Abstract
Previous studies which used intertypic reassortants of the wild-type reovirus serotype 1 Lang and the temperature-sensitive (ts) serotype 3 mutant clone tsA279 identified two ts lesions; one lesion, in the M2 gene segment, was associated with defective transmembrane transport of restrictively assembled virions (P. R. Hazelton and K. M. Coombs, Virology 207:46–58, 1995). In the present study we show that the second lesion, in the L2 gene segment, which encodes the λ2 protein, is associated with the accumulation of a core-like particle defective for the λ2 pentameric spike. Physicochemical, biochemical, and immunological studies showed that these structures were deficient for genomic double-stranded RNA, the core spike protein λ2, and the minor core protein μ2. Core particles with the λ2 spike structure accumulated after temperature shift-down from a restrictive to a permissive temperature in the presence of cycloheximide. These data suggest the spike-deficient, core-like particle is an assembly intermediate in reovirus morphogenesis. The existence of this naturally occurring primary core structure suggests that the core proteins λ1, λ3, and ς2 interact to initiate the process of virion capsid assembly through a dodecahedral mechanism. The next step in the proposed capsid assembly model would be the association of the minor core protein μ2, either preceding or collateral to the condensation of the λ2 pentameric spike at the apices of the primary core structure. The assembly pathway of the reovirus double capsid is further elaborated when these observations are combined with structures identified in other studies.
Virus assembly is an important late step in viral replication that is poorly understood in animal viruses. The mechanisms of viral assembly, release, extracellular transport, attachment, penetration, and uncoating determine the size and shape of the “packing crate” which must carry the viral genome between replicative cycles (40, 70). This packing crate must be a meta-stable structure to protect the genome during transport but also capable of releasing the genome and necessary associated replicative enzymes once it enters a new host cell. X-ray crystallography has provided valuable insight into the overall structure and putative protein interactions in a number of viruses at a macromolecular level (47, 82). Recent studies have elucidated structural interactions of components such as the G-H loop of foot-and-mouth disease virion capsid protein VP1 (24) and the hemagglutinin of influenza virus expressed in a bacterial vector (46). Electron cryomicroscopic studies have utilized image averaging and various planes of focus to provide three-dimensional and, to a degree, internal imaging of whole-virus structures (6, 75, 77, 86), a limited number of subviral particles (28, 103), reassortant viruses (85), capsids assembled from expressed proteins (45), proteolytically degraded reovirus intermediate structures (60), capsid-like structures produced by engineered and expressed rotavirus capsid protein VP2 (54), and reovirus and rotavirus undergoing in vitro transcription (53, 104). These studies showed complete structures and some assembly and disassembly intermediate structures, permit the localization of some proteins, and may identify regions of interactions between some proteins (60) and nucleic acids (77). Unfortunately, physical determinations of static structures may not define the dynamic processes and pathways by which viral proteins interact to assemble the packing crate needed to carry the viral genomic cargo safely through a hostile environment to its next address. For example, recent evidence suggests that the conformation of flock house virus in solution may differ dramatically from that predicted by X-ray crystallography (12).
An alternative approach to study macromolecular assembly processes is to use assembly-defective systems. The assembly pathways of some bacteriophages have been successfully studied in detail by using conditionally lethal amber mutations of bacteriophage T4 (10, 11, 101) and P22 (78). However, the many different strategies for biochemical regulation and interaction with host cells, genome organization, and assembly and structural designs employed by the eucaryotic viruses have mitigated against similar success in virus-eucaryote systems. The success in using bacteriophage conditionally lethal mutants (29, 30) to deduce procaryotic virus assembly pathways suggests that the Fields panel of conditionally lethal reovirus temperature-sensitive (ts) mutants (19, 32, 79) (reviewed in reference 21) may be elegant tools for dissecting the assembly pathways of a eucaryotic virus.
The complete mammalian reovirus particle contains nonequivalent amounts of eight different structural proteins assembled into a double-shelled particle approximately 80 to 85 nm in diameter. The outer capsid is made up of the major proteins μ1 and ς3 (28) and the minor protein ς1 (91). The inner core structure, a protein shell approximately 60 nm in diameter, is composed of the major core proteins λ1 and ς2, core spike protein λ2, and minor core proteins λ3 and μ2 and encases the 10 segments of double-stranded RNA (dsRNA) which comprise the genome (28). The segmented nature of the viral genome allows for the generation of intertypic reassortants which may be exploited to assign biologic functions to individual genes and their protein products (26, 44, 57, 95, 105). (For recent reviews of the structures and properties of the mammalian reoviruses, see references 68 and 71). The morphological variants produced at restrictive temperatures have been reported for only a few of the ts mutants of the Fields panel, either in thin-sectioned infected cells (33), by negative stain electron microscopy of gradient fractions of cell extracts (62, 65), or by both methods (23). Therefore, the identification of additional assembly-defective ts mutants might help elucidate eucaryotic virus assembly. The ts mutants of recombination group A (32) contain one or more lesions in the monocistronic M2 gene segment (67), which encodes the reovirus μ1 protein, a major component of the outer shell of the complete virion. Previous studies with the prototype clone tsA201 showed mild expression of the conditionally lethal phenotype (32). This clone did not produce aberrant particles when cultured under restrictive conditions (33). However, recombination group A contains more than 20 different mutant clones, all of which are believed to contain lesions in the M2 gene segment (21, 32).
Using intertypic reassortant analysis, we previously reported that the mutant clone tsA279 contains two ts mutations. One lesion is in the M2 gene and is associated with strong expression of the ts phenotype at elevated temperatures due to a blockade in the transmembrane transport of restrictively assembled virions. The second lesion, associated with mild expression of the ts phenotype at elevated temperatures, is in the L2 gene segment (44), which encodes the core spike protein λ2. In this study we report a blockade in assembly as a second mechanism for the expression of the ts phenotype by the mutant clone tsA279. The blockade in assembly is associated with the ts lesion in the L2 gene segment, and results in the accumulation of primary core particles which are defective for proteins λ2 and μ2. Correlating the previously reported assembly intermediates produced by other members of the Fields panel with the pattern of assembly intermediates produced under restrictive growth conditions by the mutant tsA279 further elaborates the assembly pathways of mammalian reoviruses.
MATERIALS AND METHODS
Cells, viruses, and immunologic reagents.
Mouse L929 cells, reovirus type 1 Lang (T1L), type 3 Dearing (T3D), and the ts mutant clone tsA279, derived from T3D, are laboratory stocks. tsA279 is a double ts mutant, with lesions mapping to the L2 and M2 gene segments (proteins λ2 and μ1, respectively) (44). The panel of T1L×tsA279 reassortants, the plaque assay to determine virus titrations, and the preparation of polyvalent rabbit anti-whole reovirus antiserum were as previously described (44). Polyvalent rabbit anti μ2 antiserum was a gift kindly provided by E. Brown, University of Ottawa. Hybridoma cell line 7F4, which produces a monoclonal antibody that recognizes λ2, was kindly provided by H. Virgin, Washington University. Hybridomas were maintained and monoclonal antibody 7F4 was purified as described previously (93).
Negative-stain-electron microscopy of infected cell extracts.
Preliminary analyses of different negative stains and stain conditions indicated that 1.2 mM phosphotungstic acid, pH 7.0 (PTA), gave the best overall quality of staining (43a). Subconfluent L929 monolayers in 24-well culture dishes were infected with either T1L, T3D, tsA279, or various T1L×tsA279 reassortants at a multiplicity of infection (MOI) of 5 PFU/cell. After attachment for 1 h at 4°C, the infections were overlaid with prewarmed Joklik’s modified minimal essential medium (S-MEM) supplemented with 2.5% fetal calf serum–2.5% VSP agammaglobulin neonatal bovine serum–2 mM l-glutamine and containing 20% preadapted medium and then incubated for 36 h at either 32 or 40°C. In some experiments, cycloheximide was added to the cultures at various times postattachment to a final concentration of 100 μg/ml, and the incubation temperature was changed 15 min later to ensure that no permissively translated viral protein was available for assembly onto structures produced under restrictive conditions (74); the infections were then cultured an additional 6 h (4). The cultures were freeze-thawed three times, and the cell debris was cleared by centrifugation for 10 s at 15,000 × g in an Eppendorf model 5412 benchtop centrifuge. Then, 50-μl aliquots of the supernatants were layered over 100-μl cushions of 30% potassium tartrate (pH 7.2), and viral and subviral particles were pelleted by centrifugation in a Beckman Airfuge (A100 rotor; 26 lb/in2, 1 h). Pellets were resuspended in 50 μl of 0.1% glutaraldehyde in S-MEM (pH 7.2), allowed to fix for 10 min at 4°C, centrifuged directly onto Formvar-coated, carbon-stabilized 400 mesh copper electron microscopy grids (Beckman Airfuge, EM-90 Rotor; 26 lb/in2, 30 min) as previously described (38), and stained with 1.2 mM PTA. Samples were viewed and photographed at machine magnifications of ×30,000 and ×70,000. The relative proportions and types of particles produced in each infection were determined by direct counting of all particles in each of five randomly selected, nonadjacent grid squares from the four outer quadrants and the central area of the grid (38). The infectious titer produced by each culture was determined by standard plaque assay at 32°C as previously described (44). Statistical probabilities were determined by using a χ2 test with the Yates correction (41).
Immunoelectron microscopy of virions and assembly intermediate structures.
Except where otherwise indicated, all preparative and conjugation steps were conducted at 20°C. Gold probes (12 nm) were prepared from chloroauric acid as described earlier (89) and filtered through 0.2-μm pore filters to remove aggregates. Polyvalent rabbit anti-reovirus immunoglobulin G (IgG) was purified by protein A-Sepharose affinity chromatography essentially as described previously (76). Briefly, antireovirus antisera was preabsorbed against an L929 monolayer, heat inactivated at 56°C for 30 min, passed through a protein A-Sepharose column (Sigma, St. Louis, Mo.), and washed with 50 mM Tris (pH 8.0)–150 mM sodium chloride (wash buffer), and the bound IgG was then eluted directly into 0.5 M phosphate buffer (pH 8.0) with elution buffer (100 mM sodium acetate, 150 mM sodium chloride [pH 4.0]). UV absorbance was determined at 280 nm, and the peak fractions were pooled. After extensive dialysis against 2 mM sodium borate (pH 8.0), the pooled fractions were concentrated with a Minicon B-15 concentrator (Millipore, Bedford, Mass.). The concentration of IgG needed to stabilize 10 ml of gold probe was determined by adsorption isotherm assay essentially as described earlier (36), except that IgG was diluted in 2 mM sodium borate (pH 8.0) for assay and conjugation to gold (48, 58, 94). After conjugation the probe was washed, resuspended in 10 mM Tris (pH 8.0)–150 mM NaCl–1% bovine serum albumin (BSA)–0.1% Carbowax M20–0.2% low bloom gelatin (Sigma) (stabilizing buffer) as described before (9), briefly sonicated at low energy to disrupt loose aggregates, filtered, and stored at 4°C until use (94). Viral lysates were prepared from T3D- and tsA279-infected cells, cleared, and pelleted directly onto Formvar-carbon-coated 400-mesh nickel electron microscope grids as described above. The grids were washed successively in HEPES-buffered MEM (pH 8.0), HEPES-buffered MEM with 1% BSA, and stabilization buffer and then incubated in a humid chamber for 30 min at 20°C on 5-μl aliquots of IgG labeled gold probes. The grids were washed with successive changes of phosphate-buffered saline (pH 8.0) and stained with PTA (81). The samples were evaluated and photographed as described above.
Isolation and identification of labeled virus and intermediate assembly structures.
L929 cells were infected with either tsA279 or T3D at an MOI of 5 PFU/cell. After attachment for 1 h at 4°C, prewarmed supplemented S-MEM was added to provide for a final concentration of 5 × 105 cells/ml, and the infections were incubated in spinner cultures for 36 h at 40°C. Control infections were incubated at 32°C for an equivalent number of replicative cycles (54 h). Viral proteins were labeled 3 h postattachment by the addition of [35S]methionine-[35S]cysteine (NEN, Boston, Mass.) to a final concentration of 25 μCi/ml. Infected cells were harvested and extracted with 1,1,2-trichloro-1,2,2-trifluoroethane (Freon 113) as described earlier (35). Viral structures were then purified by buoyant density centrifugation on preformed 1.2- to 1.55-gm/ml cesium chloride (CsCl) gradients (7.5 h at 6°C at 35,000 rpm in an SW41 Rotor (Spinco, Palo Alto, Calif.), 300-μl fractions were collected by bottom puncture, and the fraction density was determined by refractive index (Abbe 3-L Refractometer; Bausch and Lomb, Rochester, N.Y.). Aliquots from each fraction were evaluated for the presence of labeled material with a model LS 5000CE Scintillation Counter (Beckman Scientific, Palo Alto, Calif.), and for the presence of viral material by electron microscopy after direct centrifugation as described above. Aliquots of each fraction were diluted threefold in sterile distilled water, precipitated at −20°C overnight in 70% ethanol–300 mM sodium acetate, and resuspended in 0.24 M Tris (pH 6.8)–1.5% dithiothreitol–1% sodium dodecyl sulfate (SDS) (electrophoresis sample buffer) (51).
Protein separation with the Tris-glycine-urea (TGU)-SDS-polyacrylamide gel system.
Proteins were separated in the Laemmli discontinuous Tris-glycine buffering gel system (51), modified by the inclusion of urea (TGU gel) as previously described (20). Briefly, the TGU resolving gel consisted of a 5 to 16% polyacrylamide exponential gradient covered with a 4 to 5% polyacrylamide linear gradient, both containing 43% urea. A 4% polyacrylamide step which did not contain urea was layered over the upper gradient. The resolving gel was in 375 mM Tris (pH 8.8). Gels were allowed to polymerize for between 6 and 10 h, a 3.75% polyacrylamide stacking gel in 125 mM Tris (pH 6.8) was layered over the resolving gel, and proteins were resolved by electrophoresis at either 9 mA for 9.5 h or 7.5 mA for 16 h. The gels were fixed and impregnated with Enlightning (DuPont, Boston, Mass.) and fluorographed by exposure to X-ray film (Kodak X-AR, Rochester, N.Y.). Multiple film exposures of the gels were made and scanned with an LKB Ultroscan XL laser densitometer as described earlier (44), and the amount of each protein present was determined. The densities of major core protein λ1 bands were used to standardize the relative proportions of other viral proteins in each evaluated fraction.
Immunoblot analysis of protein composition of assembly intermediate structures.
Proteins were separated by using the TGU acrylamide gel system, and selected lanes were transferred to nylon membranes (Immobilon; Millipore) by using a Bio-Rad Semi-Dry Transblot transfer system (Bio-Rad, Richmond, Va.) at a 20-V constant voltage for 35 min. Nonspecific binding was blocked by washing the membranes in TBS-T (10 mM Tris [pH 7.5]–100 mM NaCl–0.1% Tween 20 containing 5% skim milk proteins; the membranes were then reacted with a mixture of antireovirus antiserum and antireovirus μ2 antiserum in TBS-T for 90 min at room temperature and washed with fresh TBS-T, and the blots were probed with goat anti-rabbit IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, West Grove, Pa.) in TBS-T containing 0.1% BSA for 90 min at room temperature. After a washing with TBS-T, the immune complexes were detected with diamino benzoate (39). Primary and secondary antibodies were stripped from the membranes by being washed in 100 mM 2-mercaptoethanol–2% SDS–62.5 mM Tris-HCl (pH 6.7) for 30 min at 50°C (1) and then reprobed with the anti-λ2 monoclonal antibody 7F4 (93). After treatment with goat anti-mouse IgG conjugated to horseradish peroxidase and a washing as above, the immune complexes were detected by using the Amersham enhanced chemiluminescence system (Amersham Life Sciences, Little Chalfont, United Kingdom) in accordance with the manufacturer’s instructions.
RESULTS
tsA279 selectively produces a novel subviral particle at the restrictive temperature.
Previous experiments that used thin-section electron microscopy to examine group A mutants suggested there was no interference in assembly at 39°C in tsA201 infections (33) and that both core-like and top-component particles were present in 39.5°C tsA279 cultures (43, 44). Thin-section electron microscopy provides insufficient resolution to distinguish between structures with subtle differences. In addition to difficulties in identifying particle type, analyses of random cell sections may not include the population of particles released by cell lysis. An ideal way to determine the nature of various products is negative-stain electron microscopy because of the higher resolution of this technique (13, 42, 64). However, when the production of virally encoded products is reduced, as may occur in restrictive mutant infections, total populations of intermediates may be missed when determinations are made from gradient-purified fractions. To determine the exact nature and quantity of the different structures present, direct particle counting of negative-stained whole-cell cytoplasmic extracts was conducted after direct centrifugation onto an electron microscope grid (38, 63).
The relative proportions of structures produced by tsA279 and T3D at both restrictive and permissive temperatures, as determined from direct particle counts of whole-cell lysates, are shown in Table 1. Cells infected with tsA279 and grown at a restrictive temperature produced approximately 50-fold fewer viral particles than those grown at permissive temperatures. The reduction in particle yield is consistent with previous reports of reduced viral protein production by this mutant under restrictive growth conditions (44). The particle/PFU ratio did not differ significantly between permissive and restrictive cultures of both T3D and tsA279 and ranged from 9 to 14 particles per PFU (data not shown). These values are similar to those in previous reports (80).
TABLE 1.
Distribution of species of particles produced by T3D and tsA279 at permissive and restrictive temperaturesa
| Clone | Temp (°C) | Time (h) | Total no. of particlesb | Total no. of particles/mlc | Proportion of particle (% [SEM])d
|
|||
|---|---|---|---|---|---|---|---|---|
| Virion | Top component | Outer shell | Core | |||||
| T3D | 32 | 36 | 1,773 | 1.7 × 108 | 85.7 (4.3) | 6.5 (0.7) | 2.7 (0.8) | 5.2 (3.0) |
| 72 | 24,682 | 2.9 × 109 | 78.6 (<0.1) | 15.4 (0.3) | 5.9 (0.3) | 0.1 (<0.1) | ||
| 40 | 36 | 1,632 | 1.3 × 108 | 64.1 (2.4) | 22.4 (2.8) | 9.1 (1.8) | 1.7 (0.3) | |
| 72 | 4,904 | 4.4 × 108 | 56.1 (8.5) | 22.5 (5.4) | 9.9 (0.7) | 11.5 (4.0) | ||
| tsA279 | 32 | 36 | 12,546 | 1.3 × 109 | 76.9 (3.8) | 19.0 (3.2) | 2.1 (0.6) | 1.9 (1.1) |
| 72 | 31,659 | 1.3 × 109 | 79.0 (1.0) | 18.8 (1.8) | 2.0 (1.8) | 0.2 (0.2) | ||
| 40 | 36 | 3,401 | 3.2 × 107 | 60.6 (2.7) | 19.6 (3.9) | 1.0 (0.4) | 18.8 (5.9) | |
| 72 | 1,209 | 3.6 × 107 | 50.2 (7.5) | 18.6 (7.3) | 0.7 (0.7) | 30.5 (15.1) | ||
Calculated from three to four separate experiments.
Total number of viral and subviral particles counted.
Total number of particles in the cell lysates, calculated as previously described (38).
Proportion of indicated particle type, expressed as the percentage of the total number of particles produced, with the standard error of the mean of replicate experiments shown in parentheses.
The predominant structural form produced at 32°C by both tsA279 and T3D was the genome complete whole virion (diameter of approximately 80 nm) after both 36 and 72 h of incubation (between 77 and 86% of the total particles produced) (Table 1). The next most common type of particle produced was the top component (6 to 19%, diameter of approximately 80 nm). Outer shell structures (diameter of approximately 90 nm) and core particles (approximately 50 nm in diameter) each comprised less than 6% of the particles produced. When infected cells were incubated at a restrictive temperature of 40°C, the distribution of particles produced was different. The proportion of whole virions was reduced to approximately 50 to 60% of all observed particles for both parental T3D and the ts mutant. With T3D there was a trend toward increased proportions of top component and outer shells at the restrictive temperature at both 36 and 72 h postattachment. In addition, there was a trend toward an increased proportion of complete core particles, which contained both genome and the characteristic λ2 apical spike, at 72 h postinfection. The mutant produced similar proportions of top-component structures and about 10-fold fewer outer-shell structures than did T3D at a restrictive temperature. However, the proportion of core-like particles produced by the mutant clone increased from less than 2% at the permissive temperature to 18 and 31% at the nonpermissive temperature for the two different times tested (P < 0.0001 by χ2 test with the Yates correction) (Table 1).
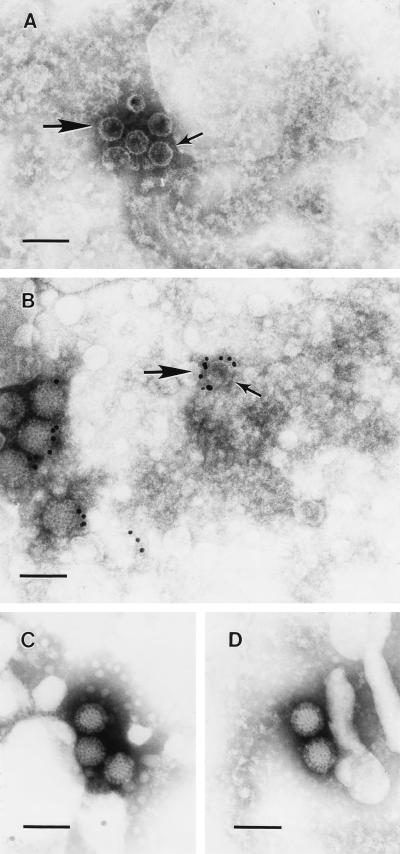
Core-like particles produced by tsA279 at a restrictive temperature (diameter of approximately 50 nm) appeared to be defective for the λ2 pentameric spike structure (Fig. 1A), whereas core particles produced under permissive growth conditions for both T3D and tsA279 and at a restrictive temperature for T3D clearly demonstrated the presence of the λ2 spike structure (data not shown). In addition, mutant core-like particles which were restrictively assembled (cultured and assembled under restrictive growth conditions) were penetrated by the negative stain, indicating that they were deficient for genomic dsRNA, while core particles produced under permissive and restrictive conditions by T3D and under permissive conditions by tsA279 were not penetrated by the negative stain, indicating that these particles contained genomic dsRNA. The identity of the core-like particles produced by tsA279 under restrictive culture conditions was confirmed with polyclonal antireovirus IgG conjugated to a 12-nm colloidal gold probe (Fig. 1B, large arrow). The distributions of particles produced under the different temperature conditions were not significantly altered by input MOIs of either 5 or 50 PFU/cell (data not shown).
FIG. 1.
Negative-stain and immunogold-labeled λ2 spike-defective core particles produced at restrictive temperature. (A) Restrictive temperature cytoplasmic extracts of tsA279-infected cells were centrifuged directly onto electron microscope grids and negatively stained with PTA as described in Materials and Methods. (B) Same as in panel A except the grid samples were immunolabeled (12-nm gold) before negative staining. The core-like particles in both panels (large arrows) demonstrated apical depressions where the λ2 pentameric core spike structure would be expected (small arrows). (C) Permissive-temperature cytoplasmic extracts of tsA279-infected cells. (D) Restrictive-temperature cytoplasmic extracts of T3D-infected cells. Magnification, ×100,000. The bars represent 100 nm.
The production of λ2 deficient core particles maps to the tsA279 L2 gene.
To determine which tsA279 gene(s) was associated with the spike-defective phenotype, we analyzed the distribution of particles produced at the restrictive temperature by various reassortant clones which segregated the mutant L2 and M2 gene segments (Table 2). Each of the six clones that contain the mutant L2 gene produced significant amounts of spike-defective particles, whereas each clone with the T1L L2 gene produced insignificant amounts of spike-defective particles. All of the remaining gene segments were randomly associated with respect to the presence or absence of the spike.
TABLE 2.
Intertypic reassortant mapping of the particle species produced by tsA279 restrictive cultures
| Clone | Gene segmenta
|
Total no. of particlesb | Proportion of particle (%)c
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virion | Top component | Outer shell | Core particles
|
|||||||||||||
| L1 | L2 | L3 | M1 | M2 | M3 | S1 | S2 | S3 | S4 | With λ2 | Without λ2 | |||||
| tsA279 | A | A | A | A | A | A | A | A | A | A | 1,209 | 50.2 | 18.6 | 0.7 | 0.1 | 30.5 |
| LA279.08 | 1 | A | 1 | A | A | A | A | A | A | A | 5,768 | 50.9 | 20.1 | 1.6 | 0.1 | 27.4 |
| LA279.11 | A | A | A | A | 1 | A | A | A | A | 1 | 1,144 | 56.2 | 27.1 | 0.1 | 1.7 | 15.0 |
| LA279.28 | A | A | A | 1 | A | A | 1 | A | 1 | 1 | 1,241 | 9.1 | 29.5 | 10.3 | 2.2 | 48.9 |
| LA279.39 | A | A | 1 | 1 | 1 | A | 1 | 1 | 1 | 1 | 2,112 | 19.0 | 27.1 | 5.2 | 0.6 | 48.1 |
| LA279.76 | A | A | 1 | 1 | 1 | 1 | 1 | 1 | A | A | 6,415 | 15.0 | 38.9 | 0.0 | 0.0 | 46.1 |
| LA279.10 | A | 1 | 1 | 1 | A | A | 1 | A | A | A | 1,603 | 26.5 | 67.4 | 1.1 | 3.9 | 1.1 |
| LA279.15 | 1 | 1 | 1 | 1 | 1 | A | 1 | 1 | 1 | 1 | 10,554 | 82.8 | 11.7 | 1.3 | 4.1 | 0.2 |
| LA279.56 | 1 | 1 | 1 | 1 | A | 1 | 1 | 1 | 1 | 1 | 2,689 | 38.6 | 53.9 | 1.5 | 5.8 | 0.3 |
| T1L | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4,965 | 72.0 | 23.2 | 1.9 | 0.6 | 2.4 |
Denotes parental origin of gene: 1, T1 Lang; A, tsA279. Bold type indicates the gene segment to which the phenotype maps.
Total number of viral and subviral particles counted.
Proportion of indicated particles, expressed as percentages of the total number of particles produced.
tsA279 produces structures which do not contain the λ2 core spike protein nor the minor core protein μ2.
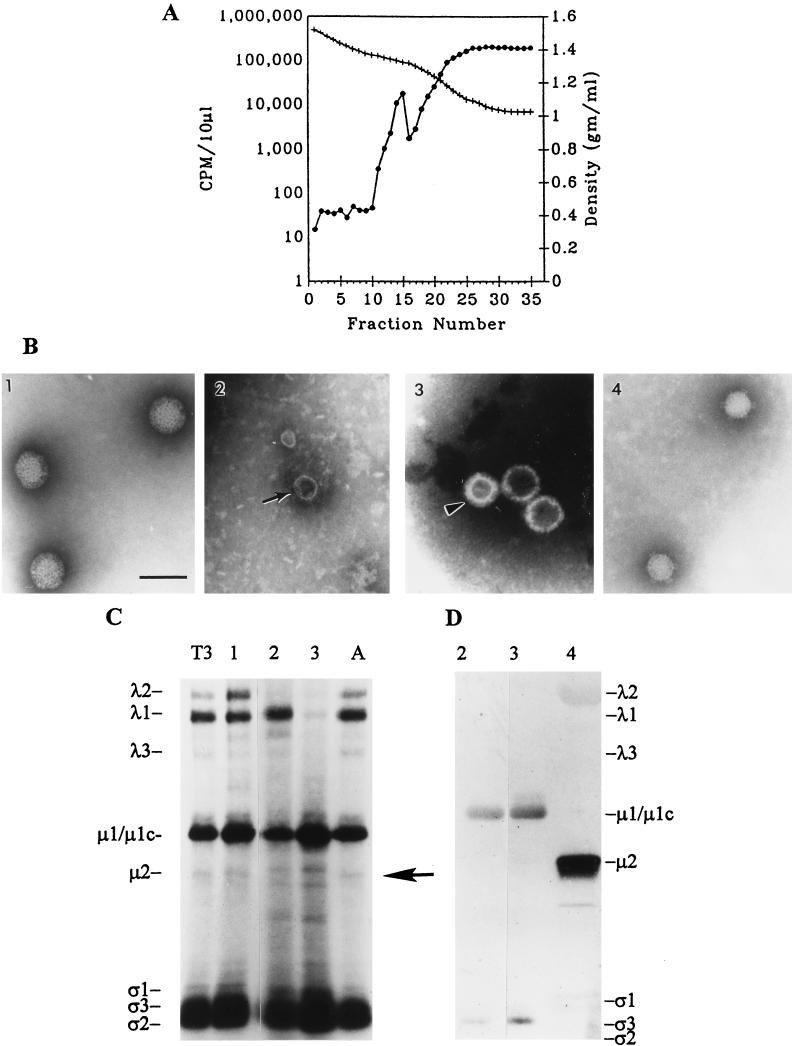
The above results indicated that the tsA279 L2 gene was associated with the nonpermissive production of a λ2 defective core-like structure. To determine whether the core-like particles contained λ2, but in a conformation which was not resolved, or were devoid of λ2, we purified them and analyzed their protein content. Restrictively assembled tsA279 assembly intermediate structures labeled with [35S]methionine-[35S]cysteine were isolated by using cesium chloride buoyant density centrifugation as detailed in Materials and Methods. Scintillation counting of fraction aliquots identified a single peak of activity at a density of 1.33 to 1.35 gm/ml (Fig. 2A). Electron microscopic examination of this fraction revealed complete virions at this density (Fig. 2B1). No individual peaks of radioactivity could be distinguished in fractions of lower density. Core-like particles which were deficient for both genome and the λ2 spike structure were present at a density of 1.28 to 1.29 gm/ml (Fig. 2B2), and outer-capsid structures were isolated at a density range of 1.26 to 1.28 gm/ml (Fig. 2B3). No core-like particles were observed at higher buoyant densities. It was not possible to cleanly separate the λ2 protein-deficient core particles from the outer-shell structures by rate zonal centrifugation due to their similar sedimentation characteristics and the small amount of material present in the fractions after sucrose gradient centrifugation (data not shown).
FIG. 2.
Purification of restrictively assembled core-like particles. L929 cells were infected with tsA279, viral proteins were labeled with [35S]methionine-[35S]cysteine during culture at 40°C, cell pellets were collected, the cytoplasmic membranes were solubilized with desoxycholate, and the samples were extracted two times with Freon 113. (A) Viral components from 6 × 108 cells were separated on preformed CsCl gradients, fractionated, and the density of each fraction determined from the refractive index; aliquots were then were counted for radioactivity as described in Materials and Methods. ●, Radioactivity in counts per minute/10 μl; +, fraction density in grams/cubic centimeter. (B) Electron micrographs of representative particles found in separate gradient fractions from the experiment shown in panel A. Panels: 1, whole virus (fraction 15); 2, λ2 spike-defective core particles (fraction 19); 3, outer-shell structures (gradient fraction 19); 4, separately prepared and purified T3D core particles prepared from permissively assembled whole T3D virions. A small amount of top component was present in each of fractions 17 through 20 (arrowhead, panel 3). Note the apical cavity present in the spike-defective core (small arrow, panel 2) (bar = 100 nm). (C) Fluorogram of a TGU-SDS-PAGE gel loaded with equivalent scintillation counts of permissively assembled T3D and tsA279 whole virions and from the same samples shown in panel B. Note the absence of λ2 in lanes 2 and 3, which are comprised primarily of core particles (lane 2) and outer shells (lane 3), with a small amount of top component in each fraction. T3, permissively assembled, purified T3D virions. Lanes 1 to 3 correspond to the samples represented in panel B and lane A shows permissively assembled, purified tsA279 virions. (D) Immunoblot of a TGU-SDS-PAGE gel run in parallel with and with the same samples as shown in panel C, with the addition of lane 4 which represents permissively assembled cores (same as panel B4). The gel was probed simultaneously by both anti-whole reovirus antiserum and anti-μ2 antiserum. Note the intense reaction of anti-μ2 in lane 4 and absence of reaction in the remaining lanes. The anti-whole reovirus antiserum shows weak reaction to λ2 in lane 4 at the concentration used but no reaction to λ2 in the remaining lanes. Similar results were obtained in each of four other experiments.
TGU-SDS-polyacrylamide gel electrophoresis (PAGE) protein analysis of the above fractions indicated that both the spike-defective cores and the outer-shell structures contained little, if any, protein λ2 (Fig. 2C). Shorter exposures confirmed that these particles were also devoid of the ς1 protein (data not shown). Parallel TGU gels which were immunoprobed with the anti-λ2 monoclonal antibody 7F4 (93) confirmed the presence of λ2 in intact core samples and the absence of λ2 in the spike-less particle fractions (data not shown). No fractions which contained structures deficient for the λ2 protein were identified electron microscopically or by electrophoretic protein profile in the permissive cultures of either T3D or tsA279. To measure the copy numbers of various proteins present in the core-like particles, laser densitometric scans of fluorograms representing multiple samples were analyzed. The number of cysteines and methionines contributed by each structural protein to whole virions and core particles was determined by multiplying the copy number of the protein in the structure by the number of cysteines and methionines present in one copy of the relevant protein. The copy number of residues contributed by each protein was then standardized to the proportion of residues contributed by the major core protein λ1 (Table 3). The signal for λ1 was determined for each fraction and used to determine the predicted signal for the remaining structural proteins in that fraction. The actual signal for each protein was then expressed as a proportion of the predicted signal (Table 4).
TABLE 3.
Relative copy numbers and cysteine and methionine content of the structural proteins in reovirus virions and core particles
| Protein | Mol wt (kDa) | No. of copies of proteina
|
No. of copies of cysteine and methionine
|
Relative proportion of cysteine and methionineb
|
||||
|---|---|---|---|---|---|---|---|---|
| Virus | Core | Moleculec | Viriond | Core | Virion | Core | ||
| λ1 | 137.4 | 120 | 120 | 55 | 6,600 | 6,600 | 1.0 | 1.0 |
| λ2 | 144.0 | 60 | 60 | 43 | 2,580 | 2,580 | 0.4 | 0.4 |
| λ3 | 142.4 | 12 | 12 | 65 | 780 | 780 | 0.1 | 0.1 |
| μ1c | 76.3 | 600 | 20 | 12,000 | 1.8 | |||
| μ2 | 83.3 | 21e | 14 | 39 | 819 | 546 | 0.1 | 0.1 |
| ς1 | 51.4 | 36 | 9 | 324 | 0.1 | |||
| ς2 | 47.1 | 120e | 120 | 18 | 2,160 | 2,160 | 0.3 | 0.3 |
| ς3 | 41.2 | 600 | 27 | 16,200 | 2.5 | |||
Dryden et al. (28).
The relative proportion of cysteine and methionine in each structure is normalized to the total copy number of cysteine (C) and methionine (M) in the λ1 protein by the following formula: [(copies of C and M for the protein) × (copies of the protein)] ÷ [(copies of C and M in λ1) × (copies of λ1)].
From published sequence data as follows: λ1 (7), λ2 (84), λ3 (99), μ1 (49, 98), μ1c (69), μ2 (97), ς1 (8), ς2 (25, 100), and ς3 (37).
The total cysteine and methionine in the structure for each protein was calculated by using the following formula: (total C and M in the protein) × (copies of protein).
The copy numbers of μ2 are from reference 20.
TABLE 4.
Relative amounts of proteins present in permissively and restrictively assembled tsA279 virions and intermediate structuresa
| Protein | Permissive
|
Restrictive
|
|||
|---|---|---|---|---|---|
| T3D virions | tsA279 virions | T3D virionsb |
tsA279
|
||
| Virions | Core fractions | ||||
| λ1 | 100 (0) | 100 (0) | 100 | 100 (0) | 100 (0) |
| λ2 | 112 (11) | 107 (4) | 108 | 109 (5) | 2 (1) |
| λ3 | 93 (10) | 95 (2)c | 102 | 98 (9) | 91 (12) |
| μ1/1c | 142 (34) | 146 (31) | 130 | 178 (41) | 88 (28) |
| μ2 | 100 (5) | 79 (10) | 100 | 97 (5) | 5 (3) |
| ς1 | ND | ND | ND | ND | ND |
| ς2 and ς3d | 156 (30) | 124 (10) | 88 | 201 (47) | 109 (33) |
All values were calculated from three to four separate infections except as noted. All values are expressed as the percentage of the predicted quantity, with the standard error of the mean of replicate cultures shown in parentheses. Integration values for λ1 are expressed as 100% of the expected value. Values for the other proteins are scaled to the λ1 values. Values for ς1 were not determined (ND).
Calculated from one experiment.
Calculated from two separate experiments.
ς2 and ς3 were analyzed jointly due to insufficient resolution in the TGU gel system for scan analysis.
When these densitometric calculations were applied to T3D and tsA279 virions which had been permissively assembled (cultured and assembled under permissive growth conditions), there was no significant difference between the wild type and the mutant. Restrictively assembled T3D and tsA279 virions also contained essentially the same number of copies of all structural proteins as permissively assembled virions (Table 4). However, there were significant differences in the relative amounts of core proteins λ2 and μ2 in the restrictive mutant core-like particle fractions; these structures contained approximately 2% of the expected amount of λ2, representing less than two copies per particle (Table 4). These fractions also contained outer-capsid proteins μ1/μ1c and ς3, which we assume to have been contributed by the contaminating outer-shell structures.
Gel analyses also indicated the minor protein μ2 produced by the mutant tsA279 at restrictive temperatures was either absent from the λ2-deficient core particles or migrated differently from μ2 produced at the permissive temperature (Fig. 2C, arrow). Laser densitometric scans indicated there were approximately normal amounts of the minor core protein λ3 but that there were about 5% the expected levels of μ2 (less than one copy per particle) in the core-like particles produced by the mutant at the nonpermissive temperature. Western blot evaluation of parallel TGU gels with polyclonal anti-μ2 antiserum confirmed that the core-like particles were deficient for this protein (Fig. 2D).
To determine whether the spike-deficient particles may represent normal assembly intermediates or “dead-end” products, we next determined the capacity of restrictively assembled spike-less core particles to accumulate spikes after temperature downshift experiments. After temperature downshift from 40 to 32°C in the presence of cycloheximide, the distribution of particles produced by the mutant was changed, with the proportion of core-like particles produced at a restrictive temperature being reduced from over 22 to below 2%, and the proportion of core particles which contained λ2 spikes increased from below 1 to approximately 20% (Table 5). There was little difference between the proportion of other species of particles produced after temperature downshift. The proportion of various particles that accumulated in the presence of cycloheximide after 6 h in each of the cultures maintained at a constant temperature did not differ dramatically from the proportion of particles produced in comparable non-cycloheximide-treated cultures (Tables 1 and 5).
TABLE 5.
Distribution of species of particles produced by tsA279 after temperature downshifta
| Temp (°C)b | Total no. of particlesc | Proportion of particle (% produced [SEM])d
|
||||
|---|---|---|---|---|---|---|
| Virion | Top component | Outer shell | Cores
|
|||
| With λ2 | No λ2 | |||||
| 32 | 2,464 | 79.6 (1.7) | 16.1 (0.8) | 1.4 (0.3) | 2.9 (0.8) | 0.1 (0.1) |
| 40/32 | 2,358 | 47.2 (1.1) | 29.3 (2.2) | 1.7 (0.4) | 20.1 (2.4) | 1.7 (0.8) |
| 40 | 3,206 | 43.9 (0.8) | 30.0 (1.8) | 2.3 (0.6) | 0.7 (0.3) | 22.5 (0.8) |
Calculated from three separate experiments. Infections were incubated for 36 h, with the addition of cycloheximide at 29.75 h postattachment.
32, incubation at 32°C for 36 h; 40/32, temperature downshift from 40 to 32°C at 30 h postattachment; 40, incubation at 40°C for 36 h.
The total number of viral and subviral particles counted.
Proportion of indicated particle type, expressed as the percentage of the total number of particles produced, with the standard error of the mean of replicate experiments shown in parentheses.
DISCUSSION
A mutant L2 gene (λ2 protein) is associated with the accumulation of spike-deficient core particles.
The assembly pathways of simple helical plant viruses have been extrapolated from disassembly and reassembly studies of tobacco mosaic virus (15, 17, 34, 92). While it has been possible to perform disassembly studies with complete reovirus virions (68) and to assemble capsids of this (102) and related viruses from expressed proteins (45, 91), sequential disassembly and reassembly has been marginally successful when applied to this (5) or other moderately complicated animal virus systems. Bacteriophage conditionally lethal mutants have been successfully used to derive assembly pathways of structurally complicated bacteriophage (29, 30). That success suggested that the Fields panel of reovirus conditionally lethal ts mutants (19, 32, 79) (reviewed in reference 21) could be used to determine the assembly pathways of a eucaryotic virus.
Direct particle counting of cytoplasmic extracts from restrictive tsA279-infected cultures confirmed the presence of and provided further insight into the nature of the core-like particles observed by thin-section electron microscopy (44). First, the proportion of core particles produced by the mutant under restrictive conditions was significantly greater than the proportion of cores produced under any of the permissive conditions (Table 1). Second, permissively assembled core particles observed in cultures of both T3D and tsA279 and restrictively assembled core particles from T3D demonstrated the λ2 apical spike and contained genomic material. Third, the core particles produced at restrictive temperature were deficient for both the λ2 and μ2 proteins and contained reduced levels of genomic material. Finally, when culture temperatures were shifted from 40 to 32°C during culture and maintained in the presence of cycloheximide to ensure the unavailability of permissibly transcribed viral protein, the proportion of spike-defective core-like particles declined, while the proportion of particles demonstrating spikes increased. This is the first report that core-like particles deficient for core proteins λ2 and μ2 (Table 4) may accumulate in the assembly pathway. Analysis of a panel of T1L×tsA279 intertypic reassortants indicated the production of the λ2 spike-deficient core particles was associated with the mutant L2 gene segment (Table 2).
The apparent inability to isolate comparable spike-less particles from wild-type infected cell extracts, coupled with the ability to readily disassemble virions and create spiked core particles (22, 28, 59, 70), has led to the assumption that the core particles usually seen in infected cells by thin-section electron microscopy are complete core particles. This premise is also supported by the accumulation of core particles which contain the λ2 pentameric spike in restrictive cultures from representatives of recombination groups B (65) and G (23, 65, 88), which code for the core spike protein λ2 and major outer capsid protein ς3, respectively (21, 67). Spike-less core particles have been prepared in laboratory settings from top component and whole virus (90, 96, 106). The core particles generated by White and Zweerink demonstrated apical cavities after the removal of the λ2 pentameric structure by high-pH treatment (96), as do those produced by treatment at high temperature (106). The core particles produced by tsA279 also demonstrated such cavities (Fig. 1A and 2B2, small arrow). Similarities in particle morphology of the spike-deficient core particle produced by tsA279 and the spike-deficient core particles produced in vitro (96, 106), coupled with the protein profiles of previously prepared spikeless cores (96, 106), suggested that all other core proteins were present. The unexpected observation that the μ2 protein was also not present in the core-like particles identified in this study provides further insight into the assembly pathway of reovirus.
Assembly pathway of reovirus.
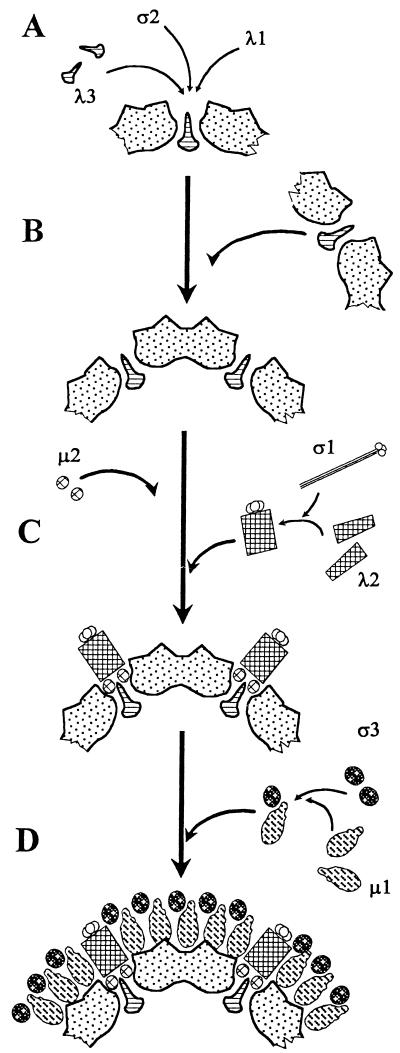
The accumulation of λ2 spike-deficient cores in the restrictively grown tsA279 cultures implies that the assembly pathway is blocked as a result of the altered λ2 product. The identification of this structure in vivo suggests that initiation of capsid assembly may involve interactions between the core proteins λ1, λ3, and ς2 to form a “primary core particle” (Fig. 3B). This notion is supported by coexpression studies that show that λ1 and ς2 together, but not alone, are minimally required for core capsid assembly, and that coexpression of these two major proteins with any of the other core proteins resulted in incorporation of each protein into a genome-deficient core-like particle (102). The inclusion of λ3 in the primary core identified in the current study implies that this minor protein is incorporated into the structure at an early point in assembly, while the concomitant lack of both λ2 and μ2 in the primary core suggests that neither protein is necessary for the initiation of assembly.
FIG. 3.
The proposed model for the assembly of reovirus. (A) Assembly of the dodecahedral base unit. Five dimers of the core protein λ1, ten units of ς2, and one copy of λ3 associate to form the dodecahedral base structure. (B) Assembly of the primary core particle. Twelve dodecahedral base units assemble to form the primary capsid structure. (C) Assembly of the intermediate particle. Two copies of μ2 associate with each apex of the primary core particles immediately prior to or collateral with assembly of the twelve λ2 pentameric spikes. The trimeric ς1 attachment protein is most likely coassembled with the λ2 spike, with the amino termini of the ς1 fibers interacting with the carboxy termini of the λ2 spike proteins. (D) The outer capsid proteins ς3 and μ1 associate with each other and then condense around the λ2 pentameric spike to form the complete virion.
Previous reports have identified l3, m3, s3, and s4 as the first viral mRNAs present in reovirus-infected cultures (52). The encoded proteins for three of these—μNS, ςNS, and ς3—are the first viral proteins to associate with viral single-stranded RNA (ssRNA) to form ssRNA-containing complexes (ssRCCs) in restrictive cultures of T3D and the dsRNA-negative ts mutants tsC447, tsD357, and tsE320, which are defective for ς2, λ3, and ςNS, respectively (3). Small quantities of λ1 (encoded by L3) and λ2 were the next constituents identified in the ssRCC complexes. The major core protein ς2 was not identified in these complexes from T3D or in the different ts mutants examined. These complexes were not examined for the presence of λ3 nor μ2 (3). The next structures identified in that study were dsRNA-containing complexes (dsRCCs), which were comprised of the proteins μNS, ςNS, ς3, and λ2. Coprecipitation studies with antibodies directed against the major core proteins ς2 and λ1 also precipitated dsRCCs from restrictive infections with T3D (3). The results from that study are consistent with previous reports of an ssRNA transcriptase particle comprised of the proteins λ1, ς2, μNS, and reduced quantities of λ2 (66, 107, 108). Review of the data in those reports does not preclude the presence of μ2 or λ3, which comigrate with other proteins in the gel system used. While the primary core particle identified in the present study differs from the dsRCCs reported with other ts mutants (3) in that it lacks the nonstructural proteins, it bears some similarity to the previously identified transcriptase particle (66). This 40-nm particle has been reported to contain all core structural proteins plus the nonstructural protein μ0 (now called μNS) (66).
These studies are consistent with a dodecahedral assembly model based upon a five-sided apical complex which contains 1 central copy of the minor core protein λ3, 5 dimers of the major core protein λ1 and 10 copies of ς2, in which λ3 interacts with the λ1 amino termini (27, 68) to form the dodecahedral base unit (Fig. 3A) from which the primary core particle is subsequently formed. Nonstructural proteins would be excluded as base units assemble, resulting in the primary core particle identified in the current studies (Fig. 3B). Dodecahedral structures constructed from penton base units have been identified in adenovirus type 3 infections (73). Coexpression of the adenovirus 3 penton base and fiber proteins also produce dodecahedral particles comprised of 12 penton and fiber base units (31, 83). Since each multimeric penton base unit is comprised of 5 penton protein monomers, these particles are made up of 60 penton monomers, plus 12 trimeric apical fibers, making them analogous to miniature capsids (83), with a triangulation lattice T=1 (16, 50). Dodecahedral models have also been proposed for the assembly of small capsids with triangulation lattice numbers equal to 1, such as the picornaviruses (reviewed in reference 2), bacteriophage φ6 (14), and the yeast virus L-A (18). Bacteriophage φ6 and yeast virus L-A are other dsRNA viruses constructed from 120 copies of a major capsid protein (14, 18). The core particle of reovirus shares similar copy numbers of major core proteins and an apparent triangulation lattice value of 1 (20, 68).
Subsequent assembly of the complete core particle is proposed to involve association of the minor core protein μ2 with the apices of the primary core particle either immediately prior to or collaterally with λ2 (Fig. 3C). Electron cryomicroscope reconstructions of T1L top-component cores further suggest that λ2 and μ2 interact with λ3 at the apices of the primary core particle (27). While the precise location of the two minor proteins in the apical complex have not been established, all three proteins have been associated with functions of the RNA-dependent RNA polymerase either separately or in different combinations (72, 87, 106). The finding that expressed λ2 exists as a monomer (61) suggests that λ2 condenses into pentamers during association with the primary core particle. Mutation(s) in the λ2 protein could affect the appearance of the primary core particle in two ways: by either preventing pentamerization of the protein or blocking the interactions of the protein and/or its pentamer with the core to produce the complete core particle. In addition, the accumulation of outer-shell structures comprised of proteins μ1 and ς3, but clearly lacking proteins λ2 and ς1 (Fig. 2), suggests that the mutant λ2 protein or its pentamer is not able to interact with the outer capsid proteins when assembled at restrictive temperatures, in contrast to wild-type λ2 protein, which can interact with the outer capsid proteins (28, 62). The observation that temperature shiftdown experiments (Table 5) lead to condensation of spikes onto the previously spike-less restrictive particles is consistent with the proposed condensation of these proteins to form the complete core particle. In the proposed dodecahedral assembly path for the reovirus primary core particle, the replication of the minus sense viral RNA to complete the genome at this stage could also result in a maturational event similar to that reported for φ6 (14), where the dodecahedral structure of the base unit would be displaced radially from the pentameric faces during assembly to provide a triangulation lattice T=1 icosahedral structure.
The precise nature of the interactions of ς1 with the λ2 spike is not clear (27, 68). This protein was not examined during previous coexpression and assembly studies. However, incorporation of the trimeric ς1 attachment fiber with the λ2 pentamer most likely occurs at the time the λ2 spike condenses and associates with the primary core, with the amino terminus of the ς1 fibers interacting with the carboxy terminus of the λ2 proteins (56, 60), thereby forming an intermediate particle.
The final stage of reovirus capsid assembly involves condensation of the outer capsid around the core lattice (Fig. 3D). Previous reports had identified the presence of outer-shell structures (L top component) in restrictive cultures of tsC447 that contained λ2, μ1, ς1, and ς3 (62). Electron cryomicroscope examination of whole virions and ISVPs have demonstrated interactions between λ2 and both μ1 and ς3. The same study showed little contact between μ1 and ς3 and the core shell (28). Those reports suggested that the λ2 complex may serve as a nucleation site for condensation of μ1 into the outer capsid lattice (28). However, the presence of μ1/ς3 outer-shell complexes that clearly lack both λ2 and ς1 in tsA279 restrictive cultures indicates that these proteins are not required for condensation of an outer-capsid-like structure. Coprecipitation studies indicated that the outer capsid proteins μ1 and ς3 form complexes in permissive cultures of T3D (55) and tsG453 (88). The inability to coprecipitate these outer capsid proteins in restrictive cultures of tsG453, combined with the observation that the restrictive product of tsG453 infections is a core-like particle (23, 88) rather than an intermediate subviral particle, indicate that the outer capsid proteins μ1 and ς3 must interact with each other before they can condense to form the outer capsid (88).
In conclusion, investigations into the intermediate assembly structures that accumulate in restrictive cultures of tsA279, a member of the Fields panel of reovirus ts mutants, have identified a novel primary core capsid comprised of λ1, λ3, and ς2. This structure may represent the transitional stage between the RNA-containing complexes described by Antczak and Joklik (3) and the λ2 spike-containing core structure identified in restrictive cultures of tsB352 (65) and tsG453 (23, 65, 88). We propose that these complexes combine through a dodecahedral mechanism of assembly to form the primary core capsid as the first capsid structure in the reovirus assembly pathway. The remaining core proteins λ2 and μ2 then combine with the primary core capsid, along with the ς1 trimeric attachment complex, to form the more complete intermediate particle. Finally, the outer capsid proteins μ1 and ς3 interact and coprecipitate around this intermediate particle to form the complete reovirus capsid.
ACKNOWLEDGMENTS
We thank R. C. Brunham for his ongoing support of this work; C. Power and J. N. Simonsen for critical review of this study; L. Petrycky-Cox for stock cell maintenance; and G. Dow, J. D. Berry, M. Makarovsky, N. Keirstead, M. Patrick, G. Wong, and P. Yin for helpful discussion.
This research was supported by grant MT-11630 from the Medical Research Council of Canada.
ADDENDUM IN PROOF
Recent crystallography data of the bluetongue virus core structure by Grimes et al. (J. M. Grimes, J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. C. Mertens, and D. I. Stuart, Nature 395:470–478, 1998) suggest that the VP3 scaffold assembles by a dodecahedral assembly mechanism.
REFERENCES
- 1.Amersham Life Science. ECL Western blotting protocols. PI/341/95/04. Buckinghamshire, England: Amersham International plc.; 1995. [Google Scholar]
- 2.Ansardi D C, Porter D C, Anderson M J, Morrow C D. Poliovirus assembly and encapsidation of genomic RNA. Adv Virus Res. 1996;46:1–68. [PubMed] [Google Scholar]
- 3.Antczak J B, Joklik W K. Reovirus genome segment assortment into progeny genomes studied by the use of monoclonal antibodies directed against reovirus proteins. Virology. 1992;187:760–776. doi: 10.1016/0042-6822(92)90478-8. [DOI] [PubMed] [Google Scholar]
- 4.Arsenakis M, Roizman B. A post-alpha gene function turns off the capacity of a host protein to bind DNA in cells infected with herpes simplex virus 1. J Virol. 1984;49:813–818. doi: 10.1128/jvi.49.3.813-818.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astell C, Silverstein S C, Levin D H, Acs G. Regulation of the reovirus RNA transcriptase by a viral capsomere protein. Virology. 1972;48:648–654. doi: 10.1016/0042-6822(72)90149-3. [DOI] [PubMed] [Google Scholar]
- 6.Baker T S, Drak J, Bina M. Reconstruction of the three-dimensional structure of simian virus 40 and visualization of the chromatin core. Proc Natl Acad Sci USA. 1988;85:422–426. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartlett J A, Joklik W K. The sequence of the reovirus serotype 3 L3 genome segment which encodes the major core protein lambda 1. Virology. 1988;167:31–37. doi: 10.1016/0042-6822(88)90051-7. [DOI] [PubMed] [Google Scholar]
- 8.Bassel-Duby R, Jayasuriya A, Chatterjee D, Sonenberg N, Maizel J V, Jr, Fields B N. Sequence of reovirus haemagglutinin predicts a coiled-coil structure. Nature. 1985;315:421–423. doi: 10.1038/315421a0. [DOI] [PubMed] [Google Scholar]
- 9.Behnke O, Ammitzboll T, Jessen H, Klokker M, Nilausen K, Tranum-Jensen J, Olsson L. Non-specific binding of protein-stabilized gold sols as a source of error in immunocytochemistry. Eur J Cell Biol. 1986;41:326–338. [PubMed] [Google Scholar]
- 10.Berget P B, King J. T4 tail morphogenesis. In: Mathews C, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. Washington, D.C: American Society for Microbiology; 1983. pp. 246–258. [Google Scholar]
- 11.Black L W, Showe M K. Morphogenesis of the T4 head. In: Matthews C, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. Washington, D.C: American Society for Microbiology; 1983. pp. 219–245. [Google Scholar]
- 12.Bothner B, Dong X F, Bibbs L, Johnson J E, Siuzdak G. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J Biol Chem. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- 13.Brenner S, Horne R W. Negative staining method for high-resolution electron microscopy of viruses. Biochem Biophys Acta. 1959;34:103–110. doi: 10.1016/0006-3002(59)90237-9. [DOI] [PubMed] [Google Scholar]
- 14.Butcher S J, Dokland T, Ojala P M, Bamford D H, Fuller S D. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. EMBO J. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butler P J. The current picture of the structure and assembly of tobacco mosaic virus. J Gen Virol. 1984;65:253–279. doi: 10.1099/0022-1317-65-2-253. [DOI] [PubMed] [Google Scholar]
- 16.Caspar D L D, Klug A. Physical properties in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–32. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 17.Caspar D L, Namba K. Switching in the self-assembly of tobacco mosaic virus. Adv Biophys. 1990;26:157–185. doi: 10.1016/0065-227x(90)90011-h. [DOI] [PubMed] [Google Scholar]
- 18.Cheng R H, Caston J R, Wang G J, Gu F, Smith T J, Baker T S, Bozarth R F, Trus B L, Cheng N, Wickner R B, Steven A C. Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric Gag dimers. J Mol Biol. 1994;244:255–258. doi: 10.1006/jmbi.1994.1726. [DOI] [PubMed] [Google Scholar]
- 19.Coombs K M. Identification and characterization of a double-stranded RNA- reovirus temperature-sensitive mutant defective in minor core protein μ2. J Virol. 1996;70:4237–4245. doi: 10.1128/jvi.70.7.4237-4245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coombs K M. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology. 1998;243:218–228. doi: 10.1006/viro.1998.9061. [DOI] [PubMed] [Google Scholar]
- 21.Coombs K M. Temperature-sensitive mutants of reovirus. Curr Top Microbiol Immunol. 1998;233:69–107. doi: 10.1007/978-3-642-72092-5_4. [DOI] [PubMed] [Google Scholar]
- 22.Coombs K M, Fields B N, Harrison S C. Crystallization of the reovirus type 3 Dearing core. Crystal packing is determined by the lambda 2 protein. J Mol Biol. 1990;215:1–5. doi: 10.1016/s0022-2836(05)80089-0. [DOI] [PubMed] [Google Scholar]
- 23.Danis C, Garzon S, Lemay G. Further characterization of the ts453 mutant of mammalian orthoreovirus serotype 3 and nucleotide sequence of the mutated S4 gene. Virology. 1992;190:494–498. doi: 10.1016/0042-6822(92)91241-l. [DOI] [PubMed] [Google Scholar]
- 24.de Prat-Gay G. Conformational preferences of a peptide corresponding to the major antigenic determinant of foot-and-mouth disease virus: implications for peptide-vaccine approaches. Arch Biochem Biophys. 1997;341:360–369. doi: 10.1006/abbi.1997.9982. [DOI] [PubMed] [Google Scholar]
- 25.Dermody T S, Schiff L A, Nibert M L, Coombs K M, Fields B N. The S2 gene nucleotide sequences of prototype strains of the three reovirus serotypes: characterization of reovirus core protein ς2. J Virol. 1991;65:5721–5731. doi: 10.1128/jvi.65.11.5721-5731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drayna D, Fields B N. Activation and characterization of the reovirus transcriptase: genetic analysis. J Virol. 1982;41:110–118. doi: 10.1128/jvi.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dryden K A, Farsetta D L, Wang G, Keegan J M, Fields B N, Baker T S, Nibert M. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology. 1998;245:33–46. doi: 10.1006/viro.1998.9146. [DOI] [PubMed] [Google Scholar]
- 28.Dryden K A, Wang G, Yeager M, Nibert M L, Coombs K M, Furlong D B, Fields B N, Baker T S. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fane B, King J. Identification of sites influencing the folding and subunit assembly of the P22 tailspike polypeptide chain using nonsense mutations. Genetics. 1987;117:157–171. doi: 10.1093/genetics/117.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fane B, Villafane R, Mitraki A, King J. Identification of global suppressors for temperature-sensitive folding mutations of the P22 tailspike protein. J Biol Chem. 1991;266:11640–11648. [PubMed] [Google Scholar]
- 31.Fender P, Ruigrok R W H, Gout E, Buffet S, Chroboczek J. Adenovirus dodecahedron, a new vector for human gene transfer. Nat Biotechnol. 1997;15:52–56. doi: 10.1038/nbt0197-52. [DOI] [PubMed] [Google Scholar]
- 32.Fields B N, Joklik W K. Isolation and preliminary genetic and biochemical characterization of temperature-sensitive mutants of reovirus. Virology. 1969;37:335–342. doi: 10.1016/0042-6822(69)90217-7. [DOI] [PubMed] [Google Scholar]
- 33.Fields B N, Raine C S, Baum S G. Temperature-sensitive mutants of reovirus type 3: defects in viral maturation as studied by immunofluorescence and electron microscopy. Virology. 1971;43:569–578. doi: 10.1016/0042-6822(71)90282-0. [DOI] [PubMed] [Google Scholar]
- 34.Fraenkel-Conrat H, Williams R C. Reconstitution of active tobacco mosaic virus from its inactive protein and nucleic acid components. Proc Natl Acad Sci USA. 1955;41:690–698. doi: 10.1073/pnas.41.10.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furlong D B, Nibert M L, Fields B N. ς1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geoghegan W D, Ackerman G A. Adsorption of horseradish peroxidase, ovomucoid and anti-immunoglobulin to colloidal gold for the indirect detection of concanavalin A, wheat germ agglutinin and goat anti-human immunoglobulin G on cell surfaces at the electron microscopic level: a new method, theory and application. J Histochem Cytochem. 1977;25:1187–1200. doi: 10.1177/25.11.21217. [DOI] [PubMed] [Google Scholar]
- 37.Giantini M, Seliger L S, Furuichi Y, Shatkin A J. Reovirus type 3 genome segment S4: nucleotide sequence of the gene encoding a major virion surface protein. J Virol. 1984;52:984–987. doi: 10.1128/jvi.52.3.984-987.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammond G W, Hazelton P R, Cheung I, Klisko B. Improved detection of viruses by electron microscopy after direct ultracentrifuge preparation of specimens. J Clin Microbiol. 1981;14:210–221. doi: 10.1128/jcm.14.2.210-221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harlow E, Lane D, editors. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 40.Harrison S C. Virus structure. In: Fields B N, Knipe D M, editors. Fields virology. 2nd ed. Vol. 1. New York, N.Y: Raven Press; 1990. pp. 37–61. [Google Scholar]
- 41.Hassard T H. Understanding biostatistics. St. Louis, Mo: Mosby; 1991. [Google Scholar]
- 42.Hayat M A. Basic techniques for transmission electron microscopy. London, United Kingdom: Academic Press; 1985. [Google Scholar]
- 43.Hazelton P R. Morphogenesis of reovirus as defined by the Fields panel of temperature sensitive mutants. Ph.D. thesis. Winnipeg, Manitoba, Canada: University of Manitoba; 1998. [Google Scholar]
- 43a.Hazelton, P. R. Unpublished data.
- 44.Hazelton P R, Coombs K M. The reovirus mutant tsA279 has temperature-sensitive lesions in the M2 and L2 genes: the M2 gene is associated with decreased viral protein production and blockade in transmembrane transport. Virology. 1995;207:46–58. doi: 10.1006/viro.1995.1050. [DOI] [PubMed] [Google Scholar]
- 45.Hewat E A, Booth T F, Loudon P T, Roy P. Three-dimensional reconstruction of baculovirus expressed bluetongue virus core-like particles by cryo-electron microscopy. Virology. 1992;189:10–20. doi: 10.1016/0042-6822(92)90676-g. [DOI] [PubMed] [Google Scholar]
- 46.Hoffman L R, Kuntz I D, White J M. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: irreversible inhibition of infectivity. J Virol. 1997;71:8808–8820. doi: 10.1128/jvi.71.11.8808-8820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogle J M, Chow M, Filman D J. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 48.Horisberger M. Quantitative aspects of labelling colloidal gold with proteins. In: Verkleij A J, Leunissen J L M, editors. Immuno-gold labelling in cell biology. Boca Raton, Fla: CRC Press; 1989. pp. 49–61. [Google Scholar]
- 49.Jayasuriya A K, Nibert M L, Fields B N. Complete nucleotide sequence of the M2 gene segment of reovirus type 3 dearing and analysis of its protein product mu 1. Virology. 1988;163:591–602. doi: 10.1016/0042-6822(88)90300-5. [DOI] [PubMed] [Google Scholar]
- 50.Klug A, Caspar D L D. The structure of small viruses. Adv Virus Res. 1960;7:225–325. doi: 10.1016/s0065-3527(08)60012-3. [DOI] [PubMed] [Google Scholar]
- 51.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 52.Lau R Y, Van Alstyne D, Berckmans R, Graham A F. Synthesis of reovirus-specific polypeptides in cells pretreated with cyclohexamide. J Virol. 1975;16:470–478. doi: 10.1128/jvi.16.3.470-478.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawton J A, Estes M K, Prasad B V. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 54.Lawton J A, Zeng C Q, Mukherjee S K, Cohen J, Estes M K, Prasad B V. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee P W K, Hayes E C, Joklik W K. Characterization of antireovirus immunoglobulins secreted by cloned hybridoma cell lines. Virology. 1981;108:134–146. doi: 10.1016/0042-6822(81)90533-x. [DOI] [PubMed] [Google Scholar]
- 56.Leone G, Mah D C, Lee P W. The incorporation of reovirus cell attachment protein sigma 1 into virions requires the N-terminal hydrophobic tail and the adjacent heptad repeat region. Virology. 1991;182:346–350. doi: 10.1016/0042-6822(91)90678-5. [DOI] [PubMed] [Google Scholar]
- 57.Lucia-Jandris P, Hooper J W, Fields B N. Reovirus M2 gene is associated with chromium release from mouse L cells. J Virol. 1993;67:5339–5345. doi: 10.1128/jvi.67.9.5339-5345.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lucocq J M, Baschong W. Preparation of protein colloidal gold complexes in the presence of commonly used buffers. Eur J Cell Biol. 1986;42:332–337. [PubMed] [Google Scholar]
- 59.Luftig R B, Kilham S S, Hay A J, Zweerink H J, Joklik W K. An ultrastructure study of virions and cores of reovirus type 3. Virology. 1972;48:170–181. doi: 10.1016/0042-6822(72)90124-9. [DOI] [PubMed] [Google Scholar]
- 60.Luongo C L, Dryden K A, Farsetta D L, Margraf R L, Severson T F, Olson N H, Fields B N, Baker T S, Nibert M L. Localization of a C-terminal region of λ2 protein in reovirus cores. J Virol. 1997;71:8035–8040. doi: 10.1128/jvi.71.10.8035-8040.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao Z X, Joklik W K. Isolation and enzymatic characterization of protein lambda 2, the reovirus guanylyltransferase. Virology. 1991;185:377–386. doi: 10.1016/0042-6822(91)90785-a. [DOI] [PubMed] [Google Scholar]
- 62.Matsuhisa T, Joklik W K. Temperature-sensitive mutants of reovirus. V. Studies on the nature of the temperature-sensitive lesion of the group C mutant ts447. Virology. 1974;60:380–389. doi: 10.1016/0042-6822(74)90333-x. [DOI] [PubMed] [Google Scholar]
- 63.Miller M F. A universal sedimentation virus particle counting procedure. In: Bailey G W, editor. 37th Annual Proceedings of the Electron Microscopy Society of America. San Francisco, Calif: San Francisco Press; 1979. pp. 56–57. [Google Scholar]
- 64.Miller S E. Diagnosis of viral infections by electron microscopy. In: Lennette E H, Lennette D A, Lennette E T, editors. Diagnostic procedures for viral, rickettsial and chlamydial infections. Washington, D.C: American Public Health Association; 1995. [Google Scholar]
- 65.Morgan E M, Zweerink H J. Reovirus morphogenesis. Corelike particles in cells infected at 39° with wild-type reovirus and temperature sensitive mutants of groups B and G. Virology. 1974;59:556–565. doi: 10.1016/0042-6822(74)90465-6. [DOI] [PubMed] [Google Scholar]
- 66.Morgan E M, Zweerink H J. Characterization of transcriptase and replicase particles isolated from reovirus-infected cells. Virology. 1975;68:455–466. doi: 10.1016/0042-6822(75)90286-x. [DOI] [PubMed] [Google Scholar]
- 67.Mustoe T A, Ramig R F, Sharpe A H, Fields B N. A genetic map of reovirus. III. Assignment of the double-stranded RNA mutant groups A, B, and G to genome segments. Virology. 1978;85:545–556. doi: 10.1016/0042-6822(78)90460-9. [DOI] [PubMed] [Google Scholar]
- 68.Nibert M L. Structure of mammalian orthoreovirus particles. Curr Top Microbiol Immunol. 1998;233:1–30. doi: 10.1007/978-3-642-72092-5_1. [DOI] [PubMed] [Google Scholar]
- 69.Nibert M L, Fields B N. A carboxy-terminal fragment of protein mu 1/mu 1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J Virol. 1992;66:6408–6418. doi: 10.1128/jvi.66.11.6408-6418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nibert M L, Furlong D B, Fields B N. Mechanisms of viral pathogenesis. Distinct forms of reoviruses and their roles during replication in cells and host. J Clin Invest. 1991;88:727–734. doi: 10.1172/JCI115369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nibert M L, Schiff L A, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1557–1596. [Google Scholar]
- 72.Noble S, Nibert M L. Core protein mu2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J Virol. 1997;71:7728–7735. doi: 10.1128/jvi.71.10.7728-7735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Norrby E. The relationship between the soluble antigens and the virion of adenovirus type 3. Virology. 1964;28:236–248. doi: 10.1016/0042-6822(66)90148-6. [DOI] [PubMed] [Google Scholar]
- 74.Ono K, Dubois-Dalcq M E, Schubert M, Lazzarini R A. A mutated membrane protein of vesicular stomatitis virus has an abnormal distribution within the infected cell and causes defective budding. J Virol. 1987;61:1332–1341. doi: 10.1128/jvi.61.5.1332-1341.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paredes A M, Brown D T, Rothnagel R, Chiu W, Schoepp R J, Johnston R E, Prasad B V. Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peeling R, MacLean I, Brunham R C. In vitro neutralization of Chlamydia trachomatous with monoclonal antibody to an epitope on the major outer membrane protein. Infect Immun. 1984;46:484–488. doi: 10.1128/iai.46.2.484-488.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prasad B V, Rothnagel R, Zeng C Q, Jakana J, Lawton J A, Chiu W, Estes M K. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–473. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 78.Prevelige P E, Jr, King J. Assembly of bacteriophage P22: A model for ds-DNA virus assembly. Prog Med Virol. 1993;40:206–221. [PubMed] [Google Scholar]
- 79.Ramig R F, Ahmed R, Fields B N. A genetic map of reovirus: assignment of the newly defined mutant groups H, I, and J to genome segments. Virology. 1983;125:299–313. doi: 10.1016/0042-6822(83)90203-9. [DOI] [PubMed] [Google Scholar]
- 80.Rhim J S, Smith K O, Melnick J L. Complete and coreless forms of reovirus (ECHO 10): ratio of number of virus particles to infective units in the one-step growth cycle. Virology. 1961;15:428–435. doi: 10.1016/0042-6822(61)90110-6. [DOI] [PubMed] [Google Scholar]
- 81.Robinson E N, Jr, Clemens C M, Schoolnik G K, McGee Z A. Probing the surface of Neisseria gonorrhoeae: immunoelectron microscopic studies to localize cyanogen bromide fragment 2 in gonococcal pili. Mol Microbiol. 1989;3:57–64. doi: 10.1111/j.1365-2958.1989.tb00104.x. [DOI] [PubMed] [Google Scholar]
- 82.Rossmann M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H J, Johnson J E, Kamer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 83.Schoehn G, Fender P, Chroboczek J, Hewat E A. Adenovirus 3 penton dodecahedron exhibits structural changes of the base on fibre binding. EMBO J. 1996;15:6841–6846. [PMC free article] [PubMed] [Google Scholar]
- 84.Seliger L S, Zheng K, Shatkin A J. Complete nucleotide sequence of reovirus L2 gene and deduced amino acid sequence of viral mRNA guanylyltransferase. J Biol Chem. 1987;262:16289–16293. [PubMed] [Google Scholar]
- 85.Shaw A L, Rothnagel R, Chen D, Ramig R F, Chiu W, Prasad B V. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell. 1993;74:693–701. doi: 10.1016/0092-8674(93)90516-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shaw A L, Samal S K, Subramanian K, Prasad B V. The structure of aquareovirus shows how the different geometries of the two layers of the capsid are reconciled to provide symmetrical interactions and stabilization. Structure. 1996;4:957–967. doi: 10.1016/s0969-2126(96)00102-5. [DOI] [PubMed] [Google Scholar]
- 87.Sherry B, Blum M A. Multiple viral core proteins are determinants of reovirus-induced acute myocarditis. J Virol. 1994;68:8461–8465. doi: 10.1128/jvi.68.12.8461-8465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shing M, Coombs K M. Assembly of the reovirus outer capsid requires mu 1/sigma 3 interactions which are prevented by misfolded sigma 3 protein in temperature-sensitive mutant tsG453. Virus Res. 1996;46:19–29. doi: 10.1016/s0168-1702(96)01372-x. [DOI] [PubMed] [Google Scholar]
- 89.Slot J W, Geuze H J. A new method of preparing gold probes for multiple-labeling cytochemistry. Eur J Cell Biol. 1985;38:87–93. [PubMed] [Google Scholar]
- 90.Smith R E, Zweerink H J, Joklik W K. Polypeptide components of virions, top component and cores of reovirus type 3. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 91.Strong J E, Leone G, Duncan R, Sharma R K, Lee P W. Biochemical and biophysical characterization of the reovirus cell attachment protein sigma 1: evidence that it is a homotrimer. Virology. 1991;184:23–32. doi: 10.1016/0042-6822(91)90818-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turner D R, McGuigan C J, Butler P J. Assembly of hybrid RNAs with tobacco mosaic virus coat protein. Evidence for incorporation of disks in 5′-elongation along the major RNA tail. J Mol Biol. 1989;209:407–422. doi: 10.1016/0022-2836(89)90006-5. [DOI] [PubMed] [Google Scholar]
- 93.Virgin H W, IV, Mann M A, Fields B N, Tyler K L. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J Virol. 1991;65:6772–6781. doi: 10.1128/jvi.65.12.6772-6781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vreeswijk J, Folkers E, Wagenaar F, Kapsenberg J G. The use of colloidal gold immunoelectron microscopy to diagnose varicella-zoster virus (VZV) infections by rapid discrimination between VZV, HSV-1 and HSV-2. J Virol Methods. 1988;22:255–271. doi: 10.1016/0166-0934(88)90108-5. [DOI] [PubMed] [Google Scholar]
- 95.Weiner H L, Powers M L, Fields B N. Absolute linkage of virulence and central nervous system tropism of reovirus to viral hemagglutinin. J Infect Dis. 1980;141:609–616. doi: 10.1093/infdis/141.5.609. [DOI] [PubMed] [Google Scholar]
- 96.White C K, Zweerink H J. Studies on the structure of reovirus cores: selective removal of polypeptide λ2. Virology. 1976;70:171–180. doi: 10.1016/0042-6822(76)90247-6. [DOI] [PubMed] [Google Scholar]
- 97.Wiener J R, Bartlett J A, Joklik W K. The sequences of reovirus serotype 3 genome segments M1 and M3 encoding the minor protein mu 2 and the major nonstructural protein mu NS, respectively. Virology. 1989;169:293–304. doi: 10.1016/0042-6822(89)90154-2. [DOI] [PubMed] [Google Scholar]
- 98.Wiener J R, Joklik W K. Evolution of reovirus genes: a comparison of serotype 1, 2, and 3 M2 genome segments, which encode the major structural capsid protein mu 1C. Virology. 1988;163:603–613. doi: 10.1016/0042-6822(88)90301-7. [DOI] [PubMed] [Google Scholar]
- 99.Wiener J R, Joklik W K. The sequences of the reovirus serotype 1, 2, and 3 L1 genome segments and analysis of the mode of divergence of the reovirus serotypes. Virology. 1989;169:194–203. doi: 10.1016/0042-6822(89)90055-x. [DOI] [PubMed] [Google Scholar]
- 100.Wiener J R, McLaughlin T, Joklik W K. The sequences of the S2 genome segments of reovirus serotype 3 and of the dsRNA-negative mutant ts447. Virology. 1989;170:340–341. doi: 10.1016/0042-6822(89)90392-9. [DOI] [PubMed] [Google Scholar]
- 101.Wood W B, Crowther R A. Long tail fibers: genes, proteins, assembly, and structure. In: Mathews C, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. Washington, D.C: American Society for Microbiology; 1983. pp. 259–269. [Google Scholar]
- 102.Xu P, Miller S E, Joklik W K. Generation of reovirus core-like particles in cells infected with hybrid vaccinia viruses that express genome segments L1, L2, L3, and S2. Virology. 1993;197:726–731. doi: 10.1006/viro.1993.1648. [DOI] [PubMed] [Google Scholar]
- 103.Yeager M, Berriman J A, Baker T S, Bellamy A R. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 1994;13:1011–1018. doi: 10.1002/j.1460-2075.1994.tb06349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yeager M, Weiner S C, Coombs K M. Transcriptionally active reovirus core particles visualized by electron cryo-microscopy and image reconstruction. Biophys J. 1996;70:A116. [Google Scholar]
- 105.Yin P, Cheang M, Coombs K M. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J Virol. 1996;70:1223–1227. doi: 10.1128/jvi.70.2.1223-1227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yin, P., P. R. Hazelton, and K. M. Coombs. Unpublished data.
- 107.Zweerink H J, Ito Y, Matsuhisa T. Synthesis of reovirus double-stranded RNA within virion-like particles. Virology. 1972;50:349–358. doi: 10.1016/0042-6822(72)90386-8. [DOI] [PubMed] [Google Scholar]
- 108.Zweerink H J, Morgan E M, Skyler J S. Reovirus morphogenesis: characterization of subviral particles in infected cells. Virology. 1976;73:442–453. doi: 10.1016/0042-6822(76)90405-0. [DOI] [PubMed] [Google Scholar]