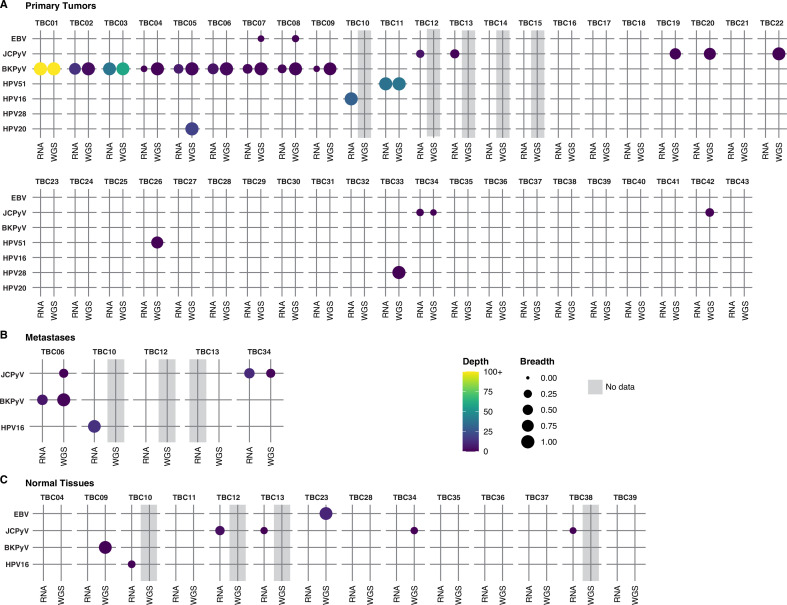
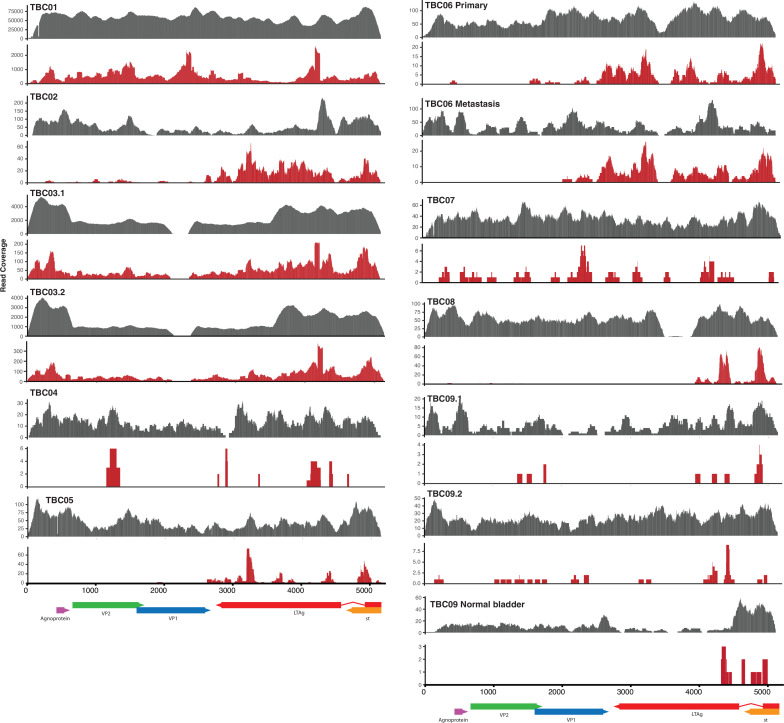
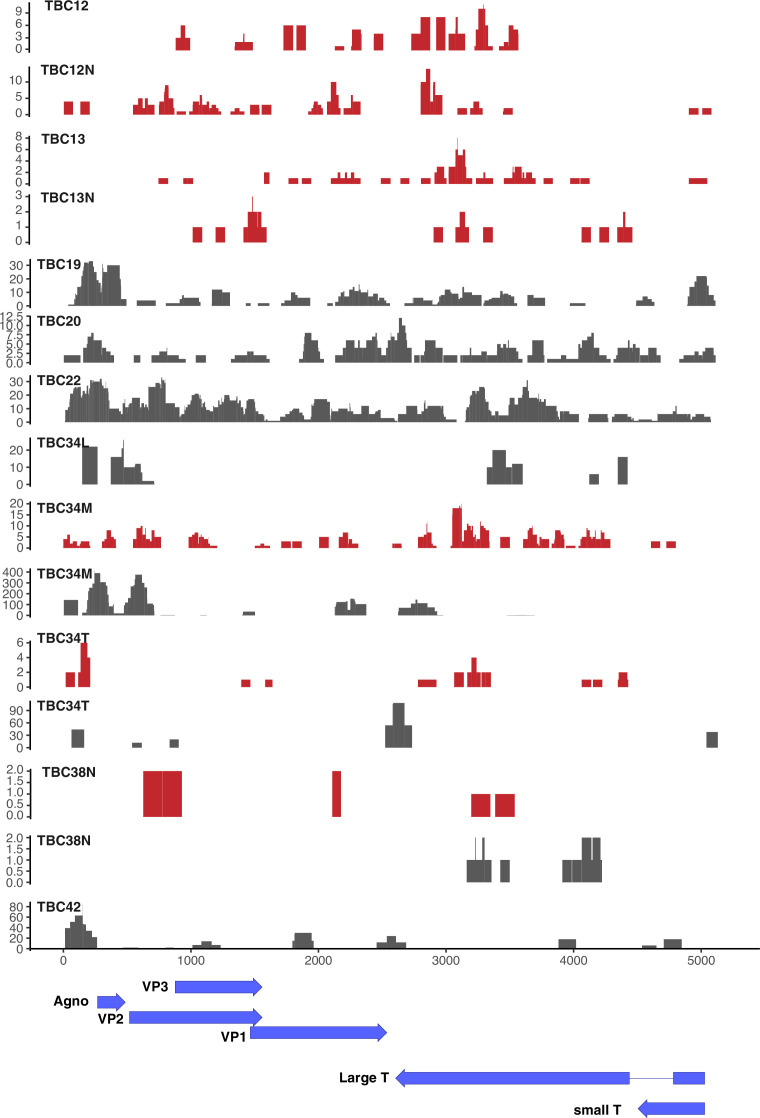
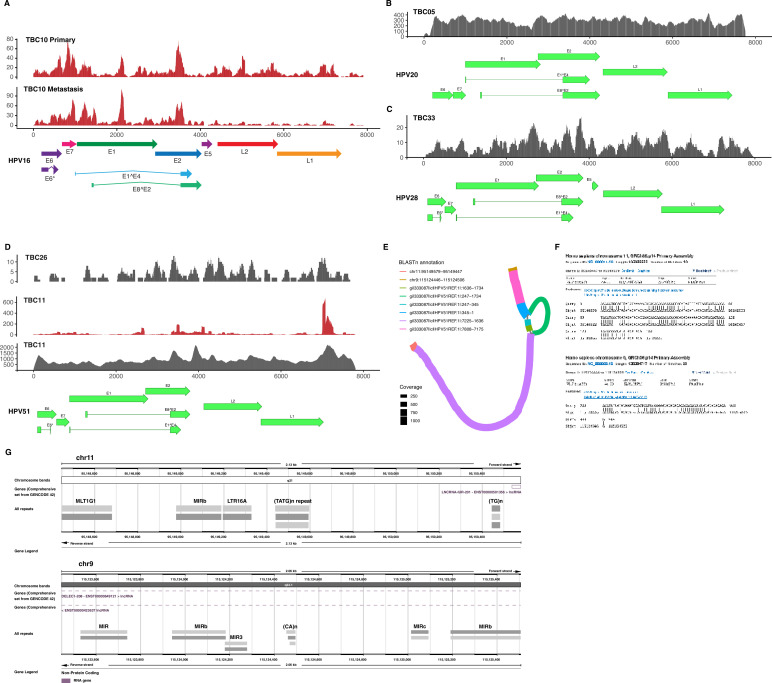
Figure 1. Detection of viral sequences.
(A) Primary tumors. (B) Metastatic tumors. (C) Normal tissues. Viral species are shown on the rows, and each case in the cohort (represented with a TBC number) is a column. TBC numbers represent a single case and are consistent across primary, metastatic, and normal tissues. Circle size represents the breadth or fraction of the viral genome covered, and color represents the average depth of coverage of the viral k-mers with all coverages over 100 binned together. Specimens without sequencing data have a gray background.