Abstract
Advanced strategies to interconvert cell types provide promising avenues to model cellular pathologies and to develop therapies for neurological disorders. Yet, methods to directly transdifferentiate somatic cells into multipotent induced neural stem cells (iNSCs) are slow and inefficient, and it is unclear whether cells pass through a pluripotent state with full epigenetic reset. We report iNSC reprogramming from embryonic and aged mouse fibroblasts as well as from human blood using an engineered Sox17 (eSox17FNV). eSox17FNV efficiently drives iNSC reprogramming while Sox2 or Sox17 fail. eSox17FNV acquires the capacity to bind different protein partners on regulatory DNA to scan the genome more efficiently and has a more potent transactivation domain than Sox2. Lineage tracing and time-resolved transcriptomics show that emerging iNSCs do not transit through a pluripotent state. Our work distinguishes lineage from pluripotency reprogramming with the potential to generate more authentic cell models for aging-associated neurodegenerative diseases.
Reprogramming with engineered SOX17 bypasses pluripotency to induce neural stem cells offering models for aging and disease.
INTRODUCTION
The discovery of induced pluripotent stem cells (iPSCs) reprogramming via the overexpression of Oct4, Sox2, Klf4, and c-Myc (OSKM) (1, 2) opened up opportunities for the patient-specific modeling of diseases, personalized drug evaluation, and cell-replacement therapies (3). Controlled differentiation of PSCs led to functional neuronal and gliogenic lineages suitable for cell replacement therapies (4–6). Likewise, patient-specific iPSCs can be established from patients with Alzheimer’s disease, Huntington’s disease, and Parkinson’s disease (7, 8) and further used to model diseases and test drugs. However, there are several shortcomings associated with iPSC-derived neural cells. First, the generation of defined neural cells from iPSCs requires cumbersome and lengthy protocols and quality control procedures during both the iPSC generation and differentiation stages. Second, the tumorigenic risk of remnant iPSCs in the differentiated cell population complicates clinical application (9). Third, iPSCs represent a rejuvenated fetal-like state even from centenarian donors limiting their use to model age-associated neurodegenerative diseases (10).
As a potential solution to these problems, several studies have described the direct conversion of mouse and human somatic cells into induced neurons (iNs) (11, 12). Unlike iPSC-derived neurons, iNs are barely rejuvenated (13–15). Thus, they can closely reflect the transcriptional and epigenetic signatures of aging. These properties make iNs a suitable cell model for aging-related neurodegenerative diseases (10, 16). A shortcoming of postmitotic iNs is their inability to divide and lack of multipotency. Therefore, they cannot be produced at scale or used as starting materials for organoids.
Induced neural stem cells (iNSCs) could potentially take the best parts of iPSCs and iN methods while avoiding their shortcomings of lengthy procedures, tumorigenicity, rejuvenation as well as lack of self-renewal and multipotency. Expandable iNSCs that can differentiate into neurons, astrocytes, and oligodendrocytes were reported in several seminal studies (17–26). Most of the iNSC induction methods depend on the ectopic expression of multiple transcription factors (TFs), among which Sox2 is the most commonly used factor. Refined composition of TF cocktails (27), small molecules (28), and special culture conditions (29) have also been applied to enhance the iNSC reprogramming. Yet, the reprogramming mechanism and roadmap has not been clearly defined for any reprogramming cocktails. Lineage-tracing analysis suggested that methods that rely on the short-term expression of OSKM or the continuous expression of BSKM (Brn4 replaces Oct4 along with SKM) transit through a pluripotent state (30, 31). However, the precise roadmap depends on the transgene delivery method and the design of the reprogramming cassette (32). Overall, none of the iNSCs generation methods has found widespread use and is often hampered by rather low efficiency and protocols that can consume several months (33). We therefore investigated whether iNSC reprogramming could be enhanced with engineered proteins and engaged in a systematic comparison of iNSC reprogramming vis-à-vis pluripotency reprogramming.
Natural mammalian TFs have not evolved to direct non-natural cell fate conversions in cultured cells. We have previously applied rational protein engineering techniques and directed molecular evolution in mammalian cells to identify variants of SRY-related high-mobility group box (SOX) and Pit-Oct-Unc (POU) family TFs that enhance speed and efficiency of pluripotency reprogramming and allow the omission of otherwise essential factors (34–36). For example, pooled saturation mutagenesis library screens led to the identification of eSox17 variants with high performance in pluripotency reprogramming that dwarf the conventionally used Sox2 (36). This led us to test whether eSox17 efficiently substitutes for the commonly used Sox2 to enhance iNSC reprogramming and reduce the requirement for otherwise essential components.
Here, we present a robust protocol which relies on eSox17FNV to efficiently generate self-renewable and tripotent iNSCs from mouse and human somatic cells. Unexpectedly, only eSox17FNV can support iNSC reprogramming, while wild-type Sox factors cannot. POU factors such as Oct4 or Brn4 are not necessary for iNSC reprogramming. The two-factor (2F) cocktails support iNSCs generation, while they are unable to generate iPSCs. We further evaluate the role of metabolism, transgene silencing, chromatin opening, time-course transcriptomics, chromatin scanning, transactivation, and partner factor interaction in iNSC reprogramming to define molecular and physiological cornerstones. A sensitive lineage-tracing system shows that iNSCs reprogrammed with eSox17FNV within 2F and three-factor (3F) cocktails do not transit pluripotency. Mouse iNSCs (miNSCs) generated from aged fibroblasts retain aging-related epigenetic profiles. This indicates the possibility of decoupling reprogramming and rejuvenation and designates iNSCs as a more suitable cell source than iPSCs to model neurodegenerative diseases.
RESULTS
eSox17FNV enables rapid and efficient lineage reprogramming in mouse
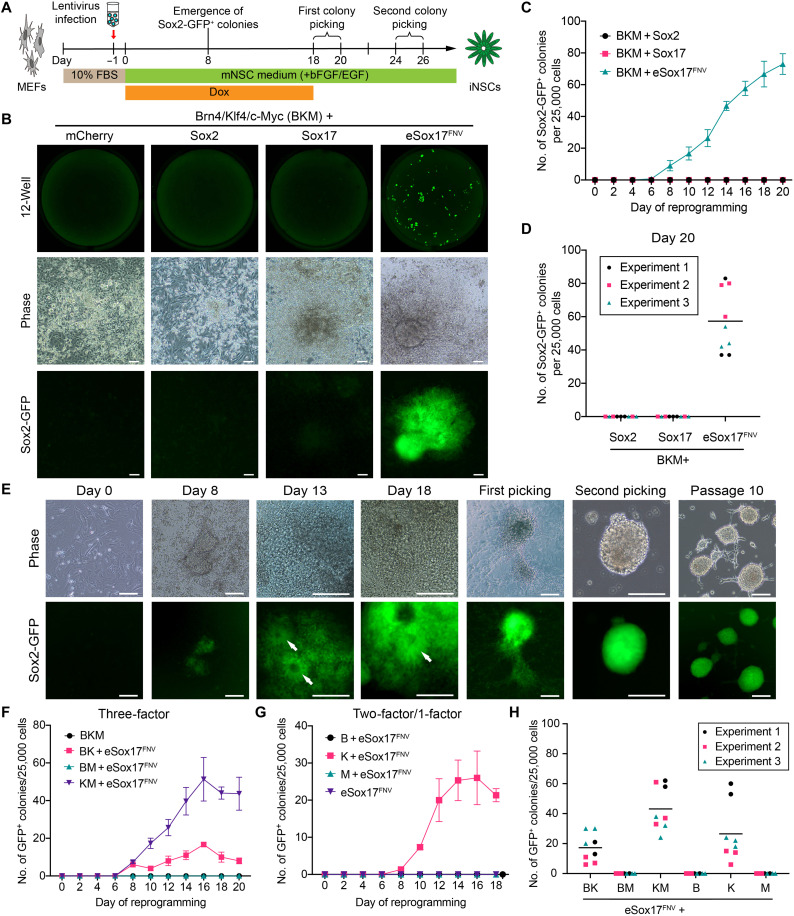
Han et al. (20) reported the intriguing finding that the replacement of Oct4 with Brn4 in a four-factor (4F) cocktail [Brn4 (B, also known as Pou3f4), Klf4 (K), c-Myc (M) and Sox2] leads to the direct reprogramming of fibroblast to a neural stem cell (NSC) state. However, only two to five colonies emerge 4 to 5 weeks after transduction followed by another 2 months to ensure transgene silencing and the acquisition of multipotency (33). We selected this miNSC reprogramming method to evaluate whether engineered TFs (eTFs) can enhance efficiency and increase the kinetics of neural reprogramming. We used an established mouse NSC medium supplemented with basic fibroblast growth factor (bFGF) and epidermal growth factor (EGF) (Fig. 1A), a Sox2-GFP reporter to identify miNSCs (fig. S1A) (37) and doxycycline (Dox)–inducible lentiviral vectors. Mouse embryonic fibroblasts (MEFs) were isolated from Sox2-GFP+ embryos (fig. S1B), and the contaminant GFP+ neural cells were removed by fluorescence-activated cell sorting (FACS) (fig. S1, C and D). We initially performed pooled screens in which Sox2 was replaced by site-saturation mutagenesis libraries of Sox17, and identified several eSox17 variants that could generate Sox2-GFP+ cell clusters (fig. S1E). However, none of them outperformed our previously identified Sox17L111F/V118N/E122V triple mutant (henceforth called eSox17FNV) (36) with regard to reprogramming speed and efficiency (fig. S1F). We thus chose the eSox17FNV variant for further iNSC reprogramming experiments. We then performed time-course experiments to determine the optimal duration of reprogramming factor overexpression (fig. S1G). While B/K/M/eSox17FNV efficiently generated Sox2-GFP+ colonies, we were unable to obtain any Sox2-GFP+ colonies with the NSC factor Sox2 or the endodermal gene Sox17 even if we extended the reprogramming experiments up to 35 days (Fig. 1, B and C, and fig. S1, H and I). The minimal time period of Dox treatment to induce GFP+ colonies that can be stably expanded was 12 days, and the number of GFP+ colonies increased substantially with prolonged Dox treatment (fig. S1I).
Fig. 1. eSox17FNV rapidly and efficiently generates mouse Sox2-positive cells.
(A) Schematic illustration of the neural reprogramming experiment. (B) Whole-well scans (top) of wells from 12-well plate at reprogramming day 20 using a GFP channel; representative phase-contrast (middle) and corresponding Sox2-GFP fluorescence (bottom) images of reprogramming cells in indicated conditions at day 20. Scale bar, 80 μm. (C) Number of Sox2-GFP+ colonies in indicated conditions from day 0 to day 20. (D) Counts of Sox2-GFP+ colonies at day 20 of reprogramming in indicated 4F conditions. (E) Representative microscope images of miNSC reprogramming with B/K/M/eSox17FNV from day 0 to day 18 and picked miNSC colonies at passages 1, 2, and 10. Neurorosette-like structures are indicated by white arrows. Scale bar, 160 μm. (F and G) The number of Sox2-GFP+ colonies in indicated 3F (F) and 2F/one-factor (G) conditions from day 0 to day 20 of reprogramming. (H) Counts of GFP+ colonies at day 20 of reprogramming in indicated 3F and 2F conditions containing eSox17FNV. (C), (F), and (G) show a representative reprogramming time course performed in three technical replicates as mean ± SEM, and (D) and (H) show data points from three independent experiments done in triplicate, the black bar denotes the mean. Source data are provided in data S1. B, Brn4; K, Klf4; M, c-Myc.
In the eSox17FNV condition, we observed the first Sox2-GFP+ colonies as early as day 8, and the number of GFP+ colonies increased rapidly after day 8 to 40 to 80 GFP+ colonies per 25,000 starting cells by day 20 (Fig. 1, C and D). The Sox2-GFP+ cells formed neurosette-like structures after day 13 and could be maintained as individual clonal lines for more than 20 passages in the absence of Dox while retaining the ability to form neurospheres on gelatin (Fig. 1E). These results show that eSox17FNV can rapidly and efficiently generate Sox2+ cells from fibroblasts while the wild-type Sox genes fail.
eSox17FNV and Klf4 are sufficient for mouse lineage reprogramming
To evaluate the requirement for each factor of the 4F cocktail to generate Sox2+ cells, we omitted individual TFs in a systematic manner. In 3F conditions, B/K/M and B/M/eSox17FNV cocktails could not convert MEFs into Sox2+ cells. Conversely, a K/M/eSox17FNV cocktail without Brn4 can generate Sox2+ cells with efficiency and kinetics reminiscent of the 4F cocktail. A B/K/eSox17FNV cocktail lacking cMyc can also generate Sox2+ cells although with decreased efficiency (Fig. 1, F and H, and fig. S2A). Among all of the 2F combinations, only eSox17FNV alongside Klf4 could generate Sox2+ cells, while eSox17FNV alone could not (Fig. 1, G and H, and fig. S2B). These Sox2+ cells generated with eSox17FNV 3F and 2F cocktails could be expanded for more than 20 passages (fig. S2, C and D). Together, these results suggest that POU and Myc factors are dispensable for the generation of Sox2+ cells while the eSox17FNV/Klf4 duo is essential.
eSox17FNV enables lineage reprogramming of adult and old fibroblasts
Adult, aged, and senescent cells are typically more recalcitrant and difficult to reprogram compared to embryonic cells. To address whether eSox17FNV-driven reprogramming depends on specific starting cell populations, we tested reprogramming of Sox2-GFP adult ear fibroblasts (aEFs, 8-week-old) and old tail-tip fibroblasts (oTTFs, 12-month-old) (fig. S2E). aEFs and oTTFs could be effectively reprogrammed into Sox2+ cells with eSox17FNV containing 4F and 3F cocktails (fig. S2, F and G). These results suggest that eSox17FNV is capable to reprogram not only fetal cells but also aged mouse cells from different tissues toward Sox2+ cells.
eSox17FNV-reprogrammed Sox2-positive mouse cells can differentiate into neurons, astrocytes, and oligodendrocytes
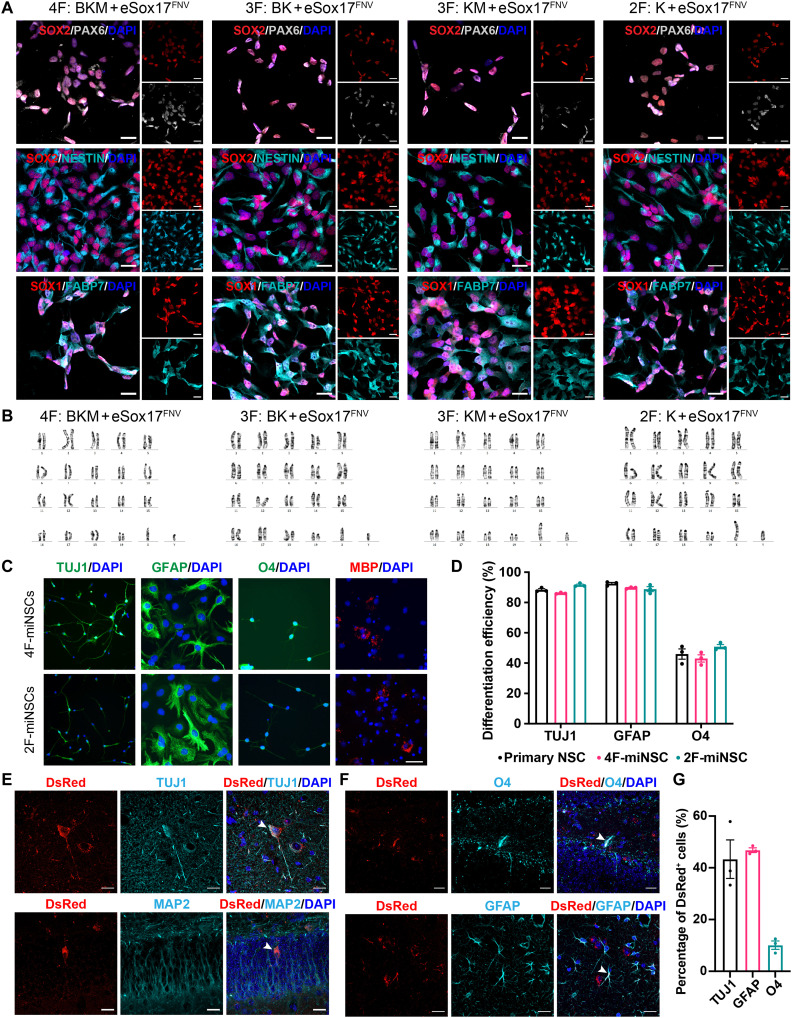
We next characterized eSox17FNV-reprogrammed Sox2+ cells. These Sox2+ cells generated from embryonic, adult, and aged mouse fibroblasts with different factor combinations express the NSC markers Nestin, Sox1, Sox2, Pax6, Olig1, Olig2, and Fabp7 at protein and transcript levels (Fig. 2A and fig. S3, A and B). By contrast, they did not express fibroblast genes Thy1, Col1a1, and Pdgfr (fig. S3C) or pluripotency genes Pou5f1 and Nanog (fig. S3D). Clonal cell lines derived from these Sox2+ colonies showed lentiviral transgene silencing after three passages in the absence of Dox (fig. S3E). These Sox2+ cells could be stably expanded on gelatin and Matrigel in mNSC medium for more than 20 passages without acquiring karyotypic abnormalities (Fig. 2B).
Fig. 2. miNSCs generated with eSox17FNV self-renew and are tripotent.
(A) Immunocytochemistry of MEF-derived miNSCs using antibodies against NESTIN, SOX1, SOX2, PAX6, and FABP7. Scale bar, 20 μm. (B) Representative karyogram of MEF-derived miNSCs at passage 15. (C and D) In vitro differentiation of MEF-derived 4F and 2F miNSCs into Tuj1+ neurons, GFAP+ astrocytes, and O4+ or MBP+ oligodendrocytes determined by immunostaining (C). Differentiation efficiency was quantified by the ratio of the number of immunoreactive cells to the number of DAPI+ cells (D). Scale bar, 40 μm (C). D, n = 3, mean ± SEM. (E to G) In vivo differentiation of miNSCs. DsRed-tagged MEF-derived 2F miNSCs transplanted into the mouse hippocampus differentiated into Tuj1+ or MAP2+ neurons, GFAP+ astrocytes and O4+ oligodendrocytes. Arrows point to representative colocalized cells. Scale bar, 20 μm. (G) Quantification of DsRed+ cells that are immunoreactive for Tuj1, GFAP, or O4 (n = 3 mice, mean ± SEM).
To evaluate the multipotency of these Sox2+ cells, we first differentiated them in vitro into Tuj1+ neurons, glial fibrillary acidic protein–positive (GFAP+) astrocytes, and oligodendrocytes immuoreactive for O4 and myelin basic protein (MBP) (Fig. 2C and fig. S3, F and G). These Sox2+ cells showed an efficient differentiation potential reminiscent to primary NSCs (Fig. 2D). We further investigated their differentiation potential in vivo. DsRed-labeled 2F-reprogrammed Sox2+ cells derived from MEFs were transplanted into the hippocampus of adult mice and analyzed after 2 months. They survived, did not form tumors, and were able to differentiate into Tuj1- and MAP2-positive neurons (Fig. 2E), O4-positive oligodendrocytes, and GFAP-positive astrocytes (Fig. 2, F and G). In sum, eSox17FNV-reprogrammed Sox2+ cells express NSC markers and self-renew. They are tripotent with the ability to generate neurons, astrocytes, and oligodendrocytes in vitro and in vivo. Therefore, we term them miNSCs.
eSox17FNV-reprogrammed miNSCs exhibit a gene expression profile reminiscent of primary NSCs
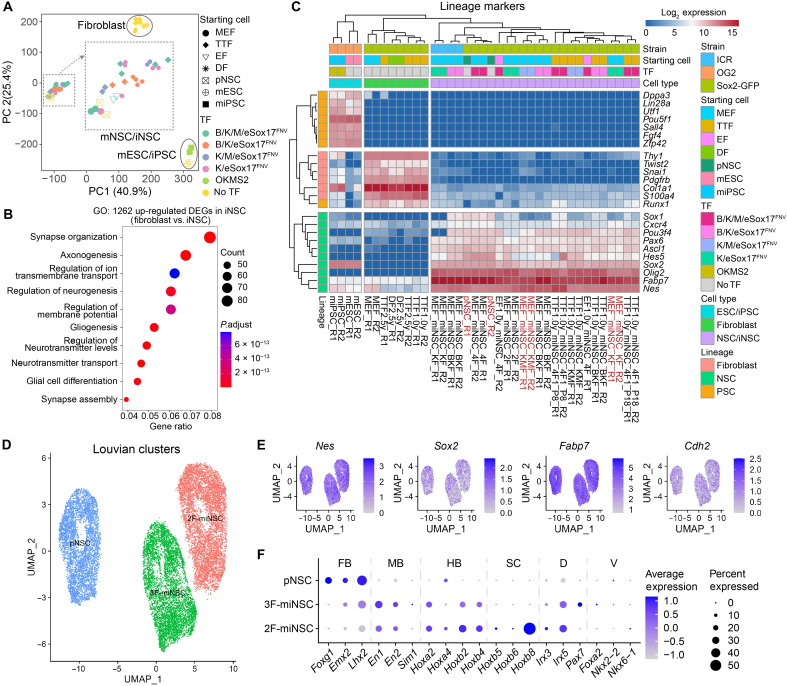
To further verify the identity of eSox17FNV-reprogrammed miNSCs, we next profiled their gene expression by bulk RNA sequencing (RNA-seq). Principal components (PC) analysis (PCA) showed that all miNSCs closely cluster with primary mouse NSCs but clearly differ from the parental fibroblasts and mouse PSCs including embryonic stem cells (ESCs) and iPSCs (Fig. 3A and fig. S4A). Differential gene expression analysis showed 1262 and 4202 differentially expressed genes (DEGs) in miNSCs compared with fibroblasts or PSCs, respectively (fig. S4, B and C). Gene ontology (GO) analysis of the 1262 up-regulated DEGs in miNSCs compared with fibroblasts revealed an enrichment for neuronal functions such as synapse organization, axonogenesis, and regulation of neurogenesis (Fig. 3B). Consistent with immunostaining and quantitative real-time polymerase chain reaction (qRT-PCR) results, miNSCs and primary NSCs showed high expression levels of NSC markers but not pluripotency markers or fibroblast markers (Fig. 3C). We observed some variability across miNSC lines generated with 2F or 4F cocktails. For example, miNSC lines obtained by 2F reprogramming showed a lower expression of some NSC markers such as Sox1 and Pou3f4 compared to 4F-generated iNSC lines (Fig. 3C). Yet, in 2F miNSCs, the expression of these two markers is substantially up-regulated compared with fibroblasts (fig. S4D).
Fig. 3. Expression profile and positional identity of eSox17FNV-reprogrammed miNSCs.
(A) Principal component (PC) analysis (PCA) of global gene expression profiles determined by bulk RNA-seq with two technical replicates. Mouse ESC and iPSC data are from GSE93029. (B) Gene ontology (GO) analysis performed using up-regulated DEGs in miNSCs compared with fibroblasts. (C) Clustered heatmap based on the expression of selected fibroblast, NSC and PSC lineage markers represented as log2-transformed read counts. Cells used for single-cell RNA-seq (scRNA-seq) were highlighted in red. (D) Uniform manifold approximation and projection (UMAP) plots of Louvain clusters of scRNA-seq data of primary mouse NSCs (pNSC), K/M/eSox17FNV-reprogrammed 3F-miNSCs and K/eSox17FNV-reprogrammed 2F-miNSCs. (E) UMAP plots of NSC genes in scRNA-seq data. (F) Dot plots of regional identity genes for forebrain (FB), midbrain (MB), hindbrain (HB) and spinal cord (SC) as well as dorsal (D) and ventral (V) axis in scRNA-seq data. TTF, tail-tip fibroblast; EF, ear fibroblast; DF, dermal fibroblast; pNSC, primary Sox2-GFP NSC; mESC, OG2 mouse ESC; miPSC, OG2 mouse iPSCs; OKMS2, Oct4/Klf4/c-Myc/Sox2.
We next inspected the expression of regional markers for the antero-posterior and dorso-ventral axes in the RNA-seq data. Most miNSCs lines express forebrain markers (Emx2 and Lhx2), midbrain markers (En1 and En2), and hindbrain markers (Gbx2, Hoxa1, Hoxa2, Hoxa4, Hoxb2, and Hoxb4) (38). The forebrain marker Foxg1 is only expressed in one 4F-miNSC line as well as in the primary NSC derived from embryonic mouse brain (fig. S4E). Most of the MEF- and EF-derived miNSCs express dorsal markers (Irx3 and Irx5) but not ventral markers (Nkx2-2, Nkx6-2, and Shh) (39) or spinal cord markers (Hoxb5, Hoxb6, Hoxb7, and Hoxb8), suggesting a dorso-anterior identity of these miNSCs (fig. S4E). By contrast, the tail-tip fibroblast (TTF)–derived miNSCs and one 2F MEF-derived miNSC line express spinal cord markers (Hoxb5, Hoxb6, and Hoxb8) and dorsal marker (Irx3 and Irx5) but not ventral markers, suggesting a dorso-posterior identity of these miNSCs (fig. S4E).
To evaluate the homogeneity of miNSC lines, we performed single-cell RNA-seq (scRNA-seq) for a K/M/eSOX17FNV-reprogrammed 3F-miNSC line and a K/eSOX17FNV-reprogrammed 2F-miNSC line as well as a primary NSC line. These three cell lines show similar expression of NSC markers (Fig. 3, D and E). The three cell lines form different clusters as they differentially express regional identity markers in line with bulk RNA-seq analysis. The forebrain markers Foxg1, Emx2, and Lhx2 are highly expressed in the primary NSCs, the spinal cord marker Hoxb8 is highly expressed in the 2F-minNSCs, and dorsal markers Irx3 and Irx5 are expressed in all these cell lines (Fig. 3F, and data S2). scRNA-seq analysis also identifies cell cycle stages in all these three clusters (fig. S4F), which underscores the ability of these cells to proliferate and self-renew. Together, these results show that miNSCs generated with eSox17FNV form homogeneous cultures resembling primary NSCs. Yet, both the reprogramming factor cocktail as well as the cell source for the reprogramming experiments affect the regional identities of miNSC lines.
miNSCs derived from old fibroblasts resist a full epigenetic reset
iPSC induction is accompanied by a drastic epigenetic reset and the rejuvenation of old cells (40). Therefore, iPSC-derived neural lineages are also rejuvenated even if the somatic cells used for reprogramming were derived from aged donors (16, 22). To evaluate whether our miNSC reprogramming method also leads to rejuvenation, we generated miNSCs from 2.5-year-old mouse TTFs and performed reduced representation bisulfite sequencing (RRBS). Age prediction with a Mouse Epigenetic Clock (41) revealed that the biological ages of two of five miNSC lines are close to their chronological ages (fig. S5A). PCA of the methylation patterns of 20 aging-related hypermethylated regions (42) indicated that miNSCs from old TTF show a methylation pattern that is distinct from young primary NSCs and MEFs (fig. S5B). In particular, miNSCs from old TTFs show high methylation levels at nine CpG sites of Hsf4 promoter (fig. S5, C and D). These results indicate that miNSCs derived from old fibroblasts do not experience the full epigenetic reset encountered during iPSC generation and suggest that it is possible to decouple iNSC reprogramming from rejuvenation.
A glycolytic switch is required for miNSC reprogramming
We next set out to investigate the roles of Klf4 and eSox17FNV factors in neural transdifferentiation. We noted a color change of the reprogramming medium that depends on the used TF combinations. The NSC medium of cells transduced with K/eSox17FNV turned yellow faster than that of cells transduced with B/eSox17FNV or M/eSox17FNV (fig. S6A). This might indicate lactic acidosis caused by enhanced glycolysis, a metabolic state reminiscent of the Warburg effect in cancer cells (43). Thus, we evaluated the effect of metabolic change during miNSC reprogramming. The treatment of a glycolytic inhibitor dichloroacetic acid markedly reduced the number of Sox2-GFP+ miNSC colonies in both B/K/M/eSox17FNV and K/eSox17FNV conditions (fig. S6B) without affecting cell growth (fig. S6C). This indicates that glycolysis is crucially required for iNSC generation.
We found that in 2F conditions, overexpression of Klf4, compared with Brn4 and c-Myc, decreased mitochondrial content (fig. S6D) and increased glucose uptake (fig. S6E). We found that the glucose transporter genes Slc2a1 [encoding glucose transporter 1 (GLUT1)] and Slc2a3 [encoding glucose transporter 3 (GLUT3)] (fig. S6F) were selectively up-regulated under conditions that support miNSC generation. In miNSC reprogramming condition, overexpression of Klf4 led to substantially increased glycolysis and glycolytic capacity. By contrast, overexpression of Brn4, c-Myc, or eSox17FNV did not affect the metabolic phenotype of MEFs (fig. S6, G to L). These results suggest that Klf4 plays a key role in miNSC reprogramming by up-regulating glycolytic activity.
Transgene silencing permits miNSC maturation after eSox17FNV priming
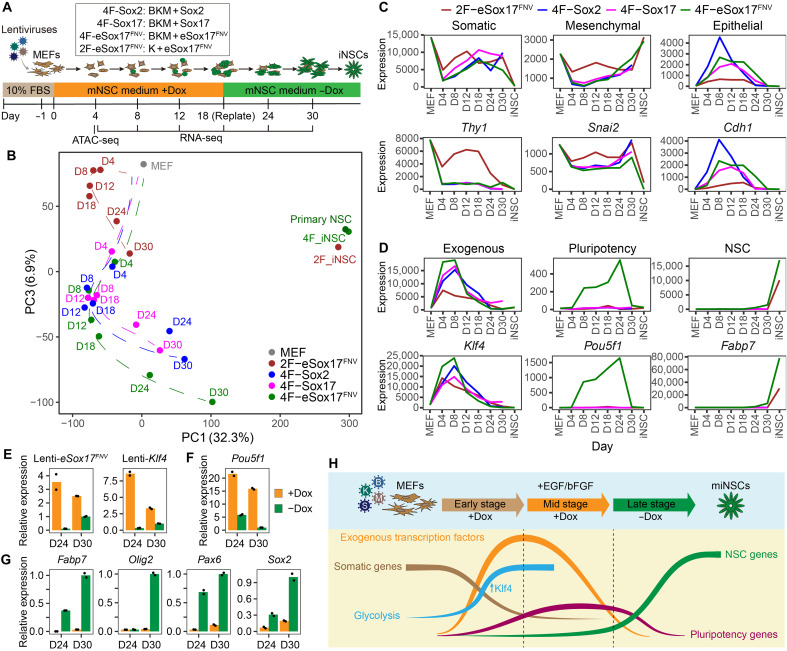
To understand the unique role of eSox17FNV in iNSC generation, we measured gene expression of cell intermediates from reprogramming day 4 to day 30 by bulk RNA-seq (Fig. 4A). We included 4F conditions transduced with B/K/M/Sox2 (4F-Sox2), B/K/M/Sox17 (4F-Sox17), and B/K/M/eSox17FNV (4F-eSox17FNV) and a 2F condition transduced with K/eSox17FNV (2F-eSox17FNV) (fig. S7, A and B). The expression profiles of the 2F-eSox17FNV condition were distinct from the 4F conditions at all time points and showed a separated route (Fig. 4B and fig. S7C). At the early stage of reprogramming (day 4 and day 8), all 4F conditions showed rapid loss of somatic MEF identity, silencing of mesenchymal genes, and activation of epithelial genes (Fig. 4C and fig. S7, D to F), indicating a mesenchymal-to-epithelial transition (MET) (44). However, compared with the 4F condition, the K/eSox17FNV 2F condition showed mitigated silencing of mesenchymal genes and reduced up-regulation of epithelial genes (Fig. 4C and fig. S7, D to F). These results suggest that cells in the K/eSox17FNV 2F condition go through a different trajectory during reprogramming.
Fig. 4. Transgene silencing is critical for the maturation toward miNSCs.
(A) Schematic illustration of miNSC reprogramming and RNA-seq, ATAC-seq experiments. (B) PCA of mean expression profiles of two RNA-seq replicates in indicated conditions determined by RNA-seq. D, day. (C and D) Mean expression of genes for indicated categories (top) and a representative gene from each category (bottom) in indicated conditions. Dash line indicates the time point to withdraw Dox at day 18. See fig. S7 (D to H) for more genes in each category. (E to G) Expression of lentiviral transgenes (E), pluripotency gene (F), and NSC genes (G) in cells cultured in NSC medium at day 24 and day 30 with or without Dox detected by qRT-PCR (n = 2, mean). (H) Schematic diagram demonstrating the sequential events occurring during miNSC reprogramming with 4F cocktail.
Before day 18 (Dox-on stage), the three 4F conditions showed overall similar global expression profiles at each time point (Fig. 4B and fig. S7C), indicating that global expression changes progress similarly regardless of whether reprogramming competent or incompetent Sox factors were transduced. At this stage, NSC markers are still barely expressed in all conditions and only very slowly emerge in the eSox17FNV condition (Fig. 4D and fig. S7G). Notably, in the 4F-eSox17FNV condition, pluripotency genes such as Pou5f1 (encoding OCT4) were selectively up-regulated (Fig. 4D and fig. S7H). When Dox was removed after day 18 (Dox-off stage), the expression of exogenous factors was rapidly silenced (Fig. 4D). Then, the 4F-eSox17FNV condition started to show distinct expression profiles and gradually transits toward NSCs in stark contrast to Sox2 or Sox17-expressing cells (Fig. 4D). Pluripotency genes markedly decreased, and NSC genes such as Fabp7 were prominently up-regulated in this condition (Fig. 4D and fig. S7, G and H). At day 30, the 4F-eSox17FNV condition showed a clear up-regulation of NSC markers over the 4F-Sox2 and 4F-Sox17 conditions (fig. S7I). These results indicate gradual maturation of 4F-eSox17FNV-reprogrammed cells into NSCs after exogenous gene silencing. After manual picking and expansion, the established 4F-eSox17FNV and 2F-eSox17FNV miNSC clonal lines became similar to the primary NSCs (Fig. 4B), and GO enrichment analysis of the top 200 PC1 genes showed a prevalence of neural-related functions (fig. S7J).
To evaluate the effect of exogenous factors at the late stage of reprogramming, we extended Dox treatment in the 4F-eSox17FNV condition. We found that continuous Dox treatment led to the failure of complete exogenous gene silencing (Fig. 4E), high expression levels of pluripotency genes (Fig. 4F), and inhibition of NSC gene activation (Fig. 4G). By contrast, after withdrawing Dox, exogenous genes were rapidly silenced (Fig. 4E), pluripotency gene expression decreased (Fig. 4F), and NSC genes started to express (Fig. 4G). Collectively, results suggest that the exogenous 4F-eSox17FNV cocktail initially reprograms the cells to a plastic intermediate stage necessary for the subsequent maturation toward miNSCs. Exogenous factor silencing is required for the cells to mature into authentic miNSCs (Fig. 4H).
The chromatin opening in miNSC reprogramming is muted compared to iPSC reprogramming
The ability to access and open silent chromatin and initiate cell fate conversion is a key feature of iPSC reprogramming factors, and widespread opening and closing of chromatin is apparent from day 1 in pluripotency medium (45). To evaluate the chromatin accessibility at the onset of iNSC reprogramming and to ask whether the three 4F cocktails engage and remodel the chromatin of MEFs differentially, we performed ATAC-seq (assay for transposase-accessible chromatin using sequencing) (46). PCA of ATAC-seq peaks revealed a cluster of 4F conditions distinct from the 2F condition and MEFs (fig. S8A). We next defined categories of ATAC-seq peaks with regard to their status in MEFs to identify closing or opening sites at this early stage of reprogramming. Most of the open chromatin loci were already open in MEFs (OO) in all samples. Around 8.2% of the sites that were open in MEFs got closed in all the reprogramming conditions (OC). About 4.6% of the sites were closed in MEFs but commonly open in all reprogramming conditions (CO), and only 1.5% of the sites were specifically opened in different reprogramming conditions (condition-specific CO) (fig. S8B). Overall, the data showed similar global chromatin opening associated with TF induction, and only a small portion of genomic loci were uniquely opened by each Sox factor at the early stage of reprogramming. Among the open or closed genomic loci, we identified somatic genes specific to OC group, such as Cdh2 and Twist2, and epithelial genes specific to the CO group, such as Cdh1 and Krt78 (fig. S8C), indicating the occurrence of MET.
We compared our ATAC-seq data of miNSC reprogramming with the public ATAC-seq data of mouse iPSC reprogramming (45, 47). iPSC reprogramming using Klf4/Sox2 or Oct4/Klf4/Sox2 at day 1 showed three times more CO peaks than miNSC reprogramming with B/K/M/Sox cocktails at day 4, while the number of OC peaks were similar (fig. S8D). This global analysis suggests that the pioneer activity of eSox17FNV, Sox2, and Sox17 is broadly similar at the onset of reprogramming. Further, the chromatin opening in miNSC reprogramming conditions is overall more muted compared to the rapid and profound changes seen in iPSC reprogramming as early as day 1 (45, 47).
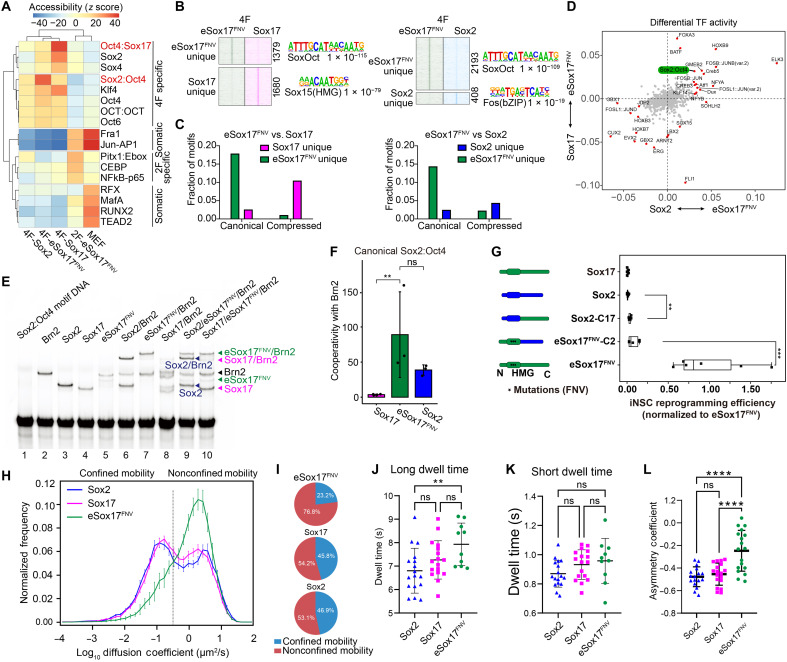
eSox17FNV dimerize with neural POU factors on the canonical SoxOct motif more tightly than Sox2
We next analyzed the ATAC-seq data in a more quantitative manner to contrast the chromatin landscape formed by the three Sox factors. We used the sensitive chromVAR method (48) to ask whether peaks annotated with specific TF motifs exhibit a detectable variation in chromatin accessibility across all ATAC-seq samples. We found that peaks containing the canonical Sox2:Oct4 motifs showed increased accessibility specifically in the 4F-eSox17FNV condition (Fig. 5A). By contrast, loci with the compressed Oct4:Sox17 motifs, single Sox motifs (Sox2 and Sox4), and Klf4 motifs showed increased accessibility in the 4F-Sox17 condition (Fig. 5A). Both canonical and compressed SoxOct motifs are composed of directly adjacent Sox and Oct half sites that differ in a single base-pair spacer (canonical: CATTGTcATGCAAAT versus compressed CATTGTATGCAAAT; octamer sequences underlined, spacer in small cap). Using MAnorm (49), we defined 1379 and 2193 peaks unique for eSox17FNV when compared with Sox17 or Sox2, respectively, many of which are marked by the canonical SoxOct motif (Fig. 5, B and C). Likewise, ATAC-seq footprinting analysis using hmm-based identification of transcription factor footprints (HINT) (50) revealed a specific binding activity of eSox17FNV at loci with the canonical SoxOct motifs (Fig. 5D). Reanalysis of chromatin immunoprecipitation sequencing data during miPSC reprogramming (36) verified that eSox17FNV acquired a strong preference for the canonical Sox2:Oct4 motif while Sox17 prefers the compressed Sox17:Oct4 motif (fig. S8E). Consistently, electrophoretic mobility shift assays (EMSAs) using purified proteins of the Sox high mobility group domain (HMG) and Brn2 POU domains with fluorescent DNA probes showed that eSox17FNV cooperatively dimerizes with the neural POU factor Brn2 on the canonical SoxOct motif reminiscent to Sox2 and in stark contrast to wild-type Sox17 (Fig. 5, E and F, and fig. S8, F and G). We also performed EMSAs using full-length proteins extracted from whole-cell lysates and verified the preference of Sox17/Brn2 dimers for the compressed and of Sox2/Brn2 as well as eSox17FNV/Brn2 dimers for the canonical element (fig. S8H). In single-tube competition assays, eSox17FNV outcompeted Sox2 in the dimerization with Brn2 on the canonical DNA element, suggesting that it dimerizes with critical partner factors more tightly (fig. S8H). This may contribute to the outperformance of eSox17FNV over Sox2 in miNSC reprogramming. Collectively, these results indicate that eSox17FNV acquired fundamentally different preference to associate with partner factors to open chromatin in comparison to Sox17. This suggests that altered protein-protein interactions in a chromatin context are key to direct the reprogramming process.
Fig. 5. DNA engagement, dimerization and chromatin mobility of eSox17FNV is profoundly different from Sox17.
(A) Accessibility variation of ATAC-seq peaks annotated by the presence of indicated TF motifs determined by chromVAR (48). (B) Heatmaps for ATAC-seq signals around unique peaks defined by MAnorm (49) with a peak number and top known motif of each cluster. (C) Occurrences of the canonical Sox2:Oct4 motifs and the compressed Oct4:Sox17 motifs mapping to ATAC-seq peaks in each of the four clusters in (B). (D) Motifs with differential TF binding activity in ATAC-seq footprints analyzed by HINT-ATAC (50). P < 0.05, t test. (E) EMSA using indicated mixtures of purified Sox HMG proteins with the Brn2 POU protein and Cy5-labeled canonical Sox2:Oct4 DNA element. Brn2 POU, amino acids 264 to 421; Sox2 HMG, amino acids 41 to 119; Sox17 HMG, amino acids 66 to 144; eSox17FNV, amino acids 58 to 166. (F) Estimation of cooperativity factors for the three Sox factors and Brn2 on the canonical Sox2:Oct4 DNA element. n = 3 to 5, mean ± SD, **P < 0.01 from Tukey HSD. (G) miNSC reprogramming efficiency using Sox2, Sox17, and eSox17FNV domain swap constructs co-expressed with Brn4, Klf4, and c-Myc. n = 2 biological replicates with three technical replicates. t test, ***P < 0.001, **P < 0.01. (H to K) SMT assay of Sox2, Sox17, and eSox17FNV. (H) Population mobility profile based on mean squared displacement. (I) Nonconfined mobility versus confined mobility population based on the mobility profile. Long-lived dwell time (J) and short-lived dwell time (K). (J and K) n ≥ 10 cells. (L) AC calculated by the frequency of forward angles (0° to 30°) and backward angles (150° to 180°) (51). n ≥ 17 cells. (J to L) Mean ± SD, Tukey-corrected analysis of variance (ANOVA) post hoc test, **P < 0.005, ****P < 0.0001.
The C terminus of eSox17FNV is critical for high-performance miNSC reprogramming
We next wondered whether the poorly conserved C termini of both proteins distinguish their function in iNSC generation. A Sox2 construct with the Sox17 C terminus (Sox2-C17) bestowed some iNSC reprogramming capacity to the otherwise inactive Sox2 (Fig. 5G). Conversely, replacing the C terminus of Sox17 with the C terminus of Sox2 (eSox17FNV-C2) led to a marked impairment of the reprogramming capabilities of eSox17FNV (Fig. 5G). This shows that the HMG box alone cannot explain the unique ability of eSox17FNV to drive iNSC reprogramming but its C-terminal transactivation domain critically contributes to this function and the outperformance of Sox2.
Single-molecule tracking reveals that Sox17 and eSox17FNV scan chromatin differently
Previous studies have used single-molecule tracking (SMT) to reveal the search pattern of TFs on chromatin and its effect on gene regulation (51–54). We used SMT to investigate how eSox17FNV navigates the human genome in comparison to both Sox2 and Sox17. To do this a Halo-tagged version of each TF was used (51, 53, 54). This method provides molecular mobility profiles based on mean squared displacement, the temporal binding dynamics, and distribution of angles between consecutive frames to distinguish confined or nonrestricted motions of the TFs. Compared to wild-type Sox17 and Sox2, eSox17FNV molecules have a greater nonconfined mobility fraction of the population (Fig. 5, H and I). Despite an overall more mobile behavior, the long dwell time of eSox17FNV was increased compared to Sox2 (Fig. 5, J and K). The long dwell time signifies specific binding to functional binding sites likely involved with transcriptional modulation per se, while the short dwell time is considered to represent states where a TF is associated with nonspecific binding sites or scanning behavior (52, 53). The increase in diffusion of eSox17FNV is also observed in the distribution of angles between consecutive frames. Typically, small-angle trajectories (0° to 30°) denote a more linear trajectory and have a more diffusive behavior, whereas trajectories with large angles (150° to 180°) correspond to more confined and restricted mobility (51). The eSox17FNV molecules show an enlarged proportion of smaller angles compared to wild-type Sox17 and Sox2 (fig. S9, A to D). This observation is further mirrored by the asymmetry coefficient (AC) with eSox17FNV having a more positive AC compared to the other Sox2 and Sox17 (Fig. 5L). A negative AC represents the frequency at which molecules encounter obstacles, are caught in confined regions, or transiently interact with other proteins (51, 55).
Together, these data imply that eSox17FNV navigates the genome in an altered pattern compared to its wild-type counterpart. Subtle mutations at the DNA binding domain that affects protein partnerships and cobinding to DNA profoundly change diffusion, chromatin scanning, and dwell time which in aggregate facilitates the efficiency to search for target genes.
eSox17FNV-mediated miNSC reprogramming sidesteps a pluripotent state
Lineage tracing with Nanog or Oct4 reporters suggested that transdifferentiation of somatic cells into iNSC driven by the BSKM or restricted expression of OSKM may pass through a transient pluripotent stage (30, 31). Our goal, however, is to establish an iNSC reprogramming method that exclusively directs cells toward a neural lineage to avoid the shortcomings of iPSCs. We first used Oct4-GFP reporter MEFs to monitor the pluripotency status during miNSC reprogramming (fig. S10A). Unexpectedly, the Oct4-GFP reporter was occasionally activated in K/M/eSox17FNV conditions even when cells were cultured in mNSC medium (fig. S10, B to D). However, in four independent experiments, we only once observed a single GFP+ colony from Oct4-GFP MEFs with K/eSox17FNV, but it was impossible to pick and maintain these cells.
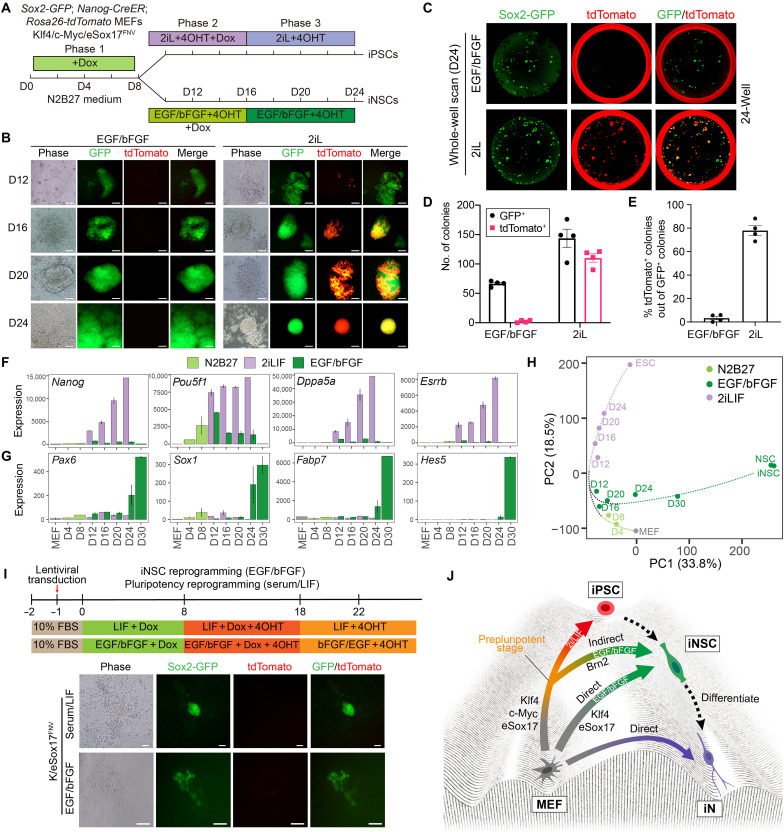
We next used a more stringent Nanog lineage tracing system developed by the Hanna laboratory (30) to unequivocally track the pluripotency acquisition during eSox17FNV-mediated reprogramming. For this purpose, we introduced Nanog-CreER:Rosa26-loxP-STOP-loxP-tdTomato reporter into ESCs derived from Sox2-GFP mice to generate double-reporter MEFs after blastocyst injection (fig. S10E). To further dissect the routes of eSox17FNV-mediated reprogramming in an unbiased manner, we performed miPSC and miNSC reprogramming experiments in parallel using a feeder-free and chemically defined media and the “POU free” 3F cocktail consisting of K/M/eSox17FNV (Fig. 6A). We first induced transgene expression with Dox in serum-free N2B27 medium from days 0 to 8 without growth factors (phase 1). Next, we split cultures and transferred the cells to either N2B27 2i/LIF medium for miPSC induction [phase 2 (2iLIF)] or bFGF/EGF NSC medium [phase 2 (EGF/bFGF)] for miNSC induction. We withdrew Dox at day 16 [phase 3 (2iLIF or EGF/bFGF)] (Fig. 6A). After day 8, Sox2-GFP+/tdTomato− cell colonies emerged in EGF/bFGF conditions and Sox2-GFP+/tdTomato+ colonies in 2iLIF condition (Fig. 6, B and C). At day 24, more than 50 Sox2-GFP+/tdTomato− miNSC colonies or more than 100 double-positive miPSC colonies were obtained (Fig. 6D). Notably, around 80% of Sox2-GFP+ miPSC colonies were also tdTomato+, whereas less than 5% of Sox2-GFP+ miNSC colonies were tdTomato+ (Fig. 6E). This confirms a recent finding by Velychko et al. (56) that POU factors are not required for effective pluripotency reprogramming. It further indicates that NSC conditions strongly drive neural reprogramming and only rare cell populations pass to a pluripotent state. Time-course RNA-seq analysis showed the progressive activation of pluripotency genes in the 2iLIF condition starting when transgenes are active (Fig. 6F). By contrast, the effective activation of NSC genes in the EGF/bFGF condition is delayed and requires transgene silencing (Fig. 6G). PCA indicates the reprogramming roadmap for these two conditions (Fig. 6H). Within the first 8 days when Klf4/c-Myc/eSox17FNV was induced, gene expression profiles gradually departed from MEFs. After the split of induction conditions, the separation of transcriptional states became apparent, indicating the acquisition of divergent cellular fates dictated by extrinsic factors. Cells in 2iLIF condition rapidly developed a gene expression pattern reminiscent of ESC. By contrast, cells in EGF/bFGF condition more slowly approach expression profiles resembling NSCs (Fig. 6H).
Fig. 6. The eSox17FNV/Klf4/c-Myc direct nonoverlapping reprogramming routes toward miPSC and miNSCs driven by distinct signaling cues.
(A) Schematic illustration of iNSC and iPSC reprogramming experiments. 2iL: CHIR99021, PD0325901, and LIF; 4OHT, 4-hydroxytamoxifen. (B) Representative microscope images of colonies in indicated conditions. Scale bar, 80 μm. (C) Whole-well scans of 24-well plates at day 24 of reprogramming using green and red fluorescence channels. (D and E) Counts of Sox2-GFP+ colonies and tdTomato+ colonies in indicated conditions (D) and percentage of tdTomato+ colonies out of Sox2-GFP+ colonies at reprogramming day 24 (E) (n = 4, mean ± SEM). (F and G) Mean RNA-seq expression of selected pluripotency genes (F) and NSC marker genes (G). (H) PCA of transcriptomes of bulk cell populations at different time points with ESCs and NSCs/iNSC as references. (I) Klf4/eSox17FNV combination is not able to activate Nanog during reprogramming. Representative microscope images of cells at day 22 of iNSC and pluripotency reprogramming with Klf4/eSox17FNV. Scale bar, 80 μm. (J) Possible routes of eSox17FNV-mediated reprogramming toward iNSCs and iPSCs.
Next, we tested the dual-reporter MEFs in pluripotency and miNSC reprogramming conditions with the 2F Klf4/eSox17FNV combination. The tdTomato reporter was never activated by Klf4/eSox17FNV transduction in neither Serum/LIF miPSC reprogramming medium nor EGF/bFGF miNSC reprogramming medium (Fig. 6I). This indicates that overexpression of Klf4 and eSox17FNV does not lead to the acquisition of pluripotency. We conclude that eSox17FNV can drive the fibroblasts to alternative cell fates upon exposure to extrinsic factors for pluripotency (LIF) and NSCs (EGF and bFGF). miNSC reprogramming with Klf4/c-Myc/eSox17FNV 3F cocktail typically goes through an Oct4+/Nanog− prepluripotent stage. This state can be avoided when c-Myc is eliminated from the reprogramming cocktail as the Klf4/eSox17FNV combination directly converts fibroblasts into miNSCs in a lineage-exclusive fashion (Fig. 6J).
eSOX17FNV enables reprogramming of human blood cells into iNSCs
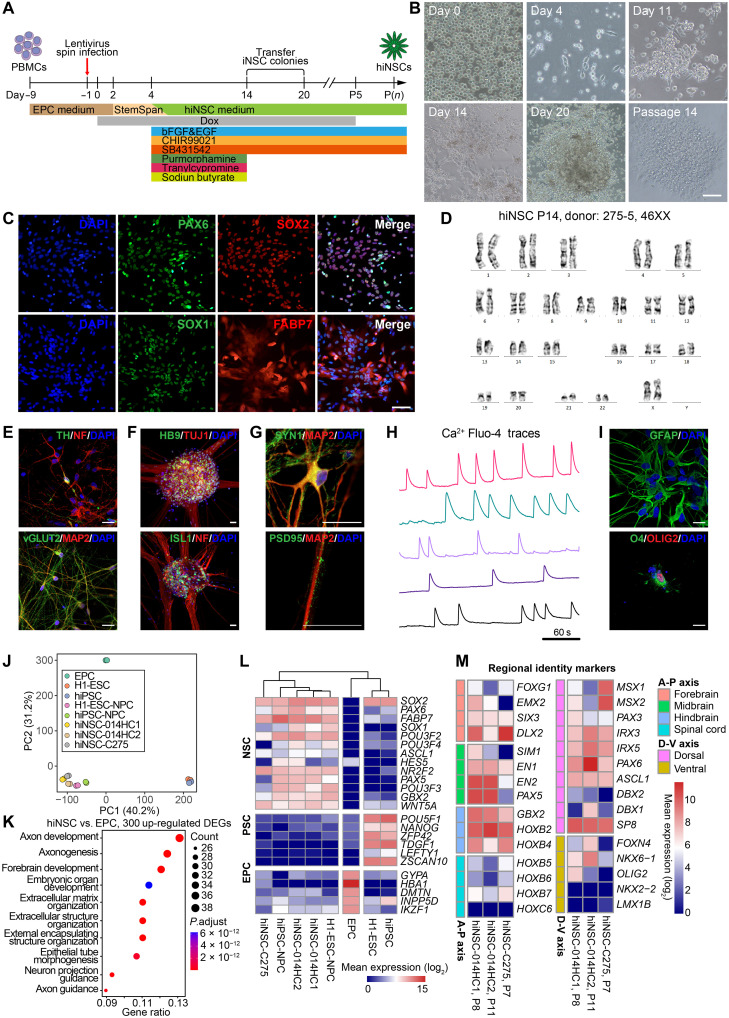
To apply eSox17FNV-driven iNSC reprogramming to human somatic cells, peripheral blood mononuclear cells (PBMCs) were used instead of fibroblasts because of their availability via noninvasive means and a lower mutation burden compared to skin fibroblasts. To avoid chromosome rearrangement frequently observed in lymphocytes, erythroid progenitor cells (EPCs) were expanded and used as starting cells to generate human iNSCs (hiNSCs). EPCs were transduced with Dox-inducible lentiviral particles encoding human KLF4, C-MYC, and eSOX17FNV (K/M/eSOX17FNV). We found that overexpression of K/M/eSOX17FNV and culture in an optimized human NSC medium with bFGF, EGF, CHIR99021 (glycogen synthase kinase-3 inhibitor), SB431542 (transforming growth factor–β receptor type I/ALK5 inhibitor), purmorphamine (PMA) [hedgehog-smoothened (Shh) agonist], tranylcypromine (monoamine oxidase inhibitor), and sodium butyrate (histone deacetylase inhibitor) allows the conversion of EPCs into iNSCs (Fig. 7A and fig. S11A). After omitting individual factors from this 3F cocktail, we found that C-MYC and eSOX17FNV (M/eSOX17FNV) are sufficient for iNSC conversion (fig. S11A). Transduced EPCs formed adherent, elongated cells within 4 days, and NSC-like colonies within 11 to 20 days (Fig. 7B). These colonies can be picked and expanded in hNSC medium supplemented with bFGF, EGF, CHIR99021, and SB431542. Stable hiNSCs independent of transgenes could be generated after five passages (Fig. 7B). Immunostaining and quantitative real-time PCR (qRT-PCR) revealed that hiNSCs express NSC markers SOX1, SOX2, PAX6, and FABP7 (BLBP) (Fig 7C and fig. S11B), but EPC markers CD71 (TFRC) and CD117 (KIT) or pluripotency genes POU5F1 and NANOG could not be detected (fig. S11, C and D). hiNSCs could be stably expanded on Matrigel in hNSC medium for more than 20 passages with a normal karyotype (Fig. 7D).
Fig. 7. Generation of iNSCs with eSOX17FNV from human blood cells.
(A) Schematic illustration of reprogramming of human EPCs into hiNSCs. (B) EPCs were converted to hiNSCs with C-MYC and eSOX17FNV (M/eSOX17FNV) combination. hiNSC colonies emerging at days 11 to 20 were picked and expanded on Matrigel for 14 passages. Scale bar, 160 μm. (C) Immunocytochemistry of hiNSCs using antibodies against SOX2, PAX6, SOX1, and FABP7. Scale bar, 80 μm. (D) Representative karyograms of hiNSCs from EPCs of donor 275-5 at passage 14. (E) After 3 weeks of in vitro spontaneous differentiation, hiNSCs gave rise to NF+/TH+ dopaminergic neurons and MAP2+/vGLUT2+ glutamatergic neurons. (F) After 2 weeks of in vitro motor neuron differentiation, hiNSCs gave rise to TUJ1+/HB9+ and NF+/ISL1+ motor neurons. (G) Neurons spontaneously differentiated from hiNSCs were immunoreactive for MAP2, SYN1, and PSD95. (H) Normalized Ca2+ Fluo-4 traces of fluorescence live-cell imaging in five individual spontaneously differentiated neurons from hiNSCs reveals spontaneous neural network activity. One region of interest (ROI)/cell. (I) In vitro differentiation of hiNSCs into GFAP+ astrocytes and O4+/OLIG2+ oligodendrocytes. (J) PCA of global gene expression profiles determined by RNA-seq. (K) GO analysis performed using top 300 up-regulated DEGs in hiNSCs compared with EPCs. (L) Mean expression of selected NSC, PSC and EPC lineage markers represented as log2-transformed read counts. (M) Mean expression of selected forebrain, midbrain, hindbrain, spinal cord, dorsal and ventral regional identity genes represented as log2-transformed read counts. P, passage. Scale bars, 80 μm (B and C) and 20 μm (E to H). (J and L) H1-ESC data are from GSE75748, hiPSC data are from GSE79636, H1-ESC–derived neural progenitor cell (H1-ESC-NPC) data are from GSE56785. A-P, antero-posterior; D-V, dorso-ventral.
To evaluate the multipotency of M/ eSOX17FNV-reprogrammed hiNSCs, we differentiated hiNSCs in various conditions. In a spontaneous neuronal differentiation condition within 3 weeks, hiNSCs gave rise to NF+/TH+ dopaminergic neurons and MAP2+/vGLUT2+ glutamatergic neurons, respectively (Fig. 7E). hiNSCs could also quickly differentiate into TUJ1+/HB9+ and NF+/ISL1+ motor neurons within 2 weeks (Fig. 7F). Notably, hiNSC-derived neurons were immunoreactive for presynaptic marker Synapsin-1 (SYN1) and postsynaptic marker postsynaptic density protein 95 (PSD95) (Fig. 7G). These neurons (12 of 34) displayed frequent and spontaneous neuronal activity detected by calcium imaging (Fig. 7H and movie S1), indicating the generation of mature and functional neurons. In addition to neuronal lineages, hiNSCs were also able to differentiate into GFAP+ astrocytes and O4+/OLIG2+ oligodendrocytes (Fig. 7I). These results indicated that EPC-hiNSCs can generate multiple neuronal subtypes and glial cells.
To further characterize the hiNSCs, we analyze the global gene expression of EPCs, three hiNSCs lines from two donors, ESCs, hiPSCs, and ESC/hiPSC-derived neural progenitor cells (NPCs). PCA of bulk RNA-seq revealed that hiNSCs clustered closely with ESC/hiPSC-derived NPCs but are distinct from EPC and PSC clusters (Fig. 7J). GO analysis of the top 300 up-regulated genes in hiNSCs compared with EPCs unveiled an enrichment for neural-related biological processes, such as axon development, axonogenesis, and forebrain development (Fig. 7K). In hiNSCs, many NSC markers such as SOX1, SOX2, PAX6, FABP7, and HES5 were highly expressed, while EPC markers such as GYPA, HBA1, and IKZF1 and pluripotency markers such as POU5F1, NANOG, and ZFP42 were repressed (Fig. 7L). We also investigated the regional identity of hiNSCs. hiNSCs expressed forebrain markers (SIX3 and DLX2), midbrain marker (PAX5, EN1, and EN2), hindbrain markers (GBX2, HOXB2, and HOXB4) but not spinal cord markers (HOXB5, HOXB6, HOXB7, and HOXC6). Along the dorso-ventral axis, hiNSCs expressed several dorsal markers such as PAX3, IRX3, IRX5, and SP8, while ventral markers were poorly expressed (Fig. 7M) (39). These results suggest that hiNSCs have a dorso-anterior identity. All together, we demonstrated that eSOX17FNV can generate self-renewing and multipotent iNSCs directly from human blood.
DISCUSSION
iNSCs might combine the benefits of iPSCs and postmitotic iNs while avoiding their limitations to establish authentic, safe, and accessible cell models to study and treat neurological disorders. A problem with past protocols of iNSC generation has been low efficiency and long conversion times that precluded its widespread use across laboratories akin to the now omnipresent iPSC technology (57, 58). Here, we present a fast and efficient approach to generate iNSCs from murine and human cells with eSox17FNV, which comes with several advantages: (i) Compared with somatic-iPSC-NSC pipelines and previous iNSC reprogramming protocols (33) (which may take up to 2 to 3 months), the direct reprogramming of fibroblasts into iNSCs with eSox17FNV is markedly faster with fewer purification steps. The first initial iNSC colonies emerge within 2 weeks of reprogramming and clonal iNSC lines can be established within less than 4 weeks. (ii) Our approach can efficiently generate iNSC colonies from a low number of starting cells. The robust and reproducible iNSC generation with eSox17FNV opens up the opportunity for nonspecialized laboratories to take advantage of iNSCs. (iii) iNSCs generated with eSox17FNV are almost identical to primary NSC in morphology, global gene expression profile, and tripotency, indicating their bona fide NSC identity. iNSCs exhibit vigorous self-renewal in long-term culture, which bestows them clonogenicity and potential for large-scale production and genome editing. iNSCs can be differentiated into diverse neuronal subtypes and gliogenic lineages. This multipotency enables the study of various neurological disorders as well as the generation of multitissue organoid models. When starting from iNSCs, the differentiation into mature neuronal subtypes is substantially faster compared to starting from iPSCs. (iv) Direct reprogramming bypasses the oncogenic iPSC state and generates iPSC-free iNSCs, which could substantially reduce the risk of tumorigenesis when applied for tissue regeneration in vivo. (v) NSCs exhibit molecular aging in the mature brain, which may impair their normal function during aging and brain injury (59, 60). It was reported that iNSCs generated by direct reprogramming fully or partially preserved age-related epigenetic signatures of the parental cells (22, 61). In contrast, the age of iPSCs and iPSC-derived NSCs were epigenetically reset to an embryonic state (22). We show that iNSCs avoid a full epigenetic reset and retain at least some aging-related epigenetic profiles of the starting cells, indicating directly reprogrammed iNSCs could be better cell sources than iPSC and iPSC-derived NSC to model aging-related neurological diseases. The degree of rejuvenation associated with iNSC vis-à-vis iPSC generation in mouse and human cells should be a focus of future research.
In our approach, ectopic eSox17FNV is essential to generate not only miNSCs but also hiNSCs. However, in both systems, an additional accessory factor is needed. In iNSC reprogramming from mouse fibroblasts, Klf4 is required while the reprogramming of human blood requires C-MYC. We reasoned that the species and starting cell types may affect the dependency on accessory reprogramming factors.
Direct reprogramming of somatic cells into iNSCs is a progressive conversion passing through plastic intermediate stages that can also be achieved with factor combinations allowing iPSC generation (62). We used fluorescent reporters, lineage tracing system, and time-course global gene expression analysis to dissect cellular trajectories during iNSC reprogramming. Our results show that Klf4/c-Myc/eSox17FNV efficiently reprograms mouse fibroblasts to iNSCs without maturing into full-blown Nanog-positive pluripotent cells. Cells depart from this intermediate stage once transgenes are switched off guided by extrinsic factors (bFGF and EGF) (Fig. 6J). Yet, in culture conditions favouring pluripotent cells, the 3F Klf4/c-Myc/eSox17FNV cocktail can also effectively generate iPSCs. By contrast, the 2F Klf4/eSox17FNV combination does not activate pluripotency genes throughout the reprogramming period regardless of the culture medium, indicating direct conversion from fibroblasts toward iNSCs (Fig. 6J). This shows that the reprogramming toward pluripotency and multipotency can be achieved using mutually incompatible factor cocktails.
We demonstrate that eTFs can facilitate the direct conversion of somatic cells into iNSCs. Why is only eSox17FNV able to drive fast and efficient generation of iNSCs whereas wild-type Sox2 and Sox17 fail? The inability of Sox2 to promote reprogramming contrasts to previous reports where Sox2-based cocktails led to successful iNSC generation (20). These outcomes can be rationalized by key differences between our method and the pioneering findings from Han et al. First, we used Dox-inducible lentivirus instead of a constitutive retrovirus to control exogenous gene expression. Second, we used a much smaller number of starting cells and shorter reprogramming time frame. Third, we used purified Sox2-GFP MEFs rather than bulk cells without reporter as starting material. To ascertain that engineered Sox17 is indeed critical for our direct lineage reprogramming approach and to work out the mechanistic underpinnings for its function, we used Sox2 and Sox17 as controls in most experiments. Clearly, the changed capacity to bind partner factors including but not limited to Brn2 affects DNA and chromatin interactions and alters the mode as to how eSox17FNV explores genomic DNA as shown by SMT. Compared to Sox2, Sox17 has a unique C-terminal domain that potently promotes iNSC generation (Fig. 5G). How this domain affects epigenetic processes and protein-protein interaction networks remains to be investigated. During transgene overexpression, we only observe subtly different gene expression profiles and chromatin opening between eSox17FNV and Sox2/Sox17. Yet, as soon as transgenes are silenced, profound differences emerge and only cells previously exposed to eSox17FNV mature toward a NSC state. This suggests that eSox17FNV creates a poised state through altered genome engagement and cofactor recruitment but needs to give way to endogenous factors for the lineage commitment to take place. This is in stark contrast to iPSC reprogramming where active transgenes do not impede the activation of pluripotency network. Yet, even for iPSCs, prolonged transgene activation leads to a “fuzzy” state of pluripotency (63).
Together, this study presents a robust approach to reprogram murine and human somatic cells into iNSCs with eSox17FNV. We anticipate that the application of engineered and enhanced TFs (eTFs) could also benefit a wide range of cell fate conversions for basic research and translational purposes. The translation of eTF-driven direct lineage reprogramming of human iNSCs could lead to more authentic cell models for age-associated neurodegenerative by advancing speed of cell model generation, eliminating the safety risk of oncogenes, and reducing the rejuvenation associated with the iPSC technology while retaining the scalability and multilineage potential of stem cells.
MATERIALS AND METHODS
Ethics statement
PBMCs were obtained from healthy volunteers with informed consent. The precision medicine study and stem cell platform was approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (study approval no.: UW 21-640).
Animals
Sox2-GFP mice (Mutant Mouse Resource & Research Centers, no. 037525-UNC) and OG2 mice (Jackson Laboratory, no. 004654) were maintained in the Centre for Comparative Medicine Research (CCMR) at the University of Hong Kong. All animal experiments were authorized by licenses from the Hong Kong Government Department of Health and University of Hong Kong Committee on the Use of Live Animals in Teaching and Research (CULATR No. 4855-18, No. 5278-19, and No. 5510-20).
Cell culture
Human embryonic kidney (HEK) 293T cells were cultured in 293T medium: high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco, 12100046) supplemented with 10% fetal bovine serum (FBS, Gibco, no. 10270106). All MEFs were maintained in MEF medium: DMEM supplemented with 10% FBS, 1% GlutaMAX (Gibco, no. 35050061), 1% nonessential amino acids (NEAA, Gibco, no. 11140050), 1% penicillin-streptomycin (Gibco, no. 15140122).
Primary mouse NSCs and miNSCs were plated in 0.2% gelatin (Sigma-Aldrich, no. G9391) or 1% Matrigel (Corning, no. 354234)–coated cell culture plates and cultured in mNSC medium: N2B27 medium supplemented with recombinant human bFGF (10 ng/ml; PeproTech, no. 10018B), recombinant human EGF (10 ng/ml; PeproTech, no. AF10015). The N2B27 medium contains DMEM/F-12 (Gibco, no. 12500062) supplemented with 1% N2 (Gibco, no. 17502048), 2% B27 (Gibco, no. 17504044), 1% GlutaMax, and 1% penicillin-streptomycin.
hiNSCs were plated in Matrige-coated plates and cultured in hiNSC medium: hN2B27 medium [DMEM/F12:Neurobasal (Gibco, no. 21103049) (1:1), 1% N2, 2% B27 (minus vitamin A, Gibco, no. 12587010), 1% GlutaMAX, and 1% penicillin-streptomycin], bFGF (20 ng/ml), EGF (20 ng/ml), 3 μM CHIR99021 (SelleckChem, no. S2924), 5 μM SB431542 (SelleckChem, no. S1067), and l-ascorbic-acid-2-phosphate (LAAP; 64 μg/ml; Sigma-Aldrich, no. 49752).
Primary mouse NSCs, miNSCs, and hiNSCs were routinely passaged every 4 to 5 days by treatment with Accutase (Gibco, A1110501). All cells were cultured at 37°C, 21% O2, and 5% CO2 and tested free of mycoplasma before they were used for experiments.
Plasmid construction and lentivirus production
The pHAGE2-TetO-mBrn4, pHAGE2-TetO-mKlf4 (Addgene, no. 136613), pHAGE2-TetO-mcMyc (Addgene, no. 136614), and pHAGE2-TetO-mSox2 (Addgene, no. 136612) lentiviral vectors were provided by S. Velychko (56). The pHAGE2-TetO-hBKSZ lentiviral vector was provided by M. Thier (26). Age I and Sal I restriction sites were added to this backbone by PCR. The coding DNA sequencing of mouse Sox17, eSox17FNV, Sox2-C17, and eSox17FNV-C2 and human KLF4, C-MYC, eSOX17FNV, and SOX2-C17 were cloned into this backbone by restriction digestion and ligation. The constructed plasmids will be available via Addgene (www.addgene.org/Ralf_Jauch/) or by request. The FUW-M2rtTA plasmid was from Addgene (no. 20342). The lentiviruses were produced in HEK293T cells. Briefly, HEK293T cells were transfected with pHAGE2-TetO lentiviral plasmid and packaging plasmids psPAX2 (Addgene, no. 12260) and pMD2.G (Addgene, no. 12259) using polyethyleneimine (Polysciences, no. 23966). Culture medium was changed 12 hours after transfection. Supernatant of each virus was collected three times 48, 72, and 96 hours after transfection, pooled, and filtered through a 0.45 μm filter (Millipore) to remove cells debris. Viral titer was measured with the qPCR Lentivirus Titer Kit (Abm, no. LV900). Filtered lentivirus supernatant of mouse TFs was used directly for mouse cell reprogramming. For human iNSC reprogramming from blood cells, lentiviruses were concentrated by 100-fold by ultracentrifugation at 60,000g, 4°C for 120 min followed by resuspension in Dulbecco’s phosphate-buffered saline (DPBS).
Preparation of MEFs
MEFs were isolated from day 13.5 mouse embryos. Briefly. the embryos were isolated from the uteri and washed with DPBS (Gibco, no. 21600044). The head and red tissue (heart and liver) of each embryo were removed. The remaining tissues were washed with DPBS and minced into small pieces using razor blades and trypsinized with 3 ml 0.25% trypsin (Gibco, no. 25200072) per embryo for 15 min at 37°C. MEF medium was added to stop trypsinization, and the dissociated cells were collected by centrifugation at 300g for 10 min. The cells from every three embryos were resuspended in 25 ml MEF medium, seeded on a 150 mm tissue culture dish, and incubated in a humidified 5% CO2 incubator at 37°C. For MEFs from Sox2-GFP–positive embryos, GFP-positive cells were removed by FACS. For reprogramming experiments, MEFs within three passages were used.
Preparation of ear and tail-tip fibroblasts from adult and old mice
Ears and 5 cm of tail of a mouse were cut into small pieces (less than 5 mm in size) with scissors. The cut tissues were then transferred to cryotubes containing collagenase D-pronase solution (Sigma-Aldrich, no. 10165921001) and digested at 37°C for 90 min with horizontal shaking at 200 rpm. The digested tissues were then neutralized with complete medium: RPMI 1640 medium (Gibco, no. 31800022) supplemented with 10% FBS, 0.05 mM β-mercaptoethanol, and 1% penicillin-streptomycin. Cell debris was removed by passing through 70 μm cell strainer. The resultant cell suspension was spun down by centrifugation for 7 min at 500g. Supernatant was removed, and cell pellet was washed with complete medium by centrifugation. Cell pellets from ear tissues and tails were resuspended in complete medium and incubated at 37°C in a humidified 5% CO2 incubator.
Derivation of primary mouse NSCs
Primary (NSCs) were derived from a Sox2-GFP embryo at day 13.5. Briefly, the head of a Sox2-GFP embryo was collected during MEF preparation and minced with a razor blade into small pieces. After trypsinization with 0.25% trypsin for 15 min at 37°C, MEF medium was added to stop trypsinization and the cells were collected by centrifugation at 300g for 10 min. The cells were washed with DPBS, resuspended in NSC medium, and seeded on a 100 mm tissue culture dish. After incubation in a humidified 5% CO2 incubator at 37°C for 2 to 4 days, GFP-positive neurospheres were picked and seeded directly on a Matrigel-coated 24-well plate in NSC medium. After culture for 2 to 4 days, the cells were digested with Accutase. Single-cell suspension in NSC medium was seeded on a 96-well plate at the density of one cell per well. The cells were cultured for another 6 to 10 days. Each GFP-positive neurosphere with a size >200 μm in diameter was transferred to one well of a Matrigel-coated 48-well plate and expanded as clonal lines.
Generation of miNSCs from mouse fibroblasts
To generate miNSCs, mouse fibroblasts (including MEFs, aEFs, and oTTFs) were seeded in MEF medium into 12-well plates at a density of 2.5 × 104 cells per well (~6.5 × 103 cells/cm2) 12 to 16 hours before transduction. On the day of lentiviral transduction, 1 ml mixture containing 100 μl of lentivirus supernatant of each TFs, 200 μl M2rtTA lentivirus, MEF medium, and polybrene (8 μg/ml) was prepared and added into each well. Twenty-four hours after transduction, lentiviruses were removed. The cells were washed with DPBS once and cultured in NSC medium containing Dox (1 μg/ml; Sigma-Aldrich, no. D9891) to initiate reprogramming. The day of adding NSC medium containing Dox was considered as reprogramming day 0. NSC medium containing Dox was changed every other day for around 21 days. The reprogramming cells were monitored using a CKX53 inverted fluorescence microscope (Olympus) during reprogramming experiments. Distinct colonies were manually picked at days 20 and 21 of reprogramming and expanded in NSC medium as clonal lines. Whole-well scans were taken at day 20 of reprogramming from 12-well plate using an ImageXpress Micro XLS confocal High-Content Imaging System (Molecular Devices) or a Typhoon5 Biomolecular Imager (GE Healthcare).
Generation of human iNSCs from blood cells
EPCs were expanded from human PBMCs as previously reported (64). Briefly, PMBCs were seeded at 2 × 106 to 3 × 106 cells/ml and cultured in EPC medium: StemSpan SFEM (Stem Cell Technologies, no. 09600) supplemented with SCF (300 ng/ml; PeproTech, no. 300-07), erythropoietin (10 U/ml) (MedChem Express, no. HY-P7164), IL-3 (50 ng/ml; Gibco, no. PHC0034), 1 μM dexamethasone (Sigma-Aldrich, no. D4902), 1% Insulin-Transferrin-Selenium-Ethanolamine (ITS -X) (Gibco, no. 51500056) for 1 day. Floating cells were transferred to empty cell culture plates to remove the attached cells and cultured for one more day without medium change. Then, the cells was adjusted to 5 × 105 cells/ml and cultured in EPC medium for another 6 days with medium change every other day.
To initiate hiNSC reprogramming, at day −1, 10 μl of concentrated lentiviruses of each TFs and M2rtTA were added to 1 × 105 EPCs resuspended in 0.5 ml of EPC medium in a 24-well plates. Spin infection was performed at 900g, 32°C for 60 min using an Eppendorf 5910R centrifuge with swing-out buckets for plates. Cells were then re-suspended without medium change and cultured for another 1 day. At day 0, the medium was changed to fresh EPC medium containing Dox (1 μg/ml) and incubated for another 2 days. At day 2, the transduced cells were replated into Matrigel-coated 12-well plates in StemSpan SFEM containing Dox and cultured for another 2 days. After day 4, the medium was half changed to fresh hiNSC medium supplemented with 0.5 μM PMA (SelleckChem, no. S3042), 100 μM sodium butyrate (NaB, Sigma-Aldrich, no. B5887), and 5 μM tranylcypromine (Sigma-Aldrich, no. P8511) every 2 days. PMA, NaB and tranylcypromine were only present for 10 days, Dox was present from day 0 to passage 5. hiNSC colonies were picked at day 20 and further expanded in Matrigel-coated plates in hiNSC medium.
Pluripotency reprogramming of mouse cells
MEFs were seeded on a 12-well plate at a density of 1.5 × 104 cells per well (~4 × 103 cells/cm2). After 12 to 16 hours of incubation, MEFs were transduced with Dox-inducible lentiviruses and rtTA lentivirus. Twenty-four hours after transduction, medium was changed to mESC medium containing Dox (1 μg/ml). mESC medium contains high-glucose DMEM supplemented with 15% FBS, 1% GlutaMAX, 1% NEAA, 1% sodium pyruvate (Gibco, no. 11360070), 0.05 mM β-mercaptoethanol (Gibco, no. 31350010), 2-phospho-l-ascorbic acid trisodium salt (LAAP; 50 μg/ml; Sigma-Aldrich, no. 49752), 1% penicillin-streptomycin, and human LIF (10 ng/ml; prepared in house). Reprogramming cells were fed with fresh mESC medium containing Dox every day for 2 to 3 weeks. Whole-well scans were taken at day 20 of reprogramming.
Estimation of cell proliferation
At day 0, iNSCs were dissociated with Accutase into single cells and resuspended in NSC medium. Cells were counted with a LUNA-II Automated Cell Counter (Logos Biosystems, no. L40002) and seeded at a density of 1 × 104 cells on each well of Matrigel-coated 24-well plates. After every 4 days, cells were dissociated and counted for 8 days. Cumulative cell number was calculated to estimate cell proliferation.
Differentiation of miNSCs in vitro
To differentiate iNSC toward neurons, iNSCs were seeded at a density of 1 × 105 cells per well on Matrigel-coated 12-well plates in NSC medium. After 24 hours, the medium was switched to neuronal differentiation medium: Neurobasal medium (Gibco, no. 21103049) supplemented with 1% N2, 2% B27, 1% GlutaMAX, 1% NEAA, and 1% penicillin-streptomycin. The medium was changed every other day. After 6 to 8 days, the cells were stained for Tuj1 to detect neurons. Neuronal differentiation efficiency was determined by the percentage of Tuj1+/DAPI+ (4′,6-diamidino-2-phenylindole–positive) cells out of total DAPI+ cells.
To differentiate iNSCs toward astrocytes, iNSCs were seeded at a density of 5 × 104 cells per well on Matrigel-coated 12-well plates and incubated in NSC medium. After 24 hours, the medium was replaced with astrocytic differentiation medium: DMEM/F-12 supplemented with 1% N2, 2% B27, 1% GlutaMAX, 1% NEAA, 2% FBS, and 1% penicillin-streptomycin. After medium change every 2 days for 6 to 8 days, the cells were stained for GFAP to detect astrocytes. Astrocyte differentiation efficiency was determined by the percentage of GFAP+/DAPI+ cells out of total DAPI+ cells.
To derive oligodendrocytes, iNSCs were seeded at a density of 3 × 104 cells per well on Matrigel-coated 12-well plates and incubated in NSC medium for 24 hours. Oligodendrocyte differentiation was induced in two steps. For the first 4 days, the medium was switched to oligodendrocyte differentiation medium for step one: DMEM/F-12 supplemented with 1% B27, bFGF (20 ng/ml), recombinant human PDGF-AA (20 ng/ml; PeproTech, no. 10013A) and 1% penicillin/streptomycin. After 4 days, the medium was changed to oligodendrocyte differentiation medium for step 2: DMEM/F-12 supplemented with 1% B27, PDGF-AA (5 ng/ml), 15 nM T3 (Sigma-Aldrich, no. T6397) and 1% penicillin-streptomycin. After another 5 to 7 days, the cells were stained for O4 to detect oligodendrocytes. Oligodendrocyte differentiation efficiency was determined by the percentage of O4+/DAPI+ cells out of total DAPI+ cells.
Differentiation of human iNSCs in vitro
For spontaneous hiNSC differentiation, cells were seeded on PDL-coated (Gibco, no. A3890401)/laminin-coated (Sigma-Aldrich, no. 11243217001) glass coverslip and cultured in hN2B27 medium supplemented with LAAP (64 μg/ml), BDNF (10 ng/ml; PeproTech, no. 450-02), GDNF (10 ng/mg; PeproTech, no. 450-10), and 0.5 mM dbcAMP (Sigma-Aldrich, no. D0627) for 21 days. For induction of motor neurons, hiNSCs were cultured in hN2B27 medium supplemented with LAAP (64 μg/ml), 10 μM DAPT (GSI-IX, PeproTech, no. 2088055), 0.5 μM retinoic acid (RA, Sigma-Aldrich, no. R2625), 1 μM PMA, 10 μM forskolin for 2 days and then BDNF (10 ng/ml), GDNF (10 ng/ml), and NT-3 (10 ng/ml; PeproTech, no. 450-03) were added to the medium for another 8 to 10 days.
For astrocyte differentiation, hiNSCs were seeded on a Matrigel-coated glass coverslip and cultured in N2B27 medium supplemented with CNTF (20 ng/ml; PeproTech, no. 450-13) and BMP-4 (10 ng/ml; PeproTech, no. AF-120-05ET) for 21 days. Oligodendrocytes were generated in three steps as previously described (65). First, hiNSCs were seede on a Matrigel-coated glass coverslip and cultured in N2 medium (DMEM/F12, 1% N2, 1.6 g/l D-glucose, insulin (0.02 mg/ml; Sigma-Aldrich, no. 91077C), 1% penicillin/streptomycin) supplemented with EGF (10 ng/ml), PDGF-AA (10 ng/ml), and 10 μM forskolin for 2 weeks. Second, the medium were switch to N2 medium supplemented with PDGF-AA (10 ng/ml), 45 nM T3, noggin (200 ng/ml; PeproTech, no. 120-10C), and LAAP (64 μg/ml) for 1 week. Third, for terminal differentiation, the cells were cultured in N2 medium supplemented with 45 nM T3, LAAP (64 μg/ml), and laminin (1 μg/ml) for 4 weeks. For each differentiation, half of the medium was changed every 2 days before the cells were collected for immunocytochemistry.
Transplantation of iNSCs and immunohistochemistry
miNSCs and primary mouse NSCs were labeled with pMX-DsRed retrovirus and were purified by FACS. DsRed-tagged miNSCs were expanded and dissociated into single cells with Accutase and resupended and diluted to a final concentration of 1 × 105 cells/μl. The transplantation was performed on 2-month-old female wild-type C57BL/6 J mice. For surgery, the animals were deeply anesthetized with ketamine/xylazine. Before surgery, the hair was removed from the surgical site, and the skull surface was exposed by making a midline incision. Using X and Y stereotaxic coordinates, the injection skull sites were thinned with microdrill, 3 μl of the cell suspension was injected into the hippocampus using a Hamilton 7105KH 5- μl syringe at a rate of 1 μl/min. After injection, the skin was sutured with nylon suture and returned to the home cage. After holding for 2 months, the injected animals were deeply anesthetized and intracardially perfused with 30 ml 4% PFA in DPBS. The brains were collected and postfixed with 4% PFA overnight at 4°C followed by rinsing three times with DPBS. Coronal sections (50 μm) were cut with a Vibratome (Leica VT 1000 S). Free-floating sections were permeabilized in PBST (DPBS with 0.3% Triton X-100) for 30 min. Sections were blocked with blocking buffer (PBST with 5% BSA, 1.5% goat serum, 2.5% and 1.5% donkey serum) for 1 hour, and then incubated with primary antibodies diluted in blocking buffer overnight at 4°C followed by incubation with Alexa-fluorophore conjugated secondary antibodies at room temperature for 2 hours and with DAPI for 5 min. Sections were analyzed with a Zeiss LSM 900 confocal microscope.
Calcium imaging
Calcium imaging of iNSC-derived neurons was performed as previously reported (66). In brief, cells cultured on PDL/laminin-coated glass coverslip were incubated with 2 μM Fluo-4-AM (Thermo Fisher Scientific, no. F14201) at 37°C, 5% CO2 for 30 min. The cells were washed three times with artificial cerebrospinal fluid (ACSF) (125 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, 25 mM, d-glucose in Deionized water) and incubated at room temperature in the dark for additional 20 to 25 min to allow for complete dye de-esterification. Then the cells were placed in a 30° to 32°C heated adaptor and live-cell imaging was performed by a Nikon ECLIPSE Ti Inverted Microscope with 20X objective and excitation wavelength of 485 nm and emission wavelength of 525 nm. Images were taken every second for 5 min using a ZEISS Axiocam 702 mono camera and analyzed with ZEISS ZEN 3.3 (blue edition). Data were exported and transferred to GraphPad Prism 8 for further analysis.
Karyotyping
More than 2 × 106 mouse or human iNSCs at 90% confluence were treated with colcemid (0.025 μg/ml; Gibco, no. 15212012) at 37°C for 45 min. The cells were subsequently washed with DPBS and dissociated into single cells with Accutase. The cells were resuspended in 6 ml of 0.075 M KCl and incubated at 37°C for 15 min. Then 0.5 ml of fixative solution (acetic acid and methanol at 1:3) were gently added, mixed and incubated at 37°C for 5 min. The cells were fixed three times in 5 ml of fixative solution at 37°C for 40 min. The fixed cells were gently resuspended in 300 μl of ice-cold fixative solution, dropped onto pre-cleaned cold glass slice and incubated at 65°C overnight. After trypsin treatment and Giemsa staining with.
Karyomax Giemsa Stain Solution (Gibco, no. 10092013), metaphase spreads were screened by a Zeiss Axioscope inverted microscope with 100X objective and analyzed with Ikaros software (MetaSystems). At least 20 metaphase spreads were analyzed for karyograms.
RNA isolation and qRT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen, no. 15596018) following the user guide quantified using a Nanodrop One spectrophotometer (Thermo Fisher Scientific). 1.5 μg total mRNA from each sample was used to synthesize the cDNA with a ReverTra Ace qPCR RT master mix with gDNA remover (TOYOBO, no. FSQ-301). qRT-PCR was performed with iTaq Universal SYBR Green Supermix (Bio-Rad, no. 1725122) on a CFX-96 thermocycler (Bio-Rad) in technical triplicates. PCR primers used for qRT-PCR are listed in table S1. The 2-∆∆CT method was used to calculate relative gene expression with GAPDH as reference gene. The R package ggplot2 was use to plot the results.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (PFA; Sigma-Aldrich, no. P6148) at room temperature for 15 min, washed three times with DPBS, permeabilized with 0.2% Triton X-100 (Sigma-Aldrich, no. T8787) at room temperature for 20 min, blocked in 5% bovine serum albumin (BSA; United States Biological, no. A1311) at room temperature for 1 to 2 hours and incubated in primary antibodies diluted in 1% BSA at 4°C overnight or at room temperature for 2 hours. After three-times rinse with DPBS, the cells were incubated in secondary antibodies diluted in 1% BSA in dark at room temperature for 2 hours followed by three times washing with DPBS. After DAPI staining at room temperature for 15 min, the cells were washed three times and imaged with a CKX53 inverted fluorescence microscope (Olympus) or an LSM 900 confocal microscope (Zeiss). All antibodies used in this study are listed in table S2.
Generation of Sox2-GFP/Nanog-CreER/Rosa26-tdTomato reporter cells
Sox2-EGFP+/−/Rosa26-tdTomato+/− double knock-in mouse ESC (mESC) lines were derived as described before (67). Briefly, homozygous Rosa26 LoxP-stop-LoxP tdTomato+/+ mice (Jackson Laboratory, no. 007905) were crossbred with heterozygous Sox2-EGFP+/− mice. At day E3.5, blastocysts were extracted and individually seeded on a flat-bottom 12-well plate on inactivated MEF feeder cells in KSR-mESC medium: high-glucose DMEM supplemented with 15% knockout serum replacement (KSR, Gibco, no. 10828028), 1% GlutaMAX, 1% NEAA, 1% penicillin/streptomycin, 0.1 mM β-mercaptoethanol, and LIF (10 ng/ml). Developed double knock-in mESC were expanded as single clonal lines.
Nanog-CreER cassette was knocked into the double knock-in mESC to generate Sox2-GFP/Nanog-CreER/Rosa26-tdTomato reporter cells as described previously (30). Briefly, Nanog-CreER-pgk-Hygro targeting construct, a gift from J. Hanna (30) (Addgene, no. 59720) was linearized with Sal1 restriction enzyme (NEB, no. R3138), transfected into the double knock-in mESCs with Lipofectamine Stem Transfection Reagent (Invitrogen, no. STEM00001). Twenty-four hours after transfection, the cells were dissociated with 0.05% trypsin into single cells, seeded on Matrigel-coated six-well plates in serum/2iLIF medium containing Hygromycin (100 μg/ml; GoldBio) for 7 to 10 days. Serum/2iLIF medium contains high-glucose DMEM supplemented with 15% FBS, 1% GlutaMAX, 1% NEAA, 1% sodium pyruvate, 0.05 mM β-mercaptoethanol, LAAP (50 μg/ml), 1% penicillin-streptomycin, LIF (10 ng/ml), 3 μM CHIR99021 (SelleckChem, no. S2924), and 1 μM PD0325901 (SelleckChem, no. S1036). Hygromycin-resistant Sox2-EGFP+/−/Nanog-CreER+/−/Rosa26-tdTomato+/− triple knock-in mESCs was confirmed by genotyping PCR using primers in table S1 and 4-OHT treatment test (PeproTech, no. 6833585). Correctly targeted clonal triple knock-in mESCs were microinjected into blastocysts and transferred into pseudopregnant host mice, which was done in CCMR. At day E13.5, chimeric embryos were collected and used to prepare reporter MEFs as described above. To remove the cells from the host embryos, MEFs at passage 0 were cultured in MEF medium containing Hygromycin (100 μg/ml) for 4 to 6 days, followed by FACS to remove GFP-positive cells. GFP-negative reporter MEFs used for reprogramming experiments within three passages.
Mitochondrial staining assay
Cells were washed once with DPBS, trypsinized into single cells with 0.05% trypsin (Gibco, no. 25300062) and centrifuged for 5 min at 300g. After removing the supernatant, the cells were resuspended to 5 × 105 cells/ml with NSC medium. In a 96-well cell culture plate, 200 μl of cells (1 × 105 cells) were added per well, and then 20 μl of MitoTracker Green FM (Invitrogen, no. M7514) was added to each well (final concentration 50 nM) and incubated for 30 min at 37°C. After incubation, the cells from each well were washed once with 1 ml of DPBS, centrifuged for 5 min at 300g. After removing supernatant, the cells were resuspended in FACS buffer and filtered through 40 μm strainers. Mean fluorescence intensities (MFI) of the green fluorescent probe in stained live single cells were quantified using NovoCyte Advanteon flow cytometer (Agilent) with a 488 nm laser.
Glucose uptake assay
Single-cell suspension was prepared as described in mitochondrial staining assay. Cells (100 μl; 5 × 104 cells) were added to each well in a 96-well cell culture plate, then 100 μl of 2-NBDG (Invitrogen, no. N13195) was added to each well (final concentration 100 μM) and incubated for 30 min at 37°C. After incubation, the cells were washed once with DPBS and resuspended in FACS buffer and filtered through 40 μm strainers. MFIs of the green fluorescent probe in stained live single cells were quantified using NovoCyte Advanteon flow cytometer with a 488 nm laser.
Extracellular metabolic flux analysis
Glycolysis stress tests were performed with the Seahorse XF Glycolysis Stress Test Kit (Agilent, no. 103020100) following instructions by the manufacture. In brief, the day before measurement, the XFe96 Sensor Cartridge (Agilent, no. 102418100) was hydrated in Seahorse XF Calibrant (Agilent, no. 100840000) at 37°C in a non-CO2 incubator overnight. Cells were seeded onto a XF96 plate (Agilent) precoated with Matrigel at 2 × 104 cells per well in NSC medium and incubate at 37°C in a CO2 incubator overnight. On the day of measurement, the medium was changed to XF DMEM (Agilent, no. 103575100) supplement with 2 mM XF glutamine (Agilent, no. 103579100) and was equilibrated for 1 hour at 37°C in a non-CO2 incubator. Then, the extracellular acidification rate (ECAR) was measured in four steps before and after sequential injections of 10 mM glucose, 1 μM oligomycin, and 50 mM 2-deoxyglucose (2-DG) using the calibrated sensor cartridge and a Seahorse Xfe96 Analyzer (Agilent). ECAR was repeatedly measured in at least three mix-wait-measure cycles in each step for the duration of the experiment (90 to 120 min).
Protein purification
The constructs of pETG20a-Sox were used to express the fusion protein containing an N-terminal His6-TrxA tag were expressed in Escherichia coli Rosetta (DE3) competent cells grown in LB medium supplemented with 0.2% Glucose and ampicillin (100 μg/ml) at 37°C to OD600 of ~0.6 to 0.8 before adding 0.5 mM isopropyl-β-thio galactoside for induction at 16 to 18°C for 18 to 22 hours. Cell pellet was resuspended in [Sox lysis: 20 mM tris-HCl (pH 8.0), 500 mM NaCl, and 20 mM imidazole] and incubated for 30 min on ice before freezing at −80°C. Cells are thawed and disrupted by sonification. The fusion proteins in the soluble fraction were first captured using HisPur Ni-NTA Superflow Agarose washed with 30 mM imidazole buffer and eluted with elution buffer [Sox: 20 mM tris-HCl (pH 8.0), 500 mM NaCl, and 300 mM imidazole]. Fusion proteins are subsequently cleaved using tobacco etch virus protease at 4°C overnight to remove the Trx tags. The cleaved protein was further purified using a 1 ml HiTrap SP HP (Cytiva) column equilibrated in buffer A [Sox: 20 mM tris-HCl (pH 8.0), and 250 mM NaCl] connected to an ÄKTA pure protein purification system (GE Healthcare) and eluted with a linear NaCl gradient using buffer B [Sox: 20 mM tris-HCl (pH 8.0) and 1 M NaCl]. Fractions containing the desired protein were pooled, exchanged into a buffer A using HiLoad 16/600 Superdex 75 pg (Cytiva), and frozen in liquid nitrogen and stored in aliquots at −80°C.
EMSAs using purified proteins
Native 12% polyacrylamide gel electrophoresis (PAGE) gels were prepared with tris/glycine (TG) buffer [25 mM tris (pH 8.3); 192 mm glycine] and cast as mini gels. Double-stranded DNA probes with 5′ cy5 or cy3 dyes at forward strand were prepared using an annealing buffer [20 mM tris/HCl, 50 mM MgCl2, and 50 mM KCl (pH 8.0)] and heated to 95°C for 5 min and subsequent cooling at 1°C/min to 16°C in a T100 Thermo Cycler (Bio-Rad). Proteins were thawed and filtered through 0.45 μm centrifugal filters. Protein samples and fluorescently labeled DNA are incubated for ~1 to 2 hours in EMSA buffer [10 mM tris/HCl (pH 8.0), BSA (0.1 mg/ml), 50 μM ZnCl2, KCl 100 mM, 10% glycerol, 0.10% Igepal CA630, and 2 mM βME]. Gels were first to pre-ran using 1 × TG buffer [0.25 mM tris and 192 mM glycine (pH 8.0)] at 200 V for 30 min, and then 10 μl samples were loaded and gels run for 30 min at 200 V at 4°C. Images are captured using an Amersham Typhoon 5 Biomolecular Imager and quantified using ImageQuantTL 7.0.
Having two different proteins, hence two separate monomeric binding states. There were four microstates defined as f0, f1, f2, and f3, which correspond to the fractional concentrations of the free DNA (f0), two alternative monomer-DNA complexes (f1,2), and the heterodimer-DNA complex (f3), respectively. The heterodimer cooperativity factor ω can be calculated from these values through equation in (68).
EMSA using whole-cell protein extraction from mammalian cells
The coding sequences of full-length TFs were amplified by PCR and inserted into pLVTHM-3xflag vector [1] between Pme I and Mlu I sites by restriction digestion and ligation. Plasmids were transfected into HEK293T cells to overexpress the proteins of interest for 72 hours. Cells were collected, washed with DPBS and then re-suspended in lysis buffer [20 mM Hepes-KOH (pH 7.8), 150 mM NaCl, 0.2 mM EDTA (pH 8.0), 25% glycerol, and 1 mM dithiothreitol (DTT) with cOmplete protease inhibitor cocktail (Roche, no. 11836145001) added freshly]. To lyse the cells, 4 cycles of freeze-thaw with liquid nitrogen were used followed by centrifugation at 14,000g for 10 min at 4°C. Supernatant was collected and total protein concentration was estimated with a Nanodrop One spectrophotometer followed by quantification by Western blot analysis with anti-flag antibody. When loading onto the gels, the SOXs concentrations were adjusted to the same level and the total protein levels between different samples were adjusted by adding nontransduced lysate. To resolve protein/DNA complexes, 6% native 18.5 cm by 20 cm PAGE gels [cast with tris/glycine buffer (Bio-Rad, no. 1610771)] were used. DNA probes and proteins mixtures were incubated for 2 hours on ice in binding buffer [25 mM Hepes-KOH (pH 8.0), 50 mM NaCl, 0.5 mM EDTA, 0.07% Triton X-100, BSA (4 mg/ml), 7 mM DTT, and 10% glycerol]. Gels were pre-run at 300 V for 2 hours with 1 × tris/glycine buffer at 4°C. Then, samples were loaded and run for 2.5 hours at 300 V at 4°C. Images were scanned with a GE Typhoon 5 Biomolecular Imager.
SMT assay
HeLa cells were cultured in DMEM supplemented with 10% FBS, 5% GlutaMAX, and 1% NEAA and maintained at 37°C with 5% CO2. Cells were seeded at a density of 20,000 cells per well in eight-well chamber glass slides (Ibidi) coated with 0.5% gelatin 24 hours before transfection. Transfections were performed using the X-tremeGENE 9 Transfection Reagent kit (Roche, Basel, Switzerland) to introduce 500 ng of plasmid DNA using FluoroBrite DMEM (Gibco) as the low serum transfection media. After 24-hour growth, cells were washed twice and cultured in Fluorobrite DMEM (Gibco) imaging media with 1.5 nM JF549 Halo-tag ligand for 10 min. Following incubation, cells were washed twice and transferred to Fluorobrite DMEM (Gibco) supplemented with 10% FBS, 5% GlutaMAX, and Hepes.
Images were acquired on a Nikon total internal reflection fluorescence (TIRF) microscope at a TIRF angle of 60.18° to achieve HILO illumination, an iXon Ultra 888 EMCCD camera with filter cube TRF49909-ET-561 laser bandpass filter and a 100 X oil 1.49 NA TIRF objective. Cells were imaged using a 561 nm excitation laser at a power density of 10.3 μW to perform two different acquisition techniques: a fast frame rate that uses 50 Hz (20 ms frame rate) to acquire 6000 frames without intervals to measure displacement distribution and fraction bound, and a slow frame rate that uses 2 Hz (500 ms frame rate) to acquire 500 frames without intervals to measure residence times.
All images were cropped using ImageJ (fast and slow tracking). Molecules were identified using a custom-written MATLAB implementation of the MTT algorithm, known as SPT_LocAndTrack (69). Parameters used for mobility and fraction bound analysis: Localization error: 10−6.5; Blinking (frames): 1; max number of competitors: 3; max expected diffusion coefficient (μm2/s): 2. Time points = 7, clipfactor = 4. Cells with less than 700 trajectories based on above parameters are excluded from analysis. All trajectories that have less than 7 (time points) tracks are removed from further analysis. Mean squared displacement calculations use the first four frames of each trajectory. Parameters used for Residence time tracking: Localization error: 10−7; Blinking (frames): 1; max number of competitors: 3; max expected diffusion coefficient (μm2/s): 0.31. Residence time analysis was performed using MATLAB code (Calculatelength_2fitting_v3) provided by Zhe Liu.
Angle and AC: Angles and AC were calculated using the same method and formulas as described by Izeddin et al. (51).
AC formula
where FWD = 0° to 30° and BWD = 150° to 180°.
Data were plotted and statistically tested using GraphPad Prism 9.1 (GraphPad Software) and Python. Results are displayed as means ± the SD unless indicated otherwise. Statistical significance was determined by analysis of variance (ANOVA) followed by Tukey post hoc test.
RNA sequencing
Cells were cultured in 6-well or 12-well culture plates in replicates, washed once with DPBS, and collected by adding 500 μl Trizol (Invitrogen). Total RNA was isolated using Trizol reagent as described above and dissolved in nuclease-free water. More than 1.5 μg total RNA of each sample was submitted to Novogene (Beijing, China) for 150 bp paired-end RNA-seq. Raw sequencing data per sample (6 Gb) in fastq format were provided for analysis.
RNA-seq data analysis
Raw RNA-seq reads in fastq format were aligned to the reference transcriptome (mm10 for mouse and hg38 for human) using RSEM (70) and bowtie 2.0 (71) (with options: rsem-calculate-expression --bowtie2 --bowtie2-sensitivity-level very_sensitive --no-bam-output --estimate-rspd --paired-end). Gene level alignment results of samples were merged and annotated using glbase3 (72). Read counts per genes were calculated using RSEM and normalized to GC content using EDAseq (73). GC-normalized read counts ≥20 in at least two samples were kept, leading to a final read count table. This table was used to inspect individual gene expression, generate correlation heatmaps, perform PCA and analyze DEGs. Correlation heatmaps were generated using the correlation_heatmap function of glbase3 with option mode = “r2,” or using the cor function of R with the option method = “pearson.” PCA was generated using the get_pca function of glbase3 or using the rpca function of R. DEG analysis was performed using the DEseq function with the option fitType = “local” and the results function of DESeq2. The R packages data.table, ggplot2, and pheatmap were used for data analysis and visualization. GO analysis was performed using enrichGO function of clusterProfiler for DEGs or top 200 gene loadings in PCA.
Single-cell RNA-seq
Mouse primary NSCs and iNSCs at passage 15 were dissociated into single cells using Accutase and resuspended in DPBS containing 0.04% BSA with more than 90% viability. scRNA-seq library preparation and Illumina sequencing (Pair-End sequencing of 151 bp) were done at The University of Hong Kong, LKS Faculty of Medicine, Centre for PanorOmic Sciences (CPOS), Genomics Core. Single cell encapsulation and cDNA library was prepared from each sample using the 10X Genomic Chromium Single Cell Controller (10X Genomics) and Chromium Next GEM Single Cell 3′ Reagent Kit v3.1 and Chromium Next GEM Chip G Single Cell Kit according to manufacturer’s instructions. In brief, single cells in suspension were counted and loaded into individual wells of 10X Chromium Single Cell chip. Single cells were encapsulated into Gel Beads-in-emulsion (GEM) by 10X Chromium Single Cell Controller. Single Cell 3’ Reagent Kit v3.1 was used to perform reverse transcription on GEMs, followed by cDNA purification and amplification. The double-stranded cDNA then went through enzymatic fragmentation, adapter ligation, index PCR and SPRIselect size selection following manufacturer’s protocol. Library size and concentration were determined by Qubit, qPCR and Bioanalyzer assays. Pooled libraries were then sequenced on Illumina NovaSeq 6000 platform using 151 bp paired-end sequencing. Raw sequencing reads were assigned into individual samples using bcl2fastq (Illumina) with each sample having an average throughput of 60.9 Gb of raw data (406 million reads) followed by alignment to mm10 mouse genome using Cell Ranger analysis pipeline (10X Genomics) with default parameters. A total of 18,143 cells were profiled to an average depth of 66,713 reads per cell.
scRNA-seq data analysis
scRNA-seq data analysis and visualization were performed using Seurat R package (version 4.3.0, https://satijalab.org/seurat/) (74). A Seurat object was created for each sample using CreateSeuratObject function after reading in the gene-by-cell unique molecular identifier (UMI) count matrix from Cell Ranger analysis using Read10X function. All Seurat objects were merged using merge function for standard pre-processing workflow. Cells were filtered using subset function and cells with 2000 to 30,000 UMIs and with 2000 to 6000 genes expressed and with less than 15% mitochondria UMI rate were kept, leaving 16,953 genes across 16,478 cells for further analysis. The expression matrix was then normalized by LogNormalize method to 10,000 UMIs per cell using NormalizeData function. Two thousand variable genes were identified by vst method using FindVariableFeatures function. The data was scaled using ScaleData function and PCA was performed using RunPCA function based on the scaled data using the variable genes as input. Cells were clustered using the FindNeighbors and FindClusters function with the first 10 PCs and resolution = 0.1 and Louvain algorithm. The first 10 PCs were used for Uniform Manifold Approximation and Projection (UMAP) construction using RunUMAP function. The marker genes were calculated by FindAllMarkers function with Wilcoxon Rank Sum test algorithm and logfc.threshold = 0.25 and min.pct = 0.1. Marker genes were shown in UMAP plots using FeaturePlot function and DotPlot function. Cell cycle scores were calculated using the CellCycleScoring function.
Reduced representation bisulfite sequencing
Genomic DNA (gDNA) of cells was isolated using Quick-DNA Microprep Kit (Zymo, no. D3021). More than 1.5 μg gDNA of each sample was submitted to Novogene (Beijing, China) for RRBS. RRBS libraries were prepared as previously reported (75). Briefly, the gDNA was digested using Msp I, followed by end repair, and dA-tailing. The adapter ligation was performed by using adapters with all cytosines being methylated. After size selection, DNA fragments were bisulfite treated. After the bisulfite conversion, unmethylated cytosines were converted to U and further to T after PCR amplification, while methylated cytosines remained unchanged. After PCR amplification, the libraries were checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries were pooled and sequenced on Illumina NovaSeq 6000 platform using 150 bp paired-end sequencing with more than 10 Gb of raw data (67 million reads) per sample in fastq format.
RRBS data analysis and methylation age prediction
RRBS data were processed for mouse methylation age prediction as previously described (41). RRBS raw reads were trimmed with Trim Galore (version 0.6.6, −rrbs) followed by alignment to the mouse genome (GRCm38/mm10) with Bismark (version 0.23.1).Then Bismark coverage files were generated from the deduplicated bam files using Bismark Methylation Extractor (−gzip –bedGraph). The bedGraph files were convertecd to BigWig files using bedGraphToBigWig. The Bismark coverage files were used to calculate the methylation age using mouse epigenetic clock with an imputation step (https://github.com/EpigenomeClock/MouseEpigeneticClock). The Bismark coverage results were filtered to exclude CpG sites which had less than 5X coverage in more than 90% of samples, resulting more than 2.2 million CpG sites included in all samples.
Assay for transposase-accessible chromatin using sequencing
Sox2-GFP MEFs at reprogramming day 4 were washed once with DPBS, trypsinized with 0.05% trypsin into single cells, and cryopreserved in MEF medium containing 10% DMSO. The cryopreserved cell samples were submitted to Novogene (Beijing, China) for ATAC-seq. Raw sequencing data per sample (12 Gb) in fastq format were provided for analysis.
ATAC-seq data analysis
ATAC-seq data were trimmed to 35 bp length to remove Tn5 transposase sequence and then mapped to mouse genome assembly mm10 using bowtie2 (−-very-sensitive --end-to-end --no-unal --no-mixed -X 2000). Mapped reads were filtered to removed low quality reads and sorted using samtools (76) (samtools view -q 30 | sort). Mitochondrial reads were removed afte alignment (grep -v chrM). PCR duplicates were removed using samtools (rmdup) and only uniquely mapped reads were kept. The BAM files of replicates of each sample were merged with samtools (merge). The merged BAM files were converted to BigWig files using bedtools (77) (bamtobed), genomeCoverageBed from bedtools and bedGraphToBigWig. Peaks were called with MACS2 (78) (with option -g mm -f BAMPE --call-summits). Enriched motifs in 200 bp regions of ATAC-seq peaks was analyzed using the findMotifsGenome.pl function of HOMER (79) (mm10 -size -100,100). Nearest genes were assigned to the ATAC-seq peaks using the annotatePeak.pl function of HOMER.
chromVAR analysis
For ATAC-seq data, chromVAR (48) was used to compare the chromatin accessibility variation for peaks marked by different TF motifs. Peaks of all ATAC-seq samples were combined into a read count matrix. For overlapping peaks, the peaks with higher peak scores or overall higher read counts were kept. Motif position weight matrices (PWMs) from the HOMER database were obtained with the chromVARmotifs R package. Motif matches in peak regions were assigned using the motifmatchr package. GC bias corrected accessibility deviations were calculated based on merged reads from ATAC-seq replicates. For better comprehension, the two motifs from HOMER database are renamed as canonical SoxOct [OCT4-SOX2-TCF-NANOG (POU,Homeobox,HMG)] and compressed Sox17:Oct4 [Oct4:Sox17 (POU,Homeobox,HMG)]. Accessibility variability data are presented as a heatmap using the pheatmap R package.
Footprint analysis
HINT was used to identify the active TF binding sites for ATAC-seq data as described previously (50). In brief, Tn5 insertion bias was corrected and footprints were called using the rgt-hint footprinting function. Motifs overlapping with the footprints were found with the rgt-motifanalysis matching function using motifs from JASPAR database. Average ATAC-seq profiles around binding sites of motifs and Differential TF binding were analyzed using the rgt-hint differential function.
Acknowledgments
We would like to thank P. F. Wong for administrative support. We thank V. S. F. Chan, S. O. Chung, and C. S. Lau for providing human PBMCs from healthy donors, A. Lee from CCMR, HKU for help in blastocyst collection and microinjection; D. Han, G. Wu, H. R. Schöler, V. Veerapandian, and Y. Sivastava for help in the design of reprogramming experiments; V. Malik for help in bioinformatics analysis; and Image and Flow Cytometry Core of CPOS at University of Hong Kong for instrument support. We thank Y. Huang, J. Wong, and G. Lau for providing access to tools, equipment, and intellectual input.
Funding: This study was supported by Health and Medical Research Fund (06174006 and 08192886), the National Natural Science Foundation of China (grant no. 31771454), Research Grants Council of Hong Kong General Research Fund (RGC/GRF, grant. nos. 17128918, 17101120, and 17106622), Collaborative Research Fund (CRF, grant no. C7064-22G), Germany/Hong Kong Joint Research Scheme sponsored by the Research Grants Council of Hong Kong and the German Academic Exchange Service (reference no. G-HKU701/18), Liu Po Shan/Dr. Vincent Liu Endowment Fund for Motor Neurone Disease, and Innovation Technology Commission Funding Scheme Health@InnoHK.
Author contributions: M.W. contributed to study design, performed most of the experiments, and analyzed data. H.H. and D.S.T. performed cloning, protein purification, and EMSA assay. J.L., M.H., and S.R. contributed to reprogramming experiments. D.H.H.H. prepared adult and old mouse fibroblasts. Y.G. performed cloning and karyotyping. M.F. supervised the SMT assay of M.S.G. and analyzed data. Y.H. and C.S.W.L. contributed to the design and analysis of in vivo differentiation experiments. G.S.L. contributed to the design and analysis of metabolic assays. R.J. designed and supervised the study, acquired funding, and analyzed data. M.W. and R.J. wrote the manuscript with input from all authors.
Competing interests: M.W., D.H.H.H., H.H., and R.J. will file a nonprovisional patent application for reprogramming methods with engineered SOX17. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The next-generation sequencing data underlying this article are available in the Gene Expression Omnibus (GEO) with the accession numbers GSE195556 (bulk RNA-seq: GSE195551, GSE225829, and GSE225830; scRNA-seq: GSE225306; ATAC-seq: GSE195554; and RRBS: GSE225305). SMT data are available via the BioImage Archive with the accession number S-BIAD703. The materials can be provided by R.J. pending scientific review and a completed material transfer agreement. Requests for the code and materials should be submitted to R.J.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Tables S1 and S2
Legend for movie S1
Legends for data S1 and S2
Other Supplementary Material for this manuscript includes the following:
Movie S1
Data S1 and S2
REFERENCES AND NOTES
- 1.K. Takahashi, S. Yamanaka, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 2.K. Takahashi, K. Tanabe, M. Ohnuki, M. Narita, T. Ichisaka, K. Tomoda, S. Yamanaka, Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- 3.S. Yamanaka, Pluripotent stem cell-based cell therapy-promise and challenges. Cell Stem Cell 27, 523–531 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Y. Fan, J. Hackland, A. Baggiolini, L. Y. Hung, H. Zhao, P. Zumbo, P. Oberst, A. P. Minotti, E. Hergenreder, S. Najjar, Z. Huang, N. M. Cruz, A. Zhong, M. Sidharta, T. Zhou, E. de Stanchina, D. Betel, R. M. White, M. Gershon, K. G. Margolis, L. Studer, hPSC-derived sacral neural crest enables rescue in a severe model of Hirschsprung's disease. Cell Stem Cell 30, 264–282.e9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D. Jgamadze, J. T. Lim, Z. Zhang, P. M. Harary, J. Germi, K. Mensah-Brown, C. D. Adam, E. Mirzakhalili, S. Singh, J. B. Gu, R. Blue, M. Dedhia, M. Fu, F. Jacob, X. Qian, K. Gagnon, M. Sergison, O. Fruchet, I. Rahaman, H. Wang, F. Xu, R. Xiao, D. Contreras, J. A. Wolf, H. Song, G. L. Ming, H. I. Chen, Structural and functional integration of human forebrain organoids with the injured adult rat visual system. Cell Stem Cell 30, 137–152.e7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.T. W. Kim, J. Piao, S. Y. Koo, S. Kriks, S. Y. Chung, D. Betel, N. D. Socci, S. J. Choi, S. Zabierowski, B. N. Dubose, E. J. Hill, E. V. Mosharov, S. Irion, M. J. Tomishima, V. Tabar, L. Studer, Biphasic activation of WNT signaling facilitates the derivation of midbrain dopamine neurons from hESCs for translational use. Cell Stem Cell 28, 343–355.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.P. R. di Val Cervo, D. Besusso, P. Conforti, E. Cattaneo, hiPSCs for predictive modelling of neurodegenerative diseases: Dreaming the possible. Nat. Rev. Neurol. 17, 381–392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A. Bose, G. A. Petsko, L. Studer, Induced pluripotent stem cells: A tool for modeling Parkinson's disease. Trends Neurosci. 45, 608–620 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. Piao, S. Zabierowski, B. N. Dubose, E. J. Hill, M. Navare, N. Claros, S. Rosen, K. Ramnarine, C. Horn, C. Fredrickson, K. Wong, B. Safford, S. Kriks, A. El Maarouf, U. Rutishauser, C. Henchcliffe, Y. Wang, I. Riviere, S. Mann, V. Bermudez, S. Irion, L. Studer, M. Tomishima, V. Tabar, Preclinical efficacy and safety of a human embryonic stem cell-derived midbrain dopamine progenitor product, MSK-DA01. Cell Stem Cell 28, 217–229.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.V. A. Church, K. Cates, L. Capano, S. Aryal, W. K. Kim, A. S. Yoo, Generation of human neurons by microRNA-mediated direct conversion of dermal fibroblasts. Methods Mol. Biol. 2239, 77–100 (2021). [DOI] [PubMed] [Google Scholar]
- 11.A. S. Yoo, A. X. Sun, L. Li, A. Shcheglovitov, T. Portmann, Y. Li, C. Lee-Messer, R. E. Dolmetsch, R. W. Tsien, G. R. Crabtree, MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Z. P. Pang, N. Yang, T. Vierbuchen, A. Ostermeier, D. R. Fuentes, T. Q. Yang, A. Citri, V. Sebastiano, S. Marro, T. C. Sudhof, M. Wernig, Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. B. Victor, M. Richner, H. E. Olsen, S. W. Lee, A. M. Monteys, C. Ma, C. J. Huh, B. Zhang, B. L. Davidson, X. W. Yang, A. S. Yoo, Striatal neurons directly converted from Huntington's disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat. Neurosci. 21, 341–352 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J. R. Herdy, L. Traxler, R. K. Agarwal, L. Karbacher, J. C. M. Schlachetzki, L. Boehnke, D. Zangwill, D. Galasko, C. K. Glass, J. Mertens, F. H. Gage, Increased post-mitotic senescence in aged human neurons is a pathological feature of Alzheimer's disease. Cell Stem Cell 29, 1637–1652.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. M. Oh, S. W. Lee, W. K. Kim, S. Chen, V. A. Church, K. Cates, T. Li, B. Zhang, R. E. Dolle, S. Dahiya, S. C. Pak, G. A. Silverman, D. H. Perlmutter, A. S. Yoo, Age-related Huntington's disease progression modeled in directly reprogrammed patient-derived striatal neurons highlights impaired autophagy. Nat. Neurosci. 25, 1420–1433 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. Mertens, J. R. Herdy, L. Traxler, S. T. Schafer, J. C. M. Schlachetzki, L. Bohnke, D. A. Reid, H. Lee, D. Zangwill, D. P. Fernandes, R. K. Agarwal, R. Lucciola, L. Zhou-Yang, L. Karbacher, F. Edenhofer, S. Stern, S. Horvath, A. C. M. Paquola, C. K. Glass, S. H. Yuan, M. Ku, A. Szucs, L. S. B. Goldstein, D. Galasko, F. H. Gage, Age-dependent instability of mature neuronal fate in induced neurons from Alzheimer's patients. Cell Stem Cell 28, 1533–1548.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Thier, P. Worsdorfer, Y. B. Lakes, R. Gorris, S. Herms, T. Opitz, D. Seiferling, T. Quandel, P. Hoffmann, M. M. Nothen, O. Brustle, F. Edenhofer, Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 10, 473–479 (2012). [DOI] [PubMed] [Google Scholar]
- 18.K. L. Ring, L. M. Tong, M. E. Balestra, R. Javier, Y. Andrews-Zwilling, G. Li, D. Walker, W. R. Zhang, A. C. Kreitzer, Y. D. Huang, Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11, 100–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.E. Lujan, S. Chanda, H. Ahlenius, T. C. Sudhof, M. Wernig, Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. U.S.A. 109, 2527–2532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D. W. Han, N. Tapia, A. Hermann, K. Hemmer, S. Hoing, M. J. Arauzo-Bravo, H. Zaehres, G. M. Wu, S. Frank, S. Moritz, B. Greber, J. H. Yang, H. T. Lee, J. C. Schwamborn, A. Storch, H. R. Scholer, Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 10, 465–472 (2012). [DOI] [PubMed] [Google Scholar]
- 21.J. Kim, J. A. Efe, S. Zhu, M. Talantova, X. Yuan, S. Wang, S. A. Lipton, K. Zhang, S. Ding, Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 108, 7838–7843 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Sheng, J. Jungverdorben, H. Wiethoff, Q. Lin, L. J. Flitsch, D. Eckert, M. Hebisch, J. Fischer, J. Kesavan, B. Weykopf, L. Schneider, D. Holtkamp, H. Beck, A. Till, U. Wullner, M. J. Ziller, W. Wagner, M. Peitz, O. Brustle, A stably self-renewing adult blood-derived induced neural stem cell exhibiting patternability and epigenetic rejuvenation. Nat. Commun. 9, 4047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D. Xiao, X. Liu, M. Zhang, M. Zou, Q. Deng, D. Sun, X. Bian, Y. Cai, Y. Guo, S. Liu, S. Li, E. Shiang, H. Zhong, L. Cheng, H. Xu, K. Jin, M. Xiang, Direct reprogramming of fibroblasts into neural stem cells by single non-neural progenitor transcription factor Ptf1a. Nat. Commun. 9, 2865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K. R. Yu, J. H. Shin, J. J. Kim, M. G. Koog, J. Y. Lee, S. W. Choi, H. S. Kim, Y. Seo, S. Lee, T. H. Shin, M. K. Jee, D. W. Kim, S. J. Jung, S. Shin, D. W. Han, K. S. Kang, Rapid and efficient direct conversion of human adult somatic cells into neural stem cells by HMGA2/let-7b. Cell Rep. 10, 441–452 (2015). [DOI] [PubMed] [Google Scholar]
- 25.E. Shahbazi, S. Moradi, S. Nemati, L. Satarian, M. Basiri, H. Gourabi, N. Zare Mehrjardi, P. Gunther, A. Lampert, K. Handler, F. F. Hatay, D. Schmidt, M. Molcanyi, J. Hescheler, J. L. Schultze, T. Saric, H. Baharvand, Conversion of human fibroblasts to stably self-renewing neural stem cells with a single zinc-finger transcription factor. Stem Cell Rep. 6, 539–551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M. C. Thier, O. Hommerding, J. Panten, R. Pinna, D. García-González, T. Berger, P. Wörsdörfer, Y. Assenov, R. Scognamiglio, A. Przybylla, P. Kaschutnig, L. Becker, M. D. Milsom, A. Jauch, J. Utikal, C. Herrmann, H. Monyer, F. Edenhofer, A. Trumpp, Identification of embryonic neural plate border stem cells and their generation by direct reprogramming from adult human blood cells. Cell Stem Cell 24, 166–182.e13 (2019). [DOI] [PubMed] [Google Scholar]
- 27.P. S. Hou, C. Y. Chuang, C. H. Yeh, W. Chiang, H. J. Liu, T. N. Lin, H. C. Kuo, Direct conversion of human fibroblasts into neural progenitors using transcription factors enriched in human ESC-derived neural progenitors. Stem Cell Rep. 8, 54–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Zhu, R. Ambasudhan, W. Sun, H. J. Kim, M. Talantova, X. J. Wang, M. L. Zhang, Y. Zhang, T. Laurent, J. Parker, H. S. Kim, J. D. Zaremba, S. Saleem, S. Sanz-Blasco, E. Masliah, S. R. McKercher, Y. S. Cho, S. A. Lipton, J. Kim, S. Ding, Small molecules enable OCT4-mediated direct reprogramming into expandable human neural stem cells. Cell Res. 24, 126–129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.L. Cheng, W. Hu, B. Qiu, J. Zhao, Y. Yu, W. Guan, M. Wang, W. Yang, G. Pei, Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 24, 665–679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.I. Maza, I. Caspi, A. Zviran, E. Chomsky, Y. Rais, S. Viukov, S. Geula, J. D. Buenrostro, L. Weinberger, V. Krupalnik, S. Hanna, M. Zerbib, J. R. Dutton, W. J. Greenleaf, R. Massarwa, N. Novershtern, J. H. Hanna, Transient acquisition of pluripotency during somatic cell transdifferentiation with iPSC reprogramming factors. Nat. Biotechnol. 33, 769–774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O. Bar-Nur, C. Verheul, A. G. Sommer, J. Brumbaugh, B. A. Schwarz, I. Lipchina, A. J. Huebner, G. Mostoslavsky, K. Hochedlinger, Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat. Biotechnol. 33, 761–768 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. Velychko, K. Kang, S. M. Kim, T. H. Kwak, K. P. Kim, C. Park, K. Hong, C. Chung, J. K. Hyun, C. M. MacCarthy, G. Wu, H. R. Scholer, D. W. Han, Fusion of reprogramming factors alters the trajectory of somatic lineage conversion. Cell Rep. 27, 30–39.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 33.S. M. Kim, H. Flasskamp, A. Hermann, M. J. Arauzo-Bravo, S. C. Lee, S. H. Lee, E. H. Seo, S. H. Lee, A. Storch, H. T. Lee, H. R. Scholer, N. Tapia, D. W. Han, Direct conversion of mouse fibroblasts into induced neural stem cells. Nat. Protoc. 9, 871–881 (2014). [DOI] [PubMed] [Google Scholar]
- 34.D. S. Tan, Y. Chen, Y. Gao, A. Bednarz, Y. Wei, V. Malik, D. H. Ho, M. Weng, S. Y. Ho, Y. Srivastava, S. Velychko, X. Yang, L. Fan, J. Kim, J. Graumann, G. D. Stormo, T. Braun, J. Yan, H. R. Scholer, R. Jauch, Directed evolution of an enhanced POU reprogramming factor for cell fate engineering. Mol. Biol. Evol. 38, 2854–2868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R. Jauch, I. Aksoy, A. P. Hutchins, C. K. Ng, X. F. Tian, J. Chen, P. Palasingam, P. Robson, L. W. Stanton, P. R. Kolatkar, Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells 29, 940–951 (2011). [DOI] [PubMed] [Google Scholar]
- 36.V. Veerapandian, J. O. Ackermann, Y. Srivastava, V. Malik, M. Weng, X. Yang, R. Jauch, Directed evolution of reprogramming factors by cell selection and sequencing. Stem Cell Rep. 11, 593–606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.P. Ellis, B. M. Fagan, S. T. Magness, S. Hutton, O. Taranova, S. Hayashi, A. McMahon, M. Rao, L. Pevny, SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 26, 148–165 (2004). [DOI] [PubMed] [Google Scholar]
- 38.G. La Manno, K. Siletti, A. Furlan, D. Gyllborg, E. Vinsland, A. Mossi Albiach, C. Mattsson Langseth, I. Khven, A. R. Lederer, L. M. Dratva, A. Johnsson, M. Nilsson, P. Lonnerberg, S. Linnarsson, Molecular architecture of the developing mouse brain. Nature 596, 92–96 (2021). [DOI] [PubMed] [Google Scholar]
- 39.T. Rayon, R. J. Maizels, C. Barrington, J. Briscoe, Single-cell transcriptome profiling of the human developing spinal cord reveals a conserved genetic programme with human-specific features. Development 148, dev199711 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.C. Lopez-Otin, M. A. Blasco, L. Partridge, M. Serrano, G. Kroemer, Hallmarks of aging: An expanding universe. Cell 186, 243–278 (2023). [DOI] [PubMed] [Google Scholar]
- 41.T. M. Stubbs, M. J. Bonder, A. K. Stark, F. Krueger, B. I. A. C. Team, F. von Meyenn, O. Stegle, W. Reik, Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 18, 68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D. Chondronasiou, D. Gill, L. Mosteiro, R. G. Urdinguio, A. Berenguer-Llergo, M. Aguilera, S. Durand, F. Aprahamian, N. Nirmalathasan, M. Abad, D. E. Martin-Herranz, C. Stephan-Otto Attolini, N. Prats, G. Kroemer, M. F. Fraga, W. Reik, M. Serrano, Multi-omic rejuvenation of naturally aged tissues by a single cycle of transient reprogramming. Aging Cell 21, e13578 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D. Hanahan, R. A. Weinberg, Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 44.P. Samavarchi-Tehrani, A. Golipour, L. David, H. K. Sung, T. A. Beyer, A. Datti, K. Woltjen, A. Nagy, J. L. Wrana, Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 (2010). [DOI] [PubMed] [Google Scholar]
- 45.D. Li, J. Liu, X. Yang, C. Zhou, J. Guo, C. Wu, Y. Qin, L. Guo, J. He, S. Yu, H. Liu, X. Wang, F. Wu, J. Kuang, A. P. Hutchins, J. Chen, D. Pei, Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell 21, 819–833.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 46.J. D. Buenrostro, P. G. Giresi, L. C. Zaba, H. Y. Chang, W. J. Greenleaf, Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.V. Malik, L. V. Glaser, D. Zimmer, S. Velychko, M. Weng, M. Holzner, M. Arend, Y. Chen, Y. Srivastava, V. Veerapandian, Z. Shah, M. A. Esteban, H. Wang, J. Chen, H. R. Scholer, A. P. Hutchins, S. H. Meijsing, S. Pott, R. Jauch, Pluripotency reprogramming by competent and incompetent POU factors uncovers temporal dependency for Oct4 and Sox2. Nat. Commun. 10, 3477 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. N. Schep, B. Wu, J. D. Buenrostro, W. J. Greenleaf, chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Z. Shao, Y. Zhang, G. C. Yuan, S. H. Orkin, D. J. Waxman, MAnorm: A robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 13, R16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Z. Li, M. H. Schulz, T. Look, M. Begemann, M. Zenke, I. G. Costa, Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 20, 45 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.I. Izeddin, V. Récamier, L. Bosanac, I. I. Cissé II, L. Boudarene, C. Dugast-Darzacq, F. Proux, O. Bénichou, R. Voituriez, O. Bensaude, M. Dahan, X. Darzacq, Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. eLife 3, e02230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. Chen, Z. Zhang, L. Li, B. C. Chen, A. Revyakin, B. Hajj, W. Legant, M. Dahan, T. Lionnet, E. Betzig, R. Tjian, Z. Liu, Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Z. Liu, W. R. Legant, B. C. Chen, L. Li, J. B. Grimm, L. D. Lavis, E. Betzig, R. Tjian, 3D imaging of Sox2 enhancer clusters in embryonic stem cells. eLife 3, e04236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.A. J. McCann, J. Lou, M. Moustaqil, M. S. Graus, A. Blum, F. Fontaine, H. Liu, W. Luu, P. Rudolffi-Soto, P. Koopman, E. Sierecki, Y. Gambin, F. A. Meunier, Z. Liu, E. Hinde, M. Francois, A dominant-negative SOX18 mutant disrupts multiple regulatory layers essential to transcription factor activity. Nucleic Acids Res. 49, 10931–10955 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.S. Condamin, V. Tejedor, R. Voituriez, O. Benichou, J. Klafter, Probing microscopic origins of confined subdiffusion by first-passage observables. Proc. Natl. Acad. Sci. U.S.A. 105, 5675–5680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Velychko, K. Adachi, K.-P. Kim, Y. Hou, C. M. MacCarthy, G. Wu, H. R. Schöler, Excluding Oct4 from Yamanaka Cocktail Unleashes the Developmental Potential of iPSCs. Cell Stem Cell 25, 737–753.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.L. J. Flitsch, O. Brüstle, Evolving principles underlying neural lineage conversion and their relevance for biomedical translation. F1000Research 8, Rev-1548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.A. Erharter, S. Rizzi, J. Mertens, F. Edenhofer, Take the shortcut: Direct conversion of somatic cells into induced neural stem cells and their biomedical applications. FEBS Let. 593, 3353–3369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.A. Ibrayeva, M. Bay, E. Pu, D. J. Jorg, L. Peng, H. Jun, N. Zhang, D. Aaron, C. Lin, G. Resler, A. Hidalgo, M. H. Jang, B. D. Simons, M. A. Bonaguidi, Early stem cell aging in the mature brain. Cell Stem Cell 28, 955–966.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.P. Navarro Negredo, R. W. Yeo, A. Brunet, Aging and rejuvenation of neural stem cells and their niches. Cell Stem Cell 27, 202–223 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.N. Gatto, C. Dos Santos Souza, A. C. Shaw, S. M. Bell, M. A. Myszczynska, S. Powers, K. Meyer, L. M. Castelli, E. Karyka, H. Mortiboys, M. Azzouz, G. M. Hautbergue, N. M. Markus, P. J. Shaw, L. Ferraiuolo, Directly converted astrocytes retain the ageing features of the donor fibroblasts and elucidate the astrocytic contribution to human CNS health and disease. Aging Cell 20, e13281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.J. Ha, B. S. Kim, B. Min, J. Nam, J. G. Lee, M. Lee, B. H. Yoon, Y. H. Choi, I. Im, J. S. Park, H. Choi, A. Baek, S. M. Cho, M. O. Lee, K. H. Nam, J. Y. Mun, M. Kim, S. Y. Kim, M. Y. Son, Y. K. Kang, J. S. Lee, J. K. Kim, J. Kim, Intermediate cells of in vitro cellular reprogramming and in vivo tissue regeneration require desmoplakin. Sci. Adv. 8, eabk1239 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.P. D. Tonge, A. J. Corso, C. Monetti, S. M. I. Hussein, M. C. Puri, I. P. Michael, M. Li, D.-S. Lee, J. C. Mar, N. Cloonan, D. L. Wood, M. E. Gauthier, O. Korn, J. L. Clancy, T. Preiss, S. M. Grimmond, J.-Y. Shin, J.-S. Seo, C. A. Wells, I. M. Rogers, A. Nagy, Divergent reprogramming routes lead to alternative stem-cell states. Nature 516, 192–197 (2014). [DOI] [PubMed] [Google Scholar]
- 64.C. Skorik, N. K. Mullin, M. Shi, Y. Zhang, P. Hunter, Y. Tang, B. Hilton, T. M. Schlaeger, Xeno-free reprogramming of peripheral blood mononuclear erythroblasts on laminin-521. Curr. Protoc. Stem Cell Biol. 52, e103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.R. Gorris, J. Fischer, K. L. Erwes, J. Kesavan, D. A. Peterson, M. Alexander, M. M. Nothen, M. Peitz, T. Quandel, M. Karus, O. Brustle, Pluripotent stem cell-derived radial glia-like cells as stable intermediate for efficient generation of human oligodendrocytes. Glia 63, 2152–2167 (2015). [DOI] [PubMed] [Google Scholar]
- 66.X. Yin, J. W. Kim, S. Liu, T. M. Dawson, V. L. Dawson, Protocol for measurement of calcium dysregulation in human induced pluripotent stem cell-derived dopaminergic neurons. STAR Protoc. 2, 100405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.V. Bryja, S. Bonilla, L. Cajanek, C. L. Parish, C. M. Schwartz, Y. Luo, M. S. Rao, E. Arenas, An efficient method for the derivation of mouse embryonic stem cells. Stem Cells 24, 844–849 (2006). [DOI] [PubMed] [Google Scholar]
- 68.C. K. Ng, N. X. Li, S. Chee, S. Prabhakar, P. R. Kolatkar, R. Jauch, Deciphering the Sox-Oct partner code by quantitative cooperativity measurements. Nucleic Acids Res. 40, 4933–4941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.D. T. McSwiggen, A. S. Hansen, S. S. Teves, H. Marie-Nelly, Y. Hao, A. B. Heckert, K. K. Umemoto, C. Dugast-Darzacq, R. Tjian, X. Darzacq, Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. eLife 8, e47098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.B. Li, C. N. Dewey, RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.B. Langmead, S. L. Salzberg, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.A. P. Hutchins, R. Jauch, M. Dyla, D. Miranda-Saavedra, glbase: A framework for combining, analyzing and displaying heterogeneous genomic and high-throughput sequencing data. Cell Regen 3, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D. Risso, K. Schwartz, G. Sherlock, S. Dudoit, GC-content normalization for RNA-Seq data. BMC Bioinformatics 12, 480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.A. Butler, P. Hoffman, P. Smibert, E. Papalexi, R. Satija, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.H. Gu, Z. D. Smith, C. Bock, P. Boyle, A. Gnirke, A. Meissner, Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat. Protoc. 6, 468–481 (2011). [DOI] [PubMed] [Google Scholar]
- 76.P. Danecek, J. K. Bonfield, J. Liddle, J. Marshall, V. Ohan, M. O. Pollard, A. Whitwham, T. Keane, S. A. McCarthy, R. M. Davies, H. Li, Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.A. R. Quinlan, I. M. Hall, BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Y. Zhang, T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nusbaum, R. M. Myers, M. Brown, W. Li, X. S. Liu, Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.S. Heinz, C. Benner, N. Spann, E. Bertolino, Y. C. Lin, P. Laslo, J. X. Cheng, C. Murre, H. Singh, C. K. Glass, Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 and S2
Legend for movie S1
Legends for data S1 and S2
Movie S1
Data S1 and S2