Abstract
Solid-state nuclear magnetic resonance (ssNMR) methods can probe the motions of membrane proteins in liposomes at the atomic level and propel the understanding of biomolecular processes for which static structures cannot provide a satisfactory description. In this work, we report our study on the fluoride channel Fluc-Ec1 in phospholipid bilayers based on ssNMR and molecular dynamics simulations. Previously unidentified fluoride binding sites in the aqueous vestibules were experimentally verified by 19F-detected ssNMR. One of the two fluoride binding sites in the polar track was identified as a water molecule by 1H-detected ssNMR. Meanwhile, a dynamic hotspot at loop 1 was observed by comparing the spectra of wild-type Fluc-Ec1 in variant buffer conditions or with its mutants. Therefore, we propose that fluoride conduction in the Fluc channel occurs via a “water-mediated knock-on” permeation mechanism and that loop 1 is a key molecular determinant for channel gating.
Fluoride permeation in Fluc channels occurs by water-mediated knock-on.
INTRODUCTION
Fluoride (F−) is ubiquitous in the aquatic biosphere and acts as an exogenous inhibitor of metalloenzymes essential for fundamental biological processes (1, 2). Microorganisms protect themselves against external fluoride through membrane transporters. The CLCF family of F−/H+ antiporters and the Fluc (fluoride channel) family of fluoride ion channels, derived from two evolutionally independent protein branches, are responsible for exporting F− and thereby undermining the cytoplasmic accumulation of this toxic halide (3–6). Fluoride channels encoded by the crcB genes are referred to as Fluc in bacteria and FEX (fluoride exporter) in eukaryotes (e.g., fungi, plants, and filter-feeding ocean animals such as sponges and sea anemones) (3, 5).
Crystal structures of representative Fluc channels from an Escherichia coli virulence plasmid (Fluc-Ec2) and Bordetella pertussis (Fluc-Bpe) with monobodies provide detailed insights into the molecular basis of how fluoride ions are transported in Flucs (7–9). Sequence alignment of Fluc-Ec1 (investigated in this study), Fluc-Ec2, and Fluc-Bpe is provided in fig. S1. Fluc channels are characterized by a unique structural architecture in that two subunits are arranged in a dimeric antiparallel orientation with two fluoride permeation pathways, such that the intracellular and extracellular ion entry pathways are structurally identical (7, 10, 11). Although assembled as a dual-topology architecture, the two permeation pathways work independently (12). Five disconnected electron densities observed in the Fluc crystal structure were interpreted as four fluoride ions along transmembrane helix TM4 and one sodium ion (Na+) in the center of the plug separating the vestibules (7, 13). The interfacial nontransported sodium is required to stabilize the active state of the channel (14). A ladder of modestly conserved polar residues acting as H-bond donors (Ser102, His106, Ser110, Thr114 on TM4, Asn41 on TM2, and Ser84 on TM3b) constitutes a “polar track” demarking the ion conduction pathway in the crystal structure of the Ec2-S9 complex [Protein Data Bank (PDB) ID: 5A43] (15). Four fluoride ions, labeled as F1 and F2 in noncrystallographic symmetry–related pairs, are coordinated by dipolar H-bond donors in the polar track and the electropositive edges of centrally located Phe rings (Phe80 and Phe83) (12, 15). Recent x-ray crystallographic studies revealed that bromide (Br−) resides in the funnel-shaped, electropositive vestibule, demonstrating that this position (hereafter, F0) may function as the entrance of the permeation pathway (9).
The Fluc channels are remarkable for this idiosyncratic dual-topology architecture and for exceedingly high F− selectivity (>104 fold) over the close analog chloride (Cl−) (10, 11). Although mutations were designed to investigate the correlation between structure and function, no modification has been found that allows for the transportation of any other anion (7, 15). Meanwhile, more inexplicable phenomena were observed during these mutation analyses. For instance, the highly similar crystal structures of the F80M and F83M mutants offer no explanation for why F80M is functionally active and F83M is not (15). The assignment of fluoride ions in the polar track aroused controversy as well. Ariz-Extreme and Hub (16) challenged the assignment of the fluoride ions in the crystal structures based on molecular dynamics (MD) simulations, and Yue et al. (17) observed stable binding of fluoride at the F1 site using polarizable force fields. In addition, both McIlwain et al. (9) and Yue et al. (17) proposed that side-chain rotamerization accompanies fluoride movement to the F1 site and suggested that this precludes simultaneous occupancy of both F1 and F2. Thus, the ion permeation and regulatory mechanism of Fluc channels remain under debate.
In this study, we combined solid-state nuclear magnetic resonance (ssNMR) spectroscopy and MD simulations to investigate the detailed permeation mechanism of CrcB (Fluc-Ec1, from E. coli K12 strains) without monobodies. Contrary to other methods in structural biology, ssNMR can be used to study the structural dynamics of membrane proteins in native-like lipid bilayers at room temperature and under physiological buffer conditions (18–24). The additional fluoride binding sites F0 in the vestibules were confirmed by 13C chemical shift perturbation and 19F spin diffusion experiments. MD simulations starting from the Fluc-Ec2 crystal structures (PDB ID: 6BX4 and 7KKR) suggested that both fluoride and chloride could transiently explore the vestibular binding sites, indicating the nonselective anion binding features of F0. With a series of 1H-detected and 19F-detected ssNMR data, we found that fluoride ions reside in F2 sites and water molecules occupy the F1 sites of Fluc-Ec1 in a high F− concentration environment. Meanwhile, the structural plasticity of loop 1 was identified as one key molecular determinant for Fluc channel gating by analyzing ssNMR spectra of Fluc-Ec1 in an F−-free environment for wild-type (WT) and F80M/F83M mutants. On the basis of our combined experimental and computational results, we propose a “water-mediated knock-on” model in which water molecules are cotransported with fluoride ions and conduction through the pore occurs by alternating water molecules and fluoride ions. These results not only provide valuable insights into the fluoride conduction mechanism of Fluc-Ec1 but also have important implications for our deeper understanding of fluoride channels.
RESULTS
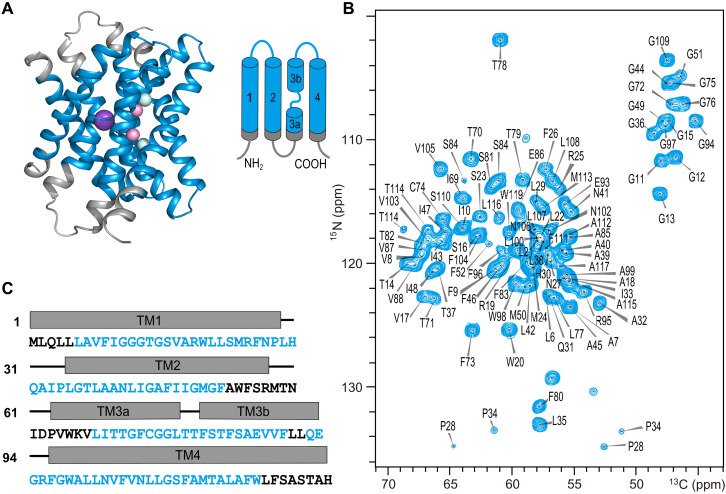
Sequential assignments of Fluc-Ec1 in liposomes were obtained by ssNMR
The Fluc-Ec1 protein was overexpressed in E. coli, purified by immobilized metal affinity chromatography, and reconstituted into E. coli total lipid extract liposomes. Fluoride efflux from liposomes measurements using 19F-detected solution NMR verified that reconstituted Fluc-Ec1 in the lipid bilayer is functional (fig. S2) (6). To investigate the conformation of Fluc-Ec1 in lipid bilayers, we recorded a series of 13C-detected ssNMR spectra of Fluc-Ec1. De novo sequential backbone and side-chain chemical shift assignments were obtained from two-dimensional (2D) and 3D correlation spectra (25) of the uniformly [13C, 15N]-labeled Fluc-Ec1 in liposomes under high F− concentration conditions (150 mM NaF). For illustration, a sequential walk from Ala18 to Gly13 is shown in fig. S3. All visible signals in the 3D spectra could be assigned to 97 residues, which are highlighted in the structure model and amino acid sequence of Fluc-Ec1 (Fig. 1). Unassigned residues can be segmented into three nearby regions on the Fluc-Ec1 structure: (i) A53-V67, the loop connecting TM2 and TM3; (ii) the N terminus up to L5; and (iii) residues L120 up to the C terminus. The signals from these regions cannot be detected in dipolar coupling–based experiments, most likely due to conformational heterogeneity caused by strong structural dynamics. The signals from flexible residues, as observed in scalar coupling–based experiments, strongly correlate with the residue types found on the loop between TM2 and TM3, C- and N-termini (fig. S4). Secondary chemical shifts confirm that the protein is predominantly α helical (fig. S5) (26), in line with the secondary structure of the x-ray–determined Fluc channel structures from E. coli or B. pertussis (7).
Fig. 1. High-resolution ssNMR spectrum of Fluc-Ec1.
(A) Structural model of Fluc-Ec1 based on the crystal structure of the Fluc channel (PDB ID: 5NKQ). Fluoride ions are shown as pink (F1 sites) or cyan (F2 sites) spheres, and sodium as a purple sphere. (B) Assigned 2D NiCAi correlation spectrum of Fluc-Ec1 in the presence of 150 mM F−. (C) Amino acid sequence of Fluc-Ec1 with transmembrane (rectangle) and loop (short line) annotation based on the crystal structure indicated above the sequence. Assigned residues are highlighted in blue, and unassigned residues are shown in black. ppm, parts per million.
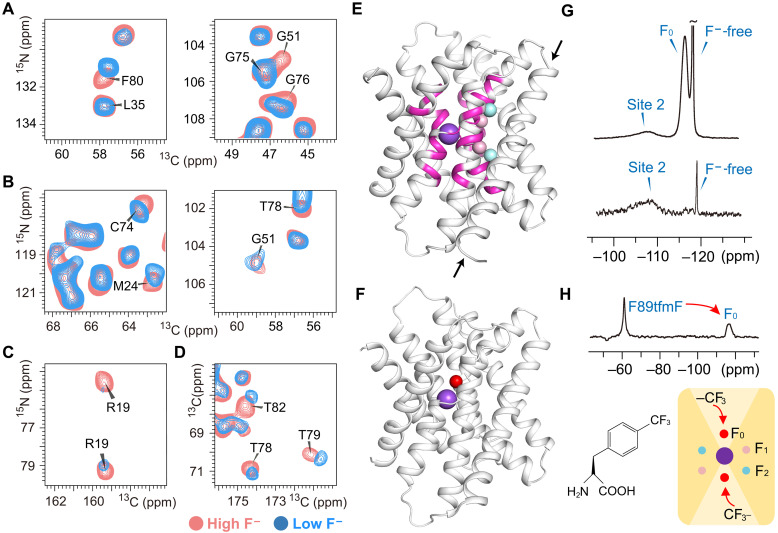
Previously unidentified fluoride ion binding sites (F0 sites) were characterized in the aqueous vestibules
To discriminate the fluoride binding sites, we tried to compare the Fluc-Ec1 spectra in the absence of F− with the spectra recorded in the presence of F− ions. However, the purified Fluc-Ec1 could not be successfully reconstituted into liposomes without F−. As an alternative, a “low-F− sample” was made by washing the Fluc-Ec1 proteoliposome sample with 150 mM NaCl buffer for approximately 1 week. Two-dimensional correlation spectra of Fluc-Ec1 in different F− environments with high resolution were recorded, including NiCAi, NiCOCAi-1, NiCOi-1, and 13C–13C proton-driven spin diffusion (PDSD) spectra (figs. S6 to S9) (27). Chemical shift perturbation data show that residues with notable chemical shift changes are located in the vestibules instead of the TM4 polar face which contributes to the fluoride permeating polar track (Fig. 2, A to E). Peak intensity of the conserved vestibule arginine (Arg19) side chain decreased (Fig. 2C), indicating increased structural dynamics of this site in the low-F− sample. Thus, there should be F− ions binding in the vestibule in high-F− concentration solution, and these F− ions will be released easily in low-F− concentration solutions.
Fig. 2. Previously unidentified fluoride ion binding sites (F0 sites) in the aqueous vestibules.
Comparison of 2D NCA (A), NCOCA (B), NCO (C), and PDSD (D) spectral close-ups of residues with remarkable chemical shift changes in the low-F− sample (blue) and high-F− sample (light coral). (E) Residues with remarkable chemical shift changes (magenta) labeled on the structure model. These residues reside near the central Na+. Fluoride ions assigned in the crystal structure (PDB ID: 5NKQ) are shown as pink (F1 sites) or cyan (F2 sites) spheres, and the sodium ion as a purple sphere. Notably, the entire amino acids located on TM4 (indicated by black arrows), encompassing the polar track residues, have remained immune to any chemical shift perturbations. (F) Fluoride (red sphere) bound in the vestibular site. This is a snapshot of the MD simulations starting from the crystal structure with PDB ID 7KKR, after the replacement of bromide by fluoride. (G) Direct 19F-detected ssNMR spectra of the high-F− sample (top) and low-F− sample (bottom). (H) 19F spin diffusion from labeled unnatural amino acid 4-(trifluoromethyl)-l-phenylalanine (tfmF) to fluoride in the vestibule (F0 sites).
A previous x-ray crystallography study of Fluc-Ec2 showed that bromide ions (Br−) could reside in the bottom of vestibules coordinating Ser81 (9). The authors proposed that this anion-binding site is part of the fluoride permeation pathway. To investigate whether fluoride and/or chloride can reside in the vestibule, we carried out an MD simulation of Fluc embedded into a lipid bilayer. The simulation was based on the Fluc-Ec2 crystal structure with PDB ID 7KKR (9), which contains bromide ions bound in the two F0 sites, as well as a central sodium ion. The halide anions were replaced by fluoride (Fig. 2F), and the simulated system additionally contained five chloride counterions to make it neutral. During the simulation, one of the fluoride ions leaves the vestibular site, whereas the other remains bound. The vestibular binding site devoid of F− is transiently explored by chloride ions, which are coordinated by four water molecules, Arg19 and Thr82. In contrast to the Br− binding residues in the crystal structure (9), Ser81 does not seem to be part of the coordination sphere of Cl−, in line with chloride being able to accommodate lower coordination numbers (28). Similar transient exploration of the vestibule by chloride ions was observed in another MD simulation started from the Fluc-Ec2 crystal structure with PDB ID 6BX4 (29) by replacing the four assigned fluoride ions with water molecules (see Materials and Methods) and including seven chloride ions in the solution to achieve a neutral system. In contrast, when F− is bound in the vestibule, its position is only ~4.5 Å from the central sodium ion, which is deeper inside the vestibule compared to bromide (9) or chloride ions (fig. S10). We surmise that this might be due to fluoride being the smallest halide ion and thus able to move further toward the electropositive sodium ion and Arg19 side chain. Although the well-known limitations of the fluoride force field (17) preclude an accurate quantification of the energetics or kinetics of fluoride binding to F0, the vestibular fluoride site identified in our simulation (fig. S10C) is consistent with the conservation of Ser81 across Flucs and mutagenesis data (9).
19F magic-angle spinning (MAS) NMR spectroscopy has emerged as a powerful means for investigating protein structure and dynamics (30). To directly observe the bound fluoride, we measured the 19F direct polarization ssNMR spectrum under a 35-kHz MAS rate. The spectrum can be deconvoluted into three components. A large sharp peak [−118.3 parts per million (ppm), 0.22-ppm linewidth] can be attributed to free F− ions in solution. The broad peak (−116.4 ppm) disappeared in the 19F spectrum of the low-F− sample, which can be attributed to the vestibule binding F− ions that can be washed out in a relatively short period of time (Fig. 2G). This is in line with the aforementioned observation that one fluoride ion leaves the vestibular site within the 500-ns time scale of our MD simulation. The other broad peak (−108.0 ppm) representing the stable binding F− ions may correspond to the fluoride in the polar track. 19F-19F distances up to ∼2 nm can be measurable in proteins via fast MAS 19F spin-exchange NMR (31). To site-specifically introduce other 19F spins, the F89 residue that points into the vestibule space was replaced by 4-(trifluoromethyl)-l-phenylalanine (tfmF) by the codon extension method (fig. S11) (32). The F− permeation function of the F89tfmF mutant was verified by fluoride efflux from liposomes (fig. S12). Successful 19F-19F spin diffusion transfer from tfmF to the F− ions with −116.4-ppm chemical shift indicated F− ion binding in the vestibules in high-F− ion solutions (Fig. 2H).
Fluoride resides in F2 sites, while water resides in F1 sites in a high F− environment
To investigate the fluoride binding sites in the polar track, we attempted to wash out the stable binding sites with long-term dialysis in F−-free buffer. After approximately 3 months of dialysis, no 19F signal was detected in the Fluc-Ec1 proteoliposome sample, named the “F−-free sample” (Fig. 3A). Two-dimensional carbon-detected spectra of the F−-free Fluc-Ec1 sample with high resolution were recorded for backbone resonance analysis (figs. S13 and S14). Chemical shifts of the backbone residues remain unchanged in the helix region compared to the low-F− sample. However, the peak intensity of the residues located on loop 1 between TM1 and TM2 decreased (Fig. 3, B to D). This indicates that loop 1 has high mobility, given that we used dipolar ssNMR experiments, in which signal sensitivity decreases with increased dynamics. Meanwhile, the cross peaks of Asn102 and Asn106 side chains oriented toward F2 sites on TM4 were also undetectable (Fig. 3C). We believe that this may be due to the washed-out fluoride leading to structural rearrangement in the vicinity of F2 sites, which altered the flexibility of loop 1 and the Asn102/Asn106 side chains (Fig. 3E). Therefore, fluoride ions reside in the F2 sites of Fluc-Ec1 only in a high F− environment.
Fig. 3. Spectral difference between the high-F− sample and the F−-free sample.
(A) Direct 19F-detected ssNMR spectra of high-F− sample (black) and F−-free sample (red). Comparison of 2D NCA (B) and NCOCA (C) spectral close-ups of residues (marked as a black font) with a notable signal intensity decrease in the F−-free sample compared to the high-F− sample. (D) The signal-to-noise ratio of the NCA spectrum is shown as a histogram, and loop 1 (R25FNPLHQAIPL35) is indicated. A.U., arbitrary unit. (E) Residues with notably decreased signals are shown in magenta on the structure model.
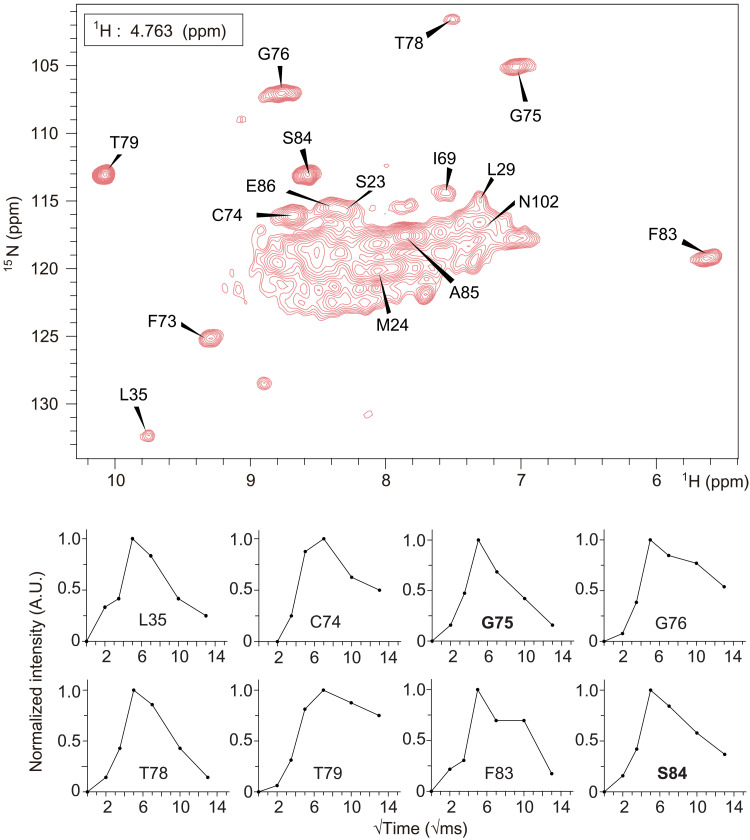
Since no notable peak shift or peak intensity change was observed in the vicinity of F1 sites of the F−-free sample compared to the other two samples, we assume that the density of F1 sites observed in the crystal structure may correspond to water molecules. To directly determine the spin type (19F or H2O) at the F1 site, we detected water-protein interactions directly through a 1H-1H spin diffusion experiment in which magnetization from bound water could be transferred to the protein (Fig. 4, top). Deuterium substitution is necessary to decrease the incoherent background from hydrogen atoms to achieve hydrogen detection in Fluc-Ec1 (33). Within a certain short mixing time, the distance between bound water and amide protons directly determines the efficiency of the magnetization transfer through 1H-1H spin diffusion. This method has been used, for example, to detect bound water in the potassium selective ion channel KcsA and other membrane proteins (23, 34–36). The magnetization transfer efficiency between bound water and nearby residues can be seen by the amide proton build-up rate. The amide protons of residues G75 and S84 build up relatively fast, and then decay with a relaxation time that corresponds to rigid protons in the protein. A similar buildup rate and decay rate indicate comparable short-distance contact with the bound water of these two residues (Fig. 4, bottom). In the Fluc channel crystal structure with the highest resolution (7) (PDB ID: 5NKQ, B. pertussis), residues S86 and G77 (corresponding to S84 and G75 in Fluc-Ec1 used here) are 6.9 and 3.7 Å away from the nearest stably bound water, respectively (fig. S15). Because of the strong dependence of dipolar coupling on distance (as 1/r3), the long distance between the S84 amide proton and bound water should not result in a buildup rate comparable to that of G75. Considering that the distance from the F1 site to the amide proton of S84 is 4.6 Å (fig. S15A), we propose that the molecule at the F1 site may be 1H2O. Although the waters captured in the crystal structure are not necessarily reflective of stable waters in solution at ambient temperature, these water-HN build-up data are valuable supporting data, complementary to the 13C-detected ssNMR data.
Fig. 4. Water-edited hNH spectrum of the high-F− sample.
The 2D HN slice taken at the chemical shift of bound water (4.763 ppm) from the 3D H(H)NH spin diffusion spectrum is shown in the top panel. Spin diffusion buildup plots from bound water to amide protons for the indicated residues are shown in the bottom panel with Gly75 and Ser84 marked in bold font. The distances of each highlighted residue to F1 and F2 and the closest crystallographic water are annotated in table S2.
To investigate water occupancy of the F1 site, we set up another MD simulation, starting from the Fluc-Ec2 crystal structure with PDB ID 6BX4 (29), replacing fluoride ions bound in sites F1 and F2 by water molecules. These water molecules present in the F1 binding sites in the starting conformation stay there during the whole simulation time of 500 ns. Instead, water exchange in the F2 sites is much faster, with a time scale of several nanoseconds, in line with the F2 sites being more exposed to the bulk solvent. To neutralize the +7e charge of the protein and sodium ion, the simulated system contains seven chloride counterions. During the simulation, we observed that one of these chloride counterions explores transiently the vestibule, indicating that the electropositive nature of this site attracts halide ions in a nonselective manner.
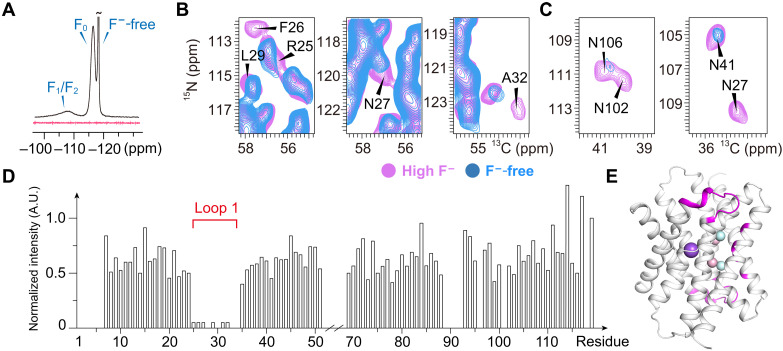
Loop 1 plays a critical role in F− permeation
On the basis of the previous study, two phenylalanine residues in the polar track (Phe80 and Phe83) directly coordinate with fluoride through a quadrupolar-ion interaction and can only be functionally replaced by methionine. When mutated with methionine, F80M led to a still active fluoride channel, while the F83M led to a loss of fluoride conductivity. However, the crystal structures of both Met mutants are virtually identical to WT, and all residues surrounding the substitutions are unmoved (15). The broad peak in the 19F spectra corresponding to a stably bound fluoride is visible for both mutants, indicating that fluoride ions remain in the polar track (fig. S16).
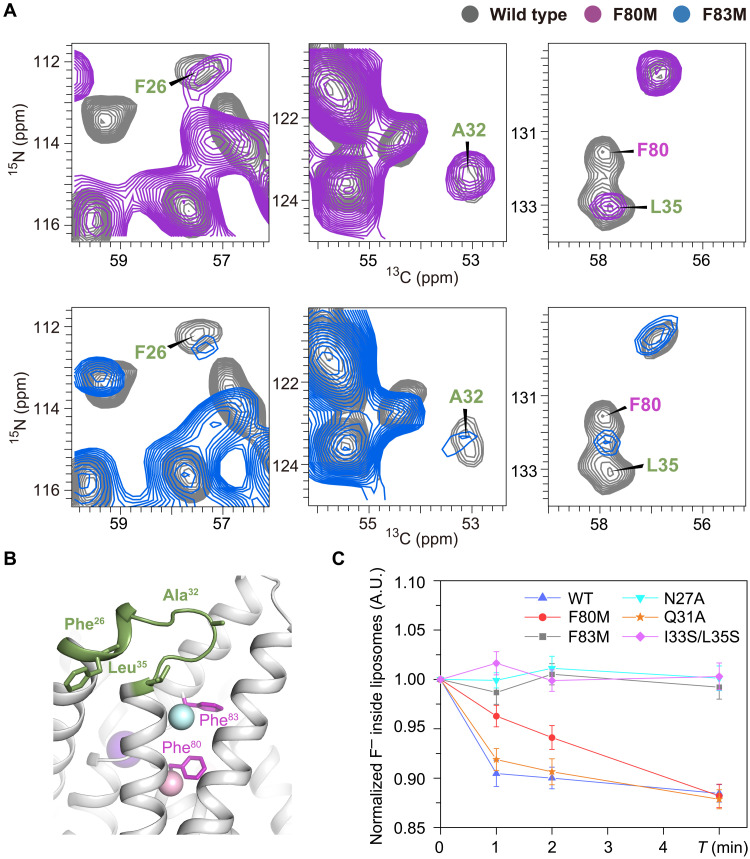
To investigate why F80M is functionally active and F83M is not, we prepared uniformly [13C,15N]-labeled Fluc-Ec1 F80M and F83M proteoliposome samples in the presence of a high concentration of F− and collected 2D 13C-detected ssNMR spectra. The spectral resolution of both mutants is slightly decreased, indicating increased structural dynamics or inhomogeneity (fig. S17). Residues surrounding the mutated F80 or F83 show notable peak drift on the spectra. However, the chemical shift difference may result from the absence of the benzene ring on the phenylalanine side chain. Comparing the spectra of these two mutants (F80M and F83M) with those of WT, we observed a remarkable signal decrease of F83M at three isolated residues (Phe26, Ala32, and Leu35), while these three residues in F80M maintained a signal intensity similar to that of WT (Fig. 5A). The decreased signal intensity in dipolar coupling–based experiments is most likely due to increased structural dynamics. These data indicate increased flexibility of loop 1, which includes Phe26, Ala32, and Leu35 (Fig. 5B). Together with the spectra of WT Fluc-Ec1 in F−-free buffer (Fig. 3, B and C), the increased flexibility of loop 1 may provide a possible explanation for the loss of fluoride transport ability of the F83M mutant. To explore whether the flexibility of loop 1 is involved in ion permeation, we constructed three mutations, N27A, Q31A, and I33S/L35S, which may weaken the interaction between loop 1 (see fig. S1) and TM3b/TM4 based on the crystal structures. We verified the biochemical properties of those mutants via size exclusion chromatography and collected 1D 19F spectra to investigate whether they are able to bind F− ions and performed 19F NMR transport assays (6) to assess channel function. Although all the mutants are biochemically well-behaved and able to bind to fluoride ions in the vestibule and polar track (figs. S18 and S19), they show variant functional behaviors. Fluoride transport was inhibited in N27A and I33S/L35S mutants compared to WT, while the mutant of unconserved Q31 residue exhibited a similar conducting phenotype of WT (Fig. 5C). These data further support the importance of loop 1 in the fluoride permeation mechanism.
Fig. 5. Importance of loop 1 structural dynamics observed in NMR.
(A) Spectral difference of the F80M and F83M mutants compared to WT Fluc-Ec1. The NCA spectrum of WT is shown in gray, F80M in magenta, and F83M in blue. (B) Residues labeled on the spectra shown in the Fluc-Ec1 structure model as green sticks. (C) Fluoride transport of variant mutants compared to WT Fluc-Ec1 in liposomes revealed by solution NMR.
DISCUSSION
Microbes are protected from the cytoplasmic accumulation of environmental fluoride ions by exporting the toxic anions via fluoride channels such as Fluc channels (5). A series of electrophysiology, x-ray crystallography, and liposome flux assays have been used to identify the fluoride conduction mechanism of the Fluc channel homologs Fluc-Bpe and Fluc-Ec2 (4, 7–9, 11–13, 15, 37). Although mutation analysis accompanied by crystal structures revealed two antiparallel and independent pores, the four disconnected electron densities in the polar track interpreted as fluoride ions were challenged by MD simulation data (16). Meanwhile, functionally important structural dynamics may be altered by detergent micelles and additional monobodies used for protein crystallization (7–9); the latter block the entrance and exit of the permeation pathway. In the present study, the structural dynamics of the Fluc channel in phospholipid bilayers were investigated by the combined use of ssNMR and MD simulations.
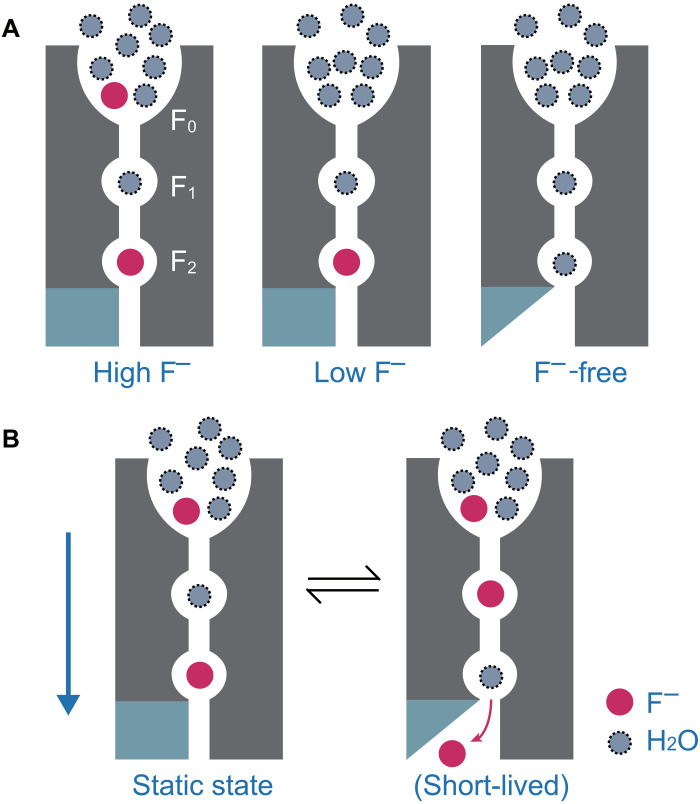
To observe the fluoride binding sites of Fluc-Ec1, we attempted to titrate F− into apo Fluc-Ec1 in liposomes and map the fluoride binding interfaces by chemical shift perturbation. However, Fluc-Ec1 cannot be efficiently reconstituted into liposomes in the absence of F−. As an alternative, a low-F− sample and an F−-free sample were made by washing the Fluc-Ec1 proteoliposome sample with 150 mM NaCl buffer for 1 week and more than 3 months, respectively. According to the chemical shift perturbation data and the spin diffusion data, we determined the nature of the molecules residing in the ion permeation pathway of Fluc-Ec1 in varying F− environments. In the presence of a high concentration of F−, fluoride ions bind to F2 sites and nonspecific anion binding sites (F0) in the aqueous vestibule, while water molecules reside in F1 sites (Fig. 6A). The fluoride ions in F0 sites could be easily washed out by 1-week dialysis in NaCl buffer, in line with the MD simulations showing that fluoride can be exchanged by water molecules or chloride ions. Furthermore, the stable binding fluoride in F2 sites could be washed out by 3 months of dialysis, leading to a more flexible loop 1 near the F2 sites (Fig. 6A). Note that low abundant states may not be detectable with our NMR experiments due to limited sensitivity or conformation exchange. The broadened peak of the F1/F2 site on the 19F spectrum may be due to the two fluorides at F1 and F2 exchanging with each other on an intermediate time scale. We propose that, in the resting state of the channel, fluoride ions occupy the F0 and F2 sites, while water molecules reside in the F1 site (Fig. 6B, left). The fluoride ion in the vestibule then settles into the F1 site, and the water molecule moves farther to the F2 site (Fig. 6B, right). Although the distances among the F0, F1, and F2 sites in Fluc channels are longer than that of K+ channels, that water/F− in the selectivity filter may still be electrostatically pushed by the incoming F−, similar to the ryanodine receptor (38) and the transient receptor potential vanilloid (TRPV) channel (39).
Fig. 6. Schematic model of the fluoride conduction mechanism in the Fluc channel.
(A) Equilibrium of Fluc-Ec1 conformations in the presence of variant F− concentration environment showing fluoride binding sites F0, F1 and F2. For clarity, only one pore is shown. Rigid loop 1 is shown as a cyan rectangle, and flexible loop 1 is shown as a cyan triangle. Fluoride ions are shown in red, and water molecules are shown as dashed gray circles. (B) The multi-ion permeation mechanism for the Fluc channel is proposed. The proposed movement of ions between binding sites is indicated by arrows.
The channel is nonfunctional if loop 1 is enforced by monobodies or loop 1 flexibility is enhanced by point mutations. The previous study showed that the T37-E86-N102 triad in Fluc-Ec1 [corresponding to the T39-E88-Y104 triad in Fluc-Bpe (9)] is important for ion permeation. A conserved residue N27 on loop 1 forms a hydrogen bond with E86, so loop 1 conformational changes may involve in the E86 side-chain rotation for fluoride-releasing from the F2 site. In a functional channel, the F− ion located at the F2 site generates electrostatic repulsion on the triad E86 carboxylate, which triggers the rotamerization of E86 with the assistance of loop 1 conformation change, disrupting the stable conformation of the T-E-N triad and ultimately facilitating the release of fluoride at F2. Within this framework, the fluoride ion bound at F2 couples loop 1 and E86 movement. Instead, in the absence of fluoride at the F2 site, loop 1 flexibility is uncoupled from E86. Previous crystallographic studies on the Fluc channel did not reveal the same degree of conformational plasticity of loop 1 because of the presence of blocking monobodies (5).
The water-mediated knock-on mechanism proposed here for fluoride conduction in Flucs is similar to the initially proposed K+ permeation mechanism for potassium channels (40), compatible with the multi-ion permeation mechanism proposed by the group of Stockbridge (9). In conclusion, our model complements the previously proposed kinetic binding mechanisms in Fluc and thereby provides a more thorough understanding of fluoride ion permeation and gating mechanism in Fluc channels.
MATERIALS AND METHODS
Sample preparation
The DNA coding region for full-length Fluc-Ec1 was amplified from the E. coli BL21(DE3) genome and cloned into a pET22b vector with a C-terminal hexahistidine (His6) sequence. For structure and function determination of Fluc-Ec1, F80M, F83M, F89tfmF, N27A, Q31A, and I33S/L35S were constructed by mutagenesis experiments. All the primers used for construction in this study are listed in table S3. WT Fluc-Ec1 and those constructs were overexpressed in E. coli C41 (DE3). At an OD600 (optical density at 600 nm) of 1.0, the cultures were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 6 hours at 37°C. Cells were harvested and resuspended in lysis buffer [20 mM tris-HCl, 150 mM NaCl, and 5 mM NaF (pH 7.5)] and lysed by a high-pressure homogenizer (Union-Biotech Ltd., China) with 68.9 MPa working pressure. Channels were extracted with 2% (w/v) n-decyl-β-d-maltopyranoside (DM; Anatrace, USA) for 2 hours at 4°C. The solubilized lysate was clarified by centrifugation at 20,000g for 40 min, and then purification was performed using Talon cobalt resin (Takara Biotechnology, USA) equilibrated with 20 mM tris-HCl, 150 mM NaCl, and 5 mM NaF (pH 7.5). The concentration of purified Fluc-Ec1 was estimated using a Bradford assay. The purity of the sample was confirmed by SDS– polyacrylamide gel electrophoresis.
To produce the uniformly [13C, 15N]-labeled samples, the protein was expressed in C41(DE3) cells in an M9 medium with 13C-glucose as the sole carbon source and 15NH4Cl as the sole nitrogen source. The [2H,13C,15N]-labeled Fluc-Ec1 sample was expressed in E. coli C41(DE3) using a deuterated M9 medium (D2O, 99.9% D) supplemented with d-glucose (U-13C6, 99%; 1,2,3,4,5,6,6-D7, 97 to 98%; catalog no. CDLM-3813-10) as the sole carbon source and lyophilized deuterated 15NH4Cl as the sole nitrogen source. H/D back exchange was performed by purification and the procedure was the same as that described for the aforementioned [13C, 15N]-labeled sample.
Purified WT Fluc-Ec1 and other mutants were mixed with E. coli total lipid extract (Avanti Polar Lipids, USA) at a lipid/protein ratio of 1.5:1 (w/w). For this purpose, the E. coli total lipid extract was first dissolved in 5% DM detergent buffer. Subsequently, detergent was removed and proteoliposome formation was performed by dialysis for 1 week at 100× dilution against 1 liter of 20 mM tris-HCl and 150 mM NaF (pH 7.5) to obtain “high-F− sample”, while dialysis for 1 week or at least 3 months at 100× dilution against 1 liter of 20 mM tris-HCl and 150 mM NaCl (pH 7.5) to obtain a low-F− sample and an F−-free sample, respectively. The reconstituted proteoliposomes were collected by ultracentrifugation at 180,000g for 4 hours at 4°C.
Preparation of F89tfmF labeled sample
19F-labeled protein was expressed and purified by incorporating the unnatural amino acid tfmF into the specific site as previously described (41). The Fluc-Ec1 F89tfmF mutant was performed by site-directed mutagenesis. The pDule-tfmF plasmid-coding tRNACUA/tfmF-specific–aminoacyl–tRNA synthetase pair can specifically insert the tfmF into polypeptide chains in response to the amber codon UAG. On the basis of this fact, the Fluc-Ec1 F89tfmF mutant plasmid and pDule-tfmF plasmid were cotransformed into E. coli C41(DE3) cells. A single colony was grown on LB medium containing ampicillin (50 μg/ml) and tetracycline (2 μg/ml) at 37°C. Cells were induced with 0.02% arabinose when OD600 reached 0.8 in the presence of 1 mM unnatural amino acid tfmF. The purification and reconstitution processes were both consistent with those described above.
19F-detected solution NMR transport assay
To establish whether reconstituted Fluc-Ec1 in liposomes is functional, we used 19F solution NMR to monitor F− transmembrane export. For 19F solution NMR analysis, liposomes were formed at a concentration of 5 mg of 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC)/ml, 10 μg of protein/ml and loaded with 300 mM potassium fluoride(KF), 20 mM Hepes-KOH (pH 7.0). After reconstitution, the protein/POPC mixture was extruded 25 times through a 400-nm membrane filter (Whatman, Newton, MA) with an Avanti mini-extruder to make unilamellar liposome vesicles. Immediately before functional assays, the extra liposomal solution was exchanged for 300 mM SHES and 20 mM Hepes-NaOH (pH 7.0) via dialysis. This procedure resulted in approximately 3 nM F− remaining in the external solution. Each sample was prepared with 5% D2O to enable deuterium locking and 10 μM trifluoroacetic acid (TFA) as an internal reference standard. One-dimensional 19F solution NMR experiments were performed on a Bruker 600 MHz spectrometer equipped with a TCI H&F/C/N-D probe at 25°C. Each dataset was recorded with an interscan delay of 1.5 s along with eight scans per transient, generating a net acquisition time of roughly 12 s per spectrum. After recording the first spectrum to quantify the initial inside/outside F− signals, F− transport was initiated by adding 1.5 μM valinomycin to the NMR tube with brief agitation. After a dead time of 20 to 30 s necessary for reinserting the sample into the spectrometer, F− efflux was followed by recording four spectra over 5 min to measure channel conductance in POPC. The Bruker Topspin software was used to process the NMR data and quantify fluoride efflux from liposomes via the intraliposomal peak integration.
SsNMR spectroscopy and analysis
The proteoliposome pellets were transferred into MAS rotors and used for ssNMR analysis. SsNMR spectra required for chemical shift assignment were recorded on a 600-MHz wide-bore NMR spectrometer (Bruker BioSpin) equipped with a 3.2-mm triple-resonance EFree MAS probe. Spectra were recorded at 20-kHz MAS rate and calibrated with external DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) as a proton chemical shift reference. Two-dimensional NCA, NCACO, NCO, NCOCA, and CANCOCA and 3D CANCO, NCACB, NCACO, NCACX, and NCOCA spectra were recorded for obtaining unambiguous resonance assignments. A 13C-13C PDSD spectrum was recorded at 11-kHz MAS with 20-ms mixing time to assist the side-chain assignment (fig. S9). An Insensitive Nuclei Enhanced by Polarization Transfer (INEPT)-based spectrum and an INEPT-total through bond correlation spectroscopy (TOBSY) 1H-13C correlation spectrum were recorded to detect the flexible region of Fluc-Ec1. The complete chemical shift list for Fluc-Ec1 in the presence of 150 mM NaF is provided in table S1. Two-dimensional NCA, NCOCA, NCO, and PDSD spectra were recorded to detect chemical shift perturbation and signal intensity change of Fluc-Ec1 mutants under different fluoride conditions.
Three-dimensional H(H)NH spectra with 4, 12.25, 25, 49, 100, and 169 ms proton-proton mixing times were recorded at 16.4 T and 40-kHz MAS with the [2H,13C,15N]-labeled Fluc-Ec1 sample. A recycle delay of 1 s and 90° pulses of 83.3, 50, and 35.7 kHz for 1H, 13C, and 15N were used, respectively. After the 1H-1H spin diffusion build block, a 35.7-kHz pulse on the 15N channel and an 84.11-kHz 100% to 80% ramp pulse on the 1H channel were used for 1H-15N CP or 15N-1H CP transfer. A short contact time of 0.4 ms was used in the CP transfer to eliminate long-distance transfer. The acquisition times were 25 ms for direct 1H acquisition, 7.1 ms for indirect 1H evolution, and 11.1 ms for indirect 15N evolution, respectively.
For 1D 19F signals assignment, 19F-NMR spectra were obtained on a Bruker 600 MHz spectrometer equipped with an HFX probe at 35-kHz MAS rate. A 1D 19F spectrum was acquired with a standard pulse program of “zgbs”. 19F chemical shifts were referenced to an external standard TFA. The 19F-19F spin diffusion experiment that was used to identify fluoride binding sites F0 in Fluc-Ec1 was recorded with 1-ms mixing time.
The sample temperature was kept constant at ~4°C for carbon-detected ssNMR experiments, while the sample temperature was ~15°C for the proton-detected experiments as measured by the temperature-dependent position of the water proton resonance. All spectral processing was performed with Topspin 4.1 and SPARKY software.
MD simulations
We ran MD simulations, starting from two different high-resolution crystal structures of Fluc-Ec2. One was solved with F− bound in sites F1 and F2 (PDB ID: 6BX4) (29), while the other was obtained using Br− (PDB ID: 7KKR), which turned out to bind in the vestibule (9). The blocking monobodies were removed from both crystal structures and the Fluc channel was embedded in a POPC lipid bilayer and solvated with water. In the 6BX4 structure, the four fluoride ions bound in sites F1 and F2 were replaced by water molecules to model the fluoride-free state. In the 7KKR structure, the two bromide ions bound in the vestibules were exchanged with fluoride to investigate the behavior of F− at the F0 site, compared to Br− captured in the x-ray structure. Additional chloride counterions (seven for 6BX4 and five for 7KKR) were added to neutralize the systems. All atoms, including those in the lipids and water molecules, were represented explicitly. MD simulations were carried out using GROMACS 2020 (42), the CHARMM36 force field for protein and lipids (43, 44), and the TIP3P water model (45). Parameters for fluoride were taken from (46). Short-range electrostatic interactions were calculated with a cutoff of 1.2 nm, whereas long-range electrostatic interactions were treated by the particle mesh Ewald method. The cutoff for van der Waals interactions was set to 1.2 nm. The default protocol of CHARMM-GUI (47) was used to equilibrate each of the two above-described systems. The simulations were performed at 300 K with a Nosé-Hoover thermostat (48). The pressure was kept constant at 1 bar by means of a semi-isotropic Parrinello-Rahman barostat (49). Bonds containing hydrogen atoms were constrained with the LINCS algorithm, allowing for a 2-fs integration time step. The production simulation for each of the two systems was run for 500 ns. For the 6BX4 structure, we ran three replicas, by varying the protonation states of the two titratable residues lining the permeation pathway (H106 and E86): (i) neutral H106 and negatively charged E86, (ii) positively charged H106 and negatively charged E86, and (iii) neutral H106 and neutral E86.
Acknowledgments
Funding: This work was supported by the Chinese Ministry of Science and Technology (2021YFA0909400 and 2019YFA0904100) and the National Natural Science Foundation of China (22077117) (to C.S.), as well as the Deutsche Forschungsgemeinschaft (DFG) via the Research Unit FOR2518 “Functional Dynamics of Ion Channels and Transporters-DynIon” (project P6 to P.C. and M.A.-P.) and the Research Training Group RTG2416 MultiSenses-MultiScales (368482240/GRK2416 to P.C.). This work was also supported by “USTC Research Funds of the Double First-Class Initiative” and “The Strategic Priority Research Program of the Chinese Academy of Science (XDB37040200).”
Author contributions: J.Z. and D.S. prepared the Fluc-Ec1 NMR samples. J.Z., J.L., and C.S. performed solid-state NMR experiments and analyzed NMR data. J.Z. and C.S. performed 19F-detected solution NMR transport assay. F.K.S. performed MD simulations, and F.K.S. and M.A.-P. analyzed MD data. J.Z., C.S., P.C., and M.A.-P. wrote the paper. All authors discussed the results and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The Fluc-Ec1 chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) under accession code 51680. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S19
Tables S1 to S3
REFERENCES AND NOTES
- 1.R. E. Marquis, S. A. Clock, M. Mota-Meira, Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiol. Rev. 26, 493–510 (2003). [DOI] [PubMed] [Google Scholar]
- 2.E. Adamek, K. Pawlowska-Goral, K. Bober, In vitro and in vivo effects of fluoride ions on enzyme activity. Ann. Acad. Med. Stetin. 51, 69–85 (2005). [PubMed] [Google Scholar]
- 3.J. L. Baker, N. Sudarsan, Z. Weinberg, A. Roth, R. B. Stockbridge, R. R. Breaker, Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335, 233–235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.C. Ji, R. B. Stockbridge, C. Miller, Bacterial fluoride resistance, Fluc channels, and the weak acid accumulation effect. J. Gen. Physiol. 144, 257–261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.B. C. McIlwain, M. T. Ruprecht, R. B. Stockbridge, Membrane exporters of fluoride ion, in Annual Review of Biochemistry, R. D. Kornberg, Ed. (Annual Review of Biochemistry, 2021), vol. 90, pp. 559–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R. B. Stockbridge, H.-H. Lim, R. Otten, C. Williams, T. Shane, Z. Weinberg, C. Miller, Fluoride resistance and transport by riboswitch-controlled CLC antiporters. Proc. Natl. Acad. Sci. U.S.A. 109, 15289–15294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R. B. Stockbridge, L. Kolmakova-Partensky, T. Shane, A. Koide, S. Koide, C. Miller, S. Newstead, Crystal structures of a double-barrelled fluoride ion channel. Nature 525, 548–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.B. C. McIlwain, S. Newstead, R. B. Stockbridge, Cork-in-bottle occlusion of fluoride ion channels by crystallization chaperones. Structure 26, 635–639.e1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.B. C. McIlwain, R. Gundepudi, B. B. Koff, R. B. Stockbridge, The fluoride permeation pathway and anion recognition in Fluc family fluoride channels. eLife 10, e69482 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.R. B. Stockbridge, A. Koide, C. Miller, S. Koide, Proof of dual-topology architecture of Fluc F− channels with monobody blockers. Nat. Commun. 5, 5120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R. B. Stockbridge, J. L. Robertson, L. Kolmakova-Partensky, C. Miller, A family of fluoride-specific ion channels with dual-topology architecture. eLife 2, e01084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.N. B. Last, L. Kolmakova-Partensky, T. Shane, C. Miller, Mechanistic signs of double-barreled structure in a fluoride ion channel. eLife 5, e18767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D. L. Turman, R. B. Stockbridge, Mechanism of single- and double-sided inhibition of dual topology fluoride channels by synthetic monobodies. J. Gen. Physiol. 149, 511–522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.B. C. McIlwain, K. Martin, E. A. Hayter, R. B. Stockbridge, An interfacial sodium ion is an essential structural feature of fluc family fluoride channels. J. Mol. Biol. 432, 1098–1108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.N. B. Last, S. Sun, M. C. Pham, C. Miller, Molecular determinants of permeation in a fluoride-specific ion channel. eLife 6, e31259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.I. Ariz-Extreme, J. S. Hub, Assigning crystallographic electron densities with free energy calculations-The case of the fluoride channel Fluc. PLOS ONE 13, e0196751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Z. Yue, Z. Wang, G. A. Voth, Ion permeation, selectivity, and electronic polarization in fluoride channels. Biophys. J. 121, 1336–1347 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. A. Shcherbakov, G. Hisao, V. S. Mandala, N. E. Thomas, M. Soltani, E. A. Salter, J. H. Davis Jr., K. A. Henzler-Wildman, M. Hong, Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers. Nat. Commun. 12, 172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. A. Shcherbakov, P. J. Spreacker, A. J. Dregni, K. A. Henzler-Wildman, M. Hong, High-pH structure of EmrE reveals the mechanism of proton-coupled substrate transport. Nat. Commun. 13, 991 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R. Shukla, F. Lavore, S. Maity, M. G. N. Derks, C. R. Jones, B. J. A. Vermeulen, A. Melcrova, M. A. Morris, L. M. Becker, X. Wang, R. Kumar, J. Medeiros-Silva, R. A. M. van Beekveld, A. Bonvin, J. H. Lorent, M. Lelli, J. S. Nowick, H. D. MacGillavry, A. J. Peoples, A. L. Spoering, L. L. Ling, D. E. Hughes, W. H. Roos, E. Breukink, K. Lewis, M. Weingarth, Teixobactin kills bacteria by a two-pronged attack on the cell envelope. Nature 608, 390–396 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.S. Jekhmane, J. Medeiros-Silva, J. Li, F. Kümmerer, C. Müller-Hermes, M. Baldus, B. Roux, M. Weingarth, Shifts in the selectivity filter dynamics cause modal gating in K+ channels. Nat. Commun. 10, 123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. Kaplan, S. Narasimhan, C. de Heus, D. Mance, S. van Doorn, K. Houben, D. Popov-Celeketic, R. Damman, E. A. Katrukha, P. Jain, W. J. C. Geerts, A. J. R. Heck, G. E. Folkers, L. C. Kapitein, S. Lemeer, P. M. P. van Bergen En Henegouwen, M. Baldus, EGFR dynamics change during activation in native membranes as revealed by NMR. Cell 167, 1241–1251.e11 (2016). [DOI] [PubMed] [Google Scholar]
- 23.C. Oster, K. Hendriks, W. Kopec, V. Chevelkov, C. Shi, D. Michl, S. Lange, H. Sun, B. L. de Groot, A. Lange, The conduction pathway of potassium channels is water free under physiological conditions. Sci. Adv. 5, eaaw6756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D. Lacabanne, T. Wiegand, M. Di Cesare, C. Orelle, M. Ernst, J.-M. Jault, B. H. Meier, A. Boeckmann, Solid-State NMR reveals asymmetric ATP hydrolysis in the multidrug ABC transporter BmrA. J. Am. Chem. Soc. 144, 12431–12442 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.C. Shi, H. K. Fasshuber, V. Chevelkov, S. Xiang, B. Habenstein, S. K. Vasa, S. Becker, A. Lange, BSH-CP based 3D solid-state NMR experiments for protein resonance assignment. J. Biomol. NMR 59, 15–22 (2014). [DOI] [PubMed] [Google Scholar]
- 26.S. Luca, D. V. Filippov, J. H. van Boom, H. Oschkinat, H. J. M. de Groot, M. Baldus, Secondary chemical shifts in immobilized peptides and proteins: A qualitative basis for structure refinement under magic angle spinning. J. Biomol. NMR 20, 325–331 (2001). [DOI] [PubMed] [Google Scholar]
- 27.N. M. Szeverenyi, M. J. Sullivan, G. E. Maciel, observation of spin exchange by two-dimensional Fourier-transform 13C cross polarization-magic-angle spinning. J. Magn. Reson. 47, 462–475 (1982). [Google Scholar]
- 28.L. Ge, L. Bernasconi, P. Hunt, Linking electronic and molecular structure: Insight into aqueous chloride solvation. Phys. Chem. Chem. Phys. 15, 13169–13183 (2013). [DOI] [PubMed] [Google Scholar]
- 29.D. L. Turman, A. Z. Cheloff, A. D. Corrado, J. T. Nathanson, C. Miller, Molecular interactions between a fluoride ion channel and synthetic protein blockers. Biochemistry 57, 1212–1218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Wang, M. Lu, M. P. Fritz, C. M. Quinn, I.-J. L. Byeon, C.-H. Byeon, J. Struppe, W. Maas, A. M. Gronenborn, T. Polenova, Fast magic-angle spinning 19F NMR spectroscopy of HIV-1 capsid protein assemblies. Angew. Chem. Int. Ed. 57, 16375–16379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. Roos, T. Wang, A. A. Shcherbakov, M. Hong, Fast magic-angle-spinning 19F spin exchange NMR for determining nanometer 19F-19F distances in proteins and pharmaceutical compounds. J. Phys. Chem. B 122, 2900–2911 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. C. Jackson, J. T. Hammill, R. A. Mehl, Site-specific incorporation of a 19F-amino acid into proteins as an NMR probe for characterizing protein structure and reactivity. J. Am. Chem. Soc. 129, 1160–1166 (2007). [DOI] [PubMed] [Google Scholar]
- 33.L. C. Shi, I. Kawamura, K. H. Jung, L. S. Brown, V. Ladizhansky, Conformation of a seven-helical transmembrane photosensor in the lipid environment. Angew. Chem. Int. Ed. 50, 1302–1305 (2011). [DOI] [PubMed] [Google Scholar]
- 34.R. Linser, M. Dasari, M. Hiller, V. Higman, U. Fink, J. M. L. del Amo, S. Markovic, L. Handel, B. Kessler, P. Schmieder, D. Oesterhelt, H. Oschkinat, B. Reif, Proton-detected solid-state NMR spectroscopy of fibrillar and membrane proteins. Angew. Chem. Int. Ed. 50, 4508–4512 (2011). [DOI] [PubMed] [Google Scholar]
- 35.T. Schubeis, T. Le Marchand, L. B. Andreas, G. Pintacuda, 1H magic-angle spinning NMR evolves as a powerful new tool for membrane proteins. J. Magn. Reson. 287, 140–152 (2018). [DOI] [PubMed] [Google Scholar]
- 36.L. B. Andreas, M. Reese, M. T. Eddy, V. Gelev, Q. Z. Ni, E. A. Miller, L. Emsley, G. Pintacuda, J. J. Chou, R. G. Griffin, Structure and mechanism of the influenza A M218–60 Dimer of dimers. J. Am. Chem. Soc. 137, 14877–14886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D. L. Turman, J. T. Nathanson, R. B. Stockbridge, T. O. Street, C. Miller, Two-sided block of a dual-topology F- channel. Proc. Natl. Acad. Sci. U.S.A. 112, 5697–5701 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. Zhang, H. Yu, C. Liu, C. Song, The Ca2+ permeation mechanism of the ryanodine receptor revealed by a multi-site ion model. Nat. Commun. 11, 922 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.C. M. Ives, N. J. Thomson, U. Zachariae, A cooperative knock-on mechanism underpins Ca2+-selective cation permeation in TRPV channels. J. Gen. Physiol. 155, e202213226 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J. H. Morais-Cabral, Y. F. Zhou, R. MacKinnon, Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature 414, 37–42 (2001). [DOI] [PubMed] [Google Scholar]
- 41.L. Li, J. Zhang, W. Sun, W. Gong, C. Tian, P. Shi, C. Shi, Allosteric conformational changes of G proteins upon its interaction with membrane and GPCR. Chin. Chem. Lett. 33, 747–750 (2022). [Google Scholar]
- 42.M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess, E. Lindahl, GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015). [Google Scholar]
- 43.A. D. Mackerell Jr., M. Feig, C. L. Brooks III, Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 (2004). [DOI] [PubMed] [Google Scholar]
- 44.J. B. Klauda, R. M. Venable, J. A. Freites, J. W. O'Connor, D. J. Tobias, C. Mondragon-Ramirez, I. Vorobyov, A. D. MacKerell Jr., R. W. Pastor, Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey, M. L. Klein, Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 46.H. M. Senn, D. O'Hagan, W. Thiel, Insight into enzymatic C-F bond formation from QM and QM/MM calculations. J. Am. Chem. Soc. 127, 13643–13655 (2005). [DOI] [PubMed] [Google Scholar]
- 47.J. Lee, X. Cheng, J. M. Swails, M. S. Yeom, P. K. Eastman, J. A. Lemkul, S. Wei, J. Buckner, J. C. Jeong, Y. Qi, S. Jo, V. S. Pande, D. A. Case, C. L. Brooks III, A. D. MacKerell Jr., J. B. Klauda, W. Im, CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D. J. Evans, B. L. Holian, The Nosé-Hoover thermostat. J. Chem. Phys. 83, 4069–4074 (1985). [Google Scholar]
- 49.M. Parrinello, A. Rahman, Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S19
Tables S1 to S3