Abstract
Herpesvirus gene expression can be classified into four distinct kinetic stages: latent, immediate early, early, and late. Here we characterize the kinetic class of a group of 16 Kaposi’s sarcoma-associated herpesvirus (KSHV)/human herpesvirus 8 genes in a cultured primary effusion cell line and examine the expression of a subset of these genes in KS biopsies. Expression of two latent genes, LANA and vFLIP, was constitutive and was not induced by chemicals that induce the lytic cycle in primary effusion lymphoma (PEL) cell lines. An immediate-early gene, Rta (open reading frame 50 [ORF50]), was induced within 4 h of the addition of n-butyrate, and its 3.6-kb mRNA was resistant to inhibition by cycloheximide. Early genes, including K3 and K5 that are homologues of the “immediate-early” gene of bovine herpesvirus 4, K8 that is a positional homologue of Epstein-Barr virus BZLF1, vMIP II, vIL-6, and polyadenylated nuclear (PAN) RNA, appeared 8 to 13 h after chemical induction. A second group of early genes that were slightly delayed in their appearance included viral DHFR, thymidylate synthase, vMIP I, G protein-coupled receptor, K12, vBcl2, and a lytic transcript that overlapped LANA. The transcript of sVCA (ORF65), a late gene whose expression was abolished by Phosphonoacetic acid, an inhibitor of KSHV DNA replication, did not appear until 30 h after induction. Single-cell assays indicated that the induction of lytic cycle transcripts resulted from the recruitment of additional cells into the lytic cycle. In situ hybridization of KS biopsies showed that about 3% of spindle-shaped tumor cells expressed Rta, ORF K8, vIL-6, vMIP I, vBcl-2, PAN RNA, and sVCA. Our study shows that several KSHV-encoded homologues of cellular cytokines, chemokines, and antiapoptotic factors are expressed during the viral lytic cycle in PEL cell lines and in KS biopsies. The lytic cycle of KSHV, probably under the initial control of the KSHV/Rta gene, may directly contribute to tumor pathogenesis.
The newly identified herpesvirus, Kaposi’s sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8 (HHV8), is found in Kaposi’s sarcoma (KS) and in another AIDS-associated malignancy, primary effusion lymphoma (PEL) (8, 10). KSHV sequences have been detected in nearly all biopsies from HIV-related, classic, renal transplant-related, and endemic forms of KS, as well as in PEL. Moreover, the presence of antibodies to KSHV-encoded antigens strongly correlates with clinical KS (1, 12, 19, 20, 24, 25, 28, 34, 41). KSHV DNA and RNA are detected in tumor cells of KS lesions by in situ hybridization (5, 18, 27, 47, 57). All cell lines derived from PEL contain the KSHV genome, and these cells express both latent and lytic-cycle products of KSHV (2, 9, 33, 34, 42, 45).
At least three scenarios could account for the strong association of KSHV with these tumors. First, the virus may be the etiologic agent, perhaps by encoding classical immortalizing or transforming functions. Second, the virus may play an accessory role in pathogenesis by, for example, stimulating the production of cytokines which enhance tumor cell growth. Third, the virus may be a passenger that readily infects the two types of tumor cells associated with KSHV.
KS and PEL differ from each other in their association with the KSHV genome. While the genome is found in all PEL cell lines studied so far, KSHV sequences are rapidly lost during culture of primary KS tumors and are not detected in any long-term cell cultures established from KS lesions (11, 17, 37). This raises the possibility that KSHV is not required to maintain proliferation of spindle cells in vitro. Furthermore, the development of KS is associated with many cytokine abnormalities (13, 30, 48). High-level expression of interleukin-6 (IL-6), basic fibroblast growth factor (FGF), platelet-derived growth factor (PDGF), and their receptors is detected in KS lesions (6, 7, 14, 16, 31, 32, 36, 49, 55). Cell cultures composed of characteristic spindle-shaped tumor cells have been established from KS lesions by the addition of cytokines (37). The required components identified so far include tumor necrosis factor alpha (TNF-α) and TNF-β, gamma interferon (IFN-γ), IL-1, IL-6, granulocyte-macrophage colony-stimulating factor (GM-CSF), and oncostatin M (13, 30, 48). Inflammatory cytokines induce KS cells to produce bFGF and enhance KS-like lesion formation in nude mice (14, 15). These findings are consistent with the hypothesis that the proliferation of KS tumors is driven by cellular cytokines.
One observation that seems to unify the pathogenetic concepts of viral etiology and proliferation driven by cytokines is that KSHV itself encodes an array of homologues of proinflammatory cytokines such as IL-6 and macrophage inflammatory protein (MIP) (35, 38–40, 44). Moreover, the virus encodes homologues of antiapoptotic factors such as vBcl2 and vFLIP (44).
As a step in unraveling the role of KSHV in pathogenesis of KS and PEL, we characterized the kinetics of expression of a group of KSHV genes, including a number of cellular homologues, in PEL cell lines and in KS biopsies. Previous studies showed that in PEL cell lines and in KS biopsies a restricted region of the viral genome was expressed (33, 46, 47, 57). Two highly expressed RNAs were designated T0.7 and T1.1 (57). If the PEL cell lines were treated with tetradecanoyl phorbol acetate (TPA) or n-butyrate, agents that are known to induce lytic cycle expression of other gammaherpesviruses, there was more extensive transcription of the viral genome (33, 34, 42). T0.7, a transcript expressed in the majority of cells in KS biopsies, was considered to be a latent product, whereas T1.1 (also known as polyadenylated nuclear [PAN] RNA or nut-1 RNA) was expressed in only a few cells in KS biopsies (51, 47, 57). Studies in PEL cell lines showed that PAN or T1.1 RNA was strongly induced by chemical treatments that activated the lytic cycle (51, 56). Therefore, PAN RNA was considered to be a lytic-cycle product. Expression of several other genes, including homologues of bovine immediate-early genes, viral chemokines, and viral cytokines were enhanced when PEL cell lines were treated with TPA or n-butyrate (35, 39, 40, 46). Thus, PEL cells provide an experimental system in which to analyze the kinetic class of KSHV genes and to study the control of the lytic cascade. Our experiments extend these observations by identifying a KSHV immediate-early gene and by demonstrating that key viral cytokines such as vIL-6 and vMIP I, as well as vBcl2 and vGPCR, which are expressed in the early lytic phase of the viral life cycle in PEL cell lines behave similarly in KS biopsies. These observations point to the possibility that activation of the lytic cycle accompanied by expression of virus-encoded cytokines may be involved in KSHV-driven proliferation of neighboring cells.
MATERIALS AND METHODS
Cell culture.
PEL cell lines containing KSHV DNA, BC-1, or BCBL-1 (9, 42) were grown in RPMI 1640 supplemented with 15% fetal bovine serum at 37°C in the presence of 5% CO2. The HH514-16 cell line is an Epstein-Barr virus (EBV)-infected derivative of P3J-HR1(HR-1) that was used as a negative control for the expression of KSHV genes. Cells that were chemically induced into the lytic cycle were harvested at intervals after exposure to 3 mM sodium butyrate or 20 ng of TPA (phorbol-12-myristate-13-acetate [PMA]) per ml.
Cloning of KSHV genomic DNA and DNA sequence analysis.
A cosmid library was constructed in the Supercos-1 vector (Stratagene) with total genomic DNA from BC-1 cells. Genomic walking on KSHV DNA was initiated with the KS Bam330 and KS Bam631 fragments (10). DNA probes labeled by the random-primed method or RNA probes labeled during in vitro transcription were used for colony hybridization screening. The sequences of viral genes were determined in both directions via primer walking. DNA sequence data were compiled and analyzed with GELASSEMBLE, BLAST, and FRAMES of the Wisconsin Sequence Analysis Package, version 8 (Genetics Computer Group, Madison, Wis.). The sequence data of four contiguous sequences containing 13 genes analyzed here were deposited on Sept. 19, 1996, under the GenBank accession numbers U71365, U71366, U71367, and U71368. This sequence data was used to identify viral genes and to prepare probes for KSHV transcripts. The sequence data for the three viral genes LANA, vFLIP, and K12 was obtained from published sources (44, 57).
RNA preparation and Northern blot analysis.
Total cellular RNAs were prepared with the RNeasy kit (Qiagen), fractionated on 1% formaldehyde agarose gels, and transferred to nylon membranes (Nytran; Schleicher & Schuell) by standard procedures. Two types of probes were used on Northern blots: (i) single-stranded antisense oligonucleotides beginning about 50 nucleotides downstream of the 5′ ends of each open reading frame (ORF) and (ii) double-stranded DNA probes prepared by PCR and containing portions of PAN RNA (51), ORF65 (28), and ORF50 (52). PCRs contained 20 ng of each primer. PCR was performed for 30 cycles at 94°C for 30 s, 55°C for 30 s, and 74°C for 60 s. The sequences of the oligonucleotides and PCR primers are available upon request. Hybridization was carried out overnight in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt’s–1% sodium dodecyl sulfate (SDS)–100 μg of ssDNA per ml at 50°C for oligonucleotides and 65°C for double-stranded probes. Filters were washed with 6× SSC–0.1% SDS twice for 15 min each and in 1× SSC–0.1% SDS once for 30 min at 45°C for oligonucleotide probes and at 65°C for double-stranded DNA probes. For quantitation of RNA loading, blots were reprobed with the gene for H1 RNA of RNase P (3); the band intensity was quantitated with a PhosphorImager (Molecular Dynamics).
In situ hybridization of PAN RNA in PEL cells.
In situ hybridization procedures for detection of RNA were modified from published methods (22, 23). Cells were pelleted and resuspended in phosphate-buffered saline (PBS) to a density of 3 × 106 cells/ml. Then, 10 μl of cell suspension was placed onto fluorescent antibody slides (Bellco). Air-dried cells were fixed with freshly made 4% paraformaldehyde in PBS (pH 7.4) for 15 min at room temperature. Biotin-11-dUTP labeled probes were prepared by nick translation of 2 μg of pBKCMV (Stratagene) or pBKCMV-PAN plasmid DNA (51). Products with an average length of 200 bp were purified through G-50 Sephadex. Next, 10 μg each of Escherichia coli tRNA and denatured salmon sperm DNA were added to 150 ng of nick-translated biotin-11-dUTP labeled probe and then dried in a Speed Vac. The dried pellet was resuspended in 10 μl of formamide, denatured by heating for 10 min at 70°C, and immediately chilled on ice. The probe was resuspended in 20 μl of hybridization mixture containing 10% dextran, 0.2% bovine serum albumin, 2× SSC, and 0.2 mM vanadyl adenosine. The hybridization mixture was layered onto slides which were covered with a coverslip, sealed with mineral oil, and incubated in a humid chamber at 37°C for 3 to 18 h. Slides were washed at 37°C for 30 min each in 50% formamide in 2× SSC, 2× SSC, and 1× SSC in succession. The hybridization signal was detected by incubation for 1 h at 37°C with fluorescein isothiocyanate (FITC)-conjugated avidin DN (Vector Laboratories) at 2 μg/ml in 4× SSC containing 1% bovine serum albumin. Slides were rinsed at room temperature for 10 min each in 4× SSC, 4× SSC with 0.1% Triton X-100, and again with 4× SSC. Cells were examined in antifade medium containing propidium iodide.
Immunofluorescent detection of sVCA in PEL cells.
PEL cells were fixed in acetone-methanol and incubated for 1 h at 37°C with rabbit antiserum to sVCA or, as a control, preimmune rabbit serum (28). The primary antibody was removed by two washes with PBS; the slides were incubated with FITC-conjugated goat antibodies against rabbit immunoglobulin G (IgG). The primary antibody was used at a dilution of 1:10; the secondary antibody was used at a dilution of 1:40.
In situ hybridization of a KS lesion.
Viral genes were amplified by PCR and cloned in both orientations into the pcDNA3.1 expression vector (Invitrogen) between the BamHI and EcoRI sites adjacent to the T7 promoter. Plasmid DNA was linearized at the distal end of the viral DNA insert and transcribed with T7 RNA polymerase in the presence of [35S]UTP to yield RNA probes with a specific activity of ∼2.4 × 109 dpm/μg. In situ hybridization was performed as previously described (47). Briefly, 6-μm thin sections of formalin-fixed paraffin-embedded tissue were deparaffinized, rehydrated, and pretreated with an antigen retrieval protocol by using microwave irradation. After acetylation, the specimens were hybridized for 16 to 18 h at 45°C with a hybridization mixture containing 105 cpm riboprobe per μl. The slides were washed, dehydrated, coated with Kodak NTB-2 photographic emulsion, exposed for variable periods at 4°C, developed, and stained with hematoxylin and eosin.
Double-label in situ hybridization and immunohistochemistry.
Double labeling was performed as previously described (47). Briefly, tissue sections were deparaffinized, pretreated, and hybridized as described above with 35S-labeled riboprobe. After the posthybridization wash, the slides were blocked for immunochemistry with 5% (wt/vol) nonfat dry milk in PBS, reacted for 16 to 18 h at 4°C with a 1:50 dilution of monoclonal antibody to human CD34 (QB-END/10; Vector Laboratories), and developed with a peroxidase-conjugated secondary antibody and 3,3′-diaminobenzidine. Slides were dehydrated, coated with Kodak NTB-2 photographic emulsion, exposed for variable periods at 4°C, developed, and counterstained briefly with hematoxylin. For colocalization of two different viral transcripts, a hybridization mixture containing both 35S-labeled and digoxigenin-labeled riboprobes was applied to pretreated slides. After hybridization and posthybridization washes, the slides were processed to detect the digoxigenin-labeled probe, dehydrated through graded alcohols, and then coated with photographic emulsion.
RESULTS
Experimental design.
We examined the expression of 16 KSHV-encoded genes in a PEL cell line, BC-1, in which the KSHV/HHV8 lytic cycle can be induced by treatment with sodium butyrate (33). Although this cell line is also infected with EBV, butyrate treatment does not alter the abundance of EBV mRNA, proteins, or viral DNA (reference 33 and data not shown). Total cellular RNA was harvested from untreated cells and from chemically induced cells at intervals from 4 to 48 h after the addition of n-butyrate. Gene expression was monitored by hybridizing Northern blots with probes specific for different ORFs of KSHV. RNA loading was standardized by measuring the amount of H1 RNA of RNase P, an abundant stable small RNA involved in tRNA processing. The abundance of the mRNAs was quantitated by phosphorimaging and standardized for the level of H1 RNA.
A latent transcript was defined as a constitutively expressed mRNA. A lytic-cycle transcript markedly increased in abundance after the addition of the chemical inducing stimulus. Cell-by-cell assays, including in situ hybridization and immunofluorescence on untreated and chemically induced cell populations, were employed to further distinguish latent from lytic-cycle products. Latent genes were expressed in the majority of cells before the addition of an inducing stimulus. Lytic-cycle genes were expressed in a subpopulation of cells that increased in number after the addition of an inducing stimulus. A lytic-cycle gene was categorized as immediate early if the abundance of the mRNA after chemical induction was resistant to the action of cycloheximide, an inhibitor of protein synthesis. Early genes were defined as lytic-cycle transcripts whose expression was abolished by cycloheximide but was not blocked by phosphonoacetic acid (PAA), an inhibitor of herpesvirus-encoded DNA polymerase (43, 50). Late lytic-cycle transcripts were inhibited by PAA. Figure 1 shows representative latent, immediate-early, and early KSHV genes.
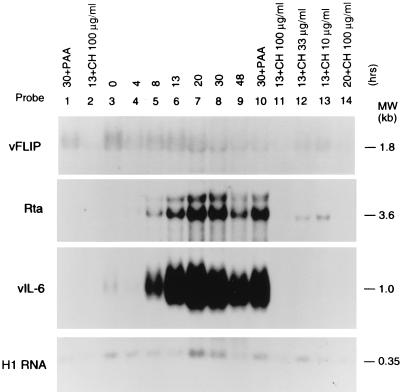
FIG. 1.
Examples of KSHV latent, immediate-early, and early genes. The BC-1 cell samples in lanes 1 to 3 were not exposed to a chemical agent that induces the KSHV lytic cycle. The samples in lanes 4 to 14 were treated with 3 mM n-butyrate for the time periods indicated (in hours) at the top of each lane. The cell samples in lanes 1 and 10 were treated with 0.5 mM PAA for 30 h to inhibit viral DNA synthesis. The samples in lanes 2 and 11 to 14 received cycloheximide in the indicated dose for 13 or 20 h to inhibit protein synthesis. The same total cellular RNA samples were hybridized with probes specific for vFLIP, a latent product; for Rta, an immediate-early product; and vIL-6, an early product. To control for RNA loading, the Northern blots were probed for cellular H1 RNA, a component of RNase P (3).
Latent gene expression.
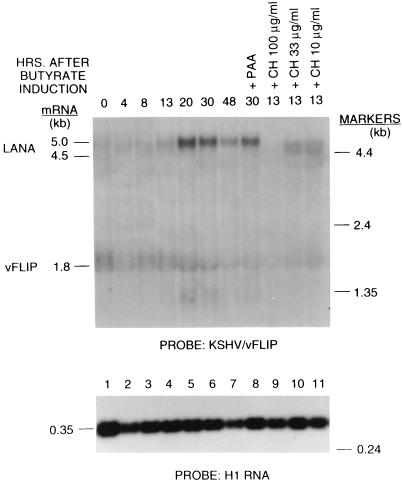
The KSHV gene (ORF71) encoding a homologue of the antiapoptotic factor vFLIP was an example of a latent gene (Fig. 1 and 2) (54). Its 1.8-kb transcript was present in untreated cells, was not increased by chemical induction, and remained at approximately the same level for at least 30 h after chemical induction. This mRNA was resistant to inhibition by cycloheximide. Another latency gene was present in ORF73, encoding the latent nuclear antigen (LANA) (25, 41). Rainbow et al. and Kedes et al. have shown by immunostaining that LANA is expressed in nearly all cells. The LANA transcript of 4.5 kb was present in untreated cells and was also cycloheximide resistant. The transcripts for LANA and vFLIP overlapped. Figure 2 shows that a probe for vFLIP detected both the 4.5-kb LANA mRNA and the 1.8-kb vFLIP transcript. However, a probe for LANA did not detect vFLIP (data not shown). The probes for LANA and vFLIP also detected a 5.0-kb early lytic cycle transcript which appeared between 13 and 20 h after chemical induction. The probe for vFLIP also detected a 1.4-kb lytic-cycle transcript. Thus, the locus of the KSHV genome containing the LANA and vFLIP genes is transcribed both during latency and during the lytic cycle.
FIG. 2.
Expression of two KSHV latency genes, latent nuclear antigen (LANA) and vFLIP. The conditions for butyrate induction and treatment with PAA and cycloheximide are as described in the legend to Fig. 1. The probe was an oligonucleotide in the vFLIP ORF.
Immediate-early gene expression.
We recently described an immediate-early gene, namely, the KSHV homologue of the EBV BRLF1 gene that encodes Rta (R transcriptional activator) which functions in the earliest phases of reactivation from latency into the lytic cycle (52). The KSHV homologue of Rta, encoded mainly in ORF50, has an analogous function. Transfection of KSHV/Rta drives early and late KSHV gene expression (52). Treatment of BC-1 cells with n-butyrate was followed within 4 h by expression of three transcripts, which were 4.0, 3.6, and 3.2 kb in size, that were detected with a probe for ORF 50 (Fig. 1). Of these three transcripts the 3.6-kb mRNA was most abundant; the appearance of the 3.2-kb transcript was faint and variable. Although the 3.6-kb ORF50 transcript was only faintly detectable on the autoradiograph at 4 h after n-butyrate treatment, phosphorimaging showed that it was induced 3- to 7.5-fold above background levels; at 4 h several abundant early-cycle transcripts, including those for vIL6 and PAN RNA, were not detectable. The 3.6-kb KSHV/Rta mRNA reached its maximal abundance (from 21- to 70-fold above background in three experiments) at 20 h after chemical treatment.
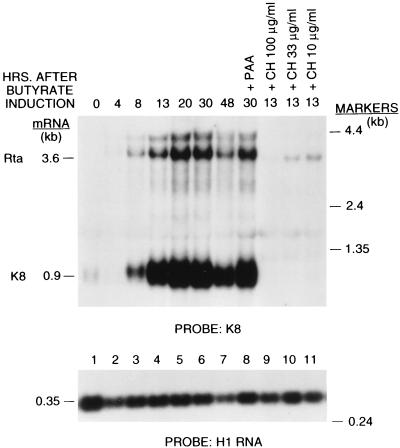
The 3.6-kb KSHV/Rta mRNA was partially resistant to inhibition by cycloheximide. In three replicate experiments, the level of ORF50 mRNA in the presence of 100 μg of cycloheximide per ml was 9 to 32% of the level in the absence of cycloheximide treatment. When 33 μg of cycloheximide per ml was used, 22 to 60% of the KSHV/Rta mRNA remained after 13 h. By contrast, only 2 to 6% of the level of three early lytic cycle transcripts, i.e., K8, vIL6, and PAN RNA, remained after cyclohexamide treatment (a representative experiment is shown in Table 1). The relative resistance of KSHV/Rta to inhibitors of protein synthesis classified it as an immediate-early gene (53). In previous related work using a probe from ORF50, we showed that the 3.6-kb mRNA was resistant to cycloheximide (52). In the current work we show that the same mRNA is detected with a probe for K8, the downstream gene in a bicistronic transcript and still appears to be cycloheximide resistant (Fig. 3).
TABLE 1.
Effect of cycloheximide treatment on the abundance of KSHV lytic-cycle mRNAs
| Cycloheximide dosea (μg/ml) | % of KSHV gene:
|
|||
|---|---|---|---|---|
| Rta | K8 | vIL-6 | PAN | |
| 0 | 100 | 100 | 100 | 100 |
| 10 | 28 | 4 | 4 | 3 |
| 33 | 23 | 3 | 3 | 2 |
| 100 | 24 | 6 | 4 | 3 |
Cycloheximide included from 0 to 13 h during n-butyrate treatment.
FIG. 3.
Comparison of the expression of KSHV/Rta and KSHV/K8. The single probe for this Northern blot was derived from the K8 ORF.
Other candidate immediate-early genes.
Besides KSHV/Rta, the KSHV genome contains at least three additional genes, K8, K3, and K5, that could represent immediate-early genes. KSHV ORF K8 is an analogue of EBV BZLF1, the immediate-early gene that encodes ZEBRA. Its location on the genome relative to Rta is similar in KSHV and EBV. K8 is weakly homologous to exons I and II of ZEBRA. The KSHV Rta and K8 transcription units overlap in a manner similar to EBV BRLF1 and BZLF1 (29). A K8 probe detects the Rta mRNAs and the prominent K8 mRNA of 0.9 kb. The KSHV/Rta probe, however, does not detect K8 transcripts (52). When the probe for K8 is used, the kinetics of expression of Rta and K8 can be compared on the same Northern blot (Fig. 3). K8 expression was delayed relative to expression of KSHV/Rta. At 4 h after chemical treatment, KSHV/Rta was induced sevenfold, while K8 was unaffected. At 8 h postinduction, KSHV/Rta was stimulated 18-fold and K8 was stimulated 4.6-fold. Unlike KSHV/Rta, K8 expression was abolished by cyclohexamide, even though the K8 transcript was more abundant than that of KSHV/Rta (Fig. 3). Its kinetics of expression and its resistance to PAA classify K8 as an early gene. K8 expression was induced by transfection of KSHV/Rta (52). Therefore, K8 is downstream in the lytic cycle from KSHV/Rta.
The kinetics of expression of two KSHV genes, K3 and K5, that are homologues of immediate-early gene 1 of BHV4 mirrored that of K8. The transcripts were first detectable 8 h after butyrate treatment and reached a maximum 20 h after induction. These transcripts, while resistant to PAA, were completely inhibited by cycloheximide; therefore, like K8, K3 and K5 are early genes.
Early lytic cycle gene expression.
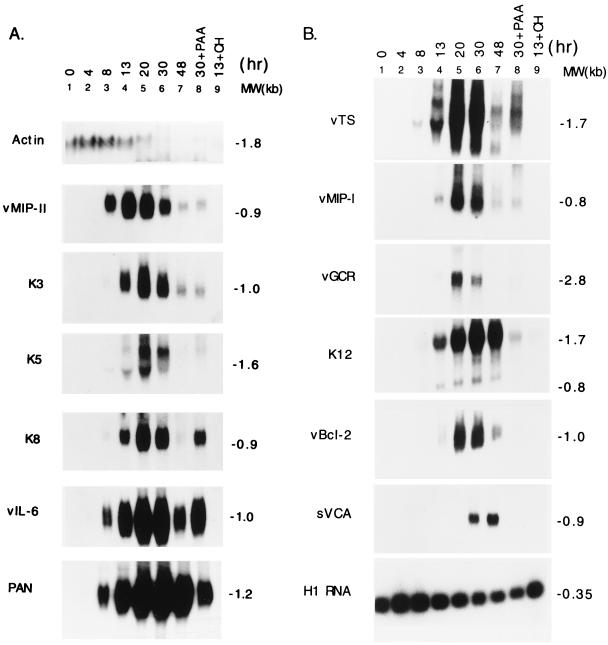
Figure 4 illustrates 12 genes that were classified as early on the basis of their resistance to PAA and their sensitivity to cycloheximide. In addition to K8, K3, and K5, the group of early genes included classical components of herpesvirus early lytic cycle gene expression, such as enzymes involved in nucleotide biosynthesis, thymidylate synthase (vTS), and viral dihydrofolate reductase (vDHFR) (data not shown). Surprisingly, however, several chemokines, proinflammatory cytokines, and putative cytokine receptors also behaved as early lytic cycle genes. These included vIL-6 (Fig. 1 and Fig. 4A), vMIP I and vMIPII (Fig. 4), and the G-protein coupled receptor (vGCR). This group of early genes differed somewhat in their kinetics. Those illustrated in Fig. 4A, including vMIP II and vIL-6, appeared earlier and reached a peak earlier than those shown in Fig. 4B, including vMIPI and vGCR.
FIG. 4.
Kinetics of early lytic gene expression of KSHV in the BC-1 PEL cell line. BC-1 cells were treated with 3 mM n-butyrate for the time indicated before total RNA was extracted. In lane 8, an RNA sample was prepared from cells that were treated with 0.5 mM PAA and 3 mM n-butyrate from time zero to 30 h. In lane 9, an RNA sample was prepared from cells that were treated with 3 mM n-butyrate and 100 μg of cycloheximide per ml from time zero to 13 h. The same RNA preparation was used for the hybridization with different probes, as indicated on the left of the blots. (A) Expression of the cellular β-actin gene and the viral early lytic genes, vMIP II gene, ORF K3, ORF K5, ORF K8, vIL-6, and PAN RNA. (B) Expression of cellular H1 RNA and viral delayed early and late lytic genes, viral thymidylate synthase (vTS), vMIP I, G-protein coupled receptor (vGCR), ORF K12, vBcl2, and sVCA gene.
T0.7 has previously been classified as a latency transcript on the basis of its expression in the majority of cells in KS lesions (47, 57). K12 is a small ORF (60 amino acids) that is contained within T0.7. By using an antisense oligonucleotide probe within the K12 ORF, two lytic cycle transcripts of 1.7 and 0.8 kb were detected in the BC-1 PEL cell line (Fig. 4B). This is another locus that may be transcribed during latency and the lytic cycle. The pattern of transcription may also vary with the cell type.
PAN RNA expression.
The 1.1-kb PAN RNA is the most abundant lytic cycle transcript of KSHV (51, 56). PAN RNA behaved as an early gene; its expression was induced 7-fold 8 h after chemical treatment and reached a maximal level 30 h after induction when it was 48-fold above the background level. PAN RNA was stable; high levels remained 48 h after n-butyrate treatment (Fig. 4A). More than 97% of its expression was eliminated by cycloheximide. Although PAN RNA was resistant to PAA, its expression was more sensitive to PAA than other early-lytic-cycle genes. For example, 27% of PAN RNA remained after PAA treatment; by comparison 82% of vIL-6 remained after PAA. Thus, some late-cycle expression of PAN RNA may be dependent on viral DNA synthesis.
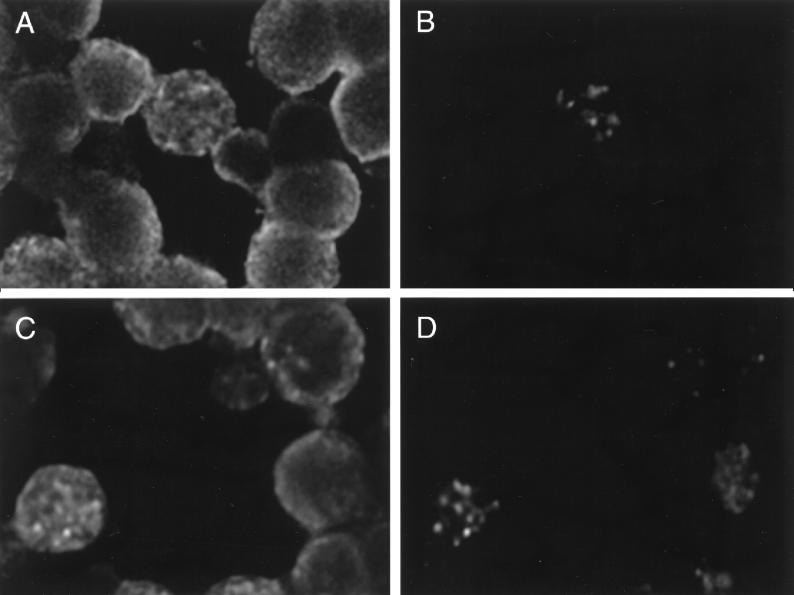
For some highly abundant and stable-lytic-cycle transcripts, such as PAN RNA, a faint signal was detectable on a Northern blot containing RNA prepared from untreated cells. To determine whether this signal was produced by all the cells, thus representing a latency transcript, or by a subpopulation of cells, as would be expected of a lytic-cycle transcript, we carried out in situ hybridization. About 1% of the untreated BC-1 cells expressed PAN RNA (Fig. 5A and B). Therefore, the signal of PAN RNA in untreated BC-1 cells was due to the rare cell that spontaneously entered the lytic cycle. By 24 h after chemical induction, up to 40% of the BC-1 cells expressed PAN RNA (Fig. 5C and D).
FIG. 5.
In situ hybridization of PAN RNA in BC-1 cells. BC-1 cells were hybridized with a FITC-labeled DNA probe specific for PAN RNA. Panels A and B show untreated cells. Panels C and D show cells treated with n-butyrate for 24 h. In panels A and C, the cells were counterstained with propidium iodide. A specific hybridization signal for PAN RNA is seen in 1 of 17 untreated cells and in 4 of 10 n-butyrate-treated cells in the illustrated field.
Expression of KSHV-encoded chemokines.
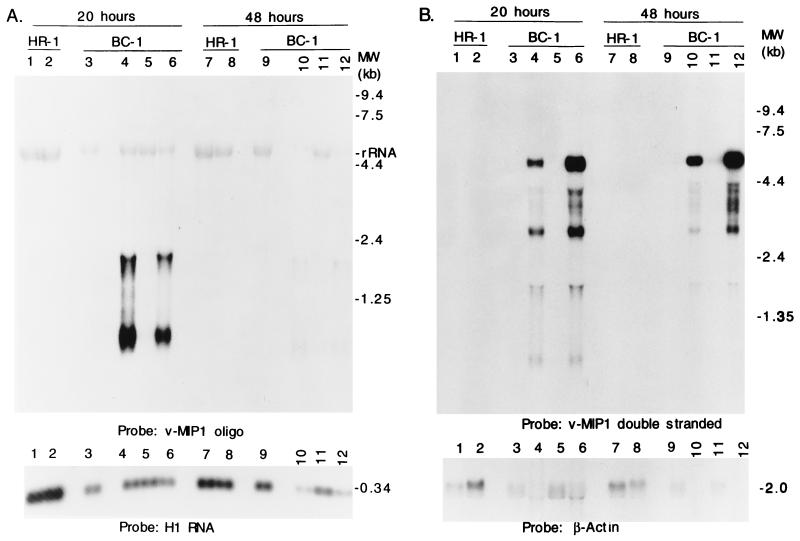
The two viral homologues of the cellular chemokine MIP1α both behaved as early lytic cycle genes but differed from each other in abundance and the kinetics of expression. The more abundant 0.9-kb vMIP II transcript appeared at 8 h and peaked at 13 h after induction (Fig. 4A). Transcription from vMIP I was first detected at 13 h and peaked at 30 h after the addition of n-butyrate (Fig. 4B).
Lytic-cycle transcripts were identified from both strands of the region encoding vMIP I (Fig. 6). When double-stranded probes were used in Northern analysis four distinct transcripts of 6.6, 3.2, 1.8, and 0.8 kb were detected, as well as a heterogeneous group of less-abundant transcripts that varied in size from 4.0 to 5.0 kb. Single-stranded oligonucleotide probes specific for vMIP I detected only the two smaller transcripts of 1.8 and 0.8 kb. These vMIP I-specific transcripts markedly decreased in abundance at 48 h after induction, while the larger transcripts from the opposite strand remained at high levels at this late time. Thus, the locus of vMIP I is transcriptionally complex and is likely to encode both early and late products.
FIG. 6.
Transcription at the viral MIP I locus. Northern blots hybridized with a single-stranded oligonucleotide representing the antisense strand at the 5′ end of the vMIP I ORF (A) or with a double-stranded probe generated by PCR (B) are shown. Aliquots of the same RNA preparations were electrophoresed on two agarose gels. The samples were prepared from BC-1 cells and KSHV-negative, EBV-positive B cells, the HH514-16 subclone of P3J-HR1K (HR-1) cells. The samples in lanes 1, 3, 7, and 9 were untreated; the samples in the even-numbered lanes were treated with n-butyrate for the indicated times. The samples in lanes 5 and 11 were treated with 20 ng of TPA per ml. The samples in lanes 6 and 12 received both butyrate and TPA.
In situ hybridization with 35S-labeled riboprobes for vMIP I detected RNA in only a rare cell of the PEL line BCBL-1 (data not shown); however, riboprobes in both the sense and antisense orientations spanning the vMIP I gene detected an RNA signal in these cells. This result is consistent with Northern analyses showing that both strands of the vMIP I locus encode lytic-cycle mRNAs.
Late gene expression.
A representative viral late gene is the small viral capsid antigen (sVCA) encoded in KSHV ORF65 (Fig. 2B) (28). The behavior of this mRNA was distinctly different from that of early genes in three respects: (i) it was not detected until 30 h after chemical induction, (ii) it reached a maximal level 48 h after the addition of n-butyrate, and (iii) its expression was completely inhibited by PAA.
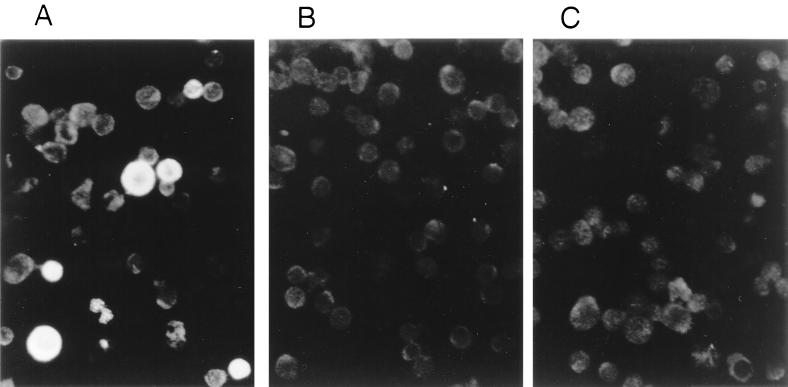
The increase in abundance of sVCA mRNA resulted from recruitment of a significant proportion of BC-1 cells into the late lytic cycle. Using a monospecific antibody to sVCA in an indirect immunofluorescence assay, we found that 0.5% of untreated BC-1 cells expressed sVCA. The proportion of the BC-1 cell population expressing sVCA rose to 20% 48 h after chemical induction (Fig. 7).
FIG. 7.
Detection of sVCA in PEL cells by indirect immunofluorescence. BC-1 cells were treated (A) with n-butyrate or left untreated (B) and reacted with rabbit antiserum to recombinant sVCA and then FITC-conjugated goat anti-rabbit IgG. In panel C, the n-butyrate-treated cells were incubated with preimmune rabbit serum. Specific staining for sVCA in the field illustrated in panel A was seen in 8 of 35 (23%) of cells.
In summary, from these kinetics experiments we found that immediate-early gene expression in BC-1 cells is first detectable by 4 h, and early gene expression is first detectable by 8 to 13 h after the induction of the lytic cycle. Early gene expression peaks at 20 h after induction. Late gene expression is initiated between 20 and 30 h after chemical induction. By inference, lytic viral DNA synthesis must occur in this interval between 20 and 30 h after the addition of the inducing agent.
In situ detection of KSHV-HHV8 genes in KS biopsies.
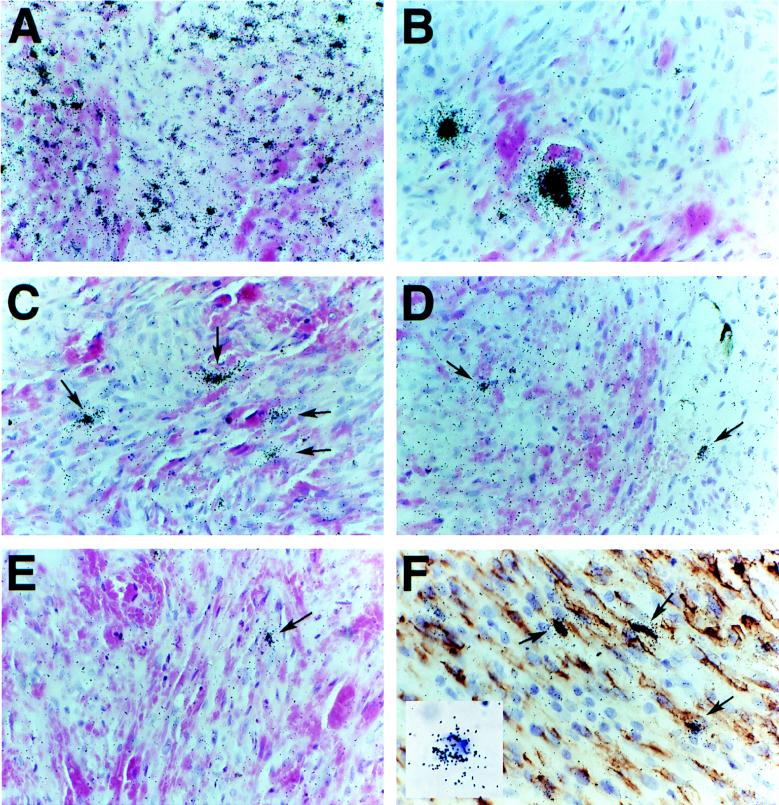
We used in situ hybridization of 35S-labeled riboprobes to determine the distribution and relative abundance of viral transcripts in a typical dermal KS lesion. In this lesion, as previously reported for an oral KS lesion (47), we found that the majority of the KS spindle cells are infected and transcribing viral T0.7 RNA (Fig. 8A) but that only a minor subset of these cells, 1 to 3%, contain the lytic transcript PAN (Fig. 8B). Riboprobes specific for transcripts of KSHV Rta (Fig. 8D), K8 (Fig. 8C), vBcl2 (Fig. 8E), vMIP I (Fig. 8F), and vIL-6 (data not shown) detected transcripts in a subpopulation of 1 to 3% of spindle-shaped cells similar in size to the subpopulation that contained PAN RNA. The intensity of the hybridization signal varied in rough correspondence to the relative abundance of transcript detected in Northern blots of lytically induced PEL cell lines; the most abundant RNA was PAN (T1.1), followed by vMIP I and K8. Double labeling with antibody to CD34 clearly showed that the vMIP I signal was present in the CD34+ spindle cells. There did not appear to be any increase in the inflammatory cells in the vicinity of the cells expressing vMIP I (Fig. 8F). Simultaneous hybridization with 35S-labeled probe for vMIP I and digoxigenin-labeled probe for PAN RNA showed colocalization of these transcripts to the same subset of spindle cells (Fig. 8F, inset). We have previously shown that PAN RNA colocalizes to cells in KS lesions that are transcribing the late lytic gene MCP (ORF25) (47), and in the current study we found that PAN RNA colocalizes to cells in PEL lines that react with specific antibody to another late lytic gene product, sVCA (ORF65) (not shown). Thus, spontaneous early and late lytic genes appear to be expressed in the same subpopulation of cells in KS lesions and PEL cell lines.
FIG. 8.
Expression of KSHV genes in a KS tumor lesion. 35S-labeled antisense riboprobes specific for KSHV genes were hybridized to subjacent sections of a dermal KS lesion. Panels: A, T0.7, 3-day exposure; B, PAN RNA, 18-h exposure; C, K8, 7-day exposure; D, Rta, 7-day exposure; E, vBcl2, 7-day exposure; F, vMIP I, 2-day exposure. Panels A to E were counterstained with hematoxylin and eosin. In situ hybridization was combined with immunochemistry with monoclonal antibody to human CD34 and horseradish peroxidase-conjugated secondary antibody. CD34 reactivity results in brown staining of the spindle-shaped tumor cells. Nuclei, purple in color, were counterstained with hematoxylin. (Inset) Double in situ hybridization with digoxigenin-labeled probe for PAN RNA (purple nucleus) and [35S]vMIP I probe (silver grains).
DISCUSSION
In this study of the kinetics of expression of KSHV genes we provide significant new observations pertinent to the molecular pathogenesis of KSHV. Detailed kinetic studies identify KSHV/Rta as an immediate-early gene product. This observation is consistent with functional experiments showing that KSHV/Rta is an initiator of the lytic cascade (52). Other genes having homology with immediate-early genes of other herpesviruses including K8, the homologue of the EBV ZEBRA gene, are kinetically downstream of KSHV/Rta. A number of virally encoded cellular homologues, such as the proinflammatory cytokines vIL-6, vMIP I, and vMIP II, and the antiapoptotic gene vBcl2 are expressed as early lytic cycle products (Fig. 4 and Fig. 6). The pattern of expression of these early lytic cycle genes in a cultured PEL cell line is consistent with the expression of these genes in a subpopulation of cells in KS biopsies in which the virus has undergone lytic cycle activation (Fig. 8). Thus, activation of the KSHV lytic cascade under the control of the KSHV/Rta gene is likely to be a key element of pathogenesis.
Behavior of gammaherpesvirus immediate-early genes in activation of the lytic cascade.
Based on functional assays with the transfection of plasmids, the KSHV/Rta gene has been found to be competent to initiate reactivation of the lytic cascade (52). In this study we show that KSHV/Rta behaves as an immediate-early gene (Fig. 1 and 3) and that it is expressed in KS biopsies in the same proportion of cells that manifest other lytic-cycle products, including the viral homologues of cytokines and chemokines (Fig. 8). All these data are consistent with the idea that KSHV/Rta is the principal driver of the lytic cascade and functions as a switch gene in the disruption of latency. The definition of an immediate-early gene in the alpha- and betaherpesvirus systems requires that the initial expression of the gene following de novo infection be resistant to the action of inhibitors of protein synthesis. This can occur if the virion contains a potent viral transactivator which, upon de novo infection, activates expression of one or more target genes, or if expression of the immediate-early gene is under the control of preexisting host cell activators. However, in the analysis of reactivation of gammaherpesviruses from latency the definition requires that the expression of the gene be resistant to protein synthesis inhibitors after induction of the switch between latency and the lytic cycle. In the analysis of reactivation of the gammaherpesvirus EBV from latency, we have found that both BZLF1 and BRLF1 are quite sensitive to inhibition by cycloheximide (data not shown). The inhibition of EBV immediate-early gene expression during reactivation from latency may occur as the result of a requirement for de novo cell protein synthesis to activate the promoters of the BZLF1 and BRLF1 genes. Alternatively, large amounts of EBV ZEBRA and Rta proteins may be needed to activate their own expression by an autostimulatory mechanism. By contrast, transcription of the HHV8/Rta gene is significantly resistant to the action of cycloheximide (Table 1), an observation consistent with its control by preexisting cell proteins or latent viral proteins. Meanwhile, K8 the homologue of EBV BZLF1 is clearly sensitive to cycloheximide (Fig. 3) and in transfection experiments is activated by KSHV/Rta (52). Therefore, the K8 gene is clearly downstream both kinetically and functionally from KSHV/Rta. Similarly, other candidate immediate-early genes, such as K3 and K5, behave as early genes, are downstream of KSHV/Rta, and do not synergize with KSHV/Rta (data not shown).
The complex transcription program of KSHV.
Sarid et al. have provided a general description of the transcription program in BC-1 cells by using relatively large double-stranded DNA probes (46). Our studies, using shorter, strand-specific oligonucleotide probes refine these studies for several selected groups of genes. Sarid et al. only studied one time point: 48 h after induction of the lytic cycle. At this time point the abundance of many early gene transcripts has decreased substantially (Fig. 4). Our studies provide kinetic information and use classical inhibitors to subdivide the lytic-cycle genes into immediate-early, early, and late classes. Sarid et al. defined one class of transcripts (class II) that were expressed during latency and increased in abundance during the lytic cycle. Our studies provide at least two explanations for this group of transcripts. Some of the class II genes, for example, PAN RNA and vIL-6, may represent abundant transcripts from a small percentage of cells that spontaneously enter lytic-cycle replication (Fig. 5). Other class II transcripts may result when the same locus is transcribed during latency and overlapping transcripts are expressed during the lytic cycle (Fig. 2).
Our studies describe several aspects of the complexity of the KSHV transcriptional program. First, there appears to be overlapping monocistronic and bi- or polycistronic mRNAs. For example, the LANA and vFLIP ORFs (Fig. 2) and the K8 and ORF50 transcripts are related in this manner (Fig. 3). There are at least two sets of overlapping transcripts in the vMIP I locus (Fig. 6). Second, there is bidirectional transcription clearly evident at the vMIP I locus (compare Fig. 6A and B). Third, the same locus can be expressed during latency and the lytic cycle. For example, lytic-cycle mRNAs are detected with probes for LANA and vFLIP (Fig. 2), as well as T0.7 (Fig. 4B).
Expression of vIL-6, vBcl2, vMIP I, and vGCR as early lytic cycle transcripts.
The studies presented here provide several lines of evidence that several KSHV encoded homologues of cellular genes, including vIL-6, vBcl2, vMIP I, and vGCR, are expressed as viral lytic cycle products in cultured cell lines derived from body cavity lymphoma and in vivo in biopsies of KS. In PEL cell lines transcripts of these genes are generally undetectable in untreated cells but are markedly induced by the addition of chemicals such as n-butyrate that are known to trigger the viral lytic cycle (33). In related experiments we have found that transfection of PEL lines with KSHV/Rta also causes an increase in the abundance of vIL-6, vMIP I, and vMIP II transcripts. Based on similar studies with the related gamma herpesvirus EBV, the response of these genes to Rta is behavior that would be expected of early lytic cycle genes (21).
In PEL cell lines and in KS biopsies a small fraction (from less than 1 to 3%) of cells spontaneously express classical viral lytic cycle products, such as the mRNA of the immediate-early gene KSHV/Rta and the small viral capsid protein encoded in ORF65. The fraction of cells in PEL lines and in KS biopsies that express these classical lytic cycle products is almost identical to the fraction that express vIL-6, vBcl2, and vMIP homologues. This finding is consistent with the notion that many of the cellular homologues are lytic cycle products in vivo. Moreover, there is good correlation between the kinetic behavior of many genes in cultured PEL cells and the transcript patterns observed in KS biopsies. The single exception so far is T0.7. This locus is transcribed as an early lytic gene in the BC-1 PEL cell line (Fig. 4B, K12), but is evidently transcribed as a latency mRNA in KS biopsies (Fig. 8A). This potential discrepancy requires further investigation. It could represent the complexity of overlapping latent and lytic transcription units, it could result from cell type-specific gene expression, or it could be influenced by the microenvironment in the host. Differences in the kinetic behavior of T0.7 RNA may also occur among different PEL cell lines. Using in situ hybridization, Sturzl et al. have found that T0.7 RNA is expressed in most cells of the BCBL-1 line (49a).
Possible functions of KSHV vIL-6, vBcl2, vMIP, and vGCR as lytic cycle products in the pathogenesis of KS.
One obvious role for lytic-cycle reactivation is to facilitate virus production and thereby transmission of the virus to additional cells. It is not likely that the KS lesions themselves would represent the major site of virus production for person-to-person transmission. Although KSHV has been detected in saliva and the prostate, the principal sites and modes of transmission may be multiple and may vary from one population group to another (26, 47). Lytic-cycle expression of nonstructural viral genes, homologues of cellular IL-6, Bcl2, MIP, and GCR, may play a specific role in viral transmission, both from person to person and within an infected host. By serving an antiapoptotic role vIL-6 and vBcl2 may allow lytically infected cells to survive long enough to produce infectious virus. IL-6 may also be secreted and spread to adjacent cells. This may provide a stimulus for the growth of cells latently infected with virus. Alternatively, secreted viral IL-6 may recruit uninfected cells that now become suitable targets for de novo viral infection. This view is consistent with the notion that the KS lesion is cytokine dependent. vMIP might play an analogous role by acting as a chemoattractant for cells that can be infected by KSHV or for cells that produce cytokines that are favorable to the growth and persistence of virally infected cells. Recent evidence suggests that viral MIP II binds to cellular chemokine receptors and blocks the response to cellular chemokines (4). Thus, viral MIP may also play a role in protecting the virus-infected population from an immune response. The role of vGCR in pathogenesis is not clear. Since it is expressed as a lytic-cycle product, it is unlikely to be a direct transforming product.
These observations suggest a novel paradigm for pathogenesis and tumorigenesis by an oncogenic herpesvirus. In most other models tumorigenesis results from the direct transforming functions of certain gene products expressed during latency by the majority of cells. While this scenario may ultimately also prove true for KSHV, there is the additional possibility that the growth of latently infected cells is enhanced by the creation of a favorable milieu resulting from the production of viral chemokines or cytokines by adjacent infected cells undergoing a switch from latency to the lytic cycle. Thus, understanding the detailed mechanism of lytic-cycle activation could suggest avenues for intervention.
ACKNOWLEDGMENT
R.S., S.-F.L., and K.S. all contributed equally to this study.
This work was supported by NIH grants CA70036 to G.M., T32 CA09159 to R.S., and CA75172 to A.H.
REFERENCES
- 1.Andre S, Schatz O, Bogner J R, Zeichhardt H, Stoffler-Meilicke M, Jahn H U, Ullrich R, Sonntag A K, Kehm R, Haas J. Detection of antibodies against viral capsid proteins of human herpesvirus 8 in AIDS-associated Kaposi’s sarcoma. J Mol Med. 1997;75:145–152. doi: 10.1007/s001090050099. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Mesri E A, Nador R G, Said J W, Asch A S, Knowles D M, Cesarman E. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 3.Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989;3:488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O’Leary J J. Kaposi’s sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 6.Bussolino F, Arese M, Montrucchio G, Barra L, Primo L, Benelli R, Sanavio F, Aglietta M, Ghigo D, Rola P M, et al. Platelet activating factor produced in vitro by Kaposi’s sarcoma cells induces and sustains in vivo angiogenesis. J Clin Invest. 1995;96:940–952. doi: 10.1172/JCI118142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai J, Gill P S, Masood R, Chandrasoma P, Jung B, Law R E, Radka S F. Oncostatin-M is an autocrine growth factor in Kaposi’s sarcoma. Am J Pathol. 1994;145:74–79. [PMC free article] [PubMed] [Google Scholar]
- 8.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman E, Moore P S, Rao P H, Inghirami G, Knowles D M, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 10.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 11.Corbeil J, Evans L A, Vasak E, Cooper D A, Penny R. Culture and properties of cells derived from Kaposi sarcoma. J Immunol. 1991;146:2972–2976. [PubMed] [Google Scholar]
- 12.Davis D A, Humphrey R W, Newcomb F M, O’Brien T R, Goedert J J, Straus S E, Yarchoan R. Detection of serum antibodies to a Kaposi’s sarcoma-associated herpesvirus-specific peptide. J Infect Dis. 1997;175:1071–1079. doi: 10.1086/516444. [DOI] [PubMed] [Google Scholar]
- 13.Ensoli B, Barillari G, Gallo R C. Cytokines and growth factors in the pathogenesis of AIDS-associated Kaposi’s sarcoma. Immunol Rev. 1992;127:147–155. doi: 10.1111/j.1600-065x.1992.tb01412.x. [DOI] [PubMed] [Google Scholar]
- 14.Ensoli B, Gendelman R, Markham P, Fiorelli V, Colombini S, Raffeld M, Cafaro A, Chang H K, Brady J N, Gallo R C. Synergy between basic fibroblast growth factor and HIV-1 Tat protein in induction of Kaposi’s sarcoma. Nature. 1994;371:674–680. doi: 10.1038/371674a0. [DOI] [PubMed] [Google Scholar]
- 15.Ensoli B, Markham P, Kao V, Barillari G, Fiorelli V, Gendelman R, Raffeld M, Zon G, Gallo R C. Block of AIDS-Kaposi’s sarcoma (KS) cell growth, angiogenesis, and lesion formation in nude mice by antisense oligonucleotide targeting basic fibroblast growth factor. A novel strategy for the therapy of KS. J Clin Invest. 1994;94:1736–1746. doi: 10.1172/JCI117521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ensoli B, Nakamura S, Salahuddin S Z, Biberfeld P, Larsson L, Beaver B, Wong-Staal F, Gallo R C. AIDS-Kaposi’s sarcoma-derived cells express cytokines with autocrine and paracrine growth effects. Science. 1989;243:223–226. doi: 10.1126/science.2643161. [DOI] [PubMed] [Google Scholar]
- 17.Flamand L, Zeman R A, Bryant J L, Lunardi-Iskandar Y, Gallo R C. Absence of human herpesvirus 8 DNA sequences in neoplastic Kaposi’s sarcoma cell lines. J Acquired Immune Defic Syndr Hum Retrovirol. 1996;13:194–197. doi: 10.1097/00042560-199610010-00011. [DOI] [PubMed] [Google Scholar]
- 18.Foreman K E, Bacon P E, Hsi E D, Nickoloff B J. In situ polymerase chain reaction-based localization studies support role of human herpesvirus-8 as the cause of two AIDS-related neoplasms: Kaposi’s sarcoma and body cavity lymphoma. J Clin Invest. 1997;99:2971–2978. doi: 10.1172/JCI119492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao S J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 20.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 21.Hardwick J M, Lieberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez G L, Spector D L. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- 23.Johnson C V, Singer R H, Lawrence J B. Fluorescent detection of nuclear RNA and DNA: implications for genome organization. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- 24.Kedes D H, Ganem D, Ameli N, Bacchetti P, Greenblatt R. The prevalence of serum antibody to human herpesvirus 8 (Kaposi sarcoma-associated herpesvirus) among HIV-seropositive and high-risk HIV-seronegative women. JAMA. 1997;277:478–481. [PubMed] [Google Scholar]
- 25.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Invest. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koelle D M, Huang M L, Chandran B, Vieira J, Piepkorn M, Corey L. Frequent detection of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in saliva of human immunodeficiency virus-infected men: clinical and immunologic correlates. J Infect Dis. 1997;176:94–102. doi: 10.1086/514045. [DOI] [PubMed] [Google Scholar]
- 27.Li J J, Huang Y Q, Cockerell C J, Friedman-Kien A E. Localization of human herpes-like virus type 8 in vascular endothelial cells and perivascular spindle-shaped cells of Kaposi’s sarcoma lesions by in situ hybridization. Am J Pathol. 1996;148:1741–1748. [PMC free article] [PubMed] [Google Scholar]
- 28.Lin S F, Sun R, Heston L, Gradoville L, Shedd D, Haglund K, Rigsby M, Miller G. Identification, expression, and immunogenicity of Kaposi’s sarcoma-associated herpesvirus-encoded small viral capsid antigen. J Virol. 1997;71:3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manet E, Gruffat H, Trescol B M C, Moreno N, Chambard P, Giot J F, Sergeant A. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 1989;8:1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miles S A. Kaposi sarcoma: a cytokine-responsive neoplasia? Cancer Treat Res. 1992;63:129–140. doi: 10.1007/978-1-4615-3088-6_6. [DOI] [PubMed] [Google Scholar]
- 31.Miles S A, Martinez M O, Rezai A, Magpantay L, Kishimoto T, Nakamura S, Radka S F, Linsley P S. Oncostatin M as a potent mitogen for AIDS-Kaposi’s sarcoma-derived cells. Science. 1992;255:1432–1434. doi: 10.1126/science.1542793. [DOI] [PubMed] [Google Scholar]
- 32.Miles S A, Rezai A R, Salazar-Gonzalez J F, Vander Meyden M, Stevens R H, Logan D M, Mitsuyasu R T, Taga T, Hirano T, Kishimoto T, et al. AIDS Kaposi sarcoma-derived cells produce and respond to interleukin 6. Proc Natl Acad Sci USA. 1990;87:4068–4072. doi: 10.1073/pnas.87.11.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller G, Rigsby M O, Heston L, Grogan E, Sun R, Metroka C, Levy J A, Gao S J, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi’s sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 35.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 36.Nair B C, DeVico A L, Nakamura S, Copeland T D, Chen Y, Patel A, O’Neil T, Oroszlan S, Gallo R C, Sarngadharan M G. Identification of a major growth factor for AIDS-Kaposi’s sarcoma cells as oncostatin M. Science. 1992;255:1430–1432. doi: 10.1126/science.1542792. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura S, Salahuddin S Z, Biberfeld P, Ensoli B, Markham P D, Wong-Staal F, Gallo R C. Kaposi’s sarcoma cells: long-term culture with growth factor from retrovirus-infected CD4+ T cells. Science. 1988;242:426–430. doi: 10.1126/science.3262925. [DOI] [PubMed] [Google Scholar]
- 38.Neipel F, Albrecht J C, Ensser A, Huang Y Q, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo H G, Reitz M S, Hayward G S. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H G, Hayward G S, Reitz M S. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 41.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 43.Rickinson A B, Finerty S, Epstein M A. Mechanism of the establishment of Epstein-Barr virus genome-containing lymphoid cell lines from infectious mononucleosis patients: studies with phosphonoacetate. Int J Cancer. 1977;20:861–868. doi: 10.1002/ijc.2910200607. [DOI] [PubMed] [Google Scholar]
- 44.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Said W, Chien K, Takeuchi S, Tasaka T, Asou H, Cho S K, de Vos S, Cesarman E, Knowles D M, Koeffler H P. Kaposi’s sarcoma-associated herpesvirus (KSHV or HHV8) in primary effusion lymphoma: ultrastructural demonstration of herpesvirus in lymphoma cells. Blood. 1996;87:4937–4943. [PubMed] [Google Scholar]
- 46.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sturzl M, Brandstetter H, Roth W K. Kaposi’s sarcoma: a review of gene expression and ultrastructure of KS spindle cells in vivo. AIDS Res Hum Retroviruses. 1992;8:1753–1763. doi: 10.1089/aid.1992.8.1753. [DOI] [PubMed] [Google Scholar]
- 49.Sturzl M, Roth W K, Brockmeyer N H, Zietz C, Speiser B, Hofschneider P H. Expression of platelet-derived growth factor and its receptor in AIDS-related Kaposi sarcoma in vivo suggests paracrine and autocrine mechanisms of tumor maintenance. Proc Natl Acad Sci USA. 1992;89:7046–7050. doi: 10.1073/pnas.89.15.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Sturzl M, Blasig C, Schreier A, Neipel F, Hohenadl C, Cornali E, Ascherl G, Esser S, Brockmeyer N H, Ekman M, Kaaya E E, Tschachler E, Biberfeld P. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi’s sarcoma. Int J Cancer. 1997;72:68–71. doi: 10.1002/(sici)1097-0215(19970703)72:1<68::aid-ijc10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 50.Summers W C, Klein G. Inhibition of Epstein-Barr virus DNA synthesis and late gene expression by phosphonoacetic acid. J Virol. 1976;18:151–155. doi: 10.1128/jvi.18.1.151-155.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun R, Lin S F, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 55.Xerri L, Hassoun J, Planche J, Guigou V, Grob J J, Parc P, Birnbaum D, deLapeyriere O. Fibroblast growth factor gene expression in AIDS-Kaposi’s sarcoma detected by in situ hybridization. Am J Pathol. 1991;138:9–15. [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong W, Ganem D. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:1207–1212. doi: 10.1128/jvi.71.2.1207-1212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6661. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]