Abstract
The influenza virus genome is transcribed in the nuclei of infected cells but assembled into progeny virions in the cytoplasm. This is reflected in the cellular distribution of the virus nucleoprotein (NP), a protein which encapsidates genomic RNA to form ribonucleoprotein structures. At early times postinfection NP is found in the nucleus, but at later times it is found predominantly in the cytoplasm. NP contains several sequences proposed to act as nuclear localization signals (NLSs), and it is not clear how these are overridden to allow cytoplasmic accumulation of the protein. We find that NP binds tightly to filamentous actin in vitro and have identified a cluster of residues in NP essential for the interaction. Complexes containing RNA, NP, and actin could be formed, suggesting that viral ribonucleoproteins also bind actin. In cells, exogenously expressed NP when expressed at a high level partitioned to the cytoplasm, where it associated with F-actin stress fibers. In contrast, mutants unable to bind F-actin efficiently were imported into the nucleus even under conditions of high-level expression. Similarly, nuclear import of NLS-deficient NP molecules was restored by concomitant disruption of F-actin binding. We propose that the interaction of NP with F-actin causes the cytoplasmic retention of influenza virus ribonucleoproteins.
The influenza A virus genome consists of eight segments of negative-sense single-stranded RNA (vRNA) which are transcribed in infected cells to yield two types of positive-sense transcripts: capped and polyadenylated mRNAs and exact complements (cRNA) which serve as replicative intermediate RNAs for the production of further vRNAs (18). Four viral proteins are necessary and sufficient to carry out this process (16): the three subunits of an RNA-dependent RNA polymerase (PB1, PB2, and PA) and a 55-kDa single-stranded RNA-binding nucleoprotein (NP). Indeed, the v- and cRNA segments are always associated with these polypeptides to form ribonucleoprotein (RNP) structures, thought to comprise one copy of the trimeric polymerase and approximately one NP polypeptide per 20 bases of RNA (18).
Unusually for a virus with no DNA coding stage, influenza virus transcription occurs in the nuclei of infected cells (15). Thus, on initiation of infection the incoming RNPs are targeted to the nucleus, and during infection newly synthesized RNP proteins also undergo nuclear import. Consistent with this, nuclear localization signals (NLSs) have been identified in all four transcriptase proteins (9, 25, 26–28, 41, 41a), including three in NP, the major protein component of RNPs. At early times postinfection, the polymerase subunits and NP accumulate in the nucleus (reviewed in reference 18), and NP has been shown to be necessary for the nuclear import of vRNA in permeabilized cells (30). However, virion assembly, including packaging of progeny RNPs, occurs in the cytoplasm and at later times this is reflected in the cytoplasmic accumulation of the four transcriptase proteins, generally assumed to be in the form of RNPs (18). This differential localization of RNPs necessitates regulatory mechanisms, and evidence points to the involvement of at least three viral polypeptides: the virion matrix (M1) protein, the minor virion component NS2, and NP itself. M1 is capable of binding to membranes, RNA, NS2, and RNPs. In the virion, it is thought to act as the link between the lipid envelope and the packaged genome segments (44). Moreover, M1 itself can localize to the nucleus, where it is thought to promote the export of RNPs (22, 43). NS2, which binds to RNPs via the M1 protein (47), has recently been shown to possess a nuclear export signal (31). Microinjected anti-NS2 antibodies inhibit export of RNPs, and it has therefore been proposed that NS2 is the viral factor directly responsible for the export of RNPs from the nucleus (31). Accordingly, import of NP (and RNPs early in infection) can be postulated to occur because of the presence of NLSs and export can be postulated to occur because of the subsequent expression of components (M1 and NS2) that target RNPs to an export pathway (31, 44). However, this hypothesis does not fully explain why RNPs accumulate in the cytoplasm in the absence of M1 export (33, 42) or why a mutant influenza virus which synthesizes very little or no NS2 is fully replication competent (36, 45). Moreover, since vRNA synthesis is not temporally separated from RNP export, it is not clear how import of newly synthesized NP is maintained in the presence of active export of the same polypeptide in the form of RNP complexes, or why cytoplasmic RNPs are not simply reimported back into the nucleus by virtue of their polyvalent NLS sequences. Therefore, there must be other elements involved in the control of RNP localization.
NP possesses the ability to locate to both cytoplasm and nucleus. In the absence of other viral proteins, it shuttles between the two compartments (43), which suggests that it interacts with the cellular nuclear export apparatus in the absence of M1 and NS2. NP can also accumulate in the cytoplasm at “late” times postexpression (27), indicating that the activities of the NP NLSs are modulatable. Therefore, RNP localization may also be regulated by mechanisms acting through NP. One well-documented way of modulating nuclear import is through interactions with cytoplasmic anchoring proteins (29, 40), and recent work involving cell fractionation and fluorescent staining experiments has suggested that cytoplasmic NP is associated with the cytoskeleton (3, 17).
Here we demonstrate that purified NP shows high-affinity binding to filamentous actin (F-actin) in vitro with a Kd on the order of 1 μM and a stoichiometry of 1 per actin subunit. Additionally, there is a further interaction of lower affinity which may reflect oligomerization of NP itself. We also show the formation of complexes containing NP, RNA, and actin and have identified a cluster of amino acid residues in NP important for the interaction with F-actin. Localization of exogenously expressed NP was found to depend on the amount expressed, with small amounts entering the nucleus and large amounts accumulating in the cytoplasm, where the protein colocalizes with F-actin. Mutations in NP which decrease F-actin binding change the cellular distribution of the protein, favoring accumulation in the nuclei even when the expression level is high. We propose that F-actin binding acts as a cytoplasmic retention signal for NP.
MATERIALS AND METHODS
Plasmids, antisera, and viruses.
A cDNA copy of A/PR8/34 segment 5 was excised from plasmid pVB5+ (6) with EcoRI and ligated into similarly linearized plasmid pMAL-c2 (New England Biolabs). The NP and maltose binding protein (MBP) open reading frames (ORFs) were then fused by oligonucleotide-directed mutagenesis. The resulting plasmid (pMAL-NP) contains the lac promoter, the MBP gene, a factor Xa protease recognition site, an alanine codon, and the NP ORF (with an NcoI restriction enzyme site flanking the initiation codon). Point mutations were introduced into this plasmid by standard procedures. All mutations were confirmed by nucleotide sequencing of the appropriate region of the NP gene. To construct a glutathione S-transferase (GST)–NP fusion protein, plasmid pMAL-NP was digested with NcoI, end filled with the Klenow fragment of DNA polymerase, and digested with EcoRI. The DNA fragment containing the NP ORF was then ligated into SmaI-EcoRI-cut plasmid pGEX-2T (Pharmacia) to create plasmid pGEX-NP. To obtain eukaryotic expression of nonfused NP, NP genes were excised from the pMAL constructs by digestion with NcoI and XbaI and inserted into similarly digested plasmid pKT0 (39). The resulting plasmids (pKT5 series) contain the NP gene under control of a T7 RNA polymerase promoter.
Antisera against NP were raised by immunizing rabbits with a β-galactosidase fusion protein containing amino acids 340 to 498 of A/PR8/34 NP. Rabbit antiserum to RNP cores was the generous gift of S. Inglis.
Recombinant vaccinia viruses expressing the three influenza virus A/PR8/34 P proteins and NP (37) or bacteriophage T7 RNA polymerase (vTF-7 [11]) have previously been described.
Expression and purification of NP.
Exponentially growing cultures of Escherichia coli TG1 containing plasmid pMAL-NP, pGEX-NP, or pGEX-2T were induced by the addition of isopropyl-thiol-β-galactosidase and incubated with shaking for a further 4 h. Bacteria were pelleted by centrifugation and resuspended in a 1/5 culture volume of amylose column buffer (ACB) (500 mM NaCl, 20 mM Tris-Cl [pH 7.6], 1 mM EDTA, 0.02% sodium azide) (pMAL) or phosphate-buffered saline (PBS) (pGEX), containing 0.25 μg of lysozyme per ml. After incubation for 30 min on ice and overnight at −20°C, the suspension was thawed and sonicated before centrifugation at an average of 9,000 × g for 30 min. The supernatants were sequentially filtered through Whatman no. 1 filter paper and a 0.45-μm-pore-size cellulose acetate filter before chromatography over amylose resin (New England Biolabs) or glutathione Sepharose (Pharmacia) as appropriate. Columns were washed sequentially with 10 volumes of ACB or PBS, 10 volumes of buffer containing 2 M NaCl, and another 10 volumes of ACB or PBS. Bound protein was eluted with buffer supplemented with 10 mM maltose or 50 mM reduced glutathione.
The MBP moiety of MBP-NP was removed by the addition of 2 mM CaCl2 and 0.5% (wt/wt) factor Xa protease (New England Biolabs) in an overnight incubation at 14°C. All polypeptides displayed the expected mobilities on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (see Fig. 1) and reacted as predicted in Western blots using anti-MBP, -GST, and -NP sera (data not shown). Protein microsequencing confirmed the predicted N-terminal sequence for NP cleaved from MBP. Proteins were either dialyzed or gel filtered into 50 mM NaCl–10 mM HEPES (pH 8)–0.1 mM EDTA–10% glycerol (storage buffer) and kept at 4 or −70°C. Protein concentrations were determined by the Bradford method (7) or by absorbance at 280 nm with a molar extinction coefficient of 119,000 M−1 · cm−1 calculated according to the formula of Gill and von Hippel (13). All samples were clarified by centrifugation at an average of 390,000 × g for 15 min before use.
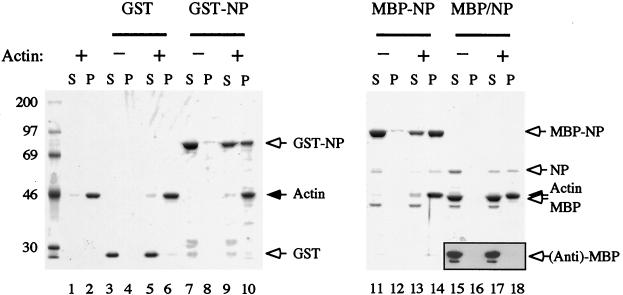
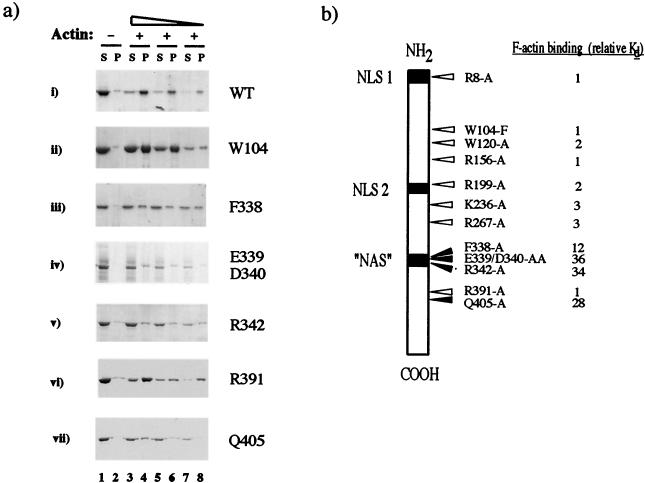
FIG. 1.
F-actin-binding activities of GST, GST-NP, and MBP-NP. The proteins (MBP/NP corresponds to MBP-NP digested with factor Xa protease) were centrifuged in the presence (+) or absence (−) of 3 μM F-actin. Supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE and staining with Coomassie blue or by Western blotting with anti-MBP serum (inset, lanes 15 to 18). Arrows indicate the named polypeptides (right), and molecular mass markers (in kilodaltons) are indicated on the left.
Actin-binding assays.
Purified rabbit muscle actin (38) was polymerized in 100 mM NaCl–10 mM Tris-Cl (pH 8.0)–1 mM MgCl2–0.1 mM ATP–0.2 mM EGTA–1 mM sodium azide (F buffer). Sedimentation assay mixtures contained 3 μM actin in 50 mM NaCl–10 mM HEPES (pH 7.6)–1 mM MgCl2–10% glycerol–0.1 mM EDTA–0.1 mM ATP in a final volume of 100 μl. After being mixed, the samples were centrifuged at an average of 200,000 × g for 20 min at 20°C before separation into pellet and supernatant fractions. Equivalent amounts of the two fractions were analyzed by SDS-PAGE and stained with Coomassie brilliant blue dye. Quantitation was performed by using a Molecular Dynamics computing densitometer (32). For electron microscopy, actin (1 μM) and MBP or MBP-NP were mixed in F buffer and left on ice for 20 min. Drops (30 μl) were applied to carbon-coated grids for 1 min and then grids were rinsed with 5 drops of F buffer and 5 drops of 1% uranyl acetate before being blotted dry and viewed in a Philips 208S electron microscope at a magnification of 20,000 or 30,000.
RNA transcription and binding assays.
For in vitro RNA binding assays, a radiolabelled 178-nucleotide synthetic RNA target was generated by in vitro transcription of plasmid pKT8-Δ3′5′ (generous gift of K. Tibbles) with bacteriophage T7 RNA polymerase in the presence of α-[32P]CTP (100 μM; specific activity, 2 Ci/mmol) according to standard procedures. The transcript corresponds to influenza virus A/PR8/34 segment 8 but with nucleotides 84 to 795 (inclusive) deleted and a C-to-A transversion of the penultimate base. Shorter transcripts corresponding to the minimal terminal repeat (“panhandle”) sequences or only the 3′ end of segment 8 were generated by in vitro transcription of the appropriate oligonucleotide templates cloned into plasmid pNEB193 (New England Biolabs). For in vivo RNA transcription assays, a synthetic influenza virus genome segment containing an antisense chloramphenicol acetyltransferase (CAT) gene was produced by in vitro transcription of plasmid pPB2CAT (generous gift of Mark Krystal). Filter binding assays were performed by incubating protein samples with 20 fmol of RNA (around 5,000 cpm) in 25 mM Tris-Cl (pH 7.6)–50 mM NaCl–5 mM MgCl2–0.5 mM dithiothreitol–5% glycerol at room temperature for 20 min. The reaction mixtures were passed through nitrocellulose filters equilibrated in 20 mM Tris-Cl (pH 7.6)–50 mM NaCl and washed three times with 200 μl of the same buffer. Bound radioactivity was quantified by liquid scintillation counting.
Transfection of tissue culture cells, transient replication assays, and indirect immunofluorescence.
BHK cells (in 35-mm-diameter dishes) were infected with recombinant vaccinia viruses at a multiplicity of infection of 5 of each virus per cell for 2 h at 37°C. The cells were washed three times with serum-free medium before transfection with 1 μg of plasmid DNA encoding NP and 0.5 μg of pPB2CAT9 in vitro-transcribed RNA and 10 μg of a cationic liposome mixture (Lipofectin [GIBCO-BRL] or Escort [Sigma-Aldrich]), according to the manufacturers’ instructions. The cells were incubated at 37°C for 20 h, washed three times with ice-cold PBS, and solubilized in 1 ml of CAT ELISA lysis buffer (Boehringer Mannheim). The lysate was clarified by centrifugation at 14,000 × g for 10 min at 4°C, and CAT expression was quantified relative to known standards by a commercial enzyme-linked immunosorbent assay (Boehringer Mannheim). Replication activities of mutants were calculated as percentages of the CAT synthesis seen with the wild-type (WT) gene after subtraction of the background obtained in the absence of NP. Values were calculated from a minimum of three separate experiments and two independent plasmid DNA preparations.
For immunofluorescence analysis, BHK or HeLa cells on coverslips were infected and transfected with plasmid DNA as described above. After 4 h of incubation at 37°C, the cells were washed three times with PBS containing 1% newborn calf serum and fixed in PBS containing 4% formaldehyde for 20 min at room temperature before being washed as before. The cells were permeabilized with 0.2% TX-100 in PBS for 5 min at room temperature, washed, and incubated for 1 h at room temperature with 200 μl of a 1-in-250 dilution of anti-RNP serum. After cells had been washed, bound immunoglobulin G was stained with swine anti-rabbit immunoglobulin G–fluorescein isothiocyanate conjugate (DAKO). Coverslips were applied in the presence of an antiphotobleaching compound (Citifluor). To preextract cells before fixation, cells were washed with 60 mM PIPES (pH 6.8)–20 mM HEPES–1 mM MgSO4–0.1 mM EGTA at 37°C, incubated with the same buffer supplemented with 0.1% TX-100 and 5 μM phallacidin for 2 min at room temperature, washed twice more, and then fixed and processed as described above. F-actin was detected by using 5 nM BODIPY-FL phallacidin (Molecular Probes), and β-actin was detected by using monoclonal antibody clone AC-74 (Sigma) at a dilution of 1:50. Fluorescence was viewed with a Leitz Orthoplan microscope or an MRC 1024 confocal microscope. Control experiments in which cells including untransfected cells were stained with individual fluorescent reagents confirmed that the labelling was channel specific (data not shown).
RESULTS
Binding of NP to F-actin.
We tested whether purified NP bound F-actin in vitro by a cosedimentation assay, in which F-actin readily separates from monomeric protein by centrifugation. As shown in Fig. 1, most of the actin sedimented after centrifugation (lane 2) and only the expected critical actin monomer concentration (0.1 to 0.2 μM) remained in the supernatant (lane 1). Control assays showed that only trace amounts of the expressed proteins pelleted in the absence of actin (Fig. 1, lanes 4, 8, and 12). However, when GST, GST-NP, or MBP-NP was sedimented after mixing with F-actin, the NP-containing polypeptides partitioned approximately evenly between the supernatant and pellet fractions (Fig. 1, lanes 9, 10, 13, 14) while GST did not (lanes 5 and 6), demonstrating association of NP with actin filaments. To test the ability of nonfused NP to bind F-actin, MBP-NP was cleaved into MBP and NP moieties by digestion with factor Xa protease and sedimented with or without F-actin. Since MBP and actin comigrate on SDS-PAGE, the samples were analyzed in parallel by Western blotting with an anti-MBP serum, confirming that MBP alone did not pellet whether or not actin was present (Fig. 1, inset, lanes 15 to 18). However, the NP polypeptide pelleted only in the presence of actin (Fig. 1, lanes 15 to 18). Thus, NP binds F-actin in vitro.
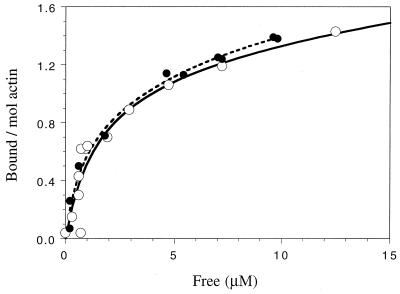
To quantitate the interaction between NP and F-actin, we titrated increasing concentrations of MBP-NP with a constant amount of F-actin in the sedimentation assay and determined the amount bound. Figure 2 shows the molar ratio of MBP-NP bound to F-actin as a function of the free MBP-NP. Saturation occurred in excess of two NP molecules per actin subunit, indicating more than one binding site for NP in the complex. The curve could best be described (based on nonlinear least-squares fitting) as resulting from a single high-affinity site per actin subunit (approximate Kd, 1 μM) and a lower-affinity site(s) having a Kd in excess of 40 μM. While we cannot exclude the possibility that a second NP binding site is present on F-actin, this weaker binding phase may be due to self-association with already bound NP.
FIG. 2.
Titration binding curves of WT and R156-A MBP-NP for F-actin. Sedimentation assays were performed by using 3 μM F-actin and WT (open circles) or mutant R156-A (closed circles) MBP-NP. The amount of bound MBP-NP (expressed as the molar ratio relative to actin) is plotted against the concentration of free MBP-NP.
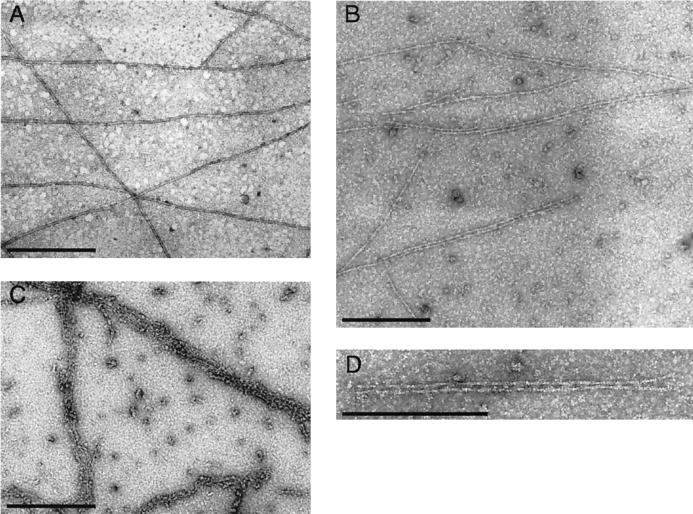
The binding of NP to actin filaments was also visualized directly by electron microscopy. Actin filaments incubated with an equimolar amount of MBP formed an essentially random distribution of fibers (Fig. 3A). However, the addition of MBP-NP to actin in an equimolar ratio induced a predominantly pairwise association of filaments (Fig. 3B), with a regular alternation of electron-dense and transparent regions between the filaments suggestive of cross-bridges (Fig. 3D). Addition of higher ratios of NP to actin induced the bundling of fibers into arrays containing multiple strands (Fig. 3C). The spacing between the cross-bridge arrangements at equimolar ratios of NP/actin was measured to be 332 ± 39.5 Å, coincident with every half-twist of the actin filament giving a ratio of 1 cross-bridge to every 13 actins. The formation of ordered bundles of fibers confirms that NP binds F-actin and is also consistent with the formation of NP-NP contacts.
FIG. 3.
Electron microscopy of actin filaments mixed with equimolar concentrations of MBP (A) and MBP-NP (B and D) and a threefold excess of MBP-NP (C). Scale bars, 300 nm.
RNA and the NP-actin interaction.
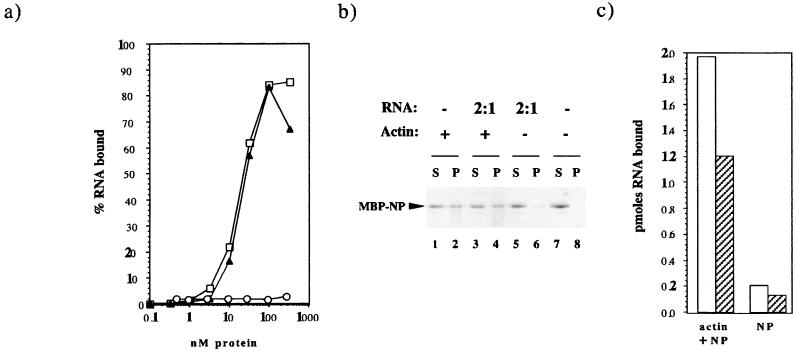
Since NP is a single-stranded-RNA-binding protein, we investigated the relationship between actin binding and RNA binding. First, we examined the effect of actin on the ability of NP to bind RNA in solution by titrating a radiolabelled 178-nucleotide RNA corresponding to the 5′ and 3′ noncoding sequences from vRNA segment 8 with increasing concentrations of MBP, MBP-NP, and MBP-NP plus 3 μM actin. Addition of MBP up to a concentration of 300 nM did not result in significant retention of the labelled probe, indicating that MBP itself does not bind RNA (Fig. 4a). However, titration with increasing amounts of MBP-NP resulted in the retention of 50% of the input RNA with around 20 nM protein (Fig. 4a). In replicate experiments, the affinity of MBP-NP for RNA was determined to be 16.3 ± 3.8 nM−1 (n = 5), which is in good agreement with that estimated for NP purified from influenza virus virions (20 nM−1 [4]). Therefore, the bacterially expressed NP behaved similarly to native NP and was fully competent to bind RNA. Moreover, addition of F-actin to the reaction mixtures had no significant effect on this RNA-binding activity (Fig. 4a).
FIG. 4.
RNA and the interaction between NP and F-actin. (a) Effect of actin on the RNA-binding activity of NP. The amounts of radiolabelled RNA (1 nM) bound by MBP (circles), MBP-NP (squares), and MBP-NP plus 3 μM F-actin (triangles) were determined by a filter assay. (b) Effect of RNA on actin binding by NP. MBP-NP (0.75 μM) was cosedimented in the presence (+) or absence (−) of 3 μM F-actin with the indicated molar ratio of a 34-mer RNA. S, supernatant; P, pellet. (c) Cosedimentation of RNA with actin and NP. Radiolabelled 34-mer (100 nM in 100 μl; open bars) or 54-mer (50 nM; shaded bars) was cosedimented in the presence of 3 μM MBP-NP with or without 3 μM F-actin.
Since NP binds to RNA with higher affinity than it does to actin (Kd ∼16 nM versus 1 μM [Fig. 2 and 3]), we examined the effect of a molar excess of RNA on actin binding by NP. We used a 34-mer transcript corresponding to the 3′ end of segment 8, which is long enough to bind efficiently to NP (47) but is not sufficiently large to bind multiple copies of NP and thus form RNP structures which would sediment irrespective of any association with actin fibers. In control reactions, this RNA did not cause a significant increase in the quantity of MBP-NP that pelleted in the absence of actin (Fig. 4b, lanes 5 to 8). However, the extent of sedimentation of MBP-NP with F-actin was unaffected by the presence of a twofold molar excess of RNA transcript relative to NP (Fig. 4b, compare lanes 1 and 2 with lanes 3 and 4).
To test whether NP was capable of binding both actin and RNA simultaneously, we assayed the sedimentation of radiolabelled RNAs (either the 3′ end [34-mer] or the 5′ and 3′ panhandle sequences [56-mer] of segment 8) with actin and MBP-NP. Mixtures of RNA, actin, and MBP-NP were subjected to centrifugation, and the pellet fractions were examined for their RNA content. Figure 4c shows that substantial quantities of both radiolabelled RNAs associated with the MBP-NP and the actin pellet (Fig. 4c). By contrast, there was no binding to actin alone (Fig. 4a) and minimal binding to MBP-NP alone (Fig. 4c), presumably reflecting the presence of oligomerized MBP-NP. Thus, NP, RNA, and F-actin can exist in termolecular complexes.
Mutational analysis of actin binding.
To further delineate the NP–F-actin interaction, we performed a mutational analysis of NP. On the grounds that an interaction with a relatively invariant protein such as actin might be reflected by sequence conservation within NP, we altered residues in regions conserved among the NPs of influenza A, B, C, and Dhori viruses (12) (summarized in Fig. 5b) and tested the effects of the mutations on actin-binding activity. Many of the mutations had no effect on the interaction. Quantitative analysis of the R156-A mutant produced a binding curve indistinguishable from that of the WT polypeptide (Fig. 2). Similarly, when tested at concentrations in the low micromolar range, the W104-F and R391-A mutants bound efficiently to F-actin (Fig. 5a, panels ii and vi, respectively; cf. panel i [WT]). The binding affinities for F-actin of these and the R8-A, W120-A, R199-A, K236-A, and R267-A mutants were found to vary from that of the WT protein by less than threefold (Fig. 5b). However, the F338-A, E339/D340-AA, R342-A, and Q405-A polypeptides showed markedly different behavior, with the majority of each protein remaining in the supernatant fractions at each concentration tested (Fig. 5a, panels iii, iv, v, and vii). Quantification produced estimated binding constants for F-actin some 10- to 35-fold lower than for the WT protein (Fig. 5b). Thus, four mutations that disrupted the ability of the protein to bind F-actin were identified in the C-terminal one-third of NP.
FIG. 5.
Identification of NP point mutations which disrupt actin binding. (a) WT or mutant MBP-NP polypeptides were cosedimented in the absence (−) or presence (+) of 3 μM F-actin, and supernatant (S) and pellet (P) fractions were examined by SDS-PAGE. (b) Summary of point mutations and cellular localization signals in NP. Arrows indicate the positions of amino acid changes introduced into NP. White arrows indicate mutations with no major effect on F-actin binding and black arrows indicate those that substantially reduced binding. Tabulated values are the fold decrease in binding affinity for F-actin relative to values for the WT protein. Also shown are the positions of the three postulated cellular localization signals that have been identified in NP.
Ability of actin-binding mutants to support viral transcription.
Although we had identified mutations that reduced the ability of NP to bind actin, it was necessary to examine whether the mutations affected other functional properties of NP. Many lines of evidence indicate that NP is essential for virus RNA synthesis (18). This has been confirmed in a recombinant system where NP and the three subunits of the influenza virus RNA polymerase are necessary and sufficient to drive transcription and replication of a synthetic influenza virus genome segment containing an antisense CAT (flu-CAT) reporter gene (16). We used an adaptation of this assay to apply the stringent test of whether the mutants defective in actin binding could support virus RNA synthesis.
The WT and mutant NP genes were subcloned into plasmid pKT0 (as described in Materials and Methods) to allow T7 RNA polymerase-driven expression of native NP. BHK cells were multiply infected with vaccinia viruses expressing the influenza virus RNA polymerase and bacteriophage T7 RNA polymerase (11, 37) and transfected with flu-CAT RNA and plasmids encoding WT or mutant NP, and the accumulated CAT polypeptide levels were measured 20 h later. Control experiments established that, as expected, the flu-CAT gene was transcribed and replicated to produce CAT polypeptide in the presence of the influenza virus RNA polymerase and NP and that NP was essential for this process (data not shown). The activities of the mutants were then quantified relative to that of the WT gene. The E339/D340-AA mutant failed to support the accumulation of CAT polypeptide (0.1% ± 0.3%). However, the Q405-A mutant supported CAT synthesis to a degree indistinguishable from that of the WT protein (98.2% ± 10.1%), while the F338-A and R342-A mutants possessed somewhat diminished (61.0% ± 20.3% and 23.9% ± 6.5%, respectively) activity. Hence, the last three mutants are capable of successfully participating in the multiple protein-protein and protein-RNA interactions necessary for the formation of a transcriptionally active influenza virus RNP core.
Actin and the intracellular localization of NP.
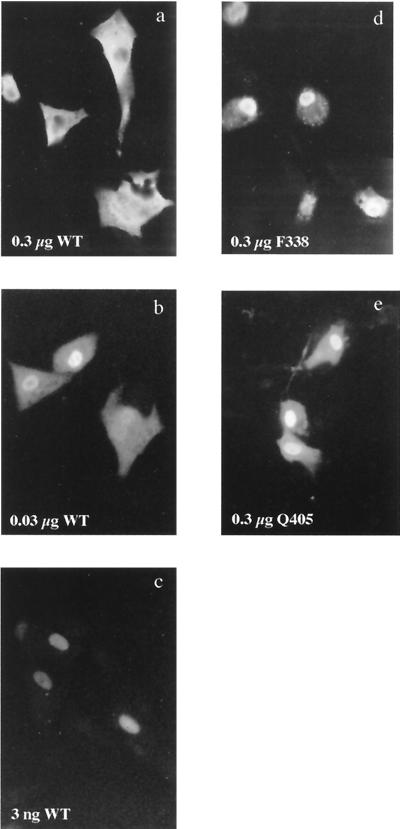
Since F-actin is cytoplasmic and NP can also accumulate in the cytoplasm, we examined the relevance of actin binding to the localization of NP. Because exogenously expressed NP shuttles between nucleus and cytoplasm and has been shown to accumulate in the cytoplasm under certain circumstances (27, 43), we first established the localization parameters of the WT polypeptide in our expression system by testing the effect of expression levels on the compartmentalization of NP. BHK cells were infected with a recombinant vaccinia virus expressing T7 RNA polymerase and transfected with various amounts of plasmid pKT5, and the subsequent expression and cellular localization of NP were examined by indirect immunofluorescence at 4 h posttransfection. Nontransfected cells revealed only a diffuse, low-level fluorescence (data not shown). In contrast, cells transfected with a plasmid containing the WT NP gene exhibited strong fluorescence, the location of which depended on the amount of plasmid transfected. In cells transfected with a high dose of plasmid (0.3 μg/105 cells), virtually all the fluorescence was cytoplasmic, often with apparent sparing of the nucleus (Fig. 6a). However, cells transfected with a low dose of plasmid (3 ng/106 cells) showed almost exclusively nuclear fluorescence (Fig. 6c). Cells transfected with an intermediate amount of plasmid (30 ng/105 cells) showed an intermediate pattern, with many cells containing substantial amounts of cytoplasmic NP but with nuclear accumulation as well (Fig. 6b). When lysates from cells transfected in parallel were subjected to SDS-PAGE and Western blot analysis to assess the quantity of NP expressed, the amount correlated with the plasmid dose (data not shown). The transfection efficiency varied by less than twofold (generally ranging between 20 and 40%; data not shown) over the range of plasmid concentrations tested. This indicates that the expression levels depended primarily on the amount of plasmid received per cell and not the number of cells transfected. Thus, the cellular distribution of NP depended on the amount (or rate) of synthesis of protein within any one cell.
FIG. 6.
Cellular localization of WT and mutant NP polypeptides. BHK cells (105) were infected with a recombinant vaccinia virus expressing T7 RNA polymerase, transfected with the indicated amounts of plasmids, and examined 4 h later by indirect immunofluorescence with anti-NP serum.
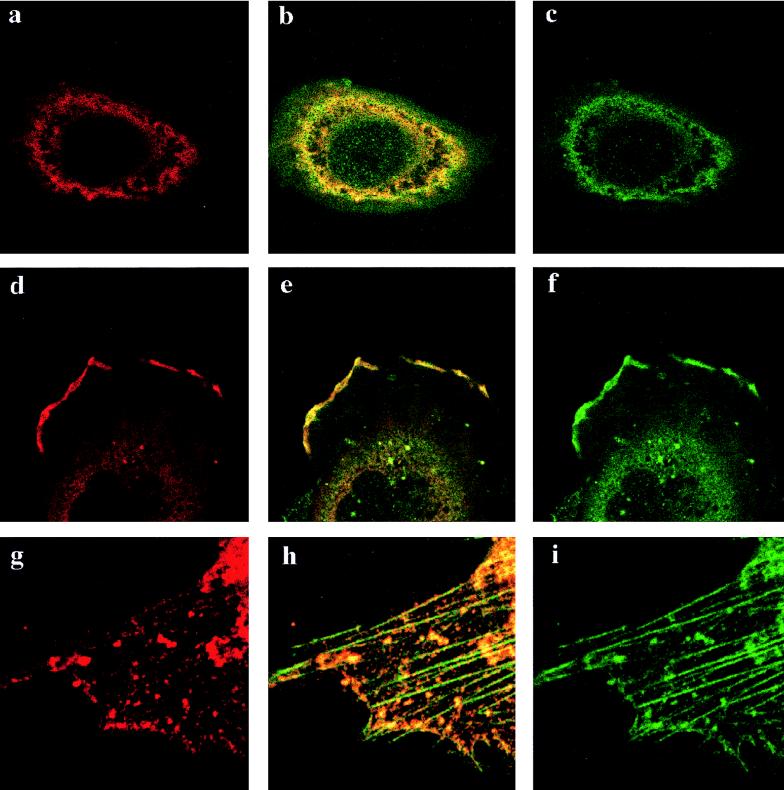
We next tested whether cytoplasmic NP associated with F-actin in vivo. The perinuclear actin network in cells was visualized by staining with anti-β-actin as an irregular ring structure that at most but not all points coincided with anti-NP fluorescence (Fig. 7a to c). Striking colocalization of NP and β-actin was also observed at the periphery of cells, particularly at the leading edges of lamellipodia (Fig. 7d to f). We also saw colocalization of NP with β-actin that had been incorporated into stress fibers (data not shown). To examine this further, we extracted cells with nonionic detergent before fixation, to remove soluble cytosolic proteins, and stained them with anti-NP and fluorescent phallacidin, a reagent that preferentially binds to actin in the form of stress fibers. Under these conditions, substantial quantities of NP remained cell associated, but in a granular pattern, often denser in the perinuclear area (data not shown). Moreover, many of the granules were arranged in linear arrays coincident with the phallacidin-stained F-actin stress fibers (Fig. 7g to i). In addition, colocalization between NP and actin was observed in a variety of cell types, including HeLa (Fig. 7a to f), BHK (Fig. 7g to i), and CV1 cells (data not shown). Therefore, exogenously expressed NP interacts with actin in vivo.
FIG. 7.
Colocalization of NP and F-actin. Cells were transfected with 0.3 μg of pKT5 and examined 4 h later by confocal laser microscopy after staining for NP (red) or actin (green). Panels b, e, and h are images resulting from the merger of the red and green channels. Panels a through f are from HeLa cells fixed before detergent extraction. Panels g through i are from a BHK cell extracted with detergent before fixation.
We next examined the intracellular localization of mutant NP molecules defective for actin binding. Significantly, even when cells were transfected with high doses of plasmids encoding these mutants, the fluorescence was predominantly nuclear (Fig. 6d and e). This increased nuclear accumulation was not simply the result of lower expression of the mutants, since the amounts of polypeptide detected by SDS-PAGE and Western blot analysis of parallel transfections were comparable with those of the WT polypeptide (relative densitometric scores of 1.1 ± 0.1 and 1.0 ± 0.2 for F338-A and Q405-A, respectively; data not shown). This suggests that the F338-A and Q405 polypeptides are more karyophilic than the WT protein.
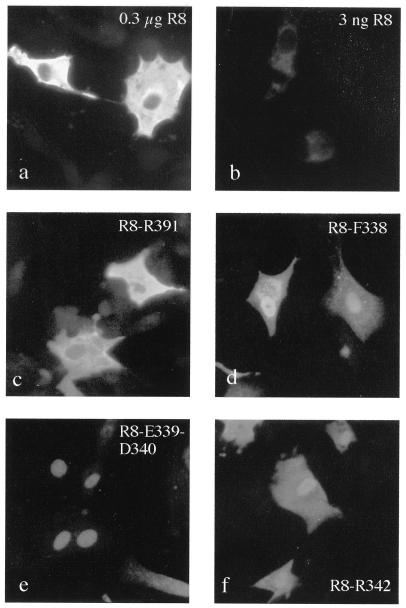
To examine further the effect of mutations affecting actin binding on the cellular localization of NP, we tested the effects of combining them with a mutation (R8-A [Fig. 5b]) that disrupted the N-terminal mammalian NLS (41). In BHK cells, the R8-A mutation rendered NP almost exclusively cytoplasmic at 4 h posttransfection regardless of the amount of plasmid transfected (Fig. 8a and b). In contrast to the situation for WT NP, even levels of R8-A expression that were barely detectable by immunofluorescence resulted in cytoplasmic staining and apparent sparing of the nucleus (Fig. 8b; cf Fig. 6c). This is consistent with the importance of the N-terminal NLS for nuclear import in mammalian cells (Fig. 5, NLS-1) (41). Combining the R8-A mutation with others that did not affect actin binding also resulted in a staining pattern where the nuclei emitted lower-intensity fluorescence than did the cytoplasm (Fig. 8c [R8-R391] and data not shown [R8-K236]). However, when either of the F338-A, E339/D340-AA, or R342-A mutations (which disrupt actin binding) were introduced into R8-A, the ability of the polypeptide to accumulate in the nuclei of cells was partially restored (Fig. 8d to f). Therefore, mutations which weaken the affinity of NP for actin can compensate for a mutation which disrupts a nuclear import signal. Thus, the NP-actin interaction causes cytoplasmic retention of NP.
FIG. 8.
Cellular location of NP mutants bearing amino acid substitutions in NLS I. Cells transfected with 0.3 μg (except for those shown in panel b, which were transfected with 3 ng) of plasmids encoding the indicated mutants were examined by indirect immunofluorescence with anti-NP serum.
DISCUSSION
Two recent studies suggested that influenza virus NP interacts with microfilaments of the host cell cytoskeleton late in infection (3, 17). Both reports found that NP partitioned into cytoskeleton-containing pools during biochemical fractionation of infected cells; that NP colocalized with microfilaments, especially at the cell periphery; and that disruption of microfilaments with cytochalasins altered the distribution of NP. Here, we confirm and extend these observations by using two methods to show that NP binds directly to F-actin in vitro (Fig. 1 to 3) with an affinity comparable to that of many cellular F-actin-binding proteins (35). We also show that NP associates with actin filaments in vivo in the absence of other influenza virus proteins (Fig. 7). Furthermore, complexes containing F-actin, NP, and RNA could be formed (Fig. 4). As the in vitro actin-binding assay employed high-speed centrifugation, we could not directly test whether authentic RNP particles bound F-actin because they sediment under these conditions, but we predict that RNP structures bind F-actin.
We have identified point mutations in NP which weaken its affinity for actin in vitro (Fig. 5) and decrease the extent of colocalization of the proteins in vivo (Fig. 6 and 8). These mutations were largely localized to a region of NP previously identified as important for the accumulation of the protein in the nuclei of Xenopus oocytes (9). The signal mapped by Davey and colleagues was one of the first NLSs proposed, but as subsequent NLSs are identified, it appears increasingly atypical. In sequence composition, it does not possess the small stretch of basic amino acids found in other viral proteins such as simian virus 40 large T antigen, or the bipartite basic NLS contained in many cellular proteins (10). In oocytes it acts as a sequence which causes the retention of NP that has diffused into the nucleus, rather than as a signal for the active uptake of the protein; hence it was called a “nuclear accumulation” signal (NAS) (9). Subsequently, the study of Wang et al. (41) found that this NAS was not essential for nuclear localization of NP in mammalian cells. Here we show that mutation of residues within the NAS actually increases the ability of NP to localize to the nuclei of mammalian cells (Fig. 6 and 8). The reason for the discrepancy between the activities of the NAS in mammalian and amphibian cells is not clear, but the fact that, unlike most somatic cells, the nuclei of amphibian oocytes have been shown to contain actin may be relevant (8, 34). Nevertheless, the region of NP defined as the NAS, which we find important for actin binding, is one of the most highly conserved blocks of amino acids between influenza A, B, C, and Dhori virus NPs (12), suggesting that it is functionally important.
The most plausible role for the NP-actin interaction is during the late stages of viral infection, when NP is also found in the cytoplasm. In this study, we show that the cellular localization of exogenously expressed NP depends on the amount of protein synthesized. Low levels of NP were targeted efficiently to the nucleus, but in a dramatic switch, large amounts localized almost exclusively to the cytoplasm (Fig. 6), where in part NP associated with actin (Fig. 7). These phenomena could perhaps be explained by saturation of the nuclear import machinery leading to cytoplasmic accumulation of NP and subsequent F-actin binding. However, the fact that all the mutations that weakened F-actin binding restored nuclear import, even under conditions of high-level expression (Fig. 6) or in the absence of a functional N-terminal NLS (Fig. 8), strongly implies a causative link between F-actin binding and cytoplasmic retention of NP. We propose that the interaction between NP and actin plays a role in regulating the localization of RNPs by causing their cytoplasmic retention late in infection. Previous work has shown that nuclear import of NP and RNPs is driven by NLSs in NP and that export of RNPs depends at least in part on the subsequent expression of M1 and NS2. Mechanistically, retention of RNPs on cytosolic actin filaments would act in opposition to the import pathway, but in concert with the export pathway, to prevent reimport of exported RNPs destined for packaging into progeny virions.
Cytosolic anchoring as a means of regulating nuclear localization is well established for several cellular proteins, such as the transcription factors of the NF-κB family, or Xenopus xnf7 (29, 40). In the case of NF-κB, retention is thought to result from masking of the NLS by the cytoplasmic anchor, since the addition of an extra NLS overrides the retention mechanism (5). In contrast, Xenopus xnf7 remains cytoplasmic even after addition of another NLS, leading to the proposal that cytoplasmic retention results from a physical interaction with a fixed anchor, possibly the cytoskeleton (20). Here we show that direct interaction with actin microfilaments mediates cytoplasmic retention of an NLS-containing protein. This represents a novel mechanism for controlling the cellular location of a ribonucleoprotein complex and may be applicable to a wider range of cellular proteins and RNAs.
It seems likely that the interaction between NP and actin is subject to regulation. The quantity of actin in the cell is such that the switch in NP localization between low- and high-dose transfections cannot be explained purely in terms of the protein’s affinity for actin. In addition, the nonuniform colocalization of NP with actin (Fig. 7) also suggests a regulated process. Since the microfilament network in cells consists of a polarized distribution of the various actin isoforms in complex with many different cellular actin-binding proteins (14), this could reflect a largely passive process, resulting from the occlusion of potential actin-binding sites and/or preferential binding to others. However, influenza virus may also have evolved positive mechanisms for regulating the NP-actin interaction, a likely candidate being phosphorylation of NP (2, 27). This is consistent with the effect of protein kinase inhibitors on NP localization noted by Neumann et al. (27), while phosphorylation is a well-documented mechanism for the control of the subcellular localization of cellular proteins, by modulation of both nuclear import (19, 21, 23) and actin binding (1, 24).
ACKNOWLEDGMENTS
We thank Mark Krystal for the gift of plasmid pPB2CAT, Geoff Smith for the gift of recombinant vaccinia viruses, Keff Tibbles for the gift of plasmids pKT8-Δ3′5′ and pKT0, and Sabine Gonsior for help with confocal microscopy. We also thank Ian Brierley, John McCauley, and Laurence Tiley for helpful criticism.
This work was supported by grants from the Royal Society, the Medical Research Council (grant G9232370), and the Wellcome Trust (grant 048911) to P.D. P.D. is a Royal Society University research fellow.
REFERENCES
- 1.Agnew B J, Minamide L S, Bamburg J R. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- 2.Almond J W, Felsenreich V. Phosphorylation of the nucleoprotein of an avian influenza virus. J Gen Virol. 1982;60:295–305. doi: 10.1099/0022-1317-60-2-295. [DOI] [PubMed] [Google Scholar]
- 3.Avalos R T, Yu Z, Nayak D P. Association of influenza virus NP and M1 proteins with cellular cytoskeletal elements in influenza virus-infected cells. J Virol. 1997;71:2947–2958. doi: 10.1128/jvi.71.4.2947-2958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baudin F, Bach C, Cusak S, Ruigrok R W H. Structure of influenza RNP I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beg A A, Ruben S M, Scheinmann R I, Haskil S, Rosen C A, Baldwin A S. IκB interacts with the nuclear localization sequences of the subunits of NFκB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 6.Blok V C. Studies of the influenza virus RNA polymerase. Ph.D. thesis. Cambridge, United Kingdom: University of Cambridge; 1988. [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Clark T G, Merriam R W. Diffusible and bound actin in nuclei of Xenopus laevis oocytes. Cell. 1977;12:883–891. doi: 10.1016/0092-8674(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 9.Davey J, Dimmock N J, Colman A. Identification of the sequence responsible for the nuclear accumulation of the influenza virus nucleoprotein in Xenopus oocytes. Cell. 1985;40:667–675. doi: 10.1016/0092-8674(85)90215-6. [DOI] [PubMed] [Google Scholar]
- 10.Dingwall C, Laskey R. Nuclear targetting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 11.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller F J, Freedman-Faulstich E Z, Barnes J A. Complete nucleotide sequence of the tick-borne, orthomyxo-like Dhori/Indian/1313/61 virus nucleoprotein gene. Virology. 1987;160:81–87. doi: 10.1016/0042-6822(87)90047-x. [DOI] [PubMed] [Google Scholar]
- 13.Gill S C, von Hippel P H. Calculation of protein extinction coefficients from amino-acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 14.Herman I M. Actin isoforms. Curr Opin Cell Biol. 1993;5:48–55. doi: 10.1016/s0955-0674(05)80007-9. [DOI] [PubMed] [Google Scholar]
- 15.Herz C, Stavnezer E, Krug R M. Influenza virus, an RNA virus, synthesises its messenger RNA in the nucleus of infected cells. Cell. 1981;26:391–400. doi: 10.1016/0092-8674(81)90208-7. [DOI] [PubMed] [Google Scholar]
- 16.Huang T-S, Palese P, Krystal M. Determination of influenza virus proteins required for genome replication. J Virol. 1990;64:5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husain M, Gupta C M. Interactions of viral matrix protein and nucleoprotein with the host cell cytoskeletal actin in influenza viral infection. Curr Sci. 1997;73:40–47. [Google Scholar]
- 18.Lamb R A, Krug R M. Orthomyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1353–1396. [Google Scholar]
- 19.Leukel M, Jost E. Two conserved serines in the nuclear localisation signal flanking region are involved in the nuclear targetting of human Iamin A. Eur J Cell Biol. 1995;68:133–142. [PubMed] [Google Scholar]
- 20.Li X, Shou W, Kloc M, Reddy B A, Etkin L D. Cytoplasmic retention of Xenopus nuclear factor 7 before the mid blastula transition uses a unique anchoring mechanism involving a retention domain and several phosphorylation sites. J Cell Biol. 1994;124:7–17. doi: 10.1083/jcb.124.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao W, Ou J H. Phosphorylation and nuclear localisation of the hepatitis B core protein: significance of serine in the three repeated SPRRR motifs. J Virol. 1995;69:1025–1029. doi: 10.1128/jvi.69.2.1025-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 23.Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SW15. Cell. 1991;68:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- 24.Morgan T E, Lockerbie R O, Minamide L S, Browning M D, Bamburg J R. Isolation and characterisation of a regulated form of actin depolymerising factor. J Cell Biol. 1993;122:623–633. doi: 10.1083/jcb.122.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukaigawa J, Nayak D P. Two signals mediate nuclear localization of influenza virus (A/WSN/33) polymerase basic protein 2. J Virol. 1991;65:245–253. doi: 10.1128/jvi.65.1.245-253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nath S T, Nayak D P. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1N1) Mol. Cell Biol. 1990;10:4139–4145. doi: 10.1128/mcb.10.8.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann G, Castrucci M R, Kawaoka Y. Nuclear import and export of influenza virus nucleoprotein. J Virol. 1997;71:9690–9700. doi: 10.1128/jvi.71.12.9690-9700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieto A, de la Luna S, Barcena J, Portela A, Ortin J. Complex structure of the nuclear translocation signal of influenza virus polymerase PA subunit. J Gen Virol. 1994;75:29–36. doi: 10.1099/0022-1317-75-1-29. [DOI] [PubMed] [Google Scholar]
- 29.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 30.O’Neill R E, Jaskunas R, Blobel G, Palese P, Moroianu J. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem. 1995;270:22701–22704. doi: 10.1074/jbc.270.39.22701. [DOI] [PubMed] [Google Scholar]
- 31.O’Neill R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pope B, Way M, Matsudaira P T, Weeds A. Characterization of the F-actin binding domains of villin: classification of F-actin binding-proteins into 2 groups according to their binding-sites on actin. FEBS Lett. 1994;338:58–62. doi: 10.1016/0014-5793(94)80116-9. [DOI] [PubMed] [Google Scholar]
- 33.Rey O, Nayak D P. Nuclear retention of M1 protein in a temperature-sensitive mutant of influenza (A/WSN/33) virus does not affect nuclear export of viral ribonucleoproteins. J Virol. 1992;66:5815–5824. doi: 10.1128/jvi.66.10.5815-5824.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheer U, Hinnsen H, Franke W W, Jockusch B M. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 1984;39:111–122. doi: 10.1016/0092-8674(84)90196-x. [DOI] [PubMed] [Google Scholar]
- 35.Sheterline P, Clayton J, Sparrow J. Actin. Protein Profile. 1995;2:1–103. [PubMed] [Google Scholar]
- 36.Smith D B, Inglis S C. Regulated production of an influenza virus spliced mRNA mediated by virus-specific products. EMBO J. 1985;4:2313–2319. doi: 10.1002/j.1460-2075.1985.tb03932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith G L, Levin J Z, Palese P, Moss B. Synthesis and cellular location of the ten influenza polypeptides individually expressed by recombinant vaccinia viruses. Virology. 1987;160:336–345. doi: 10.1016/0042-6822(87)90004-3. [DOI] [PubMed] [Google Scholar]
- 38.Taylor R S, Weeds A G. The magnesium-ion-dependent adenosine triphosphatase of bovine cardiac myosin and its subfragment-1. Biochem J. 1976;159:301–315. doi: 10.1042/bj1590301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tibbles K W, Brierley I, Cavanagh D, Brown T D K. A region of the coronavirus infectious bronchitis virus 1a polyprotein encoding the 3C-like protease domain is subject to rapid turnover when expressed in rabbit reticulocyte lysate. J Gen Virol. 1995;76:3059–3070. doi: 10.1099/0022-1317-76-12-3059. [DOI] [PubMed] [Google Scholar]
- 40.Vandromme M, Gauthier-Rouvière C, Lamb N, Fernandez A. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- 41.Wang P, Palese P, O’Neill R E. The NPI-1/NPI-3 (karyopherin α) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Weber F, Kochs G, Gruber S, Haller O. A classical bipartite nuclear localization signal on Thogoto and influenza A virus nucleoproteins. Virology. 1998;250:9–18. doi: 10.1006/viro.1998.9329. [DOI] [PubMed] [Google Scholar]
- 42.Whittaker G, Kemler I, Helenius A. Hyperphosphorylation of mutant influenza virus matrix protein, M1, causes its retention in the nucleus. J Virol. 1995;69:439–445. doi: 10.1128/jvi.69.1.439-445.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whittaker G, Bui M, Helenius A. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J Virol. 1996;70:2743–2756. doi: 10.1128/jvi.70.5.2743-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whittaker G, Bui M, Helenius A. The role of nuclear import and export in influenza virus infection. Trends Cell Biol. 1996;6:67–71. doi: 10.1016/0962-8924(96)81017-8. [DOI] [PubMed] [Google Scholar]
- 45.Wolstenholme A J, Barret T, Nichol S T, Mahy B W J. Influenza virus-specific RNA and protein syntheses in cells infected with temperature-sensitive mutants defective in the genome segment encoding nonstructural proteins. J Virol. 1980;35:1–7. doi: 10.1128/jvi.35.1.1-7.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamanaka K, Ishihama A, Nagata K. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J Biol Chem. 1990;265:11151–11155. [PubMed] [Google Scholar]
- 47.Yasuda J, Nakada S, Kato A, Toyoda T, Ishihama A. Molecular assembly of influenza virus: association of the NS2 protein with virion matrix. Virology. 1993;196:249–255. doi: 10.1006/viro.1993.1473. [DOI] [PubMed] [Google Scholar]