Abstract
Determining cellular DNA content is valuable in study of numerous biological processes, including organ development and injury repair. While FACS analysis of dissociated cells is a widely used method for assaying ploidy in a tissue cell population, for many tissue samples it is possible and convenient to measure ploidy in situ using light microscopy. Here, we present two protocols for measuring cellular ploidy in tissues. These protocols are based on our studies in Drosophila melanogaster, but are applicable to other settings as well. We present example results from Drosophila hindgut, midgut, and wing imaginal disc as examples. The first protocol focuses on measuring DNA content from decondensed interphase nuclei, while the second protocol details the visualization of condensed chromosomes for ploidy determination, either from mitotic cells or from interphase cells with drug-induced chromosome condensation. These techniques can be completed in one day and require readily available supplies as well as a fluorescence light microscope.
1. Introduction
Polyploidy, or whole genome duplication, is a widespread tissue property throughout evolution. As an example of its prevalence, polyploidy is now appreciated to occur in at least nine out of eleven human organ systems[1]. Polyploidy often arises in specific cell types during tissue development, or following tissue injury. Cellular ploidy levels can differ within an organism by over 1000-fold, and the degree of polyploidy can have important biological consequences[2, 3].
Here, we present two protocols for measuring cellular ploidy in situ. Both protocols use physical force-based tissue squashing of DAPI-stained nuclei to facilitate consistent and accurate DNA content measurements by fluorescent light microscopy. These protocols should be applicable to any tissue sample that does not require sectioning and can be flattened using physical pressure as described here. This protocol is adapted from other long-used protocols in Drosophila for visualizing polytene chromosomes or mitotic neuroblast metaphase spreads[4-6]. In general, we find that tissues of 50-100um thickness can be compressed to 10-15 um thickness using this protocol. This reduction of thickness both minimizes and evens out the distance of each nucleus to the objective, which is critical when using light intensity to determine DNA content. Siliconized coverslips are used, which cause liquid to bead easily on the coverslip. This facilitates transfer of the tissue to a positively charged slide during the squash process. Protocol 3.1 is for measuring the DNA content of populations of interphase nuclei. This protocol requires a known tissue ploidy standard such as haploid sperm. Protocol 3.2 is for visualizing condensed chromosomes, either from preparations containing mitotic chromosomes, or for interphase cells expressing the phosphatases PP2A and PP1, which can be inhibited by the drug Calyculin A to cause premature chromosome condensation[7, 8]. We have used both protocols to assay ploidy previously[8-12]. Both protocols can be completed in a short amount of time and rely on light microscopy to determine DNA content. These protocols should be of interest to anyone interested in measuring ploidy in situ, in samples that are amenable to physical flattening using the procedures described here.
2. Materials
2.1. Siliconized coverslips
Pour several ml of Sigmacote (Sigma) into a small plastic container, such as the lid of a pipette tip container. Using forceps, transfer several 18 x 18 cm coverslips into the Sigmacote solution. Leave for 5 min. Set the coverslips to dry by leaning them at a 45 degree angle against the side of an Eppendorf tube rack. Dried siliconized coverslips can be stored for several months.
2.2. Solutions
For all protocols:
0.5% Na Citrate
1XPBS (diluted in H2O from 10X stock[13])
95% EtOh in Coplin jar at −20C
For protocol 3.1:
3.7% Paraformaldehyde (diluted from 37% stock using phosphate buffered saline, PBS)
For protocol 3.2:
11:11:2 MeOH:Acetic Acid:H2O
45% Acetic Acid
200 nM Calyculin A (Cell Signaling Technology, diluted in 1XPBS, for interphase chromosome visualization only)
50 ug/ml Colcemid (Sigma, diluted in 1X PBS, if an enrichment in metaphase cells is desired)
2.3. Squashing supplies
Dissection supplies as needed (e.g. forceps, dissection dish)
Platform shaker
Dewar of liquid nitrogen
Tongs for handling slides
Positively charged slides (superfrost plus, VWR)
Standard and gel loading pipette tips
Metal dissection probe (Fisher)
Paper towels
Vise (Avenger Gold, Penn Tool Co)
Razor blade
2.4. Staining supplies
5 ug/ml DAPI
1X PBS
Mounting media (Vectashield, Vector Labs)
Clear Nail polish
22 x 22 coverslips
3. Methods
3.1. Tissue squash protocol for measuring nuclear ploidy from decondensed interphase nuclei
Tissue preparation
Dissect tissue of interest with forceps in 1X PBS. Remove any excess tissue not needed for ploidy measurements, as is possible. If the ploidy of your tissue sample is unknown, also include a known ploidy standard. A convenient standard for Drosophila tissue is the testis, which contains elongated haploid sperm nuclei (Fig1).
Transfer tissue to a siliconized coverslip with 50 ul of Na citrate for 10 min. This serves to swell the tissue and can impact the final size of the nuclei in your sample.
While viewing your tissue under a dissecting microscope, remove as much of the Na Citrate as possible without drying out the tissue, using a pipette with a gel loading tip (Fig2A).
Quickly add 0.5ml 37% paraformaldehyde fix solution. Leave tissue in fix for 15 min.
While viewing your tissue under a dissecting microscope, remove most of the fix, using a pipette with a standard pipette tip.
Replace with 50 ul 1X PBS as a quick wash.
While viewing your tissue under a dissecting microscope, remove as much of the 1X PBS as possible using a pipette with a gel loading tip.
Replace with 7-10ul of fresh 1X PBS.
Gently place a charged slide (charged slide facing down) over the coverslip with the drop of PBS. The coverslip should adhere to the slide, and you can then flip the slide/coverslip over (Fig2B).
With one hand, apply very gentle pressure to opposite corners of the coverslip using your index and middle fingers.
Use the needle of the probe to trace a grid-like pattern first horizontally and then vertically across the coverslip. Apply gentle pressure to the coverslip with the probe needle as you do this step (Fig2C).
Flip the slide over onto a paper towel. Place one finger at opposite ends of the slide and apply gentle pressure. Next, through the paper towel, take the handle end of the dissecting probe and again trace a grid-like pattern over the portion of the slide bottom that is opposite the coverslip on the other side. These steps (10-13) prevent slippage of the coverslip during squashing (Fig2D).
Fold the paper towel in half and place in the vise. The coverslip should be facing away from the handle side of the vise (Fig2E).
Gradually apply increased pressure to the slide. As a general rule, you should continue to increase pressure until it no longer becomes possible to easily tighten the vise handle. Leave the slide for 1-2 minutes in the vise.
Release the vise pressure and remove the slide.
Using tongs, submerge the slide in liquid nitrogen. After about 5 seconds, the bubbling of the nitrogen will stop.
Remove the slide and place coverslide up on a paper towel.
Quickly pick up the razor blade. While holding the slide down, pry one corner of the coverslide off with the razor blade using a flicking motion (flicking the blade away from you and your fingers (Fig2F).
Quickly transfer the slide to the Coplin jar containing −20C EtOH. The coverslip can be reused if still in one piece (remove all pieces of the coverslip if shattered during removal).
Slides can be stored in EtOH for one hour to several days. Upon removal, allow the slide to air dry.
Rinse the tissue on the slide by pipetting 1ml of 1XPBS and then quickly removing. Repeat 2-3 times.
Add 1ml of 1XPBS + 5ug/ml DAPI for 5 min to the slide. Incubate in the dark for 15 min. To minimize the chance of the solution/sample from drying out, enclose the slide in a sealed Tupperware container with a moist paper towel.
Rinse the tissue on the slide by pipetting 1ml of 1XPBS and then quickly removing. Repeat 2-3 times.
Apply a drop of mounting media to the tissue and place a 22 x 22 cm coverslip over the drop.
Seal the coverslip with nail polish.
Figure 1-. DAPI stained haploid sperm nuclei.
Scale bar= 20 microns.
Figure 2-. The Squash procedure.
A. Aspirating a bead of liquid on a siliconized coverslip with a gel loading tip. B. Picking up the coverslip with a positively charged slide. C. Tracing a grid over the coverslip with a metal probe. D. Tracing a grid on the bottom of the slide (covered with a paper towel) with a probe handle. E. Placing the slide in the vise. F. Removing the coverslip with a razor blade.
Imaging and Ploidy Measurement
-
26.
For each individual slide, image on an epifluorescence or confocal microscope. We typically use a 40X or 60X oil objective. Image a few candidate polyploid cells to determine an exposure setting that does not contain any saturated pixels. Set this exposure setting to be constant while imaging each slide. Re-measure again for each new slide.
-
27.
Image several candidate polyploid cells, taking Z-stacks as needed to capture the entire nucleus. Due to the squash, your sample thickness should not be greater than 10-15 microns. We recommend a Z-stack step size of 1 micron.
-
28.
Using the same exposure settings, image your internal ploidy control (e.g. elongated spermatid nuclei, Fig1) as well as nuclei in your tissue of interest.
-
29.
Continue steps 26-28 for all slides in an experiment.
-
30.
Once imaging for an experiment is complete, open each Z-stack in ImageJ (ImageJ.nih.gov). Create a Z-projection for the DAPI channel using the “Sum-slices” option.
-
31.
Within each projection, use the free-hand ROI indicator to outline each nucleus (Fig3A). Only measure nuclei which are not on top/underneath a neighboring nucleus.
-
32.
After drawing an ROI for an individual nucleus, use the “measure” function. Make sure that “Integrated Density” is being recorded.
-
33.
Repeat steps 31-32 for all nuclei that can be possibly measured in each Z-stack image. Measure both candidate polyploid cells and your known ploidy standard. Keep track of which nuclear measurements are for each cell type.
-
34.
When you’ve completed imaging for a given slide, export the measurements into a database program (e.g. Excel). Transfer your notes about which measurements go with which cell type.
-
35.
For the ploidy standard nuclei from each slide, calculate the median integrated density value. Set the value for this internal standard to reflect the C value of the standard population (e.g., for haploid cells, set a value of 1.0, reflecting 1C). Divide all other nuclei measurements for the standard sample by the C value for the median value (Fig3B). This should give you a fairly narrow range of values. For a 1C cell population, we aim to have a range of values all between 0.5-1.5, though even narrower is better.
-
36.
For the candidate polyploid nuclei, normalize all nuclei to the value of the median integrated density value of the ploidy standard.
-
37.
Repeat steps 34-36 for all slides in an experiment. This will now give you a set of values normalized to C, which enables you to combine data from different slides into your final dataset.
-
38.
When your data analysis is complete, create a histogram from your data. Try several binning ranges to generate your desired plot (Fig3C).
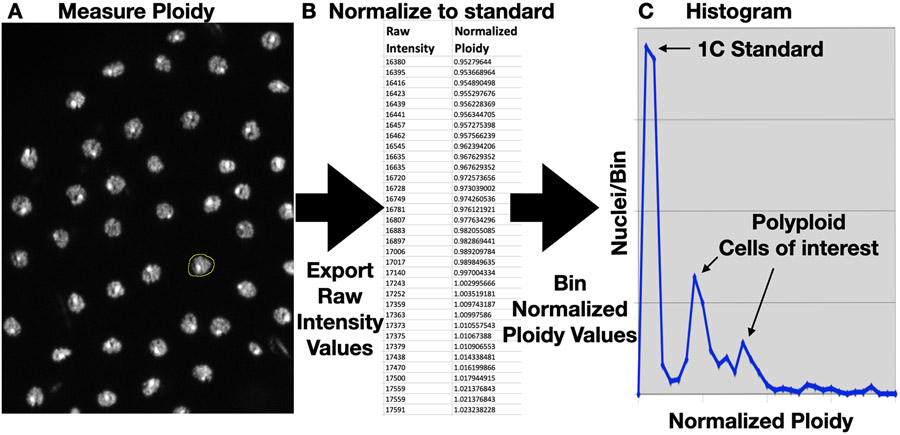
Figure 3-. Producing ploidy histogram data from samples prepared using Protocol 3.1.
A. ROI selection (yellow) of a DAPI-stained nucleus of interest in ImageJ. B. Example range of raw integrated density and converted ploidy data (C), normalized to a haploid standard. C. An example histogram plot that includes both a 1C standard and polyploid cells of interest.
3.2. Tissue squash protocol for measuring nuclear ploidy from condensed chromosomes
Tissue preparation
Dissect tissue as in protocol 3.1 step 1.
If inducing premature chromosome condensation of interphase nuclei is desired, incubate tissue in CalyculinA for 30 minutes in a dissecting dish on a platform shaker for 30 min. If an enrichment of metaphase cells is desired, incubate tissue in Colchicine for 30 minutes in a dissecting dish on a platform shaker for 30 min. Shake at a speed that gently circulates the solution. If premature chromosome condensation or enrichment of metaphases is not needed, skip to the next step.
Transfer tissue to a siliconized coverslip with 50 ul of Na citrate for 10 minutes. This serves to swell the tissue. Longer incubations in Na citrate can experimentally induce sister chromatid separation.
While viewing your tissue under a dissecting microscope, remove as much of the Na Citrate as possible without drying out the tissue, using a pipette with a gel loading tip (Fig2A).
Quickly add 11:11:2 MeOH:Acetic Acid:H2O fix solution. Fix for 10-20 seconds.
While viewing your tissue under a dissecting microscope, remove most of the fix, using a pipette with a standard pipette tip.
While viewing your tissue under a dissecting microscope, remove as much of the fix as possible without drying out the tissue, using a pipette with a gel loading tip.
Add 7 microliters 45% Acetic Acid. Wait 1-2 minutes.
Repeat steps 9-25 of Protocols 3.1 to squash the tissue (Fig2B-F).
Imaging and Chromosome Visualization
-
10.
Image as described in Protocols 3.1, with the exception that Z-stacks may not be necessary, as one should focus on taking single images of each nucleus of interest where all chromosomes are in focus. The above protocol should enable visualization of condensed chromosomes. Ploidy analysis from such images is done simply by counting chromosome number.
-
11.
Counting of individual chromosomes can be done from images to score for degree of polyploidy (Fig4) and/or aneuploidy. Each chromosome in a species typically has a distinct morphology[4]. N value can be determined by counting the number of chromosomes of each type, while C value can be determined by the number of distinct chromatids that are visible for each chromosome type. Drosophila melanogaster has a haploid number of 4 chromosomes and is a diploid organism. Due to both homologous chromosome pairing and the polytene nature of chromosomes produced by endocycles, it is possible for a cell of high ploidy to have a low N value. This is due to all chromosomes being part of a single giant polytene chromosome [14].
Figure 4-. Condensed chromosome imaging of Drosophila melanogaster cells, prepared using protocol 3.2.
A. A metaphase spread from a 2N/4C XY mitotic rectal epithelial cell. B. A metaphase spread from a 2N/4C XX mitotic midgut intestinal stem cell. C. A metaphase spread from an 8N/16C XY hindgut rectal papillar cell. D. An interphase wing imaginal disc cell without Calyculin A. E. An interphase XX wing imaginal disc cell treated with Calyculin A to reveal 2N/2C ploidy (note- the small 4th chromosomes are not present in this image). F. An interphase hindgut rectal papillar cell of similar ploidy to the cell in C, treated with Calyculin A. The centromeric regions on each chromosome become un-paired in this assay[8]. For all images where distinct chromosomes are seen, the chromosome type is labeled. Chromosome types are pseudo-colored as follows: X/Y: purple, 2nd or 3rd autosomes: green or red, autosome 4: blue. Scale bar= 25 microns.
4. Notes
If you do not have a vise, you may apply strong pressure using your thumbs. However, in our experience, this results in more variability in tissue appearance between tissue preparations and between users.
It is generally more effective to use a smaller amount of tissue on many coverslips, rather than a large amount of tissue on a single coverslip. As a general guideline, we place 200 x 200 micron-sized tissue dissections from 2-3 Drosophila melanogaster per coverslide along with one adult testis for an internal ploidy control.
Cracking the slide, either during vise tightening or following submersion into liquid nitrogen, is a sign that you are tightening the vise too much.
Frequent re-use of siliconized coverslips (more than 3-4 times) usually leads to poor transfer of tissue to the slide and/or coverslip cracking. Make new siliconized coverslips as needed.
In our experience, it is easy to see the area where the 18 x 18 coverslip was after its removal. However, if this is not obvious to the experimenter, you may consider drawing the location of the coverslip on the bottom of the slide opposite the coverslip with a wax pencil.
Please be advised that any slide labeling should be done in pencil prior to removal from EtOH.
For Protocols 3.1, if the range of measured values for your ploidy standard exceeds +/− 50% of the median, this is a sign that your squash protocol did not work well and that the tissue is likely under-squashed. Try increasing the pressure on the vise.
For Protocols 3.1, in our experience, N values of 100 nuclei per tissue type are required to accurately reflect tissue ploidy distribution as visualized in a histogram.
For Protocols 3.1, in our experience, GFP fluorescence from a transgenic animal, expressed in a tissue of interest, generally survives the 3.7% fixation and squash protocol. This is not the case for Protocol 3.2, however.
For Protocol 3.2, we note that for Drosophila samples with prematurely condensed chromosomes, chromosome pairing may prevent resolution of N value, but will enable visualization of each chromosome type, including in cells with polytene chromosomes (Fig4D-F).
For Protocol 3.2, we caution against relying solely on absence of chromosomes from mitotic preparations as a sign of aneuploidy, as chromosomes can physically dissociate from nuclei during the squash procedure. Chromosome gains, in contrast, can be reliably scored as a readout of aneuploidy, and if the frequency of perceived chromosome losses is similar to that of chromosome gains, then the chromosome losses may then be reliably scored as a sign of aneuploidy.
Protocols 3.1 and 3.2 can be adapted for antibody staining. The success of antibody staining in visualizing your protein of interest is highly dependent on the antibody. Many epitopes do not survive the freezing protocol or the MeOH fixation step in particular. We recommend trying antibody staining both before and after the squash procedure to identify optimal conditions for antibody signal. Optimal concentrations of primary antibody may differ from standard (non-squash) tissue preparations.
Protocols 3.1 and 3.2 can be adapted for in situ hybridization. This may require different fixation conditions.
Acknowledgments
Study of polyploidy in the Fox laboratory is supported by a National Institutes of Health (NIH) grant R01GM118447. Delisa Clay is supported by a GRFP Fellowship from the National Science Foundation (NSF). We thank Archan Chakraborty for comments on the manuscript.
Literature Cited
- 1.Peterson NG & Fox DT (2021) Communal living: the role of polyploidy and syncytia in tissue biology. Chromosome Research, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Øvrebø JI, Edgar BA (2018) Polyploidy in tissue homeostasis and regeneration. Development 145: dev.156034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox DT, Soltis DE, Soltis PS, et al. (2020) Polyploidy: A Biological Force From Cells to Ecosystems. Trends Cell Biology 30: 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatti M, Bonaccorsi S, Pimpinelli S (1994) Looking at Drosophila Mitotic Chromosomes. Methods Cell Biol 44:371–391. [DOI] [PubMed] [Google Scholar]
- 5.Kennison JA (2008) Dissection of larval salivary glands and polytene chromosomes preparation. Cold Spring Harb Protoc 3:pdb.prot4708. [DOI] [PubMed] [Google Scholar]
- 6.Johansen KM, Cai W, Deng H, et al. (2009) Polytene chromosome squash methods for studying transcription and epigenetic chromatin modification in Drosophila using antibodies. Methods 48:387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotoh E, Asakawa Y, Gotoh E, Kosaka H (1995) Inhibition of protein serine/threonine phosphatases directly induces premature chromosome condensation in mammalian somatic cells. Biomed Res 16:63–68. [Google Scholar]
- 8.Stormo BM, Fox DT (2016) Distinct responses to reduplicated chromosomes require distinct Mad2 responses. eLife 5:e15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox DT, Gall JG, Spradling AC (2010) Error-prone polyploid mitosis during normal Drosophila development. Genes Dev 24:2294–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoenfelder KP, Montague RA, Paramore SV, et al. (2014) Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development 141:3551–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stormo BM, Fox DT (2019) Interphase cohesin regulation ensures mitotic fidelity after genome reduplication. Mol Biol Cell 30:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen E, Allen SR, Sawyer JK, Fox DT (2018) Fizzy-related dictates a cell cycle switch during organ repair and tissue growth responses in the Drosophila hindgut. eLife 7:e38327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(2007) 10X PBS. Cold Spring Harb Protoc. 10.1101/pdb.rec10768 [DOI] [Google Scholar]
- 14.Stormo BM, Fox DT (2017) Polyteny: still a giant player in chromosome research. Chromosome Research 25: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]