Abstract
There is significant evidence that the etiology of chronic otorhinolaryngology infections such as chronic rhinosinusitis, adenotonsillitis, and otitis media depends on biofilms. As biofilm-forming bacteria can be resistant to the immune system, antibiotics, and other treatments, biofilm infections are often chronic. To identify the genus and species of the clinical isolates obtained from the swabs collected from the patients with chronic infections of the nasal and paranasal sinus, nasopharynx, and oropharynx and to evaluate phenotypic and genotypic methods for the detection of biofilms and antimicrobial resistance among the isolated organisms. A total of 100 patients with chronic rhinosinusitis and adenotonsillitis participated in this study. Various clinical samples from the nasal cavity, nasopharynx, and oropharynx were obtained and subjected to microbiological analysis and biofilm-forming capacity by three methods: tube methods, Congo red staining, and microtiter plate method. The various specific genes were amplified by polymerase chain reaction. The amplified gene products were separated by gel electrophoresis. This was a prospective cohort study conducted on a total of 100 patients with chronic rhinosinusitis and adenotonsillitis. The age of the study participants was between 7 and 53 years with a mean age of 29.22 ± 15.03. This study included 54 (54%) nasal tissue samples and 46 (46%) adenotonsillar tissue. The frequently cultured organisms are coagulase-negative staphylococci (17%), E. coli (10%), Citrobacter (10%), and Klebsiella (7%). Staphylococcus aureus (4), and Methicillin-resistant Staphylococcus aureus (3) produced strong biofilm. Acenobacter (3), Citrobacter (4), and E. coli (4) showed moderate biofilm production. Coagulase-negative Staphylococcus aureus (11), E. coli (6), and Klebsiella (7) showed weak biofilm formation. Citrobacter (6), and Coagulase negative Staphylococcus aureus (6) were negative for biofilm production. Staphylococcus aureus expressed mecA gene (3) and Panton-Valentine Leukocidin gene (2), Pseudomonas expressed mucA gene (2), Citrobacter expressed blaCARB-2 (4) qnrA gene (2), E. coli expressed bla SHV (2) and bla TEM1 gene (2) and Klebsiella expressed Kfu (2) and uge (1). Acenobacter was negative for blaIMP1, blaVIM2 genes. This study adds to the information on the common pathogens-forming biofilms in various nasal pathologies and adenotonsillitis. The knowledge that a particular organism has a higher biofilm-forming capacity will help to sensitize the physician that factors such as biofilms may be at play and take appropriate measures.
Keywords: Biofilm, Adenotonsillitis, Chronic rhinosinusitis, Microbiota
Introduction
Infections of the upper respiratory tract (URT) include inflammation of the nose (rhinitis), sinuses (sinusitis), the middle ear (otitis media), pharynx (pharyngitis), tonsils (tonsillitis), and larynx (laryngitis) [1]. Inflammation of the nasal and paranasal sinus mucosa for at least 12 consecutive weeks is called chronic rhinosinusitis (CRS), which is frequently encountered in everyday practice [2]. There must be congestion in at least one nostril, discharge from at least one nostril, postnasal drip, hyposmia, a headache, or pain in at least one facial region for a diagnosis of rhinitis to be made. The pathophysiology of chronic rhino-sinusitis (CRS) is poorly understood despite extensive research efforts. It is challenging for doctors to manage patients because it is difficult to differentiate and determine the contribution of anatomic, environmental, and host immunological factors implicated in the etiology [3].
The Waldeyer’s lymphatic ring, which consists of the adenoid, which is present in the nasopharynx at the junction of the roof and posterior wall, the tubal tonsils, which are close to the pharyngeal openings of the Eustachian tubes, the palatine tonsils, which are in the oropharynx, and the lingual tonsil, which is present on the posterior one-third of the tongue, acts as a barrier to the microorganisms [4]. Multiple, recurrent infections of the tonsils result in the development of chronic tonsillitis. The disease's etiology results from numerous consecutive episodes of acute tonsillitis or from a persistent infection that causes chronic inflammation that lasts for a long time and advances gradually. Every lymphatic component of the Waldeyer’s ring is susceptible to inflammation. Inflammation most frequently affects the palatine tonsils and adenoids [5].
According to the Centers for Disease Control and Prevention (CDC), biofilms are believed to be the main cause of at least 65% of all human illnesses [6]. A biofilm is a collection of microorganisms in which cells adhere to one another on a surface and build a matrix of extracellular polymeric substances that are controlled by the expression of the cell-to-cell adhesion mediator polysaccharide intracellular adhesion molecule [7]. They may damage local tissue by remaining dormant and hidden from the immune system and subsequently result in an acute infection. The bacteria within the biofilm alter their metabolism, gene expression, and protein synthesis to adapt to the anoxic environment and nutritional deficiency, which can result in a slower metabolic rate and a slower rate of cell division and make the bacteria more resistant to antimicrobial therapy [8]. The biofilm serves like a physical barrier that keeps antibiotics from penetrating.
Multiple different types of bacteria can always be found in the microbiota of the various URT locations. The genera Streptococcus, Neisseria, Haemophilus, Moraxella, Staphylococcus, Corynebacterium, Propionibacterium, Prevotella, and Porphyromonas are the most frequently isolated bacteria. The most prevalent among them are Haemophilus influenzae, Moraxella catarrhalis, Streptococcus pneumoniae, and Staphylococcus aureus [9].
This study aimed to identify the genus and species of the clinical isolates obtained from the swabs collected from the patients with chronic infections of the nasal and paranasal sinus, nasopharynx, and oropharynx and to evaluate phenotypic and genotypic methods for the detection of biofilms and antimicrobial resistance among the isolated organisms.
Materials and Methodology
Study Design
The study was a prospective cohort study conducted in the department of Otorhinolaryngology at Chettinad hospital and research institute, Chettinad academy of research and education (tertiary care hospital), Chengalpet district.
Study Duration
One year.
Sample Size Calculation
Sample size calculation was done based on the incidence rate, n = z2p (1 − p)/d2 where n = sample size Z = Z statistic for a level of confidence (1.96) P = expected prevalence of proportion (expected prevalence was 5%) d = precision (if precision was 5%, then d = 0.05).
Subject Selection
Patients with Chronic Rhinosinusitis and Adenotonsillitis infections, who were planned for tonsillectomy, adenoidectomy, and Functional Endoscopic Sinus Surgery (FESS) procedure.
Inclusion Criteria
Patient between belonging to both sexes and age group of 6–55 years of age.
Patient with either chronic tonsillitis, tonsillar hypertrophy, adenoid hypertrophy, chronic rhinosinusitis with or without nasal polyps.
Exclusion Criteria
Patients below the age group of 6 years.
Patients above the age group of 55 years.
Patients with signs of acute tonsillitis, acute pharyngeal infection, peritonsillitis, peritonsillar abscess, acute sinusitis, and suspected malignant neoplasm.
Method
The study was performed on a total of 100 patients, in the department of Otorhinolaryngology, at Chettinad Hospital and Research institute, Chettinad academy of research and education, after getting approved by the Ethical Committee of the Chettinad University. A standard anaesthetic protocol was used for all the patients. After General anaesthesia, the usual physiologic parameters (respiratory rate, pulse rate, blood pressure, and temperature) were monitored during surgery. Standard surgical techniques were performed. Intra-operatively mucosa from nasal and paranasal sinuses, tonsils, and adenoid tissue was collected aseptically for bacteriological investigation (Figs. 1, 2, 3). Patients were extubated and shifted to the postoperative ward.
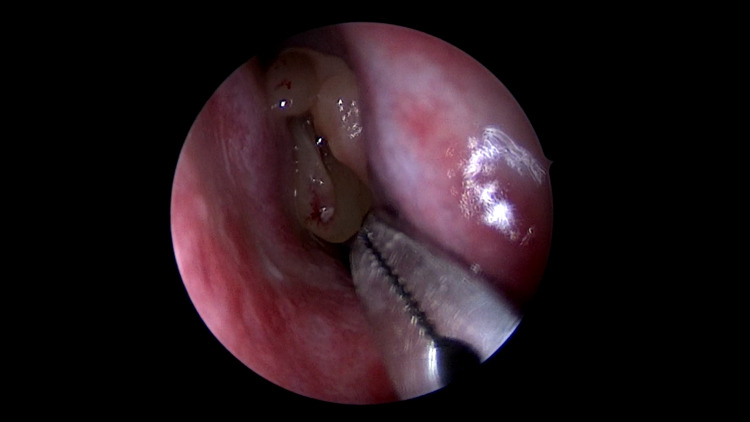
Fig. 1.
Nasal polyp involving left nasal cavity
Fig. 2.
Bilateral tonsils in the oropharynx
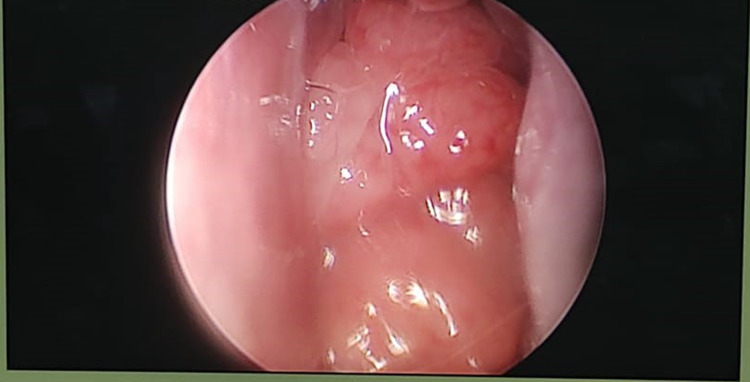
Fig. 3.
Endoscopy image showing adenoid
Specimen Collection
Using proper aseptic precautions, specimen were collected in a saline container. These clinical specimens were transported to the laboratory within the 2 h of collection for culture.
Phenotyping
The samples were inoculated onto primary plating media like blood agar, chocolate agar, MacConkey agar, thioglycolate broth and Sabouraud Dextrose agar incubated at 37 °C overnight and were studied at 24 and 48 h. The cultured colonies will be evaluated using Gram staining. These colonies were further subjected to various biochemical tests and also antimicrobial susceptibility testing by disc diffusion method.
Biofilm Production
The isolates' capacity to produce biofilms was examined using three different techniques: congo red staining, microtiter plate analysis, and tube procedures (Figs. 4, 5, 6) [10–12]. The optical density at 595 nm (OD595 nm) of a well containing stained adherent bacteria and CW, the OD595 nm of the stained control wells containing bacterium-free media, were used to determine the biofilm formation (BF) using the formula: BF = AB − CW.
Fig. 4.

Congo red staining showing biofilm production [10]
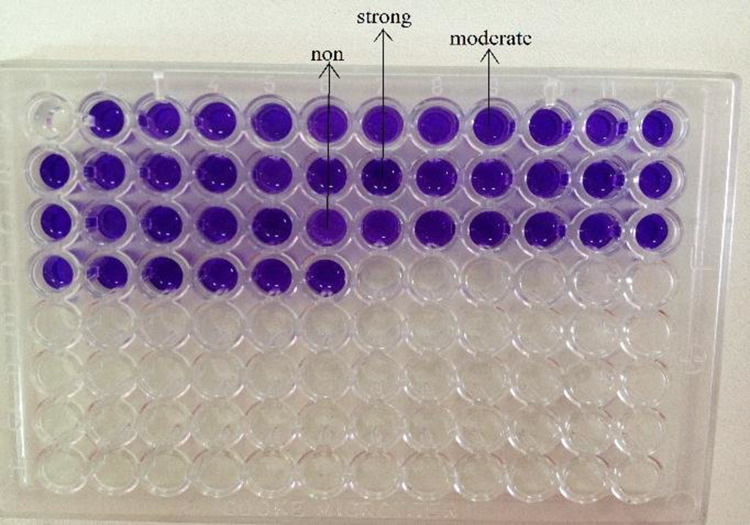
Fig. 5.
Microtitre platting technique [11]
Fig. 6.

Tube method for production of biofilm [12]
The samples were divided into four categories as follows: strong: greater than or equal to 0.300; moderate: between 0.22 and 0.29; weak: between 0.100 and 0.199; and negative: less than or equal to 0.100 [7].
Based on the antibiogram created for the isolates, oral antibiotics were prescribed postoperatively. Patients who received negative culture results also received and managed with broad-spectrum antibiotics.
Genotypic Methods
The bacterial genomes were extracted from the culture using the boiling method. The isolates were subjected to polymerase chain reaction (PCR) to identify the specific genes for various cultured organisms. This is done by using various primers specific to each gene. mecA and Panton-Valentine Leukocidin gene were specific to Staphylococcus aureus [13]. Pseudomonas isolates were analysed to detect mucA and lasR genes and Citrobacter for blaCARB-2, qnrA genes [14, 15]. bla TEM1, bla SHV and bla CTX-M genes for E. coli, mag A, Kfu and Uge genes for Klebsiella and blaIMP1, blaVIM2 for Acenobacter [16–18]. The PCR products were separated by gel electrophoresis.
Statistical Analysis
The Statistical Package for Social Sciences (SPSS)-Version 19 was used to evaluate the data, which was put into an Excel spreadsheet. For quantitative variables, descriptive statistics were generated with mean, standard deviation, and proportions (%). Chi Square and cross table were employed to examine the claim.
Ethical Consideration
Institutional ethical committee approval was obtained before starting of the study. Confidentiality of study participants was maintained in all the phases of study.
Results
This study was conducted on a total of 100 patients, between the age of 7 and 53 years with a mean age of 29.22 and a standard deviation was 15.03. About 57 (57%) were males and 43 (43%) were females in this study. 11 (11%) patients had diabetes mellitus, 9 (9%) patients had systemic hypertension, and 7 (7%) patients had ischaemic heart disease (Table 1).
Table 1.
Distribution of demographical distribution
| Age | Minimum (years) | Maximum (years) | Mean | SD |
|---|---|---|---|---|
| N = 100 | 7 | 53 | 29.22 | 15.03 |
| Gender | Frequency (N = 100) | Percentage (%) | ||
| Male | 57 | 57 | ||
| Female | 43 | 43 | ||
| Co-morbidities | Frequency (N = 100) | Percentage (%) | ||
| DM | 11 | 11 | ||
| SHTN | 9 | 9 | ||
| IHD | 7 | 7 | ||
The specimen for analysis were taken from nasal tissue in 54 (54%) patients and from adenotonsillar tissue in 46 (46%) patients (Fig. 7).
Fig. 7.
Nature of specimen
During the culture of the organism, the organism cultured was coagulase-negative Staphylococcus in 17 (17%) samples. Citrobacter was found in 10 (10%) samples, Escherichia coli in 10 (10%) samples, Klebsiella in 7 (7%) samples, and Staphylococcus aureus in 4 (4%) samples. The combination of Klebsiella and Pseudomonas was 3 (3%), Klebsiella and Staphylococcus aureus is 3 (3%), Pseudomonas and coagulase-negative staphylococci is 3 (3%), methicillin-resistant Staphylococcus aureus is 3 (3%), and Acenobacter is 3 (3%) (Fig. 8).
Fig. 8.
Distribution of organism among the study population
About 58 (59%) patients were sensitive to Gentamycin and Ciprofloxacin, 47 (47%) patients were sensitive to Piperacillin-Tazobactum, 45 (45%) had Cotrimoxazole sensitivity, 40 (40%) had Amikacin sensitivity, 34 (34%) had Tobramycin sensitivity, 31 (31%) had cefepime and cefotaxime sensitivity, 23 (23%) had Meropenem and Vancomycin sensitivity, 20 (20%) had Imepenem and Clindamycin sensitivity, 19 (19%) had Cefazolin sensitivity, 17 (17%) had Tetracyclin sensitivity and only 7 (7%) patients were sensitivity to Ampicillin (Table 2).
Table 2.
Antimicrobial sensitivity of microorganism
| Drug sensitivity | Frequency (N = 100) | Percentage (%) |
|---|---|---|
| Amikacin | 40 | 40 |
| Cefepime | 31 | 31 |
| Cefotaxim | 31 | 31 |
| Cotrimoxazole | 45 | 45 |
| Gentamycin | 58 | 58 |
| Piperacillin–Tazobactum | 47 | 47 |
| Tobramycin | 34 | 34 |
| Ciprofloxacin | 58 | 58 |
| Ampicillin | 7 | 7 |
| Cefazolin | 19 | 19 |
| Imepenem | 20 | 20 |
| Meropenem | 23 | 23 |
| Tetracyclin | 17 | 17 |
| Clindamycin | 20 | 20 |
| Vancomycin | 23 | 23 |
Among the cultured organisms, 13 (13%) became negative for biofilm formation, 13 (13%) showed strong biofilm formation, 14 (14%) showed moderate biofilm formation and 24 (24%) showed weak biofilm formation (Fig. 9).
Fig. 9.
Grading of biofilm production among organism
Based on biofilm-forming capacity, the organisms were classified into four categories: Staphylococcus aureus (N = 4), Methicillin-resistant Staphylococcus aureus (N = 3), and combination of Klebsiella with Staphylococcus aureus (N = 3), and Klebsiella with Pseudomonas (N = 3) produced strong biofilm. Acenobacter (N = 3), Citrobacter (N = 4), and E. coli (N = 4), and the combination of Pseudomonas with coagulase-negative Staphylococci (N = 3) showed moderate biofilm production. Coagulase-negative Staphylococcus aureus (N = 11), E. coli (N = 6), and Klebsiella (N = 7) showed weak biofilm formation. Citrobacter (N = 6), Coagulase negative Staphylococcus aureus (N = 6), and Enterobacter (N = 1) were negative for biofilm production (Fig. 10).
Fig. 10.
Biofilm forming capacity of the organisms
The amplified PCR products were sequenced using gel electrophoresis. Among Staphylococcus aureus, three strains were positive for mecA gene and two strains were positive for Panton-Valentine Leukocidin gene. mucA gene was found in two isolates of Pseudomonas and lasR gene was not found any isolates of Pseudomonas. blaCARB-2 was seen in four strains of Citrobacter and qnrA gene was seen in two strains of Citrobacter. Among E. coli, two strains were positive for bla SHV, two strains was positive for bla TEM1 gene and negative for bla CTX-M gene. Kfu was found in two strains of Klebsiella, uge in one strains of Klebsiella and mag A was not found. Acenobacter was negative for blaIMP1, blaVIM2 genes (Table 3).
Table 3.
Various genotypes of bacterial isolates
| Organism | Gene | No. of expressed strains |
|---|---|---|
| Staphylococcus aureus | mecA | 3 |
| Panton-Valentine Leukocidin | 2 | |
| Pseudomonas | mucA | 2 |
| lasR | 0 | |
| Citrobacter | blaCARB-2 | 4 |
| qnrA | 2 | |
| E. coli | bla SHV | 2 |
| bla TEM1 | 2 | |
| bla CTX-M | 0 | |
| Klebsiella | Kfu | 2 |
| mag A | 0 | |
| Uge | 1 | |
| Acenobacter | blaIMP1 | 0 |
| blaVIM2 | 0 |
Discussion
There are two different types of bacteria: planktonic and biofilm. Almost all bacteria prefer to exist in biofilm, which represents their preferred state. In comparison to their planktonic counterparts, the bacteria that form a biofilm exhibit several significant changes in terms of growth dynamics and genetic expression [19]. The steps in production of biofilm are initial attachment, development of three dimentional structure, biofilm development, maturity and detachment (Fig. 11) [20]. Cholesteatoma, chronic tonsillitis, and otitis media with effusion have all been linked to biofilms. Less is known about how biofilms affect the etiology of chronic rhinosinusitis.
Fig. 11.
Production of biofilm [20]
This study is a prospective cohort study conducted on a total of 100 patients with chronic rhinosinusitis and adenotonsillitis. The age of the study participants was between 7 and 53 years with a mean age of 29.22 ± 15.03. This study included 54 (54%) nasal tissue samples and 46 (46%) adenotonsillar tissue. The frequently cultured organisms are coagulase-negative staphylococci (17%), E. coli (10%), Citrobacter (10%), and Klebsiella (7%). Staphylococcus aureus (4), and Methicillin-resistant Staphylococcus aureus (3) produced strong biofilm. Acenobacter (3), Citrobacter (4), and E. coli (4) showed moderate biofilm production. Coagulase-negative Staphylococcus aureus (11), E. coli (6), and Klebsiella (7) showed weak biofilm formation. Citrobacter (6), and Coagulase negative Staphylococcus aureus (6) were negative for biofilm production. Staphylococcus aureus expressed mecA gene (3) and Panton-Valentine Leukocidin gene (2), Pseudomonas expressed mucA gene (2), Citrobacter expressed blaCARB-2 (4) qnrA gene (2), E. coli expressed bla SHV (2) and bla TEM1 gene (2) and Klebsiella expressed Kfu (2) and uge (1). Acenobacter was negative for blaIMP1, blaVIM2 genes.
Kostic et al. [1], analyzed 79 tonsil tissue obtained from patients diagnosed with Chronic Tonsillitis and studied the microbiological profile, they found Streptococcus parasanquinis, Streptococcus oralis, and S. aureus were discovered to be the most prevalent bacteria, and the existence of biofilm are the causes infections to recur. The current study showed the highest prevalence of coagulase-negative Staphylococcus, E. coli, and Citrobacter.
In 50 cases with chronic rhino sinusitis, Karunasagar et al. [3], evaluated the existence of bacteria and their capacity to generate biofilm. Staphylococcus was the most prevalent organism identified, and 11 of the 16 Staphylococcus developed biofilm. The present study showed a higher incidence of coagulase-negative Staphylococcus (17) in the culture, among which 11 produced biofilm.
When Healy et al. examined mucosa samples from 11 individuals having sinus surgery in 2008, they discovered bacterial and fungal biofilms coexisting [21]. This study analyzed 54 nasal samples and all of them showed bacterial biofilm.
In 2009, in a study by Foreman et al. [22], fungal biofilms were discovered in 11/50 (22%) of the CRS patients, with associated S. aureus infection in 7 of these cases. The current study showed coagulase-negative S. aureus in 17 samples, Methicillin-resistant S. aureus in 3 samples, and S. aureus in 4 samples.
In 2011, Boase et al. [23] showed that fungal inoculation of sheep with obstructed frontal sinuses resulted in robust biofilm formation, but only when S. aureus co-inoculation was present.
Malik et al. [24], analyzed biofilm-forming bacteria in clinical isolates of CRS patients presenting with or without nasal polyposis in 60 patients and concluded that the prevalence of bacterial Biofilms in sinonasal mucosa of CRS patients as Biofilms existed in 53% of the patients. This current study showed the production of biofilm in 51 (51%) patients out of 100 patients.
Al-Mazrou et al. [25], assessed the presence of adherent biofilms on the surface epithelium of tonsils and adenoids among 76 patients. Adherent biofilm formation was demonstrated in 46 patients (61%). This present study demonstrated the production of biofilm in 51 (51%) patients out of 100 patients.
In 2007, Swidsinski et al. [26] examined tissues from 70 patients’ tonsils and adenoids that had been removed during surgery and found that H. influenzae had invaded the tissue, Fusobacteria, Pseudomonas, and Burkholderia were only present in adherent bacterial infiltrates and layers.
Both microbiologists and doctors find it difficult to detect biofilms. Many techniques have been used successfully, but their usage in resource-poor nations is constrained by cost and accessibility. The majority of investigations rely on expensive and time-consuming techniques for direct visualization of biofilms [27].
Conclusion
A biofilm is a community of multiple microorganisms responsible for resistance to antibiotic treatment and the host immune system which acts as a reservoir that causes recurrent and chronic infections. This produces a negative impact on the quality of life and public health. It is becoming increasingly evident how bacterial biofilms contribute to the persistence and recalcitrance of CRS and chronic adenotonsillitis.
This study adds to the information on the common pathogens-forming biofilms in nasal pathologies and adenotonsillar tissue. The knowledge that a particular organism has a higher biofilm-forming capacity will help to sensitize the physician that factors such as biofilms may be at play and take appropriate measures.
Future therapies are more likely to target the disruption of their adherence and proliferation through biochemical or electromechanical mechanisms. Pulsed laser therapy has been demonstrated to remove the biofilm. Drug delivery mechanisms like electromagnetic, ultrasonic, and phototherapy need to be explored in the future.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
The approval for the study was obtained from the Human ethical committee of the Chettinad University, Chennai.
Informed Consent
Written and informed consent was taken from all patient for participation in the study. Confidentiality of patients maintained.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kostić M, Ivanov M, Babić SS, Tepavčević Z, Radanović O, Soković M, Ćirić A. Analysis of tonsil tissues from patients diagnosed with chronic tonsillitis—microbiological profile, biofilm-forming capacity and histology. Antibiotics. 2022;11(12):1747. doi: 10.3390/antibiotics11121747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fokkens WJ, Lund VJ, Hopkins C, Hellings PW, Kern R, Reitsma S, et al. European position paper on rhinosinusitis and nasal polyps 2020. Rhinology. 2020;58(Suppl S29):1–464. doi: 10.4193/Rhin20.600. [DOI] [PubMed] [Google Scholar]
- 3.Karunasagar A, Garag SS, Appannavar SB, Kulkarni RD, Naik AS. Bacterial biofilms in chronic rhinosinusitis and their implications for clinical management. Indian J Otolaryngol Head Neck Surg. 2018;70(1):43–48. doi: 10.1007/s12070-017-1208-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry M, Whyte A. Immunology of the tonsils. Immunol Today. 1998;19(9):414–421. doi: 10.1016/S0167-5699(98)01307-3. [DOI] [PubMed] [Google Scholar]
- 5.Plank L. Tonsillitis, chronic. In: Volavšek M, editor. Head and neck pathology. Encyclopedia of pathology. Cham: Springer; 2016. pp. 497–501. [Google Scholar]
- 6.Potera C. Forging a link between biofilms and disease. Science. 1999;283:1837–1839. doi: 10.1126/science.283.5409.1837. [DOI] [PubMed] [Google Scholar]
- 7.Vasanthi R, Karthikeyan D, Jeya M. Study of biofilm production and antimicrobial resistance pattern of the bacterial isolates from invasive devices. Int J Res Health Sci. 2014;31:274–281. [Google Scholar]
- 8.Vestby LK, Grønseth T, Simm R, Nesse LL. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics. 2020;9(2):59. doi: 10.3390/antibiotics9020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark SE. Commensal bacteria in the upper respiratory tract regulate susceptibility to infection. Curr Opin Immunol. 2020;66:42–49. doi: 10.1016/j.coi.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdulrahim U, Kachallah M, Rabiu M, Usman NA, Adeshina GO, Olayinka BO. Molecular detection of biofilm-producing Staphylococcus aureus isolates from national orthopaedic Hospital Dala, Kano State, Nigeria. Open J Med Microbiol. 2019;9(3):116–126. doi: 10.4236/ojmm.2019.93012. [DOI] [Google Scholar]
- 11.Al-Hadban W, Kandala N, Mohmmed H. Detection of biofilm formation and icaA gene among Iraqi clinical isolates of both methicillin resistant and susceptible Staphylococcus aureus. Curr Res Microbiol Biotechnol. 2017;5(2):1031–1037. [Google Scholar]
- 12.Eladawy M, El-Mowafy M, El-Sokkary MM, Barwa R. Effects of lysozyme, proteinase K, and cephalosporins on biofilm formation by clinical isolates of Pseudomonas aeruginosa. Interdiscip Perspect Infect Dis. 2020;2020:1–9. doi: 10.1155/2020/6156720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavalcanti VP, Camargo LA, Moura FS, Melo Fernandes EJ, Lamaro-Cardoso J, Braga CA, André MC. Staphylococcus aureus in tonsils of patients with recurrent tonsillitis: prevalence, susceptibility profile, and genotypic characterization. Braz JInfect Diseases. 2019;20(23):8–14. doi: 10.1016/j.bjid.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen SK, Rau MH, Johansen HK, Ciofu O, Jelsbak L, Yang L, Folkesson A, Jarmer HØ, Aanæs K, Von Buchwald C, Høiby N. Evolution and diversification of Pseudomonas aeruginosa in the paranasal sinuses of cystic fibrosis children have implications for chronic lung infection. ISME J. 2012;6(1):31–45. doi: 10.1038/ismej.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brecher SM, Tickler IA, Tenover FC. Phenotypic and genotypic discrepancies for carbapenemase-producing Citrobacter freundii in multiple isolates from a single patient. Ann Clin Microbiol Antimicrob. 2023;22(1):1–7. doi: 10.1186/s12941-023-00579-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnamurthy V, Vijaykumar GS, Kumar S, Prashanth HV, Prakash R, Nagaraj ER. Phenotypic and genotypic methods for detection of extended spectrum β lactamase producing Escherichia coli and Klebsiella pneumoniae isolated from ventilator associated pneumonia. J Clin Diagn Res: JCDR. 2013;7(9):1975. doi: 10.7860/JCDR/2013/6544.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massar AM, Al-Khafaji JK, Al-Shibly IK. Genotypic and phenotypic characterization of some virulence genes of Klebsiella pneumoniae in Hilla city
- 18.Uma Karthika R, Srinivasa Rao R, Sahoo S, Shashikala P, Kanungo R, Jayachandran S, Prashanth K. Phenotypic and genotypic assays for detecting the prevalence of metallo-β-lactamases in clinical isolates of Acinetobacter baumannii from a South Indian tertiary care hospital. J Med Microbiol. 2009;58(4):430–435. doi: 10.1099/jmm.0.002105-0. [DOI] [PubMed] [Google Scholar]
- 19.Fastenberg JH, Hsueh WD, Mustafa A, Akbar NA, Abuzeid WM. Biofilms in chronic rhinosinusitis: pathophysiology and therapeutic strategies. World J Otorhinolaryngol-Head Neck Surg. 2016;2(4):219–229. doi: 10.1016/j.wjorl.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Microbewiki.kenyon.edu (2022) https://microbewiki.kenyon.edu/images/thumb/8/84/Biofilm.png/800px-Biofilm.png
- 21.Healy DY, Leid JG, Sanderson AR, Hunsaker DH. Biofilms with fungi in chronic rhinosinusitis. Otolaryngol Head Neck Surg. 2008;138(5):641–647. doi: 10.1016/j.otohns.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Foreman A, Psaltis AJ, Tan LW, Wormald PJ. Characterization of bacterial and fungal biofilms in chronic rhinosinusitis. Am J Rhinol Allergy. 2009;23(6):556–561. doi: 10.2500/ajra.2009.23.3413. [DOI] [PubMed] [Google Scholar]
- 23.Boase S, Valentine R, Singhal D, Tan LW, Wormald PJ (2011) A sheep model to investigate the role of fungal biofilms in sinusitis: fungal and bacterial synergy. In: International forum of allergy & rhinology, vol 1, no 5. Wiley, Hoboken, pp 340–347 [DOI] [PubMed]
- 24.Malik BA. Prevalence of bacterial biofilms in patients of chronic rhino sinusitis (CRS) with or without nasal polyposis. Sch J App Med Sci. 2021;11:1642–1645. doi: 10.36347/sjams.2021.v09i11.001. [DOI] [Google Scholar]
- 25.Al-Mazrou KA, Al-Khattaf AS. Adherent biofilms in adenotonsillar diseases in children. Arch Otolaryngol-Head Neck Surg. 2008;134(1):20–23. doi: 10.1001/archoto.2007.18. [DOI] [PubMed] [Google Scholar]
- 26.Swidsinski A, Göktas Ö, Bessler C, Loening-Baucke V, Hale LP, Andree H, Weizenegger M, Hölzl M, Scherer H, Lochs H. Spatial organisation of microbiota in quiescent adenoiditis and tonsillitis. J Clin Pathol. 2007;60(3):253–260. doi: 10.1136/jcp.2006.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vermee Q, Cohen R, Hays C, Varon E, Bonacorsi S, Bechet S, et al. Biofilm production by Haemophilus influenzae and Streptococcus pneumoniae isolated from the nasopharynx of children with acute otitis media. BMC Infect Dis. 2019;19:44. doi: 10.1186/s12879-018-3657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]