Mr ≃ 120,000 to 155,000, depending on the virus) make up 150 hexons and 12 pentons. The other two proteins form the triplexes, complexes that are present on the outer surface at the 320 sites of local threefold symmetry and have an α2β stoichiometry (43). Typically, the α-protein has a mass of 33 to 35 kDa, and the mass of the β-protein is either similar or in the 50-kDa range (10). Most herpesvirus capsids also contain an additional low-molecular-weight protein that binds around the rims of hexons but not to pentons (11, 65, 72, 73).
The above account owes most to observations relating to herpes simplex virus type 1 (HSV-1), the archetypal alphaherpesvirus. However, one property of herpesviruses that does not suggest an immutable capsid structure is their variation in genome size, which ranges from ∼120 to ∼230 kbp (18). Betaherpesviruses account for three of the eight herpesviruses known to cause disease in humans and include the cytomegaloviruses (CMVs), which have the largest of all known herpesvirus genomes (4). Although the overall structure of the herpesvirus capsid appears to be conserved, it has not been clear whether there are features specific to each subfamily. To investigate this possibility, we have used cryoelectron microscopy to study isolated capsids of simian CMV (SCMV), a close and experimentally more tractable relative of human CMV (HCMV) [24].
A second motivation for this study was that it might offer insight into tegumentation. Nonenveloped capsids have been observed in significant numbers in the cytoplasms of cells infected with CMV (57) and appear to be at least partly tegumented (24, 30). We have noticed that, late in the infection of cultured cells with SCMV, B-capsids appear in the cytoplasmic fraction, and these cytoplasmic B-capsids differ from nuclear B-capsids in several respects, including their state of tegumentation. Here, we characterize the structures of nuclear and cytoplasmic B-capsids at a resolution of ∼2.2 nm and, by quantitative comparison between them, identify sites of tegument attachment.
MATERIALS AND METHODS
Virus, cells, and capsid preparation.
The Colburn strain of SCMV was grown in human foreskin fibroblasts prepared and infected as described earlier (24). Much of the capsid recovery procedure has also been described (33). In brief, 5 days after infection when the cytopathic effect was extensive and many cells were beginning to detach and float, all cells (∼5 × 107) were scraped into the medium, collected by centrifugation (1,500 × g, 5 min, 4°C), and suspended in PB buffer (150 mM NaCl, 40 mM phosphate; pH 7.4) containing 0.5% Nonidet P-40 (NP-40; BDH Laboratory Supplies, Poole, United Kingdom), at 1.0 × 108 cells per ml. After 5 min on ice, the detergent-treated cells were subjected to centrifugation (1,500 × g, 5 min, 4°C), yielding a supernatant (NP-40 cytoplasmic fraction) and pellet (NP-40 nuclear fraction). The NP-40 nuclei were suspended in 1.5 ml of PB and lysed by 10 passages back-and-forth through a hypodermic syringe with a 23-gauge needle. After clearing the lysate of particulate material by centrifugation (16,000 × g, 2 min, 4°C), the resulting supernatant and the NP-40 cytoplasmic fraction were subjected to rate-velocity sedimentation in sucrose gradients (15 to 50% sucrose [wt/wt] in PB buffer; 40,000 rpm, 20 min, 4°C; Beckman SW41 rotor). After centrifugation, light scattering bands were observed in both gradients at the position corresponding to B-capsids (24). These bands were collected by aspiration through the wall of the centrifuge tube, diluted 1:1 with PB buffer, and concentrated by pelleting (35,000 rpm, 2 h, 4°C; SW41 rotor).
Protein analysis.
Capsid proteins were separated by sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE), essentially as described previously (39) but with a Tris-Tricine buffer system (54) and a 10 to 20% gradient gel (Novex, San Diego, Calif.). Samples of the same nuclear and cytoplasmic B-capsid preparations used for cryo-electron microscopy and image reconstruction were subjected to SDS-PAGE (150 V for 60 min). Proteins were stained with Coomassie brilliant blue; the gel was then destained, dried, and digitized by optical scanning. Stained proteins were quantified by using the Quant mode of MacBas 2.5 software (Fuji Film Co., Tokyo, Japan) after the linear range was established with an optical density wedge.
Cryoelectron microscopy.
Immediately prior to specimen preparation, capsid suspensions at a 1- to 2-mg/ml protein concentration were vortexed for 30 s on a G-560 Vortex (Daigger Scientific Industries, Inc.) to disperse aggregates. Thin film specimens were formed over holey carbon films and vitrified by quenching them in liquid ethane cooled by liquid nitrogen according to standard procedures (9, 16). Specimens were observed in a Philips CM120 electron microscope equipped with a liquid N2-cooled model 626 Cryoholder (Gatan, Pleasanton, Calif.) and anticontamination blades. The grids were surveyed at low magnification for areas with ice of suitable thickness and appropriate particle concentrations by using a Gatan RMC 794 slow-scan charge-coupled device camera. Micrographs were recorded on Kodak SO-163 film at a nominal magnification of ×36,000 by using low-dose procedures. For size calibration, SCMV capsids were mixed with purified T4 bacteriophages, and the axial spacing of the T4 tail sheath was used to calibrate the dimensions of capsids recorded in the same micrographs (10).
Image processing and reconstruction.
Images used in this analysis were evaluated by optical diffraction, and the best images were scanned at 0.70 nm/pixel with a Perkin-Elmer 1010MG microdensitometer. The software and procedures were as described earlier (2, 22, 23). These images had their first zeros of the contrast transfer function (CTF) at frequencies varying from (1.9 nm)−1 to (2.5 nm)−1. For those images whose first zero frequency was less than (2.2 nm)−1, we computationally inverted the phases of the Fourier terms beyond the zero, i.e., in the second lobe of the CTF. Particles were extracted from the scanned fields and preprocessed by a semiautomatic procedure (17). For the nuclear B-capsid reconstruction, initial estimates of the orientation angles were obtained for a base set of particles by the polar Fourier transform (PFT) method of Baker and Cheng (1), with an appropriately scaled map of HSV-1 B-capsids (17) taken as starting model. Using these orientations, a density map was calculated and used in the first of several subsequent cycles of PFT-based refinement of these particles’ orientations and to solve additional particles. The final reconstruction combined 350 capsids from five micrographs into a three-dimensional density map with a resolution of 2.2 nm, according to the FRC3D criterion (17).
Cytoplasmic B-capsids were divided by visual criteria into three classes: lightly, moderately, and heavily tegumented. The three classes were analyzed separately, and corresponding maps were calculated, by using the nuclear B-capsid as a starting model in each case. The lightly tegumented capsid map included 121 capsids from 4 micrographs for a resolution of 2.2 nm; the moderately tegumented map included 257 capsids from 16 micrographs for a resolution of 2.2 nm; and for the heavily tegumented capsids, 79 images from the same 16 micrographs were used for a resolution of 2.5 nm. The smaller number of the latter particles reflects both their relative rarity and a lower success rate in determining reliable orientations. To calculate the radial density profile of the inner shell, 15 particles with seemingly well preserved, i.e., round and symmetrical, cores were selected, their surface shells were stripped away (3), and their exposed cores were aligned by cross-correlation methods and then averaged. Further noise reduction was effected by azimuthal averaging, and from this projection a radial density profile was calculated (61).
To estimate the molecular weights of the two tegument proteins, their volumes were estimated and expressed as masses by using a conversion factor of 0.78 kDa/nm3. To obtain the volume estimates, portions of the difference maps (moderately tegumented minus nuclear) and (heavily tegumented minus moderately tegumented) were excised in which the respective molecules were well represented (see Fig. 5c and d). The numbers of voxels in the corresponding envelopes of continuously connected density were then calculated. For the capsomer-capping protein, the molecule overlying the penton was used. Two estimates of volume were obtained in each case and define the limits of the range quoted (see Results). They represent the use of differing contour levels and correspond respectively to the nuclear capsid containing 100 or 120% of the expected mass (17).
FIG. 5.
Stereo pairs presenting the outer surfaces of the moderately tegumented cytoplasmic B-capsid (a) and the heavily tegumented cytoplasmic B-capsid (b) of SCMV. Also shown are difference maps of the moderately tegumented cytoplasmic B-capsid minus the nuclear B-capsid (c) and the heavily tegumented cytoplasmic B-capsid minus the moderately tegumented cytoplasmic B-capsid (d). Because we wished to focus on components associated with the outer surface of the capsid, the internal density was set to zero in both maps to avoid distracting differences from the relatively noisy internal regions. Map c reveals the capsomer-capping tegument protein, and map d reveals the triplex-associated tegument protein. Bar (a), 10 nm.
RESULTS
Nuclear and cytoplasmic B-capsids: protein composition.
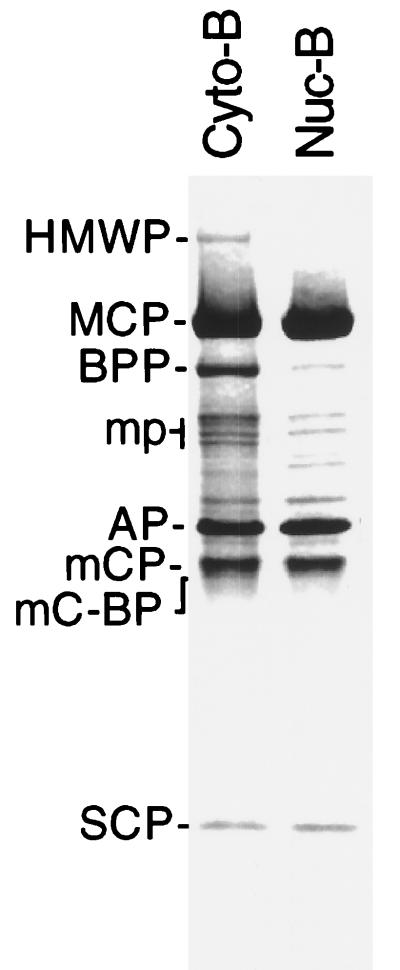
Cultured human fibroblasts were harvested 5 days after infection and separated into nuclear and cytoplasmic fractions after treatment with the detergent NP-40 to disrupt membranes. B-capsids were then isolated from both fractions by rate-velocity sedimentation through sucrose gradients. Their protein compositions were then evaluated by SDS-PAGE (Fig. 1) and are indistinguishable with respect to their contents of capsid proteins (i.e., MCP, AP, mCP, mC-BP, and SCP in Fig. 1). They differed, however, in that cytoplasmic B-capsids contain several proteins whose electrophoretic mobilities correspond to those of proteins previously classified as tegument components (24, 25, 28, 67), the strongest of which is the 119-kDa basic phosphoprotein (BPP; also called pp150 for HCMV). There are also several closely spaced bands in the size range of the 69-kDa upper-matrix protein (UM; also called pp71 for HCMV) and the 66-kDa lower-matrix protein (LM, also called pp65 for HCMV), as well as a small amount of the 205-kDa high-molecular-weight protein. None of these SCMV tegument proteins has been cloned and sequenced, but each appears to be the counterpart of an HCMV protein that has been so characterized (25).
FIG. 1.
Comparison of the protein compositions of nuclear and cytoplasmic B-capsids of SCMV (Colburn) by SDS-PAGE with protein detection by staining with Coomassie brilliant blue. Additional proteins are present in the cytoplasmic capsid preparation. Abbreviations (25, 28): HMWP, high-molecular-weight protein (205 kDa, homolog of HCMV UL48 protein); MCP, major capsid protein (151 kDa, homolog of HCMV UL86 protein); BPP, basic phosphoprotein (119 kDa, homolog of HCMV UL32 protein); “mp” refers to the several bands in the molecular-weight range of the upper- and lower matrix proteins (UM, 69 kDa, is the homolog of HCMV UL82 protein, and LM, 66 kDa, is the homolog of HCMV UL83 protein); AP, assembly protein (30 kDa, mature form, i.e., proteolytically processed; homolog of HCMV UL80.5 protein); mCP, minor capsid protein (35 kDa, homolog of HCMV UL85 protein); mC-BP, minor capsid binding protein (33 kDa, homolog of HCMV UL46 protein); SCP, smallest capsid protein (∼8 kDa, homolog of HCMV UL48/49 protein).
Nuclear and cytoplasmic B-capsids: morphology.
Typical cryomicrographs of both capsid preparations are presented in Fig. 2. Nuclear B-capsids (Fig. 2a) have a thick-walled outer shell with peripheral serrations. The angularity of a given projection and the sharpness of its serrations vary according to the viewing direction, as with equine herpesvirus 1 (EHV-1) (3) and HSV-1 (e.g., references 43 and 73). The only marked difference between nuclear B-capsids of SCMV and those of the alphaherpesviruses is their inner ring of density, which represents the projection of an internal protein shell. This ring is present in >90% of these capsids, the remainder being empty, like A-capsids. As discussed below, we interpret this internal structure to be the “small core” previously described in in situ electron microscopic observations of B-capsids (e.g., reference 47).
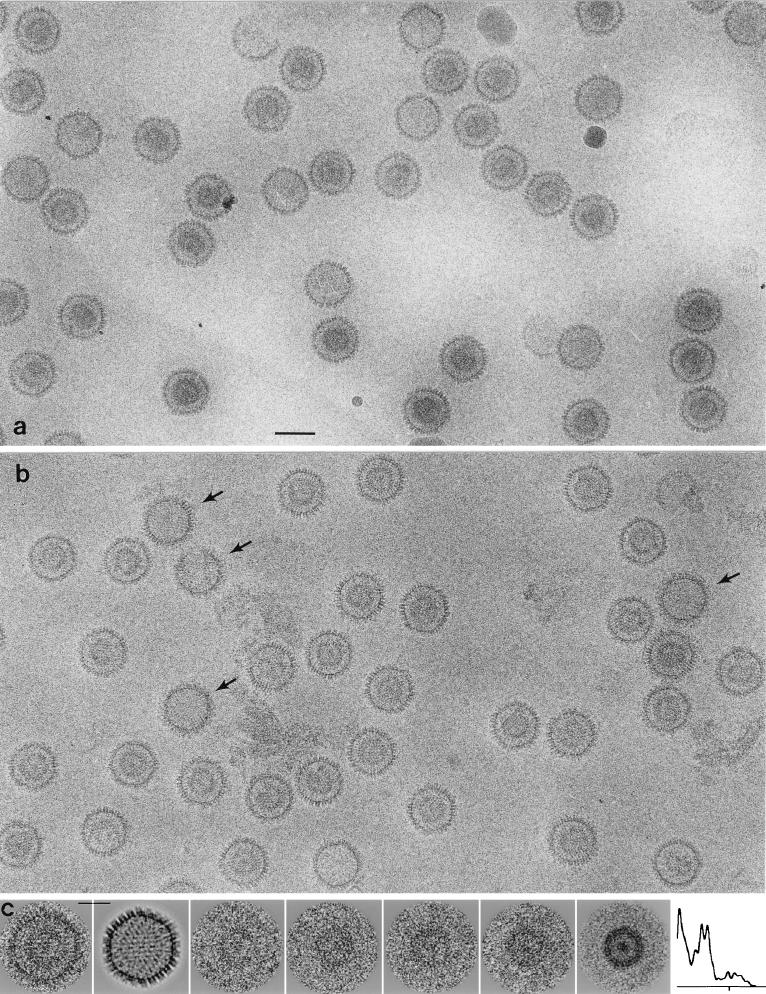
FIG. 2.
Cryoelectron micrographs of purified SCMV B-capsids (nuclear [a] and cytoplasmic [b]). (b) Several heavily tegumented capsids are marked with arrows in panel b. Also present in panel a is a phage T4 virion, which was included because its tail provided an internal magnification standard. Bar, 100 nm. (c) Gallery of cores of nuclear B-capsids. The three left panels show such a capsid and the separated images of its surface shell and core. Bar, 50 nm. The next three panels show the cores of other capsids. The last two panels show an averaged image of the core and a radial density profile calculated by rotational averaging of this image. They disclose a double ring surrounding some central density. The tick mark on the x axis of the density profile denotes a radius of 50 nm.
Cytoplasmic B-capsids are less uniform in appearance (Fig. 2b). Many of them (40 to 50%) resemble nuclear B-capsids. The remainder have additional material attached to their outer surface: of these, a subset of ∼8% carry markedly greater amounts of this material (Fig. 2b, arrows). Consistent with its location on the outer surface of the capsid and the presence of specific tegument proteins in this preparation (Fig. 1), we interpret this material as tegument and view the three classes of particles as varying in the extent to which they have acquired or retained such proteins. We refer to them as lightly, moderately, and heavily tegumented cytoplasmic B-capsids, respectively, noting that these are relative terms and that, in virions, the capsid may be still more heavily tegumented. Most of the heavily tegumented B-capsids (55 to 60%) had little or no internal density: in contrast, only ∼8% of the remaining cytoplasmic B-capsids were empty, a finding similar to the proportion of empty nuclear B-capsids (∼9%).
To obtain unobstructed views of the inner shell, the coprojected surface shell was computationally erased from a set of particles by subtracting reprojections of a three-dimensional density map of the capsid (see below) that had been emptied by setting its internal densities to the background value. A gallery of cores is shown in Fig. 2c: some are more circularly symmetric than others, and it is likely that they represent cores in a relatively good state of preservation (see Discussion). The averaged core image shows two closely spaced concentric rings and a central patch of density (Fig. 2c, rightmost panels).
Nuclear B-capsids of SCMV and HSV-1 compared.
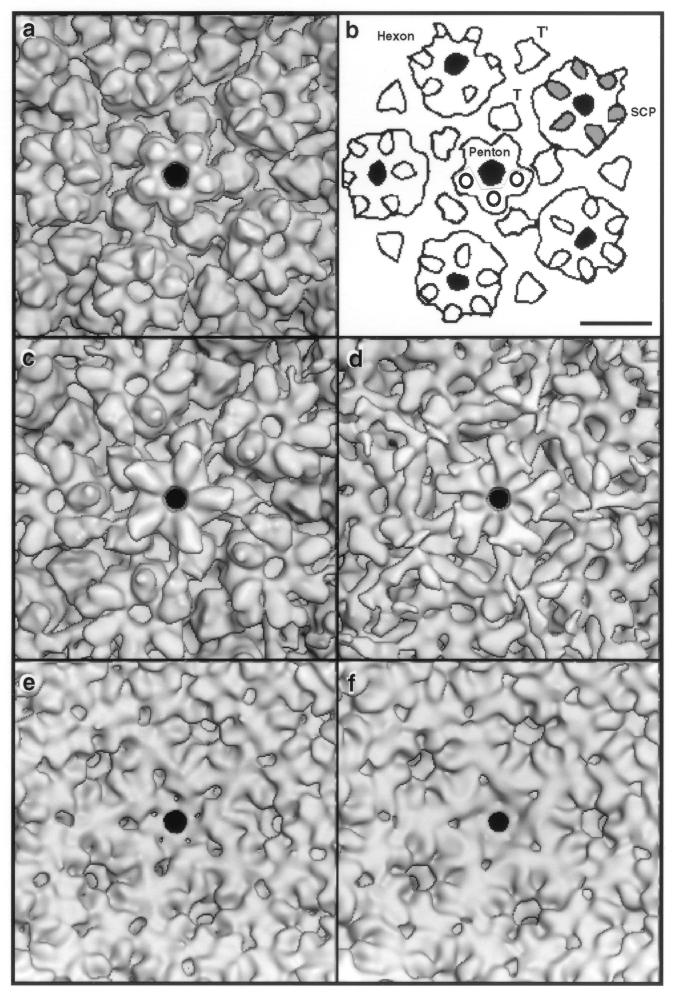
To examine the structure of the nuclear B-capsid, we calculated a three-dimensional density map to a 2.2-nm resolution (Fig. 3a and d). The molecular architecture of the capsid is largely conserved between SCMV and other previously characterized herpesviruses, i.e., in the T=16 triangulation geometry; in the overall shape of the large and elaborate capsomers, whose columnar protrusions extend ∼10 nm out from the contiguous “floor”; and in the presence of triplexes at the threefold sites between capsomer protrusions. On the HSV-1 capsid, the 12-kDa VP26 (UL35) protein (20, 41) is present around the outer rims of the hexons but absent from the pentons (11, 65, 72, 73). We infer that this property also is reproduced in SCMV because the protuberances around the rims of its hexons are considerably larger than those on the pentons (cf. Fig. 3a and b and 6a). Thus, the additional outer part of the SCMV hexon protuberance represents a monomer of SCP (∼8 kDa [32]), its counterpart to VP26 (see Fig. 6b).
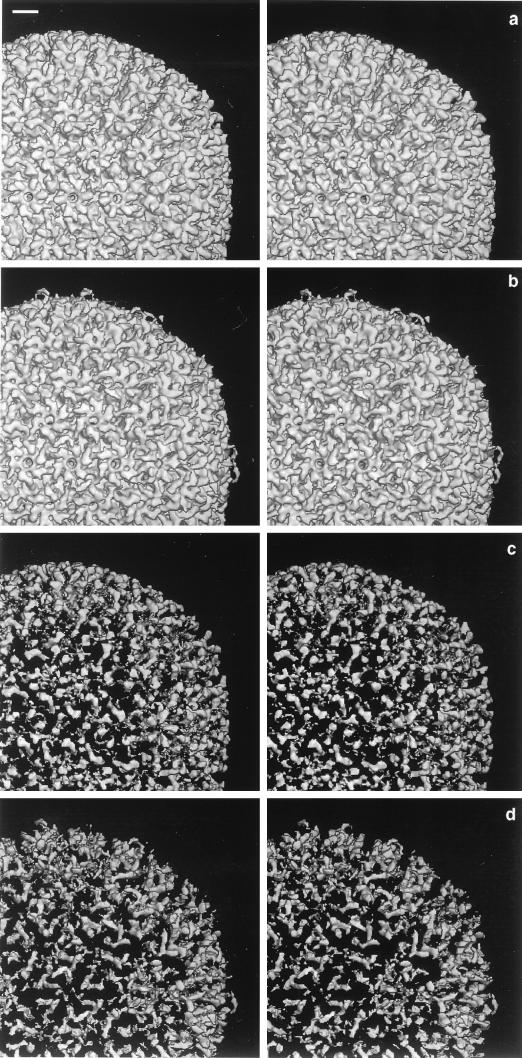
FIG. 3.
Reconstructed density map of nuclear B-capsids of SCMV at 2.2-nm resolution, viewed along an axis of twofold symmetry showing the outer surface (a) and the inner surface (d). Bar (d), 10 nm. To allow comparison with the HSV-1 capsid (at 2.4-nm resolution, reproduced from reference 17), corresponding features centered on the E-hexon, midway between two pentons, are compared in panels b and c and panels e and f: b and e, SCMV; c and f, HSV-1; b and c, outer surfaces; e and f, inner surfaces. Bar (b), 10 nm. On the outer surface, the orientations of the capsomer protrusions differ between SCMV and HSV-1 (cf. panels b and c). In principle, this might reflect an arbitrarily different assignment of handedness to the two T=16 density maps (handedness is a subtle feature in nonskewed icosahedral lattices, such as T=16). However, the relative handedness of the capsids was determined by maximizing the correlation between the two maps, and they match well in respect to the handedness on the inner surface (cf. panels e and f) and elsewhere. Thus, we consider that the differing orientations of their external protrusions is a genuine distinction, although the absolute handedness of either capsid has yet to be determined.
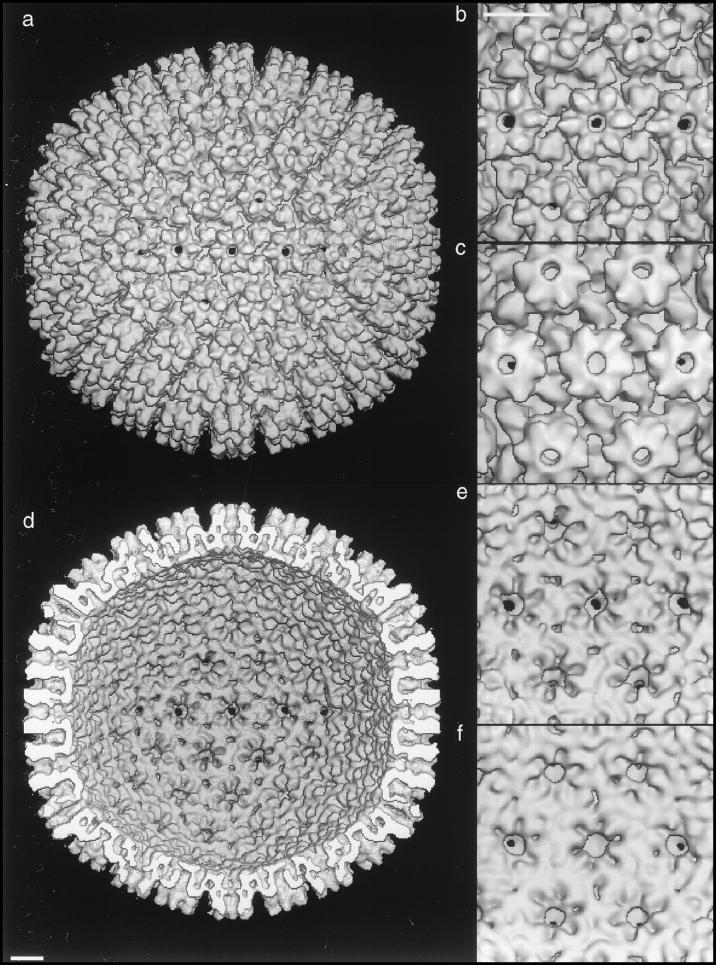
FIG. 6.
Peripentonal regions of the outer (a to d) and inner (e and f) surfaces of SCMV capsids. Panels: a, nuclear B-capsid; b, diagram indexing features of note in panel a, where SCP is the smallest capsid protein present on hexons (gray outer portion of protuberance on top of the hexon subunits) but not on pentons, where the corresponding protuberance is markedly smaller and rounder; c, moderately tegumented cytoplasmic capsid; d, heavily tegumented cytoplasmic capsid. Also shown are the inner surfaces of the nuclear B-capsid (e) and the moderately tegumented cytoplasmic B-capsid (f). Bar, 10 nm.
At a more detailed level, differences are apparent between the outer surfaces of the HSV-1 and SCMV capsomers (cf. Fig. 3b and c), whereas the inner surfaces are almost superimposable (cf. Fig. 3e and f). The main structural distinctions relate to the outer parts of the respective hexon protrusions, which lie in different orientations relative to the surface lattice. Moreover, they have different shapes: in SCMV, unlike HSV-1, there are deep notches between the protuberances around the outer rims of the capsomer protrusions (cf. Fig. 3b and c), suggesting that the corresponding portions of their major capsid proteins are folded differently. Their respective triplexes also have somewhat different shapes.
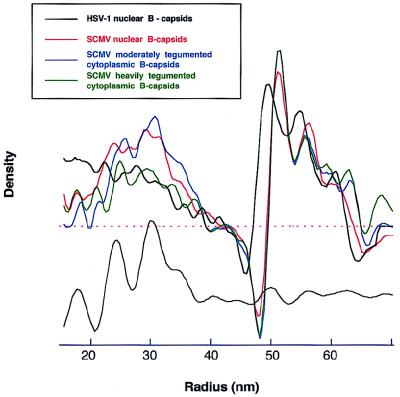
The SCMV capsid is slightly larger than the HSV-1 capsid, as assessed in terms of spherically averaged radial density profiles (Fig. 4). In outer radius, it is ∼2% larger but the inner radius is ∼4% larger, the SCMV shell being slightly thinner. Both radial profiles show three peaks in the surface shell, between ∼50 and 65 nm. For SCMV, the second peak is lower relative to the first peak than for HSV-1.
FIG. 4.
Radial density profiles of SCMV capsids: nuclear B-capsids (red), lightly tegumented cytoplasmic B-capsids (blue), and heavily tegumented cytoplasmic B-capsids (green). Also shown for comparison (in black) is the profile of HSV-1 nuclear B-capsids (reproduced from reference 17). The curves were obtained by spherical averaging of the corresponding three-dimensional density maps. The inner part of these curves (r < 40 nm, relating to the core) becomes progressively noisier towards the lower radii, both because of sparser sampling and because some distorted cores were included in the calculation. An optimized rendition of this part of the profile, calculated from nuclear B-capsids with well-preserved cores (see Materials and Methods), is shown below (in gray) offset, relative to the other curves, for clarity.
Radial organization of nuclear and cytoplasmic B-capsids.
The material adhering to heavily tegumented cytoplasmic B-capsids appears not to be arranged with perfect icosahedral symmetry, to judge by its irregular appearance. We found it more difficult to determine their orientations than for other capsids in the same micrographs, presumably because of this material. Nevertheless, enough particles (79 in all) were solved to yield a well defined map at 2.5-nm resolution. We also generated maps at a 2.2-nm resolution from larger numbers of moderately and lightly tegumented B-capsids.
Averaged radial density profiles were calculated from these maps, and those of the lightly and heavily tegumented B-capsids are compared in Fig. 4 with that of nuclear B-capsids. In the inner part of the capsid shell, i.e., between radii of ∼48 and ∼58 nm, the curves of the lightly and heavily tegumented B-capsids are superimposable and differ slightly in relative peak heights from that of the nuclear B-capsid, which may reflect slight conformational differences between them. From ∼58 to ∼65 nm, the profile of the heavily tegumented B-capsid overlies that of the lightly tegumented B-capsid by a small margin, and both have more density in this region than the nuclear B-capsid. Between 65 and 70 nm, the additional outer density of heavily tegumented B-capsids becomes more evident.
SCMV B-capsids contain an inner shell, which lies between radii of ∼20 and ∼40 nm and is separated from the surface shell by a gap of ∼10 nm (Fig. 4). In contrast, the internal proteins of HSV-1 B-capsids, which are not regularly ordered in a shell but are icosahedrally averaged in the reconstruction, register no inner edge (cf. Fig. 4). At lower radii, these profiles become progressively noisier but nevertheless reveal the following features. Their overall shapes are generally similar for all kinds of SCMV B-capsids, although the profile of heavily tegumented B-capsids is ∼50% lower, reflecting their higher proportion of empty capsids. The curves all show a minimum at a radius of ∼28 nm and an outer shoulder located between ∼33 and ∼40 nm.
The inner part of the profile (i.e., at radii of <40 nm) is smeared because not all particles included in the reconstruction have well-preserved cores. Some have lesions that suggest incipient degeneration and other cores are off-centered (Fig. 2a). However, by selecting particles with round, seemingly well preserved cores and correcting for off-centering, we calculated an optimized averaged image (Fig. 2c). After rotational averaging to obtain a projection profile, two density peaks are clearly resolved at radii of ∼25 and 30 nm, respectively (Fig. 2c, rightmost panel). From this curve, an average radial density profile was calculated (Fig. 4, bottom curve), which shows more clearly the strong minimum between them at the 28-nm radius.
Visualization of capsid-tegument interactions.
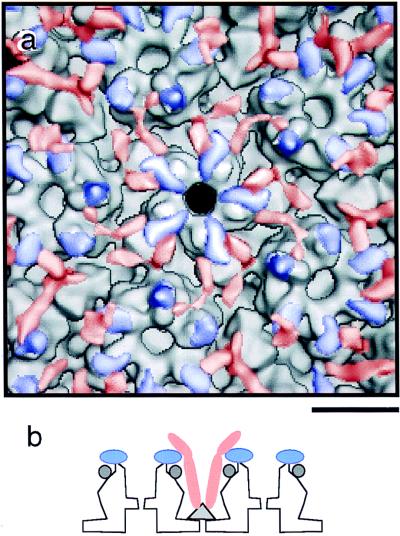
The outer surfaces of moderately and heavily tegumented cytoplasmic B-capsids are presented in stereo in Fig. 5a and b. Figure 5c is a difference map calculated between the moderately tegumented cytoplasmic B-capsid and the nuclear B-capsid. The two maps are at the same resolution (2.2 nm), and this difference map is intended to highlight any additional components present on cytoplasmic B-capsids. Although there is some noise that we attribute mainly to minor conformational differences between the respective shells, the most pronounced difference is the presence of additional density capping each subunit of major capsid protein. This density is carrot-shaped, ∼6 nm long by ∼2.5 nm wide at its midpoint, and oriented with its long axis tangential to the capsid surface and its tapered end pointing in towards the symmetry axis of the capsomer (Fig. 6c; see also Fig. 5c, lower right quadrant). Similarly shaped features overlie both the hexons and the pentons (see also Fig. 8a). Presumably, they represent protein subunits, and we refer to them as the capsomer-capping protein(s). Consistent results were obtained with a difference map calculated between lightly tegumented cytoplasmic B-capsids and nuclear B-capsids (data not shown).
FIG. 8.
(a) Region surrounding the fivefold axis showing the interactions of the two major tegument proteins with the SCMV capsid. The capsomer-capping tegument protein is shown as blue, and the triplex-associated tegument protein is shown as red. The segmentation was achieved by removing noise from the two difference maps (Fig. 5c and d) and then recombining them with the nuclear B-capsid image. Bar, 10 nm. (b) Schematic summary of the interactions that link the tegument to the capsid. The capsomer-capping tegument protein (blue) binds to the major capsid protein on top of the capsomer protrusions. Two copies of the triplex-binding tegument protein (red) diverge from each triplex (the triplex is shown in panel b as a triangle). This elongated protein contacts the capsomer-capping protein (blue) on two different capsomers and then extend outwards. This protein may have an additional domain or domains that extend beyond the portions sketched in panel b, which are not well visualized in our density map (panels b and c) on account of flexibility or otherwise poor ordering.
We also calculated a difference map between the heavily and moderately tegumented cytoplasmic B-capsids (Fig. 5d) to accentuate components that are present in higher occupancy on the former. The predominant features of this difference map are V-shaped densities extending from the triplexes. The tip of the V makes contact with the triplex, and its legs are ∼10 nm long and 2 to 2.5 nm thick. We infer that the two halves of the V represent two copies of the triplex-associated protein.
The molecular weights of these proteins were estimated from their apparent volumes, applying a partial specific volume typical for protein. We gauged the reliability of the method by using it to measure the mass of a triplex in a density map of HSV-1 capsid, contoured to contain 100% of the total mass as determined by scanning transmission electron microscopy (43). The value obtained was 130 kDa, ∼8% higher than the theoretical value of 119 kDa (one copy of VP19c plus two copies of VP23). In the same way, the mass of the penton-capping protein was estimated from the SCMV density maps as 50 to 60 kDa and that of the triplex-associated protein as 100 to 120 kDa. We note that, notwithstanding our control experiment, such estimates may be biased downwards by substoichiometric occupancy or local disordering.
The outer surfaces of these capsids in the region around a penton are shown at higher magnification in Fig. 6. The nuclear B-capsid is shown in Fig. 6a, a diagram marking its salient features in Fig. 6b, and the moderately and heavily tegumented cytoplasmic B-capsids in Fig. 6c and d, respectively. The inner surfaces of all three classes of cytoplasmic B-capsids are essentially identical when depicted at the same resolution. As a representative, that of the moderately tegumented cytoplasmic B-capsid (Fig. 6f) is compared with that of the nuclear B-capsid (Fig. 6e). Minor differences between them (cf. Fig. 6e and f) suggest that the surface shells of nuclear and cytoplasmic B-capsids may be in slightly different conformations.
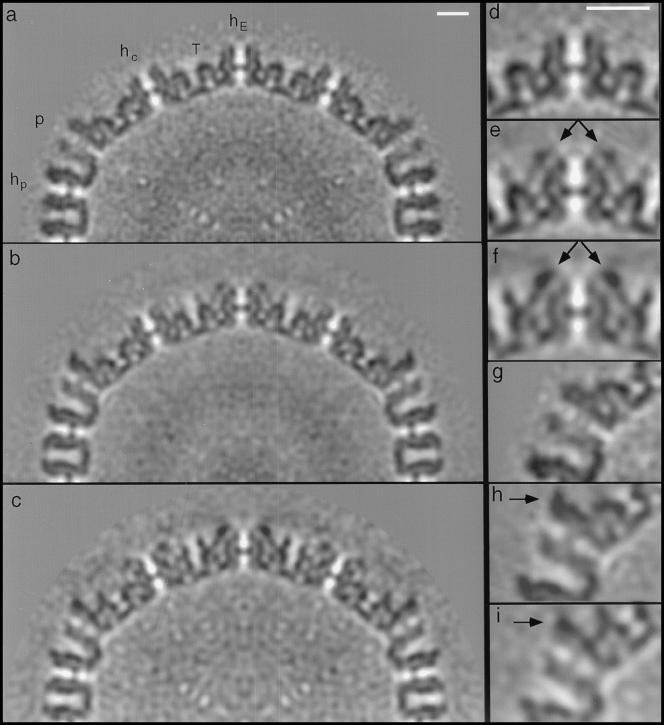
To visualize components that, although not stoichiometric, are present in sufficient quantity to be discernible in a reconstruction, it is informative to examine sections through the maps (Fig. 7). The core is visible in our maps of both the nuclear B-capsid (Fig. 7a) and the moderately tegumented cytoplasmic B-capsid (Fig. 7b). The triplex-associated protein is faintly visible in the moderately tegumented B-capsid (Fig. 7e), whereas in the heavily tegumented B-capsid (Fig. 7f), it is as strongly represented as the capsid proteins, implying full occupancy. This protein is anchored on the triplex, passes along the wall of the capsomer protrusion, makes contact with the capsomer-capping protein (see Fig. 6c and 8a), and then extends out beyond the protrusions. In all three cytoplasmic B-capsids, the density associated with the capsomer-capping protein (Fig. 7h and i, arrows) is similar to that of the major capsid protein, suggesting full occupancy.
FIG. 7.
Central sections (half-plane) through density maps of the nuclear B-capsid (a), moderately tegumented cytoplasmic B-capsid (b), and heavily tegumented cytoplasmic B-capsid (c) of SCMV. The plane of the 0.4-nm-thick section is perpendicular to an axis of twofold icosahedral symmetry and passes centrally through pentons (p), peripentonal hexons (hp), edge hexons (hE), and central hexons (hc) (see Fig. 2 of reference 60 for nomenclature) and triplexes (T). The regions centered on the hE hexon and the penton from all three maps are compared at higher magnification in panels d to i. The arrows in panels e and f indicate the additional density on cytoplasmic B-capsids, which is attributed to the triplex-associated tegument protein. This density is faint in panel e and strong in panel f. The arrows in panels h and i indicate the additional density attributed to the capsomer-capping protein (here, on the penton) on both kinds of cytoplasmic B-capsids. The difference in this density on either side of the penton occurs because the section plane passes centrally through this protein only on one side of the penton. The distribution of capsomer-capping protein subunits over the capsid surface is shown in Fig. 5c. Bars (a and d), 10 nm.
The components of the heavily tegumented cytoplasmic B-capsid, i.e., its capsid shell, the capsomer-capping protein, and the triplex-associated protein, are distinguished by color coding in Fig. 8a. The apparent connectivity of the interactions that link them together is shown schematically in Fig. 8b.
DISCUSSION
We have investigated the capsid structure of SCMV, viewed as a representative betaherpesvirus. To date, the only other herpesviruses whose capsids have been characterized in comparable detail are the alphaherpesviruses, EHV-1 (3) and HSV-1 (e.g., references 17 and 73), and channel catfish virus (10), a distant and still-uncategorized relative (19). The molecular architecture of the nuclear B-capsid of SCMV turned out to be quite similar to those already described, a finding consistent with its protein composition being conserved (25, 28). Nevertheless, several differences came to light in the present study.
Limited variability in capsid size among herpesviruses.
Since CMVs have the largest genomes of all herpesviruses, one might expect corresponding adaptations in capsid structure. However, the average inner radius of SCMV (49.5 nm) is only slightly greater than those of HSV-1 (47.2 nm) and channel catfish virus (45.9 nm), whose packing densities are quite similar (9). With the same packing density as HSV-1, the SCMV capsid would accommodate a genome of ∼171 kbp. Although the SCMV genome has not yet been completely sequenced, estimates from restriction enzyme digests indicate that its size is higher at ∼209 kbp (33a). The additional DNA could be accommodated by slightly tighter packing, i.e., reducing the interduplex spacing of HSV-1 (9) by 7% would give a spacing of 2.57 nm, which remains reasonable for packaged viral DNA (cf. 2.54 nm for bacteriophage T7 [15]). In summary, we expect that other herpesvirus genomes are packed in much the same “liquid crystalline” fashion as in HSV-1 (9), whereby substantial variations in genome size may be accommodated by proportionately much smaller changes in capsid dimensions and/or packing density.
The small core of isolated SCMV B-capsids.
Most of these capsids contain an inner shell composed mainly of the assembly protein in its mature, proteolytically processed form (31, 69). Presumably, these structures were originally assembled as procapsid scaffolds (64) and subsequently contracted upon proteolytic processing and maturation of the procapsid, in their transition from large-cored to small-cored B-capsids (49, 62). The large gap between the inner and outer shells (Fig. 4) supports this explanation. Although isolated B-capsids of EHV-1 (3) and HSV-1 (44) were found to contain irregular coagulates of their assembly proteins, their appearance as small cores in thin sections of infected cells (see, for example, reference 47) implies that this material was originally organized like the inner shell of SCMV B-capsids but became disordered during isolation.
Domain structure of assembly protein.
There is strong physical (3, 50) and biochemical (43) evidence that the small cores of herpesvirus B-capsids are composed primarily of the scaffolding-assembly protein in its proteolytically cleaved form. Accordingly, examination of the spherical small core of SCMV may yield some insight into the structure of this key morphogenic protein. We confine this discussion to its optimized radial density profile (Fig. 4, lower curve), which describes the distribution of mass along this molecule. The profile discloses, in addition to some material at the center, a shell that is ∼16 nm thick, with peaks separated by a minimum at a radius of ∼28 nm. Deferring interpretation of the material at the center, this profile implies that the 30-kDa assembly protein is an elongated molecule (∼16 nm) and consists of two domains separated by a low-density linker.
The outer domain is presumably the C-terminal moiety, since the C terminus of the precursor should interact with the surface shell (7, 21, 36, 44, 45, 69, 71). We tentatively suggest that the linker may be an α-helical coiled coil, about four heptads (4 nm) long, based on scanning the SCMV assembly protein sequence (68) with a coiled-coil detection algorithm (40) (unpublished results). Such a coiled coil has been reported for the assembly protein of HSV-1 (45), and we note that the radial profile of the inner shell of the HSV-1 procapsid also exhibits a pronounced minimum (cf. Fig. 6 of reference 64).
Variable tegumentation of cytoplasmic B-capsids.
The origins of cytoplasmic B-capsids and their relationship to the export pathway of maturing virions are not clear. We obtained these capsids in significant amounts only at very late times after infection, when their appearance in the cytoplasm may reflect deteriorating nuclear and/or cytoplasmic compartmentation. We consider it unlikely that they derive from infecting virions, since the majority retain assembly protein, which is absent from virions. However, about one-half of the heavily tegumented B-capsids (∼5% of the total) are coreless, and it is not ruled out that they represent infecting virions that have discharged their genomes. Heavily tegumented cytoplasmic B-capsids may represent a more advanced stage of tegumentation or a subpopulation that fortuitously retained more of its tegument through the isolation procedure. Notwithstanding their origins or maturation prospects, cytoplasmic B-capsids afford the possibility of insight into capsid-tegument interactions.
Interactions between capsid and tegument.
The three classes of cytoplasmic B-capsids were reconstructed separately. Comparing these reconstructions with each other and with the nuclear B-capsid revealed two sites of tegument attachment. On all cytoplasmic B-capsids, each capsomer subunit is capped with an additional protein (Fig. 5c, 7h, and 7i), i.e., this protein is approximately equimolar with the major capsid protein. Although the proteins that overlie hexons and pentons, respectively, are similar in size and shape and binding site on the capsomer protrusions, the penton-associated protein may be slightly longer, and it is not ruled out that more than one species is involved. Although the smallest capsid protein (SCP) is present around the outer rim of hexons, the capsomer-capping protein(s) presumably binds to the major capsid protein, since it is also present on pentons, which lack the SCP (cf. Fig. 6a to c).
The second protein that we have detected has a V-shaped structure, whose tip is anchored on a triplex and whose “legs” diverge outwards. This protein makes contact with the capsomer-capping protein at the tips of neighboring capsomer protrusions, before it extends into the outer region of diffuse density (Fig. 5d and 7f). This triplex-associated protein is faintly visible in the moderately tegumented B-capsid, reflecting low occupancy, and is strongly visible in the heavily tegumented B-capsid, reflecting nearly complete occupancy (cf. Fig. 7e and f). As noted above, it is likely to be a dimer, in which case it should be bound to the minor capsid protein (the counterpart of VP23 of HSV-1 [43]), which is present in two copies per triplex (6). At full occupancy, there would be 640 copies of this protein per capsid.
Candidates for the capsid-binding tegument proteins.
Current evidence does not allow conclusive identification of either protein. However, by correlating their molecular sizes and abundances, as estimated by electron microscopy and with corresponding data from SDS-PAGE, we can identify possible candidates.
The triplex-associated protein Mr value was estimated to be 100,000 to 120,000, which tallies with BPP (∼119,000). Its molar ratio relative to the major capsid protein in this preparation was approximated from the observed proportions of lightly, moderately, and heavily tegumented B-capsids, and its amount on each, as given by its map density relative to that of the major capsid protein and a stoichiometry of two copies per triplex. This calculation yielded a figure of ∼0.15, which is in reasonable agreement with the value obtained for BPP by quantitating the stained gel bands (i.e., 0.13). At full occupancy, the triplex-associated protein would have a molar ratio of 0.67 relative to the major capsid protein, which is close to the value of 0.8 estimated for BPP in virions (37). These correlations suggest BPP as a plausible candidate for this protein.
We estimate the capsomer-capping protein(s) to be ∼50 to 60 kDa and close to equimolar with the major capsid protein. Cytoplasmic B-capsids contain proteins in this size range (cf. Fig. 1), but the complexity and close spacing of the protein bands in this region of the gel precluded reliable quantitation. The only known tegument proteins in this size range that are of appropriate abundance in virions are the matrix proteins, UM and LM (26, 37). UM has been reported to be equimolar with the major capsid protein (25, 37), whereas LM has been found to vary considerably in amount among the different strains of CMV (38) and is not required for virion assembly (55). These considerations may favor UM as a candidate. However, further data are required to test this hypothesis.
Binding of other tegument proteins.
Our reconstructions assume icosahedral symmetry and thus reveal only the most highly ordered part of the tegument, the part in direct contact with the capsid shell. As such, these observations represent only a first step towards an account of the molecular organization of the tegument. Nevertheless, they imply that the coupling of the SCMV tegument to its capsid is not confined to pentons (66) but is instead distributed all over the capsid surface.
Each copy of the major capsid protein, in hexons and pentons alike, is capped with an additional protein. If this capsomer-capping protein is a single species, it differs in this respect from the SCP which binds only to hexons (cf. Fig. 6a) as a consequence of differences between the penton and hexon conformations of the major capsid protein (70). We have conjectured (70) that the absence of SCP from pentons may confer the advantage of diversifying the binding sites presented on the capsid surface, which could facilitate the binding of different tegument proteins. However, if the same capsomer-capping protein binds to both hexons and pentons, this opportunity is not exploited in tegumentation, at least, for SCMV. It may, however, be relevant to interactions in which the capsid engages at other stages of the replication cycle (cf. references 5, 34, 35, 46, and 58).
A diffuse outer halo of density is seen in the maps of moderately and heavily tegumented B-capsids (Fig. 2b, Fig. 4, and Fig. 7b and c), indicating that some proteins are present in this region, albeit in a disordered state. The triplex-associated protein extends into this region, where it may serve as a platform for other tegument components. However, the mechanisms whereby they are selected and bound, and how they may function in envelopment or in other viral functions, remain to be addressed.
ACKNOWLEDGMENTS
We thank T. Baker and J. Conway for software, D. Belnap for helpful discussions, and S. Butcher for dialog on CMV structure.
This work was supported in part by USPHS grant AI13718 to W.G.
REFERENCES
- 1.Baker T S, Cheng R H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J Struct Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 2.Baker T S, Drak J, Bina M. Reconstruction of the three-dimensional structure of simian virus 40 and visualization of the chromatin core. Proc Natl Acad Sci USA. 1988;85:422–426. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker T S, Newcomb W W, Booy F P, Brown J C, Steven A C. Three-dimensional structures of maturable and abortive capsids of equine herpesvirus 1 from cryoelectron microscopy. J Virol. 1990;64:563–573. doi: 10.1128/jvi.64.2.563-573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Chee M S, Hutchison C A, Kouzarides T, Martignetti J A, Preddie E. The DNA sequence of the human cytomegalovirus genome. DNA Seq. 1991;2:1–12. doi: 10.3109/10425179109008433. [DOI] [PubMed] [Google Scholar]
- 5.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral replicative cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baxter M, Gibson W. The putative human cytomegalovirus triplex proteins, minor capsid protein (mCP) and mCP-binding protein (mC-BP), form a heterotrimeric complex that localizes to the cell nucleus in the absence of other viral proteins. 22nd International Herpesvirus Workshop, La Jolla, Calif. 1997. [Google Scholar]
- 7.Beaudet-Miller M, Zhang R, Durkin J, Gibson W, Kwong A D, Hong Z. Viral specific interaction between the human cytomegalovirus major capsid protein and the C terminus of the precursor assembly protein. J Virol. 1996;70:8081–8088. doi: 10.1128/jvi.70.11.8081-8088.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black L W, Showe M K, Steven A C. Morphogenesis of the T4 head. In: Karam J, editor. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 218–258. [Google Scholar]
- 9.Booy F P, Newcomb W W, Trus B L, Brown J C, Baker T S, Steven A C. Liquid-crystalline, phage-like, packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booy F P, Trus B L, Davison A J, Steven A C. The capsid architecture of channel catfish virus, an evolutionarily distant herpesvirus, is largely conserved in the absence of discernible sequence homology with herpes simplex virus. Virology. 1996;215:134–141. doi: 10.1006/viro.1996.0016. [DOI] [PubMed] [Google Scholar]
- 11.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12 kDa VP26) in a large complex (the 200 MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown J H, Cohen C, Parry D A. Heptad breaks in alpha-helical coiled coils: stutters and stammers. Proteins. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 13.Campadelli-Fiume G, Farabegoli F, Di Gaeta S, Roizman B. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J Virol. 1991;65:1589–1595. doi: 10.1128/jvi.65.3.1589-1595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Publishing Corp.; 1988. pp. 15–91. [Google Scholar]
- 15.Cerritelli M E, Cheng N, Rosenberg A H, McPherson C E, Booy F P, Steven A C. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 16.Conway J F, Duda R L, Cheng N, Hendrix R W, Steven A C. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- 17.Conway J F, Trus B L, Booy F P, Newcomb W W, Brown J C, Steven A C. Visualization of three-dimensional density maps reconstructed from cryoelectron micrographs of viral capsids. J Struct Biol. 1996;116:200–208. doi: 10.1006/jsbi.1996.0031. [DOI] [PubMed] [Google Scholar]
- 18.Davison A. Herpesvirus genes. Rev Med Virol. 1998;3:237–244. [Google Scholar]
- 19.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 20.Davison M D, Rixon F J, Davison A J. Identification of genes encoding two capsid proteins (VP24 and VP26) of herpes simplex virus type 1. J Gen Virol. 1992;73:2709–2713. doi: 10.1099/0022-1317-73-10-2709. [DOI] [PubMed] [Google Scholar]
- 21.Desai P, Person S. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology. 1996;220:516–521. doi: 10.1006/viro.1996.0341. [DOI] [PubMed] [Google Scholar]
- 22.Fuller S D. The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 23.Fuller S D, Butcher S J, Cheng R H, Baker T S. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J Struct Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 24.Gibson W. Structural and nonstructural proteins of strain Colburn cytomegalovirus. Virology. 1981;111:516–537. doi: 10.1016/0042-6822(81)90354-8. [DOI] [PubMed] [Google Scholar]
- 25.Gibson W. Protein counterparts of human and simian cytomegaloviruses. Virology. 1983;128:391–406. doi: 10.1016/0042-6822(83)90265-9. [DOI] [PubMed] [Google Scholar]
- 26.Gibson W. Cytomegalovirus protein structure and function. Amsterdam, The Netherlands: Elsevier Science Publishers B. V.; 1991. [Google Scholar]
- 27.Gibson W. Molecular biology of human cytomegalovirus. New York, N.Y: Springer-Verlag; 1993. [Google Scholar]
- 28.Gibson W. Structure and assembly of the virion. Intervirology. 1996;39:389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- 29.Gibson W, Roizman B. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J Virol. 1972;10:1044–1052. doi: 10.1128/jvi.10.5.1044-1052.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson W, van Breemen R, Fields A, LaFemina R, Irmiere A. d,l-alpha-difluoromethylornithine inhibits human cytomegalovirus replication. J Virol. 1984;50:145–154. doi: 10.1128/jvi.50.1.145-154.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson W, Marcy A I, Comolli J C, Lee J. Identification of precursor to cytomegalovirus capsid assembly protein and evidence that processing results in loss of its carboxy-terminal end. J Virol. 1990;64:1241–1249. doi: 10.1128/jvi.64.3.1241-1249.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson W, Clopper K S, Britt W J, Baxter M K. Human cytomegalovirus (HCMV) smallest capsid protein identified as product of short open reading frame located between HCMV UL48 and UL49. J Virol. 1996;70:5680–5683. doi: 10.1128/jvi.70.8.5680-5683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson W, Baxter M K, Clopper K S. Cytomegalovirus “missing” capsid protein identified as heat-aggregable product of human cytomegalovirus UL46. J Virol. 1996;70:7454–7461. doi: 10.1128/jvi.70.11.7454-7461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Hayward, G. Personal communication.
- 34.Hensel G, Meyer H, Gärtner S, Brand G, Kern H F. Nuclear localization of the human cytomegalovirus protein pp150 (ppUL32) J Gen Virol. 1995;76:1591–1601. doi: 10.1099/0022-1317-76-7-1591. [DOI] [PubMed] [Google Scholar]
- 35.Hensel G M, Meyer H H, Buchmann I, Pommerehne D, Schmolke S, Plachter B, Radsak K, Gärtner S, Brand G, Kern H F. Intracellular localization and expression of the human cytomegalovirus matrix phosphoprotein pp71 (ppUL82): evidence for its translocation into the nucleus. J Gen Virol. 1996;77:3087–3097. doi: 10.1099/0022-1317-77-12-3087. [DOI] [PubMed] [Google Scholar]
- 36.Hong Z, Beaudet-Miller M, Burkin J, Zhang R, Kwong A D. Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein. J Virol. 1996;70:533–540. doi: 10.1128/jvi.70.1.533-540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irmiere A, Gibson W. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology. 1983;130:118–133. doi: 10.1016/0042-6822(83)90122-8. [DOI] [PubMed] [Google Scholar]
- 38.Jahn G, Scholl B-C, Traupe B, Fleckenstein B. The two major structural phosphoproteins (pp65 and pp150) of human cytomegalovirus and their antigenic properties. J Gen Virol. 1987;68:1327–1337. doi: 10.1099/0022-1317-68-5-1327. [DOI] [PubMed] [Google Scholar]
- 39.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 41.McNabb D S, Courtney R J. Identification and characterization of the herpes simplex virus type 1 virion protein encoded by the UL35 open reading frame. J Virol. 1994;66:2653–2663. doi: 10.1128/jvi.66.5.2653-2663.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newcomb W W, Homa F L, Booy F P, Thomsen D R, Trus B L, Steven A C, Spencer J V, Brown J C. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J Mol Biol. 1996;263:432–446. doi: 10.1006/jmbi.1996.0587. [DOI] [PubMed] [Google Scholar]
- 43.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 44.Oien N L, Thomsen D R, Wathen M W, Newcomb W W, Brown J C, Homa F L. Assembly of herpes simplex virus capsids using the human cytomegalovirus scaffold protein: critical role of the C terminus. J Virol. 1997;71:1281–1291. doi: 10.1128/jvi.71.2.1281-1291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelletier A, Do F, Brisebois J J, Lagace L, Cordingley M G. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. J Virol. 1997;71:5197–5208. doi: 10.1128/jvi.71.7.5197-5208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penfold M E, Armati P, Cunningham A L. Axonal transport of herpes simplex virions to epidermal cells: evidence for a specialized mode of virus transport and assembly. Proc Natl Acad Sci USA. 1994;91:6529–6533. doi: 10.1073/pnas.91.14.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preston V G, Coates J A, Rixon F J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol. 1983;45:1056–1064. doi: 10.1128/jvi.45.3.1056-1064.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Preston V G, Rixon F J, McDougall I M, McGregor M, Alkobaisi M F. Processing of the herpes simplex virus assembly protein-icp35 near its carboxy terminal end requires the product of the whole of the ul26 reading frame. Virology. 1992;186:87–98. doi: 10.1016/0042-6822(92)90063-u. [DOI] [PubMed] [Google Scholar]
- 49.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 50.Rixon F J, Cross A M, Addison C, Preston V G. The products of herpes simplex virus type 1 gene UL26 which are involved in DNA packaging are strongly associated with empty but not with full capsids. J Gen Virol. 1988;69:2879–2891. doi: 10.1099/0022-1317-69-11-2879. [DOI] [PubMed] [Google Scholar]
- 51.Roffman E, Albert J P, Goff J P, Frenkel N. Putative site for the acquisition of human herpesvirus 6 virion tegument. J Virol. 1990;64:6308–6313. doi: 10.1128/jvi.64.12.6308-6313.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2221–2230. [Google Scholar]
- 53.Roizman B, Furlong D. The replication of herpes viruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. New York, N.Y: Plenum Press; 1974. pp. 229–403. [Google Scholar]
- 54.Schagger H, Von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 55.Schmolke S, Kern H G, Drescher P, Jahn G, Plachter B. The dominant phosphoprotein pp65 of human cytomegalovirus. Virus Res. 1995;69:5959–5968. doi: 10.1128/jvi.69.10.5959-5968.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sherman G, Bachenheimer S L. Characterization of intranuclear capsids made by morphogenic mutants of HSV-1. Virology. 1988;163:471–480. doi: 10.1016/0042-6822(88)90288-7. [DOI] [PubMed] [Google Scholar]
- 57.Smith J D, Harven E. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. II. An ultrastructural study of viral penetration. J Virol. 1974;14:945–956. doi: 10.1128/jvi.14.4.945-956.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spencer J V, Newcomb W W, Thomsen D R, Homa F L, Brown J C. Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid. J Virol. 1998;72:3944–3951. doi: 10.1128/jvi.72.5.3944-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steven A C, Roberts C R, Hay J, Bisher M E, Pun T, Trus B L. Hexavalent capsomers of herpes simplex virus type 2: symmetry, shape, dimensions, and oligomeric status. J Virol. 1986;57:578–584. doi: 10.1128/jvi.57.2.578-584.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steven A C, Hainfeld J F, Trus B L, Steinert P M, Wall J S. Radial distributions of density within macromolecular complexes determined from dark-field electron micrographs. Proc Natl Acad Sci USA. 1984;81:6363–6367. doi: 10.1073/pnas.81.20.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steven A C, Spear P G. Herpesvirus capsid assembly and envelopment. In: Chiu W, Burnett R M, Garcea R L, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 312–351. [Google Scholar]
- 63.Tooze J, Hollinshead M, Reis K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- 64.Trus B L, Booy F P, Newcomb W W, Brown J C, Homa F L, Thomsen D R, Steven A C. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J Mol Biol. 1996;263:447–462. doi: 10.1016/s0022-2836(96)80018-0. [DOI] [PubMed] [Google Scholar]
- 65.Trus B L, Homa F L, Booy F P, Newcomb W W, Thomsen D R, Cheng N, Brown J C, Steven A C. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vernon S K, Lawrence W C, Long C A, Rubin B A, Sheffield J B. Morphological components of herpesvirus. IV. Ultrastructural features of the envelope and tegument. J Ultrastruct Res. 1982;81:163–171. doi: 10.1016/s0022-5320(82)90072-7. [DOI] [PubMed] [Google Scholar]
- 67.Weiner D, Gibson W. Phosphorylation, maturational processing, and relatedness of strain Colburn matrix proteins. Virology. 1983;129:155–169. doi: 10.1016/0042-6822(83)90403-8. [DOI] [PubMed] [Google Scholar]
- 68.Welch A R, McNally L M, Gibson W. Cytomegalovirus assembly protein nested gene family: four 3′-coterminal transcripts encode four in-frame, overlapping proteins. J Virol. 1991;65:4091–4100. doi: 10.1128/jvi.65.8.4091-4100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welch A R, Woods A S, McNally L M, Cotter R J, Gibson W. A herpesvirus maturational protease, assemblin: identification of its gene, putative active site domain, and cleavage site. Proc Natl Acad Sci USA. 1995;88:10792–10796. doi: 10.1073/pnas.88.23.10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wingfield P T, Stahl S J, Thomsen D R, Homa F L, Booy F P, Trus B L, Steven A C. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol. 1997;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wood L J, Baxter M K, Plafker S M, Gibson W. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pUL86) through two different domains. J Virol. 1997;71:179–190. doi: 10.1128/jvi.71.1.179-190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Z H, Prasad B V V, Jakana J, Rixon F J, Chiu W. Protein subunit structures in the herpes simplex virus capsid determined from 400-kV spot-scan electron cryomicroscopy. J Mol Biol. 1994;242:456–469. doi: 10.1006/jmbi.1994.1594. [DOI] [PubMed] [Google Scholar]
- 73.Zhou Z H, He J, Jakana J, Tatman J D, Rixon F J, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]