ABSTRACT
Microbial cell-free DNA (mcfDNA) sequencing is an emerging infectious disease diagnostic tool which enables unbiased pathogen detection and quantification from plasma. The Karius Test, a commercial mcfDNA sequencing assay developed by and available since 2017 from Karius, Inc. (Redwood City, CA), detects and quantifies mcfDNA as molecules/μL in plasma. The commercial sample data and results for all tests conducted from April 2018 through mid-September 2021 were evaluated for laboratory quality metrics, reported pathogens, and data from test requisition forms. A total of 18,690 reports were generated from 15,165 patients in a hospital setting among 39 states and the District of Columbia. The median time from sample receipt to reported result was 26 h (interquartile range [IQR] 25 to 28), and 96% of samples had valid test results. Almost two-thirds (65%) of patients were adults, and 29% at the time of diagnostic testing had ICD-10 codes representing a diverse array of clinical scenarios. There were 10,752 (58%) reports that yielded at least one taxon for a total of 22,792 detections spanning 701 unique microbial taxa. The 50 most common taxa detected included 36 bacteria, 9 viruses, and 5 fungi. Opportunistic fungi (374 Aspergillus spp., 258 Pneumocystis jirovecii, 196 Mucorales, and 33 dematiaceous fungi) comprised 861 (4%) of all detections. Additional diagnostically challenging pathogens (247 zoonotic and vector-borne pathogens, 144 Mycobacterium spp., 80 Legionella spp., 78 systemic dimorphic fungi, 69 Nocardia spp., and 57 protozoan parasites) comprised 675 (3%) of all detections. This is the largest reported cohort of patients tested using plasma mcfDNA sequencing and represents the first report of a clinical grade metagenomic test performed at scale. Data reveal new insights into the breadth and complexity of potential pathogens identified.
KEYWORDS: microbial cell-free DNA, high-throughput nucleic acid sequencing, liquid biopsy for infectious diseases, metagenomics
INTRODUCTION
Sequencing microbial cell-free DNA (mcfDNA) in plasma represents integration of progress in genomic sequencing, computation capacity, and recognition of cell-free DNA as a clinically useful blood analyte (1–3). Over the last 4 decades, PCR-based tests, specifically multiplexed broad syndromic panels, have made welcomed contributions to infectious disease diagnostics but fall short of desired performance, including breadth of pathogen detection, and require samples of infected tissue or body fluid (4). Broad-range PCR testing arguably facilitates considering a wider range of potential pathogens but is still only limited to bacteria and fungi (5). A recent meta-analysis, which included 20 studies that satisfied the Quality Assessment of Diagnostic Accuracy Studies (6) to assess the diagnostic accuracy of next generation sequencing in distinguishing infectious diseases, concluded that this group of technologies demonstrated satisfactory diagnostic performance for infections and yielded an overall detection rate superior to conventional methods (7). Four of the studies included in this review employed plasma mcfDNA sequencing. Moreover, early experience with plasma mcfDNA sequencing suggests this new approach, especially when applied early in a patient’s clinical course and for specific use cases, has potential to improve upon the above-noted shortcomings (8, 9). Plasma mcfDNA sequencing enables unbiased pathogen detection through noninvasive sampling with rapid turnaround, creating opportunities to enhance diagnosis of bloodstream and deep-seated infections (10–12). These capabilities are most urgently needed among immunocompromised patients who often have the broadest range of pathogens and are most vulnerable to serious consequences from infections. The Karius Test is an analytically and clinically validated mcfDNA sequencing test commercially available for US inpatients since 2017 as a laboratory developed test from Karius, Inc. The test can identify and quantitate molecules/μL (MPM) mcfDNA in plasma for >1,500 bacteria, DNA viruses, fungi, and parasites. The analytical and clinical validation of the test was previously reported (12). Since the time of this study, others have reported how this unbiased test may contribute to the diagnosis and management of life-threatening infections in immunocompromised patients. These contributions include minimizing invasive procedures (13), reducing time to specific etiologic diagnosis of infections compared with standard of care (SOC) microbiological testing (14), and optimizing antimicrobial therapy (15, 16). In contrast, several retrospective, observational reviews of Karius Test utilization concluded that in routine clinical practice the diagnostic and clinical impact of the test was limited, which highlights the need for diagnostic stewardship to optimize implementation and maximize clinical utility in specific patient populations (17–19).
Plasma mcfDNA sequencing for infectious disease diagnosis performance at scale with respect to time to results, laboratory quality metrics, positivity rates, and diversity of taxa detected has not been previously reported. Here, we review the results for a large commercial laboratory testing cohort of over 18,000 plasma samples from over 15,000 patients in a hospital setting with the primary objective to provide additional insights about the breadth and complexity of microbial identifications. While doing so, we also characterize clinical use in a subset of the patients in this test cohort.
MATERIALS AND METHODS
Commercial laboratory test cohort.
The Karius Test results for patients from across the United States were evaluated for reported pathogens and patient data (including basic demographics, ordering clinician, and ICD-10 codes, if provided) obtained from the test request forms (TRF) for all samples tested from 1 April 2018 through mid-September 2021. Laboratory quality metrics were gathered for all samples collected from 1 April 2018 through the end of September 2021. Diagnosis codes submitted via TRFs were summarized at both the chapter level and Clinical Classifications Software Refined categories (20, 21). Immunocompromising conditions were then flagged using definitions published by the Agency for Healthcare Research and Quality (22–24).
The Karius Test.
Plasma mcfDNA sequencing was performed as previously described (12) in the Karius clinical laboratory, certified under the Clinical Laboratory Improvement Amendments of 1988 and accredited by the College of American Pathologists. Briefly, whole-blood samples were collected in either BD Vacutainer plasma preparation tubes (PPTs) or K2-EDTA tubes. After plasma had been separated from cells, the sample was stable at ambient temperature for 96 h and at −20°C for 6 months. Upon receipt at Karius, controls for carry-over, sequencing bias, metagenomic sequencing quality, and sample mix-ups were added to the sample.
In addition, two types of batch controls were run alongside patient samples. Four replicates of environmental control samples containing buffer instead of plasma were processed in parallel with patient samples from accessioning to report generation. Environmental controls were used to monitor microbial DNA signals arising from the background at the time of batch processing. The estimated taxon abundances from the environmental control samples within the batch were combined to parameterize a model of read abundance arising from the environment with variations driven by counting noise. Two assay controls, each containing four distinct species ranging in GC-content from 30% to 65% at high or low concentrations were included in each batch. All four spiked microorganisms and no others had to be detected within a specified MPM range in both assay controls to pass the final quality control inspection.
Proprietary chemistries were used to enrich samples for mcfDNA without preselecting pathogens to test. Automated DNA extraction and sequencing library preparation protocols were optimized for high speed and low pathogen bias. Single-end, 76- or 56-cycle sequencing was performed on NextSeq 500 instruments (Illumina, San Diego, CA) with an average of >20 million reads/sample. Double-unique dual indexes were used to ensure robust sample demultiplexing. Sequencing data were processed using a proprietary analytical pipeline, and microbial reads were aligned to a database comprising >20,000 curated assemblies from >16,000 species of which >1,500 taxa are in the clinical reportable range (CRR) of the test, including bacteria, DNA viruses, fungi, and parasites.
The selection of organisms in the CRR was performed as follows. A candidate list of human pathogens was generated by two board-certified infectious disease physicians by including: (i) DNA viruses, bacteria, mycobacteria, fungi, and parasites from the standard textbooks and other relevant references, (ii) organisms in the pathogen database referenced in published case reports, and (iii) reference genomes sequenced from human clinical isolates with publications supporting potential pathogenicity. Organisms from the above list that were associated with high-quality reference genomes, as determined by our reference database quality control process, were used to further narrow the range. Finally, organisms at risk of generating common false-positive calls because of sporadic environmental contamination were removed from the CRR. The current CRR is available at https://kariusdx.com/the-karius-test/pathogen-list/.
Statistical significance values were computed for each estimated taxon abundance in each nonenvironmental control sample. Those within the CRR at high significance levels above background comprised our candidate detections. Final detections were made after additional filtering was applied, which accounted for read location uniformity, read percent identity, and cross-reactivity originating from higher abundance detections. The microorganism detections that passed these filters were reported along with abundances in MPM, a unique, absolute quantification capability of the Karius Test shown to correlate well with single-analyte qPCR measurements of DNA viruses (25, 26).
The reports also contained median and range of MPM values observed for each microorganism reported in the last 1,000 specimens, as MPM values from different microbes are not comparable, and a reference interval determined from 675 asymptomatic donors for comparison. We routinely analyzed the raw data for mcfDNA from potential pathogens, including those present at levels below our standard laboratory report thresholds. For this study, we focused on microorganisms identified in statistically significant amounts.
Notable improvements in the test wet bench procedures and analytical pipeline, as may be anticipated, occurred during the study period, and are provided in Table S1 in the supplemental material. We used the operational classes of pathogens described by Relman, Falkow, and Ramakrishnan (27). The operational classes include obligate, commensal, zoonotic, and environmental pathogens here to categorize some the pathogens detected.
Data analytics.
Data analysis and visualization were conducted using Python v3.9.7, pandas v1.3.4, matplotlib v3.4.3, and seaborn v0.11.2 (28–30). Given the taxonomy and nomenclature for some genera continue to evolve, we selected three (Legionella, Nocardia, and Mycobacterium) to examine species detections, especially multiple species codetections, more closely. All analyses were done by Karius staff.
Ethical considerations.
Using the Department of Health and Human Services regulations found at 45 CFR 46.104(d)(4), the Advarra IRB determined that this research project was exempt from IRB oversight.
RESULTS
Test cohort.
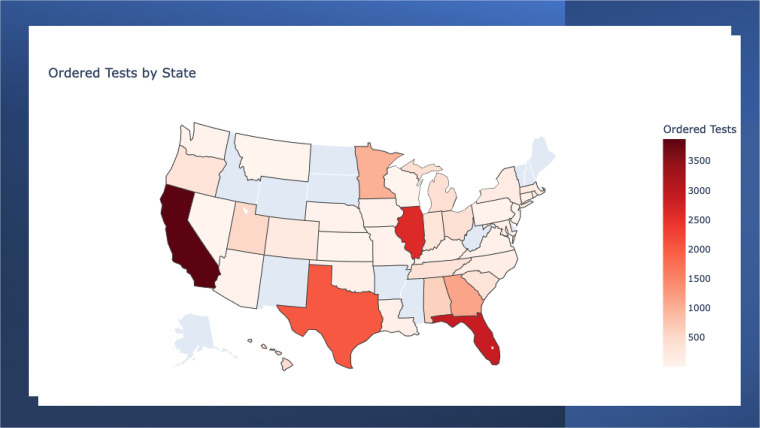
A total of 19,739 samples meeting collection and transport requirements were tested from 16,172 patients in a hospital setting in 39 states and the District of Columbia during the study period. See Fig. 1 for the number of Karius Tests ordered by state. The median time from sample receipt at the Karius laboratory to the delivery of the reported result was 26 h (interquartile range 25 to 28), and the median time from sample collection to the delivery of reported result was 63 h (interquartile range 49 to 89). Ninety-six percent of samples had valid test results. A summary of key quality metrics for the laboratory in this production data set is shown in Table 1. These metrics were not significantly different (two-sided t test P value >0.01) from those reported for the first 2,000 clinical samples run by the Karius clinical laboratory and reported in the initial validation study (12).
FIG 1.
Number of Karius Tests ordered by state from April 2018 to September 2021. No samples were received from the states shaded in blue.
TABLE 1.
Plasma mcfDNA sequencing clinical laboratory quality metrics in production from April 2018 to the end of September 2021
| Metric | Productiona | Validationb |
|---|---|---|
| Sample acceptance rate | 98% | 98% |
| Total yield | 96% | 98% |
| First pass yield | 91% | 91% |
| Results delivered next operational day | 90% | 90% |
Based on 20,087 clinical samples received during this study.
Based on 2,000 clinical samples tested as part of the initial Karius Test validation study (12).
Infectious disease and hematology/oncology providers represented most ordering clinicians, 64% (n = 9,804) and 14% (n = 2,132), respectively, for the 15,424 specimens with a National Provider Identifier indicated. We were able to capture and analyze 18,690 reports from 15,165 patients. Twelve percent (n = 1,839) of patients had at least one repeat test during the study interval. Almost two thirds (65%, n = 9,798) of patients were adults (i.e., age >18 years). More than a quarter (29%, n = 4,423) of patients at the time of diagnostic testing had ICD-10 codes representing a diverse array of clinical scenarios indicated in their TRFs (Table 2). Eighteen percent (n = 797) of these patients were indicated as immunocompromised (IC); 717 (16%) had fever; and 230 (5%) had sepsis.
TABLE 2.
ICD-10 codes by principal diagnosis type for those patients with ICD-10 codes indicated on Karius TRFs (n = 4,423) from April 2018 to September 2021a
| Principal diagnosis type (ICD-10-CM chapter) | Total | IC | Fever | Sepsis |
|---|---|---|---|---|
| (Unmappable) | 68 | |||
| Conditions not elsewhere classified | 1,437 | 722 | ||
| Respiratory system diseases | 800 | |||
| Neoplasms | 580 | 288 | ||
| Infectious/parasitic diseases | 543 | 47 | 231 | |
| Blood diseases | 485 | 320 | ||
| Circulatory system diseases | 416 | |||
| Factors influencing health status and contact with health services | 290 | 140 | ||
| Musculoskeletal system diseases | 259 | 9 | ||
| Nervous system diseases | 242 | |||
| Digestive system diseases | 162 | 21 | ||
| Skin diseases | 111 | |||
| Genitourinary system diseases | 101 | 15 | ||
| Endocrine/metabolic diseases | 91 | 4 | ||
| Injury, poisoning, external causes | 83 | 25 | ||
| Congenital malformations | 76 | |||
| Codes for special purposes | 20 | |||
| Eye/adnexa diseases | 16 | |||
| Mental/behavioral disorders | 13 | |||
| Ear/mastoid diseases | 7 | |||
| Pregnancy/childbirth | 7 | |||
| External causes of morbidity | 3 | |||
| Total no. of patients by diagnosis chapter | 5,810 | 869 | 722 | 231 |
TRF, test report form; IC, immunocompromised. Fever, any ICD-10 starting with “R50”; sepsis, any ICD-10 starting with “A41”; IC, any ICD-10 annotated as immunocompromised from the Agency for Healthcare Research and Quality (AHRQ) code list. Each TRF could contain up to two ICD-10 codes, and each patient had between 1 and 5 unique ICD-10 codes. For the study period, there were 15,165 patients with a positive or negative report (18,690 reports).
Taxa detection and quantification.
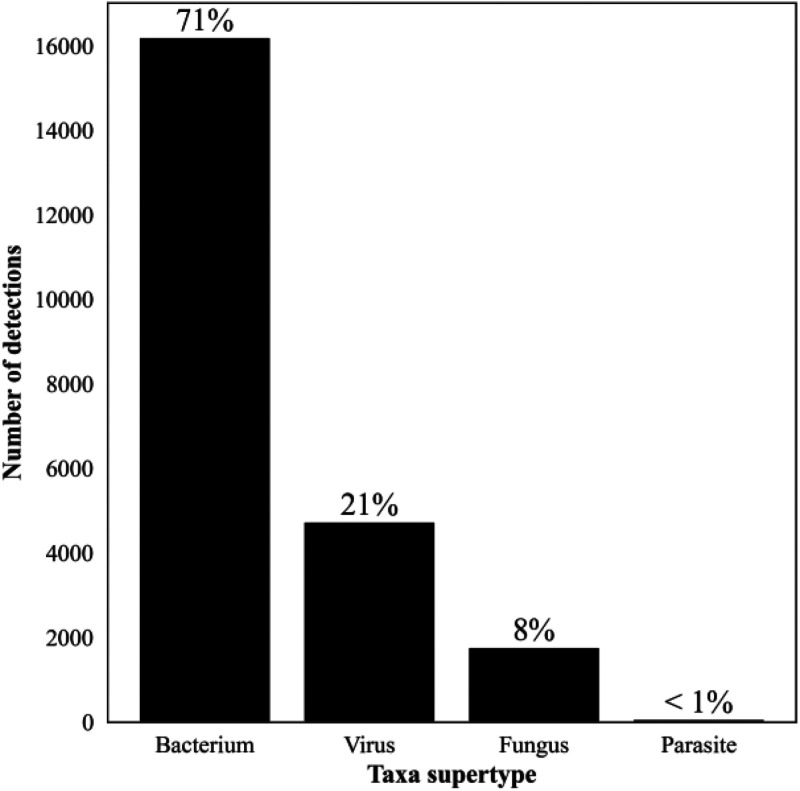
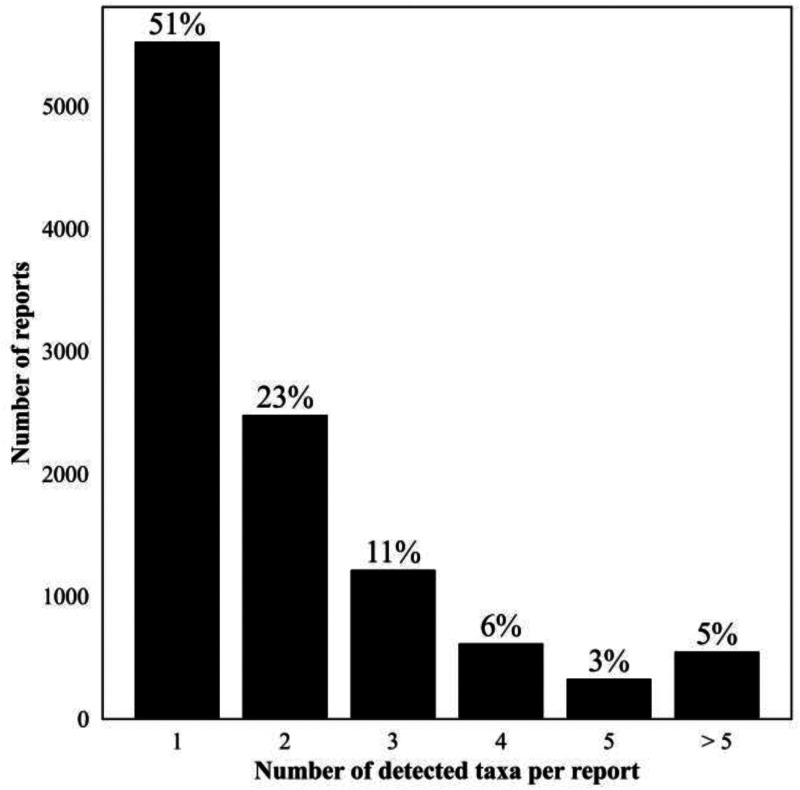
Of samples yielding a valid result, 7,938 (42%) reported a negative test with no pathogens identified. The remaining 10,752 (58%) Karius Test reports from 8,849 patients had at least one microbe identified (5,531 [30%] only one) representing 701 unique microbial taxa (526 [75%] bacteria, 103 [15%] fungi, 47 [7%] viruses, and 24 [3%] parasites) and a total of 22,792 detections. The overall frequency of detection for each of these groups is shown in Fig. 2, and the number of detected taxa counts per report for all positive reports is shown in Fig. 3. All the quality control metrics were met for taxa quantification in MPM for 9,690 (90%) samples with positive results. A complete list of all taxa reported along with their frequency of detection and median and IQR for the MPM values are shown in Table S2.
FIG 2.
Number of detections by the Karius Test of the different supergroups of taxa from April 2018 to September 2021. n = 22,792: bacteria, 16,221; viruses, 4,737; fungi, 1,758; and parasites, 70. Percentages reflect the proportion of total number of detections spanning 701 microbial taxa.
FIG 3.
Number of detected taxa counts per report for all positive reports, April 2018 to September 2021. Percentages reflect the proportion of all positive reports (n = 10,752).
Top 50 reported taxa.
The top 50 reported taxa and the median, range, and IQR of MPM for each taxon are shown in Table 3. They included 36 bacteria, 9 viruses, and 5 fungi. Together the top 50 taxa included a broad range of commensal and environmental pathogens and represented 15,692 detections (69% of all detections).
TABLE 3.
Top 50 taxa detected from April 2018 to September 2021
| Taxon namea | No. detected | Median (MPM) | IQR (MPM) |
|---|---|---|---|
| Bacteria | |||
| Acinetobacter haemolyticus | 199 | 142.97 | 192.68 |
| Bacteroides fragilis | 217 | 264.74 | 1,046.15 |
| Bacteroides ovatus | 133 | 141.43 | 476.14 |
| Bacteroides thetaiotaomicron | 146 | 221.79 | 742.60 |
| Bacteroides vulgatus | 392 | 153.01 | 413.32 |
| Enterobacter cloacae complex | 343 | 821.34 | 6,742.30 |
| Enterococcus faecalis | 582 | 617.90 | 3,285.15 |
| Enterococcus faecium | 510 | 687.63 | 3,958.33 |
| Escherichia coli | 1088 | 684.92 | 3,623.17 |
| Fusobacterium nucleatum | 299 | 460.25 | 1,922.08 |
| Haemophilus influenzae | 288 | 157.09 | 425.59 |
| Haemophilus parainfluenzae | 178 | 284.92 | 1,121.19 |
| Helicobacter pylori | 207 | 217.44 | 601.83 |
| Klebsiella pneumoniae | 501 | 910.19 | 5,889.84 |
| Lactobacillus fermentum | 165 | 256.13 | 1,058.08 |
| Lactobacillus rhamnosus | 281 | 344.33 | 1,250.14 |
| Prevotella melaninogenica | 509 | 208.52 | 545.66 |
| Prevotella oris | 170 | 517.78 | 2,094.07 |
| Pseudomonas aeruginosa | 817 | 1,005.18 | 6,874.18 |
| Rothia dentocariosa | 110 | 117.02 | 256.72 |
| Rothia mucilaginosa | 454 | 177.53 | 369.36 |
| Serratia marcescens | 99 | 492.72 | 2,956.54 |
| Staphylococcus aureus | 561 | 838.21 | 5,665.43 |
| Staphylococcus epidermidis | 716 | 579.58 | 2,689.68 |
| Staphylococcus haemolyticus | 92 | 1,137.08 | 5,450.01 |
| Stenotrophomonas maltophilia | 169 | 1,983.18 | 11,864.14 |
| Streptococcus intermedius | 100 | 354.95 | 1,048.47 |
| Streptococcus mitis | 356 | 226.15 | 897.48 |
| Streptococcus oralis | 140 | 235.54 | 730.42 |
| Streptococcus parasanguinis | 186 | 152.35 | 298.03 |
| Streptococcus pneumoniae | 214 | 975.48 | 11,933.10 |
| Streptococcus pyogenes | 95 | 1,064.08 | 9,773.54 |
| Streptococcus salivarius | 104 | 253.73 | 799.10 |
| Streptococcus thermophilus | 184 | 190.84 | 339.33 |
| Veillonella dispar | 162 | 140.88 | 269.85 |
| Veillonella parvula | 256 | 202.30 | 476.57 |
| Fungi | |||
| Aspergillus fumigatus | 186 | 300.10 | 723.60 |
| Candida albicans | 260 | 624.63 | 2,074.97 |
| Candida glabrata | 113 | 611.85 | 1,449.20 |
| Candida tropicalis | 103 | 784.49 | 3,997.85 |
| Pneumocystis jiroveci | 258 | 2,452.78 | 18,084.89 |
| Viruses | |||
| BK polyomavirus | 354 | 472.38 | 2,677.18 |
| Cytomegalovirus | 1275 | 633.93 | 3,889.67 |
| Epstein-Barr virus | 902 | 272.34 | 852.32 |
| Herpes simplex virus type 1 | 479 | 754.78 | 3,208.06 |
| Human herpesvirus 6B | 468 | 518.74 | 3,268.72 |
| Human herpesvirus 7 | 98 | 82.74 | 342.37 |
| Human adenovirus B | 100 | 3,184.75 | 69,714.71 |
| Human adenovirus C | 171 | 573.87 | 10,160.48 |
| Torque teno virus | 135 | 484.19 | 1,935.97 |
Reported taxa reflect those names used in the clinical reportable range of microbes at the time that the tests were performed.
The distribution of these taxa is as follows. There were 11,023 detections of bacteria, including 11 anaerobes (2,730, 25%), 8 Streptococcus spp. (1,379, 12%), 4 Enterobacterales (2,031, 18%), 3 Staphylococcus spp. (1,369, 12%)., 2 Rothia spp. (564, 5%), 2 Haemophilus spp. (466, 4%), 2 Enterococcus spp. (1,092, 10%), and 1 each of Acinetobacter haemolyticus (199, 2%), Pseudomonas aeruginosa (817, 7%), Stenotrophomonas maltophilia (169, 1%), and Helicobacter pylori (207, 2%). There were 3,982 viral detections of 9 different viruses that included 1,275 (32%) cytomegalovirus, 902 (23%) Epstein-Barr virus, 479 (12%) herpes simplex virus 1, 468 (12%) human herpesvirus 6B, 354 (9%) BK polyomavirus, 171 (4%) human adenovirus C, 135 (3%) torque teno virus (TTV), 100 (3%) human adenovirus B, and 98 (3%) human herpesvirus 7. Finally, there were 920 detections of fungi comprising 260 (28%) Candida albicans, 258 (28%) Pneumocystis jirovecii, 186 (20%) Aspergillus fumigatus, 113 (12%) Candida glabrata, and 103 (11%) Candida tropicalis.
Difficult-to-diagnose uncommon pathogens.
The SOC methods for the organisms listed below have considerable shortcomings, including, but not limited to, sensitivity and specificity, inclusivity, accuracy, time to result, and/or local availability.
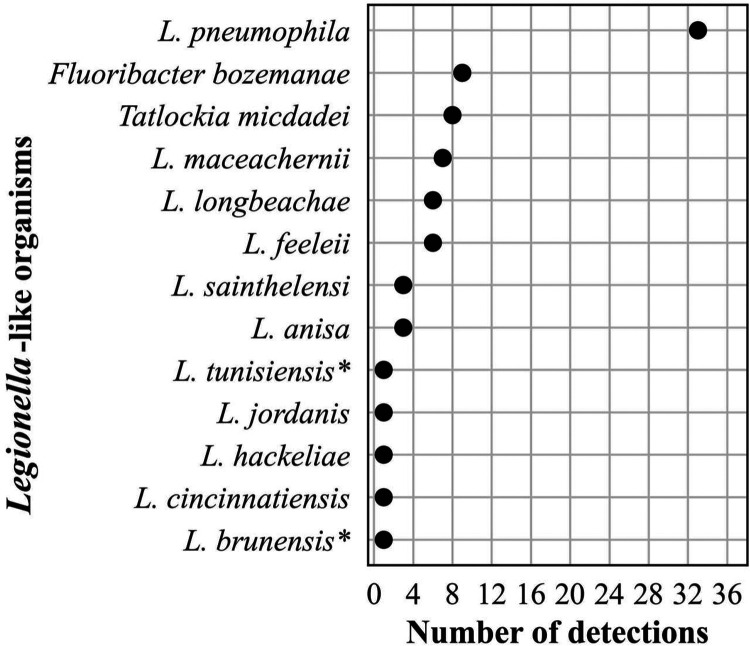
Bacteria. The frequency distribution of the number of detections of Legionella-like organisms (n = 80) is shown in Fig. 4. Forty-one percent of these detections were the most recognized pathogen, L. pneumophila. Two reports contained codetections of two different species (L. brunensis, 400 MPM and L. hackeliae, 270 MPM; and L. feeleii, 78,508 MPM and L. tunisiensis, 76,445 MPM, respectively). Neither L. brunensis nor L. tunisiensis has been associated with human disease (https://specialpathogenslab.com/legionella-species/), and the MPM values for the codetections in each report were similar.
FIG 4.
Frequency distribution of Legionella-like organisms detected, n = 80 (<1% of all bacterial detections, n = 16,203), April 2018 to September 2021. *indicates species not associated with human infections.
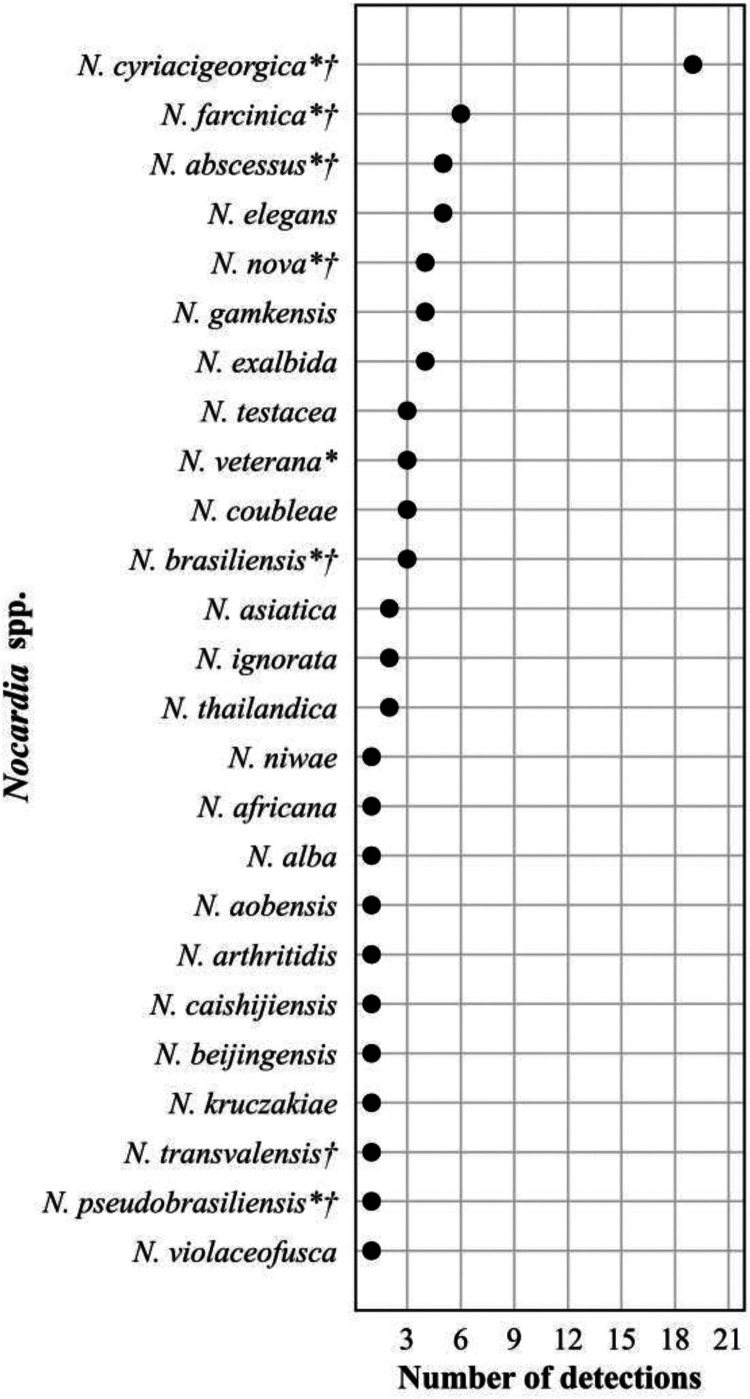
The frequency distribution of Nocardia spp. detections, n = 76, is shown in Fig. 5. Plasma mcfDNA sequencing detected 25 of the approximately 100 validly named species. Of the 8 species reported to be isolated more frequently from patients, 7 were detected. One species, N. cyriacigeorgica, dominated with 19 (25%) detections. Of the 69 patient reports represented by the Nocardia spp. detections, 12 (17%) reported ≥2 concurrent species (range 2 to 5). All the codetections were reported with similar MPM values, and 8 (67%) were codetections of closely related species (4 N. exalbida/gamkensis, 3 N. elegans/nova/africana, 1 N. kruczakiae/violaceofusca/aobensis) previously reported to be indistinguishable by mcfDNA sequencing (31, 32).
FIG 5.
Frequency distribution of Nocardia spp. detections, n = 76 (<1% bacterial detections, n = 16,203), April 2018 to September 2021. An asterisk (*) indicates species more frequently isolated from clinical specimens (69). A dagger (†) indicates species-specific susceptibility patterns (51).
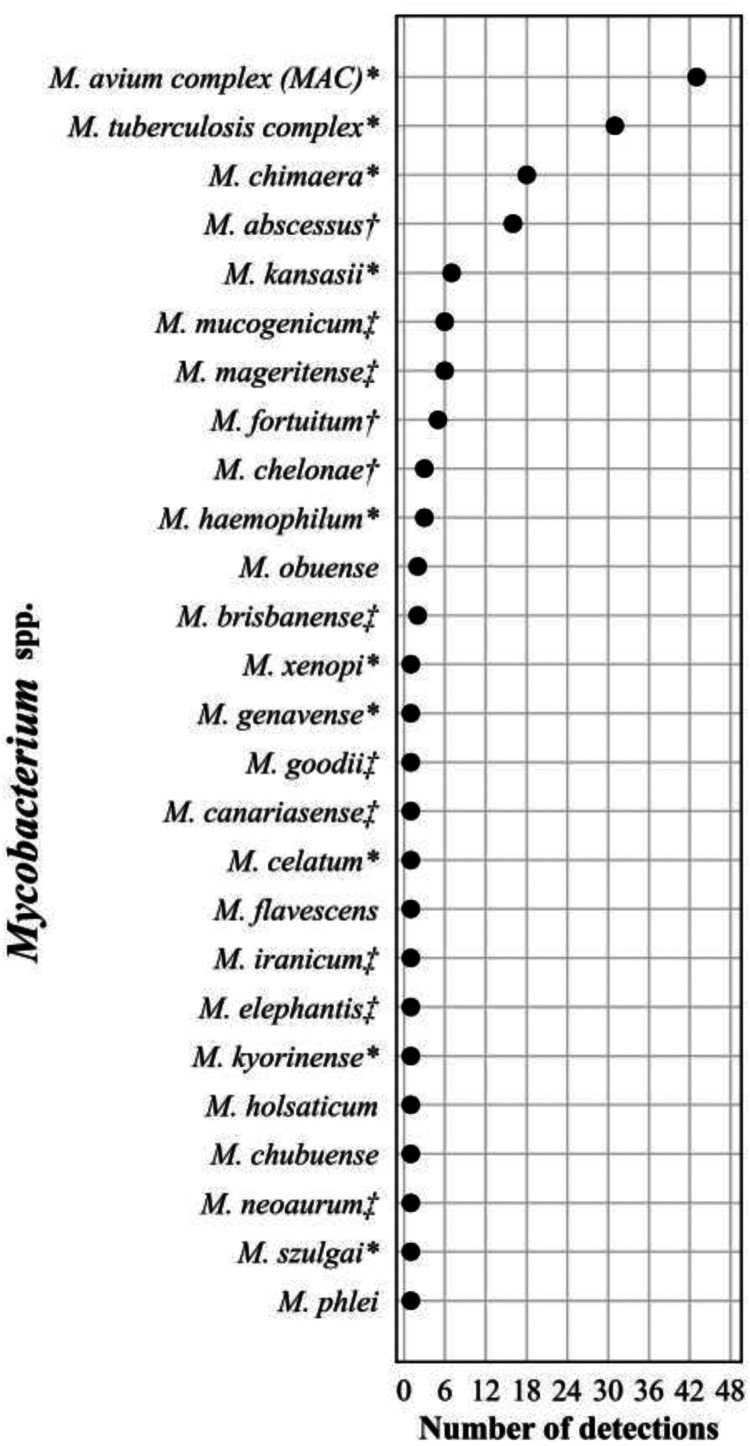
The frequency distribution of the 156 Mycobacterium spp. detections is shown in Fig. 6. Plasma mcfDNA sequencing detected 107 (69%) slowly growing mycobacteria (SGM) and 49 (31%) rapidly growing mycobacteria (RGM). The frequency of species distributions of these three genera showed similarities in that several species were predominant, followed by long tails of uncommon to single-species detections. We reported ≥2 concurrent species (range 2 to 6) in 6 (4%) of the 144 reports, including Mycobacterium spp. All the codetections were reported with similar MPM values. Three of the reports contained M. avium complex/chimera and one each M. avium complex/celatum/kyorinense, M. brisbanense/mucogenicum/obuense, and M. chubuense/elephantis/flavescens/goodii/holsaticum/phlei.
FIG 6.
Frequency distribution of Mycobacterium spp. detections, n = 156 (1% of all bacterial detections, n = 16,203), April 2018 to September 2021. Asterisk (*) indicates slowly growing mycobacteria of established clinical significance (70). Dagger (†) indicates rapidly growing mycobacteria considered common human pathogens. Double dagger (‡) indicates rapidly growing mycobacteria considered less common or rare human pathogens (71).
The codetections of multiple Legionella, Nocardia, and Mycobacterium spp., respectively, within the same sample and their resolution are shown in Table S3. We contrasted the pattern we would expect to see in the alignments in three hypothetical scenarios: (i) a true coinfection of two or more species in the Karius database, (ii) a single species in the Karius database, and (iii) a single species not in the Karius database. In each scenario, we expected certain relative proportions of reads aligning uniquely to each species in the database: those shared among closely related species, those shared among distantly related species, and those aligning more broadly across the entire genus or family. We also considered the BLAST percent identity of the alignments for each scenario and subset of reads. In 19 cases, a single species not in the Karius database seemed to fit best, while the remaining case represented a true codetection of 3 different mycobacteria species (M. brisbanense, M. mucogenicum, and M. obuense) (data not shown).
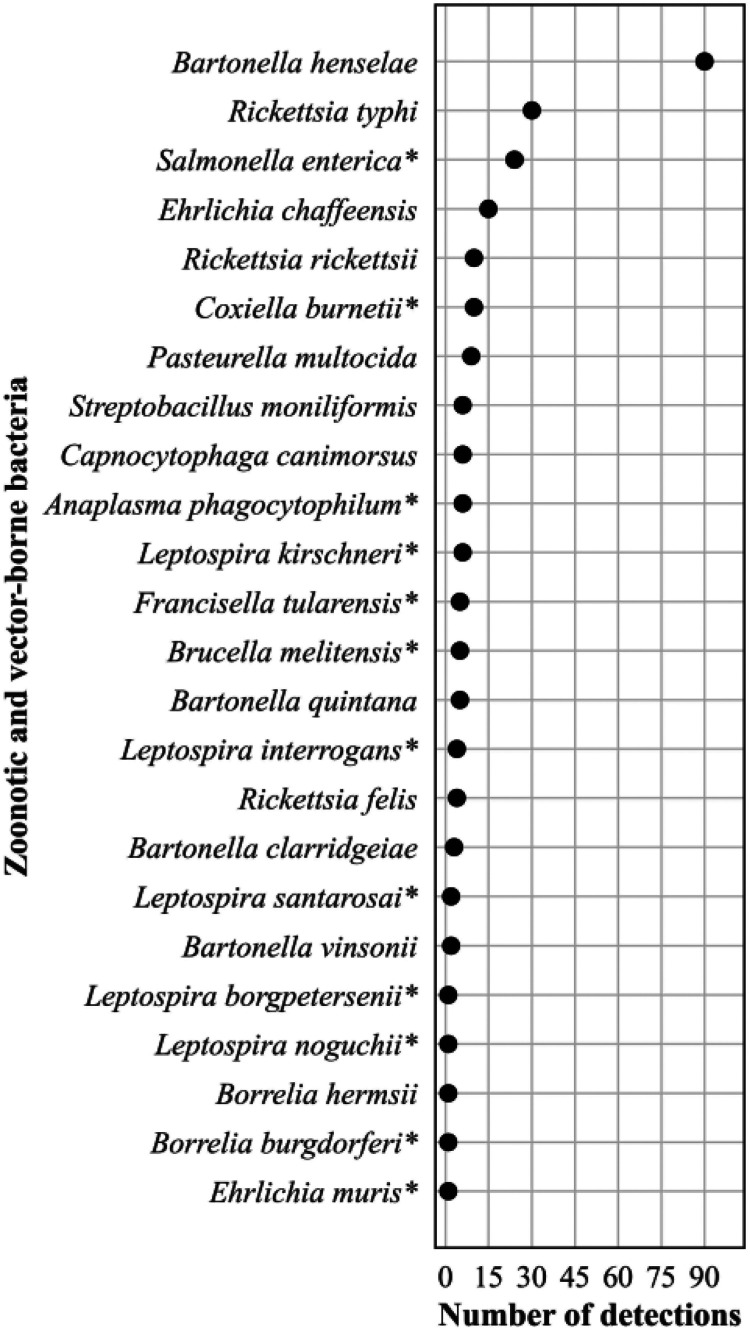
The frequency distribution of the 247 (3% of all bacterial detections) zoonotic and vector-borne bacterial detections are shown in Fig. 7. Bartonella henselae predominated with 90 (36%) detections.
FIG 7.
Frequency distribution of zoonotic and vector-borne bacteria detections, n = 247 (2% of all bacterial detections, n = 16,203), April 2018 to September 2021. Asterisk (*) indicates bacteria causing a nationally notifiable infectious disease (72).
Fungi.
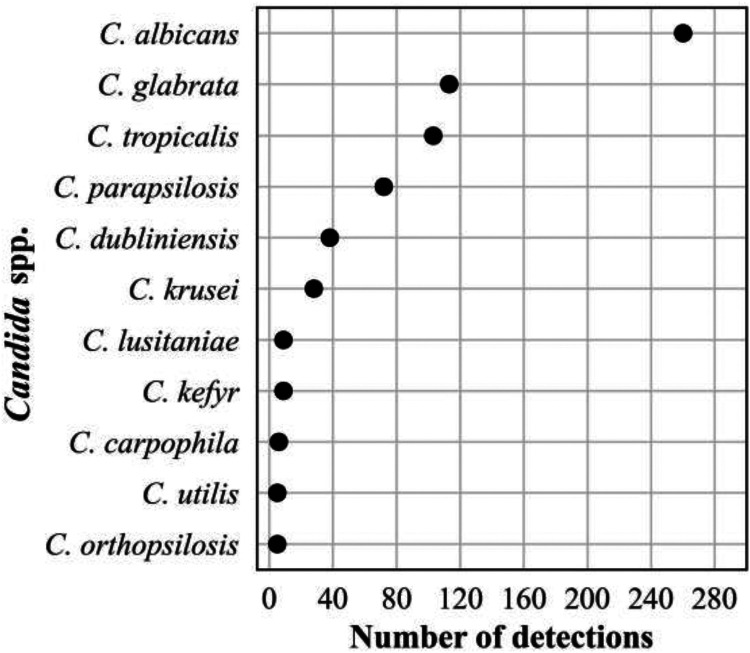
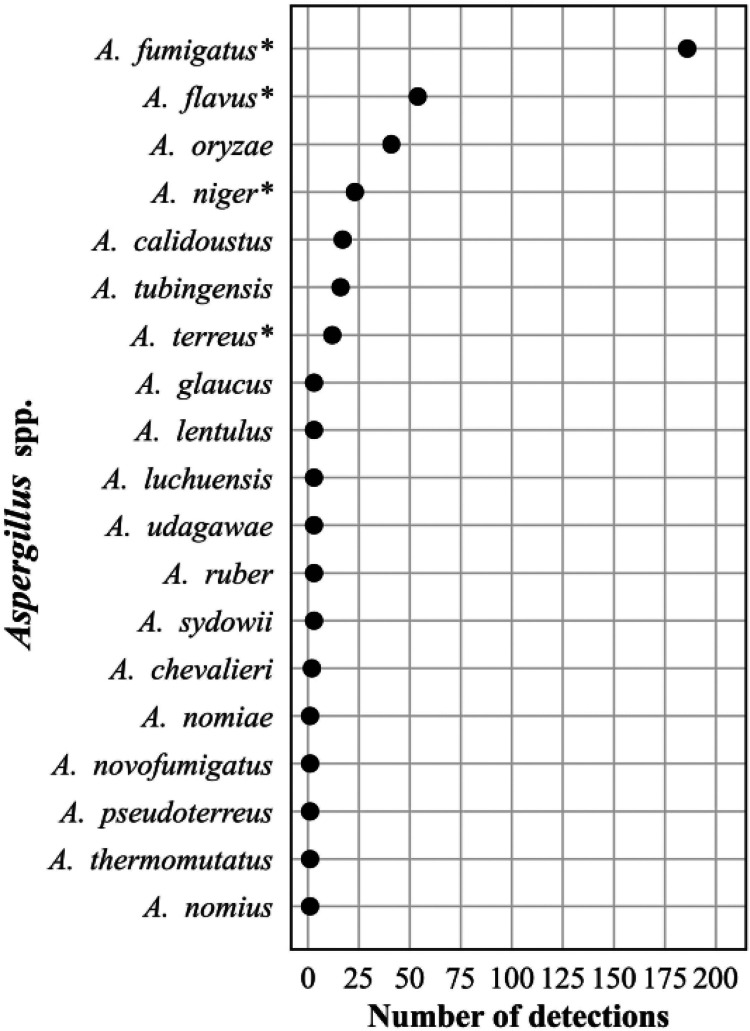
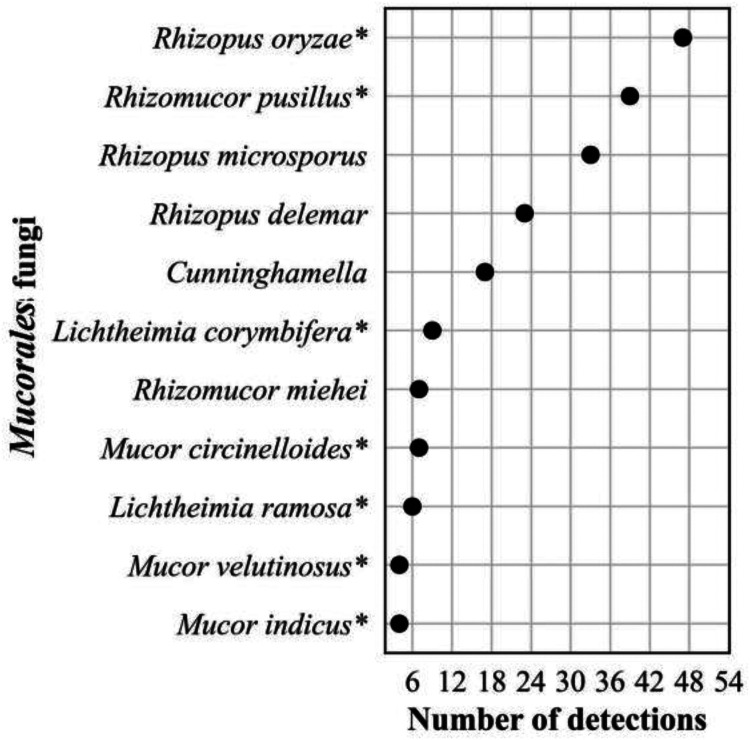
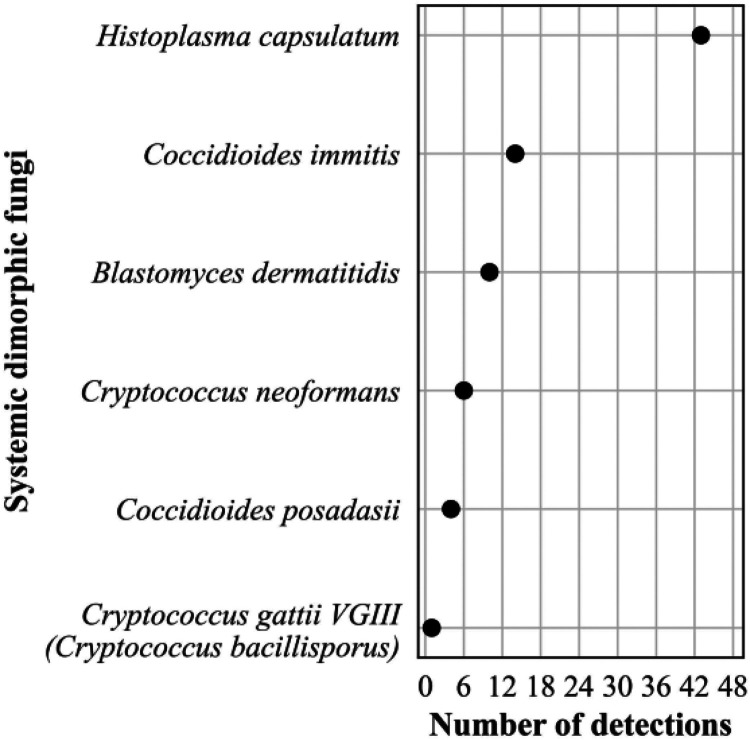
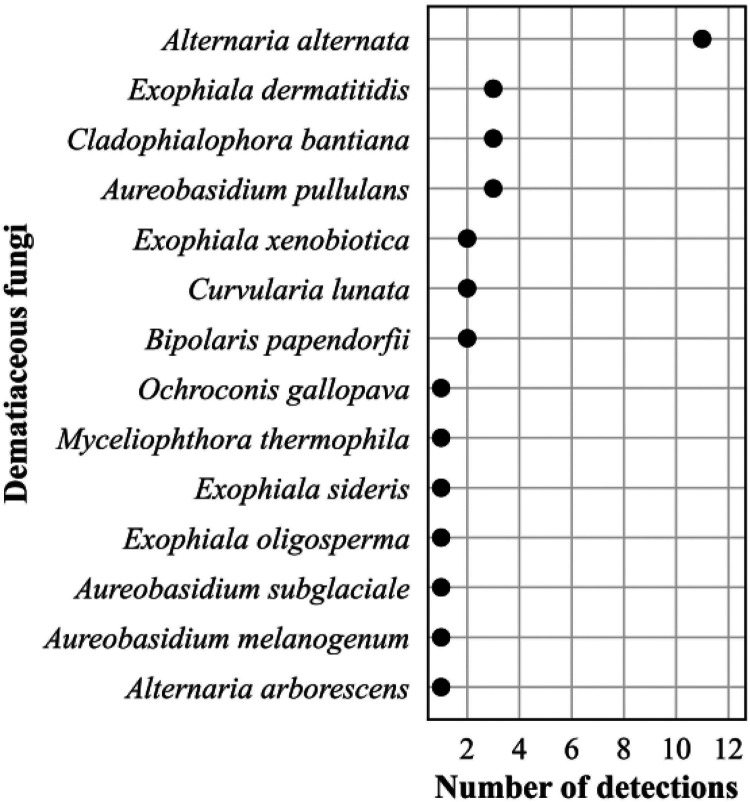
The frequency distribution of the 632 Candida spp. detections is shown in Fig. 8; 374 Aspergillus spp. detections in Fig. 9; 196 detections in the order Mucorales in Fig. 10; 78 detections of the systemic dimorphic fungi in Fig. 11; and 33 detections of dematiaceous fungi in Fig. 12. We detected 9 microsporidia, including 5 Enterocytozoon bieneusi and one each of E. cuniculi, E. hellem, Anncaliia algerae, and Vittaforma corneae. In addition, Pneumocystis jirovecii (258 detections) was among the top 50 taxa detected.
FIG 8.
Frequency distribution of Candida spp. detections, n = 648 (36% of all fungal detections, n = 1,776), April 2018 to September 2021.
FIG 9.
Frequency distribution of Aspergillus spp. detections, n = 374 (21% of all fungal detections, n = 1,776), April 2018 to September 2021. Asterisk (*) indicates most common pathogenic species (73).
FIG 10.
Frequency distribution of detections in the order Mucorales, n = 196 (11% of all fungal detections, n = 1,776), April 2018 to September 2021. Asterisk (*) indicates taxa implicated in human mucormycosis (74).
FIG 11.
Frequency distribution of detections of systemic dimorphic fungi, n = 78 (4% of all fungal detections, n = 1,776), April 2018 to September 2021.
FIG 12.
Frequency distribution of detections of dematiaceous fungi, n = 33 (2% of all fungal detections, n = 1,776), April 2018 to September 2021.
Eukaryotic parasites.
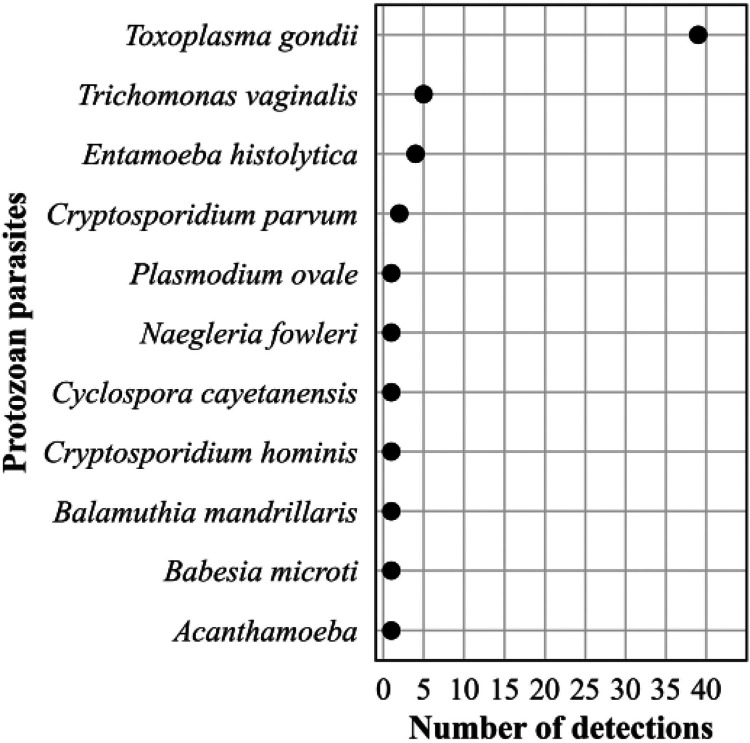
The frequency distribution of the 57 (89% of 64 parasite detections) protozoa is shown in Fig. 13. Among the protozoan parasite detections, 68% were Toxoplasma gondii, and 14% were pathogenic amoebae. Among the 7 (11%) helminthic parasites, we detected 4 nematodes (all Strongyloides stercoralis), 2 cestodes (both Echinococcus multilocularis), and 1 trematode (Schistosoma mansoni).
FIG 13.
Frequency distribution of detections of protozoan parasites, n = 57 (89% of all eukaryotic parasite detections, n = 64), April 2018 to September 2021.
DISCUSSION
We report the largest testing cohort of patients in which mcfDNA was identified and quantified. The key clinical laboratory quality metrics in this large cohort mirror what was reported with a much smaller cohort in the initial validation study (10), demonstrating that the Karius Test is robust and can be performed at scale in a clinically relevant time frame. These mcfDNA sequencing data reaffirm the ubiquity of some infections, as commonly expected microbes were detected in most patients while less common microbes were rarely detected. However, notably, the unbiased approach of the plasma mcfDNA sequencing made those “rare” detections possible, whereas conventional diagnostics require targeting specific organisms. Identifying optimal utilization, both clinical indications and timing, of the plasma mcfDNA sequencing in future studies to augment clinical decision-making and integration into current testing algorithms (17, 18, 33–35) will serve to improve the utility of the test. Examples of such studies include two recently completed prospective, observational clinical trials of the use of the Karius Test to diagnose pneumonia in immunocompromised patients (NCT04047719) (36) and infections in stem cell transplant inpatients and outpatients over time (NCT02804464) (26), respectively.
Our finding that 58% of the Karius Test reports identified ≥1 pathogen is substantially lower than the 70 to 85% reported for the test when applied to well-defined clinical uses (13, 14, 37). These findings support the need for diagnostic stewardship and additional clinical studies demonstrating how yield differs and should be carefully interpreted according to the specific patient population and disease prevalence considered.
The Karius Test provides absolute quantification of reported microbes in MPM. Generally higher concentrations have been found in confirmed infections, but since considerable overlap of MPM values exists with unlikely infections and with the asymptomatic cohort, threshold values for confirmed infections have not been established (12). However, following the decay of mcfDNA quantitatively by serial testing may have important implications for individual patient management in assessing the effectiveness of antimicrobial therapy and other medical or surgical interventions (8, 38, 39), but needs further study. MPM values can be influenced by several factors specific to the microbial genome such as turnover rate and genome size (12). Confounding patient variables (e.g., infection site, therapeutic interventions, and immune status) may also influence this measure. Lack of the clinical context prevented analyzing the data from those patients who had repeated testing to determine whether these tests were performed serially to diagnose new suspected infections, monitor therapy response, or conduct pathogen surveillance among immunocompromised patients as suggested by others in previous studies. Even with the clinical context, the data would not shed light on reproducibility of the test since plasma mcfDNA has a short half-life (16 min to 2 h) and may increase as infection progresses or decreases in response to therapeutic interventions. The precision (reproducibility) of mcfDNA sequencing was assessed in the analytical validation by testing 13 representative infections in duplicate on each of 20 days; the within run and between run coefficients of variation for MPM values were 18.2% and 19.2%, respectively (12).
Notable among the most common detections were the many commensal bacterial and fungal pathogens that cause serious and often invasive infections in patients with relevant risk factors (e.g., immunocompromised). Among the DNA viruses most detected by the Karius Test (e.g., Herpesviridae), all could represent latent infection, reactivation, or active infections depending on the respective clinical conditions; regardless, the detection of these viruses may be of particular concern for immunocompromised patients for whom they may cause considerable morbidity (40). TTV has a remarkable ability to produce chronic infections with no clearly associated clinical manifestations, so they are generally considered orphan viruses. However, detection and quantification of TTV in plasma have proven clinically useful in assessment of the kinetics of functional immune competence in a variety of solid organ transplant patients (41). Considering TTV accounts for about 70% of the total human plasma virome, their detection among the top viruses detected is not surprising. A limitation of the Karius Test is that it does not detect RNA viruses.
A key benefit of unbiased mcfDNA sequencing identified in this study is the ability to detect diagnostically challenging microbes such as opportunistic and systemic dimorphic fungi and zoonotic and vector-borne pathogens (42). These pathogens often carry a sense of urgency for the management of the individual patient and even for the public’s health, as they may be associated with considerable morbidity and potential mortality. As some are uncommonly expected pathogens, they present a major challenge for clinicians in considering and ordering appropriate SOC testing to capture all possible pathogens. For the laboratories, the microbiologic diagnosis of these infections often represents a major challenge for SOC methods, as has been described by others (43, 44).
Plasma mcfDNA sequencing has the potential to enhance disease surveillance, as illustrated by the systemic dimorphic fungi detections. At 43 detections (55% of dimorphic fungal identifications), Histoplasma capsulatum predominated, very likely related to its wide geographic range and opportunities for environmental exposure and mirrors what is known about the epidemiology and incidence of systemic dimorphic fungal infections (45). The relative frequency of detections of Coccidioides immitis and posadasii and Blastomyces dermatitidis may have been influenced by the geographic bias in this study sample cohort. Of the nine Cryptococcus spp. detected, gattii was detected only once compared with neoformans, reflecting its restricted geographic distribution and the overall rarity of infections in the United States (46).
Among the rarely detected microbes during the study period are some deserving further mention. The detection of Legionella, an obligate pathogen, signals public health concern, whether community or hospital acquired. L. pneumophila serogroup 1 (LP1) is estimated to cause 84% of community acquired Legionnaires’ disease (LD) (47); however, other serogroups of L. pneumophila and other Legionella spp. may cause 60% of hospital-acquired LD (48). Urinary antigen detection is the most common diagnostic in the United States and Europe; yet, given its specificity for LP1, as many as 40 to 50% of patients with non-LP1 legionellosis could be missed (49, 50). Plasma mcfDNA sequencing provides comprehensive testing for Legionella spp. and Legionella-like organisms in one diagnostic test and could thereby expand the known LD epidemiology, particularly in nosocomial cases.
Nocardia has 8 species-specific drug susceptibility patterns (51). While accurate species identification can predict antimicrobial susceptibility patterns, molecular methods are required for accurate identifications but are not widely available. The Karius Test detected in our cohort 7 of the 8 species with recognized susceptibility patterns and can provide results more rapidly than existing approaches to species identification.
Finally, culture for mycobacteria, while a complicated and lengthy process, is considered the gold standard, supplemented by direct detection of M. tuberculosis complex by nucleic acid amplification tests (NAATs) in many laboratories. However, NAATs for the direct detection of nontuberculous mycobacteria are not widely available, and accurate identification to species level from cultured isolates remains challenging for most laboratories. The M. chimaera may be overrepresented in our cohort since the Karius Test was optimized for its detection following reports of infections occurring after surgeries employing contaminated cardiopulmonary bypass devices (52). However, these detections demonstrate the capability of mcfDNA sequencing to provide comprehensive identification of these important obligate and commensal pathogens directly from plasma and provides additive diagnostic value to the above-mentioned SOC methods (53).
Plasma mcfDNA sequencing offers a noninvasive means of detecting microbial infection and capturing species diversity, potentially revealing new insights on genetic complexity not resolved by current taxonomic classification. Still, our findings highlight the need to expand currently available reference genomes, as many species across various genera remain undiscovered or undescribed (54, 55). The species codetections demonstrated among the genera we highlighted, Legionella, Nocardia, and Mycobacteria, were resolved as a single species not in the Karius database in all but one case, which was resolved as a true codetection of 3 different Mycobacteria species. We estimate that codetections of genetically similar microbes as described above occurred in ≤3% of all detections in the CRR. Similar challenges exist for broad-range PCR testing (5, 56) and occurred with adoption of proteomic identification by MALDI-TOF mass spectrometry (57), but the scope of the problem occurs less with next-generation sequencing (NGS) approaches.
This study has several limitations preventing us from directly comparing Karius with orthogonal SOC microbiological test results and fully elucidating its impact on patient care. While infectious disease physicians at Karius routinely communicated with ordering clinical providers regarding microbes identified as pathogens of special clinical significance (e.g., rare, and diagnostically challenging), any data collected were for the purposes of providing test interpretation support and facilitating in-house testing improvements. Karius did not have data use agreements with the over 200 referring institutions for publication purposes. However, numerous peer-reviewed publications have reported positive and negative percent agreement of plasma mcfDNA sequencing results with SOC microbiology results in identifying clinically adjudicated causes of infection across a broad range of patient populations and microbes. These studies have reported accuracy assessments on a microbe-by-microbe basis and have gone even further to report clinical validity as well. Six representative studies including 683 patients are summarized in Table S4 (8, 11, 12, 14, 58, 59).
In the studies above, the positive percent agreement (PPA) compared with SOC methods were similarly high (≥87%), but perhaps more importantly, plasma mcfDNA sequencing had higher diagnostic yields than SOC tests for diagnosis of the adjudicated causes of infections. The analytical sensitivity and breadth of microorganisms detected, combined with the diversity of the microbes across patients does present challenges in achieving high diagnostic specificity or NPA range (52.3 to 100%) compared with SOC methods. Even so, the initial analytical validation demonstrated very low levels of false reporting of mcfDNA not in the original plasma sample, consistent with high reproducibility of mcfDNA detection across independent runs (12). All samples tested in these previous publications were analyzed in the same laboratory using fundamentally the same wet bench procedures and computational algorithm as the samples reported here. Consequently, we are confident that the performance metrics in our test cohort are comparable with what has been previously reported.
The clinical context for using the Karius Test was provided in only 29% of patients and is therefore limited and likely biased as that information was voluntarily provided, may have been incomplete, and would reflect the clinician’s perspective at the time they ordered the test versus the final diagnosis. However, others have reported increased diagnostic yield of plasma mcfDNA sequencing compared with SOC tests in certain clinical scenarios (14–16, 60). Some have noted the test to be commonly applied in managing severely ill, especially IC, patients (61) who are more likely to be infected with unusual and pathogens that are difficult to diagnose (62, 63). The patient populations and disease states in these studies reflect this cohort’s most associated ICD-10 codes, thus providing insight about the real-world use of the test. In addition, infectious disease and hematology/oncology providers comprised most (78%) of the ordering clinicians, aligning with expectations that these clinicians commonly care for critically ill and IC patients with difficult-to-diagnose infections. Constraints of the underlying data structure prevented analyzing the data from the 12% of patients who had repeated testing to determine whether these tests were performed serially to diagnose new suspected infections or to monitor therapy response or conduct pathogen surveillance among immunocompromised patients as described by others (8, 26, 64–66). Further, the underlying data structure limited our ability to stratify the results by improvements to the bioinformatics pipeline over the course of the study period. However, the criteria for deciding whether a taxon was reported have only changed marginally during this study period.
Unbiased plasma mcfDNA sequencing can potentially enhance patient outcomes by direct and timely recognition of pathogens in specific clinical scenarios as well as benefit the overall public health by increasing our understanding of the epidemiology of emerging infectious diseases such as monkeypox virus (M.S. Lindner, K. Brick, N. Noll, S.Y. Park, et al., submitted for publication) or Borrelia miyamotoi (67). Further, this powerful, novel diagnostic tool may facilitate medical advances through recognizing previously unanticipated pathogens, as noted when mcfDNA sequencing was leveraged in a research-use-only modality to identify porcine cytomegalovirus infection in a patient who had received a genetically modified porcine-to-human cardiac transplant (68). As with any advanced diagnostic tool, careful, timely clinical application with expert guidance and interpretation of its results as well as appropriate diagnostic stewardship will optimize its application to offer even greater clinical impact. In addition, the development of robust clinical outcomes studies to evaluate the clinical impact and cost effectiveness of plasma mcfDNA sequencing for specific clinical indications to guide use remains a top diagnostic stewardship priority. Leading test use clinical indications reported in the literature include pneumonia in immunocompromised patients, fever of unknown origin, febrile neutropenia, invasive fungal infections, endocarditis, and complicated community acquired pneumonia. Also, the breath of pathogens detected in this study suggest mcfDNA sequencing may be useful as a single test for comprehensive diagnosis of zoonotic and vector-borne infections, systemic dimorphic fungal infections, legionellosis, and infections caused by other diagnostically challenging microbes.
This is the largest reported cohort of patients tested using plasma mcfDNA sequencing. As such, it represents the first report of a clinical grade metagenomic test performed at scale, with laboratory quality metrics that mirror what was reported in the initial validation study and demonstrates that plasma mcfDNA sequencing is robust and can be performed in a clinically relevant time frame. The large number of samples included also provides new insights into the breadth and complexity of pathogens identified with this test, not revealed in previously published studies.
Data availability.
The Karius Test is a proprietary laboratory-developed test, and thus there are several limitations on data availability that we declare here. Karius is unable to share the raw sequencing data underlying the test results evaluated here due to issues relating to patient privacy and consent. Additionally, the methodological details that can be provided to others to replicate the findings are limited due to intellectual property concerns. Taking these two limitations into account, summarized data used to reach the conclusions in this study are included in the manuscript and supplemental material.
ACKNOWLEDGMENTS
We thank the talented, experienced, and dedicated clinical laboratory operations and analytics teams at Karius for generating the test results and sequencing analyses, respectively, that were foundational to this study. We also thank T. Matthew Hill, PharmD, PhD, formerly with Karius, Inc., and Asim A. Ahmed, MD for their early contributions to the paper.
The following authors declare a conflict of interest: Sarah Y. Park, Eliza J. Chang, Martin S. Lindner, Shivkumar Venkatasubrahmanyam, Judith C. Wilber, Marla Lay Vaughn, Sivan Bercovici, Bradley A. Perkins, and Frederick S. Nolte are all Karius employees, which markets the commercial test evaluated in this study who shared responsibilities for design of the study, analysis of the data, and writing and editing of the manuscript. Nathan Ledeboer and Kevin Messacar both served as uncompensated consultants for Karius on the manuscript and participated in writing and editing of the manuscript. Their relationship with Karius has been reviewed and approved by the Medical College of Wisconsin and the University of Colorado, Children’s Hospital of Colorado, respectively, in accordance with their conflict-of-interest policies.
This study was solely supported by Karius.
Footnotes
Supplemental material is available online only.
Contributor Information
Frederick S. Nolte, Email: rick.nolte@kariusdx.com.
John P. Dekker, National Institute of Allergy and Infectious Diseases
REFERENCES
- 1.Levy SE, Myers RM. 2016. Advancements in next-generation sequencing. Annu Rev Genomics Hum Genet 17:95–115. doi: 10.1146/annurev-genom-083115-022413. [DOI] [PubMed] [Google Scholar]
- 2.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Kowarsky M, Camunas-Soler J, Kertesz M, De Vlaminck I, Koh W, Pan W, Martin L, Neff NF, Okamoto J, Wong RJ, Kharbanda S, El-Sayed Y, Blumenfeld Y, Stevenson DK, Shaw GM, Wolfe ND, Quake SR. 2017. Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA. Proc Natl Acad Sci USA 114:9623–9628. doi: 10.1073/pnas.1707009114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewinski M, Alby K, Babady E, Butler-Wu S, Dien Bard J, Greninger A, Hanson K, Naccache S, Newton D, Temple-Smolkin RL, Nolte F. in press. Exploring the utility of multiplex infectious disease panel testing for diagnosis of infection in different body sites: a joint report of the association for molecular pathology, american society for microbiology, infectious diseases society of america, and pan american society for clinical virology. The J Molecular Diagnostics. [DOI] [PubMed] [Google Scholar]
- 5.Naureckas Li C, Nakamura MM. 2022. Utility of broad-range PCR sequencing for infectious diseases clinical decision making: a pediatric center experience. J Clin Microbiol 60:e0243721. doi: 10.1128/jcm.02437-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM, Group Q . 2011. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Zhang Q, Dong YQ, Yin J, Qiu YQ. 2022. Diagnostic accuracy of metagenomic next-generation sequencing in diagnosing infectious diseases: a meta-analysis. Sci Rep 12:21032. doi: 10.1038/s41598-022-25314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichenberger EM, Degner N, Scott ER, Ruffin F, Franzone J, Sharma-Kuinkel B, Shah P, Hong D, Dalai SC, Blair L, Hollemon D, Chang E, Ho C, Wanda L, de Vries C, Fowler VG, Ahmed AA. 2023. Microbial cell-free DNA identifies the causative pathogen in infective endocarditis and remains detectable longer than conventional blood culture in patients with prior antibiotic therapy. Clin Infect Dis 76:e1492–e1500. doi: 10.1093/cid/ciac426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dworsky ZD, Lee B, Ramchandar N, Rungvivatjarus T, Coufal NG, Bradley JS. 2022. Impact of cell-free next-generation sequencing on management of pediatric complicated pneumonia. Hosp Pediatr 12:377–384. doi: 10.1542/hpeds.2021-006361. [DOI] [PubMed] [Google Scholar]
- 10.Brenner T, Skarabis A, Stevens P, Axnick J, Haug P, Grumaz S, Bruckner T, Luntz S, Witzke O, Pletz MW, Ruprecht TM, Marschall U, Altin S, Greiner W, Berger MM, Group T, TIFOnet Critical Care Trials Group . 2021. Optimization of sepsis therapy based on patient-specific digital precision diagnostics using next generation sequencing (DigiSep-Trial)-study protocol for a randomized, controlled, interventional, open-label, multicenter trial. Trials 22:714. doi: 10.1186/s13063-021-05667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han D, Li R, Shi J, Tan P, Zhang R, Li J. 2020. Liquid biopsy for infectious diseases: a focus on microbial cell-free DNA sequencing. Theranostics 10:5501–5513. doi: 10.7150/thno.45554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blauwkamp TA, Thair S, Rosen MJ, Blair L, Lindner MS, Vilfan ID, Kawli T, Christians FC, Venkatasubrahmanyam S, Wall GD, Cheung A, Rogers ZN, Meshulam-Simon G, Huijse L, Balakrishnan S, Quinn JV, Hollemon D, Hong DK, Vaughn ML, Kertesz M, Bercovici S, Wilber JC, Yang S. 2019. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 4:663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 13.Rossoff J, Chaudhury S, Soneji M, Patel SJ, Kwon S, Armstrong A, Muller WJ. 2019. Noninvasive diagnosis of infection using plasma next-generation sequencing: a single-center experience. Open Forum Infect Dis 6. doi: 10.1093/ofid/ofz327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benamu E, Gajurel K, Anderson JN, Lieb T, Gomez CA, Seng H, Aquino R, Hollemon D, Hong DK, Blauwkamp TA, Kertesz M, Blair L, Bollyky PL, Medeiros BC, Coutre S, Zompi S, Montoya JG, Deresinski S. 2022. Plasma microbial cell-free DNA next generation sequencing in the diagnosis and management of febrile neutropenia. Clin Infect Dis 74:1659–1668. doi: 10.1093/cid/ciab324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Diaz JD, Goldstein SC, Patel RD, Varela JC, Reyenga C, Smith M, Smith T, Balls J, Ahmad S, Mori S. 2021. Impact of next-generation sequencing cell-free pathogen DNA test on antimicrobial management in adults with hematological malignancies and transplant recipients with suspected infections. Transplant Cell Ther doi: 10.1016/j.jtct.2021.02.025. [DOI] [PubMed] [Google Scholar]
- 16.Francisco DMA, Woc-Colburn L, Carlson TJ, Lasco T, Barrett MB, Mohajer MA. 2020. 680. The use of plasma next-generation sequencing test in the management of immunocompetent and immunocompromised patients – a single center retrospective study. Open Forum Infect Dis 7:S393–S394. doi: 10.1093/ofid/ofaa439.872. [DOI] [Google Scholar]
- 17.Hogan CA, Yang S, Garner OB, Green DA, Gomez CA, Dien Bard J, Pinsky BA, Banaei N. 2021. Clinical impact of metagenomic next-generation sequencing of plasma cell-free DNA for the diagnosis of infectious diseases: a multicenter retrospective cohort study. Clin Infect Dis 72:239–245. doi: 10.1093/cid/ciaa035. [DOI] [PubMed] [Google Scholar]
- 18.Lee RA, Al Dhaheri F, Pollock NR, Sharma TS. 2020. Assessment of the clinical utility of plasma metagenomic next-generation sequencing in a pediatric hospital population. J Clin Microbiol 58. doi: 10.1128/JCM.00419-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niles DT, Wijetunge DSS, Palazzi DL, Singh IR, Revell PA. 2020. Plasma metagenomic next-generation sequencing assay for identifying pathogens: a retrospective review of test utilization in a large children's hospital. J Clin Microbiol 58. doi: 10.1128/JCM.00794-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Center for Health Statistics (CDC). 2015. International Classification of Diseases, Tenth Revision, Clinical Modification (ICD-10-CM). https://ftp.cdc.gov/pub/Health_Statistics/NCHS/Publications/ICD10CM/2022/icd10cm-tabular-2022-April-1.pdf. [PubMed] [Google Scholar]
- 21.Agency for Healthcare Research and Quality. 2021. Clinical Classifications Software Refined (CCSR). Healthcare Cost and Utilization Project (HCUP). Rockville, MD. https://www.hcup-us.ahrq.gov/toolssoftware/ccsr/ccs_refined.jsp. [Google Scholar]
- 22.Agency for Healthcare Research and Quality. July 2021. Immunocompromised state diagnosis and procedure codes. https://qualityindicators.ahrq.gov/Downloads/Modules/PQI/V2021/TechSpecs/PQI_Appendix_C.pdf.
- 23.Agency for Healthcare Research and Quality. July 2021. Intermediate-risk immunocompromised state diagnosis codes. https://qualityindicators.ahrq.gov/Downloads/Modules/PDI/V2021/TechSpecs/PDI_Appendix_G.pdf.
- 24.Agency for Healthcare Research and Quality. July 2021. High-risk immunocompromised state diagnosis and procedure codes. https://qualityindicators.ahrq.gov/Downloads/Modules/PDI/V2021/TechSpecs/PDI_Appendix_F.pdf.
- 25.Blauwkamp TA, Ho C, Seng H, Hollemon D, Blair L, Yao JD, Warren D, Russell P, Davis T, Hong DK. 2019. Evaluation of Karius plasma next generation sequencing of microbial cell-free DNA to detect and quantitate Cytomegalovirus, Epstein-Barr Virus, and BK Virus. American Society for Microbiology (ASM) Microbe 2019, San Francisco, CA. [Google Scholar]
- 26.Fung M, Teraoka J, Lien K, Seng H, Parham A, Hollemon D, Hong DK, Blair L, Zompì S, Logan AC, Yao JD, Chin-Hong P. 2019. Use of the quantitative Karius plasma next generation sequencing cell-free pathogen DNA test to detect and monitor Cytomegalovirus infection in allogeneic stem-cell transplant recipients, abstr 2019 TCT: Transplantation & Cellular Therapy Meetings of ASBMT and CIBMTR, Houston, TX,
- 27.Relman DA, Falkow S, Ramakrishnan L. 2020. Chapter 1: a Molecular Perspective of Microbial Pathogenicity, p 1–11. In Bennett JE, Bennett JE, Dolin R, Blaser MJ (ed), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 9th edition, vol 1. Elsevier, Philadelphia, PA. [Google Scholar]
- 28.McKinney W. Data structures for statistical computing in python. 2010. abstr 9th Python in Science Conference, Austin, TX. doi: 10.25080/Majora-92bf1922-00a. [DOI] [Google Scholar]
- 29.Hunter J. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 30.Waskom M, Botvinnik O, O'Kane D, Hobson P, Lukauskas S, Gemperline DC, Augspurger T, Halchenko Y, Cole JB, Warmenhoven J, de Ruiter J, Pye C, Hoyer S, Vanderplas J, Villalba S, Kunter G, Quintero E, Bachant P, Martin M, Meyer K, Miles A, Ram Y, Yarkoni T, Williams ML, Evans C, Fitzgerald C, Fonnesbeck C, Lee A, Qalieh A. 2017. mwaskom/seaborn: v0.8.1 (September 2017), September 3, 2017 ed Zenodo. doi: 10.5281/zenodo.883859. [DOI] [Google Scholar]
- 31.Ahmed AA, Rosen M, Hong DK, Dalai SC, Macintyre A, Blair L, Lindner M, Balakrishnan S, Bercovici S. 2019. Next-generation sequencing of pathogen cell-free DNA in plasma (Karius Test) reveals Nocardia species diversity in clinical infections, abstr ASM Microbe, San Francisco, CA, June 20–24, 2019. [Google Scholar]
- 32.Smollin M, Lindner M, Blair L, Arun A, Degner N, Equils O, de Vries CR, Dalai SC, MacIntyre A, Ahmed AA. 2021. Rapid, non-invasive detection of Legionella and Nocardia and resolution of species diversity in clinical infections in immunocompromised hosts using the Karius Test, a plasma-based microbial cell-free DNA sequencing test for pathogen identification, abstr 21st ICHS Symposium, Melbourne, Australia, February 17–19, 2021. [Google Scholar]
- 33.Graff K, Dominguez SR, Messacar K. 2021. Metagenomic next-generation sequencing for diagnosis of pediatric meningitis and encephalitis: a review. J Pediatric Infect Dis Soc 10:S78–S87. doi: 10.1093/jpids/piab067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haslam DB. 2021. Future applications of metagenomic next-generation sequencing for infectious diseases diagnostics. J Pediatric Infect Dis Soc 10:S112–S117. doi: 10.1093/jpids/piab107. [DOI] [PubMed] [Google Scholar]
- 35.Huang AL, Hendren N, Carter S, Larsen C, Garg S, La Hoz R, Farr M. 2022. Biomarker-based assessment for infectious risk before and after heart transplantation. Curr Heart Fail Rep 19:236–246. doi: 10.1007/s11897-022-00556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergin SP, Chemaly R, Duttagupta R, Bigelow R, Dadwal S, Hill JA, Lee YJ, Haidar G, Luk A, Drelick A, Chin-Hong PV, Benamu E, Davis T, Wolf O, McClain M, Maziarz E, Madut D, Bedoya A, Gilstrap DL, Todd J, Barkauskas C, Spallone A, McDowell BJ, Small CB, Shariff D, Salsgiver E, Khawaja F, Papanicolaou GA, Spagnoletti J, Van Besien K, English M, Fung M, Rusell P, Ibrahimi S, Pandey S, Adams S, Liang W, Visweswaran A, Ho C, Nemirovich-Danchenko E, Braaten J, Sundermann L, Mughar M, Chavez R, Romano R, Montgomery S, Kumar S, Dalai SC, Cho Y, Ahmed AA, et al. 2022. PICKUP: pneumonia in the immunocompromised - use of the Karius test for the detection of undiagnosed pathogens abstr IDWeek 2022, Washington, D.C., October 20, 2022.
- 37.Farnaes L, Wilke J, Ryan Loker K, Bradley JS, Cannavino CR, Hong DK, Pong A, Foley J, Coufal NG. 2019. Community-acquired pneumonia in children: cell-free plasma sequencing for diagnosis and management. Diagn Microbiol Infect Dis 94:188–191. doi: 10.1016/j.diagmicrobio.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eichenberger EM, de Vries CR, Ruffin F, Sharma-Kuinkel B, Park L, Hong D, Scott ER, Blair L, Degner N, Hollemon DH, Blauwkamp TA, Ho C, Seng H, Shah P, Wanda L, Fowler VG, Ahmed AA. 2022. Microbial cell-free DNA identifies etiology of bloodstream infections, persists longer than conventional blood cultures, and its duration of detection is associated with metastatic infection in patients with staphylococcus aureus and gram-negative bacteremia. Clin Infect Dis 74:2020–2027. doi: 10.1093/cid/ciab742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.To RK, Ramchandar N, Gupta A, Pong A, Cannavino C, Foley J, Farnaes L, Coufal NG. 2021. Use of plasma metagenomic next-generation sequencing for pathogen identification in pediatric endocarditis. Pediatr Infect Dis J 40:486–488. doi: 10.1097/INF.0000000000003038. [DOI] [PubMed] [Google Scholar]
- 40.de Melo Silva J, Pinheiro-Silva R, Dhyani A, Pontes GS. 2020. Cytomegalovirus and epstein-barr infections: prevalence and impact on patients with hematological diseases. Biomed Res Int 2020:1627824. doi: 10.1155/2020/1627824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Focosi D, Antonelli G, Pistello M, Maggi F. 2016. Torquetenovirus: the human virome from bench to bedside. Clin Microbiol Infect 22:589–593. doi: 10.1016/j.cmi.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Hilt EE, Ferrieri P. 2022. Next generation and other sequencing technologies in diagnostic microbiology and infectious diseases. Genes 13:1566. doi: 10.3390/genes13091566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaca DJ, Dobler G, Fischer SF, Keller C, Konrad M, von Loewenich FD, Orenga S, Sapre SU, van Belkum A, Kempf VAJ. 2022. Contemporary diagnostics for medically relevant fastidious microorganisms belonging to the genera Anaplasma, Bartonella, Coxiella, Orientia, and Rickettsia. FEMS Microbiol Rev doi: 10.1093/femsre/fuac013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haydour Q, Hage CA, Carmona EM, Epelbaum O, Evans SE, Gabe LM, Knox KS, Kolls JK, Wengenack NL, Prokop LJ, Limper AH, Murad MH. 2019. Diagnosis of fungal infections. a systematic review and meta-analysis supporting american thoracic society practice guideline. Ann Am Thorac Soc 16:1179–1188. doi: 10.1513/AnnalsATS.201811-766OC. [DOI] [PubMed] [Google Scholar]
- 45.Lockhart SR, Toda M, Benedict K, Caceres DH, Litvintseva AP. 2021. Endemic and other dimorphic mycoses in the americas. J Fungi (Basel) 7:151. doi: 10.3390/jof7020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention (CDC). C. gattii Infection Statistics. https://www.cdc.gov/fungal/diseases/cryptococcosis-gattii/statistics.html. Accessed June 5, 2022.
- 47.Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, Summersgill J, File T, Heath CM, Paterson DL, Chereshsky A. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis 186:127–128. doi: 10.1086/341087. [DOI] [PubMed] [Google Scholar]
- 48.Helbig JH, Uldum SA, Bernander S, Luck PC, Wewalka G, Abraham B, Gaia V, Harrison TG. 2003. Clinical utility of urinary antigen detection for diagnosis of community-acquired, travel-associated, and nosocomial legionnaires' disease. J Clin Microbiol 41:838–840. doi: 10.1128/JCM.41.2.838-840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fields BS, Benson RF, Besser RE. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev 15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phin N, Parry-Ford F, Harrison T, Stagg HR, Zhang N, Kumar K, Lortholary O, Zumla A, Abubakar I. 2014. Epidemiology and clinical management of Legionnaires' disease. Lancet Infect Dis 14:1011–1021. doi: 10.1016/S1473-3099(14)70713-3. [DOI] [PubMed] [Google Scholar]
- 51.CLSI. 2018. Performance Standards for Susceptibility Testing of Mycobacteria, Nocardia spp., and Other Aerobic Actinomycetes. CLSI Supplement M62. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 52.Sax H, Bloemberg G, Hasse B, Sommerstein R, Kohler P, Achermann Y, Rossle M, Falk V, Kuster SP, Bottger EC, Weber R. 2015. Prolonged outbreak of mycobacterium chimaera infection after open-chest heart surgery. Clin Infect Dis 61:67–75. doi: 10.1093/cid/civ198. [DOI] [PubMed] [Google Scholar]
- 53.Pollock NR, MacIntyre AT, Blauwkamp TA, Blair L, Ho C, Calderon R, Franke MF. 2021. Detection of Mycobacterium tuberculosis cell-free DNA to diagnose TB in pediatric and adult patients. Int J Tuber Lung Dis 25:403–405. doi: 10.5588/ijtld.21.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goldberg B, Sichtig H, Geyer C, Ledeboer N, Weinstock GM. 2015. Making the leap from research laboratory to clinic: challenges and opportunities for next-generation sequencing in infectious disease diagnostics. mBio 6:e01888-15–e01815. doi: 10.1128/mBio.01888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simner PJ, Miller S, Carroll KC. 2018. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis 66:778–788. doi: 10.1093/cid/cix881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wright WF, Simner PJ, Carroll KC, Auwaerter PG. 2022. Progress report: next-generation sequencing, multiplex polymerase chain reaction, and broad-range molecular assays as diagnostic tools for fever of unknown origin investigations in adults. Clin Infect Dis 74:924–932. doi: 10.1093/cid/ciab155. [DOI] [PubMed] [Google Scholar]
- 57.Steensels D, Verhaegen J, Lagrou K. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of bacteria and yeasts in a clinical microbiological laboratory: a review. Acta Clin Belg 66:267–273. [DOI] [PubMed] [Google Scholar]
- 58.Wilke J, Ramchandar N, Cannavino C, Pong A, Tremoulet A, Padua LT, Harvey H, Foley J, Farnaes L, Coufal NG. 2021. Clinical application of cell-free next-generation sequencing for infectious diseases at a tertiary children's hospital. BMC Infect Dis 21:552. doi: 10.1186/s12879-021-06292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Degner N, Vissichelli NC, Berman DM, Smollin M, Morales MK. 2023. Diagnostic value of plasma microbial cell-free DNA sequencing in hematopoietic stem cell transplant recipients: a systematic review and meta-analysis. medRxiv. doi: 10.1101/2023.01.12.22280967. [DOI] [Google Scholar]
- 60.Camargo JF, Ahmed AA, Lindner MS, Morris MI, Anjan S, Anderson AD, Prado CE, Dalai SC, Martinez OV, Komanduri KV. 2019. Next-generation sequencing of microbial cell-free DNA for rapid noninvasive diagnosis of infectious diseases in immunocompromised hosts. F1000Res 8:1194. doi: 10.12688/f1000research.19766.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edward P, Handel AS. 2021. Metagenomic next-generation sequencing for infectious disease diagnosis: a review of the literature with a focus on pediatrics. J Pediatric Infect Dis Soc 10:S71–S77. doi: 10.1093/jpids/piab104. [DOI] [PubMed] [Google Scholar]
- 62.Fishman JA. 2022. Approach to the immunocompromised patient with fever and pulmonary infiltrates. In Blumberg EA, Bond S (ed), UptoDate, Waltham, MA. [Google Scholar]
- 63.Pierce KK. 2013. Chapter 40 - immunocompromised host, p 277–292. In Parsons PE, Wiener-Kronish JP (ed), Critical Care Secrets (Fifth Edition) Mosby, Maryland Heights, MO. doi: 10.1016/B978-0-323-08500-7.00041-2. [DOI] [Google Scholar]
- 64.Goggin KP, Gonzalez-Pena V, Inaba Y, Allison KJ, Hong DK, Ahmed AA, Hollemon D, Natarajan S, Mahmud O, Kuenzinger W, Youssef S, Brenner A, Maron G, Choi J, Rubnitz JE, Sun Y, Tang L, Wolf J, Gawad C. 2020. Evaluation of plasma microbial cell-free DNA sequencing to predict bloodstream infection in pediatric patients with relapsed or refractory cancer. JAMA Oncol 6:552–556. doi: 10.1001/jamaoncol.2019.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Solanky D, Ahmed AA, Fierer J, Golts E, Jones M, Mehta SR. 2022. Utility of plasma microbial cell-free DNA decay kinetics after aortic valve replacement for Bartonella endocarditis: case report. Front Trop Dis 3:842100. doi: 10.3389/fitd.2022.842100. [DOI] [Google Scholar]
- 66.Ahsouri N, Nieves D, Singh J, Arrieta A. 2022. Serial microbial cell-free DNA next generation sequencing (NGS) as a means of diagnosis and monitoring of clinical response to treatment of invasive fungal infections (IFI) in immunocompromised pediatric patients. Open Forum Infectious Diseases 9. doi: 10.1093/ofid/ofac492.1171. [DOI] [Google Scholar]
- 67.Rubio LA, Kjemtrup AM, Marx GE, Cronan S, Kilonzo C, Saunders MEM, Pawloski JL, Dietrich EA, Liebman KA, Park SY. in press. A human case of Borrelia miyamotoi infection causing relapsing fever in an immunocompromised patient from California. Emerg Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Griffith BP, Goerlich CE, Singh AK, Rothblatt M, Lau CL, Shah A, Lorber M, Grazioli A, Saharia KK, Hong SN, Joseph SM, Ayares D, Mohiuddin MM. 2022. Genetically modified porcine-to-human cardiac xenotransplantation. N Engl J Med 387:35–44. doi: 10.1056/NEJMoa2201422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conville PS, Brown-Elliott BA, Witebsky FG. 2019. Nocardia, Rhodococcus, Gordonia, Actinomadura, Streptomyces, and other aerobic actinomycetes. In Carroll KC, Pfaller MA, Landry ML, McAdam AJ, Patel R, Richter SS, Warnock DW (ed), Manual of Clinical Microbiology, 12th ed ASM Press. [Google Scholar]
- 70.Caulfield AJ, Elvira Brown-Elliott BA, Wallace J, Richard J, Wengenack NL. 2019. Mycobacterium: laboratory characteristics of slowly growing mycobacteria other than Mycobacterium tuberculosis. In Carroll KC, Pfaller MA, Landry ML, McAdam AJ, Patel R, Richter SS, Warnock DW (ed), Manual of Clinical Microbiology, 12th ed ASM Press. [Google Scholar]
- 71.Brown-Elliott BA, Wallace J, Richard J. 2019. Mycobacterium: clinical and laboratory characteristics of rapidly growing mycobacteria. In Carroll KC, Pfaller MA, Landry ML, McAdam AJ, Patel R, Richter SS, Warnock DW (ed), Manual of Clinical Microbiology, 12th ed ASM Press. [Google Scholar]
- 72.Centers for Disease Control and Prevention (CDC). 2022. National notifiable infectious diseases. https://ndc.services.cdc.gov/search-results-year/. Accessed 1 March 2022.
- 73.Chen SC-A, Meyer W, Sorrell TC, Halliday CL. 2019. Aspergillus, Talaromyces, and Penicillium. In Carroll KC, Pfaller MA, Landry ML, McAdam AJ, Patel R, Richter SS, Warnock DW (ed), Manual of Clinical Microbiology, 12th ed. ASM Press. [Google Scholar]
- 74.Hoffmann K, Pawłowska J, Walther G, Wrzosek M, de Hoog GS, Benny GL, Kirk PM, Voigt K. 2013. The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia 30:57–76. doi: 10.3767/003158513X666259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1, S3, and S4. Download jcm.01855-22-s0001.docx, DOCX file, 0.04 MB (38.6KB, docx)
Table S2. Download jcm.01855-22-s0002.xlsx, XLSX file, 0.05 MB (53KB, xlsx)
Data Availability Statement
The Karius Test is a proprietary laboratory-developed test, and thus there are several limitations on data availability that we declare here. Karius is unable to share the raw sequencing data underlying the test results evaluated here due to issues relating to patient privacy and consent. Additionally, the methodological details that can be provided to others to replicate the findings are limited due to intellectual property concerns. Taking these two limitations into account, summarized data used to reach the conclusions in this study are included in the manuscript and supplemental material.