Abstract
The E1B-55-kDa protein of adenovirus type 5 and the p53 tumor suppressor gene product form a complex that localizes to the cytoplasm, thereby downregulating p53’s transcriptional activity. The E4orf6 protein binds and relocalizes E1B-55-kDa, and the proteins act synergistically to inactivate p53. We show that another adenovirus E4 gene product, E4orf3, is also sufficient to relocalize E1B-55-kDa from the cytoplasm to the nucleus. Both proteins are then found in discrete nuclear structures (tracks) that are known to contain components of the promyelocytic leukemia-associated nuclear structure. Simultaneously, p53 is dissociated from E1B-55-kDa and is found evenly distributed over the nucleoplasm. In the presence of E4orf3, p53-dependent transcriptional activity is no longer repressed by E1B-55-kDa. When E1B-55-kDa is coexpressed with E4orf3 and E4orf6, E1B-55-kDa is found to colocalize with E4orf6 rather than E4orf3. In parallel, p53 is inhibited and degraded by the combination of E1B-55-kDa and E4orf6, regardless of coexpressed E4orf3. This suggests that the effects of E4orf6 on E1B-55-kDa overrule the actions of E4orf3. When cells are infected with virus expressing E4orf3 but not E4orf6, E1B is found in the cell nucleus and p53 enters the virus replication centers. After infection with wild-type adenovirus, E4orf3 is expressed before E4orf6 and E1B temporarily colocalizes with E4orf3 in nuclear tracks before associating with E4orf6. We propose that during adenovirus infection, the E4orf3 protein transiently liberates p53 from its association with E1B-55-kDa. Subsequently, p53 is inactivated and degraded by the combination of E1B-55-kDa and E4orf6.
The adenovirus E1B-55-kDa protein forms a specific complex with the E4orf6 (E4-34-kDa) protein during infection (31). When transiently expressed, E1B-55-kDa can be detected in characteristic clusters confined to the cytoplasm. In contrast, coexpressed E4orf6 relocalizes E1B-55-kDa from the cytoplasm to the nucleus, and both proteins are found distributed over the nucleus (16). The complex formed by E1B-55-kDa and E4orf6 shuttles between the nucleus and the cytoplasm (11), but the equilibrium between export and import results in a predominantly nuclear location of the two proteins. During infection, both proteins localize to the periphery of the virus replication centers (26). The complex of E1B-55-kDa and E4orf6 is responsible for at least two different functions. First, the two proteins are necessary for the modulation of mRNA export during adenovirus infection. When both proteins are expressed by the virus, viral mRNA is efficiently carried to the cytoplasm, while most cellular mRNA molecules are retained in the nucleus (1, 5, 27). Second, the E1B-55-kDa protein binds and inactivates the p53 tumor suppressor gene product, resulting in oncogenic transformation of cells (32, 42). In the absence of E4 proteins, E1B-55-kDa colocalizes with p53 in cytoplasmic clusters (7, 43). However, when E4orf6 is coexpressed with p53 and E1B-55-kDa, p53 is strongly destabilized, leading to severely reduced steady-state amounts of the protein (29, 35). E4orf6 has been reported to interact with p53 (12) and to enhance the E1B-55-kDa protein’s oncogenic potential (22, 24). Thus, it appears that adenovirus employs E1B-55-kDa and E4orf6 to reduce p53 abundance and activity during infection. In contrast, herpesviruses do not seem to counteract p53 during infection. Rather, cytomegalovirus IE2 protein appears to upregulate p53 (23). Further, p53 accumulates in the cellular replication areas within the nucleus during infection with cytomegalovirus (14) and herpes simplex virus (44), suggesting that these viruses may take advantage of p53 for their replication. The fact that adenovirus E1A proteins upregulate the intracellular p53 levels (8, 21) raises the possibility that p53 may also play a positive role during the adenovirus infectious cycle, especially before E1B-55-kDa/E4orf6-mediated p53 degradation reaches completion.
E4orf6 can relocalize E1B-55-kDa from the cytoplasm to the nucleus (16). However, when cells are infected with a recombinant adenovirus lacking a functional E4orf6 coding region, E1B-55-kDa is still found in the nucleus, namely, in characteristic track-like structures (26). This suggests that at least one other adenovirus protein can relocalize E1B-55-kDa from the cytoplasm. The track-like localization is reminiscent of another adenovirus E4 protein that is encoded by the E4 open reading frame 3 (E4orf3) (9, 13). Based on these observations, we suspected that E4orf3 may also be able to relocalize the E1B-55-kDa protein from the cytoplasm to the nucleus. Further, we asked if E4orf3, like E4orf6, might modulate E1B-55-kDa’s action on p53.
We show here that the presence of E4orf3 redirects E1B-55-kDa from the cytoplasm to the nuclear tracks. p53 is then liberated from E1B-55-kDa, resulting in p53-mediated transcriptional activity. However, the E4orf6 protein cooperates with E1B-55-kDa to inactivate and degrade p53, even in the presence of E4orf3. Therefore, we propose that E4orf3 temporarily permits p53 activity in adenovirus-infected cells, before E4orf6 in concert with E1B-55-kDa eliminates p53.
MATERIALS AND METHODS
Cell culture and infection.
Cells were maintained in Dulbecco modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS). For infection, 400,000 cells in a 3.5-cm dish were washed with phosphate-buffered saline (PBS) and then overlaid with 4 × 106 infectious units of virus in 500 μl of DMEM without serum. After 3 h of gentle rocking in an incubator, the cells were washed again with PBS and further incubated in DMEM–10% FBS.
Plasmids.
The expression plasmids for p53 (20), hemagglutinin HA-tagged E1B-55-kDa (termed pCGNE1B) (11) and E4orf6 (12) have been described. To obtain an expression plasmid for flag-tagged E4orf3, the corresponding coding region was PCR amplified from genomic adenovirus type 5 DNA by using the primers 5′-CGCGGATCCGCCACCATGGACTACAAAGACGATGACGATAAGATGATTCGCTGCTTGAGGCTGAAG-3′ and 5′-CGCGGATCCTTATTCCAAAAGATTATCCAAAAC-3′. The PCR product was cloned into the pCMVneoBam expression vector (4) by using BamHI to obtain pCMVflagE4orf3. The construct was verified by sequencing.
Transfections.
Cells were transfected by using a cationic lipid preparation (Superfect; Qiagen) according to the manufacturer’s instructions. A total of 3 μg of plasmid DNA was transfected into 400,000 cells on a 3.5-cm dish. Cells were harvested 20 h after transfection. For Western blot detection of p53 (see Fig. 3), Saos-2 cells were transfected with 2 μg of plasmid DNA by using Fugene 6 (Boehringer Mannheim).
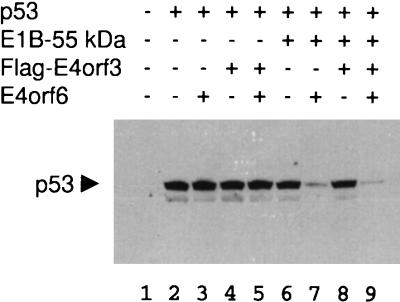
FIG. 3.
Intracellular p53 levels in the presence of E1B-55-kDa and E4 gene products. Expression plasmids for p53 (0.3 μg), E1B-55-kDa (0.3 μg), flag-tagged E4orf3 (0.6 μg) and E4orf6 (0.6 μg), or “empty” vector constructs were cotransfected as indicated into Saos-2 cells. At 24 h after transfection, the cells were harvested, subjected to SDS-polyacrylamide gel electrophoresis, and transferred to nitrocellulose. The blot was stained for p53, and the antibody was visualized by a peroxidase-mediated chemiluminescent reaction.
Immunofluorescence.
Cells were transfected as described above, but all quantities were reduced by a factor of 4, and the cells were seeded onto plastic slides (Nunc) suitable for microscopy. Transfected or infected cells were fixed with paraformaldehyde (4% in PBS, 15 min), permeabilized with Triton X-100 (0.2% in PBS, 25 min), and incubated with antibody as described earlier (10). To stain the flag tag, a mouse monoclonal antibody (M2; IBI Kodak) was used when double labeling of the flag tag with p53 was performed. Alternatively, the flag tag was stained with a polyclonal rabbit antibody (D-8; Santa Cruz) when double labeling of the flag tag with E1B-55-kDa was performed. The E2A-72-kDa DNA binding protein was stained with a polyclonal rabbit antibody (a generous gift of G. Ketner) when E1B-55-kDa was labeled simultaneously. Alternatively, the E2A-72-kDa protein was stained with a mouse monoclonal antibody (clone B8-6) when p53 was labeled simultaneously. To detect E1B-55-kDa, the murine monoclonal antibody 2A6 (33) was used. The p53 protein was stained with a polyclonal rabbit antibody (FL-393; Santa Cruz). E4orf3 in infected cells was stained with a polyclonal rabbit antibody (J. Boyer and G. Ketner). Primary mouse antibodies were visualized by secondary antibodies coupled to fluorescein isothiocyanate FITC (Jackson). Primary rabbit antibodies were detected by a Texas-Red-labeled secondary antibody (Jackson). Prior to mounting (Fluoprep, bioMérieux), the cell nuclei were briefly stained with 4,6′-diamidino-2-phenylindole (DAPI).
Immunoblots.
Proteins were separated on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels transferred to nitrocellulose; this was followed by incubation with antibodies in PBS containing 5% milk powder and Tween 20 and chemiluminescent detection (Pierce) of peroxidase-coupled secondary antibody (Jackson). The concentration of Tween 20 was 0.05% when p53 was detected and 0.5% when E4 proteins were stained. Antibody Pab1801 to p53 was from Calbiochem. Polyclonal antibodies to E4orf3 and E4orf6 were generous gifts from G. Ketner and P. Branton, respectively.
Luciferase assays.
Next, 750,000 Saos-2 cells per assay were transfected by using Superfect (Qiagen). Luciferase activities were determined by a premanufactured assay system (Promega).
RESULTS
E4orf3 is sufficient to relocalize the E1B-55-kDa protein from the cytoplasm to the nucleus.
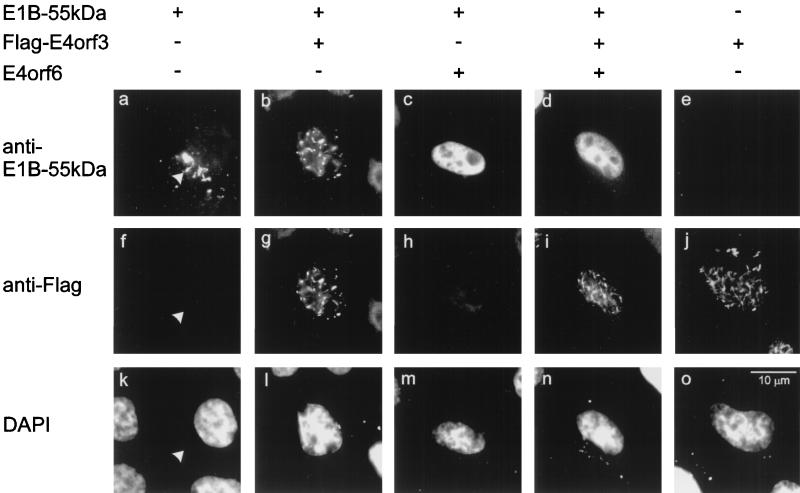
E4orf3 was transiently expressed in HeLa cells, with a flag epitope fused to the amino-terminal end of the protein. A protein of the expected size was detected by Western blot with an antibody to the tag (data not shown). By immunofluorescence, the protein was found in nuclear tracks that colocalize with the promyelocytic leukemia (PML) protein in HeLa cells and Hep-2 cells, confirming previous results (9, 13). In addition to the nuclear tracks, E4orf3 was occasionally found in filamentous structures within the cytoplasm. These cytoplasmic filaments did not detectably coincide with cytoskeletal components such as actin, tubulin, or vimentin (data not shown). Using this construct, E4orf3 was coexpressed with the adenovirus E1B-55-kDa protein, followed by simultaneous detection of the two proteins with differently labeled antibodies (Fig. 1). When E1B-55-kDa was transiently expressed in HeLa cells, it was found within characteristic cytoplasmic clusters (Fig. 1a). When E4orf3 and E1B-55-kDa were expressed together, both proteins localized to nuclear tracks (Fig. 1b and g) that were similar to the pattern observed with E4orf3 alone (Fig. 1j). The E4orf6 protein also relocalized E1B-55-kDa, resulting in an even distribution of E1B-55-kDa throughout the nucleus (Fig. 1c), as was shown previously (16). When both E4orf3 and E4orf6 were simultaneously expressed along with E1B-55-kDa (Fig. 1d and i), the pattern of E1B-55-kDa distribution was similar to the combination of E4orf6 and E1B-55-kDa. In contrast, E4orf3 remained in the nuclear tracks. Thus, the E4orf6-induced relocalization of E1B-55-kDa is dominant over the pattern achieved with E4orf3.
FIG. 1.
Relocalization of E1B-55-kDa by two E4 gene products. Expression plasmids for E1B-55-kDa, flag-tagged E4orf3, and E4orf6 (330 ng of each) or “empty” vector constructs were cotransfected into HeLa cells as indicated. At 24 h after transfection, E1B-55-kDa (panels a to e) and the tagged E4orf3 protein (panels f to j) were detected by double immunofluorescence. The location of the nuclei was visualized by DAPI stain (panels k to o). The site of cytoplasmic clusters containing E1B-55-kDa is marked by arrowheads in panels a, f, and k.
E4orf3 relieves the inhibition of p53 activity by E1B-55-kDa.
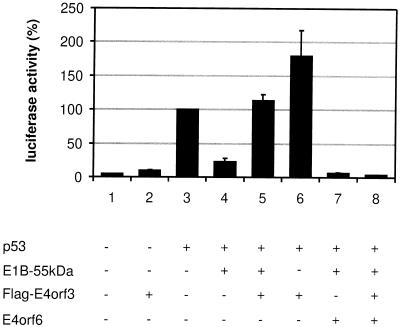
Since E1B-55-kDa is known to bind and inactivate the p53 protein, we asked if this activity might be modulated by E4orf3. To measure p53 activity, a reporter plasmid with a p53-dependent promoter (derived from the first intron of the mdm2 gene) regulating luciferase expression was transfected into Saos-2 cells, a human osteosarcoma cell line that lacks endogenous p53. At 24 h after transfection, the cells were harvested and the luciferase activity was determined (Fig. 2). While only a low level of basal activity was detectable in the absence or presence of E4orf3 (Fig. 2, lanes 1 and 2), cotransfection of a p53 expression plasmid resulted in strongly enhanced luciferase expression (Fig. 2, lane 3). The additional expression of E1B-55-kDa reduced this transcriptional activation by p53 and resulted in a low luciferase activity (Fig. 2, lane 4). However, when E4orf3 was expressed along with p53 and E1B-55-kDa, the inhibitory effect of E1B-55-kDa was abolished, and the transcriptional activity was similar to the value obtained with p53 in the absence of adenoviral proteins (Fig. 2, lane 5). When E4orf3 was coexpressed with p53, transcriptional activity was moderately enhanced compared to p53 alone (Fig. 2, lane 6). Finally, E1B-55-kDa and E4orf6 were downregulating p53 activity more profoundly than E1B-55-kDa alone (Fig. 2, lane 7), in accordance with the finding that the combination of E1B-55-kDa and E4orf6 mediates the intracellular degradation of p53 (29, 35). This effect was not reversed by the additional expression of E4orf3 (Fig. 2, lane 8). Essentially the same results were obtained when different p53-responsive promoters were used (data not shown). Taken together, these results suggest that E4orf3 can reverse the inhibitory effect of E1B-55-kDa on p53, while E4orf6 further enhances p53 inhibition in the presence of E1B-55-kDa. In the presence of all three viral proteins, the effect of E4orf6 overrules that of E4orf3.
FIG. 2.
p53 inhibition by E1B-55-kDa abolished by E4orf3. Expression plasmids for p53 (100 ng), E1B-55-kDa (400 ng), flag-tagged E4orf3 (400 ng) and E4orf6 (100 ng), or “empty” vector constructs were cotransfected along with 1 μg of a p53 responsive luciferase reporter plasmid termed pBP100luc (30) into Saos-2 cells as indicated. At 24 h after transfection, the luciferase activity was determined. The activity of p53 alone was arbitrarily set to 100%, and the activities obtained with the different plasmid combinations were normalized accordingly.
E4orf3 does not detectably influence p53 stability.
Given the fact that E1B-55-kDa and E4orf6 promote the intracellular destabilization of p53 (29, 35), we tested whether p53 stability might also be affected by E4orf3. The three adenovirus proteins in all permutations were transiently coexpressed with p53 in Saos-2 cells, followed by Western blot detection of p53 (Fig. 3). None of the adenovirus proteins affected the p53 levels when expressed alone (Fig. 3, compare lanes 3, 4, and 6 with lane 2), and combinations of E4orf3 with either E1B-55-kDa or E4orf6 did not reduce the steady-state amount of p53 (Fig. 3, lanes 5 and 8). In contrast, the p53-derived signal was strongly reduced in the presence of E1B-55-kDa and E4orf6 together (Fig. 3, lane 7), and this reduction was maintained in the presence of E4orf3 (Fig. 3, lane 9). These results argue that E1B-55-kDa and E4orf6 act in concert to destabilize p53, while E4orf3 does not significantly affect the intracellular amount of p53 protein.
E4orf3 abolishes the colocalization of p53 with E1B-55 kDa.
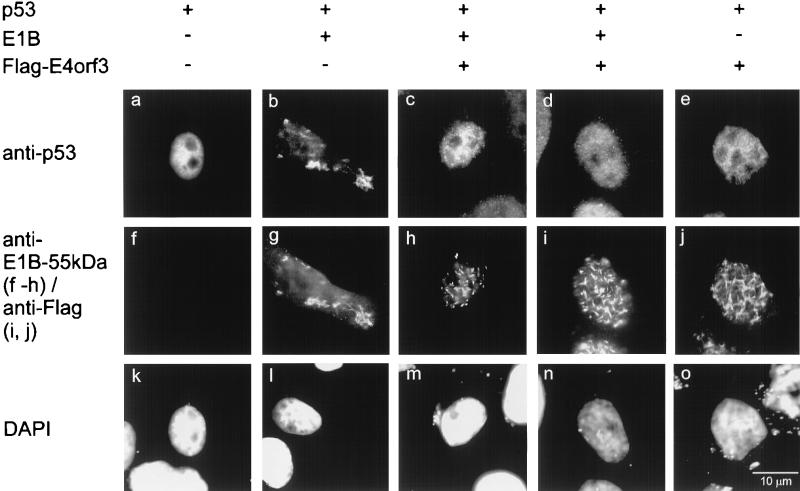
Since E4orf3 apparently reverses the inhibitory effect of E1B-55-kDa on p53 activity (Fig. 2), we asked if E4orf3 might modulate the intracellular association of E1B-55-kDa with p53. To this end, the three proteins were coexpressed in HeLa cells in different combinations and detected by immunofluorescence (Fig. 4). Transfection of Saos-2 cells yielded essentially the same results (data not shown). While p53 is found predominantly in a diffusely nuclear distribution when expressed in the absence of virus proteins (Fig. 4a), it is relocalized to cytoplasmic clusters in the presence of E1B-55-kDa (Fig. 4b and g), as described previously (7, 43). When E4orf3 was coexpressed with p53 in addition to E1B-55-kDa, the two adenovirus proteins were found in nuclear tracks (Fig. 4h and i), whereas p53 was distributed over the nucleus (Fig. 4c and d) in a pattern similar to cells expressing p53 alone. When p53 was coexpressed with E4orf3, neither of the proteins affected the other’s location (Fig. 4e and j). We conclude that while E4orf3 apparently has little if any direct effect on p53’s location or activity, it separates the E1B-55-kDa protein from p53. As a result, the two adenovirus proteins colocalize, whereas p53 is apparently “liberated” from its association with E1B-55-kDa and regains its transcriptional activity.
FIG. 4.
Dissociation of p53 from E1B-55-kDa in the presence of E4orf3. Expression plasmids for p53 (0.5 μg), E1B-55-kDa (0.5 μg) and flag-tagged E4orf3 (1 μg), or “empty” vector constructs were cotransfected as indicated into HeLa cells. The p53 protein (panels a to e), E1B-55-kDa (panels f to h), and flag-E4orf3 (panels i and j) were detected by double immunofluorescence, and the nuclei were visualized by DAPI staining (panels k to o).
E4orf3 mediates nuclear localization of E1B-55-kDa in infected cells.
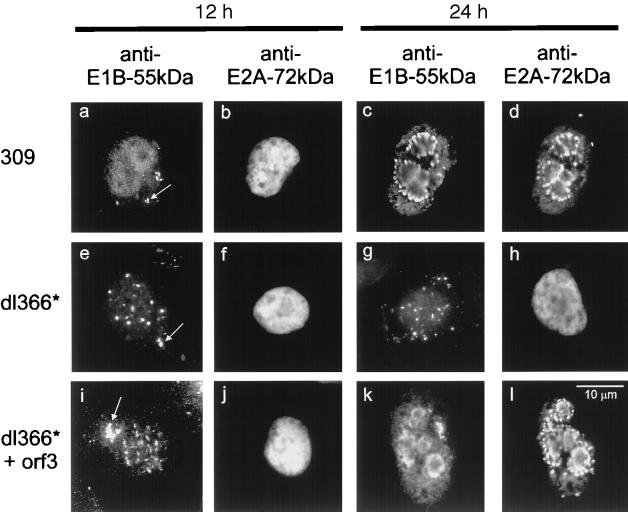
Next, we hypothesized that the E4orf3 protein might relocalize E1B-55-kDa in the context of virus infection. To test this, A549 cells were infected with different recombinant adenoviruses (Fig. 5, Table 1). Virus 309 is essentially wild type (except for the E3 region that does not detectably affect virus replication in vitro) and expresses the entire E4 region. Virus 355 (17) is derived from virus 309 but it does not express functional E4orf6. Virus 366* (18) is derived from wild-type adenovirus (termed virus 300) but it lacks the entire E4 region. Virus 366*+orf3 (18) is derived from virus 366*. It expresses E4orf3 but none of the other E4 gene products. At various times after infection, the cells were fixed and stained for E1B-55-kDa and the E2A-72-kDa DNA binding protein (Fig. 5) that is known to localize within the virus replication centers (28, 37). In cells infected with wild-type adenovirus 309, E1B-55-kDa was initially found in cytoplasmic clusters (Fig. 5a) and E2A was evenly nuclear (Fig. 5b). Later during infection, both E1B-55-kDa and E2A-72-kDa concentrated predominantly in or near the replication centers within the nucleus (Fig. 5c and d; Table 1). This location of E1B-55-kDa has been reported to depend on E4orf6 in HeLa cells (26). When the entire E4 region was absent, E1B localized to the cytoplasmic clusters throughout the observation time (Fig. 5e and g). Simultaneously, the E2A protein remained diffusely nuclear and did not concentrate in replication centers (Fig. 5f and h), a finding consistent with the reported failure of this recombinant virus to replicate in HeLa cells (18). When the cells were infected with virus 366*+orf3 lacking the E4 region but expressing E4orf3, E1B moved from the cytoplasmic clusters to the nuclear tracks (Fig. 5i; Table 1) that are characteristic for the location of E4orf3 (9, 13). Later during infection, E1B-55-kDa and E2A-72-kDa were both found to move to the nuclear centers associated with virus replication. This is in contrast with an earlier report indicating that the E4orf6 protein is required for this location of E1B-55-kDa (26). The discrepancy can possibly be explained by the fact that HeLa cells were used in the earlier studies, while A549 cells were used here. In HeLa cells, little if any E1B-55-kDa was seen in the periphery of the replication centers when cells were infected with virus lacking E4orf6 (unpublished observations). Our data strongly suggest that besides E4orf6 the E4orf3 protein also can relocalize E1B-55-kDa from the cytoplasm to the nucleus during virus infection.
FIG. 5.
E1B-55-kDa location during infection. A549 cells were infected with wild-type or recombinant adenovirus. At two different time points after infection, the cells were fixed and stained with antibodies against E1B-55-kDa and E2A-72-kDa as indicated. Cytoplasmic clusters containing E1B-55-kDa are marked with arrows in panels a, e, and i. The locations of the proteins at more time points after infection are given in Table 1.
TABLE 1.
Intracellular localization of the E1B-55-kDa protein in A549 cells infected with recombinant adenovirus
| Time (h) postinfection | Intracellular localization of E1B-55-kDa witha:
|
||||
|---|---|---|---|---|---|
| Virus 309 (+/+) | Virus 355 (+/−) | E4 in orf3 (−/+) | dl366* (−/−) | dl366*+orf3 (+/−) | |
| 6 | Cytoplasmic clusters; low levels of nucleoplasmic staining in some cells | Cytoplasmic clusters | Cytoplasmic clusters; low levels of nucleoplasmic staining in some cells | Cytoplasmic clusters | Cytoplasmic clusters |
| 9 | Cytoplasmic clusters; nuclear tracks; some diffusely nucleoplasmic staining | Cytoplasmic clusters; nuclear tracks | Cytoplasmic clusters; some diffusely nucleoplasmic staining | Cytoplasmic clusters | Cytoplasmic clusters; nuclear tracks |
| 12 | Diffusely nucleoplasmic staining; some cytoplasmic clusters | Cytoplasmic clusters; nuclear tracks | Diffusely nucleoplasmic staining; some cytoplasmic clusters | Cytoplasmic clusters | Cytoplasmic clusters; nuclear tracks |
| 18 | In the nucleus, mostly around the replication centers | In the nucleus, mostly around the replication centers | Not assayed | Cytoplasmic clusters | In the nucleus, mostly around the replication centers |
| 24 | In the nucleus, almost exclusively around the replication centers | In the nucleus, almost exclusively around the replication centers | Not assayed | Cytoplasmic clusters | In the nucleus, almost exclusively around the replication centers |
The presence (+) or absence (−) of E4orf3/E4orf6 is indicated in parentheses.
E4orf3 allows nuclear accumulation of p53 during infection when E4orf6 is absent.
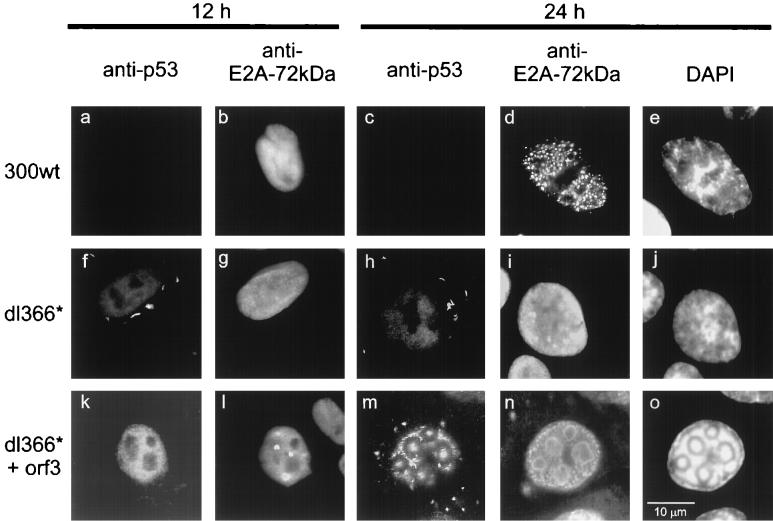
Given our findings that E4orf3 dissociates p53 from E1B-55-kDa during transient expression of all three proteins, we tested whether the same might be true during infection (Fig. 6; Table 2). The p53 protein was localized by immunofluorescence in infected A549 cells, along with E2A-72-kDa as a marker for replication centers. When wild-type adenovirus was examined, little p53 was detected in the nucleus of some cells 8 h after infection, and p53 was undetectable later during infection (Fig. 6a and c; Table 2). The same result was obtained with a recombinant virus lacking E4orf3 but expressing all other E4 gene products (termed E4 in orf3 [18]; Table 2) was used. This is consistent with reports that p53 is rapidly degraded during adenovirus infection when E1B-55-kDa and E4orf6 are expressed (29, 35). When cells were infected with adenovirus lacking the entire E4 region, p53 was mainly found in cytoplasmic clusters that are characteristic for E1B-55-kDa (Fig. 6f and h; Table 2). In contrast, when E4orf3 was expressed from the same background, p53 was localized predominantly to the nucleus (Fig. 6k and m; Table 2). Later during infection, a large proportion of p53 moved to the virus replication centers. These findings are consistent with the concept that E4orf3 liberates p53 from its association with E1B-55-kDa during infection and that p53 associates with the virus replication centers under these circumstances.
FIG. 6.
p53 location during infection. A549 cells were infected with wild-type or recombinant adenovirus. At two different time points after infection, the cells were fixed and stained with antibodies to p53 and E2A-72-kDa as indicated. The locations of the proteins at more time points after infection are given in Table 2.
TABLE 2.
Intracellular localization of p53 in A549 cells infected with recombinant adenovirus
| Time (h) postinfection | Intracellular localization of p53 witha:
|
|||
|---|---|---|---|---|
| Virus 300 wt (+/+) | E4 in orf3 (−/+) | dl366* (−/−) | dl366*+orf3 (+/−) | |
| 8 | Detectable only in a small proportion of the cells, diffusely in the nucleus | Detectable only in a small proportion of the cells, diffusely in the nucleus | Almost exclusively in the cytoplasmic clusters | Diffusely in the nucleus |
| 12 | ND | ND | Almost exclusively in cytoplasmic clusters | In the nucleus, with some concentration in emerging replication centers |
| 18 | ND | ND | Almost exclusively in cytoplasmic clusters; in the replication centers of a few cells | In the nucleus, mostly in replication centers |
| 24 | ND | ND | Almost exclusively in cytoplasmic clusters; in the replication centers of a few cells | In the nucleus, mostly in replication centers |
The presence (+) or absence (−) of E4orf3/E4orf6 is indicated in parentheses. wt, wild type; ND, not detectable.
p53 localizes within the virus replication centers, while E1B-55-kDa accumulates in the periphery of these centers.
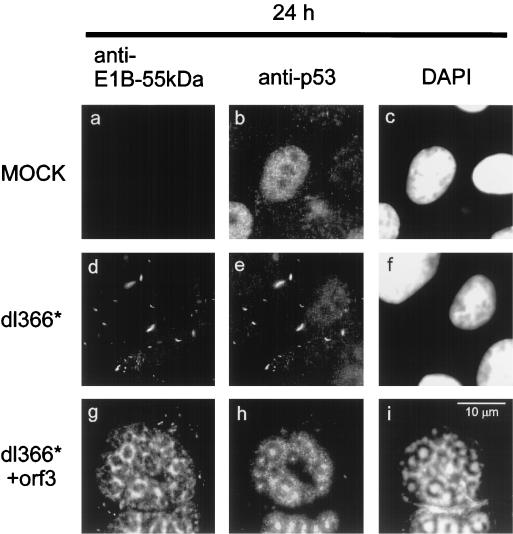
To decide whether p53 and E1B-55-kDa still colocalize in infected cells, both proteins were stained simultaneously (Fig. 7). The p53 protein was evenly distributed in the nucleus when cells were mock infected (Fig. 7b). After infection with virus lacking the E4 region, E1B-55-kDa and p53 colocalized in cytoplasmic clusters (Fig. 7d and e). However, when E4orf3 was expressed from a recombinant virus that lacks all other E4 proteins, p53 was found within the replication centers (Fig. 7h). This location did not coincide with the pattern of E1B-55-kDa; while E1B-55-kDa localized in the periphery of the virus replication centers (Fig. 7g), as reported earlier (26), the p53 protein was found in the middle of these centers (Fig. 7h, compare with Fig. 6m). At this location, DNA was highly concentrated as revealed by DAPI staining (Fig. 7i, compare with Fig. 6o). Therefore, we propose that E4orf3 dissociates p53 from its association with E1B-55-kDa not only after transfection but also in infected cells. Apparently, p53 is then enriched within the virus replication centers, colocalizing with virus DNA.
FIG. 7.
Distinct locations of p53 and E1B-55-kDa in the presence of E4orf3. A549 cells were infected with wild-type or recombinant adenovirus. At 24 h after infection, the cells were fixed and stained with antibodies to p53 and E1B-55-kDa as indicated. The location of DNA is visualized by DAPI staining (panels c, f, and i).
Expression of E4orf3 precedes expression of E4orf6 during infection.
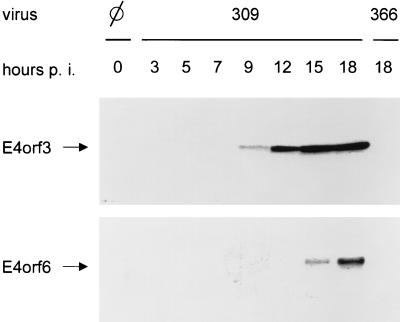
The data presented above suggest that E4orf3 can affect E1B-55-kDa’s localization, as well as E1B-55-kDa’s action on p53. However, in the presence of E4orf6, these effects are overruled. This prompted us to address the question of whether E4orf3 might be expressed at earlier time points postinfection than E4orf6, thus acting transiently on E1B-55-kDa. To test this, A549 cells were infected with wild-type adenovirus (309). At different times postinfection, the cells were harvested and the proteins were electrophoretically separated, followed by transfer onto nitrocellulose. E4orf3 and E4orf6 were visualized by using polyclonal antibodies (Fig. 8). Cells infected with virus 366 (lacking both E4orf3 and E4orf6) were processed in parallel to control for the specificity of the signal. E4orf3 was detected at 9 h postinfection, and longer exposures revealed some E4orf3 expression even after 7 h (data not shown). The amount of E4orf3 did not further increase after the 15 h time point. In contrast, E4orf6 was detectable only at 15 h postinfection, and it reached its peak later than 18 h after infection. This indicates that E4orf3 is expressed earlier than E4orf6 during infection with wild-type adenovirus.
FIG. 8.
E4orf3 is expressed earlier than E4orf6 during infection with adenovirus. HeLa cells were infected with either wild-type (virus 309) or recombinant (virus 366) adenovirus as indicated. At time points between 0 and 18 h, the cells were harvested and the lysate was subjected to SDS-polyacrylamide gel electrophoresis and Western blotting. E4orf3 (upper panel) and E4orf6 (lower panel) were detected with specific antibodies and visualized by chemiluminescence.
E1B transiently localizes in nuclear tracks in cells infected with wild-type adenovirus.
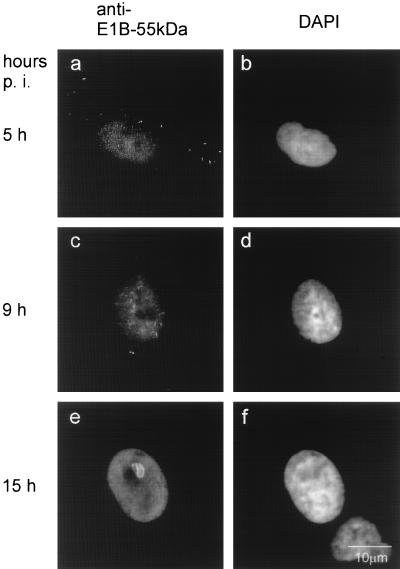
The apparent delay of E4orf6 expression in comparison to E4orf3 synthesis raised the question of where E1B would localize at early time points during infection with wild-type adenovirus. To study this, A549 cells were infected with this virus (virus 309) and fixed after 5, 9, and 15 h (Fig. 9; Table 1). As early as 5 h after infection, some E1B-55-kDa was observed in the nucleus in addition to the cytoplasmic clusters (Fig. 9a). This nuclear staining increased at 9 h postinfection, and track-like structures became visible when staining for E1B-55-kDa (Fig. 9c). At 15 h postinfection, this pattern changed again. The nuclear staining pattern of E1B-55 kDa now became more even and was only interrupted by the formation of viral replication centers (Fig. 9e). In addition, the intracellular localization of E1B-55-kDa was assessed at early time points by using recombinant viruses lacking either E4orf3 and E4orf6 (Table 1). The results show that the diffuse nuclear staining depends on the presence of E4orf6, whereas the location of E1B-55-kDa in track-like structures strictly depends on E4orf3. Taken together, these data suggest that in the early phase of infection with wild-type adenovirus, the majority of nuclear E1B-55-kDa first associates with E4orf3 in track-like structures before associating with E4orf6 and assuming a diffusely nuclear distribution.
FIG. 9.
E1B-55-kDa transiently associates with nuclear tracks during infection with adenovirus. A549 cells were infected with wild-type adenovirus (virus 309). At the indicated time points after infection, the cells were fixed and stained with an antibody to E1B-55-kDa (panels a, c, and e). The location of DNA was visualized with DAPI (panels b, d, and f).
E1B colocalizes with E4orf3 in cells infected with wild-type adenovirus.
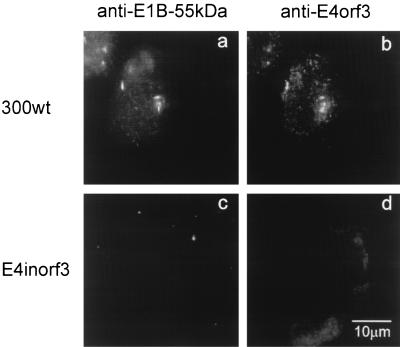
To directly visualize the association between E4orf3 and E1B-55-kDa during adenovirus infection, cells were infected for 9 h, followed by simultaneous antibody staining for E4orf3 and E1B-55-kDa. It was observed that E1B-55-kDa and E4orf3 colocalize in nuclear tracks (Fig. 10a and b). In contrast, when using a recombinant adenovirus lacking E4orf3, the antibody against E4orf3 failed to detect these structures (Fig. 10d), and E1B-55-kDa was detected predominantly in cytoplasmic clusters at this time after infection (Fig. 10c). These results confirm that E1B-55-kDa is relocalized from the cytoplasm to the nucleus by E4orf3 at early time points after infection with wild-type adenovirus.
FIG. 10.
E1B-55-kDa transiently colocalizes with E4orf3 in nuclear tracks during adenovirus infection. A549 cells were infected with wild-type adenovirus (virus 309; panels a and b) or recombinant adenovirus lacking the E4orf3 coding region (E4inorf3; panels c and d). At 9 h after infection, the cells were fixed and stained with antibodies to E1B-55-kDa (panels a and c) and E4orf3 (panels b and d).
DISCUSSION
We have shown that the adenovirus type 5 E4orf3 protein is sufficient to relocalize the E1B-55-kDa protein from the cytoplasm to the nucleus. Thus, two E4 gene products, orf3 and orf6, can each move E1B-55-kDa to the nucleus. The two associations are mutually exclusive, and complex formation between E4orf6 with E1B-55-kDa is dominant. With respect to p53, the two E4 proteins have opposite effects. While E4orf6 acts synergistically with E1B-55-kDa to inhibit and destabilize p53, the E4orf3 protein does not affect p53 stability. Moreover, it abolishes the inhibitory effect of E1B-55-kDa and restores p53 function. Similar effects were observed after transient transfection and during virus infection. During infection with wild-type adenovirus, E4orf3 is expressed considerably earlier than E4orf6, suggesting that E4orf3 acts transiently on E1B-55-kDa, before E4orf6 is present in quantities sufficient to redirect E1B-55-kDa’s localization and activities.
The E1B-55-kDa protein is found in nuclear tracks (also termed spicules) that are most obvious after infection with virus lacking E4orf6 but which also appear when cells are infected with wild-type adenovirus (Fig. 9 and 10; reference 26). Our data strongly suggest that this location is induced by the E4orf3 protein. The fact that this pattern is observed during infection with wild-type virus argues that the relocalization of E1B-55-kDa by E4orf3 plays a physiological role in this context. However, the precise function of the two viral proteins in this location is presently unknown. This function may involve components of the PML-associated nuclear structures that are known to coincide with these tracks (9, 13). Our data suggest that the association of E1B-55-kDa with the nuclear tracks shifts to a localization at the periphery of the virus replication centers later during infection. This new location does not require the presence of E4orf6, at least in the A549 cells under study. Thus, both E4orf3 and E4orf6 can induce the accumulation of E1B-55-kDa near the replication centers. This may explain at least in part why both E4 proteins can contribute to the virus’ ability to replicate (18, 19). Even though E4orf3 is apparently less efficient than E4orf6 in complementing virus replication, it is conceivable that it may cooperate with E1B-55-kDa and modulate splicing (25) or even transport (1, 27) of viral mRNA to some extent.
It remains to be determined by which mechanism E4orf3 relocalizes E1B-55-kDa from the cytoplasm to the nucleus. The most obvious possibility is that E4orf3 might directly associate with E1B-55-kDa. However, we did not detect a complex containing both proteins in vitro or in vivo. E4orf3 is strongly associated with the nuclear matrix and can therefore be fully solubilized only in the presence of strong detergents (unpublished observations) that may disrupt the putative complex. Alternatively, the two adenovirus proteins may associate only in the presence of additional proteins. In parallel, E4orf3 and the PML protein do not detectably associate with each other despite their intracellular colocalization (13).
The fact that E4orf3 abolishes the inhibitory effect of E1B-55-kDa on p53 suggests that p53’s activity is temporarily enhanced by E4orf3 during virus infection. Later during infection, p53 is apparently eliminated by the synergistic action of E1B-55-kDa and E4orf6, which was shown to be dominant over E4orf3’s effect. It was reported earlier that singly spliced E4 mRNA capable of encoding E4orf3 accumulates in infected cells earlier than multiply spliced E4 mRNA that could be translated to yield the E4orf6 protein (36). Our data support this concept by showing that E4orf3 is expressed earlier than E4orf6 during infection (Fig. 8). This is consistent with the model that during adenovirus infection E4orf3 induces a transient increase in p53 activity, before E4orf6 promotes p53’s inactivation and degradation.
When cells were infected with virus expressing E4orf3 but lacking E4orf6, p53 accumulated in the middle of the virus replication centers. It remains an open question whether p53 in these centers plays a beneficial or a detrimental role in virus infection. Herpesviruses are known to accumulate p53 in their areas of DNA replication (14, 44) and apparently did not evolve to eliminate p53 activity. This argues that p53 may support some aspects of virus DNA replication. The mechanism that recruits p53 to the sites of adenovirus DNA is unknown. Since the concentration of DNA ends and single-stranded DNA is strongly increased when compared with the cellular chromosomes, it is possible that p53 might directly interact with the viral DNA through its C-terminal portion (2, 3). This way, p53 may contribute to the formation of the panhandle structure that joins the DNA ends during DNA replication, through its reported ability to anneal DNA strands (41). Alternatively, components of the DNA repair machinery, such as TFIIH (38, 39), may relocate to the large amount of single-stranded DNA within the replication centers and recruit p53 to the sites of virus DNA. The identification of the exact mechanism responsible for this p53 location may also shed new light on the unsolved question how p53 “senses” the presence of unusual DNA forms that are the result of DNA damage.
On the other hand, an excess of p53 activity is likely to have an inhibitory effect on virus production. First, the induction of apoptosis would limit the synthesis of new virus particles. Further, viruses that lack the E4 region are known to accumulate large concatemers of virus DNA (40). Possibly, an excess of p53 tends to join single-stranded DNA ends from several virus genomes rather than the two ends of the same DNA molecule. Alternatively, excess p53 may trigger the activation of DNA repair mechanisms that result in the ligation of virus DNA. At least in some cases, viruses lacking the ability to control p53 due to a loss of E1B-55-kDa were shown to selectively replicate in tumor cells without wild-type p53 (6). Also, viruses lacking E1B-55-kDa can replicate almost exclusively when infecting the cells during S phase (15). For these reasons, it is conceivable that the virus replicates more efficiently by tightly limiting the activity of p53 by expressing E4orf6 in addition to E1B-55-kDa later during infection.
Several gene products within adenovirus are known to regulate p53 directly or indirectly. First, E1A proteins are known to enhance p53 expression (8, 21). E1B-55-kDa directly binds and inactivates p53 (32, 42). The E1B-19-kDa gene product was reported to act as an inhibitor of p53-induced transcriptional repression and apoptosis (34). The E4orf3 protein abolishes E1B-55-kDa-mediated inhibition of p53 (this study). In contrast, E4orf6 enhances p53 inhibition, and it acts in concert with E1B-55-kDa to destabilize the p53 protein (29, 35). Some of these viral gene products regulate p53 activity and abundance in opposite directions. This argues that the virus evolved to adjust p53 activity in infected cells as a function of time after infection, by the differential expression of several virus proteins. According to this model, the adenovirus E4orf3 protein is a novel internal regulator of p53 activity.
ACKNOWLEDGMENTS
We thank H.-D. Klenk and R. Arnold for their generous support. We are indebted to J. Boyer and G. Ketner, as well as E. Querido and P. Branton, for their generous supply of antibodies to E4orf3 and E4orf6, respectively. We thank P. Hearing and T. Shenk for recombinant viruses; J. Flint, G. Ketner, and T. Shenk for antibodies; K. Leppard for helpful discussions; and D. Gemsa for generous permission to use his luminometer.
This work was supported by the German Research Foundation and the P. E. Kempkes foundation. C.K. received a fellowship from the Fazit foundation, and M.D. was a recipient of the Stipendium für Infektionsbiologie of the German Cancer Research Center.
REFERENCES
- 1.Babiss L E, Ginsberg H S, Darnell J E., Jr Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakalkin G, Selivanova G, Yakovleva T, Kiseleva E, Kashuba E, Magnusson K P, Szekely L, Klein G, Terenius L, Wiman K G. p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res. 1995;23:362–369. doi: 10.1093/nar/23.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakalkin G, Yakovleva T, Selivanova G, Magnusson K P, Szekely L, Kiseleva E, Klein G, Terenius L, Wiman K G. p53 binds single-stranded DNA ends and catalyzes DNA renaturation and strand transfer. Proc Natl Acad Sci USA. 1994;91:413–417. doi: 10.1073/pnas.91.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 5.Beltz G A, Flint S J. Inhibition of HeLa cell protein synthesis during adenovirus infection. Restriction of cellular messenger RNA sequences to the nucleus. J Mol Biol. 1979;131:353–373. doi: 10.1016/0022-2836(79)90081-0. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 7.Blair Zajdel M E, Blair G E. The intracellular distribution of the transformation-associated protein p53 in adenovirus-transformed rodent cells. Oncogene. 1988;2:579–584. [PubMed] [Google Scholar]
- 8.Braithwaite A, Nelson C, Skulimowski A, McGovern J, Pigott D, Jenkins J. Transactivation of the p53 oncogene by E1a gene products. Virology. 1990;177:595–605. doi: 10.1016/0042-6822(90)90525-v. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho T, Seeler J S, Ohman K, Jordan P, Petterson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobbelstein M, Arthur A K, Dehde S, van Zee K, Dickmanns A, Fanning E. Intracistronic complementation reveals a new function of SV40 T antigen that co-operates with Rb and p53 binding to stimulate DNA synthesis in quiescent cells. Oncogene. 1992;7:837–847. [PubMed] [Google Scholar]
- 11.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 13.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 14.Fortunato E A, Spector D H. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J Virol. 1998;72:2033–2039. doi: 10.1128/jvi.72.3.2033-2039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrum F D, Ornelles D A. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J Virol. 1997;71:548–561. doi: 10.1128/jvi.71.1.548-561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodrum F D, Shenk T, Ornelles D A. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol. 1996;70:6323–6335. doi: 10.1128/jvi.70.9.6323-6335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang M M, Hearing P. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol. 1989;63:2605–2615. doi: 10.1128/jvi.63.6.2605-2615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ketner G, Bridge E, Virtanen A, Hemström C, Pettersson U. Complementation of adenovirus E4 mutants by transient expression of E4 cDNA and deletion plasmids. Nucleic Acids Res. 1989;17:3037–3048. doi: 10.1093/nar/17.8.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 21.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 22.Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muganda P, Carrasco R, Qian Q. The human cytomegalovirus IE2 86 kDa protein elevates p53 levels and transactivates the p53 promoter in human fibroblasts. Cell Mol Biol (Noisy-Le-Grand) 1998;44:321–331. [PubMed] [Google Scholar]
- 24.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordqvist K, Ohman K, Akusjarvi G. Human adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol Cell Biol. 1994;14:437–445. doi: 10.1128/mcb.14.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ornelles D A, Shenk T. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J Virol. 1991;65:424–429. doi: 10.1128/jvi.65.1.424-429.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puvion-Dutilleul F, Pedron J, Cajean-Feroldi C. Identification of intranuclear structures containing the 72K DNA- binding protein of human adenovirus type 5. Eur J Cell Biol. 1984;34:313–322. [PubMed] [Google Scholar]
- 29.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth J, Dobbelstein M, Freedman D A, Shenk T, Levine A J. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 1998;17:554–564. doi: 10.1093/emboj/17.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarnow P, Hearing P, Anderson C W, Halbert D N, Shenk T, Levine A J. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 33.Sarnow P, Sullivan C A, Levine A J. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology. 1982;120:510–517. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y, Shenk T. Relief of p53-mediated transcriptional repression by the adenovirus E1B 19-kDa protein or the cellular Bcl-2 protein. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steegenga W T, Riteco N, Jochemsen A G, Fallaux F J, Bos J L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus-infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- 36.Tigges A M, Raskas H J. Expression of adenovirus-2 early region 4: assignment of the early region 4 polypeptides to their respective mRNAs, using in vitro translation. J Virol. 1982;44:907–921. doi: 10.1128/jvi.44.3.907-921.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voelkerding K, Klessig D F. Identification of two nuclear subclasses of the adenovirus type 5-encoded DNA-binding protein. J Virol. 1986;60:353–362. doi: 10.1128/jvi.60.2.353-362.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X W, Vermeulen W, Coursen J D, Gibson M, Lupold S E, Forrester K, Xu G, Elmore L, Yeh H, Hoeijmakers J H, Harris C C. The XPB and XPD DNA helicases are components of the p53-mediated apoptosis pathway. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- 39.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G, et al. p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 40.Weiden M D, Ginsberg H S. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc Natl Acad Sci USA. 1994;91:153–157. doi: 10.1073/pnas.91.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu L, Bayle J H, Elenbaas B, Pavletich N P, Levine A J. Alternatively spliced forms in the carboxy-terminal domain of the p53 protein regulate its ability to promote annealing of complementary single strands of nucleic acids. Mol Cell Biol. 1995;15:497–504. doi: 10.1128/mcb.15.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 43.Zantema A, Fransen J A, Davis-Olivier A, Ramaekers F C, Vooijs G P, DeLeys B, Van der Eb A J. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology. 1985;142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhong L, Hayward G S. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J Virol. 1997;71:3146–3160. doi: 10.1128/jvi.71.4.3146-3160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]