Abstract
Human immunodeficiency virus type 1 (HIV-1) normally assembles into particles of 100 to 120 nm in diameter by budding through the plasma membrane of the cell. The Gag polyprotein is the only viral protein that is required for the formation of these particles. We have used an in vitro assembly system to examine the assembly properties of purified, recombinant HIV-1 Gag protein and of Gag missing the C-terminal p6 domain (Gag Δp6). This system was used previously to show that the CA-NC fragment of HIV-1 Gag assembled into cylindrical particles. We now report that both HIV-1 Gag and Gag Δp6 assemble into small, 25- to 30-nm-diameter spherical particles in vitro. The multimerization of Gag Δp6 into units larger than dimers and the formation of spherical particles required nucleic acid. Removal of the nucleic acid with NaCl or nucleases resulted in the disruption of the multimerized complexes. We conclude from these results that (i) N-terminal extension of HIV-1 CA-NC to include the MA domain results in the formation of spherical, rather than cylindrical, particles; (ii) nucleic acid is required for the assembly and maintenance of HIV-1 Gag Δp6 virus-like particles in vitro and possibly in vivo; (iii) a wide variety of RNAs or even short DNA oligonucleotides will support assembly; (iv) protein-protein interactions within the particle must be relatively weak; and (v) recombinant HIV-1 Gag Δp6 and nucleic acid are not sufficient for the formation of normal-sized particles.
The assembly of virus particles occurs through the organized multimerization of numerous protein subunits, although in some cases nucleic acid is also required (for a review, see reference 21). In retroviruses, such as human immunodeficiency virus type 1 (HIV-1), a virus-like particle can be assembled in eukaryotic cells from the viral Gag polyprotein. The first step in assembly which is visible by electron microscopy (EM) is the accumulation of Gag proteins into electron-dense patches beneath the plasma membrane of the cell. These patches enlarge and project outward from the cell to form spherical, budding particles, which pinch off and are released into the environment. Freshly budded particles have an immature morphology; the Gag proteins are located around the periphery of the particle, under the plasma membrane-derived lipid envelope, giving a doughnut-shaped appearance. Soon after budding, the viral protease (PR) is activated and some of the peripheral material condenses into a cone-shaped core at the center of the HIV-1 particle. This is a mature virus particle.
The HIV-1 Gag polyprotein is composed of separate domains, which are (from the N to the C terminus) the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains. Short spacer peptides are also present between the CA and NC domains (p2) and between the NC and p6 domains (p1). The viral protease cleaves Gag at the junctions of these domains to produce the mature structural proteins MA, CA, NC, and p6 as well as p2 and p1.
The functions of these domains during assembly are different from the functions of the mature proteins. The MA domain is important for the transport of Gag from within the cell to the plasma membrane. This domain is cotranslationally modified by the addition of myristic acid to the N terminus, and mutations or drug treatments which prevent myristylation also prevent the association of Gag with the plasma membrane. In these cases, particles assemble in the cytoplasm rather than on the plasma membrane (38, 46). The CA domain appears to guide the arrangement of the Gag molecules during assembly. Even small mutations within the CA domain can prevent particle assembly (9, 10, 58) or alter the size of the particle (9, 10). (By comparison, the CA domain of Rous sarcoma virus [RSV] can tolerate substantial deletions without preventing particle assembly [56], although the size of the particle is altered [27, 56]). As a mature protein, CA forms the shell around the viral core. The NC domain packages the viral RNA genome and promotes Gag-Gag interactions, presumably mediated by RNA binding (4). As a mature protein within the virus particle, NC protects the RNA genome at the center of the core. The function of the p6 domain, during or after assembly, remains unclear. When Gag alone is expressed, the p6 domain can be deleted without causing any significant defect in particle assembly (22, 46). However, in the context of the complete viral genome, deletion of p6 results in a late-assembly defect. In this case, particle assembly appears to proceed normally except that the particles remain tethered to the plasma membrane (18). This defect is dependent on the expression of an active PR (24).
The functions of these domains within HIV-1 Gag have primarily been identified through genetic analysis and examination of the properties of the mutant particles in vivo. The cellular environment is difficult to manipulate, and thus it is not clear whether the observed Gag mutant phenotypes are purely the result of defective Gag-Gag interactions or are due to interactions with cellular factors. Numerous cellular proteins have been confirmed (e.g., cyclophilin A) or are suspected to interact with Gag during assembly (for a review, see reference 40). Cell-free systems, involving the use of wheat germ or reticulocyte lysates, have recently been developed to study retroviral assembly (29, 47, 48, 55). These systems are more amenable to manipulation, and the results from these studies suggest that at least one cellular protein, which requires ATP for activity, is critical for HIV-1 and Mason-Pfizer monkey virus Gag assembly (29, 55). However, these systems also contain numerous cellular factors, making it difficult to identify purely Gag-Gag interactions.
A fully defined, in vitro assembly system which uses Gag protein, or fragments of Gag, expressed in Escherichia coli has been previously developed. The viral proteins can be purified in a soluble form, without denaturation, and used for in vitro assembly studies. Using this system for RSV Gag proteins, it was observed that purified proteins by themselves did not efficiently assemble into organized, virus-like particles. Efficient assembly required the addition of nucleic acid (only RNA, not DNA, was used in these studies). The CA-NC fragment of RSV Gag formed cylindrical particles with RNA (8), but when the protein was extended N terminally to also include the MA-p2-p10 domains of RSV, spherical particles were formed (7). The spherical particles that assembled in vitro were similar in appearance to authentic, immature particles which had been stripped of their lipid envelopes (49). Further analysis showed that the p10 domain was responsible for the spherical, rather than cylindrical, shape of these particles (7). The CA-NC and CA-NC-p6 fragments of HIV-1 Gag also formed cylindrical particles with RNA (8, 20).
We report here on the in vitro assembly properties of HIV-1 Gag and a Gag mutant missing the p6 domain (Gag Δp6). Both proteins formed spherical particles in the presence of nucleic acid. A wide variety of nucleic acids supported assembly, although there were upper and lower limits to the lengths of nucleic acids which could be used. However, the spherical particles which were formed in vitro were much smaller (∼30 nm in diameter) than HIV-1 particles formed in vivo (∼100 to 120 nm). These results suggest models for how nucleic acid, and possibly cellular proteins, may be used in assembly.
MATERIALS AND METHODS
Plasmids and cells.
All plasmids were constructed by using common subcloning techniques and propagated in the DH5α strain of E. coli. After confirmation of their identity by restriction digestion, the plasmids were moved into the BL21 DE3 pLys S strain of E. coli for protein expression. The plasmids pET 3xc HIV CA-NC and CA-NC-p6 were previously used for bacterial expression of the CA-NC and CA-NC-p6 fragments, respectively, of the BH10 isolate of HIV-1 Gag and have been described elsewhere (8). For expression of HIV Gag, the MA and CA segments of the gag gene of HIV-1 strain BH10 (plasmid BH10 gp II [GenBank; NIDg 326383], a generous gift from Hans-Georg Kräusslich) were amplified with a primer containing an XbaI site and an NdeI site, ATTAATCTAGACATATGGGTGCGAGAGCGTC (5′ primer at the beginning of MA, including nucleotides [nt] 112 to 128) and a primer containing sequences within CA downstream of the PstI site, TTACTTGGCTCATTGCTTCAGCCA (3′ primer within CA, nt 1199 to 1176). The amplified product was then digested with XbaI and PstI and cloned between the XbaI and PstI sites of pBluescript KS II(+) (Stratagene). The NdeI-SpeI fragment of this sequence was then substituted for the NdeI-SpeI fragment of pET 3xc HIV CA-NC-p6 to create pET 3xc HIV Gag. The pET 3xc HIV Gag Δp6 construct was created by substituting the SpeI-KpnI fragment of pET 3xc HIV CA-NC for the same fragment of pET 3xc HIV Gag. The Gag Δp6 construct contains a termination codon after the last codon in p1 (resulting in the deletion of p6). The initial Gag Δp6 clones produced by this procedure expressed high levels of protein, and this protein was used for the experiments described herein. Subsequent sequencing analysis revealed that this clone contained two point mutations, a substitution of valine for alanine at position 37 (A37V) and Q63D. The wild-type sequence was recloned into this plasmid, using a ClaI-SpeI restriction enzyme digest of BH10 gp II, and confirmed by sequence analysis. The key experiments described herein were repeated with protein expressed from the wild-type clone; the two point mutations had no effect on the results.
Protein purification.
E. coli BL21 DE3 pLys S cells were grown and induced for protein expression as described previously (7, 50, 51). HIV Gag and HIV Gag Δp6 were purified by a protocol previously described for the purification of soluble RSV proteins (7), except that 0.1% Nonidet P-40 (NP-40) was included in the initial lysis buffer.
A final purification step, involving the use of an antibody affinity column, was added to the procedure for purification of full-length Gag protein. A synthetic peptide (Chiron Mimetope, Clayton, Victoria, Australia) containing an N-terminal cysteine and the last 21 amino acids (DKELYPLTSLRSLFGNDPSSQ) of BH10 p6 was used to generate polyclonal rabbit antibodies against p6. Specific antibodies were affinity purified by using the C-terminal p6 peptide bound to a Sulfo-Link column (Pierce Chemical Company, Rockford, Ill.) according to the manufacturer’s instructions. An HIV Gag affinity column was made by conjugating these antibodies to protein A-agarose (Boehringer Mannheim, Indianapolis, Ind.) according to the manufacturer’s instructions. Partially purified HIV Gag was bound to the column at room temperature in a solution containing 20 mM Tris (pH 7.5), 0.5 M NaCl, 10 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride and then eluted with the p6 peptide (2 mg/ml) in the same buffer. Purified proteins (5 to 10 mg/ml) were stored in storage buffer (20% glycerol, 20 mM Tris [pH 7.5], 0.5 M NaCl, 10 mM DTT, and 1 mM phenylmethylsulfonyl fluoride) at −20°C.
In vitro assembly and analysis by EM.
Protein at 5 mg/ml in storage buffer was slowly diluted fivefold (to 1 mg/ml protein and 0.1 M NaCl) by dropwise addition of 20 mM Tris (pH 8.0)–10 mM DTT (0.5% NP-40 was included when indicated in the text) at room temperature. When nucleic acid was used in the assembly reactions, it was added prior to dilution and at a nucleic acid/protein ratio of 4% (wt/wt) unless otherwise specified. The reactions were routinely allowed to proceed for 2 h prior to examination by sedimentation (60 min at 21,000 × g in an Eppendorf model 5417R refrigerated microcentrifuge at 4°C) or by EM on Formvar-carbon-coated grids after negative staining with 2% uranyl acetate. E. coli cells expressing viral proteins were also examined for the presence of virus-like particles. For these assays, cells were collected by centrifugation 1 h after induction and processed for thin sectioning as described previously (7).
Nucleic acids.
Yeast tRNA, E. coli rRNA, and bacteriophage MS2 RNA were purchased from Boehringer Mannheim. Total E. coli RNA was purified as described in reference 2. DNA oligonucleotides were purchased from Life Technologies Inc. (Gaithersburg, Md.). Oligonucleotides are designated according to sequence and length; i.e., TGTGT = TG 5, TGTGTGTGTG = TG 10, etc. In vitro RNA transcripts were made by using the T7 RNA polymerase, buffers, and protocols of Promega (Madison, Wis.). The template used for transcription was pNL4-3 H3, which contains a portion of the HIV-1 pNL4-3 genomic sequence downstream from a T7 promoter in pUC 19. The transcripts initiated at nt 454 (i.e., nt 1 of the genomic RNA [1]) and terminated at various positions downstream. The longest transcript used terminated at nt 1507.
Cross-linking experiments.
For cross-linking with dimethyl suberimidate (DMS; Pierce), the assembly reactions were carried out with the dilution buffer containing 20 mM HEPES (pH 8.5) instead of Tris. After 2 h, DMS in the same buffer was added for another hour, and then the reaction was stopped with 20 mM glycine (pH 2.5) prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
RESULTS
Purification and analysis of HIV-1 Gag expressed in E. coli.
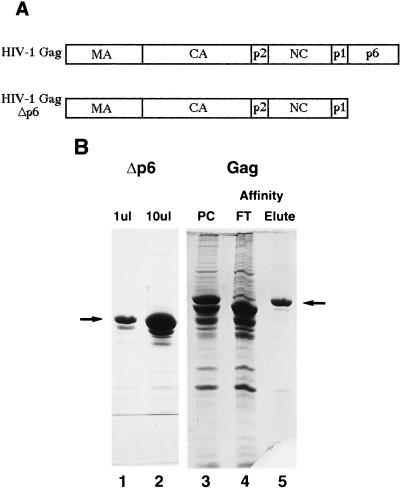
We made two constructs for the expression of HIV-1 Gag proteins in E. coli, one expressing full-length Gag and the other expressing Gag Δp6 (Gag missing the p6 domain) (Fig. 1A). Because these proteins were purified from E. coli, they were not myristylated at the amino terminus as they would have been if expressed in eukaryotic cells. When purified as described here, Gag Δp6 was soluble to about 10 mg/ml and was determined to be about 85 to 90% homogeneous by SDS-PAGE (Fig. 1B, lanes 1 and 2). A slightly smaller degradation product, presumably the result of cleavage by a bacterial protease, was also present. However, the full-length Gag protein was extensively degraded (Fig. 1B, lane 3); only about 30 to 40% of the viral protein appeared intact. This has been observed by others (6, 12, 30) and appears to be due to cleavage near the C terminus of Gag during expression in E. coli. An affinity column to which were conjugated antibodies against the C terminus of p6 was used to purify the intact Gag away from the degradation products (Fig. 1B, lane 5). The identities of all proteins were confirmed by N-terminal sequencing, migration in SDS-PAGE, and immunoblotting with anti-CA and -NC antibodies, as well as with anti-p6 antibodies in the case of Gag. Gag Δp6 protein was used in the majority of the following experiments because of the low yields of and difficulty of purifying the full-length Gag.
FIG. 1.
Schematic diagrams of proteins expressed in E. coli and purified proteins. (A) Schematic diagram of HIV-1 Gag and Gag Δp6 used for in vitro assembly. (B) Coomassie blue-stained SDS-polyacrylamide gel of purified proteins. Lanes: 1 and 2, 10 and 100 μg of purified HIV-1 Gag Δp6, respectively; 3, HIV-1 Gag purified in the same manner as Gag Δp6; 4 and 5, anti-p6 antibody affinity purification of HIV-1 Gag; 4, flow-through (FT) from the affinity column; 5, affinity-purified HIV-1 Gag (eluted with p6 peptide). Arrows indicate the protein band referred to in each lane. PC, purified on a phosphocellulose cation-exchange column.
EM analysis of particles assembled in vitro.
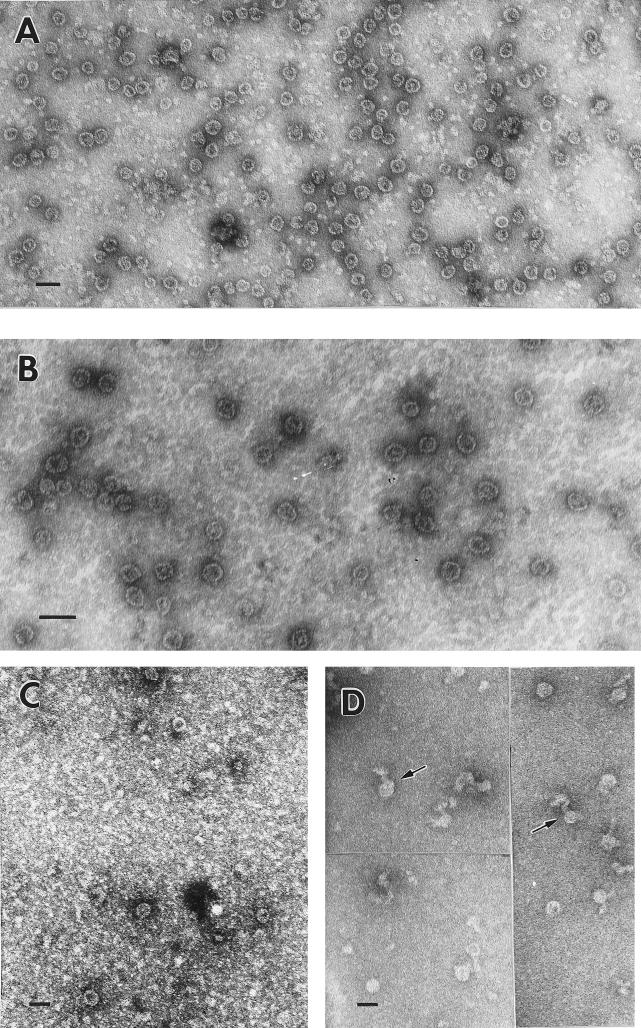
The CA-NC and CA-NC-p6 fragments of HIV-1 Gag have previously been shown to assemble in vitro into cylindrical particles under conditions of high pH (8.0) and low salt (0.1 M NaCl) in the presence of RNA (8). We used the same conditions for assembly of Gag and Gag Δp6. Protein stored at −20°C in storage buffer was routinely thawed on ice and centrifuged at 21,000 × g for 15 min prior to use. The supernatant was then diluted fivefold with 20 mM Tris (pH 8.0)–10 mM DTT. Under these conditions, EM revealed that both HIV-1 Gag and Gag Δp6 proteins formed sheets and occasional irregular round particles when incubated without nucleic acid (data not shown). These structures were similar to the ones formed by RSV Gag proteins in the absence of RNA (7, 8). When nucleic acid was included in the reaction mixture, the solution quickly became visibly turbid on dilution. The turbidity settled to the bottom of the tube within a few hours. Examination of the reactions by negative-stain EM revealed that under these conditions Gag Δp6 formed numerous round particles, 25 to 30 nm in diameter (Fig. 2A and B). Our measurements were not sufficiently precise to determine whether this size range represented two species of distinctly sized particles (25 and 30 nm) or a gradation of different sizes (25 to 30 nm). Analysis of the negative staining pattern suggested that these particles are spherical; in most cases, the stain penetrated the particles, indicating that they were hollow or at least had regions of low protein density. A central mass of protein was frequently visible within the particles which had been penetrated by the negative stain. In some cases, 16 to 20 radial striations were visible around the circumference of each particle.
FIG. 2.
Negatively stained EM images of Gag Δp6 and Gag particles assembled in vitro. (A) Gag Δp6 particles assembled with yeast tRNA; (B) Gag Δp6 particles assembled with an in vitro transcript of HIV-1 packaging sequence (∼1 kb); (C) Gag particles assembled at 4°C with E. coli rRNA; (D) Gag Δp6 particles assembled with MS2 bacteriophage RNA (∼3.5 kb); arrows indicate tails protruding from particles. All panels were negatively stained with 2% uranyl acetate. Scale bars = 50 nm.
These reactions were routinely carried out at room temperature, and Gag Δp6 produced numerous particles at that temperature. In contrast, full-length Gag formed heterogeneous, elongated structures in the presence of RNA at room temperature (data not shown). These structures were interpreted to be protein-bound RNA strands which had not completely assembled into organized particles. At 4°C, both full-length Gag (Fig. 2C) and Gag Δp6 assembled into uniform, small particles identical in appearance to those formed by Gag Δp6 at room temperature. However, the number of particles produced by full-length Gag was very small, and the majority of the protein was present in elongated structures similar to those formed at room temperature.
Normal HIV-1 virions produced from infected or transfected cells have a diameter of approximately 100 to 120 nm. The particles produced in this study in vitro had a significantly smaller diameter. To determine whether the smaller size of these particles is the result of a specific set of conditions or is an inherent property of purified HIV-1 Gag, we varied several conditions, i.e., pH, salt concentration, type of nucleic acid (see below), and Gag protein concentration. In all cases in which particles were formed, they were still 25 to 30 nm in diameter (data not shown).
Retroviral Gag proteins, or fragments of Gag, have been shown to assemble into virus-like particles within E. coli (7, 26). We performed thin-section EM on E. coli expressing HIV-1 Gag and Gag Δp6 in order to determine if HIV-1 Gag would form normal-sized particles in E. coli. As positive controls, we also examined E. coli expressing RSV Gag ΔPR, which has been shown to assemble in E. coli (7), and the CA-NC fragment of HIV-1 Gag, which forms 50-nm-diameter cylindrical particles in vitro (8). Both RSV Gag ΔPR and HIV CA-NC were observed to form the expected particles in E. coli, and these particles were readily apparent (data not shown). In contrast, HIV Gag and Gag Δp6 did not form any identifiable structures in E. coli (data not shown).
RNA requirements for in vitro assembly.
The small particles were formed by Gag Δp6 when a wide variety of nucleic acids were added: total E. coli RNA (primarily tRNA and rRNA), E. coli rRNA (a mixture of 1.6- and 3.5-kb rRNA), yeast tRNA (∼90 bases), in vitro-transcribed RNAs (up to 1 kb) containing HIV-1 packaging sequences, and short DNA oligonucleotides. No differences between the particles which were formed with these different RNAs were observed. However, bacteriophage MS2 RNA (3.5 kb) did affect the formation of spherical particles. When products of assembly reactions with MS2 RNA were examined by EM, most of the protein seemed to have formed disorganized, elongated structures; however, some spherical particles were present. These were still only 25 to 30 nm in diameter, but many of them had extra material, or tails, associated with them (Fig. 2D). We interpreted this to mean that only part of the RNA strand was used for assembly (e.g., 2 kb) and that the remainder was left as an extruded tail. This result suggests that there is an upper limit to the size of the nucleic acid which can be packaged into these particles, but determining the precise limit by EM would be difficult due to the extrusion of the extra RNA.
Particles can form from short DNA oligonucleotides.
Having determined that there was an upper limit to the length of the nucleic acid which could be incorporated into spherical particles, we decided to find out whether a lower size limit existed. Initial experiments used arbitrarily chosen DNA oligonucleotides of various lengths and sequences. Using these oligonucleotides, we found that a length of 24 nt was sufficient for efficient particle assembly, as determined by EM (data not shown). To be more systematic in our approach, we next used oligonucleotides of defined length and sequence, i.e., multiples of 5 nt in length and either poly(dA) or poly(dTG) sequence. These two particular sequences were chosen because HIV-1 NC has been shown to have a significantly higher affinity for poly(dT-G) than for poly(dA) sequences (13). We also used a precipitation assay to quantitate the multimerization efficiency. Assembly reaction mixtures were prepared with the different oligonucleotides at a fixed oligonucleotide/protein ratio. We included 0.5% NP-40 in these reactions because we had previously observed that a considerable percentage of the protein (30 to 50%) adhered to the sides of the tube in the absence of NP-40 (data not shown). Half of each assembly reaction mixture was centrifuged in a microcentrifuge for 1 h to produce a pellet and a supernatant fraction. We estimated that under these conditions, multimers of at least 30 to 50 Gag molecules would be pelleted. Figure 3 shows an example of the results of this assay for a particular set of conditions, while Fig. 4 shows a graphical summary of the results under all of the conditions tested.
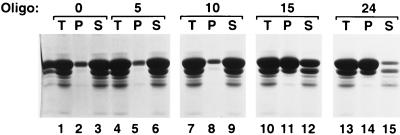
FIG. 3.
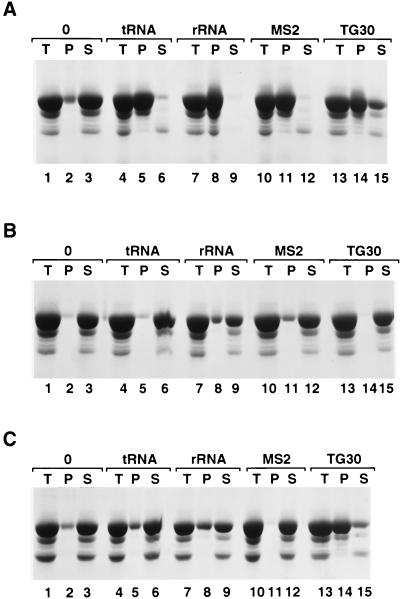
Precipitation of HIV-1 Gag Δp6 with poly(dTG) oligonucleotides (Oligo) of different lengths. HIV-1 Gag Δp6 (100 μg at 5 mg/ml in a solution containing 20 mM Tris [pH 7.5], 0.5 M NaCl, 0.5% NP-40, and 10 mM DTT) was mixed with no oligonucleotide (lanes 1 to 3) or with a 5-base (lanes 4 to 6), 10-base (lanes 7 to 9), or 15-base (lanes 10 to 12) TG oligonucleotide or the 24-base arbitrary oligonucleotide referred to in the text (lanes 13 to 15). The reaction mixtures were then diluted fivefold in pH 8.0 buffer without NaCl (final conditions, 100 μg [at 1 mg/ml] of protein, pH 8.0, 0.1 M NaCl, 0.5% NP-40, 10 mM DTT, 4 μg of oligonucleotide) and incubated for 2 h at room temperature. The precipitates were pelleted by centrifugation in a microcentrifuge for 1 h. Equal portions of the total (T), pellet (P), and supernatant (S) fractions were analyzed by SDS-PAGE and Coomassie blue staining.
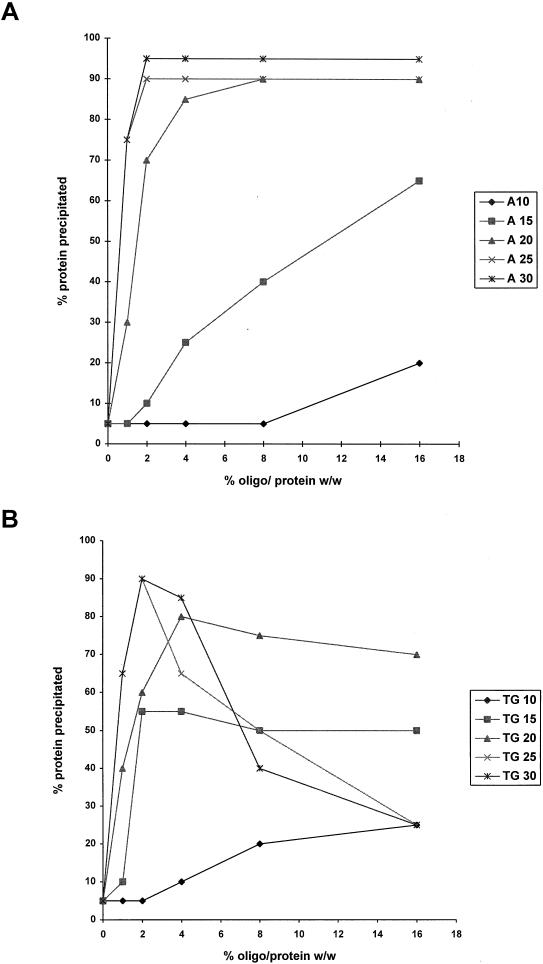
FIG. 4.
Precipitation of HIV-1 Gag Δp6 with oligonucleotides of different lengths, sequences, and oligonucleotide/protein ratios. Assembly reactions with oligonucleotides of different lengths (10, 15, 20, 25, or 30 nt long) and at different oligonucleotide/protein ratios (0, 1, 2, 4, 8, or 16% [wt/wt]) were performed and the products analyzed as described in the legend to Fig. 3. The percentage of protein in each pellet fraction was quantitated by densitometry. (A) Poly(dA) oligonucleotides; (B) poly(dTG) oligonucleotides.
Very little protein was precipitated in experiments in the absence of nucleic acid (Fig. 3, lane 2) or with poly(dTG) oligonucleotides of 5 or 10 nt at a 4% (wt/wt) oligonucleotide/protein ratio (Fig. 3, lanes 5 and 8), while the majority of the protein was multimerized with oligonucleotides of 15 nt or longer (Fig. 3, lanes 11 and 14). In this experiment, we included as a control (Fig. 3, lanes 13 to 15) the arbitrarily chosen 24-base oligonucleotide (GGGCCCCAGTACTTACCAGGAAGG) which initial EM experiments had indicated was sufficient for particle formation.
We used the precipitation assay to find out how Gag Δp6 would behave with different ratios of protein to nucleic acid and whether the behavior depended on the nucleic acid sequence as well as on its length. We used poly(dA) and poly(dTG)oligonucleotides of different lengths and at nucleic acid/protein ratios of 0, 1, 2, 4, 8, and 16% (wt/wt) for the precipitation assay and quantitated the amount of precipitated protein on SDS-PAGE gels by using a densitometer. A nucleic acid/protein mass ratio was used instead of a molar ratio to facilitate comparison of oligonucleotides of different lengths. A nucleic acid/protein ratio of 4% (wt/wt) corresponds to about 6 nt/protein molecule, approximately the binding site size for NC on nucleic acids (13, 25).
The results with poly(dA) oligonucleotides were the simplest to interpret (Fig. 4A). Short oligonucleotides (10 or 15 bases) resulted in very little precipitation, although the amount of precipitation did increase at higher nucleic acid/protein ratios. A dramatic difference was observed when 20-, 25-, or 30-base oligonucleotides were used. These three curves were very similar. A significant amount of protein was precipitated at low oligonucleotide/protein ratios (1 to 2%). The amount of precipitated protein was very large (90 to 95%) and remained constant when A 20–30 was used at higher oligonucleotide/protein ratios (8 to 16%).
The results with poly(dTG) oligonucleotides (Fig. 4B) were very different from those with poly(dA), and the oligonucleotides of different length could be placed into three groups according to their behavior. TG 10 supported very little multimerization, although it was significantly more efficient than A 10. TG 15 and TG 20 both resulted in efficient precipitation at low percentages of oligonucleotide (2 to 4%), but the amount of precipitation did not change significantly at higher oligonucleotide concentrations. TG 25 and TG 30 resulted in very efficient precipitation at low oligonucleotide percentages, but the level of precipitation declined dramatically at higher oligonucleotide percentages. Similar experiments using yeast tRNA or E. coli rRNA gave results similar to those obtained with TG 25 and TG 30 (data not shown).
Examination of the reaction products by EM confirmed these results: particles or multimeric complexes were observed only in reactions involving the use of longer oligonucleotides. Increasing the length of the oligonucleotides above the lower size limit [15 nt for poly(dTG) or 20 nt for poly(dA)] tended to increase the number of complete, spherical particles which were observed (data not shown). Inclusion of 0.5% NP-40 in the assembly reaction mixture did not noticeably affect the formation or appearance of the particles. The multimerization of Gag Δp6 was clearly affected by the length, sequence, and relative amount of nucleic acid.
Cross-linked protein complexes in the presence of nucleic acid.
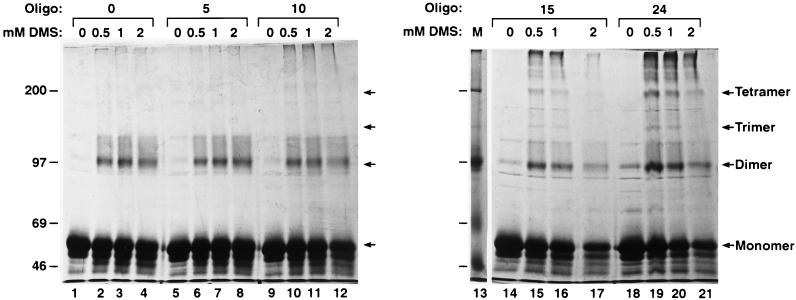
To gain a more detailed insight into how the multimerization process occurred, we performed chemical cross-linking on the products of the assembly reactions, using poly(dTG) and poly(dA) oligonucleotides (Fig. 5). DMS, which cross-links primary amines, has been used previously to cross-link MA, CA, and NC proteins in mature avian sarcoma and leukemia viruses (42–44) and in both mature and immature murine leukemia virus (41, 42). Assembly reaction mixtures with Gag Δp6 and oligonucleotides of increasing length (4% oligonucleotide/protein ratio) were incubated for 2 h prior to cross-linking with increasing concentrations of DMS (0 to 2 mM) and SDS-PAGE. Monomers and dimeric cross-linking products were present in all cross-linking reactions, even in the absence of oligonucleotides (Fig. 5, lanes 2 to 4). Larger cross-linking products (e.g., trimers or tetramers) were observed only when the oligonucleotides were longer than 15 nt for poly(dT-dG) (Fig. 5, lanes 15 to 17) or 20 nt for poly(dA) (data not shown). The dimeric and tetrameric bands were more intense than the trimeric bands, suggesting that multimerization occurs through dimers.
FIG. 5.
Cross-linking of HIV-1 Gag Δp6 with oligonucleotides (Oligo) of different lengths. The total reaction mixtures from the experiment of Fig. 3 were cross-linked with increasing amounts (0, 0.5, 1, and 2 mM) of DMS prior to SDS-PAGE and Coomassie blue staining. Lanes: 1 to 4, no oligonucleotide (0); 5 to 8, 5-base poly(dTG); 9 to 12, 10-base poly(dTG); 14 to 17, 15-base poly(dTG); 18 to 21, arbitrary 24-base oligonucleotide referred to in the text. The sizes of the molecular mass markers (M; lane 13) are indicated on the left (in kilodaltons). The positions of the Gag Δp6 cross-linking products are indicated on the right.
Gag Δp6 multimers are disassociated by NaCl or RNase.
The above results all suggest that the formation of multimeric complexes by HIV-1 Gag requires nucleic acid. However, assembly presumably involves protein-protein interactions as well as protein-nucleic acid interactions. Are these protein-protein interactions sufficient to maintain the integrity of the multimeric complex once it has formed, or is the nucleic acid still required after assembly? We addressed this question by removing the nucleic acid from the particles with nucleases or by adding NaCl to assembled particles to disrupt the protein-nucleic acid interactions. Figure 6 shows an example of the precipitation assay using yeast tRNA, E. coli rRNA, bacteriophage MS2 RNA, or the TG 30 oligodeoxynucleotide. All of these nucleic acids were quite efficient at precipitating Gag Δp6 (Fig. 6A, lanes 5, 8, 11, and 14). After assembly was allowed to proceed for 2 h, NaCl was added to these assembled complexes to a final concentration of 0.5 M. Twenty minutes later, the protein complexes were centrifuged, and very little protein was present in the pellet (Fig. 6B, lanes 5, 8, 11, and 14). When RNase A, instead of NaCl, was added at 0.1 mg/ml for 20 min, a similar result was observed, except that the complexes containing the TG 30 DNA oligonucleotide were not affected (Fig. 6C, lanes 5, 8, 11, and 14). EM also revealed that no spherical particles remained after NaCl or (in reactions with RNA) RNase A treatment (data not shown).
FIG. 6.
Precipitation of HIV-1 Gag Δp6 with RNA is reversible with RNase A or NaCl treatment. Lanes: 1 to 3, in vitro assembly reactions with HIV-1 Gag Δp6 and no oligonucleotide (0); 4 to 6, yeast tRNA, 4%; 7 to 9, E. coli rRNA, 4%; lanes 10 to 12, bacteriophage MS2, RNA 4%; 13 to 15, poly(dT-dG) 30 DNA, 4% oligonucleotide. T, total; P, pellet; S, supernatant. (A) Assembly reactions in 0.1 M NaCl; (B) assembly reactions from panel A to which NaCl was added to 0.5 M NaCl for 20 min prior to sedimentation; (C) assembly reactions from panel A to which RNase A was added (at 0.1 mg/ml) for 20 min prior to sedimentation.
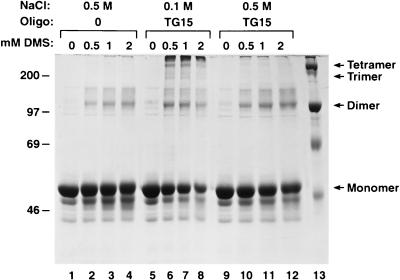
We also performed cross-linking experiments to determine the extent to which the multimeric complexes had been disassociated. After assembly was allowed to proceed in the presence of TG 15 (Fig. 7) or TG 30 (data not shown), NaCl was added to 0.5 M and the cross-linking agent was introduced. Protein in 0.5 M NaCl, without nucleic acid, was also cross-linked as a control. We found that Gag Δp6 which had assembled with TG oligonucleotides in 0.1 M NaCl was efficiently cross-linked by DMS into larger multimeric forms (Fig. 7, lanes 5 to 8), but only monomers and cross-linked dimers were present if 0.5 M NaCl had been added (Fig. 7, lanes 9 to 12). The extents of cross-linking in 0.5 M NaCl with and without nucleic acid (Fig. 7; compare lanes 1 to 4 with lanes 9 to 12) appeared to be about the same. A similar cross-linking experiment, using the RNase A-treated complexes formed with yeast tRNA (Fig. 6C), also showed a reduction in the amount of cross-linked products larger than dimers (data not shown). However, this reduction (∼50%) was not as dramatic as the above-described results obtained with NaCl. Presumably Gag Δp6 can partially protect the RNA from RNase treatment, as has been shown for HIV-1 NC (53).
FIG. 7.
Cross-linking of NaCl-treated HIV-1 Gag Δp6–TG 15 assemblies. Assembly reactions using TG 15, at a oligodeoxynucleotide/protein ratio of 4% (wt/wt), were cross-linked with increasing amounts (0, 0.5, 1, and 2 mM) of DMS prior to SDS-PAGE and Coomassie blue staining. All reaction mixtures were assembled and cross-linked for the same amount of time. Lanes: 1 to 4, no oligonucleotide (Oligo) (0), in 0.5 M NaCl; 5 to 8, TG 15, in 0.1 M NaCl; 9 to 12, TG 15 assembly reaction (in 0.1 M NaCl) to which NaCl was added to 0.5 M for 20 min prior to cross-linking. The sizes of the molecular mass markers (lane 13) are indicated on the left (in kilodaltons). The positions of the Gag Δp6 cross-linking products are indicated on the right.
DISCUSSION
We have used an in vitro assembly system to examine the properties of recombinant HIV-1 Gag and Gag Δp6 proteins. These proteins were purified from E. coli as completely soluble proteins without denaturation. Since this system uses purified viral proteins, the results of this study represent the behavior of HIV-1 Gag proteins in the absence of other viral or cellular cofactors.
To the best of our knowledge, this is the first study of the in vitro assembly properties of isolated full-length HIV-1 Gag or of Gag which is lacking only the p6 domain. Other investigators have previously used in vitro systems to study the assembly properties of fragments of HIV-1 Gag. Purified HIV-1 CA can form rod-shaped or tubular structures in vitro (11, 20) under conditions of high NaCl and protein concentrations. N-terminal extension of the CA protein to include all or part of the MA domain results in spherical particles which are heterogeneous in size (19, 54). The CA-NC and CA-NC-p6 fragments of HIV-1 Gag also form tubular structures at low NaCl and protein concentrations if RNA is also included in the assembly reaction mixture (8, 20).
The formation of HIV-1 Gag and Gag Δp6 particles in vitro occurred with a wide variety of nucleic acids. Using oligonucleotides of defined length and sequence, we have identified a minimal nucleic acid length which will support Gag Δp6 multimerization. This minimal length was found to be dependent on the nucleic acid sequence and may reflect the affinity of Gag Δp6 for the particular sequence. The results presented above are of particular importance in light of our current knowledge of how HIV-1 virions are assembled.
Retroviral Gag proteins are generally imagined to be shaped like tapered rods or cones: wide at the N terminus (MA) and narrow at the C terminus (NC). Thus, the proteins would naturally pack together to form a sphere, with the individual proteins extending radially inward from the N to the C terminus. This view is supported by EM analysis of immature particles, which reveals a striated pattern within the protein shell at the periphery of the particle (14, 23, 39, 57). It is likely that the individual striations are Gag monomers. Based on the known dimensions of the mature HIV-1 MA, CA, and NC proteins (15, 16, 32–34, 36, 37, 52), as well as cryo-EM of immature virus-like particles (14), HIV-1 Gag is believed to be about 12 to 17 nm in length. These studies, as well as others (3, 45), suggest that in a retroviral particle, Gag monomers are arranged in a roughly hexagonal network, with the centers of the hexagons being 5 to 7 nm apart. This organized packing may occur in patches or domains rather than over the entire particle (14).
Our results suggest that the protein arrangement within the HIV-1 Gag Δp6 particles assembled in vitro is similar to the protein arrangement within in vivo particles, although the in vitro particles are much smaller (Fig. 2). We also observed a striated pattern in the particles assembled in vitro. The spacing of these striations (16 to 20 striations around a 30-nm-diameter particle = 5 to 6 nm between striations) is consistent with the protein arrangement within in vivo particles. We estimate that each 30-nm particle contains approximately 80 to 120 Gag Δp6 molecules, assuming that the striations observed also represent individual Gag monomers (with each striation occupying 25 to 36 nm2 [(5 to 6 nm)2] of the surface area [∼2,800 nm2] of a 30-nm-diameter particle). The fact that Gag Δp6 particles were disrupted by RNase treatment suggests that RNase can gain access to the center of the particles, presumably through the porous packing network. The Gag Δp6 molecules should be of sufficient length (12 to 17 nm) to extend almost to the center of the 30-nm-diameter particles reported here. This may explain why particles made in vitro from full-length Gag formed only in the cold; there may not have been enough room within the particles to include the p6 domain except under conditions of low thermal vibrations. The negative staining pattern of these particles indicated that a central protein mass (presumably NC and/or CA) was separated from the surface of the particle (MA) by a less-protein-dense region which was filled with stain. This is consistent with the radial density plot for HIV-1 Gag virus-like particles observed by cryo-EM (14), in which two shells of high protein density are separated by a region of lower protein density.
In this study, both HIV-1 Gag and Gag Δp6 assembled into small (25- to 30-nm-diameter) spherical particles in vitro. This is significantly smaller than the size range (100 to 120 nm) for HIV-1 particles assembled in vivo. The small size of the in vitro particles remained constant under all of the conditions which supported assembly. However, when the same recombinant protein was incubated in the presence of cellular lysates, larger particles were formed (unpublished data). Some factor(s) present within the cell lysates must therefore be responsible for the formation of normal-sized virus-like particles. It seems likely that neither RSV nor Mason-Pfizer monkey virus requires such a factor(s), since purified Gag proteins from these viruses assemble into normal-sized particles in vitro (7, 26). We have not yet identified the responsible factor(s) or determined how it interacts with HIV-1 Gag.
The process by which retroviral Gag proteins multimerize and assemble into particles is not well understood. One of the most striking aspects of the HIV-1 Gag proteins studied here is the absolute requirement for nucleic acid in the assembly of particles in vitro. We found that assembly could occur on short oligodeoxynucleotides (Fig. 3 to 5) as well as on RNA. Furthermore, nucleic acid was required for maintaining the multimeric complex after it formed, since treatment with nuclease or NaCl disrupted preformed multimers (Fig. 6 and 7). The formation of an organized virus-like particle must also involve protein-protein interactions, in order to produce the spherical shape. Therefore, the assembly process depends on both protein-nucleic acid interactions and protein-protein interactions. However, these protein-protein interactions must be rather weak since the particles are disrupted when the protein-nucleic acid interactions are eliminated by nuclease treatment.
It is significant that oligonucleotides with as few as 10 to 15 bases are sufficient for multimerization (Fig. 3 and 4). The assembly of large numbers of Gag proteins on such a short nucleic acid implies that many nucleic acid molecules, as well as many Gag molecules, are incorporated into each multimeric complex. Thus, assembly requires that the Gag molecules not only bind to nucleic acid but also straddle or bridge the gap between individual oligonucleotide molecules. The observation that Gag Δp6 cross-linked products were produced as multiples of two (Fig. 5 and 7) suggests that multimerization occurs through the formation of Gag dimers. It is possible that 10 to 15 bases represents the minimum length to which two Gag molecules can attach, so that at each end a Gag dimer can bridge the gap to another oligonucleotide molecule. The net result would be a chain of protein dimers linked together by nucleic acid. The formation of large multimers might be more efficient with longer nucleic acids, including dimeric viral RNA, because fewer bridging events would have to occur at the ends of the nucleic acid. Additional protein-protein interactions would then cause this protein-nucleic acid chain to wind up into a spherical particle. This model is basically a refinement of one proposed earlier for the assembly of RSV particles in vitro (7, 8).
We also found that poly(dA) and poly(dTG) oligonucleotides differ in their ability to support multimerization of Gag Δp6 in vitro (Fig. 4). Since free NC protein has different affinities for these sequences (13), this difference in the ability to support multimerization strongly suggests that the Gag Δp6 molecules bind nucleic acid by their NC domains. However, the sequence-dependent differences were restricted to a very limited set of conditions, i.e., low concentrations of 10- to 15-base oligonucleotides and high concentrations of 25- to 30-base oligonucleotides.
While we do not fully understand the significance of the discrimination between dA and d(TG) oligonucleotides with respect to their ability to support multimerization in vitro, it seems likely that further experimentation in this area will provide us with important information. The d(TG) oligonucleotides, for which the NC domain of Gag presumably has a high affinity (13), were significantly more efficient than the dA oligonucleotides at the shortest lengths tested (i.e., 10 to 15 bases). This result suggests that with very short oligonucleotides, the strength of the Gag-nucleic acid interaction can be a limiting factor in the multimerization process. In contrast, at lengths of 25 bases or more, high concentrations of d(TG), but not dA, oligonucleotides inhibited multimerization. Perhaps the central portion of a relatively long d(TG) molecule can act as a sink, binding Gag proteins in a region in which they are unable to undergo bridging interactions with other oligonucleotides, or with Gag molecules on other oligonucleotides, as required for multimerization. The inhibitory effect of longer oligonucleotides on assembly was observed only with high-affinity d(TG) oligonucleotides and only when protein concentrations were not saturating, but not with lower-affinity dA oligonucleotides or, presumably, with other low-affinity oligonucleotides. Since viral genomic RNA, as well as other cellular RNAs, probably contains few high-affinity binding sites but many more lower-affinity binding sites, this inhibitory effect should not prevent assembly in vivo, where the protein concentrations may be less than saturating relative to the available RNA.
It has been known for many years that retroviral assembly in vivo is independent of the presence of genomic RNA. That is, while assembly is normally accompanied by an exquisitely specific protein-RNA interaction (i.e., packaging of genomic RNA), this interaction is completely unnecessary for assembly. Retrovirus particles are still produced from mutants which do not package genomic RNA (for a review, see reference 5) and in cells lacking packageable genomic RNA (28, 31). In these cases, the particles do not contain genomic RNA and are sometimes referred to as RNA-less or empty. However, upon closer examination, it is evident that these particles, like normal virions, still contain a substantial number of small cellular RNA molecules (17, 28, 35). We have found that nucleic acid is necessary for assembly in vitro, yet genomic RNA is clearly not required for assembly in vivo. Therefore, either (i) nucleic acid is not required for assembly in vivo or (ii) the small cellular RNAs can support assembly in vivo.
In one possible model, in every normal virus particle, the majority of the Gag proteins would use these small RNAs as structural components during the assembly of the particle. Meanwhile, a small percentage of Gag would specifically package the genomic RNA into the particle. Thus, every particle would have both genomic and nongenomic RNA, with the majority of Gag being bound to the nongenomic RNA and with the genomic RNA being essentially naked except at the specific packaging sequence(s). Alternatively, Gag might use the genomic RNA as a structural component during assembly. If Gag was produced in the cell in excess over the amount of packageable genomic RNA, the remaining Gag might then use nongenomic RNA for assembly. The resulting particles would thus consist of two populations, those containing primarily genomic RNA and those containing only nongenomic RNA. Further analysis of authentic virions will be necessary to test this hypothesis.
In summary, we have shown that HIV-1 Gag polyprotein is capable of self-assembly into regular, ordered structures. This assembly process requires the presence of nucleic acid, but we have found little evidence of specificity with respect to the identity of the nucleic acid. The spherical particles which are formed under these conditions are much smaller than authentic virions. It seems likely that some factor in vivo alters the normal radius of curvature with which the proteins assemble, but the nature of this putative factor is not yet known.
ACKNOWLEDGMENTS
This research was sponsored by the National Cancer Institute, Department of Health and Human Services, under contract with ABL.
We thank Volker Vogt and Jackson Ho for the initial construction of the HIV-1 Gag expression vector and Swati Joshi for suggesting dilution conditions for in vitro assembly. We also thank Rulong Shen and Eliana Munoz for electron microscopy technical support, Demetria Harvin for in vitro RNA transcripts, and Volker Vogt and Judith Levin for helpful comments on the manuscript.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 4.4.4–4.4.7. [Google Scholar]
- 3.Barklis E, McDermott J, Wilkins S, Schabtach E, Schmid M F, Fuller S, Karanjia S, Love Z, Jones R, Rui Y, Zhao X, Thompson D. Structural analysis of membrane-bound retrovirus capsid proteins. EMBO J. 1997;16:1199–1213. doi: 10.1093/emboj/16.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R, Fisher J, Goff S P. RNA packaging. In: Kräusslich H-G, editor. Morphogenesis and maturation of retroviruses. Heidelberg, Germany: Springer-Verlag; 1996. pp. 177–218. [Google Scholar]
- 6.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell S, Vogt V M. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chazal N, Carrière C, Gay B, Boulanger P. Phenotypic characterization of insertion mutants of the human immunodeficiency virus type 1 Gag precursor expressed in recombinant baculovirus-infected cells. J Virol. 1994;68:111–122. doi: 10.1128/jvi.68.1.111-122.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman T, Bukovsky A, Öhagen Å, Höglund S, Göttlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrlich L S, Agresta B E, Carter C A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehrlich L S, Fong S, Scarlata S, Zybarth G, Carter C. Partitioning of HIV-1 Gag and Gag-related proteins to membranes. Biochemistry. 1996;35:3933–3943. doi: 10.1021/bi952337x. [DOI] [PubMed] [Google Scholar]
- 13.Fisher R J, Rein A, Fivash M, Urbaneja M A, Cases-Finet J R, Medaglia M, Henderson L E. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller S D, Wilk T, Gowen B D, Kräusslich H-G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 15.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 16.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 17.Gorelick R J, Nigida S M, Arthur L O, Henderson L E, Rein A. Roles of nucleocapsid cysteine arrays in retroviral assembly and replication: possible mechanisms in RNA encapsidation. In: Kumar A, editor. Advances in molecular biology and targeted treatment for AIDS. New York, N.Y: Plenum Press; 1991. pp. 257–272. [Google Scholar]
- 18.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross I, Hohenberg H, Huckhagel C, Kräusslich H-G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross I, Hohenburg H, Kräusslich H-G. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249:592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 21.Harrison S C. Principles of virus structure. In: Fields B N, et al., editors. Virology. New York, N.Y: Raven Press Ltd.; 1990. pp. 37–61. [Google Scholar]
- 22.Hockley D J, Nermut M V, Grief C, Jowett J B M, Jones I M. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J Gen Virol. 1994;75:2985–2997. doi: 10.1099/0022-1317-75-11-2985. [DOI] [PubMed] [Google Scholar]
- 23.Hockley D J, Wood R D, Jacobs J P, Garrett A J. Electron microscopy of human immunodeficiency virus. J Gen Virol. 1988;69:2455–2469. doi: 10.1099/0022-1317-69-10-2455. [DOI] [PubMed] [Google Scholar]
- 24.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karpel R L, Henderson L E, Oroszlan S. Interactions of retroviral structural proteins with single-stranded nucleic acids. J Biol Chem. 1987;262:4961–4967. [PubMed] [Google Scholar]
- 26.Klikova M, Rhee S S, Hunter E, Ruml T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J Virol. 1995;69:1093–1098. doi: 10.1128/jvi.69.2.1093-1098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin J G, Grimley P M, Ramseur J M, Berezesky I K. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingappa J R, Hill R L, Wong M L, Hegde R S. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luban J, Goff S P. Binding of human immunodeficiency virus type 1 (HIV-1) RNA to recombinant HIV-1 gag polyprotein. J Virol. 1991;65:3203–3212. doi: 10.1128/jvi.65.6.3203-3212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann R, Mulligan R C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 32.Massiah M A, Starich M R, Paschall C M, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:194–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 33.Massiah M A, Worthylake D, Christensen A M, Sundquist W I, Hill C P, Summers M F. Comparison of the NMR and X-ray structures of the HIV-1 matrix protein: evidence for conformational changes during viral assembly. Protein Sci. 1996;5:2391–2398. doi: 10.1002/pro.5560051202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews S, Barlow P, Boyd J, Barton G, Russell R, Mills H, Cunningham M, Meyers N, Burns N, Clark N, Kingsman S, Kingsman A, Campbell I. Structural similarity between the p17 matrix protein of HIV-1 and interferon-γ. Nature. 1994;370:666–668. doi: 10.1038/370666a0. [DOI] [PubMed] [Google Scholar]
- 35.Méric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Erlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 37.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix J-L, Roques B P. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morikawa Y, Hinata S, Tomoda H, Goto T, Nakai M, Aizawa C, Tanaka H, Omura S. Complete inhibition of human immunodeficiency virus Gag myristoylation is necessary for inhibition of particle budding. J Biol Chem. 1996;271:2868–2873. doi: 10.1074/jbc.271.5.2868. [DOI] [PubMed] [Google Scholar]
- 39.Nermut M V, Hockley D J, Jowett J B M, Jones I M, Garreau M, Thomas D. Fullerene-like organization of HIV Gag-protein shell in virus-like particles produced by recombinant baculovirus. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 40.Ott D E. Cellular proteins in HIV virions. Rev Med Virol. 1997;7:167–180. doi: 10.1002/(sici)1099-1654(199709)7:3<167::aid-rmv199>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 41.Pepinsky R B. Localization of lipid-protein and protein-protein interactions within the murine retrovirus Gag precursor by a novel peptide-mapping technique. J Biol Chem. 1983;258:11229–11235. [PubMed] [Google Scholar]
- 42.Pepinsky R B, Cappiello D, Wilkowski C, Vogt V M. Chemical crosslinking of proteins in avian sarcoma and leukemia viruses. Virology. 1980;102:205–210. doi: 10.1016/0042-6822(80)90081-1. [DOI] [PubMed] [Google Scholar]
- 43.Pepinsky R B, Vogt V. Identification of retrovirus matrix proteins by lipid-protein cross-linking. J Mol Biol. 1979;131:819–837. doi: 10.1016/0022-2836(79)90203-1. [DOI] [PubMed] [Google Scholar]
- 44.Pepinsky R B, Vogt V M. Fine-structure analyses of lipid-protein and protein-protein interactions of gag protein p19 of the avian sarcoma and leukemia viruses by cyanogen bromide mapping. J Virol. 1984;52:145–153. doi: 10.1128/jvi.52.1.145-153.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao Z, Belyaev A S, Fry E, Roy P, Jones I M, Stuart D I. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 46.Royer M, Cerutti M, Gay B, Hong S-S, Devauchelle G, Boulanger P. Functional domains of HIV-1 Gag-polyprotein expressed in baculovirus-infected cells. Virology. 1991;184:417–422. doi: 10.1016/0042-6822(91)90861-5. [DOI] [PubMed] [Google Scholar]
- 47.Sakalian M, Parker S D, Weldon R A, Jr, Hunter E. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J Virol. 1996;70:3706–3715. doi: 10.1128/jvi.70.6.3706-3715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spearman P, Ratner L. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J Virol. 1996;70:8187–8194. doi: 10.1128/jvi.70.11.8187-8194.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Studier F W, Moffat B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 51.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 52.Summers M F, Henderson L E, Chance M R, Bess J W, Jr, South T L, Blake P R, Sagi I, Perez-Alvarado G, Sowder R C I, Hare D R, Arthur L O. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanchou V, Gabus C, Rogemond V, Darlix J-L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J Mol Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 54.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weldon R A, Jr, Parker W B, Sakalian M, Hunter E. Type D retrovirus capsid assembly and release are active events requiring ATP. J Virol. 1998;72:3098–3106. doi: 10.1128/jvi.72.4.3098-3106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y, Jones I M, Hockley D J, Nermut M V, Roy P. Complementation of human immunodeficiency virus (HIV-1) Gag particle formation. Virology. 1994;199:403–408. doi: 10.1006/viro.1994.1138. [DOI] [PubMed] [Google Scholar]