Abstract
The La autoantigen (also known as SS-B), a cellular RNA binding protein, may shuttle between the nucleus and cytoplasm, but it is mainly located in the nucleus. La protein is redistributed to the cytoplasm after poliovirus infection. An in vitro translation study demonstrated that La protein stimulated the internal initiation of poliovirus translation. In the present study, a part of the La protein was shown to be cleaved in poliovirus-infected HeLa cells, and this cleavage appeared to be mediated by poliovirus-specific protease 3C (3Cpro). Truncated La protein (dl-La) was produced in vitro from recombinant La protein by cleavage with purified 3Cpro at only one Gln358-Gly359 peptide bond in the 408-amino-acid (aa) sequence of La protein. The dl-La expressed in L cells was detected in the cytoplasm. However, green fluorescence protein linked to the C-terminal 50-aa sequence of La protein was localized in the nucleus, suggesting that this C-terminal region contributes to the steady-state nuclear localization of the intact La protein in uninfected cells. The dl-La retained the enhancing activity of translation initiation driven by poliovirus RNA in rabbit reticulocyte lysates. These results suggest that La protein is cleaved by 3Cpro in the course of poliovirus infection and that the dl-La is redistributed to the cytoplasm. dl-La, as well as La protein, may play a role in stimulating the internal initiation of poliovirus translation in the cytoplasm.
Poliovirus polypeptides are generated by cotranslational and posttranslational cleavages of the 247-kDa polyprotein precursor encoded by a unique long open reading frame of the virus RNA. Three virus-coded proteases, 2Apro (specific to Tyr-Gly), 3Cpro (Gln-Gly), and 3CDpro (Gln-Gly) are involved in protein processing during the virus replication (29, 34). 3CDpro may also function in the initiation of poliovirus plus-strand RNA synthesis via RNA-protein complex formation on the plus-strand 5′-end RNA (2, 16). These proteases also play a role in host cell alteration. It is well known that 2Apro induces the shutoff of cap-dependent translation initiation (25). 3Cpro is involved in transcriptional inhibition by the proteolytic cleavage of transcription factors, such as TATA binding protein (TBP), CREB, and Oct-1 (10, 13, 49, 50), and in changes in cell morphology by the cleavage of microtubule-associated protein (MAP-4) (22).
Translation initiation of poliovirus RNA occurs by entry of ribosomes in the internal RNA sequence called the internal ribosome entry site (6, 32). The internal initiation of poliovirus requires host cellular factors other than basic initiation factors. These host cellular factors include La protein (6, 12, 33, 44), polypyrimidine tract binding protein (19), poly(rC) binding protein 1 (PCBP-1), and PCBP-2 (7, 16). While the translation of poliovirus does not occur efficiently in a cell-free translation system prepared from rabbit reticulocyte lysates (RRL), translation is markedly improved by the addition of factors from HeLa cells (8) or recombinant La protein (33).
The La autoantigen, also called SS-B, is a cellular protein that is involved in the initiation and termination of RNA polymerase III transcription. It associates with various small RNA molecules to form La ribonucleoprotein particles (La RNPs). The RNA components of an La RNP are mostly newly synthesized RNA polymerase III transcripts, such as 7S RNA, 5S rRNA, U6 RNA, or Y RNA (42, 43, 47). In addition, some virus-coded RNA species are also bound by La (28), such as adenovirus VAI and VAII RNAs (15, 37), Epstein-Barr virus EBER1 and EBER2 RNAs (37), and the leader RNA of vesicular stomatitis virus (27). Moreover, La protein also binds to sites within the 5′ noncoding regions (NCRs) of poliovirus (32), hepatitis C virus (1), and human immunodeficiency virus (HIV) (9) mRNAs. Interaction of La with these viral mRNA 5′ NCRs stimulates translation initiation (1, 12, 33, 44).
Subcellular immunolocalization studies showed that La protein is located mainly in the nucleus is redistributed to the cytoplasm after poliovirus infection (33). Here we demonstrate that a part of the La protein is converted to a lower-molecular-weight molecule in poliovirus-infected HeLa cells and in HeLa cells expressing 3Cpro. Structural analysis of the in vitro 3Cpro-mediated cleavage product of recombinant La protein indicates that the cleavage site is between Gln358 and Gly359 in the 408-amino-acid (aa) La protein sequence. We further demonstrate that the truncated La protein, in which the C-terminal 50-aa sequence is missing, is distributed in the cytoplasm and retains the host factor activity for internal translation initiation of poliovirus in RRL.
MATERIALS AND METHODS
Cells and viruses.
Suspension-cultured HeLa S3 cells were grown in RPMI 1640 medium with 5% newborn calf serum (NCS) and used for plaque formation and the preparation of poliovirus type 1 Mahoney strain PV1(M)OM (38). Monolayer cultured HeLa S3 cells were grown in Dulbecco modified Eagle medium (DMEM) with 5% NCS. 293 cells were cultured in DMEM with 10% fetal calf serum (FCS) and used for the preparation and titration of adenovirus type 5 (Ad5), the adenovirus vector for the expression of Cre recombinase (AxCANCre) (23, 24), and control adenovirus vector (Adex1W1) (23, 24). Mouse L cells and TgSVA cells established from the kidney of a transgenic mouse carrying the human poliovirus receptor gene (38, 39) were cultured in DMEM with 5% FCS.
Antibodies.
The mouse anti-human La monoclonal antibodies (MAb) La4B6 and SW5 (36, 45) (gifts from M. Bachmann) were used. The rabbit hyperimmune sera to peptides of the C-terminal 15-aa sequence of human La protein and the C-terminal aa sequence of poliovirus 3Cpro were prepared and used as anti-La and anti-3Cpro antibodies, respectively. Rabbit hyperimmune serum to the amino-terminal (N-terminal) aa sequence of poliovirus 3Cpro was also used both as anti-3CDpro and as anti-3Cpro antibodies. The rabbit hyperimmune sera to Cre-recombinase peptides were from I. Saito. The antibody to TBP was from T. Yamamoto and M. Horikoshi.
Construction of plasmid DNAs.
Poliovirus 3Cpro gene was amplified from the cDNA corresponding to the region from nucleotides (nt) 5438 to 5986 of plasmid pOM1 (38) by PCR with the sense primer 5′-CGCGACGCGTACCCGGATGGGACCAGGGTTCGATTACGCAGTG-3′ and the antisense primer 3′-AGTATGAAGTGATGAGTCTCAGTTATTCAGCTGCGCGAG-5′, where the MluI and SalI sites are underlined and the initiation and termination codons are indicated by boldface letters. To construct pCIneo-3C, amplified products were digested with MluI and SalI, purified by gel electrophoresis, and cloned into the MluI and SalI sites of pCIneo (Promega). After confirmation of the nucleotide sequence of the 3Cpro gene, the 3Cpro gene was inserted into SwaI site of pCALNLw (23, 24), and the resultant plasmid was called pCALNLw-3C. To construct plasmid pET3C8 for the production of recombinant 3Cpro carrying a histidine tag at the C terminus, a cDNA corresponding to the nucleotide sequence from nt 5242 (HincII) to nt 6056 (HindIII) of the poliovirus RNA was inserted into the HincII and HindIII sites of pET22b (Novagen). The plasmid pGEX-La for recombinant La production is pGEX-4T-1, with La cDNA isolated from the HeLa cell cDNA library. Plasmid pCIneo-La was a gift from M. Bachmann (46). The DNA sequence corresponding to nt 861 to 1074 of La DNA was amplified by PCR with the sense primer 5′-AGATGCAAATAATGGTAACCTACAATTAAG-3′ and the antisense primer 3′-CAGACCATTTCCTTTTCATGTCAAAGTCATCACTGAGCTCTATG-5′, where the BstEII and XhoI sites are underlined and the termination codons are indicated by boldface letters, from the pCIneo-La DNA template. Plasmids pCIneo-La and pGEX-La were digested with BstEII and XhoI, and the La coding sequence was replaced by the PCR product to prepare plasmids pCIneo-dl-La and pGEX-dl-La, respectively. To construct pGEX-N′-La, DNA fragment corresponding to nt 330 to 666 of La DNA was amplified from the pGEX-La DNA template by PCR using the sense primer 5′-AGCGCAGATCTGTTTATATTAAAGGCTTCC-3′ and antisense primer 3′-TGTTTTCAATCTTCTTCTACGACTTATTATCGCCGGCGGCTGC-5′, where the BglII and NotI sites are underlined and the termination codons are indicated by boldface letters, digested with BglII and NotI, and inserted into the pGEX-La plasmid which had been digested with BglII and NotI. The EcoRI-NotI fragment from pGEX-N′-La was inserted into the EcoRI and NotI sites of pCIneo, resulting in plasmid pCIneo-N′-La. The pSV40 NLS-GFP (nuclear localization signal, green fluorescence protein) plasmid was plasmid pCE321-FL (Clontech) into which the simian virus 40 (SV40) NLS sequence-GFP fusion DNA had been inserted and which was a gift from S. Sugano. The construction of pCIneo-GFP-C′50-La and pCIneo-GFP was carried out as follows. To obtain stronger GFP fluorescence (11, 40), nucleotide substitutions were introduced into the DNA sequence of GFP by using a PCR-based method as described previously (26). The resulting plasmid, designated pGFP536, had F64L, S65T, V163A, I167T, and S175G substitutions. To make pCIneo-GFP, pGFP536 was digested with EcoRI and SalI and cloned into the EcoRI and SalI sites of pCIneo. To construct pCIneo-GFP-C′50-La, the C-terminal portion of La (aa 359 to 408) was fused to the C-terminal end of GFP536 (aa 1 to 226) as described earlier (26), and the fusion gene was cloned into the EcoRI and SalI sites of pCIneo.
Purification of 3C protease.
Escherichia coli BL21(DE3) cells were transformed with plasmid pET3C8 to express a 3Cpro-histidine tag protein and grown at 37°C. At an optical density at 600 nm of 0.5, IPTG (isopropyl-β-d-thiogalactopyranoside) was added at a final concentration of 1 mM, and the cells were further cultured at 37°C for 4 h. The pelleted cells were suspended in phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4) and then disrupted by sonication. After centrifugation at 5,000 × g for 10 min, the pellet was washed once with a binding buffer (0.5 M NaCl, 5 mM imidazole, 20 mM Tris-HCl; pH 8.0) containing 1% Triton X-100 and twice more with the binding buffer alone. The pellet was resuspended in the binding buffer containing 6 M guanidine hydrochloride and placed on ice for 60 min, and the supernatant was then applied onto an Ni-NTA agarose column (Q/Aexpress Type IV Kit; Qiagen). Recombinant 3Cpro was purified by elution with 100 mM imidazole and by dialysis against TE buffer (1 mM EDTA, 10 mM Tris-HCl; pH 8.0) containing 150 mM NaCl, 1 mM dithiothreitol (DTT), and 5% glycerol.
Purification of La protein.
E. coli BL21 cells were transformed with plasmid pGEX-La to express a glutathione S-transferase (GST)-La fusion protein and grown at 37°C. At an optical density at 600 nm of 0.5, IPTG was added to the culture at a final concentration of 1 mM and further cultured at room temperature for 20 h. The pelleted cells were suspended in PBS and disrupted by sonication. Triton X-100 at a final concentration of 1% was added and mixed at 4°C for 30 min. The fusion protein was purified by using glutathione-Sepharose 4B (Pharmacia Biotech). La protein was purified after digestion with thrombin protease (Pharmacia Biotech). Purification of dl-La and N′-La was carried out by a method similar to that for the La protein.
Immunoblot analysis.
Cells were washed in PBS, lysed in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Triton X-100; 0.1% sodium dodecyl sulfate [SDS]; 1% sodium deoxycholate; 1.5 mM phenylmethylsulfonyl fluoride; 1 μg each of aprotinin, leupeptin, and pepstatin A per ml; 1 mM Na3VO4) at 4°C and centrifuged at 15,000 rpm at 4°C for 10 min. The supernatants were heated at 100°C for 3 min in lysis buffer (2% SDS; 50 mM Tris-HCl, pH 6.8; 10% glycerol; 0.1% bromophenol blue; 50 mM DTT), separated by SDS–12% polyacrylamide gel electrophoresis (PAGE) and transferred onto a polyvinylidene difluoride membrane (Millipore). After being blocked with PBST (PBS with 0.3% Tween 20) containing 3% skim milk at 4°C overnight, the membranes were incubated with antibodies for 1 h and then with alkaline phosphatase-conjugated goat secondary antibodies to rabbit or mouse immunoglobulin G (IgG; Bio-Rad) for 1 h. Blots were visualized by enhanced chemiluminescence (ECL; Amersham).
Cell-free translation.
Cytoplasmic extract (S10) was prepared from suspension-cultured HeLa S3 cells as previously described (21, 39). RRL were purchased from Promega. Template RNA was transcribed by T7 RNA polymerase from poliovirus cDNA pOM1 cleaved by XbaI. For each reaction mixture of 12.5 μl, 0.5 μg of RNA, 170 mM potassium acetate, 1.5 mM magnesium acetate, and 10 μCi of [35S]methionine (1,000 Ci/mmol) were used. Reaction with 8.75 μl (200 μg) of RRL was carried out at 30°C for 1 h in the presence or absence of N′-La, dl-La, La, or HeLaS100 extract. Translation products were analyzed by PAGE as described above. The gels were treated with Enlightning (Dupont), dried, and subjected to autoradiography.
Immunofluorescence.
After infection or transfection, the cells were fixed in cold acetone-methanol (2:3) and subjected to indirect immunofluorescence studies. The cells were incubated with La4B6 or SW5 antibody at 37°C for 1 h. After being washed with PBS, the samples were reacted with fluorescein isothiocyanate-labeled anti-mouse IgG (Biosys, SA) at 37°C for 1 h and were then treated with propidium iodide (PI) that has a high affinity for nucleic acids at 37°C for 15 min. Fluorescence was visualized by a confocal laser scanning microscope (MRC1024 system; Bio-Rad).
RESULTS
Establishment of HeLa cells carrying Cre recombinase-inducible 3Cpro expression unit.
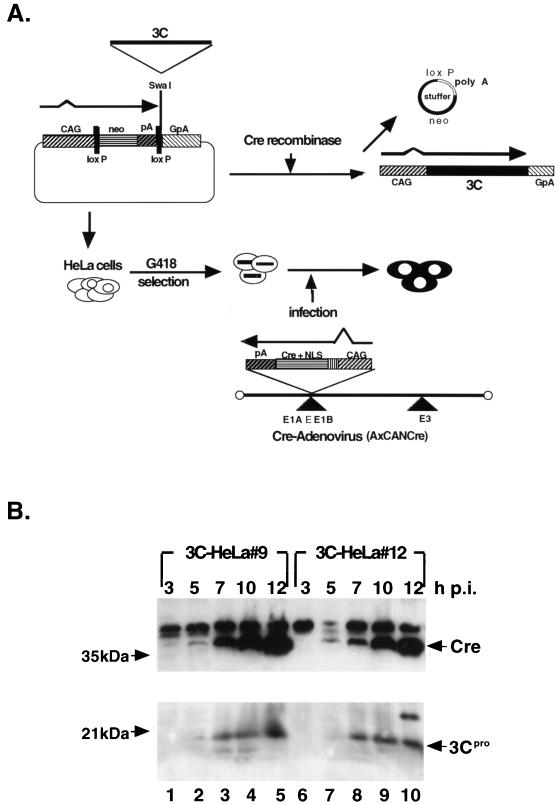
HeLa cells were transfected with plasmid pCALNLw-3C and cultured in medium containing G418. Several G418-resistant cell lines were infected with AxCANCre. 3Cpro-expressing cells were screened by immunoblot analysis with antibodies to 3Cpro. Among several cell lines thus obtained, two cell lines, 3C-HeLa9 and 3C-HeLa12 were used in the following experiments (Fig. 1A). As shown in Fig. 1B, Cre recombinase was detected 5 h after AxCANCre infection, and 3Cpro was detected 7 h after the infection. These proteins were not detected in cells infected with control virus Adex1W1 (data not shown). The cleavage of TBP was observed by immunoblot analysis in those cells infected with AxCANCre (data not shown) as reported earlier (10, 13). These data indicate that 3Cpro is expressed in 3C-HeLa9 and 3C-HeLa12 cells only when the cells are infected with AxCANCre.
FIG. 1.
Establishment of HeLa cells carrying a Cre recombinase-inducible 3Cpro expression unit. (A) Strategy for the establishment of the 3C-HeLa9 and 3C-HeLa12 cells. These cell lines produce poliovirus 3Cpro when the cells are infected with AxCANCre. (B) Expression of 3Cpro. The cells were incubated for periods indicated in the figure after AxCANCre infection. The Cre recombinase and 3Cpro were detected by immunoblot analysis of cell lysate with antibodies to Cre recombinase and to 3Cpro as described in Materials and Methods.
Cleavage of La protein by 3Cpro.
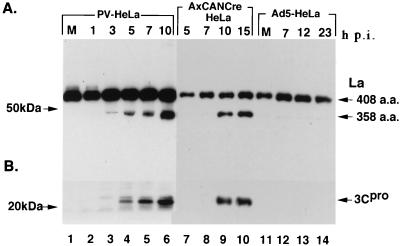
The cleavage of La protein was observed in 3C-HeLa9 and 3C-HeLa12 when 3Cpro was expressed by infection with AxCANCre (Fig. 2), but the cleavage was not detected in these cells infected with Ad5 (Fig. 2) and Adex1W1 (data not shown). During the infection of HeLa cells with poliovirus, 3Cpro was detected 3 h after the infection, and the cleavage product of La protein was slightly detected. The amount of the 3Cpro and the cleavage product of La protein increased with time after infection (Fig. 2). These results suggested that La protein was cleaved by poliovirus 3Cpro. It is possible that La protein is also cleaved by 3CDpro that can be detected slightly earlier than 3Cpro by Western blot analysis (data not shown).
FIG. 2.
Cleavage of La protein by 3Cpro. HeLa cells were incubated for 1, 3, 5, 7, and 10 h after poliovirus infection at 37°C (lanes 2 to 6). 3C-HeLa9 cells were incubated for 5, 7, 10, and 15 h after AxCANCre infection (lanes 7 to 10). HeLa cells were incubated for 7, 12, and 23 h after Ad5 infection (lanes 12 to 14). Mock-infected cells are indicated by an “M” (lanes 1 and 11). The cells were lysed in RIPA buffer and analyzed by immunoblot with MAbs La4B6 and SW5 as described in Materials and Methods.
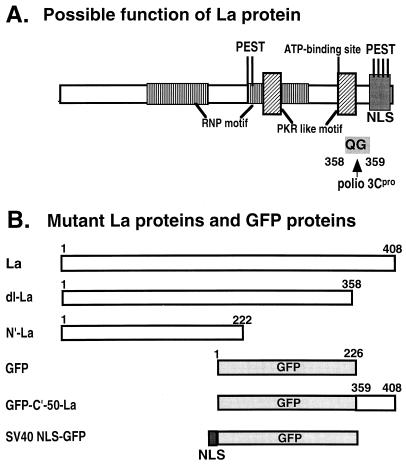
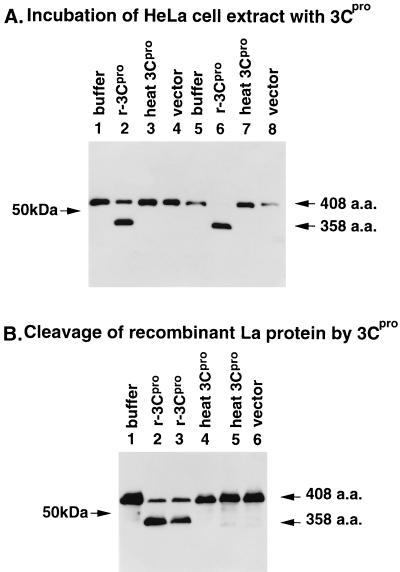
Poliovirus 3Cpro cleaves Gln-Gly bonds. As can be seen from the schematic structure of La protein (see Fig. 4A), only one Gln-Gly pair is present at aa 358 to 359 on the La protein. To determine whether 3Cpro is able to cleave La protein, HeLa cell extract or affinity-purified recombinant La protein was incubated with affinity-purified recombinant 3Cpro, and the products were analyzed by immunoblot analysis with monoclonal antibodies (MAbs) La4B6 and SW5. As shown in Fig. 3A, La protein in HeLa cell extracts was cleaved by purified 3Cpro. After incubation at 30°C for 10 h, almost all of the La protein was cleaved. La protein was not cleaved by heated 3Cpro or by the same fraction from E. coli BL21(DE3) carrying vector plasmid. Recombinant La protein was also cleaved by purified 3Cpro but not by heated 3Cpro (Fig. 3B). The lower band shown in Fig. 3A migrated to the same position as the lower band in Fig. 3B (data not shown). The lower bands in Fig. 3A and B were not detected with La antibody to the C-terminal peptide (data not shown). The C-terminal aa sequence of the lower band of recombinant La protein was determined to identify the cleavage site and was found to be Val-Gln-Phe-Gln. These results indicated that 3Cpro cleaved the Gln358-Gly359 bond in La protein both in vivo and in vitro.
FIG. 4.
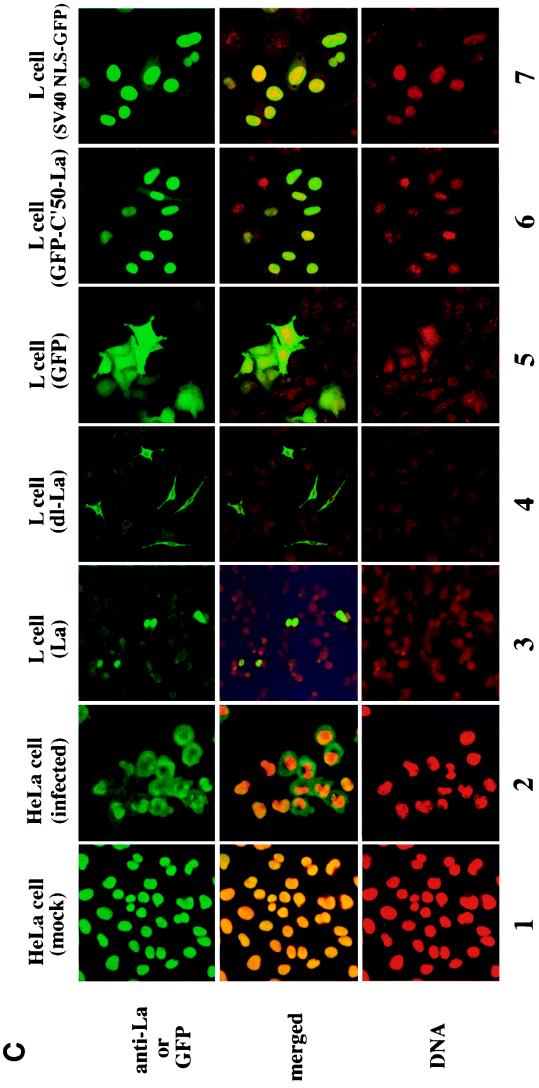
Distribution of La protein and its related proteins in HeLa cells. (A) A possible function of La protein is shown. Poliovirus 3Cpro cleaved the Gln358-Gly359 peptide bond. (B) Structures of mutant La proteins. (C) Immunostaining of La proteins in HeLa cells. Columns: 1, HeLa cells; 2, HeLa cells 6 h after poliovirus infection; 3, pCIneo-La-transfected L cells; 4, pCIneo-dl-La-transfected L cells; 5, pCIneo-GFP transfected L cells; 6, pCIneo-GFP-C′50-La-transfected L cells; 7, pCIneo-GFP-SV40 NLS-transfected L cells. MAb La4B6 was used in the immunostaining (columns 1 to 4). GFP fluorescence was detected at 480 nm with a confocal laser scanning microscope (Bio-Rad) (columns 5 to 7). Top row, immunostainings or GFP fluorescence; bottom row, stainings with PI. In the middle row the pictures are merged.
FIG. 3.
In vitro cleavage of La protein by recombinant 3Cpro. (A) HeLa cell extracts were incubated at 30°C for 5 h (lanes 1 to 4) and 10 h (lanes 5 to 8) with recombinant 3Cpro, heated 3Cpro (70°C, 10 min) and E. coli BL21(DE3) extract with vector DNA in an incubation buffer (5% glycerol, 150 mM NaCl, and 1 mM DTT). The samples were boiled in SDS sample buffer for immunoblot analysis as described in Materials and Methods. (B) Recombinant La protein was incubated at 30°C for 5 h with no protein (lane 1), recombinant 3Cpro preparations 1 (lanes 2 and 4) and 2 (lanes 3 and 5), and E. coli BL21(DE3) extract with vector DNA (lane 6) in an incubation buffer. Immunoblot analysis for the samples was performed as above.
Intracellular redistribution of 3Cpro cleavage products of La protein.
La protein was detected mainly in the nucleus by immunostaining. After poliovirus infection, La protein was redistributed to the cytoplasm (Fig. 4C2; Table 1) (33). Although only a part of the La protein was converted to the dl-La (Fig. 2), it is of interest to determine the distribution of 3Cpro-catalyzed cleavage products of La protein. Plasmids carrying cDNAs of intact La protein (aa 1 to 408), dl-La (aa 1 to 358), N′-La (aa 1 to 222), GFP, and GFP-C′50-La (Fig. 4B) were constructed and designated pCIneo-La, pCIneo-dl-La, pCIneo-N′-La, pCIneo-GFP, and pCIneo-GFP-C′50-La. Plasmid pSV40 NLS-GFP was used as a control for NLS function (Fig. 4B and C). L cells were transfected with these plasmids and, after 20 to 28 h, the cells were fixed for immunostaining with MAb La4B6 or SW5. As shown in Fig. 4C, La protein was detected in the nucleus of HeLa cells (Fig. 4C1) and L cells (Fig. 4C3), and at 6 h after poliovirus infection La protein was detected mainly in the cytoplasm of HeLa cells (Fig. 4C2). dl-La (Fig. 4C4) and N′-La (data not shown) were detected in the cytoplasm of L cells. GFP was present in both the cytoplasm and the nucleus (Fig. 4C5), and GFP-C′50-La, as well as SV40 NLS-GFP, was detected in the nucleus of L cells (Fig. 4C6 and 4C7). These results suggest that the C-terminal 50-aa sequence contributes to the nuclear localization of GFP as well as to the intact La protein. TgSVA cells carrying La, dl-La, and N′-La cDNAs, the expression of which was controlled by infection of AxCANCre, were established. The immunolocalization experiments with La, dl-La, and N′-La with these cell lines showed the same results as in the transient-transfection experiments shown in Fig. 4 (data not shown). These results suggest that the dl-La produced from La protein is distributed to the cytoplasm.
TABLE 1.
Immunofluorescence study of La protein and poliovirus antigens in poliovirus-infected HeLa cells
| Time (h) postinfection at 100 PFU/cell | % Immunofluorescence of:
|
|||||
|---|---|---|---|---|---|---|
| La proteina
|
Poliovirus antigensb
|
|||||
| Strongly in nucleus | Weakly in whole cell | Strongly in cytoplasm | Weakly in cytoplasm | Strongly near nucleus | Strongly in cytoplasm | |
| 0 | 100c | 0 | 0 | 0 | 0 | 0 |
| 1 | 100 | 0 | 0 | 0 | 0 | 0 |
| 2 | 80 | 20d | 0 | 20f | 0 | 0 |
| 3 | 40 | 50 | 10e | 70 | 10g | 0 |
| 4 | 10 | 60 | 30 | 50 | 30 | 10h |
| 5 | 5 | 5 | 90 | 10 | 30 | 50 |
| 6 | 5 | 5 | 90 | 10 | 20 | 60 |
La protein was detected with MAbs La4B6 and SW5.
Poliovirus antigens were detected with rabbit hyperimmune serum to poliovirus type 1.
La protein was detected only in the nucleus of about 100% of cells, as shown in Fig. 4C1.
La protein was distributed both in the nucleus and in the cytoplasm of about 20% of cells.
La protein was detected only in the cytoplasm of about 10% of cells, as shown in Fig. 4C2.
Poliovirus antigens were detected relatively weakly in the cytoplasm of about 20% of cells.
Poliovirus antigens were detected strongly near the nucleus of about 10% of cells.
Poliovirus antigens were detected strongly in a whole cytoplasm of about 10% of cells.
The kinetics of distribution of the La protein that might include the dl-La, in poliovirus-infected cells, demonstrated that La redistribution after poliovirus infection appeared to roughly parallel the switch from cellular translation to poliovirus-specific translation (Table 1). Subcellular localization of La protein in the cytoplasm observed about 4 h postinfection appeared to be the same as that of poliovirus antigens, that is, bright fluorescence spots in certain cytoplasmic regions near the nucleus (data not shown). A similar phenomenon has been reported by Meerovitch et al. (33) with CV-1 cells infected with poliovirus. However, it is still possible that the proteolytic processing of La is an inconsequential modification of another cellular factor.
Effect of dl-La on poliovirus translation initiation in RRL.
It is of interest to determine whether dl-La retains a stimulatory activity for poliovirus translation initiation, since the subcellular localization is mainly in the cytoplasm, the site of poliovirus replication.
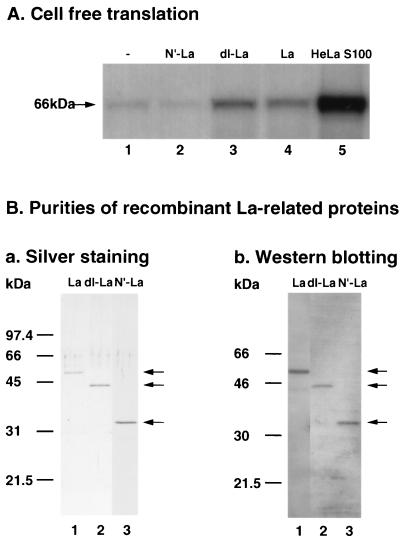
Recombinant La, dl-La, and N′-La proteins were purified as described in Materials and Methods (Fig. 5B). To examine these recombinant proteins for their stimulatory effects on poliovirus translation initiation, we employed RRL in which the efficiency of the internal translation initiation of poliovirus was very low. A 66-kDa in vitro translation product, a truncated capsid protein of poliovirus, was barely detectable in poliovirus RNA-programmed RRL (Fig. 5A, lane 1). The efficiency was enhanced by the addition of HeLa cell S100 extracts or La protein as reported previously (33, 44) (Fig. 5A, lanes 4 and 5). The dl-La, as well as La protein, stimulated the synthesis of the 66-kDa protein, but N′-La did not (Fig. 5A, lanes 2 and 3). These results suggested that the dl-La produced by 3Cpro in poliovirus-infected cells was localized in the cytoplasm and stimulated the poliovirus internal initiation of translation.
FIG. 5.
Effect of dl-La on poliovirus RNA translation in RRL. (A) Effect of recombinant La protein on cell-free translation. Template RNA (0.5 μg) was incubated in RRL containing [35S]methionine and 1 μg of N′-La, 0.7 μg of dl-La, 0.7 μg of La, or 50 μg of HeLa S100 extract as described in Materials and Methods. The mixture was boiled in SDS sample buffer for the immunoblot as described in the legend to Fig. 3. (B) Purities of recombinant La-related proteins. La-related proteins (lane 1, La; lane 2, dl-La; lane 3, N′-La) were purified as described in Materials and Methods and separated by SDS-PAGE. The results of silver staining of the gel (a) and immunoblot analysis with MAb La4B6 (b) are shown. Arrows on the right side of each figure indicate the positions of La protein, dl-La, and N′-La.
DISCUSSION
The La antigen is distributed mainly in the nucleus, but it may be shunted between the nucleus and the cytoplasm (4). A number of functions have been assigned to the La protein. In the nucleus, La protein binds the UUUOH sequence, which is the 3′ terminus of most newly synthesized polymerase III transcripts, and a part of La protein bound to RNA is exported to the cytoplasm from the nucleus (43). The La protein is involved in the initiation and termination of RNA polymerase III transcription (17, 30, 31). Fan et al. (14) have reported that RNA synthesis from isolated polymerase III transcription complex is inhibited by phosphorylation on Ser-366 in the La protein and is reversible by dephosphorylation. Goodier et al. (17) have shown by using in vitro reaction that a C-terminal basic region (aa 328 to 336) of the La protein is important for its activity as an RNA polymerase III transcription factor. The dl-La produced by poliovirus 3Cpro in the present study contained aa 328 to 336 but not Ser-366. Therefore, it is of interest to determine whether dl-La plays a role in RNA polymerase III transcription in poliovirus-infected cells, even though most dl-La is located in the cytoplasm (Fig. 4C).
La protein binds several viral RNAs, including those of poliovirus and HIV, and enhances their translation in vitro (1, 6, 9, 12, 33, 44). Svitkin et al. (44) reported that aa 1 to 194 of La protein possessed RNA-binding specificity for the 5′ NCR of poliovirus RNA but did not stimulate protein synthesis in a poliovirus RNA-programmed RRL. Recently, Craig et al. (12) reported that aa 1 to 380 of the La protein could enhance poliovirus translation but that aa 1 to 293 of the La protein could not. They concluded that aa 293 to 348 of the La protein was a functional domain that promotes homodimerization and is absolutely required for the enhancement of translation of poliovirus RNA in vitro by La. As shown in Fig. 5, dl-La (aa 1 to 358) enhanced the translation of poliovirus in vitro. Our data were consistent with the results of Craig et al. (12). The mechanism by which the La protein enhances the internal translation of poliovirus in virus-infected cells remains unclear. If La protein is necessary for poliovirus translation and replication in HeLa cells, the double-stranded RNA unwinding activity of La protein (5, 20, 48) may be important. La protein may interact with a subset of small ribosomal subunits and may directly bind to 18S ribosomal RNA (35). This interaction may also be related to the role of this protein in translational regulation.
Microinjection of mutant La proteins to Xenopus laevis oocytes indicated that the nuclear import signal of La protein probably resides in the C-terminal region between aa 382 and aa 408, a section which contains a sequence that resembles the consensus bipartite NLS (41). As shown in Fig. 4, the cis nuclear import element of the La protein may reside within aa 359 to 408. This observation is compatible with the previous report above. The dl-La was detected in the cytoplasm in this study, although Simons et al. (41, 42) reported that the sequences between aa 266 and 269 and aa 313 and 337 were the signals for nuclear retention. It is of interest to determine whether the C-terminal 50-aa sequence of La protein interacts with importines α and β (18).
Various molecular weights of proteins reactive to anti-La protein antibodies have been reported. The La protein may be easily cleaved by proteases during extraction processes from cells. Indeed, there are two PEST (Pro, Glu, Ser, and Thr)-rich regions which are putative target aa sequences of ubiquitine in the central and C-terminal regions of La protein (Fig. 4A) (41). However, cleaved La proteins may only account for a small proportion of La in cells, since an La protein of 52 kDa was obtained as a single band from uninfected cells in this study (Fig. 2 and 3). This observation suggests that the 48-kDa dl-La is not derived from random proteolysis.
After poliovirus infection, La protein in most cells is redistributed mainly to the cytoplasm. Immunoblot analysis indicated that the amount of intact 52-kDa La protein was more abundant than the 48-kDa dl-La protein in infected cells during the course of poliovirus replication, which usually came to the end 7 to 8 h after the infection began (Fig. 2). Thus, intact La protein must also be translocated to the cytoplasm in poliovirus-infected cells. The reason for this phenomenon is not clear at present. Redistribution of La protein may not be solely due to 3Cpro function but may also depend on altered cellular metabolisms caused by poliovirus replication. Indeed, the relative amount of La-related proteins redistributed in the cytoplasm in 3Cpro-expressing HeLa cells was less than in poliovirus-infected HeLa cells (data not shown). It has been reported that intracellular redistribution (cytoplasmic accumulation) of La protein also occurs under certain stress conditions, such as UV irradiation (3) and inhibition of RNA synthesis (4). Without cleavage by 3Cpro, La protein may be redistributed under certain conditions of stress induced by virus infection. The pleiotropic effects caused by virus infection are complicated, and these effects may be advantageous for virus replication.
Finally, it should be noted that the cleavage of La protein and/or redistribution of the protein to the cytoplasm may result in the inhibition of cellular reactions in the nucleus that require La protein. This would be one of several examples of how cellular functions are inhibited in poliovirus-infected cells.
ACKNOWLEDGMENTS
We are grateful to M. Bachmann for providing monoclonal antibodies to La and cDNA constructs of La. We are very grateful to N. Imamoto, E. Yoneda, K. Onodera, S. Sugano, and I. Saito for helpful comments and suggestions. We thank Nisei Sangyou Co., Ltd., for sequencing of dl-La protein C-terminal end. We thank Y. Sasaki and K. Iwasaki for expert technical assistance and E. Suzuki and M. Watanabe for help in preparation of the manuscript.
This work was supported in part by a grant-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan and the Ministry of Health and Welfare of Japan and by funds from the Science and Technology Agency of Japan.
REFERENCES
- 1.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino R, Rieckhof G E, Achacoso P L, Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann M, Chang S, Slor H, Kukulies J, Müller W E G. Shuttling of the autoantigen La between nucleus and cell surface after UV irradiation of human keratinocytes. Exp Cell Res. 1990;191:171–180. doi: 10.1016/0014-4827(90)90002-r. [DOI] [PubMed] [Google Scholar]
- 4.Bachmann M, Pfeifer K, Schröder H C, Müller W E G. The La antigen shuttles between the nucleus and the cytoplasm in CV-1 cells. Mol Cell Biochem. 1989;85:103–114. doi: 10.1007/BF00577106. [DOI] [PubMed] [Google Scholar]
- 5.Bachmann M, Pfeifer K, Schröder H C, Müller W E G. Characterization of the autoantigen La as a nucleic acid-dependent ATPase/dATPase with melting properties. Cell. 1990;60:85–93. doi: 10.1016/0092-8674(90)90718-t. [DOI] [PubMed] [Google Scholar]
- 6.Belsham G J, Sonenberg N, Svitkin Y V. The role of the La autoantigen in internal initiation. Curr Top Microbiol Immunol. 1989;203:85–98. doi: 10.1007/978-3-642-79663-0_4. [DOI] [PubMed] [Google Scholar]
- 7.Blyn L B, Towner J S, Semler B L, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown B A, Ehrenfeld E. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology. 1979;97:396–405. doi: 10.1016/0042-6822(79)90350-7. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y-N, Kenan D J, Keene J D, Gatignol A, Jeang K-T. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark M E, Lieberman P M, Berk A J, Dasgupta A. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol Cell Biol. 1993;13:1232–1237. doi: 10.1128/mcb.13.2.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 12.Craig A W B, Svitkin Y V, Lee H S, Belsham G J, Sonenberg N. The La autoantigen contains a dimerization domain that is essential for enhancing translation. Mol Cell Biol. 1997;17:163–169. doi: 10.1128/mcb.17.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das S, Dasgupta A. Identification of the cleavage site and determinants required for poliovirus 3CPro-catalyzed cleavage of human TATA-binding transcription factor TBP. J Virol. 1993;67:3326–3331. doi: 10.1128/jvi.67.6.3326-3331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan H, Sakulich A L, Goodier J L, Zhang X, Qin J, Maraia R J. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]
- 15.Francoeur A M, Mathews M B. Interaction between VA RNA and the lupus antigen La: formation of a ribonucleoprotein particle in vitro. Proc Natl Acad Sci USA. 1982;79:6772–6776. doi: 10.1073/pnas.79.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamarnik A V, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 17.Goodier J L, Fan H, Maraia R J. A carboxy-terminal basic region controls RNA polymerase III transcription factor activity of human La protein. Mol Cell Biol. 1997;17:5823–5832. doi: 10.1128/mcb.17.10.5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Görlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellen C U T, Witherell G W, Schmid M, Shin S H, Pestova T V, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hühn P, Pruijn G J M, van Venrooij W J, Bachmann M. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res. 1997;25:410–416. doi: 10.1093/nar/25.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii T, Shiroki K, Hong D-H, Aoki T, Ohta Y, Abe S, Hashizume S, Nomoto A. A new internal ribosomal entry site 5′ boundary is required for poliovirus translation initiation in a mouse system. J Virol. 1998;72:2398–2405. doi: 10.1128/jvi.72.3.2398-2405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joachims M, Harris K S, Etchison D. Poliovirus protease 3C mediates cleavage of microtubule-associated protein 4. Virology. 1995;211:451–461. doi: 10.1006/viro.1995.1427. [DOI] [PubMed] [Google Scholar]
- 23.Kanegae Y, Takamori K, Sato Y, Lee G, Nakai M, Saito I. Efficient gene activation system on mammalian cell chromosomes using recombinant adenovirus producing Cre recombinase. Gene. 1996;181:207–212. doi: 10.1016/s0378-1119(96)00516-1. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi M, Nagata S, Kita Y, Nakatsu N, Ihara S, Kaibuchi K, Kuroda S, Ui M, Iba H, Konishi H, Kikkawa U, Saitoh I, Fukui Y. Expression of a constitutively active phosphatidylinositol 3-kinase induces process formation in rat PC12 cells. J Biol Chem. 1997;272:16089–16092. doi: 10.1074/jbc.272.26.16089. [DOI] [PubMed] [Google Scholar]
- 25.Kräusslich H G, Nicklin M J H, Toyoda H, Etchison D, Wimmer E. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J Virol. 1987;61:2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuge S, Jones N, Nomoto A. Regulation of yAp-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurilla M G, Keene J D. The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1983;34:837–845. doi: 10.1016/0092-8674(83)90541-x. [DOI] [PubMed] [Google Scholar]
- 28.Lai M M C. Cellular factors in the transcription and replication of viral RNA genomes: A parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 29.Lawson M A, Semler B L. Poliovirus thiol proteinase 3C can utilize a serine nucleophile within the putative catalytic triad. Proc Natl Acad Sci USA. 1991;88:9919–9923. doi: 10.1073/pnas.88.22.9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maraia R J. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc Natl Acad Sci USA. 1996;93:3383–3387. doi: 10.1073/pnas.93.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maraia R J, Kenan D J, Keene J D. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implication for internal translation initiation. Genes Dev. 1989;3:1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 33.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicklin M J, Kräusslich H G, Toyoda H, Dunn J J, Wimmer E. Poliovirus polypeptide precursors: expression in vitro and processing by exogenous 3C and 2A proteinase. Proc Natl Acad Sci USA. 1987;84:4002–4006. doi: 10.1073/pnas.84.12.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peek R, Pruijn G J M, van Venrooij W J. Interaction of the La (SS-B) autoantigen with small ribosomal subunits. Eur J Biochem. 1996;236:649–655. doi: 10.1111/j.1432-1033.1996.0649d.x. [DOI] [PubMed] [Google Scholar]
- 36.Pruijn G J, Thijssen J P H, Smith P R, Williams D G, van Venrooij W J. Anti-La monoclonal antibodies recognizing epitopes within the RNA-binding domain of the La protein show differential capacities to immunoprecipitate RNA-associated La protein. Eur J Biochem. 1995;232:611–619. doi: 10.1111/j.1432-1033.1995.611zz.x. [DOI] [PubMed] [Google Scholar]
- 37.Rosa M D, Gottlieb E, Lerner M R, Steitz J A. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiroki K, Ishii T, Aoki T, Kobashi M, Ohka S, Nomoto A. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J Virol. 1995;69:6825–6832. doi: 10.1128/jvi.69.11.6825-6832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shiroki K, Ishii T, Aoki T, Ota Y, Yang W-X, Komatsu T, Ami Y, Arita M, Abe S, Hashizume S, Nomoto A. Host range phenotype induced by mutations in the internal ribosomal entry site of poliovirus RNA. J Virol. 1997;71:1–8. doi: 10.1128/jvi.71.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siemering K R, Golbik R, Sever R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- 41.Simons F H M, Broers F J M, van Venrooij W J, Pruijn G J M. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp Cell Res. 1996;224:224–236. doi: 10.1006/excr.1996.0132. [DOI] [PubMed] [Google Scholar]
- 42.Simons F H, Rutjes S A, van Venrooij W J, Pruijn G J. The interactions with Ro60 and La differentially affect nuclear export of hY1 RNA. RNA. 1996;2:264–273. [PMC free article] [PubMed] [Google Scholar]
- 43.Stefano J E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell. 1984;36:145–154. doi: 10.1016/0092-8674(84)90083-7. [DOI] [PubMed] [Google Scholar]
- 44.Svitkin Y V, Meerovitch K, Lee H S, Dholakia J N, Kenan D J, Agol V I, Sonenberg N. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tröster H, Bartsch H, Klein R, Metzger T E, Pollak G, Semsei I, Schwemmle M, Pruijn Ger J M, van Venrooij W J, Bachmann M. Activation of a murine autoreactive B cell by immunization with human recombinant autoantigen La/SS-B: characterization of the autoepitope. J Autoimmun. 1995;8:825–842. doi: 10.1016/s0896-8411(95)80020-4. [DOI] [PubMed] [Google Scholar]
- 46.Tröster H, Metzger T E, Semsei I, Schwemmle M, Winterpacht A, Zabel B, Bachmann M. One gene, two transcripts: isolation of an alternative transcript encoding for the autoantigen La/SS-B from a cDNA library of a patient with primary Sjögrens’ syndrome. J Exp Med. 1994;180:2059–2067. doi: 10.1084/jem.180.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willis I M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 48.Xiao Q, Sharp T V, Jeffrey I W, James M C, Pruijn G J, van Venrooij W J, Clemens M J. The La antigen inhibits the activation of the interferon-inducible protein kinase PKR by sequestering and unwinding double-stranded RNA. Nucleic Acids Res. 1994;22:2512–2518. doi: 10.1093/nar/22.13.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yalamanchili P, Datta U, Dasgupta A. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J Virol. 1997;71:1220–1226. doi: 10.1128/jvi.71.2.1220-1226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yalamanchili P, Weidman K, Dasgupta A. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology. 1997;239:176–185. doi: 10.1006/viro.1997.8862. [DOI] [PubMed] [Google Scholar]