Abstract
An outstanding question in biology is to what extent convergent evolution produces similar, but not necessarily identical, complex phenotypic solutions. The placenta is a complex organ that repeatedly evolved in the livebearing fish family Poeciliidae. Here, we apply comparative approaches to test whether evolution has produced similar or different placental phenotypes in the Poeciliidae and to what extent these phenotypes correlate with convergence at the molecular level. We show the existence of two placental phenotypes characterized by distinctly different anatomical adaptations (divergent evolution). Furthermore, each placental phenotype independently evolved multiple times across the family, providing evidence for repeated convergence. Moreover, our comparative genomic analysis revealed that the genomes of species with different placentas are evolving at a different pace. Last, we show that the two placental phenotypes correlate with two previously described contrasting life-history optima. Our results argue for high evolvability (both divergent and convergent) of the placenta within a group of closely related species in a single family.
Livebearing fishes have independently evolved different kinds of placentas multiple times.
INTRODUCTION
Phenotypic convergence, the repeated evolution of similar traits in phylogenetically independent lineages, is of interest to evolutionary biologists because of the opportunity it offers to study evolutionary processes (1, 2). Cases of convergent evolution are generally thought to arise in response to similar selective pressures driving similar adaptations in different lineages. They represent compelling natural evidence of adaptive evolution by natural selection and signals of repeatability in evolution (3–7). Examples of convergent evolution include the cranial shape of extinct marsupial thylacines and mammalian canids that evolved as an adaptation to carnivorous feeding (8, 9) and the hydrodynamic body shape in fishes (sharks), reptiles (ichthyosaurs), birds (penguins), and mammals (dolphins) that evolved in response to life in aquatic environments (10–12). However, what we can learn about the repeatability of evolution from lineages that diverged hundreds of millions of years (Ma) ago is somewhat limited because distantly related species often differ greatly in many aspects other than the convergent trait of interest, including genomic architecture, developmental programs, morphology, physiology, and ecology. Because there may be no association between phylogenetic proximity and the genetic basis of similar phenotypes (13–15), there are benefits to studying phenotypic convergence and its repeatability in a group of closely related species in which the novel trait of interest recently evolved multiple times independently.
The fish family Poeciliidae (order Cyprinodontiformes) offers a unique opportunity to study convergent evolution because here a complex organ, the placenta, has evolved repeatedly in a group of closely related species. All but one species in this family bear live young (16, 17), which means that the eggs are internally fertilized and embryos develop inside the female until birth (18). Most species in this family fully provision eggs with yolk before fertilization (lecithotrophic viviparity). Some species evolved the ability to continue to provision embryos during pregnancy via a placenta (matrotrophic viviparity) (19, 20). The level of post-fertilization maternal provisioning is represented by the matrotrophy index (MI), which quantifies how much dry weight an embryo gains from fertilization to birth (18). Lecithotrophic species produce large eggs that use yolk reserves as the embryo develops, resulting in MI indices of less than one, because they lose weight during development. In contrast, placental females produce smaller eggs, and embryos gain weight after fertilization as the female provides nutrients to the developing embryos (MI > 1). The differences between lecithotrophic (nonplacental) and matrotrophic (placental) livebearing fishes are manifested morphologically at the maternal-fetal interface, with placental species having evolved specialized anatomical adaptations to support embryo development during pregnancy. The maternal follicular wall (MFW) that surrounds each embryo (present in both lecithotrophic and matrotrophic species) has become hypertrophied and vascularized in placental lineages (21–24). Moreover, the embryos of placental species often have highly vascularized embryonic structures in the yolk sac, pericardial sac, and/or embryonic epithelium (25–27). Together, these specialized maternal and embryonic tissues form the placenta, with more complex placentas presumably allowing for more intimate physiological maternal-fetal interactions (e.g., exchange of nutrients, gasses, and waste products) throughout pregnancy. Phylogenetic and ancestral state reconstruction using more than 100 species in the family Poeciliidae revealed that the common poeciliid ancestor likely lacked placentotrophy and that the placenta evolved independently at least nine times in different lineages (20, 28, 29). These independent origins of the placenta appear in six different genera: once in Xenodexia, Phalloptychus, Phalloceros, and Heterandria; twice in the genus Poecilia; and three times within the genus Poeciliopsis (29).
Despite important efforts devoted to understanding why (30–38) and how (39–42) the placenta evolved repeatedly in this family, it is still unclear whether the independent evolutionary origins led to organs with similar morphological phenotypes and genomic signatures resulting from similar evolutionary modification to the developmental and molecular pathways. We recently found some evidence for two contrasting life-history strategies in placental poeciliids: one represented by small-bodied species that have fast life histories, and the other by large-bodied species with slow life histories (36). We hypothesized that these life-history strategies might be correlated with diversity in placental structures. In the present study, we investigated to what extent (i) evolution has produced similar (convergent evolution) or different (divergent evolution) placental phenotypes in the Poeciliidae, (ii) the evolution of different placental phenotypes is repeatable, and (iii) evolution of placental phenotypes is associated with parallel or alternative changes at the genomic level.
RESULTS
Morphological analysis reveals two types of placenta in the fish family Poeciliidae
We compared placental tissues [i.e., MFW and yolk-pericardial sac (YPS)] in species from six independent placental lineages. All studied species (Fig. 1) had a highly vascularized MFW (maternal part of the placenta) and YPS (embryonic part of the placenta), while vascularization was reduced in the embryonic dermis and epidermis (fig. S1). On the basis of the morphological details of the MFW and YPS, we identified two distinctly different types of placentas in the Poeciliidae.
Fig. 1. Phylogeny of 26 species from the family Poeciliidae.
Placental lineages are highlighted in blue lines. The tree was obtained from Kruistum et al. (42).
One type (hereafter referred to as the “villous placenta”) was characterized by the presence of an MFW with elongated villus-like structures supported by simple connective tissue (Fig. 2 and fig. S2), while the embryonic part was formed by a reduced layer of mesothelium tissue. This placental morphology was found in four of the eight studied placental species: in the southern clade of the genus Poeciliopsis (represented by Poeciliopsis turneri and Poeciliopsis presidionis) and in the subgenus Aulophallus of the genus Poeciliopsis (i.e., Poeciliopsis retropinna and Poeciliopsis paucimaculata).
Fig. 2. Morphology of villous and smooth placentas of eight poeciliid species representing six independent origins of the placenta in the family Poeciliidae.
(A) MFW and the YPS show important morphological diversity among the six placental lineages. P. turneri, P. presidionis, P. retropinna, and P. paucimaculata have a villous placenta, whereas P. prolifica, H. formosa, P. bifurca, and P. januarius have a smooth placenta. Please note that the morphology of P. presidionis (placental sister species of P. turneri) and P. paucimaculata (placental sister species of P. retropinna) is given in fig. S2). ML, mesenchymal layer; Bv, blood vessel; Ery, erythrocyte; Mbv, maternal blood vessel; Ebv, embryonic blood vessel; MeC, mesothelial cells. (B) Schematic representation of the anatomical differences between the two types of placentas: Species with a villous placenta have a MFW (maternal part of the placenta) with elongated villus-like structures (EVSs) and a YPS (embryonic part of the placenta) that consists of a reduced mesothelium layer, while species with a smooth placenta lack elongated villus-like structures on their MFW and have a more developed YPS that consists of a thicker mesothelium layer of cells supported by abundant embryonic blood vessels underlying the connective tissue.
The other type of placenta (hereafter referred to as the “smooth placenta”) was found in the other four placental species: Poeciliopsis prolifica, Phalloptychus januarius, Heterandria formosa, and Poecilia (subgenus Micropoecilia) bifurca. The placenta in these species showed a MFW that lacked elongated villus-like structures, while the YPS displayed an outer surface layer of mesothelium supported by abundant embryonic blood vessels underlying connective tissue (Fig. 2).
Because pregnancy is a dynamic process, we evaluated temporal changes in the thickness of the MFW and YPS by comparing three developmental stages (early, mid, and late). We found that in species with a villous placenta (i.e., P. turneri, P. presidionis, P. retropinna, and P. paucimaculata), the MFW was consistently thicker than the YPS at all three developmental stages (Fig. 3, A to D). On the contrary, species with a smooth placenta (i.e., P. prolifica, P. januarius, P. bifurca, and H. formosa) tended to show a thinner MFW and thicker YPS across developmental stages (Fig. 3, E to H). When comparing the MFW and YPS across species, we found that all species with villous placenta showed a thicker MFW (58.7 to 101.5 μm) while exhibiting a thinner YPS (8.5 to 14.5 μm) (Fig. 3, I and J), resulting in an MFW/YPS ratio that was consistently higher than 1 (Fig 3K). In contrast, species with smooth placenta showed a thinner MFW (8.1 to 21.4 μm) and a thicker YPS (14.7 to 29.2 μm) (Fig. 3, I and J), leading to an MFW/YPS ratio that was consistently lower than 1 (Fig 3K). These differences in the thickness of MFW and YPS between placental types were further supported when these values were corrected by the female standard length (fig. S3, A and B).
Fig. 3. Comparative analyses of placental morphology at different developmental stages in eight placental species from six independent origins of the placenta in the family Poeciliidae.
(A to H) Thickness of the MFW and YPS at three embryonic stages (early, mid, and late developmental stage) remains relatively stable within each species; there is a species-specific increase in the thickness of the MFW between early and mid embryo stages in P. turneri and a reduction in thickness of the MFW between early and mid embryo stages in P. januarius and H. formosa. Different letters indicate significant differences (P < 0.05) across developmental stages within each species in the MFW and YPS, respectively. Asterisks represent statistical differences (P < 0.05) between the tissues (i.e., MFW and YPS) at a specific developmental stage within each species. Bars represent SEs. (I to K) Comparisons of (I) the thickness of the MFW, (J) the YPS, and (K) the ratio of MFW/YPS among species at all embryonic stages. The analyses reveal two types of placental morphology: Species with a villous placenta (P. turneri, P. presidionis, P. retropinna, and P. paucimaculata) have a thicker MFW and thinner YPS, resulting in an MFW/YPS ratio > 1, while species with a smooth placenta (P. prolifica, P. bifurca, P. januarius, and H. formosa) have a thinner MFW and thicker YPS, leading to an MFW/YPS ratio < 1. Different letters indicate significant differences (P < 0.05) among species. The dotted line in (K) represents a ratio equal to 1.
The degree of placentotrophy is associated with the evolution of the maternal part of the placenta, but not with the embryonic part of the placenta
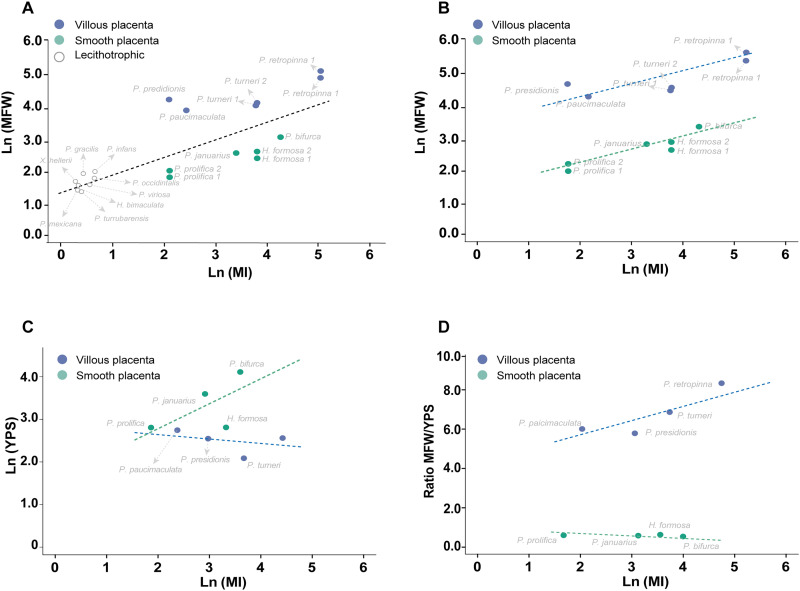
To test for associations between aspects of the morphology of “villous” and “smooth” placentas and the level of maternal provisioning, we performed phylogenetic regressions between the thickness of the MFW and YPS or their ratio and the MI. We first tested for a relationship between MFW and MI using species that represent the full range of maternal provisioning across the family (i.e., ranging from lecithotrophic to highly matrotrophic). Phylogenetic generalized least square (PGLS) analysis revealed a significant positive relationship between MFW and MI, with lecithotrophic species having a significantly thinner MFW than placental species (Fig. 4A). Next, we implemented PGLS analyses to test whether the MFW differed among species with a villous and smooth placenta while accounting for MI. We found that the two placental types significantly differed in MFW: For any given value of MI, species with a villous placenta had a thicker MFW than those with a smooth placenta (P < 0.001; Fig. 4B and fig. S4A). We then tested whether the YPS differed among species with a villous and smooth placenta and found that the two placenta types did not significantly differ in YPS and that YPS did not significantly increase with a higher MI (although a positive trend was apparent for species with “smooth placentas”; Fig. 4C and fig. S4B). However, an association between these variables should not be discarded in further studies including more species. Last, we tested whether the MFW/YPS ratio differed between species with a villous and smooth placenta. We found that the two placental types significantly differed in this ratio and that, for any given value of MI, species with a villous placenta had a substantially higher MFW/YPS ratio compared to species with a smooth placenta (Fig. 4D). This latter result was driven by the substantially thicker MFW in species with a villous placenta, as the YPS did not differ between placental types.
Fig. 4. Phylogenetic generalized least squares (PGLS) analyses between the thickness of the MFW, YPS or their ratio and the MI within the family Poeciliidae.
(A) Comparison of the MFW of poeciliid species (ranging from lecithotrophic to highly matrotrophic; table S2) reveals a significant positive relationship between thickness of the MFW and MI. (B) The thickness of the MFW shows a positive association with MI in species with both a villous (blue dashed line; P < 0.001) and a smooth placenta (green dashed line; P < 0.001). Moreover, for any given value of MI, species with a villous placenta have a significantly thicker MFW than those with a smooth placenta (P < 0.001). (C) Thickness of YPS does not differ significantly between species with a villous placenta and smooth placenta nor shows a statistically significant relationship with MI (P > 0.05). Note that the green and blue dashed lines are from the best fitting PGLS model including the interaction between MI and placenta type; these lines are included for illustration purposes only, as all terms in the model except intercept were nonsignificant (P > 0.05). (D) MFW/YPS ratio in the villous placenta (blue dashed line) is significantly higher (P < 0.001) than in the smooth placenta (green dashed line), while there is no significant association with MI (P > 0.05).
The evolution of different placental structures is accompanied by changes in the rate of evolution in genes associated with blood vessel formation and metabolism
We tested for differences in the relative evolutionary rate of 14,322 sets of orthologous genes between species with a villous placenta (P. retropinna, P. paucimaculata, P. turneri, and P. presidionis) and species with a smooth placenta (H. formosa, P. bifurca, P. januarius, and P. prolifica). When testing for the hypothesis that genes show accelerated evolution in species with a villous placenta relative to species with a smooth placenta, we found an overrepresentation of low P values in the resulting distribution (Fig. 5A and fig. S5). This suggests that a larger than expected set of genes evolves more quickly in species with a villous placenta than in species with a smooth placenta. For the opposite hypothesis, we did not find a skew toward low P values, but, instead, we found a reverse result (Fig. 5B and fig. S5). This shows that not more genes than expected by chance evolve faster in species with a smooth placenta than in species with a villous placenta.
Fig. 5. Analysis of evolutionary rate in the genome of species with different types of placenta.
(A) Distribution of P values for the test assessing that the relative evolutionary rate is significantly greater in poeciliid species with a villous placenta than in species with a smooth placenta (P = 3.56 × 10−17, Kolmogorov-Smirnov test). (B) Distribution of P values for the test assessing that the relative evolutionary rate is significantly greater in poeciliid species with a smooth placenta than in species with a villous placenta (P = 1.39× 10−16, Kolmogorov-Smirnov test). The same test with phenotypes shuffled randomly is shown in gray bars in (A) and (B). Tests were performed on 14,322 sets of orthologous genes obtained from Kruistum et al. (42). (C and D) GO enrichment analysis for the top 100 genes evolving faster in species with villous placenta. The top 15 terms for biological process (C) and top 7 terms for molecular function (D) are shown with their respective enrichment score [−log10 (P value)], which are ranked from the most significant (blue circles) to the least significant (green circles). Circle size (gene_ratio) represents the proportion of high evolving genes per total annotated genes in a GO term. Dashed line represents P = 0.05, while solid line represents P = 0.01. JNK, c-Jun N-terminal kinase; snRNA, small nuclear RNA; snRNP, small nuclear ribonucleoprotein; MAP, mitogen-activated protein.
Our Gene Ontology (GO) enrichment analysis using the top 100 most significant genes with accelerated evolution in species with villous placenta (table S5) revealed an overrepresentation of genes associated with blood vessel formation and endothelial cell regulation (Fig. 5, C and D, and fig. S6). Moreover, several GO terms related to metabolic processes were also ranked among the most significant terms (Fig. 5, C and D, and fig. S6).
Species with different placental types diverge in their life histories
Previously, we proposed that there may be two phenotypic adaptive peaks associated with placentation in the fish family Poeciliidae: one represented by large-bodied species that have few but relatively large offspring at birth, and the second by small-bodied species with many but relatively small offspring at birth (36). To assess whether the two placental types are correlated with different life histories, we tested for associations between life-history traits (i.e., female standard length, offspring size at birth, and number of embryos) and reproductive mode. For the latter, we included species with villous and smooth placentas plus lecithotrophic species for which we previously gathered life-history data (138 lecithotrophic species) (36). The PGLS showed that reproductive strategy had a significant effect on female standard length (Fig. 6A) and offspring size at birth (Fig. 6B). Females from species with a villous placenta are significantly larger than females from species with a smooth placenta (Fig. 6A). Moreover, species with a villous placenta give birth to the largest offspring, species with a smooth placenta give birth the smallest offspring, and lecithotrophic species give birth to offspring with an intermediate size (Fig. 6B). Last, we did not find differences in the number of embryos between the three reproductive strategies (Fig. 6C). Given that we identified differences in female size as a function of reproductive mode and the general association between female size and life-history traits, we then tested whether species with different reproductive strategies (i.e., lecithotrophy, smooth placenta, and villous placenta) differed in offspring size at birth and the number of embryos when accounting for female standard length (Fig. 6, D and E). We found that when accounting for female standard length, species with villous placenta have the largest offspring at birth but the lowest number of embryos, while species with a smooth placenta have smaller offspring at birth but a higher number of embryos (Fig. 6, D and E).
Fig. 6. PGLS analyses of poeciliid life histories as a function of placenta type.
In the top row, raw data plotted for (A) female standard length, (B) offspring size at birth, and (C) number of embryos, with species categorized as villous placenta, smooth placenta, or lecithotrophic. Different letters in (A) to (C) indicate statistical differences (P < 0.05) between groups. (D) Species with a villous placenta (blue dashed line) have significantly larger offspring relative to those with a smooth placenta (green dashed line) while accounting for female standard length (P < 0.001). (E) Conversely, species with a smooth placenta (green dashed line) produce significantly more embryos than species with villous placenta (blue dashed line) while accounting for female standard length (P < 0.05).
DISCUSSION
Morphological divergence in the MFW between placental types
Our comparative study includes eight species from six independent placental lineages within the family Poeciliidae. We identified two types of placentas that significantly differ in morphology. Each evolved multiple times independently across the family tree. The villous placenta, found in two placental lineages, is characterized by a relatively complex thick MFW with many elongated villi and a relatively thin/simple YPS (MFW/YPS ratio > 1). In contrast, the smooth placenta, observed in four of the six studied placental lineages, is characterized by a relatively simple thin MFW and a thick YPS that exhibits an outer surface layer of mesothelial cells upon abundant embryonic blood vessels. This is reflected by an MFW/YPS ratio ≤ 1.
The MFW of species with a villous placenta (i.e., P. retropinna, P. paucimaculata, P. turneri, and P. presidionis) shows classical features typically associated with a fish placenta, i.e., a thick follicle wall with dense villus-like structures. Notably, the elongated villus structure found in the MFW of the villous placenta resembles the microvilli organization previously described in P. prolifica (23) and H. formosa (23, 25). Nevertheless, the length of the villus structure (~86 μm) is >2 orders of magnitude greater than the length of the microvilli (~0.85 μm), indicating that while both structures might exercise similar functions (facilitation of nutrient delivery and increasing surface area), their competence to do so likely differs.
In contrast to the villous placenta, the MFW of the smooth placenta (i.e., P. prolifica, P. bifurca, H. formosa, and P. januarius) is relatively simple, being thin and lacking elongated villus-like structures. Olivera-Tlahuel et al. (23), using electron microscopy, found microvilli in the MFW of P. prolifica and H. formosa, a feature that we were not able to capture using light microscopy. It is, therefore, possible that the presence of microvilli in the MFW is another characteristic common in species with smooth placenta. The MFW of species with a smooth placenta may be relatively simple but is still more complex (i.e., thicker and with presence of blood vessels) than the MFW found in nonplacental (lecithotrophic) viviparous species in the family Poeciliidae (22–24, 41), which is to be expected as lecithotrophic species lack post-fertilization maternal-fetal nutrient provisioning [reflected by a MI value below 0.7 (19, 20)]. Together, our results suggest that there are at least three levels of complexity in the MFW within the livebearing family Poeciliidae: one in nonplacental (lecithotrophic) and two levels (the different placenta types) in placental species.
Morphological divergence in the YPS between placental types
The evolution of post-fertilization provisioning required not only adaptations on the mother’s side but also anatomical adaptations in fetal tissues (i.e., the embryonic part of the placenta) to enable nutrient acquisition by the embryo (25–27). Our comparative anatomical study reveals a general embryonic anatomy that is consistently similar among all placental species regardless of placental type, i.e., the embryos of all species evolved hypervascularized pericardial and yolk sacs, while the embryonic dermis and epidermis were not vascularized. Our results, combined with the fact that the embryonic epithelium is also present in nonplacental livebearing species (26), support the idea that pericardial and yolk sacs are the primary embryonic structures involved in the physiological exchange during maternal provisioning.
Although the thickness of the YPS did not differ greatly between species with villous and smooth placentas, these results will need further validation using more species with different placentas. Still, we identified two clear differences that likely affect their nutrient transfer efficiency from mother to fetus: (i) The YPS of the villous placenta is highly vascularized, while (ii) the YPS of the smooth placenta is also highly vascularized but, in addition, develops blood vessels that are located directly under the layer of mesothelial cells. Notably, mesothelial cells have been linked to improved nutrient acquisition in the yolk sac of primates, a key process to support embryo survival and growth between fertilization and the onset of placental function (43). Nutrients are absorbed in the yolk sac by the outer surface layer of the mesothelium and are then transported into surrounding embryonic blood vessels (44). The thickness of YPS and MI increases in synchrony only in species with a smooth placenta. These findings suggest that the presence of mesothelium and more blood vessels in the YPS of species with the smooth placenta likely enhances nutrient absorptive capacity.
Distinctive genomic changes between placental types
To test whether the two placental types that evolved within this family are associated with genomic differences, we compared the whole genomes of our study species (42). We identified a shift in the rate of gene evolution between the two placental types, with genes evolving significantly faster in species with a villous placenta than in species with a smooth placenta. The repeated evolution of each of the two types of placentas thus appears to be associated with genomic differences between them. The villous placenta is the relatively more morphologically complex placenta, typically having a thicker follicle wall with dense villous-like structures, suggesting that a higher morphological complexity is driven by a faster molecular rate evolution. Most of the fast-evolving genes in species with a villous placenta are associated with the formation of blood vessels and metabolic pathways, suggesting that these biological processes are under selection in more complex placentas. In line with this, our previous analysis of 26 poeciliid genomes revealed accelerated evolution in genes associated with metabolic transport and vesicle functioning in placental over nonplacental poeciliids (42).
Studies have shown that functionally similar complex structures or organs can result from convergent (45, 46) or divergent (47, 48) genomic changes. Our study shows the repeated occurrence of both divergent and convergent evolution of a complex organ (the placenta) in a single family (Poeciliidae).
Life-history divergence between placental types
In a previous study, we argued that there may be two phenotypic adaptive peaks, corresponding to two selective optima, associated with placentation in the fish family Poeciliidae: one represented by small-bodied species that have fast life histories, and the second by large-bodied species with slow life histories (36). Here, we extend this hypothesis by arguing that these two contrasting life-history strategies may be associated with the evolution of two distinctly different placental types. The villous placenta evolved in the largest species (P. retropinna, P. paucimaculata, P. presidionis, and P. turneri), which typically produce few but relatively large offspring at birth. It is possible that in these species, the thicker MFW facilitates maternal-fetal physiological exchange, enabling the mother to invest more in each individual offspring via an improved nutrient transfer. By contrast, species with a smooth placenta evolved in the smallest species with the fastest life histories (P. prolifica, H. formosa, P. bifurca, and P. januarius) characterized by a small adult body size, high fecundity, and relatively small offspring size at birth. For P. prolifica (smooth placenta) versus P. presidionis/P. turneri (villous placenta), we also know that P. prolifica is younger at maturity (33). Here, a relatively simple placenta may be sufficient to transfer the lower required quantities of resources to each of the developing offspring during gestation. The finding that the villous and smooth placentas are associated with these different life-history traits provides further compelling evidence for the repeated evolution of two different placental types in the Poeciliidae.
In this study, we show the occurrence of both divergent and convergent evolution of a novel complex organ in a single fish family (Poeciliidae). The evolution of two distinctly different placental types (villous versus smooth placenta) indicates the occurrence of divergent evolution in the family. Moreover, the finding that each placental type independently evolved multiple times across the family (the villous placenta at least two times and smooth placenta at least four times) provides evidence for the repeated convergent evolution of each type. These findings are supported by three lines of evidence: First, the two placental types are associated with distinctly different anatomical adaptations for maternal-fetal nutrient transfer, leading to differences in the complexity of the maternal versus embryonic parts of the placenta (morphological evidence). Second, the genomes of species with different placental phenotypes are evolving at a different pace, with genes associated with blood vessel formation and metabolic processes evolving significantly faster in species with a more complex villous placenta than in species with smooth placenta (molecular evidence). Third, the two placental types are associated with different life-history traits: The villous placenta is linked with a slow life-history with females producing a few large offspring, while a smooth placenta is associated with a fast life-history with females producing many small offspring (life-history evidence). Prior studies have shown that the poeciliid placenta evolved in a relatively short period of time (~2.3 to 0.75 Ma ago) (20, 49), arguing for a high “evolvability” of this organ. The fact that both placental types (villous versus smooth) evolved multiple times across different lineages suggests that they may be the result of adaptation to dissimilar conditions, likely modulating molecular pathways and/or underlying developmental constraints. Our study provides evidence of great flexibility in the evolutionary trajectories (both divergent and convergent) of a complex organ in a group of closely related species within a single family.
METHODS
Study species
We studied the genome and morphology of follicles and embryos from eight poeciliid species representing six of nine independent origins of the placenta in this family (Fig. 1) (29). These include P. januarius, H. formosa, P. (subgenus Micropoecilia) bifurca (representing one of two independent placental origins within the genus Poecilia), P. prolifica, P. retropinna, P. paucimaculata, P. turneri, and P. presidionis (table S1). P. prolifica represents one of the three origins of placentation in this genus. P. retropinna and P. paucimaculata represent the second origin. P. turneri and P. presidionis are sister taxa that represent the third origin. The species were obtained from stocks maintained at Wageningen University (The Netherlands) and the University of California Riverside (USA). All procedures were approved by the Animal Ethics Committee of Wageningen University (The Netherlands) and Research and the University of California Riverside (USA).
Tissue processing and imaging
After an overdose of the anesthetic tricaine methanesulphonate (MS-222), whole pregnant females or freshly dissected ovaries were fixed in 4% paraformaldehyde for at least 14 days. P. retropinna and P. paucimaculata females were first stored in 70% EtOH after capture from wild populations in Costa Rica. All ovaries were washed in water, dehydrated in graded ethanol (70, 80, 90, 96, and 100%), embedded in paraffin, cleared in xylene, and sectioned at a thickness of 5 μm in a longitudinal plane. Sections were then stained with hematoxylin and eosin and photographed using an upright DM6 B microscope (Leica) with Leica Application Suite X (LAS X) software (version 3.7.4.23463). To measure placental structures (see the “Morphological analysis of placental structures” section), the entire section was scanned at ×200 magnification with a digital camera, while details of the placental tissues were taken using ×400 magnification.
Morphological analysis of placental structures
Placentas are composed of closely apposed maternal and fetal tissues. In livebearing fishes, the maternal part of the placenta is formed by the MFW. The embryonic part consists of various structures that may be adapted for physiological exchange, i.e., the yolk sac, pericardial membranes, and/or microvillous embryonic epithelium (25, 27). We found that the yolk sac and pericardial membranes are both highly vascularized; however, the embryonic epithelium is not (fig. S1). The presence of microvillii in embryonic epithelium has furthermore been shown to occur in both placental and nonplacental lineages (26). Therefore, we considered the yolk sac and pericardial membranes (hereafter referred to as the YPS) as the embryonic part of the placenta.
To quantify the thickness of placental structures, we randomly selected nonoverlapping regions of the MFW (15 regions) or YPS (10 regions) per placenta. Using ImageJ (version 2.3.0/1.53f, National Institutes of Health, Bethesda, MD, USA; http://rsbweb.nih.gov/ij) (50), the MFW was measured from the distal part of the epithelium to the underlying connective tissues, while the YPS was measured from the connective tissue or mesothelial cells to the internal blood vessels. In addition, we studied three embryonic developmental stages (early, mid, and late based on the presence of embryonic structures and size; fig. S1). For P. paucimaculata, embryos of only two of the three stages (mid and late) were obtained from the pregnant females. To avoid analyzing the same embryo more than once, the placental structures of all embryos present in a single scanned section per ovary were analyzed.
To test for differences in the thickness of the MFW and YPS among developmental stages and among species, we implemented general linear mixed models using lme4 (v. 1.1.27.1) (51), performance (v. 0.7.3) (52) and emmeans (v. 1.6.3.9900002) (53) in R v. 4.0.4 (54). We performed two separate analyses: (i) To detect potential differences among developmental states within species, we included developmental stage (three levels: early, mid, and late stage) as a fixed effect, female as a random effect, and thickness of placental structures (in the MFW or YPS, respectively) as the response variable; (ii) to perform a cross-species comparison, species were set as fixed effects, female as a random effect, and thickness of placental structures (in the MFW or YPS, respectively) as the response variable. To control for the possible effect of female size, we performed separate analyses using the raw thickness data corrected by female length (thickness of MFW or YPS/mean female length), using the female length of each species reported by Furness et al. (36).
To test for an association between the thickness of the tissues forming the placenta (i.e., MFW and YPS) and the MI, we implemented PGLS analyses using the R package Caper (v. 1.0.1) (55). Caper estimates model parameters and lambda (phylogenetic signal) using maximum likelihood. We used Reznick et al.’s (56) well-resolved time tree of the family Poeciliidae to implement the PGLS analyses. We trimmed the tree to only include the species of interest using the function drop.tip in the R package Ape (v. 5.5) (57). The MI for each species was obtained from Pollux et al. (19).
We first tested for a relationship between MFW and MI. For this analysis, we included additional MFW data for eight lecithotrophic species and four additional previous measurements in matrotrophic species, available in the literature (table S2). Additional YPS data were unavailable, so PGLS analyses examining the relationship between YPS and MI, as well as the relationship between MFW/YPS and MI, were limited to the eight placental species for which we gathered the primary data.
Our morphological analysis revealed the presence of at least two placental conformations across the eight species from the six independent lineages studied. These two placental types differed in the complexity of the maternal and embryonic parts forming this organ (i.e., MFW and YPS). We, therefore, tested whether MFW, YPS, or the ratio MFW/YPS differed as a function of placental type (villous and smooth) and the MI. For each dependent variable (MFW, YPS, and MFW/YPS), we fit three models: (i) one that only included the MI, (ii) one that included the MI and placental type, and (iii) one that included the interaction of MI and placental type. The best model was chosen according to the Akaike information criterion (AIC), selecting the model with the lowest AIC (table S3).
Analysis of life-history traits
To test for an association between placenta type and life-history traits, we implemented PGLS analyses, as described above. For these analyses, we included previously published data on female standard length, offspring size at birth, and number of embryos for our eight placental species (four species with villous placenta and four species with smooth placenta) and for 138 lecithotrophic species (table S4) (36), allowing us to cover the entire reproductive spectrum within this fish family (from lecithotrophic to highly matrotrophic species).
We first tested whether female standard length, offspring size at birth, and number of embryos differed among species with a villous placenta, smooth placenta, and no placenta (i.e., lecithotrophic species). To statistically compare each dependent variable (female standard length, offspring size at birth, and number of embryos), we fit simple PGLS models that included only reproductive mode as the independent variable. Each PGLS model was run treating villous placenta, smooth placenta, and lecithotrophic as the reference level, which amounts to a post hoc comparison among groups.
Given that we found differences in female standard length among species with a villous placenta and smooth placenta, and the general association between female size and life-history traits, we then tested whether offspring size at birth and number of embryos differed as a function of reproductive mode (villous placenta, smooth placenta, and lecithotrophy) while accounting for female standard length. We fit four PGLS models: (i) one that included only female standard length, (ii) one that only included placental type (villous placenta, smooth placenta, and lecithotrophic species), (iii) one that included female standard length and placental type, and (iv) one that included the interaction of female standard length and placental type. The best model was chosen on the basis of the AIC, selecting the model with the lowest AIC (table S3).
Comparative genomics
To study how convergent evolution can drive genomic changes in closely related species, we performed an evolutionary rate analysis of the genome of 26 poeciliid species (8 placental and 18 lecithotrophic species) (see Fig. 1). We used Kruistum et al.’s. (42) precalculated table of relative evolutionary rates to perform our evolutionary rate analysis. First, amino acid alignments of orthologous genes were made using mafft v7.402 (58). Second, branch lengths were estimated for each branch across the reconstructed phylogeny with the AAML program of the PAML package (59) using an empirical substitution model (60). Raw branch lengths were transformed into relative rates using a projection operator method (61). The relative rates estimate how much faster or slower this gene changed on a given branch after factoring out the divergence on that branch. Therefore, this table contains the mutation rate of each gene across each branch in the poeciliid phylogeny, normalized for both gene- and branch-specific evolutionary rates (45) for 14,322 sets of orthologous genes. The resulting relative evolutionary rate resembles the deviation from the expected evolutionary rate given gene- and branch-specific averages, so that a relative evolutionary rate greater than 0 means a faster than expected evolutionary rate in the given branch, while a relative evolutionary rate lesser than 0 means a slower than expected rate of evolution.
We hypothesized that species with a seemingly morphologically more complex villous placenta would present a faster evolutionary rate than species with a smooth placenta. Therefore, we first tested the hypothesis that (i) the relative evolutionary rate is greater in phylogenetic branches leading to species with a villous placenta than in branches leading to species with a smooth placenta. To do so, a Mann-Whitney U test was performed for each set of orthologous genes. Then, a null hypothesis was developed by performing the same tests, but between randomly selected phylogenetic branches, to test whether more genes than expected show a faster evolutionary rate in species with villous placenta. To further support our finding, we second tested whether (ii) the relative evolutionary rate is greater in phylogenetic branches leading to species with a smooth placenta than in branches leading to species with a villous placenta using the same approach and null hypothesis. Then, the resulting P value distributions of both hypotheses (i and ii) were compared to the P value distribution of the control analysis using a Kolmogorov-Smirnov test. While this analysis informs about differences in the rate of evolution among lineages, it does not distinguish between higher rates of adaptation versus relaxation of selective constraints.
To functionally annotate the genes that are differentially evolving faster in species with a villous placenta over species with a smooth placenta, we conducted a GO enrichment analysis with the top 100 genes that were significant (table S5). Enriched GO terms associated with biological processes and molecular functions were determined using the weight01 algorithm and the Fisher’s test in the R package topGO (62). GO gene sets with at least three annotated genes for Poecilia reticulata were retrieved from Ensemble (www.ensembl.org) using the R package biomaRt (63, 64).
Acknowledgments
We thank H. Schipper for all the advice and support in the morphological data collection and M. Cardoso-Moreira for providing critical comments on the manuscript.
Funding: We thank the support of a scholarship from ANID Chile (74200001) awarded to D.S., the National Science Foundation grants (DEB 0416085 and DEB 1754669) awarded to D.N.R., and a VIDI grant (864.14.008) from the Netherlands Organization for Scientific Research and HORIZON-RIA grant (HLTH-2021-STAYHLTH-01: 101057390) awarded to B.J.A.P.
Author contributions: Conceptualized and designed the study: D.S. and B.J.A.P. Produced and analyzed the morphological data: D.S. and M.A. Analyzed the genomic data: H.v.K. Analyzed the phylogenetic regression analyses: A.I.F. and D.S. Provided fish species for the study: D.N.R. Drafted the manuscript: D.S. and B.J.A.P. Contributed to content and editing of the final manuscript and figures: D.S., M.A., H.v.K., A.I.F., D.N.R., G.F.W., and B.J.A.P.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper, the Supplementary Materials, and/or at Zenodo (doi:10.5281/zenodo.7620367).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Table S1 to S5
REFERENCES AND NOTES
- 1.E. B. Rosenblum, C. E. Parent, E. E. Brandt, The molecular basis of phenotypic convergence. Annu. Rev. Ecol. Evol. Syst. 45, 203–226 (2014). [Google Scholar]
- 2.T. B. Sackton, N. Clark, Convergent evolution in the genomics era: New insights and directions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D. B. Wake, M. H. Wake, C. D. Specht, Homoplasy: From detecting pattern to determining process and mechanism of evolution. Science 331, 1032–1035 (2011). [DOI] [PubMed] [Google Scholar]
- 4.J. B. Losos, Convergence, adaptation, and constraint. Evolution 65, 1827–1840 (2011). [DOI] [PubMed] [Google Scholar]
- 5.D. Schulter, E. A. Clifford, M. Nemethy, J. S. McKinnin, Parallel evolution and inheritance of quantitative traits. Am. Nat. 163, 809–822 (2004). [DOI] [PubMed] [Google Scholar]
- 6.H. D. Rundle, L. Nagel, J. W. Boughman, D. Schluter, Natural selection and parallel speciation in sympatric sticklebacks. Science 287, 306–308 (2000). [DOI] [PubMed] [Google Scholar]
- 7.F. Duponchelle, E. Paradis, A. J. Ribbink, G. F. Turner, Parallel life history evolution in mouthbrooding cichlids from the African Great Lakes. Proc. Natl. Acad. Sci. U.S.A. 105, 15475–15480 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.S. Wroe, N. Milne, Convergence and remarkably consistent constraint in the evolution of carnivore skull shape. Evolution 61, 1251–1260 (2007). [DOI] [PubMed] [Google Scholar]
- 9.C. Y. Feigin, A. H. Newton, L. Doronina, J. Schmitz, C. A. Hipsley, K. J. Mitchell, G. Gower, B. Llamas, J. Soubrier, T. N. Heider, B. R. Menzies, A. Cooper, R. J. O. Neill, A. J. Pask, Genome of the Tasmanian tiger provides insights into the evolution and demography of an extinct marsupial carnivore. Nat. Ecol. Evol. 2, 182–192 (2018). [DOI] [PubMed] [Google Scholar]
- 10.J. S. Reidenberg, Anatomical Adaptations of Aquatic Mammals. Anat. Rec. 290, 507–513 (2007). [DOI] [PubMed] [Google Scholar]
- 11.J. G. M. Thewissen, L. N. Cooper, M. T. Clementz, S. Bajpai, B. N. Tiwari, Whales originated from aquatic artiodactyls in the Eocene epoch of India. Nature 450, 1190–1194 (2007). [DOI] [PubMed] [Google Scholar]
- 12.C. Formation, O. S. Illiger, F. P. Cope, H. Member, C. Formation, The earliest known fully quadrupedal sirenian. Nature 141, 625–627 (2001). [DOI] [PubMed] [Google Scholar]
- 13.A. B. Ward, R. S. Mehta, Axial elongation in fishes: Using morphological approaches to elucidate developmental mechanisms in studying body shape. Integr. Comp. Biol 50, 1106–1119 (2010). [DOI] [PubMed] [Google Scholar]
- 14.P. J. Bergmann, G. Morinaga, The convergent evolution of snake-like forms by divergent evolutionary pathways in squamate reptiles. Evolution 73, 481–496 (2019). [DOI] [PubMed] [Google Scholar]
- 15.J. Arendt, D. Reznick, Convergence and parallelism reconsidered: What have we learned about the genetics of adaptation? Trends Ecol. Evol. 23, 26–32 (2008). [DOI] [PubMed] [Google Scholar]
- 16.P. H. F. Lucinda, Family Poeciliidae ( Livebearers )–Check List of the Freshwater Fishes of South and Central America. R. E. Reis, S. O. Kullander, C. J. J. Ferraris, Eds., (EDIPURCS, 2003), pp. 555–581. [Google Scholar]
- 17.D. Safian, G. F. Wiegertjes, B. J. A. Pollux, The fish family poeciliidae as a model to study the evolution and diversification of regenerative capacity in vertebrates. Front. Ecol. Evol. 9, 613157 (2021). [Google Scholar]
- 18.B. J. A. Pollux, M. N. Pires, A. I. Banet, D. N. Reznick, evolution of placentas in the fish family Poeciliidae: An empirical study of macroevolution. Annu. Rev. Ecol. Evol. Syst. 40, 271–289 (2009). [Google Scholar]
- 19.B. J. A. Pollux, R. W. Meredith, M. S. Springer, T. Garland, D. N. Reznick, The evolution of the placenta drives a shift in sexual selection in livebearing fish. Nature 513, 233–236 (2014). [DOI] [PubMed] [Google Scholar]
- 20.D. N. Reznick, M. Mateos, M. S. Springer, Independent origins and rapid evolution of the placenta in the fish genus Poeciliopsis. Science 298, 1018–1020 (2002). [DOI] [PubMed] [Google Scholar]
- 21.C. L. Turner, Pseudoamnion, pseudochorion, and follicular pseudoplacenta in Poeciliid fishes. J. Morphol. 67, 59–89 (1940). [Google Scholar]
- 22.L. Kwan, M. Fris, F. H. Rodd, L. Rowe, L. Tuhela, T. M. Panhuis, An examination of the variation in maternal placentae across the genus Poeciliopsis (Poeciliidae). J. Morphol. 276, 707–720 (2015). [DOI] [PubMed] [Google Scholar]
- 23.C. Olivera-Tlahuel, N. A. Moreno-Mendoza, M. Villagrán-Santa Cruz, J. J. Zúñiga-Vega, Placental structures and their association with matrotrophy and superfetation in poeciliid fishes. Acta Zool. 100, 167–181 (2019). [Google Scholar]
- 24.J. L. Ponce de León, M. C. Uribe, Morphology of yolk and pericardial sacs in lecithotrophic and matrotrophic nutrition in poeciliid fishes. J. Morphol. 282, 887–899 (2021). [DOI] [PubMed] [Google Scholar]
- 25.B. D. Grove, J. P. Wourms, follicular placenta of the viviparous fish, Heterandria formosa: II. Ultrastructure and development of the follicular epithelium. J. Morphol. 220, 167–184 (1994). [DOI] [PubMed] [Google Scholar]
- 26.T. M. Panhuis, M. Fris, L. Tuhela, L. Kwan, An examination of surface epithelium structures of the embryo across the genus Poeciliopsis (Poeciliidae). J. Morphol. 278, 1726–1738 (2017). [DOI] [PubMed] [Google Scholar]
- 27.B. D. Grove, J. P. Wourms, The follicular placenta of the viviparous fish, Heterandria formosa. 1. Ultrastructure and development of the embryonic absorptive surface. J. Morphol. 284, 265–284 (1991). [DOI] [PubMed] [Google Scholar]
- 28.R. W. Meredith, M. N. Pires, D. N. Reznick, M. S. Springer, Molecular phylogenetic relationships and the evolution of the placenta in Poecilia (Micropoecilia) (Poeciliidae: Cyprinodontiformes). Mol. Phylogenet. Evol. 55, 631–639 (2010). [DOI] [PubMed] [Google Scholar]
- 29.A. I. Furness, B. J. A. Pollux, R. W. Meredith, M. S. Springer, D. N. Reznick, How conflict shapes evolution in poeciliid fishes. Nat. Commun. 10, 3335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. C. Trexler, D. L. DeAngelis, Resource allocation in offspring provisioning: An evaluation of the conditions favoring the evolution of matrotrophy. Am. Nat. 162, 574–585 (2003). [DOI] [PubMed] [Google Scholar]
- 31.A. I. Banet, D. N. Reznick, Do placental species abort offspring? Testing an assumption of the Trexler–DeAngelis model. Funct. Ecol. 22, 323–331 (2008). [Google Scholar]
- 32.A. I. Banet, A. G. Au, D. N. Reznick, Is mom in charge? Implications of resource provisioning on the evolution of the placenta. Evolution 64, 3172–3182 (2010). [DOI] [PubMed] [Google Scholar]
- 33.R. D. Bassar, S. K. Auer, D. N. Reznick, Why do placentas evolve? A test of the life-history facilitation hypothesis in two clades in the genus Poeciliopsis representing two independent origins of placentas. Funct. Ecol. 28, 999–1010 (2014). [Google Scholar]
- 34.B. J. A. Pollux, D. N. Reznick, Matrotrophy limits a female’s ability to adaptively adjust offspring size and fecundity in fluctuating environments. Funct. Ecol. 25, 747–756 (2011). [Google Scholar]
- 35.M. Fleuren, J. L. van Leeuwen, B. J. A. Pollux, Superfetation reduces the negative effects of pregnancy on the fast-start escape performance in live-bearing fish. Proc. R. Soc. B. Biol. Sci. 286, 20192245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. I. Furness, J. C. Avise, B. J. A. Pollux, Y. Reynoso, D. N. Reznick, The evolution of the placenta in poeciliid fishes. Curr. Biol. 31, 2004–2011.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 37.A. Hagmayer, A. I. Furness, D. N. Reznick, M. L. Dekker, B. J. A. Pollux, Predation risk shapes the degree of placentation in natural populations of live-bearing fish. Ecol. Lett. 23, 831–840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. Fleuren, E. M. Quicazan-Rubio, J. L. van Leeuwen, B. J. A. Pollux, Why do placentas evolve? Evidence for a morphological advantage during pregnancy in live-bearing fish. PLOS ONE 13, e0195976 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.H. Van Kruistum, J. Van Den Heuvel, J. Travis, K. Kraaijeveld, B. J. Zwaan, M. A. M. Groenen, H. J. Megens, B. J. A. Pollux, The genome of the live-bearing fish Heterandria formosa implicates a role of conserved vertebrate genes in the evolution of placental fish. BMC Evol. Biol. 19, 156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.H. Van Kruistum, M. W. Guernsey, J. C. Baker, S. L. Kloet, M. A. M. Groenen, B. J. A. Pollux, H. J. Megens, E. Teeling, The genomes of the livebearing fish species Poeciliopsis retropinna and Poeciliopsis turrubarensis reflect their different reproductive strategies. Mol. Biol. Evol. 37, 1376–1386 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. W. Guernsey, H. van Kruistum, D. N. Reznick, B. J. A. Pollux, J. C. Baker, Molecular signatures of placentation and secretion uncovered in Poeciliopsis maternal follicles. Mol. Evol. Biol. 37, 2679–2690 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H. Van Kruistum, R. Nijland, D. N. Reznick, M. A. M. Groenen, H. Megens, B. J. A. Pollux, Parallel genomic changes drive repeated evolution of placentas in live-bearing fish. 38, 2627–2638 (2021). [DOI] [PMC free article] [PubMed]
- 43.C. Ross, T. E. Boroviak, Origin and function of the yolk sac in primate embryogenesis. Nat. Commun. 11, 3760 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.B. F. King, J. M. Wilson, A fine structural and cytochemical study of the rhesus monkey yolk sac: Endoderm and mesothelium. Anat. Rec. 205, 143–158 (1983). [DOI] [PubMed] [Google Scholar]
- 45.J. R. Gallant, L. L. Traeger, J. D. Volkening, H. Moffett, P. H. Chen, C. D. Novina, G. N. Phillips, R. Anand, G. B. Wells, M. Pinch, R. Güth, G. A. Unguez, J. S. Albert, H. H. Zakon, M. P. Samanta, M. R. Sussman, Genomic basis for the convergent evolution of electric organs. Science 344, 1522–1525 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.M. S. Pankey, V. N. Minin, G. C. Imholte, M. A. Suchard, T. H. Oakley, Predictable transcriptome evolution in the convergent and complex bioluminescent organs of squid. PNAS 111, E7436–E4742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.E. B. Rosenblum, H. Römpler, T. Schöneberg, H. E. Hoekstra, Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc. Natl. Acad. Sci. U.S.A. 107, 2113–2117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. D. Foote, Y. Liu, G. W. C. Thomas, T. Vinař, J. Alföldi, J. Deng, S. Dugan, C. E. Van Elk, M. E. Hunter, V. Joshi, Z. Khan, C. Kovar, S. L. Lee, K. Lindblad-toh, A. Mancia, R. Nielsen, X. Qin, J. Qu, B. J. Raney, N. Vijay, J. B. W. Wolf, M. W. Hahn, D. M. Muzny, K. C. Worley, M. T. P. Gilbert, R. A. Gibbs, Convergent evolution of the genomes of marine mammals. Nat. Genet. 47, 272–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R. W. Meredith, M. N. Pires, D. N. Reznick, M. S. Springer, Molecular phylogenetic relationships and the coevolution of placentotrophy and superfetation in Poecilia (Poeciliidae: Cyprinodontiformes). Mol. Phylogenet. Evol. 59, 148–157 (2011). [DOI] [PubMed] [Google Scholar]
- 50.C. A. Schneider, W. S. Rasband, K. W. Eliceiri, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D. Bates, M. Mächler, B. M. Bolker, S. C. Walker, Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 52.D. Lüdecke, M. Ben-Shachar, I. Patil, P. Waggoner, D. Makowski, Performance: An R package for assessment, comparison and testing of statistical models. J. Open Source Softw. 6, 3139 (2021). [Google Scholar]
- 53.R. V. Lenth, emmeans: Estimated Marginal Means, aka Least-Squares Means. R package (2021); https://github.com/rvlenth/emmeans.
- 54.R. C. Team, R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2021); www.R-project.org/.
- 55.D. Orme, R. Feckleton, G. Thomas, T. Petzoldt, S. Fritz, N. Isaac, W. Pearse, caper: Comparative Analysis of Phylogenetics and Evolution in R version 1. (2018); https://CRAN.R-project.org/package=caper.
- 56.D. N. Reznick, A. I. Furness, R. W. Meredith, M. S. Springer, The origin and biogeographic diversification of fishes in the family Poeciliidae. PLOS ONE 12, e0172546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.E. Paradis, K. Schliep, Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 58.K. Katoh, K. Misawa, K. Kuma, T. Miyata, MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Z. Yang, PAML 4: Phylogenetic analysis by maximum likelihood. Molec. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 60.S. Whelan, N. Goldman, A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Molec. Biol. Evol. 18, 691–699 (2001). [DOI] [PubMed] [Google Scholar]
- 61.T. Sato, Y. Yamanishi, M. Kanehisa, H. Toh, The inference of protein-protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics 21, 3482–3489 (2005). [DOI] [PubMed] [Google Scholar]
- 62.A. Alexa, J. Rahnenfuhrer, topGO: Enrichment analysis for gene ontology. R package version 2.48.0 (2022).
- 63.S. Durinck, Y. Moreau, A. Kasprzyk, S. Davis, B. De Moor, A. Brazma, W. Huber, BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 (2005). [DOI] [PubMed] [Google Scholar]
- 64.S. Durinck, P. T. Spellman, E. Birney, W. Huber, Mapping identifiers for the integration of genomic datasets with the R/ Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Table S1 to S5