Abstract
This study presents the first chromosome-level genome assembly of Hanwoo, an indigenous Korean breed of Bos taurus taurus. This is the first genome assembly of Asian taurus breed. Also, we constructed a pangenome graph of 14 B. taurus genome assemblies. The contig N50 was over 55 Mb, the scaffold N50 was over 89 Mb and a genome completeness of 95.8%, as estimated by BUSCO using the mammalian set, indicated a high-quality assembly. 48.7% of the genome comprised various repetitive elements, including DNAs, tandem repeats, long interspersed nuclear elements, and simple repeats. A total of 27,314 protein-coding genes were identified, including 25,302 proteins with inferred gene names and 2,012 unknown proteins. The pangenome graph of 14 B. taurus autosomes revealed 528.47 Mb non-reference regions in total and 61.87 Mb Hanwoo-specific regions. Our Hanwoo assembly and pangenome graph provide valuable resources for studying B. taurus populations.
Subject terms: Genomics, Genome
Background & Summary
Hanwoo is a native Korean taurine cattle breed with a 5000-year history as a draft animal for farming and transportation1. In a short period, Hanwoo underwent significant changes in its demographic history and selection. During the Korean war (1950–1953), the number of Hanwoo dropped to about 390,000, but recovered to 1.02 million by the late 1950s. With the development of the South Korean economy and agricultural industry, Hanwoo transitioned from a draft to a meat production breed in the 1960s. Modern breeding programs, including performance tests, artificial insemination and genomic selection were initiated by the South Korean government in the 1980s. These programs have improved carcass weight and meat quality of Hanwoo by increasing intramuscular fat (marbling). As a result of continuous artificial selection, Hanwoo has gained unique features both in genome and traits.
This study presents a high-quality assembly of Hanwoo which is the first chromosome-level genome assembly of Asian Bos taurus taurus using a combination of PacBio Hifi, Isoform and Illumina RNA sequencing, with scaffold N50 length of 89 Mb. The completeness of the genome was confirmed by the BUSCO score of 95.8%. The top 31 scaffolds are all greater than 17 Mb in size with a total length of 2.69 Gb. 48.7% of the Hanwoo genome is composed of various repetitive elements. The genome was annotated to contain 27,314 protein-coding genes, including 25,302 proteins with inferred gene names and 2,012 unknown proteins.
We generated a pangenome graph of 14 high-quality Bos taurus autosomes including high-quality genome assemblies of Hanwoo, Hereford, Angus, Brown Swiss, Highland, Holstein, Jersey, Original Braunvieh, Piedmontese, Simmental, Brahman, Nellore, N’Dama, and Ankole. We identified non-reference regions and breed-specific regions through the pangenome graph. In Hanwoo, 528.47 Mb of total non-reference nodes and 61.87 Mb of Hanwoo-specific nodes were identified. This pangenome graph would be used to extract structural variations and make insightful observations among various populations of Bos taurus.
Methods
Sample collection and extraction of genomic DNA and RNA
The samples used in the study of Hanwoo genome included blood, sirloin, liver, and subcutaneous fat from a steer named “bull 2050”. The samples were collected from the Experimental farm of College of Agriculture and Life Sciences at Seoul National University, Pyeongchang-gun, Gangwon-do, Republic of South Korea (Fig. 1) and were approved by the Seoul National University Institutional Animal Care and Use Committee (SNU-201129-1-1). It was castrated in 9.4 months of age, slaughtered and sampled in 32 months of age. All blood sampling was carried out by trained veterinarians, according to the approved institutional protocols. Genomic DNA were extracted from whole blood using Wizard Genomic DNA Purification kit following the manufacturer’s protocol.
Fig. 1.
Picture of Hanwoo steer used in this study and a circos plot. Shown from the outer to inner circle are the following: gene density, with the intensity of color representing the number of genes in a 10,000 bp window; N (unknown base) ratio, with the height of the bar representing the percentage of bases that are N in a 1,000,000 bp window and the overall height of the track representing from the minimum to maximum value for the whole genome which are from 0% to 0.02%, respectively; GC content, with the height of the bar representing the percentage of GC in a 10,000 bp window and the overall height of the track representing from the minimum to maximum value for the whole genome which are from 27.07% to 74.80%, respectively; and the corresponding chromosome.
Sirloin, liver and subcutaneous fat tissues of Hanwoo bull 2050 were collected immediately after slaughter and frozen using liquid nitrogen and stored in a deep freezer until RNA extraction. RNA was isolated using the RNeasy kits (Qiagen, Valencia, CA) following the manufacturer’s protocol.
DNA library construction and sequencing
DNA sequencing libraries were prepared using SMRTbell Express Template Prep kit 2.0 (Pacific Biosciences, California, USA) and libraries larger than 20 kb were used for next steps. HiFi reads were sequenced using 2 SMRT cells of 8 M Tray, Sequel II Sequencing Kit 2.0 in Pacific Biosciences (PacBio) Sequel IIe platform at NICEM in Seoul National University. Highly accurate consensus sequences were produced by PacBio CCS workflow (v 6.3.0), yielding a total of 3.5 M reads and 67.5Gbp corresponding to a genomic coverage of ~24.8X (Table 1).
Table 1.
Statistics of sequencing data.
| Platform | Tissue | Reads | Total bases (bp) | Average length (bp) | N50 length (bp) | SRA accession |
|---|---|---|---|---|---|---|
| PacBio | Blood | 3,520,375 | 67,520,132,790 | 19180 | 20224 | SRR23238456 |
| RNA-seq | Liver | 37986259 | 5773911368 | 76 | 76 | SRR23238454 |
| Subcutaneous fat | 37619668 | 5718189536 | 76 | 76 | SRR23238453 | |
| Sirloin | 40572880 | 6167077760 | 76 | 76 | SRR23238455 | |
| Iso-Seq | Sirloin | 10,054,509 | 20,639,745,850 | 2,052 | 2,268 | SRR23238452 |
RNA library construction and sequencing
For RNA-seq, paired-end libraries with insert size of 75 bp were prepared with TruSeq Stranded mRNA Sample Preparation kit (Illumina, San Diego CA USA) from total messenger RNA (mRNA) of sirloin, liver and subcutaneous fat tissues of a Hanwoo bull 2050. RNA of the three tissues were sequenced separately using Illumina NextSeq 500 with following adapters; liver: D701, D506; sirloin: D701, D507; subcutaneous fat: D701, D508. 17.65 Gb of short paired-end RNA reads were sequenced using Illumina NextSeq 500 (Table 1).
For Iso-Seq, a total of 600 ng RNA from sirloin was used for full-length transcript sequencing with Pacbio Sequel system (Pacific Biosciences, CA, USA) according to the manufacturer’s instructions. The Iso-Seq library was prepared according to the Isoform Sequencing (Iso-Seq) protocol using the NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module, PacBio SMRTbell Express Template Prep Kit 2.0 and ProNex® Size-Selective Purification System.
Total 10 μL library was prepared using PacBio SMRTbell Express Template Prep Kit 2.0. SMRTbell templates were annealed using Sequel Binding and Internal Ctrl Kit 3.0. The Sequel Sequencing Kit 3.0 and SMRT cells 1 M v3 LR Tray was used for sequencing. SMRT cells (Pacific Biosciences) using 1200 min movies were captured for each SMRT cell using the PacBio Sequel System (Pacific Biosciences).
Genome size estimation and contig assembly
Hanwoo contigs were assembled using the HiFi consensus reads and validated following the VGP (Vertebrate Genomes Project) assembly pipeline2. Adapter sequences of HiFi reads (5′–ATCTCTCTCTTTTCCTCCTCCTCCGTTGTTGTTGTTGAGAGAGAT–3′) were removed by Cutadapt (v 4.0)3. Counting k-mer and generating histogram of the k-mer count were performed on adapter trimmed sequences with k = 21 by Meryl (v 1.3.0)4. Genome properties such as genome size, maximum read depth and transition parameter were inferred using GenomeScope (v 2.0)5 from the 21-mer histogram generated by Meryl (v 1.3.0)4. Genome size of Hanwoo was estimated as 3.06 Gb based on the k-mer histogram (Fig. 2). Trimmed reads were assembled to contig level using Hifiasm (v 0.16.1)6, and the draft contig assembly consisted of 1311 contigs totaling 3.28 Gb with an N50 of 55.23 Mb (Table 2). Haplotypic duplication and low-coverage contigs of the draft contig assembly were removed using Purge_dups (v 1.2.5)7 after self-alignment using Minimap28. The primary contig assembly after removing haplotypic duplication included 603 contigs, with a size of 3.11 Gb and a contig N50 of 58.14 Mb.
Fig. 2.

Genome size estimation by GenomeScope2.
Table 2.
Statistics of contig assembly before scaffolding.
| Statistics without reference | Draft primary contig assembly | Draft alternate contig assembly | Purged primary contig assembly | Purged alternate contig assembly |
|---|---|---|---|---|
| Number of contigs | 1311 | 12491 | 603 | 8339 |
| Largest contig | 154270589 | 4548020 | 154270589 | 4727895 |
| Total length | 3278632171 | 2786845933 | 3108034269 | 2585754757 |
| N50 | 55230564 | 664031 | 58141786 | 746831 |
| N75 | 15456913 | 255609 | 23975184 | 356899 |
| L50 | 19 | 1163 | 18 | 998 |
| L75 | 44 | 2817 | 37 | 2240 |
| GC (%) | 43.93 | 43.81 | 43.44 | 43.13 |
Scaffolding and gap filling
The Hanwoo contigs after removing haplotypic duplication were scaffolded on autosome of ARS-UCD1.3, through reference-guided approach by RagTag (v 2.1.0)9. Because the Y chromosome is absent in ARS-UCD1.3, autosome and X chromosome of ARS-UCD1.3, and Y chromosome of UOA_Angus_1 were used as reference genome for scaffolding. The reference-guided scaffolding using RagTag (v 2.1.0)9 consist of ‘correct’ and ‘scaffold’ steps. The ‘correct’ step identified and corrected potential misassembly based on alignment of contig assembly to the reference genome assembly. Part of contigs were broken at points of putative misassembly, and as a result, the number of contigs increased to 1915. In the ‘scaffold’ step, these RagTag ‘corrected’ contigs were aligned to the reference genome consist of autosome and X chromosome of ARS-UCD1.3, and Y chromosome of UOA_Angus_1. As a result, there were 1598 scaffolds including 31 chromosome-level scaffolds and 1567 unplaced scaffolds.
HiFi reads used in the Hanwoo assembly were aligned using Minimap28 to perform gap filling of the chromosome-level Hanwoo genome assembly using TGS-GapCloser (v 1.0.1)10. The final 31 chromosome-level scaffolds had a total size of 2.69 Gb, which was similar to chromosome size of ARS-UCD 1.3. (Tables 3, 4). These 31 chromosome-level scaffolds composed 86.66% of the assembly, with the remaining 414.6 Mb still unanchored and requiring further investigation. Further analysis including annotation and pangenome were performed on the chromosome-level scaffolds.
Table 3.
Hanwoo genome assembly statistics.
| Assembly statistics | Value |
|---|---|
| Genome size (bp) | 3108492884 |
| Number of scaffolds | 1598 |
| Number of chromosome-scale scaffolds | 31 |
| N50 of scaffolds (bp) | 89243566 |
| L50 of scaffolds | 13 |
| Chromosome-scale scaffolds (bp) | 2693904935 |
| GC content of the genome (%) | 43.44 |
| QV score | 63.68 |
| Error rate | 4.29E-07 |
| BUSCO analysis | |
| Library | mammalia_odb10 |
| Complete | 8842 (95.8%) |
| Complete and single copy | 8664 (93.9%) |
| Complete and duplicated | 178 (1.9%) |
| Fragmented | 106 (1.1%) |
| Missing | 278 (3.1%) |
Table 4.
Length of Chromosome-level scaffolds.
| Chromosome | Length | % of assembly |
|---|---|---|
| 1 | 158347075 | 5.88 |
| 2 | 140532406 | 5.22 |
| 3 | 121557778 | 4.51 |
| 4 | 122787172 | 4.56 |
| 5 | 121175501 | 4.50 |
| 6 | 120343135 | 4.47 |
| 7 | 111272195 | 4.13 |
| 8 | 114613683 | 4.25 |
| 9 | 105990968 | 3.93 |
| 10 | 104650420 | 3.88 |
| 11 | 107792557 | 4.00 |
| 12 | 89243566 | 3.31 |
| 13 | 85553472 | 3.18 |
| 14 | 83497117 | 3.10 |
| 15 | 85308379 | 3.17 |
| 16 | 88665756 | 3.29 |
| 17 | 73790049 | 2.74 |
| 18 | 68766244 | 2.55 |
| 19 | 65427893 | 2.43 |
| 20 | 71637878 | 2.66 |
| 21 | 78435670 | 2.91 |
| 22 | 61025439 | 2.27 |
| 23 | 53933626 | 2.00 |
| 24 | 63313671 | 2.35 |
| 25 | 42768661 | 1.59 |
| 26 | 53441352 | 1.98 |
| 27 | 46802419 | 1.74 |
| 28 | 46008137 | 1.71 |
| 29 | 52029189 | 1.93 |
| X | 137682877 | 5.11 |
| Y | 17510650 | 0.65 |
| Total | 2693904935 | 100.00 |
Circos plot denoting gene density, N ratio and GC content was generated with the advanced circos function from Java-based tool TBtools11. The gene density (number of genes), N ratio (%) and GC content (%) was calculated for every 10,000 bp increment of the genome and was visualized in a heatmap format for gene density and histogram format for N ratio and GC content using BIN size 100,000.
Masking repetitive sequences
Repetitive sequences in the gap-filled Hanwoo assembly were soft-masked using RepeatMasker (v 4.1.5)12 with a known library (cow) in Dfam (v 3.7) and RepBase (v 10/26/2018) using RMBlast. Repetitive elements predicted by RepeatMasker contained 1.31 Gb of sequences, accounting for 48.7% of the genome, including 27.6%, 11.6%, 4.9%, 2.1% and 1.5% for LINEs, SINEs, LTR elements, DNA elements, and satellite repeats, respectively (Table 5).
Table 5.
Statistics of repetitive elements.
| Class | Subclass | Number | Total length (bp) | % of genome |
|---|---|---|---|---|
| SINEs: | 2083225 | 312596265 | 11.6 | |
| MIRs | 399931 | 57592626 | 2.14 | |
| LINEs: | 1318367 | 742600414 | 27.57 | |
| LINE1 | 584926 | 340390426 | 12.64 | |
| LINE2 | 255007 | 65643056 | 2.44 | |
| L3/CR1 | 34731 | 7189988 | 0.27 | |
| RTE | 442538 | 329203364 | 12.22 | |
| LTR elements: | 415490 | 131192082 | 4.87 | |
| ERVL | 75217 | 29646401 | 1.1 | |
| ERVL-MaLRs | 121580 | 39874562 | 1.48 | |
| ERV_classI | 84207 | 37072606 | 1.38 | |
| ERV_classII | 117558 | 20606823 | 0.76 | |
| DNA elements: | 289836 | 57547635 | 2.14 | |
| hAT-Charlie | 163969 | 30537889 | 1.13 | |
| TcMar-Tigger | 45005 | 11907379 | 0.44 | |
| Unclassified: | 3023 | 464793 | 0.02 | |
| Total interspersed repeats: | 1244401189 | 46.19 | ||
| Small RNA: | 254380 | 43115368 | 1.6 | |
| Satellites: | 6216 | 39399744 | 1.46 | |
| Simple repeats: | 537045 | 22458650 | 0.83 | |
| Low complexity: | 81860 | 4022678 | 0.15 | |
| Total bases masked: | 1311158349 | 48.67 | ||
Genome annotation
Illumina RNA-seq reads were trimmed to remove adapter sequences and low-quality bases using Trimmomatic (v 0.39)13. The BRAKER3 (v 3.0.3) pipeline was used for structural annotation of Hanwoo genome. The pipeline utilized three sources of extrinsic evidence; short-read RNA-seq (Illumina), protein sequences of Vertebrata in OrthoDB (v 11)14 in addition to protein sequence of ARS-UCD1.3 to train Augustus (v 3.5.0)15 for gene prediction.
The predicted gene sets were searched in 2 public functional databases, Swiss-Prot of UniProtKB16 and Pfam (v 35.0) database17 to identify the potential function with BLASTP (v 2.13.0+)18 and functional domains with InterProScan (v 5.57)19. We used scripts included in MAKER (v 3.01.03)20 to integrate functional annotations into structural annotations. The protein annotation was evaluated by analyzing amino acid sequences of protein using BUSCO (v 5.3.2)21 with the conserved core set of mammalian genes, yielding a completeness score of 87.9%. A total of 27,314 protein-coding genes were identified, including 25,302 genes with inferred names and 2,012 unknown proteins.
Assessment of the chromosome-level genome assembly
N50, L50 and lengths of the chromosome-level Hanwoo genome assembly was calculated by QUAST (v 5.0.2)22. Single copy gene completeness was assessed with BUSCO (v 5.3.2)21, using the metaeuk backend against ‘mammalia_odb10’. Quality values (QV) was calculated with Merqury (v 1.3)23, with k-mer databases (k = 21) constructed by Meryl (v 1.3)4.
Pangenome graph construction
The pangenome graph of 14 Bos taurus genomes, including the Hanwoo assembly, was generated using the Minigraph-Cactus Pangenome Pipeline (v 2.5.2)24. 14 assemblies were collected with the Hereford assembly, ARS-UCD1.325, as the reference genome. 8 haplotype-resolved assemblies of Angus (UOA_Angus_1, GCF_002263795.3), Brahman (UOA_Brahman_1)26, Simmental (ARS-Simm1.0)27, Scottish Highland bull (ARS_UNL_Btau-highland_paternal_1.0_alt, GCA_009493655.1)28, N’Dama (ROSLIN_BTT_NDA1), Ankole (ROSLIN_BTI_ANK1)29, Jersey (ARS-LIC_NZ_Jersey, GCA_021234555.1), Holstein Friesian (ARS-LIC_NZ_Holstein-Friesian_1, GCA_021347905.1) were obtained from NCBI. Original Braunvieh30, Nellore, Brown Swiss, and Piedmontese were collected from the public database (10.5281/ZENODO.5906579) and scaffolded and merged by RagTag9 following the protocol of previous article31. The repeat sequences in the genomes of Original Braunvieh, Nellore, Brown Swiss, Piedmontese and Highland were soft-masked for by RepeatMasker (v 4.1.5)12 using same parameters and repeat databases with Hanwoo. Because one sex chromosome was missing in haplotype-resolved genomes produced by trio-binning assembly, only autosomes were included in our pangenome graph.
The Minigraph-Cactus Pangenome Pipeline consisted of four steps: constructing the Minigraph GFA, mapping the genomes back to the Minigraph, creating the Cactus alignment and creating the VG indexes. The Minigraph graph was created using ARS-UCD1.3 as the reference genome, and the other 13 genomes were iteratively added. Base-level alignments of the genomes were added to the graph using Cactus24. After embedding the haplotypes into the graph, Cactus alignment were performed, resulting in variation graph (VG) and hierarchical alignment (HAL). The HAL file was converted to packed graph (PG) and chopped into 32 base pairs using ‘hal2vg’ to describe it as nodes and edges.
Non-reference nodes in pangenome graph
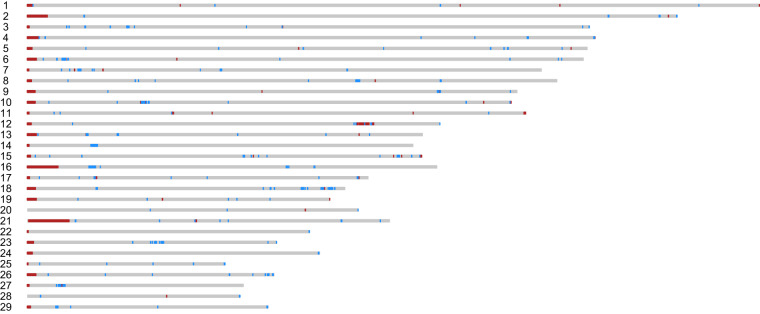
The multiple whole-genome alignments generated by CACTUS24 were transformed into the Packed Graph (PG) format by chopping into 32 base pairs using ‘hal2vg’ with the options ‘—chop 32’ and ‘—noAncestors’32. The reference nodes and non-reference nodes were separated using scripts from the Github repository (https://github.com/evotools/CattleGraphGenomePaper/tree/master/detectSequences/nf-GraphSeq) following previous research29. After excluding nodes flanking with gaps in 1 kb, the counts and lengths of the non-reference and breed-specific nodes were calculated (Table 6). Non-reference region and Hanwoo-specific regions longer and equal to 10 kb are marked in Hanwoo autosome using KaryoploteR33 (Fig. 3). The Hanwoo-specific regions are encompassed within the non-reference region, with the majority of these regions being located in the telomeric and centromeric regions. Notably, the size of satellite repeats, as identified by RepeatMasker, amounted to 39.4 Mb (Table 5). The total size of the satellite repeat, a main component of the centromere, were similar to the differences in autosome length between Hanwoo and others. This finding implies that the larger genome and specific region of Hanwoo can be attributed to expansions within repeat-rich telomeric and centromeric regions.
Table 6.
Sequence contribution of 14 bos taurus autosomes.
| Breed | Non-reference nodes | Specific nodes | Total length (autosome) | ||
|---|---|---|---|---|---|
| nodes | bp | nodes | bp | bp | |
| Hanwoo | 5644829 | 83917034 | 622052 | 61869953 | 2538711408 |
| Angus | 4876028 | 40793146 | 331609 | 23589072 | 2468157877 |
| Brown Swiss | 5135844 | 25626114 | 364958 | 8631263 | 2497220059 |
| Highland | 4917533 | 32014564 | 383674 | 14515221 | 2483452092 |
| Holstein | 5046695 | 31095517 | 434031 | 16204587 | 2468170459 |
| Jersey | 5050922 | 27795391 | 402709 | 11095169 | 2473656513 |
| Original Braunvieh | 5135877 | 27234395 | 361737 | 10537892 | 2503654516 |
| Piedmontese | 5128788 | 28520430 | 389915 | 11411557 | 2500499917 |
| Simmental | 5266669 | 40554393 | 527318 | 20773580 | 2494093306 |
| Brahman | 11480493 | 46633118 | 2650315 | 20140251 | 2478073158 |
| Nellore | 12648594 | 45129061 | 3423881 | 19092260 | 2502536439 |
| N’Dama | 7225426 | 54175845 | 1375922 | 35064951 | 2504036093 |
| Ankole | 8960222 | 44980693 | 1959559 | 23916971 | 2485084605 |
| Hereford | 2489385779 | ||||
Fig. 3.
Non-reference region and specific region in Hanwoo autosome. Non-reference regions and Hanwoo-specific regions larger than or equal to 10 kb are visualized on Hanwoo autosomes. The Hanwoo-specific regions are marked in red, while the non-reference regions shared by other Bos taurus assemblies, excluding the Hanwoo-specific regions, are marked in blue.
Furthermore, HiFi-based assemblies generally have higher telomeric completeness than Oxford nanopore- or CLR-based assemblies34. The uniqueness of origin and evolution history also supported the larger and disctinct genome of Hanwoo compared to European taurine. Mitochondrial DNA haplogroup of Hanwoo is P, which is common in European aurochs but has not been detected in modern cattle in Europe35. The haplogroup P mtDNA in Hanwoo suggested the possibility of a minor and local event of domestication or introgression of Asian aurochs36,37. Furthermore, intensive inbreeding and small effective population size of Hanwoo might facilitate fixation of these distinctive regions in Hanwoo genome38.
Data Records
The final genome assembly was deposited at DDBJ/ENA/GenBank under the accession JARDUZ00000000039.
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession SRR2323845640.
The transcriptomic Illumina sequencing data of subcutaneous fat, liver and sirloin were deposited in the SRA at NCBI SRR23238453, SRR23238454 and SRR23238455, respectively40.
The transcriptomic PacBio sequencing data of sirloin were deposited in the SRA at NCBI SRR2323845240.
The Hanwoo genome assembly which were not processed by NCBI, genome annotation, transcript sequence and protein sequence are available in figshare41.
The pangenome graph in GFA format are also available in figshare42.
Technical Validation
RNA degradation and contamination were monitored on Agilent RNA ScreenTape. The purity of RNA samples was checked using the NanoPhotometer spectrophotometer (IMPLEN, CA, USA). The integrity of RNA was assessed using the RNA ScreenTape of the Agilent 2200 TapeStation System (Agilent Technologies, CA, USA). Only RNAs with an OD260/280 ratio of 2.0–2.2, an OD260/230 ratio of 1.8–2.1, and a RIN value of ≥9.0 were considered qualified for use. RNA concentration was measured using Quant-iT™ RiboGreen™ RNA Assay Kit in Victor Nivo (PerkinElmer, Waltham, MA, USA).
The completeness of the Hanwoo genome assembly was evaluated using BUSCO21 with the mammalian data set “mammalia_odb10.” The evaluation found 95.8% (8842) of the core mammalian genes were present in the genome, including 93.9% single-copy, 1.9% duplicated, 1.9% fragmental, and 3.1% missing genes from the mammalian data set (Table 3). The k-mer databases (k = 21) constructed using HiFi reads by Meryl4, and the overall assembly quality was assessed using the k-mer databases using Merqury23. The assembly showed high quality values (QV > 63) with an error rate of 4.29 × 10−7 (Table 3). The GC content of Hanwoo (43.44%) was slightly higher than that of ARS-UCD1.3 (41.56%). These assessment results confirmed the completeness of Hanwoo genome assembly (Table 3).
Acknowledgements
This work was supported by the National Research Foundation of Korea(NRF) grant funded by the Korea government(MSIT) (No. NRF-2021R1A2C2094111).
Author contributions
H.K. conceived of the project. S.L., M.B. collected the samples and extracted the genomic DNA and RNA. J.Jang performed the data analysis and wrote the manuscript. J.Jung and Y.L. contributed to the data analyses and visualization. Y.L. revised the manuscript. All authors read and approved the final version of the manuscript.
Code availability
Parameters for all commands used to assemble the genome and construct the pangenome are available in figshare43.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee S-H, et al. Hanwoo cattle: origin, domestication, breeding strategies and genomic selection. Journal of animal science and technology. 2014;56:1–8. doi: 10.1186/2055-0391-56-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lariviere, D. et al. VGP assembly pipeline. (2022).
- 3.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 4.Meryl (GitHub, GitHub repository, 2020).
- 5.Ranallo-Benavidez, T., Jaron, K. & Schatz, M. (Nature Publishing Group, 2020). [DOI] [PMC free article] [PubMed]
- 6.Cheng H, Concepcion GT, Feng X, Zhang H, Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature methods. 2021;18:170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan D, et al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36:2896–2898. doi: 10.1093/bioinformatics/btaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alonge, M. et al. Automated assembly scaffolding elevates a new tomato system for high-throughput genome editing. BioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 10.Xu M, et al. TGS-GapCloser: a fast and accurate gap closer for large genomes with low coverage of error-prone long reads. GigaScience. 2020;9:giaa094. doi: 10.1093/gigascience/giaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 12.Chen N. Using Repeat Masker to identify repetitive elements in genomic sequences. Current protocols in bioinformatics. 2004;5:4.10. 11–14.10. 14. doi: 10.1002/0471250953.bi0410s05. [DOI] [PubMed] [Google Scholar]
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuznetsov, D. et al. OrthoDB v11: annotation of orthologs in the widest sampling of organismal diversity. Nucleic Acids Research (2022). [DOI] [PMC free article] [PubMed]
- 15.Stanke M, et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic acids research. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic acids research. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mistry J, et al. Pfam: The protein families database in 2021. Nucleic acids research. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camacho C, et al. BLAST+: architecture and applications. BMC bioinformatics. 2009;10:1–9. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell MS, Holt C, Moore B, Yandell M. Genome annotation and curation using MAKER and MAKER‐P. Current protocols in bioinformatics. 2014;48:4.11. 11–14.11. 39. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 22.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhie A, Walenz BP, Koren S, Phillippy AM. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome biology. 2020;21:1–27. doi: 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armstrong J, et al. Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature. 2020;587:246–251. doi: 10.1038/s41586-020-2871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosen BD, et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience. 2020;9:giaa021. doi: 10.1093/gigascience/giaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koren S, et al. De novo assembly of haplotype-resolved genomes with trio binning. Nature biotechnology. 2018;36:1174–1182. doi: 10.1038/nbt.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heaton MP, et al. A reference genome assembly of Simmental cattle, Bos taurus taurus. Journal of Heredity. 2021;112:184–191. doi: 10.1093/jhered/esab002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice ES, et al. Continuous chromosome-scale haplotypes assembled from a single interspecies F1 hybrid of yak and cattle. GigaScience. 2020;9:giaa029. doi: 10.1093/gigascience/giaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talenti A, et al. A cattle graph genome incorporating global breed diversity. Nature communications. 2022;13:1–14. doi: 10.1038/s41467-022-28605-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crysnanto D, Leonard AS, Fang Z-H, Pausch H. Novel functional sequences uncovered through a bovine multiassembly graph. Proceedings of the National Academy of Sciences. 2021;118:e2101056118. doi: 10.1073/pnas.2101056118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonard AS, et al. Structural variant-based pangenome construction has low sensitivity to variability of haplotype-resolved bovine assemblies. Nature Communications. 2022;13:1–13. doi: 10.1038/s41467-022-30680-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickey G, Paten B, Earl D, Zerbino D, Haussler D. HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics. 2013;29:1341–1342. doi: 10.1093/bioinformatics/btt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gel B, Serra E. karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics. 2017;33:3088–3090. doi: 10.1093/bioinformatics/btx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leonard AS, Crysnanto D, Mapel XM, Bhati M, Pausch H. Graph construction method impacts variation representation and analyses in a bovine super-pangenome. Genome Biology. 2023;24:124. doi: 10.1186/s13059-023-02969-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Achilli A, et al. Mitochondrial genomes of extinct aurochs survive in domestic cattle. Current Biology. 2008;18:R157–R158. doi: 10.1016/j.cub.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 36.Noda A, Yonesaka R, Sasazaki S, Mannen H. The mtDNA haplogroup P of modern Asian cattle: A genetic legacy of Asian aurochs? PLoS One. 2018;13:e0190937. doi: 10.1371/journal.pone.0190937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mannen H, et al. Cattle mitogenome variation reveals a post-glacial expansion of haplogroup P and an early incorporation into northeast Asian domestic herds. Scientific Reports. 2020;10:20842. doi: 10.1038/s41598-020-78040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, Y. & Kim, J.-J. Effective population size and signatures of selection using bovine 50K SNP chips in Korean native cattle (Hanwoo). Evolutionary Bioinformatics11, EBO. S24359 (2015). [DOI] [PMC free article] [PubMed]
- 39.Jang J, 2023. Bos taurus breed Hanwoo isolate HWB-2050, whole genome shotgun sequencing project. GenBank. JARDUZ000000000
- 40.2023. NCBI Sequence Read Archive. SRP419181
- 41.Jang J. 2023. Hanwoo Genome Assembly (Bos taurus) figshare. [DOI]
- 42.Jang J. 2023. Bos taurus pangenome graph. figshare. [DOI]
- 43.Jang J. 2023. Parameters for all commands used to assemble the Hanwoo genome and construct Bos taurus pangenome. figshare. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Jang J, 2023. Bos taurus breed Hanwoo isolate HWB-2050, whole genome shotgun sequencing project. GenBank. JARDUZ000000000
- 2023. NCBI Sequence Read Archive. SRP419181
- Jang J. 2023. Hanwoo Genome Assembly (Bos taurus) figshare. [DOI]
- Jang J. 2023. Bos taurus pangenome graph. figshare. [DOI]
- Jang J. 2023. Parameters for all commands used to assemble the Hanwoo genome and construct Bos taurus pangenome. figshare. [DOI]
Data Availability Statement
Parameters for all commands used to assemble the genome and construct the pangenome are available in figshare43.