Abstract
A model for the maturation of tomato spotted wilt virus (TSWV) particles is proposed, mainly based on results with a protoplast infection system, in which the chronology of different maturation events could be determined. By using specific monoclonal and polyclonal antisera in immunofluorescence and electron microscopy, the site of TSWV particle morphogenesis was determined to be the Golgi system. The viral glycoproteins G1 and G2 accumulate in the Golgi prior to a process of wrapping, by which the viral nucleocapsids obtain a double membrane. In a later stage of the maturation, these doubly enveloped particles fuse to each other and to the endoplasmic reticulum to form singly enveloped particles clustered in membranes. Similarities and differences between the maturation of animal-infecting (bunya)viruses and plant-infecting tospoviruses are discussed.
Increased knowledge about the molecular biology of tomato spotted wilt virus (TSWV), type species of the genus Tospovirus, has shed light on the replication and gene-coding strategy of this plant-infecting bunyavirus (for reviews, see references 6, 8, 9, 23, and 34). In contrast, a clear view of the TSWV particle maturation pathway in plant cells has not yet been reported, mainly because useful single-cell systems to study this process have been lacking. Early electron microscopic studies by Milne (22) and Ie (12) of infected leaf tissues summarized typical structures associated with tospovirus infections that were observed, such as the viroplasm (VP), nucleocapsid aggregates (NCA), paired parallel membranes (PPM) thought to be involved in budding events, doubly enveloped particles (DEV), and singly enveloped particles (SEV) clustered within endoplasmic reticulum (ER) membranes. It was observed that VP, NCA, PPM, and DEV were present mostly in early stages of infection whereas clustered SEV seemed to be a late or even final state in the maturation. Since TSWV is an enveloped bunyavirus, it is anticipated that preexisting intracellular membranes are used for enveloping, although it has been reported (22) that the PPM may be produced de novo since they did not seem to resemble any of the cellular membrane structures. More recently however, Kitajima et al. (14) presented three possible models for the morphogenesis of tospovirus particles, which included morphogenesis at the intracellular membranes of the ER or the Golgi system, although no conclusive choice could be made between the models proposed. While studying defective and nondefective isolates of impatiens necrotic spot virus, Lawson et al. (20) suggested that PPM, showing budding structures, could be Golgi derived based on their morphology. None of the above studies, however, presented extensive labeling data that supported either of the different models of the maturation pathway of tospoviruses in plants.
Structures possibly associated with morphogenesis of TSWV have also been observed in cells of the thrips vector Frankliniella occidentalis (32). Both N and G1/G2 proteins were immunolocalized to intracellular membranes, which were suggested to be Golgi derived (again based on their morphology), in cells of the midgut epithelium. However, no intermediate structures associated with budding virus were observed in these membranes, and no mature particles were observed in the midgut.
Since previous studies of nonsynchronous infections have hampered our understanding of the morphogenesis process of TSWV and other tospoviruses in plants, we now apply the recently developed protoplast infection system (13) to study this topic. Both cowpea (Vigna unguiculata) and Nicotiana rustica protoplast suspensions support TSWV multiplication, reaching at least 50% infection, provided that freshly prepared, highly infectious virus preparations are used as the inoculum (13). The application of a protoplast system not only has the advantage of a high level of infected cells but also allows a temporal analysis of the maturation pathway, since a high synchrony of infection is obtained. It has been shown that in cowpea protoplasts the infection cycle leads to only small numbers of enveloped particles whereas in N. rustica protoplasts large numbers of mature particles accumulate and a complete infection is produced (13). Therefore the system based on N. rustica protoplasts was used in the present studies to investigate the chronology of the TSWV maturation.
MATERIALS AND METHODS
Virus, plants, and protoplasts.
Throughout this study, a Brazilian isolate of TSWV, BR01 (2), was used, and maintained in N. rustica plants by mechanical inoculation and transmission by thrips. For (cryo)electron microscopic analysis, N. rustica plants were harvested 5 to 10 days after inoculation with TSWV and local lesions were isolated from petunia (cv. Polo Blauw) 3 to 5 days after inoculation with TSWV. N. rustica protoplasts were inoculated with freshly isolated TSWV particles as described previously (13) and harvested for immunofluorescence microscopy, (immuno)electron microscopy, or Western blotting between 0 and 40 h postinoculation (p.i.).
Antisera.
Polyclonal antisera against TSWV N protein and the hydrophilic ectodomains of G1 and G2 were raised as described previously (13). The rat monoclonal immunoglobulin M serum against the plant Golgi system (JIM84) was described previously (11), as were anti-βF1 (19) and anti-RGP1 (3) sera against the plant Golgi system.
Immunofluorescence light microscopy.
Immunofluorescence of protoplasts was performed as described previously (13). Double-labeling experiments were performed with a combination of rabbit polyclonal antiserum and rat monoclonal antiserum, with swine anti-rabbit serum conjugated to tetramethylrhodamine isothiocyanate and goat anti-rat serum conjugated to fluorescein isothiocyanate as the respective second antibodies.
(Immuno)electron microscopy.
Immunoelectron microscopy of protoplasts fixed with 3% glutaraldehyde–2% paraformaldehyde, as well as ultrastructural analysis with osmium tetroxide fixation, was performed as described previously (13).
Cryoelectron microscopy.
Aldehyde fixation, infiltration with sucrose, cryosectioning, and negative staining of infected leaf material were carried out as described by Van der Wel et al. (33).
Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western immunoblot analysis with alkaline phosphatase detection were carried out as described by Kikkert et al. (13).
RESULTS
Chronology of appearance, and labeling of structures associated with maturation.
N. rustica protoplasts were inoculated with TSWV particles by using polyethylene glycol, and samples were taken between 0 and 40 h p.i. for immunofluorescence and electron microscopy analyses. Table 1 shows the times of appearance of different structures, which have previously been reported to be associated with TSWV maturation (12, 14, 22), during the infection in protoplasts. Table 2 summarizes the reactivity of these maturation-associated structures with antisera used in electron microscopy, as discussed further throughout Results and Discussion.
TABLE 1.
Relative appearance of TSWV maturation-associated structures and intracellular membranes during TSWV infection in protoplastsa
| Time (h) p.i. | Presence of:
|
||||||
|---|---|---|---|---|---|---|---|
| VP | NCA | PPM | DEV | SEV | ER | Golgi | |
| 0 | − | − | − | − | − | + | + |
| 18 | + | + | ++ | + | − | + | + |
| 22 | + | + | ++ | + | + | + | ± |
| 26 | + | ++ | + | ++ | + | + | ± |
| 30 | + | ++ | ± | + | ++ | + | ± |
| 40 | + | ++ | − | − | ++ | + | − |
Judgment of relative presence of structures at one time point was based on at least three different specimens with 10 to 20 infected cells each.
TABLE 2.
Reactivity of TSWV maturation-associated structures and intracellular membranes with different antisera in immunogold electron microscopy, in TSWV-infected protoplastsa
| Antiserum | Reactivity of:
|
||||||
|---|---|---|---|---|---|---|---|
| VP | NCA | PPM | DEV | SEV | ER | Golgi | |
| Anti-N | + | ++ | + | + | + | − | − |
| Anti-G1-G2 | ± | ± | + | + | + | − | − |
| Anti-βF1 | − | − | + | + | + | − | + |
| Anti-RGP1 | − | − | + | − | − | − | + |
Samples were taken between 0 and 40 h p.i. as indicated in Table 1.
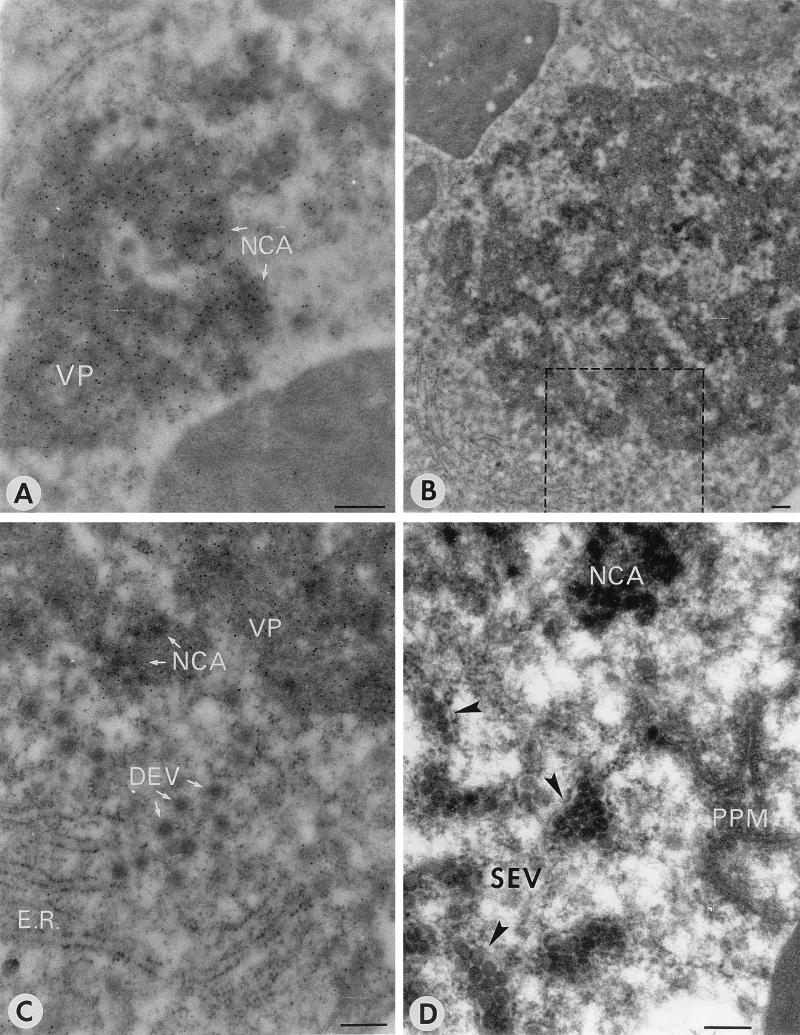
Examples of the different structures are depicted in Fig. 1. VP (Fig. 1A), characterized as amorphous medium-density material, was seen throughout the infection, very often in association with NCA, which is much more dense (Fig. 1A) and which often appears as aggregated nodules of around 60 to 80 nm in diameter embedded in the VP (Fig. 1). Both structures are strongly labeled with antiserum against the nucleocapsid protein, N (Fig. 1A and C and Table 2), suggesting that both contain viral nucleocapsid protein. They are only weakly labeled with serum against the viral glycoproteins G1 and G2 (Table 2 and data not shown). In the early stages of infection, mostly small patches of VP and NCA dispersed throughout the cytoplasm were observed (Fig. 1A), whereas in later stages, often only one or two very large areas of NCA and VP were found (Fig. 1B).
FIG. 1.
Overview of TSWV maturation-associated structures found in infected N. rustica protoplasts. (A) NCA embedded in VP, immunogold labeled with antiserum against N at 26 h p.i. (B) Large cluster of VP-NCA, as found in late stages of infection at 40 h p.i. (C) Detail of panel B (the boxed area) showing DEV and ER on the edge of the VP-NCA cluster labeled with antiserum against N. (D) SEV clusters surrounded by membrane envelopes (membranes are indicated by arrowheads) and NCA close to PPM structures at 30 h p.i. Bars, 200 nm.
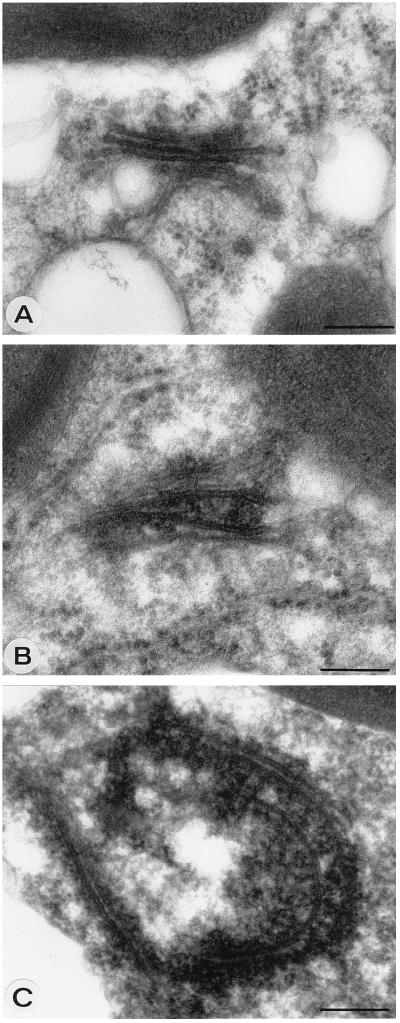
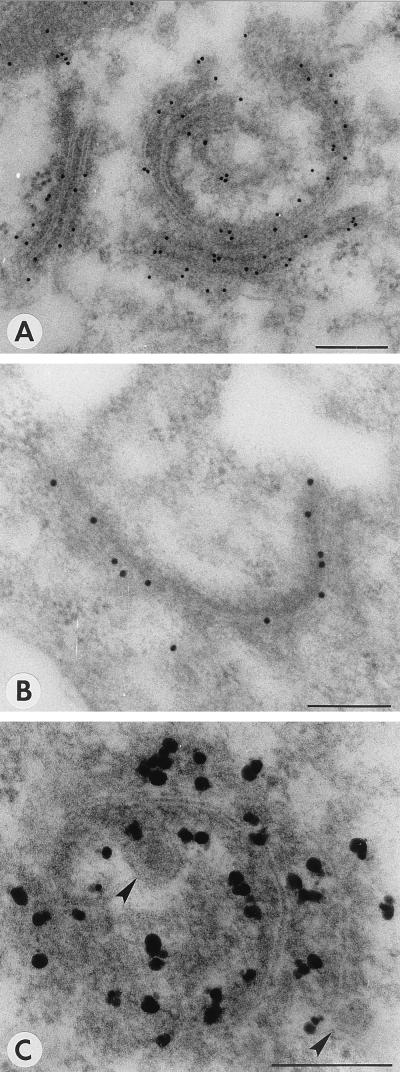
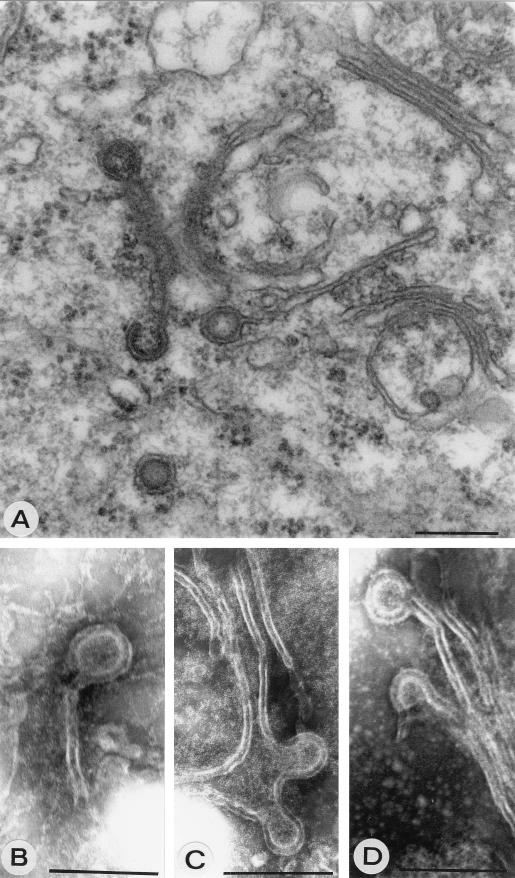
PPM are characterized as membrane cisternae that are often strongly curved, and dense material is tightly associated with them (Fig. 1D). PPM were previously suggested to be derived from Golgi stacks (20), and the modification of Golgi stacks that was observed in protoplast infections can indeed be interpreted as the formation of PPM (Fig. 2). In infected cells, the cisternae of a Golgi stack seem to drift apart and dense material accumulates between them (Fig. 2B). The cisternae become curved and often seem extended (Fig. 2C). The formation of PPM was quite obvious in protoplast infections at several time points, but a clear peak was found in the early stages of infection around 18 to 22 h p.i. (Table 1). PPM and their surroundings were labeled with antiserum against G1 (Fig. 3A), G2 (Fig. 3B), and N (Fig. 3C) proteins (Table 2) in infected protoplasts, while these antisera gave no background on healthy plant cells (data not shown). Often DEV were found in the vicinity, which appear to form at the PPM (Fig. 3C). To confirm that these structures found in protoplast infections were not artifacts of the system, early stages of systemic infections in N. rustica plants as well as TSWV local lesions in petunia were investigated. Particularly clear examples of curving and wrapping PPM were found in these plants as well (Fig. 4). The data suggest that DEV are formed by curving and wrapping of modified Golgi cisternae around dense nucleocapsid material in the cytoplasm.
FIG. 2.
Formation of PPM structures by modification of Golgi stacks. (A) Apparently unmodified Golgi stack at 18 h p.i. (B) Putative, modified Golgi stack with cisternae moving away from each other and dense material, presumably nucleocapsids, accumulating between them at 18 h p.i. (C) PPM structure showing extended and curved Golgi cisternae presumably surrounded by nucleocapsid material at 18 h p.i. Bars, 200 nm.
FIG. 3.
Immunogold labeling of PPM structures with antiserum against G1 (A), G2 (B), and N (C), all at 22 h p.i. Arrowheads indicate DEV being formed. Bars, 200 nm.
FIG. 4.
(A) PPM structures in local lesions in petunia at 4 days p.i. (B to D) PPM structures in systemically infected N. rustica plants at 5 days p.i. Formation of DEV is shown clearly. Bars, 200 nm.
From Table 1 it becomes clear that PPM largely precedes the formation of DEV and that SEV seem to represent a final stage in the infection. At late stages of infection (30 to 40 h p.i.), PPM as well as DEV were rarely found whereas SEV surrounded by membranes were abundant and were often found near the large NCA clusters (Table 1 and Fig. 1D). ER membranes were found throughout the infection, often in the vicinity of maturation-associated structures (Fig. 1A to C). Apparently unmodified Golgi stacks, however, became more scarce during the infection process and were virtually absent at late stages.
The Golgi system is the site of TSWV enveloped-particle morphogenesis.
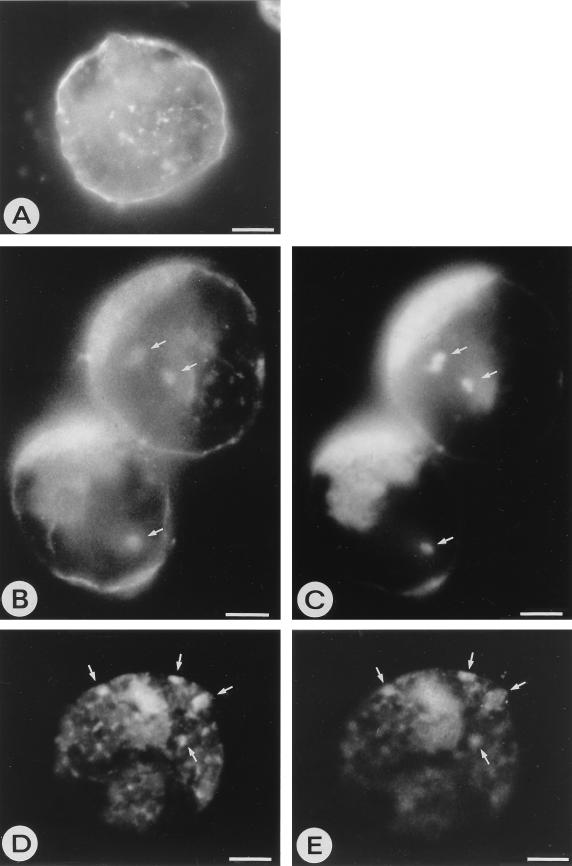
Since the morphology of PPM structures strongly suggests that they are derived from Golgi stacks, three independent plant Golgi markers were used to confirm this. JIM84, a rat monoclonal antiserum raised against carrot Golgi protein epitopes (11), anti-βF1, a polyclonal antiserum raised in carrot against complex glycans common to plant Golgi glycoproteins (19), and anti-RGP1, a polyclonal antiserum against the peptide fraction of a reversibly glycosylated trans-Golgi protein from pea (3) were used. Since JIM84 is a monoclonal antiserum, it could be used in double-label experiments together with polyclonal sera against viral proteins in immunofluorescence microscopy. JIM84 was raised against carrot Golgi epitopes, and so we verified whether it can also recognize the Golgi system of N. rustica. Indeed, in healthy N. rustica protoplasts or at 0 h p.i., JIM84 produced a typical pattern of small spots scattered throughout the cytoplasm, representing individual Golgi stacks (Fig. 5A), as was also observed in carrot, onion, and maize cells with JIM84 (11). As a result of transport of the epitope-containing protein, parts of the plasma membrane were usually labeled by JIM84 as well (Fig. 5A) (11, 27). During TSWV infection, the Golgi system did not appear dramatically different, although larger clusters were increasingly observed with JIM84, as well as the smaller ones (Fig. 5B and D). The viral glycoproteins colocalized at least partly with these Golgi structures at different time points during the protoplast infection (Fig. 5B to E). Viral N protein also colocalized with the Golgi during protoplast infections (data not shown).
FIG. 5.
Immunofluorescence images of N. rustica protoplasts. (A) Healthy protoplast labeled with JIM84 antiserum against the plant Golgi system, showing individual Golgi stacks as small clusters throughout the cytoplasm. The plasma membrane is also labeled with JIM84 due to transport of the epitope-containing Golgi proteins. (B and D) TSWV-infected protoplasts at 30 h p.i., labeled with JIM84 antiserum. (C and E) The same infected protoplasts labeled with mixed antisera against G1 and G2. Areas of clear colocalization of the viral glycoproteins with the Golgi system are indicated with arrows. Cloudy areas within cells represent the autofluorescence background. Bars, 5 μm.
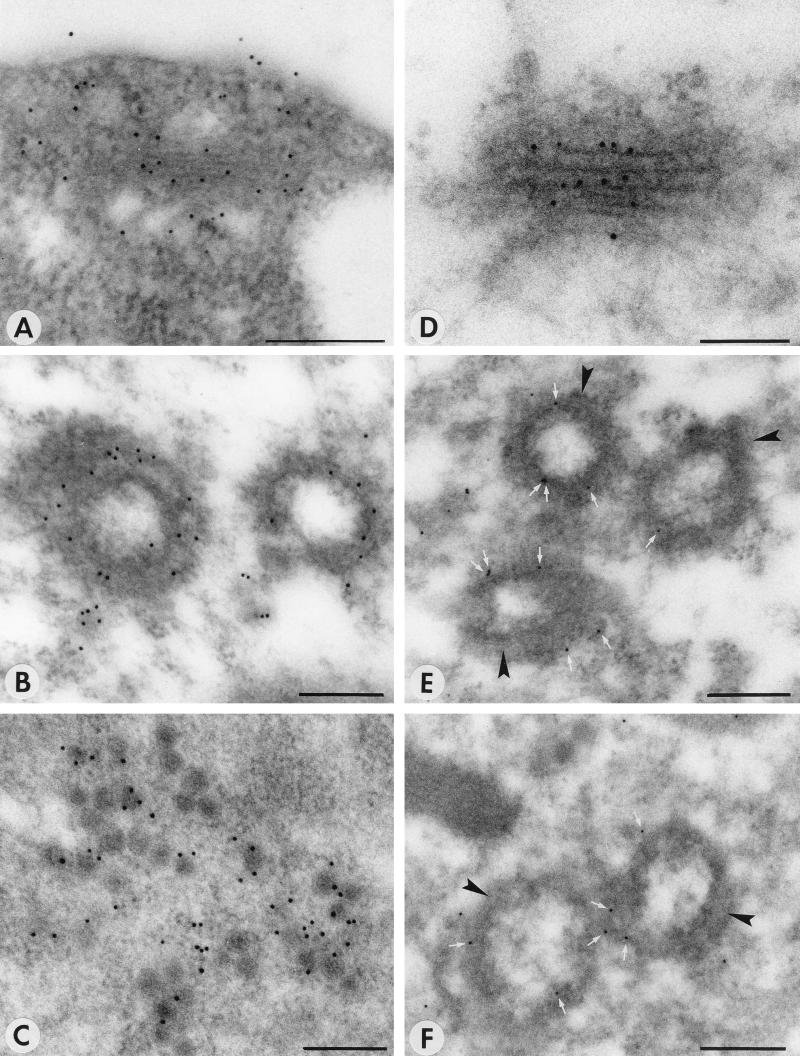
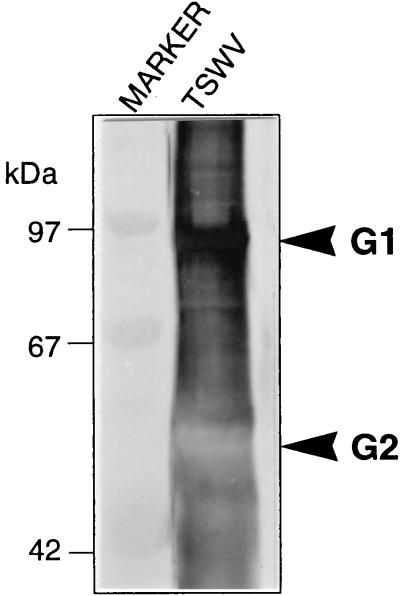
Both polyclonal anti-βF1 and anti-RGP1 sera labeled Golgi stacks in healthy N. rustica protoplasts or in protoplasts at 0 h p.i., as shown by immunogold electron microscopy (Fig. 6A and D). In infected protoplasts, anti-βF1 labeled PPM structures (Fig. 6B) as well as virus particles (Fig. 6C). Anti-RGP1 labeled PPM very specifically although not heavily (Fig. 6E and F), but it did not tag virus particles (Table 2). Figure 7 shows that in Western blots anti-βF1 cross-reacts with G1 from virus particles, which most probably explains the labeling of virus particles. The serum, however, did not react with unglycosylated G1 expressed in Escherichia coli (data not shown), indicating that the cross-reaction is an interaction with the glycans of G1. Since the anti-βF1 serum was raised against Golgi-specific glycans (19), the cross-reaction may indicate that G1 contains epitopes for anti-βF1, confirming the Golgi targeting of TSWV glycoproteins.
FIG. 6.
Immunogold labeling of maturation-associated structures with antisera against the plant Golgi system. (A) Unmodified Golgi stack in a healthy cell, labeled with anti-βF1 antiserum. (B) PPM structure labeled with anti-βF1 antiserum at 22 h p.i. (C) Virus particles labeled with anti-βF1 antiserum at 40 h p.i. (D) Unmodified Golgi stack at 0 h p.i., labeled with anti-RGP1 antiserum. (E and F) PPM structures labeled with anti-RGP1 antiserum at 26 h p.i. White arrows indicate gold particles; black arrowheads indicate PPM membranes. Bars, 200 nm.
FIG. 7.
Western blot analysis with anti-βF1 antiserum, showing cross-reaction of this serum with G1 protein from purified TSWV particles. Lanes: TSWV, purified TSWV particles; MARKER, low-molecular-mass marker proteins.
These data together indicate that PPM are in fact derived from the Golgi and form the site of particle morphogenesis.
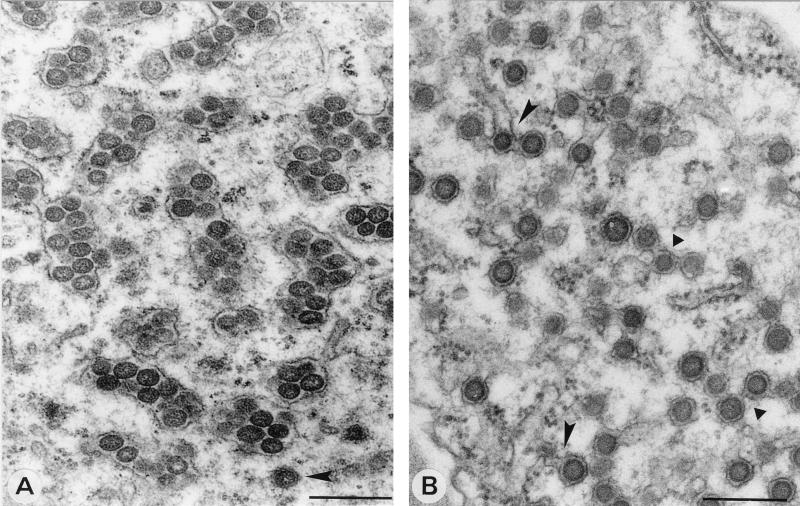
SEV are produced by fusion of DEV.
From osmium tetroxide-fixed specimens of infected protoplasts, in which DEV and SEV can be clearly distinguished, it was observed that at around 26 h p.i. SEV started to accumulate in clusters surrounded by membranes (Table 1) while DEV particles could also be observed in these areas (Fig. 8A). These kinds of structures were also found in local lesions in petunia (results not shown), along with structures suggesting the formation of these SEV accumulations in membranes (Fig. 8B). Considering the chronology of the appearance of DEV and SEV (Table 1), these images suggest that SEV is formed by fusion of DEV. In this process, the outer membranes of DEV form a tight collective smooth envelope around singly enveloped particles (Fig. 8B). DEV seem also able, either individually or fused, to fuse to (rough) ER membranes (Fig. 8B).
FIG. 8.
Formation of SEV by fusion of DEV. (A) Clustered SEV inside smooth and rough membranes, as found in late stages of TSWV infections of N. rustica protoplasts at 40 h p.i. Note the DEV at the bottom of the image (arrowhead). (B) Image from a TSWV local lesion in petunia showing DEV particles fusing with each other (triangles) and with ER membranes identified by ribosomes on the surface (arrowheads). Bars, 200 nm.
DISCUSSION
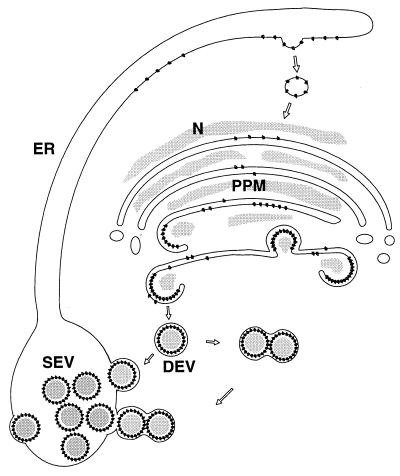
By using the recently developed N. rustica protoplast infection system, a time course of TSWV particle maturation could be produced. Analysis of the maturation intermediates in these protoplasts as well as in local lesions in petunia and systemically infected N. rustica plants leads to a TSWV particle maturation model (Fig. 9).
FIG. 9.
Model of TSWV particle morphogenesis.
The use of three independent plant-Golgi markers confirmed the suggestion that PPM are derived from Golgi stacks and form the site of doubly enveloped particle morphogenesis. The apparent retention of the viral glycoproteins G1 and G2 in the Golgi system is in agreement with the observations by immunofluorescence microscopy, which indicated that these proteins are not found on the surface of the plasma membrane (13) (Fig. 5) whereas they are assumed to enter the secretory pathway due to their N-terminal signal peptide (15). It is therefore likely that either one or both of the TSWV glycoproteins contain a Golgi retention signal, as recently documented for Uukuniemi virus (1).
In the proposed maturation model (Fig. 9), DEV is formed by wrapping of modified Golgi membranes (PPM) around nucleocapsids in the cytoplasm, and subsequently SEV is formed by fusion of DEV with each other or with ER membranes. Whether this latter process is based on an existing targeted membrane fusion mechanism in the cell or whether it is a virally induced phenomenon remains to be investigated.
Apart from the structures that now have been identified as being involved in particle morphogenesis, other virus-associated structures were also observed but not clarified. VP in association with NCA, consisting of nucleocapsid protein, does not seem to be directly involved in particle morphogenesis. However, PPM and DEV are often found in the vicinity of VP and NCA (Fig. 1B and C) or are even embedded in them, as suggested by the labeling of NCA and VP with anti-G1-G2 sera (Table 2). NCA and VP thus could be a source of nucleocapsids meant to be wrapped with Golgi membranes containing viral glycoproteins.
Still, a large part of the nucleocapsid protein that is produced during an infection is not used for producing particles but seems to be stored in large NCA clusters (Fig. 1B and C). This is also evident from Western blots of crude extracts of infected plants and purified TSWV particles, where the ratio of nucleocapsid protein to glycoproteins is much greater in crude extracts than in purified particles (data not shown). It is thought that TSWV moves from cell to cell by the action of NSm (nonstructural protein encoded by the M segment) which modifies the plasmodesmata so that infectious nucleocapsid units can pass the cell barrier (17, 31). This accounts for a part of the overproduction of nucleocapsids but most probably not for all.
The production of TSWV-encoded nonstructural (NSs) protein has not been investigated here but was observed in earlier studies (12, 14, 16, 22). So far, there is no evidence that this protein plays a role in particle morphogenesis.
Although the TSWV maturation pathway as proposed here is very different from that of the animal-infecting bunyaviruses (reviewed in 4, 10, 21, 24, 30), the assembly of particles in the Golgi system appears to be a feature of both plant- and animal-infecting bunyaviruses. Modification of the Golgi system, as observed in TSWV infections, has also been reported for Uukuniemi virus and Nairobi sheep virus (5, 18, 26); however, instead of curling and wrapping, the Golgi cisternae vacuolize extensively during these animal bunyavirus infections, thus increasing the volume of the cisternae and allowing the budding of singly enveloped particles into the lumen. The formation of doubly enveloped particles by wrapping of cisternae has never been reported for animal infecting bunyaviruses; however, it is not unknown in enveloped animal-infecting double-stranded DNA viruses. Vaccinia poxvirus is wrapped by membranes of the intermediate compartment between the ER and the Golgi and in a later stage of the maturation it is wrapped by trans-Golgi network membranes (28, 29). Varicella-zoster herpesvirus makes use of the trans-Golgi network membranes for wrapping its particles as well (7, 35), and African swine fever virus is wrapped by ER membranes (25). The wrapping phenomenon has not been reported for any other plant-infecting virus, and it is not yet precisely clear which part of the Golgi is involved in TSWV morphogenesis.
The final stages of the particle maturation also seem to be different. Animal-infecting bunyaviruses produce groups of particles inside vesicles pinched off from the Golgi (21), and the ER plays no role in particle morphogenesis. TSWV singly enveloped particles end up in large membrane envelopes as a result of self-fusion of DEV (which have two Golgi-derived membranes), or fusion with ER membranes. These envelopes surrounding SEV clusters must consequently consist of both Golgi- and ER-derived membranes.
To infect new cells, animal-infecting bunyaviruses are transported to the plasma membrane and released outside the cell via the vesicular transport pathway of the cell, whereas TSWV particles retain and accumulate in the plant cell until feeding thrips vectors ingest them for transport to other host plants.
ACKNOWLEDGMENTS
We thank Chris Hawes, Oxford School of Biological and Molecular Sciences, for kindly providing the rat monoclonal JIM84 antiserum and Maarten Chrispeels, University of California, La Jolla, for kindly providing polyclonal anti-βF1 antiserum. We also thank Kanwarpal Dhugga, Stanford University, Stanford, Calif., for sending us the anti-RGP1 antiserum. We are grateful to Wiesje Kassies for excellent technical assistance and to Beatrice Satiat-Jeunemaitre and Dick Peters for helpful discussions.
REFERENCES
- 1.Andersson A M, Melin L, Bean A, Pettersson R F. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J Virol. 1997;71:4717–4727. doi: 10.1128/jvi.71.6.4717-4727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Ávila A C, de Haan P, Smeets M L L, Resende R de O, Kormelink R, Kitajima E W, Goldbach R W, Peters D. Distinct levels of relationships between tospovirus isolates. Arch Virol. 1993;128:211–227. doi: 10.1007/BF01309435. [DOI] [PubMed] [Google Scholar]
- 3.Dhugga K S, Tiwari S C, Ray P M. A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proc Natl Acad Sci USA. 1997;94:7679–7684. doi: 10.1073/pnas.94.14.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliott R M. Molecular biology of the Bunyaviridae. J Gen Virol. 1990;71:501–522. doi: 10.1099/0022-1317-71-3-501. [DOI] [PubMed] [Google Scholar]
- 5.Gahmberg N, Kuismanen E, Keränen S, Pettersson R F. Uukuniemi virus glycoproteins accumulate in and cause morphological changes of the Golgi complex in the absence of virus maturation. J Virol. 1986;57:899–906. doi: 10.1128/jvi.57.3.899-906.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.German T L, Ullman D E, Moyer J W. Tospoviruses: diagnosis, molecular biology, phylogeny and vector relationships. Annu Rev Phytopathol. 1992;30:315–348. doi: 10.1146/annurev.py.30.090192.001531. [DOI] [PubMed] [Google Scholar]
- 7.Gershon A A, Sherman D L, Zhu Z, Gabel G A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldbach R, Peters D. Molecular and biological aspects of tospoviruses. In: Elliott R M, editor. The Bunyaviridae. New York, N.Y: Plenum Press; 1996. pp. 129–157. [Google Scholar]
- 9.Goldbach R, Kormelink R, de Haan P, Resende R de O, de Ávila A, van Poelwijk F, van Lent J, Wijkamp I, Prins M, Peters D. Tomato spotted wilt virus: genome organization, transmission and symptom induction. In: Bills D, Kung S-D, editors. Biotechnology and plant protection. Singapore: World Scientific; 1992. pp. 257–311. [Google Scholar]
- 10.Griffiths G, Rottier P. Cell biology of viruses that assemble along the biosynthetic pathway. Semin Cell Biol. 1992;3:367–381. doi: 10.1016/1043-4682(92)90022-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horsley, D., J. Coleman, D. Evans, K. Crooks, J. Peart, B. Satiat-Jeunemaitre, and C. Hawes. 1993. A monoclonal antibody, JIM84, recognizes the Golgi apparatus and plasma membrane in plant cells. J. Exp. Bot. 44(Suppl.):223–229.
- 12.Ie T S. Electron microscopy of developmental stages of tomato spotted wilt virus in plant cells. Virology. 1971;2:468–479. doi: 10.1016/0042-6822(71)90318-7. [DOI] [PubMed] [Google Scholar]
- 13.Kikkert M, Van Poelwijk F, Storms M, Kassies W, Bloksma H, van Lent J, Kormelink R, Goldbach R. A protoplast system for studying tomato spotted wilt virus infection. J Gen Virol. 1997;78:755–1763. doi: 10.1099/0022-1317-78-7-1755. [DOI] [PubMed] [Google Scholar]
- 14.Kitajima E W, de Ávila A C, Resende R O, Goldbach R W, Peters D. Comparative cytological and immunological labeling studies on different isolates of tomato spotted wilt virus. J Submicrosc Cytol Pathol. 1992;24:1–14. [Google Scholar]
- 15.Kormelink R, de Haan P, Meurs C, Peters D, Goldbach R. The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J Gen Virol. 1992;73:2795–2804. doi: 10.1099/0022-1317-73-11-2795. [DOI] [PubMed] [Google Scholar]
- 16.Kormelink R, Kitajima E W, de Haan P, Zuidema D, Peters D, Goldbach R. The nonstructural protein (NSs) encoded by the ambisense S RNA segment of tomato spotted wilt virus is associated with fibrous structures in infected plants. Virology. 1991;181:459–468. doi: 10.1016/0042-6822(91)90878-f. [DOI] [PubMed] [Google Scholar]
- 17.Kormelink R, Storms M, van Lent J, Peters D, Goldbach R. Expression and subcellular location of the NSm protein of tomato spotted wilt virus (TSWV), a putative movement protein. Virology. 1994;200:56–65. doi: 10.1006/viro.1994.1162. [DOI] [PubMed] [Google Scholar]
- 18.Kuismanen E, Hedman K, Saraste J, Pettersson R F. Uukuniemi virus maturation: accumulation of virus particles and viral antigens in the Golgi complex. Mol Cell Biol. 1982;2:1444–1458. doi: 10.1128/mcb.2.11.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurière M, Laurière C, Chrispeels M J, Johnson K D, Sturm A. Characterization of a xylose-specific antiserum that reacts with the complex asparagine-linked glycans of extracellular and vacuolar glycoproteins. Plant Physiol. 1989;90:1182–1188. doi: 10.1104/pp.90.3.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawson R H, Dienelt M M, Hsu H T. Ultrastructural comparisons of defective, partially defective, and non-defective isolates of impatiens necrotic spot virus. Phytopathology. 1996;86:650–661. [Google Scholar]
- 21.Matsouka Y, Chen S Y, Compans R W. Bunyavirus protein transport and assembly. Curr Top Microbiol Immunol. 1991;169:161–180. doi: 10.1007/978-3-642-76018-1_6. [DOI] [PubMed] [Google Scholar]
- 22.Milne R G. An electron microscope study of tomato spotted wilt virus in sections of infected cells and in negative stain preparations. J Gen Virol. 1970;6:267–276. [Google Scholar]
- 23.Mumford R A, Barker I, Wood K R. The biology of the tospoviruses. Ann Appl Biol. 1996;128:159–183. [Google Scholar]
- 24.Pettersson R F. Protein localization and virus assembly at intracellular membranes. Curr Top Microbiol Immunol. 1991;170:67–104. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 25.Rouiller I, Brookes S M, Hyatt A D, Windsor M, Wileman T. African swine fever virus is wrapped by the endoplasmic reticulum. J Virol. 1998;72:2373–2387. doi: 10.1128/jvi.72.3.2373-2387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rwambo P M, Shaw M K, Rurangirwa F R, DeMartini J C. Ultrastructural studies on the replication and morphogenesis of Nairobi sheep disease virus, a Nairovirus. Arch Virol. 1996;141:1479–1492. doi: 10.1007/BF01718249. [DOI] [PubMed] [Google Scholar]
- 27.Satiat-Jeunemaitre B, Hawes C. Redistribution of a Golgi glycoprotein in plant cells treated with brefeldin A. J Cell Sci. 1992;103:1153–1166. [Google Scholar]
- 28.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans-Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van’t Hof W, van Meer G, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens E B, Compans R W. Assembly of animal viruses at cellular membranes. Annu Rev Microbiol. 1988;42:489–516. doi: 10.1146/annurev.mi.42.100188.002421. [DOI] [PubMed] [Google Scholar]
- 31.Storms M, Kormelink R, Peters D, van Lent J, Goldbach R. The non-structural protein NSm of tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology. 1995;214:485–493. doi: 10.1006/viro.1995.0059. [DOI] [PubMed] [Google Scholar]
- 32.Ullman D E, Westcot D M, Chenault K D, Sherwood J L, German T L, Bandla M D, Cantone F A, Duer H L. Compartmentalization, intracellular transport, and autophagy of tomato spotted wilt tospovirus proteins in infected thrips cells. Phytopathology. 1995;85:644–654. [Google Scholar]
- 33.Van der Wel N N, Goldbach R W, van Lent J W M. The movement protein and coat protein of alfalfa mosaic virus accumulate in structurally modified plasmodesmata. Virology. 1998;244:322–329. doi: 10.1006/viro.1998.9117. [DOI] [PubMed] [Google Scholar]
- 34.Van Poelwijk F, Kikkert M, Prins M, Kormelink R, Storms M, van Lent J, de Haan P, Peters D, Goldbach R. Replication and expression of the tospoviral genome. Proceedings of the International Symposium on tospoviruses and thrips of floral and vegetable crops. Acta Hortic. 1996;431:201–208. [Google Scholar]
- 35.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]