Abstract
Retroviral Gag proteins, in the absence of any other viral products, induce budding and release of spherical, virus-like particles from the plasma membrane. Gag-produced particles, like those of authentic retrovirions, are not uniform in diameter but nevertheless fall within a fairly narrow distribution of sizes. For the human immunodeficiency virus type 1 (HIV-1) Gag protein, we recently reported that elements important for controlling particle size are contained within the C-terminal region of Gag, especially within the p6 sequence (L. Garnier, L. Ratner, B. Rovinski, S.-X. Cao, and J. W. Wills, J. Virol. 72:4667–4677, 1998). Deletions and substitutions throughout this sequence result in the release of very large particles. Because the size determinant could not be mapped to any one of the previously defined functions within p6, it seemed likely that its activity requires the overall proper folding of this region of Gag. This left open the possibility of the size determinant residing in a subdomain of p6, and in this study, we examined whether the late domain (the region of Gag that is critical for the virus-cell separation step) is involved in controlling particle size. We found that particles of normal size are produced when p6 is replaced with the totally unrelated late domain sequences from Rous sarcoma virus (contained in its p2b sequence) or equine infectious anemia virus (contained in p9). In addition, we found that the large particles released in the absence of p6 require the entire CA and adjacent spacer peptide sequences, whereas these internal sequences of HIV-1 Gag are not needed for budding (or proper size) when a late domain is present. Thus, it appears the requirements for budding are very different in the presence and absence of p6.
The major structural proteins of human immunodeficiency virus type 1 (HIV-1) are initially synthesized in the form of a polyprotein precursor, Pr55gag (Fig. 1). The Gag polyproteins assemble at the plasma membrane in a process that leads to the release of spherical, enveloped, and immature virions. A number of studies have demonstrated that Pr55gag is the only viral protein essential for assembly and release of viral particles (10, 17, 18, 33).
FIG. 1.
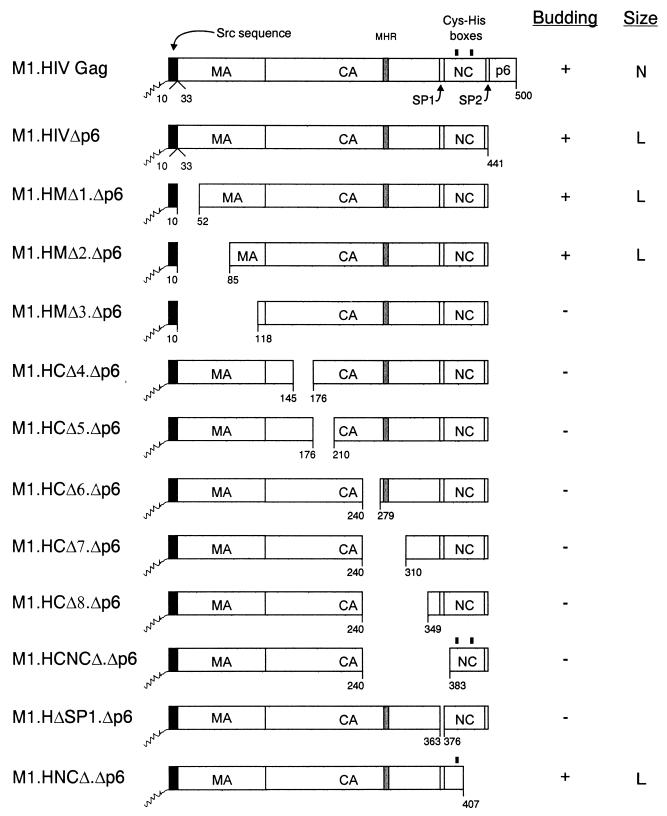
Combinations of internal Gag deletions with a terminal p6 deletion. The names of the Gag cleavage products (MA, CA, SP1, NC, SP2, and p6) are indicated. The shaded region within CA marks the MHR. The black box at the N termini of the constructs represents the first 10 amino acids from pp60v-src. The squiggle depicts the fatty acid myristate. The black rectangles above the NC sequence mark the cysteine-histidine boxes. Numbers below the Gag molecules refer to amino acid residues. The properties of the Gag mutants with regard to particle release and size distribution are summarized in columns at the right of the figure: N, homogeneous particles of normal size; L, homogeneous particles of large size.
Very late during budding or immediately after, cleavage of Pr55gag by the virally-encoded protease releases the mature products p17 (MA [matrix]), p24 (CA [capsid]), p7 (NC [nucleocapsid]), and the C-terminal peptide p6, as well as two small peptides, SP2 (spacer peptide 2) and SP1 (12, 16). However, it is the uncleaved Gag protein that directs the assembly and budding events (for a review, see reference 9). It is now clear that the MA sequence contains the M (membrane binding) domain, which specifically directs Gag proteins to the membrane. Within the NC sequence, two copies of the I (interaction) domain mediate tight interactions between Gag molecules at the membrane to give particles their proper density. Finally, the separation of the particle from the cell surface is mediated by the L (late) domain, which resides within the p6 sequence. Although the M, I, and L domains have been shown to be sufficient for the release of virus particles of normal density (for a review, see reference 9), they are insufficient for the production of normal-sized particles.
A recent study of Rous sarcoma virus (RSV [20]) has demonstrated that the size determinants map to the segment of Gag consisting of CA plus the spacer peptides located between CA and NC. Small deletions throughout CA-SP result in particles that are large and heterogeneous (20). In the case of HIV-1, the arrangement of size determinants is very different, and although the spacer peptide (SP1) following CA may be an important size determinant, the CA sequence appears to be far less critical (8, 19). Thus, while some CA mutants have been reported to have altered sizes (3, 6), many others have been shown to have normal size, although infectivity is lost (31). By employing rate-zonal gradients to systematically study the effect of deletions throughout the Gag protein, we recently reported that the HIV-1 CA sequence does not control particle size during budding. Rather, the C-terminal sequences of Gag, and especially p6, are critical for determining HIV-1 particle diameter (8). Thus, Gag proteins with deletions within MA, CA, or the N-terminal part of NC produce particles of normal size, while those with defects in the p6 sequence or the C-terminal part of NC release very large particles, even though proper buoyant density was retained. The importance of p6 is further emphasized by its ability to restore normal size to RSV CA mutants that otherwise produce large and heterogeneous particles in its absence (8).
How the p6 sequence functions to constrain the size of an emerging particle is unknown. In our previous study, the size determinant could not be mapped to motifs within p6 having known functions, including the minimal sequence that defines the L domain (8); rather, the folded structure of the entire p6 seemed to be important. Thus, it was not possible to rule out a role for the L domain in controlling particle size. To further examine this possibility, the RSV p2b and equine infectious anemia virus (EIAV) p9 sequences and their associated L domains were moved to the C terminus of HIV-1 Gag in place of p6. In both cases, the presence of the heterologous L domain was sufficient to restore normal particle size as measured by sedimentation experiments. These findings demonstrate that the L domains from three unrelated Gag proteins can function independently as determinants of HIV-1 particle size. Further evidence for this was obtained from experiments with EIAV, which showed that the C-terminal sequences of this lentiviral Gag protein, like those of HIV-1, are critical for determining particle size and can be functionally replaced with the L domains of RSV and HIV. We also uncovered evidence that the HIV-1 Gag protein utilizes a different mechanism for budding when p6 is absent, a pathway that requires the presence of an intact CA-SP1 sequence.
MATERIALS AND METHODS
All of the constructs used in this study lack viral protease activity (2, 5, 38). All DNA manipulations were carried out by using standard methods (34). Recombinant plasmids were propagated in Escherichia coli DH-5α with YT medium containing ampicillin (25 μg/ml). Each construct was sequenced, and two independent clones of each mutant were analyzed in transfection experiments to minimize possible unwanted mutations.
Construction of p6 deletion mutants.
All gag alleles were expressed by using a simian virus 40 (SV40)-based vector. Constructs pSV.M1.HIV Gag and pSV.M1.HIVΔp6 have been previously reported (8).
To combine previously described MA deletion mutations with a p6 deletion, pSV.M1.HMΔ1, pSV.M2.HMΔ2, and pSV.M1.HMΔ3 (8) were digested with MluI and SpeI, and each of the resulting small fragments was ligated into the MluI-SpeI sites of pSV.M1.HIVΔp6. The recombinants were named pSV.M1.HMΔ1.Δp6, pSV.M2.HMΔ2.Δp6, and pSV.M1.HMΔ3.Δp6, respectively (Fig. 1).
To combine previously described CA, NC, and SP1 deletion mutations with a p6 deletion, pSV.M1.HCΔ4, pSV.M1.HCΔ5, pSV.M1.HCΔ6, pSV.M1.HCΔ7, pSV.M1.HCΔ8, pSV.M1.HCNCΔ, and pSV.M1.HΔSP1 (8) were digested with BglII and BssHII and then ligated with an oligonucleotide pair containing a stop codon inserted in place of the first p6 codon (8). The resulting constructs were named pSV.M1.HCΔ4.Δp6, pSV.M1.HCΔ5.Δp6, pSV.M1.HCΔ6.Δp6, pSV.M1.HCΔ7.Δp6, pSV.M1.HCΔ8.Δp6, pSV.M1.HCNCΔ.Δp6, and pSV.M1.HΔSP1.Δp6, respectively (Fig. 1).
To combine a previously described NC deletion mutation with a p6 deletion, pSV.M1.HIVΔp6 was digested with ApaI and BglII, treated with T4 DNA polymerase, and religated. One foreign residue (Ser) was introduced at the site of the deletion. The resulting construct was named pSV.M1.HNCΔ.Δp6 (Fig. 1).
Construction of HIV-p2 deletion mutants.
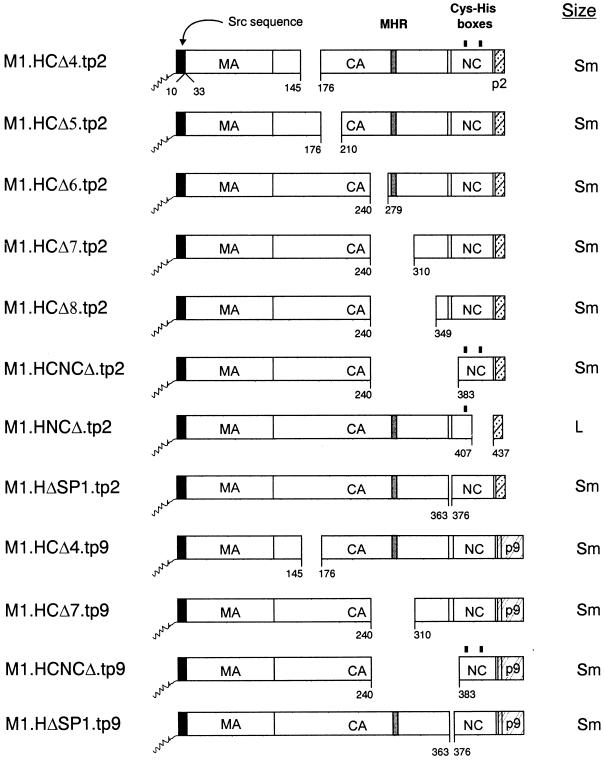
To insert the RSV p2b sequence (and its associated L domain) in the place of the HIV-p6 sequence, pSV.M1.HIV Gag and previously described CA, SP1, and NC deletion mutants (8) were digested with BglII and SpeI and ligated with the fragment containing the p2b sequence, resulting from the digestion of pSV.RHB.T10C.tp2 (25) with BglII-SpeI. The resulting constructs were named pSV.M1.HG.tp2, pSV.M1.HCΔ4.tp2, pSV.M1.HCΔ5.tp2, pSV.M1.HCΔ6.tp2, pSV.M1.HCΔ7.tp2, pSV.M1.HCΔ8.tp2, pSV.M1.HCNCΔ.tp2, and pSV.M1.HΔSP1.tp2 (Fig. 4).
FIG. 4.
Deletion of internal Gag sequences from the L domain chimeras. The names of the Gag cleavage products (MA, CA, NC, p6, p2, and p9) are indicated. The black box at the N termini of the constructs represents the first 10 amino acids from pp60v-src, and the squiggle represents the fatty acid myristate. The black rectangles above the NC sequence mark the cysteine-histidine boxes. Numbers refer to amino acid residues. The column at the right of the figure summarizes the size distribution of the mutants: Sm, particles that are uniform in size but smaller in diameter; L, homogeneous particles of large size.
To insert the RSV p2b sequence in the place of the HIV-1 p6 sequence in the NC deletion mutant M1.HNCΔ (8), pSV.M1.HG.tp2 was digested with ApaI and BglII, treated with T4 DNA polymerase, and religated. The resulting construct was named pSV.M1.HNCΔ.tp2 (Fig. 4).
Chimeric HIV-EIAV gag alleles.
To place the EIAV p9 sequence (and its associated L domain) in the place of HIV-1 p6, pSV.M1.HIV Gag, pSV.M1.HCΔ4, pSV.M1.HCΔ7, pSV.M1.HCNCΔ, and pSV.M1.HCΔSP1 were digested with ApaI-EcoRV and ligated with the fragment containing the p9 sequence, resulting from the digestion of pSV.RHE.p9 (25) with ApaI and EcoRV. The recombinants were named pSV.M1.HCΔ4.tp9, pSV.M1.HCΔ7.tp9, pSV.M1.HCNCΔ.tp9, and pSV.M1.HΔSP1.tp9 (Fig. 4).
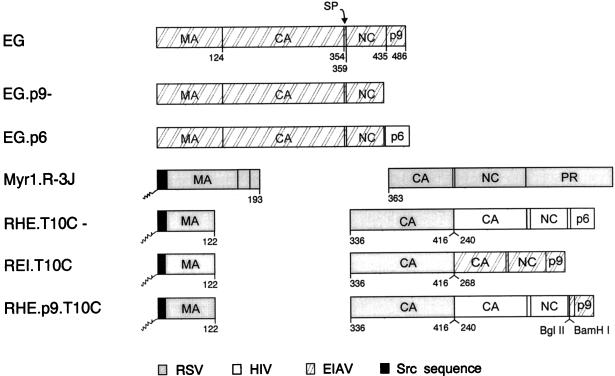
Four other HIV-EIAV chimeras used in this study (Fig. 6, pSV.EG.p6 [28], pSV.RHE.T10C− [25], pSV.REI.T10C [25], and pSV.RHE.p9.T10C [25]) as well as two EIAV constructs (pSV.EG [28] and pSV.EG.p9− [28]) have been previously reported.
FIG. 6.
Wild-type and chimeric forms of the EIAV Gag protein. The Gag protein of EIAV is illustrated at the top with the names of its cleavage products (MA, CA, SP, NC, and p9). Numbers below the Gag molecules refer to amino acid residues. Foreign sequences from RSV, HIV, and Src are indicated.
Construction of M1.HG.CY.
PCR amplification of the iso-1-cytochrome c sequence of pSV.MYCY (36) was performed to create a BglII site immediately upstream of the fourth codon and an BssHII site immediately downstream of the stop codon by using 5′-ATAGGAGGGGAGATCTGGAAGGCCGTTTCTGCTAAGA-3′ as the upstream primer and 5′-ATCCTACAGCGCGCTTACTCACAGGCTTTTT-3′ as the downstream primer. The amplified fragment was digested with BglII and BssHII (restriction endonuclease recognition sites are underlined in the primer sequences) and ligated into plasmid pSV.M1.HIV Gag (8), which had been digested with the same enzymes. The recombinant was named pSV.M1.HG.CY.
Transfection of cells and metabolic labeling.
COS-1 cells were grown in 35-mm- or 60-mm-diameter dishes in Dulbecco’s modified Eagle medium (DMEM; GIBCO BRL) supplemented with 3% fetal bovine serum and 7% bovine calf serum (HyClone, Inc.). These cells were transfected with XbaI-digested and ligated plasmids by the DEAE-dextran-chloroquine method as described previously (38). At 48 h after transfection, the COS-1 cells were labeled with [35S]methionine (10 μCi or 50 μCi; >1,000 Ci/mmol). After 2.5 h of labeling, the cells and growth medium from each labeled culture were mixed with lysis buffer containing protease inhibitors, and the Gag proteins were immunoprecipitated for 1 h at 4°C with a human HIV immunoglobulin (27), electrophoresed in sodium dodecyl sulfate-12% polyacrylamide gels, and visualized by fluorography. The autoradiograms were then quantitated by laser densitometry.
Density gradient analysis.
To ensure that the Gag deletion mutants used in this study produced dense particles, several of the key constructs (M1.HG.tp2, M1.HG.tp9, M1.HCΔ6.tp2, M1.HNCΔ.tp2, and M1.HCΔ7.tp9) were tested by centrifugation of the samples through density gradients as previously described (8). All of them released dense particles (data not shown). Many of the other constructs have been analyzed previously and found to be released into dense particles, including M1.HIV Gag (8), M1.HIV.Δp6 (8), EG.p9− (28), EG (28), EG.p6 (28), REI.T10C (25), and RHE.p9.T10C (25).
Rate-zonal gradient analysis.
Two days posttransfection, COS-1 cells were labeled in methionine-free, serum-free Dulbecco’s medium supplemented with [35S]methionine (50 μCi; >1,000 Ci/mmol) for 5 h in 0.5 ml. After the labeling period, the medium was immediately centrifuged at a low speed to remove cellular debris. Radiolabeled infectious RSV was added to each sample to provide an internal size marker. This virus, obtained from RSV-infected turkey embryo fibroblasts (TEF) which were propagated in supplemented F10 medium as described previously (15), was labeled with [35S]methionine as described above. Similarly, EIAV-infected equine dermal cells (American Type Culture Collection catalog no. CCL 57, kindly provided by Ron Montelaro and Bridget Puffer, University of Pittsburgh, Pittsburgh, Pa.) propagated in DMEM supplemented with 10% fetal bovine serum, were labeled as described above. Each mixture was layered onto 10 to 30% sucrose and centrifuged at 83,500 × g for 0.5 h in a Beckman SW41Ti. From each gradient, 0.6-ml fractions were collected, and Gag proteins were immunoprecipitated with a mixture of anti-RSV and anti-HIV antisera (27) and subjected to electrophoresis. For EIAV constructs, we used a mixture of anti-RSV and anti-EIAV antisera (a kind gift from Ron Montelaro and Bridget Puffer, University of Pittsburgh). All gradients were repeated at least once to confirm the results.
RESULTS
We recently reported that important size-controlling elements within the HIV-1 Gag protein are contained within its C-terminal sequences and especially within the entire p6 sequence (8). Moreover, we found no evidence that the HIV-1 CA sequence plays any role in determining particle size. In contrast, previous studies of RSV showed that the CA-SP sequence of its Gag protein is critical for normal size, and small deletions throughout it result in the release of large, heterogeneously-sized particles (20). These results suggested that either lentiviruses (HIV-1) and oncoviruses (RSV) have different size controlling elements or that the impact of HIV-1 CA deletions on particle size cannot be detected when the p6 sequence is present within Pr55gag. In other words, the CA sequence of HIV might be important for the mechanism of large particle production that occurs when p6 is absent.
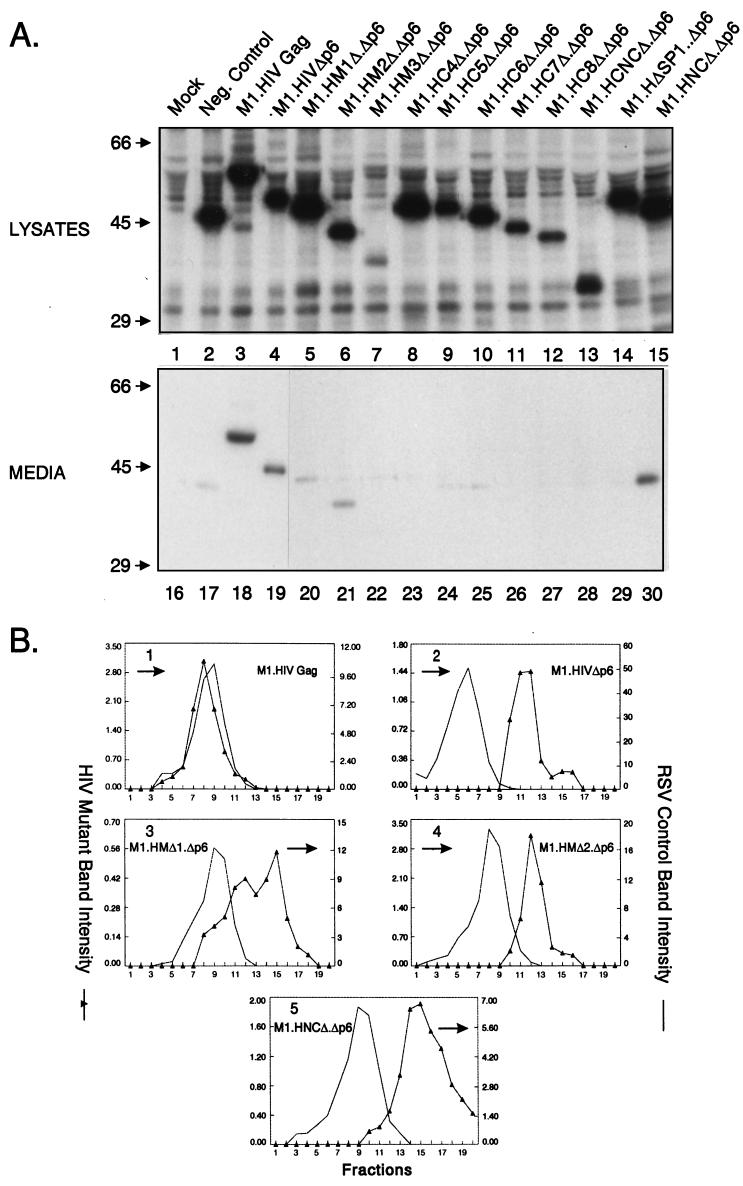
To further examine the role of the CA sequence as well as MA, SP1, and NC domains in determining particle size in the absence of p6, we removed the p6 sequence from a collection of previously described HIV-1 Gag deletion mutants (8). Each of the HIV-1 constructs has the first 10 residues of the Src protein in place of the 32-residue M domain of HIV-1 (Fig. 1). As previously reported (8), this modification has no effect on particle size but leads to enhanced production of Gag proteins and rapid release of virus-like particles into the growth medium after transfection (M1.HIV Gag, Fig. 2A, lanes 3 and 18). To assess particle size, medium samples from transfected COS-1 cells were analyzed in 10 to 30% rate-zonal sucrose gradients. We chose this method of measuring particle size because this technique, unlike electron microscopy (EM) analysis, allows the entire population of particles to be detected regardless of their morphological appearance. That is, very large particles might go unrecognized by EM methods. It should be noted that in the case of RSV, the variation in size detected by the rate-zonal gradient method was in agreement with EM results (20). The gradient analyses were performed a minimum of two times for each mutant in this study, and representative data are presented here. As previously reported (8), the parental M1.HIV particles were homogeneous and sedimented one or two fractions more slowly than authentic RSV virions (Fig. 2B, panel 1), but cosedimented with RSV particles produced by expressing RSV Gag alone in COS-1 cells (protease deletion mutant 3h [37]; data not shown). In agreement with our previous study (8), particles lacking the p6 sequence were very large but uniform in size (panel 2).
FIG. 2.
Properties of p6-deleted Gag polyproteins. (A) COS-1 cells were transfected with the indicated DNAs and labeled as described in Materials and Methods. Molecular mass standards (in kilodaltons) are indicated to the left. (B) Distribution of particle size in rate-zonal gradients. COS-1 cells were transfected with the indicated DNAs, and after 48 h were labeled with [35S]methionine for 5 h. After the labeling period, particle sizes were analyzed as described in Materials and Methods. Radiolabeled RSV from infected TEF cells was added to the gradients to provide an internal control. Arrows indicate the direction of sedimentation.
Protease treatments showed that the large sizes of these particles was not a result of changes in protein composition or their outside surface. That is, M1.HIVΔp6 particles treated with trypsin prior to centrifugation were again very large; however, they were completely susceptible to trypsin digestion in the presence of Triton X-100 (data not shown).
Deletion of p6 from MA mutants.
To address the role of MA in determining particle size in the absence of p6, COS-1 cells were transfected with M1.HMΔ1.Δp6, M1.HMΔ2.Δp6, and M1.HMΔ3.Δp6 (Fig. 1). While intracellular synthesis of Gag proteins was similar to wild-type levels for M1.HMΔ1.Δp6 and M1.HMΔ2.Δp6, Gag expression was severely reduced in M1.HMΔ3.Δp6 (Fig. 2A, compare lanes 3 and 5 to 7). Analysis of the media fractions revealed that M1.HMΔ1.Δp6 and M1.HMΔ2.Δp6 were present but greatly reduced for particle release (reduction about fivefold, Fig. 2A, compare lanes 18 and 20 to 21), while M1.HMΔ3.Δp6 was not detectable, which is due either to a lower efficiency of budding or to a lower level of expression (Fig. 2A, compare lanes 18 and 22). For comparison, an assembly-incompetent deletion mutant (RHB.T10C [25]), which lacks L domains needed for budding, is shown in lane 17. Gradient analyses revealed that both M1.HMΔ.Δp6 and M1.HMΔ2.Δp6 still produced large particles like the Δp6 parent, although those of M1.HMΔ1.Δp6 were more heterogeneous in size (Fig. 2B, panels 3 and 4). Although the efficiency of budding is reduced for the p6 deletion mutants when combined with MA deletions, these data indicate that MA does not influence particle size.
Deletion of p6 from CA, SP1 mutants.
To address the role of CA and SP1 in large-particle production, COS cells were transfected with a collection of HIV CA and SP1 deletion mutants, all of which lacked the p6 sequence (Fig. 1). While intracellular synthesis of Gag proteins was similar to wild-type levels in M1.HCΔ4.Δp6, M1.HCΔ5.Δp6, M1.HCΔ6.Δp6, and M1.HΔSP1.Δp6, the level of Gag expression was severely reduced in M1.HCΔ7.Δp6 and M1.HCΔ8.Δp6 (Fig. 2A, compare lanes 3 and 8 to 12 and 14). Unexpectedly, all of the CA and SP1 deletion mutants were defective for particle release (Fig. 2A, compare lanes 18 and 22 to 27 and 29). This was surprising because the very same CA and SP1 deletions in the presence of p6 do not greatly affect particle release or size (8). Thus, it appears that the HIV-1 CA and SP1 sequences are required for and may drive the production of large particles in the absence of p6 (see Discussion).
Deletion of p6 from NC mutants.
To determine the contribution of NC to particle size, we removed p6 from two previously described NC deletion mutants (8). M1.HCNCΔ.Δp6 contains a deletion which removes a portion of the CA upstream of the major homology region (MHR) along with SP1 and the first six residues from NC, while M1.HNCΔ.Δp6 lacks the second half of the NC, SP2, and p6 but retains I domain activity (Fig. 1). Both proteins were produced at wild-type levels (Fig. 2A, compare lanes 3 and 13 and 15). M1.HNCΔ.Δp6 was released into the medium with approximately 50% of wild-type efficiency (Fig. 2A, compare lanes 18 and 30), whereas M1.HCNCΔ.Δp6 was completely defective for particle release (Fig. 2A, compare lanes 18 and 28), again suggesting the importance of CA and SP1 in the production of large particles in the absence of p6. When analyzed for size, M1.HNCΔ.Δp6 was found to be contained in relatively homogeneous but very large particles (panel 5). This sedimentation profile was identical to that for M1.HNCΔ (8), indicating once again that an important size determinant is located within the C-terminal portion of NC.
Substitutions of p6 with RSV p2b and EIAV p9.
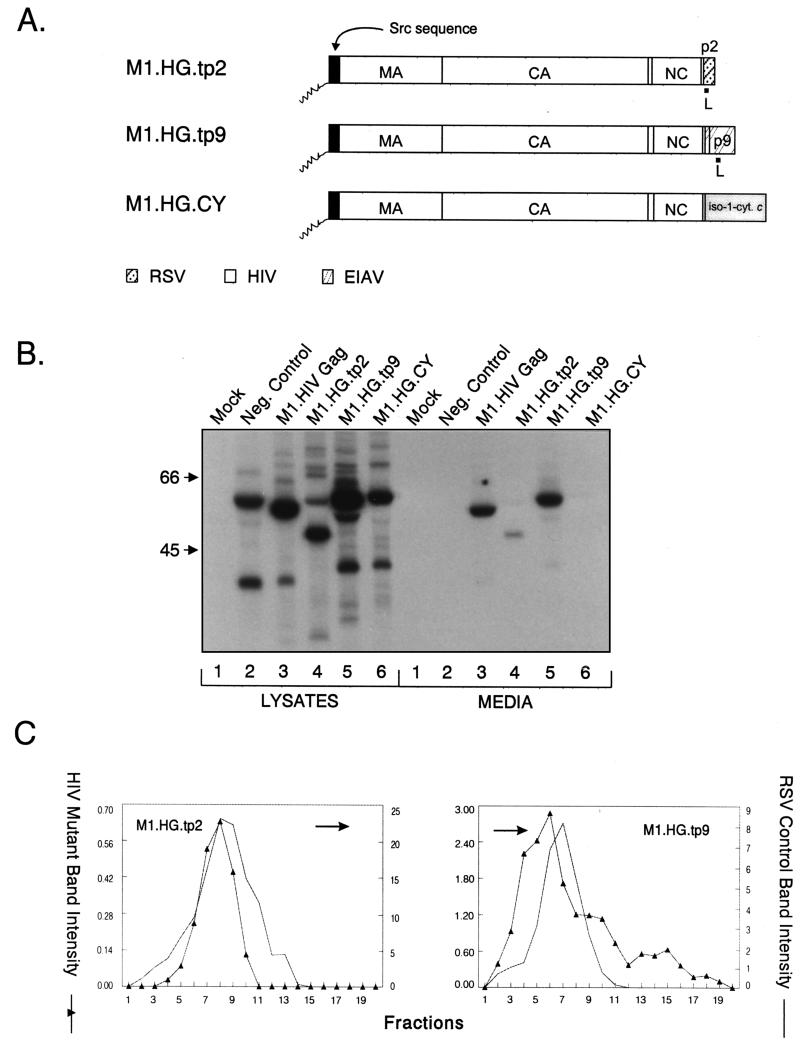
Because we were unable to analyze the impact that CA deletions might have on particle size in the absence of p6, we next attempted to restore budding to the HIV CA deletion mutants by including heterologous L domains at their C termini. We decided to use the RSV p2b and EIAV p9 sequences and their associated L domains because it has been previously shown that these sequences can provide L domain activity in the context of heterologous Gag proteins (25, 28). We hoped that they would restore budding without themselves influencing particle size, since previous studies have not implicated L domains as size determinants (8, 20). As a control, we first examined constructs with complete CA sequences. We found that the HIV-p9 chimeric particles (produced by M1.HG.tp9, Fig. 3A) were released into the medium with the same efficiency as M1.HIV Gag (Fig. 3B, compare lanes 3 and 5), whereas the level of release of HIV-p2 (produced by M1.HG.tp2, Fig. 3A) was reduced about fourfold compared to wild-type level (Fig. 3B, compare lanes 3 and 4). This decrease is likely due to suboptimal positioning of the RSV L domain at the end of HIV Gag. Consistent with this, we previously noted a 50% decrease in particle release for a RSV-HIV chimera containing the p2b sequence at this position (RHB.T10C.tp2 [25]).
FIG. 3.
Replacement of p6 with heterologous L domains. (A) M1.HG.tp2 contains the L domain of RSV Gag, while M1.HG.tp9 contains the L domain of EIAV. M1.HG.CY contains the yeast iso-1-cytochrome c sequence, which does not have an L domain. The names of the Gag cleavage products (MA, CA, NC, p6, p2, and p9) are indicated. The black box at the N termini of the constructs represent the first 10 amino acids from pp60v-src. The squiggle represents the fatty acid myristate. (B) COS-1 cells were transfected with the indicated DNAs and labeled as described in Materials and Methods. Molecular mass standards (in kilodaltons) are indicated to the left. (C) Distribution of particle size in rate-zonal gradients. COS-1 cells were transfected with the indicated DNAs and labeled with [35S]methionine for 5 h. After the labeling period, particle sizes were analyzed as described in Materials and Methods. HIV-p2 and HIV-p9 chimeras produced normal-sized particles. Arrows indicate the direction of sedimentation.
As expected, subsequent sedimentation analyses showed that both L domain chimeras produced particles of normal density when analyzed in 10 to 50% isopycnic sucrose gradients (data not shown). However, when particle size was analyzed in 10 to 30% rate zonal gradients, particles for both chimeras were not large but appeared essentially identical in size distribution to the control particles (Fig. 3C). This provided our first indication that L domains may be important for size. To address the possibility that any random sequence placed at the C terminus of HIV Gag would be sufficient to obtain normal particle size, we analyzed chimera M1.HG.CY, which contains the iso-1-cytochrome c sequence from Saccharomyces cerevisiae in place of p6 (Fig. 3A). Although this sequence does not interfere with budding when attached to the C terminus of the RSV Gag protein (36), we found that the HIV-cytochrome chimera was severely defective for particle release, although it was synthesized at levels similar to wild type (Fig. 3B, lanes 6). For comparison, an assembly-incompetent deletion mutant (T10C [37]), which lacks late domains needed for budding, is shown in lanes 2. The only known feature in common to p6, p9, and p2b is L domain activity, and thus, we conclude that the L domains play an active role in constraining particle size, at least in the context of HIV-1 Gag.
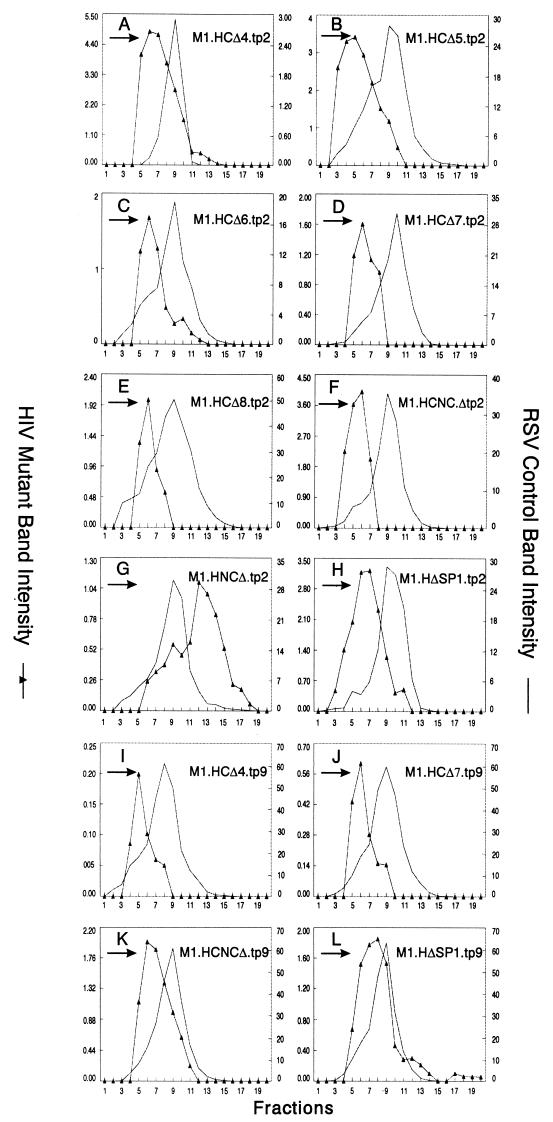
Having found that heterologous L domains do influence particle size, we could not predict what would happen when segments of CA or other regions of Gag were removed from the chimeras. For example, would a CA deletion block particle release for the p2b or the p9 chimera? If not, would particles of normal or large size be released? To answer these questions, a variety of gag deletions were inserted into the HIV-p2 and HIV-p9 chimeras (Fig. 4). As before, budding efficiencies of the p2 chimeras were lower than those observed for the HIV-p9 constructs but sufficient for size analyses (data not shown). When sedimented through sucrose, each of the CA, CA-NC, and SP1 mutants produced relatively uniform populations of particles that were not large but were somewhat smaller than the internal RSV control (curves with triangular symbols in Fig. 5A to F and H to L). For each mutant, the shift of the peak to a position four fractions or less above the control was quite reproducible and has been also seen in the case of M1.HG.tp9 (Fig. 3C). The reason for this shift toward smaller particles remains to be determined but may be due to the absence of viral components other than Gag. Only M1.HNCΔ.tp2, which lacks the second half of the NC sequence and the first four residues of SP2 (Fig. 4), produced large particles (panel G). A similar construct having p9 in the place of p2 was not tested, but when p6 is present on this mutant, large particles are also produced (8). Thus, it appears that the CA sequence, the N-terminal part of NC, and SP1 are not important for HIV-1 particle size when an L domain is present.
FIG. 5.
Heterologous L domains restore particle release and normal size to HIV Gag mutants. COS-1 cells were transfected with the indicated DNAs and labeled with [35S]methionine for 5 h. After the labeling period, particle sizes were analyzed as described in Materials and Methods. Arrows indicate the direction of sedimentation.
Analysis of size determinants of EIAV Gag.
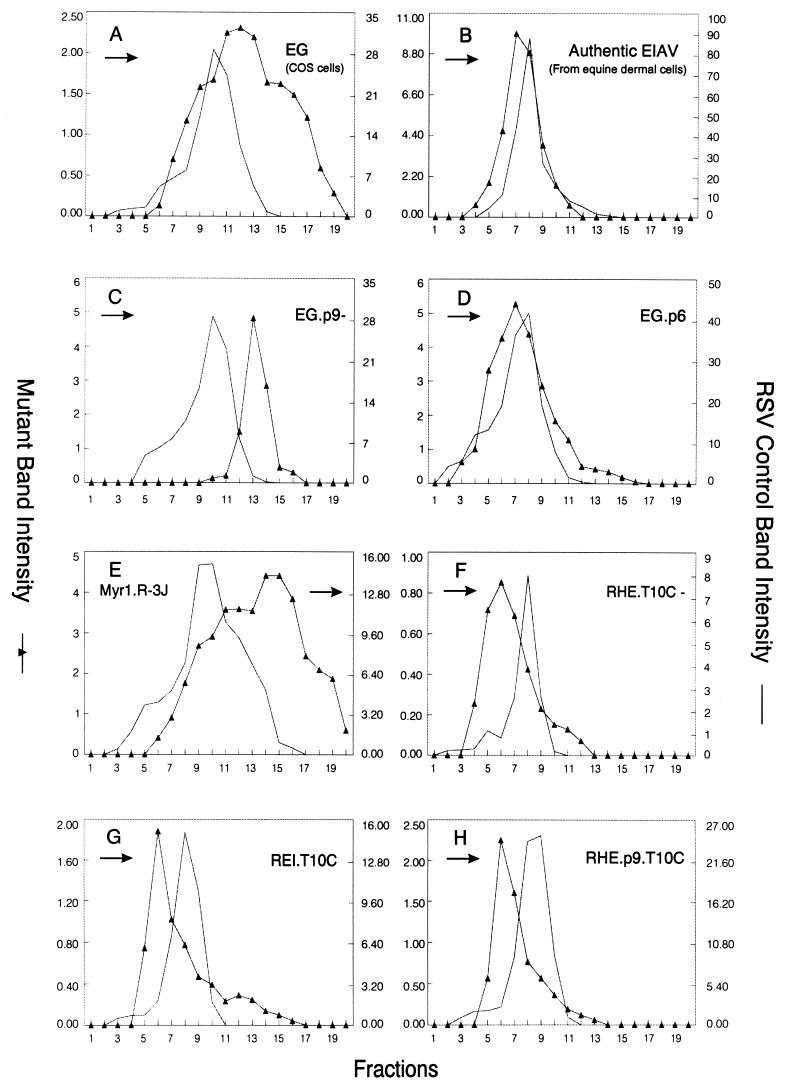
In view of the fundamental differences reported for the size determinants of RSV (20) and HIV-1 (8), we decided to examine the importance of the C terminus in another lentiviral Gag protein. In our initial experiments, particles produced by expressing the wild-type EIAV Gag protein in COS cells were subjected to rate zonal analysis (Fig. 6, construct EG). Unexpectedly, these particles were relatively heterogeneous, although they overlapped the normally-sized RSV particles (Fig. 7A). In contrast, infectious EIAV particles produced from equine dermal cells were homogeneous and beautifully sedimented one to two fractions more slowly than authentic RSV virions (panel B). These results raise the possibility that there is a cell-specific factor that contributes to the regulation of EIAV size or that an EIAV component other than Gag influences particle size. Further experiments will be required to distinguish between these two possibilities.
FIG. 7.
Distribution of wild-type and recombinant EIAV Gag particles in rate-zonal gradients. COS-1 cells were transfected with the indicated deletion mutants DNAs and labeled with [35S]methionine for 5 h. After the labeling period, particle sizes were analyzed as described in Materials and Methods. Arrows indicate the direction of sedimentation.
When the p9 sequence and its associated L domain were deleted from EIAV Gag (EG.p9−), large particles were released (panel C) in a manner similar to what we have observed with HIV p6 deletion mutants. When the HIV p6 sequence was inserted in place of the p9 sequence (EG.p6), the released particles were once again rather uniform in size but slightly smaller than RSV particles (panel D). This provides further evidence that the p6 sequence has an important role in constraining particle size and can exert this function in a heterologous system.
To examine whether p9 might be able to control the size of an oncoviral Gag protein in a manner similar to that reported for p6 (8), RSV-EIAV chimeric Gag proteins were analyzed. A RSV capsid deletion mutant which produces heterogeneously-sized particles (Myr1.R-3J [39]) is shown for comparison in Fig. 7E. Placement of C-terminal sequences from EIAV Gag onto the C terminus of such an RSV capsid mutant was suffiient to restore the production of homogeneously-sized particles (REI.T10C, panel G), just as the C-terminal sequences of HIV did (RHE.T10C, panel F). This result suggests that size determinants of EIAV Gag are indeed contained within its C-terminal sequences. The importance of the p9 sequence in determining particle size is emphasized by chimera RHE.p9.T10C (Fig. 6), a RHE.T10C construct that contains the p9 sequence in place of p6. As predicted, RHE.p9.T10C particles were found to be homogeneous (panel H). Based on our limited studies of the EIAV Gag protein, its size determinants appear to be arranged similarly to those of HIV.
DISCUSSION
In this report, we have demonstrated that three very different retroviral L domains act as major determinants of particle size, at least in the context of HIV Gag. The substitution of the HIV p6 sequence with the RSV p2b or EIAV p9 sequences and their associated L domains was sufficient to restore normal sedimentation properties to HIV-1 particles. We chose the sucrose gradient approach rather than EM methods to analyze particle size. The advantages of gradient sedimentation include the display of the entire population of particles and the standardization provided by internal markers. However, the principal disadvantage of this technique is that variations in sedimentation rate do not allow easy calculation of the actual size of the particles. Therefore, it will be interesting to analyze the particles produced by the different deletion mutants by EM in an attempt to determine their exact size and morphology. However, our transient expression systems make EM analyses difficult because most of the cells in the transfected cultures do not express Gag.
Our data suggest that the HIV-1 Gag protein can participate in two very different mechanisms of budding. One requires the L domain but not the capsid and results in the production of normally-sized particles. The second mechanism is independent of the L domain and seems to be driven by CA-SP1 but can produce only very large particles.
How do retroviral L domains influence particle size?
Although we do not yet understand how L domains orchestrate the release of normal-sized particles, we favor the idea that they recruit host proteins to the site of budding for mediating the separation of the virus from the cell. The L domain might be directly or indirectly recognized near or within the neck of the stalk by cellular proteins, thereby creating the molecular machinery needed for the release of a normal-sized particle from the plasma membrane. This model is in agreement with the finding that the Y-X-X-L sequence in the L domain of EIAV interacts in vitro and in vivo with the cellular AP-50 medium chain subunit of the plasma membrane AP-2 complex (29), which in uninfected cells mediates endocytosis (22). In the case of RSV, the P-P-P-P-Y motif in its L domain has been shown to bind to the WW domain of Yes-associated protein (Yap) in vitro (7). Although putative cellular proteins involved in budding have not yet been identified for RSV, several studies suggest that cytoskeletal proteins (actin and myosin) might participate in HIV-1 particle release (23, 26, 32, 35). Moreover, a recent study has shown that HIV virions incorporate ubiquitin and that a small amount of p6gag is covalently attached to single ubiquitin molecules inside HIV-1 virions (24). Free ubiquitin has also been detected in murine leukemia virus, simian immunodeficiency virus, and RSV virions (24, 30). These observations, coupled with the fact that several cytoskeletal proteins as well as members of the microtubule network are conjugated to single ubiquitin molecules (1, 4, 21), suggest that the presence of ubiquitin into released HIV-1 virions might be the result of an interaction between viral structural proteins and a monoubiquitinated cytoskeletal protein during assembly and/or budding. Interestingly, monoubiquitination is believed to be a signal for plasma membrane receptor internalization (13). Therefore, the identification of cellular proteins that interact specifically with different L domains may lead to a better understanding of the mechanism by which these domains define particle size.
What happens in the absence of L domains?
In the case of RSV and EIAV, when L is nonfunctional, virus particles accumulate at the cell surface but fail to be released (25, 28, 40). With regard to HIV-1, multiple lines of evidence prove that p6 contains a late budding function (11, 25), although this activity appears to be influenced by PR (14). More recently, it has been found that p6-deleted Gag particles are extremely large in size (8), suggesting that a mechanism of release independent of the L domain can take place. When p6 and its associated L domain (and any cellular proteins normally involved in virus-cell separation) are absent, the particles grow very large, perhaps as a result of many nascent particles coalescing. Unexpectedly, our results indicate that the CA-SP sequence is absolutely required for this and may drive the release of these large particles. That is, all of the CA and SP1 deletion mutants were defective for particle release in the absence of p6. Perhaps the rigid structure created by the interactions among the intact CA-SP sequences stabilizes the nascent particle long enough so that a less efficient or alternative mechanism of membrane separation can occur (one not dependent on the L domain of p6). Although in our study we were unable to map an L domain-like sequence in the capsid, it may be that deletions throughout CA alter its overall conformation and hence prevent such a sequence from being properly presented. Further experiments are necessary to test these hypotheses.
Lentiviruses and oncoviruses have different size-controlling elements.
Our results show that the size determinants of oncoviruses (RSV) and lentiviruses (HIV-1 and EIAV) are located at very different positions within Gag and raise the possibility that they work through different mechanisms. Data obtained with the EIAV chimeras indicate that the size-controlling elements, like those of HIV-1, are contained within the C-terminal sequences of Gag, and especially within the p9 sequence. Unexpectedly, we also found that EIAV Gag particles produced from COS cells were relatively heterogeneous compared to infectious EIAV particles produced from equine cells. This suggests the possibility that EIAV Gag is better adapted to host proteins present in equine cells than simian cells.
In conclusion, we have identified a novel function of the retroviral L domain. While further experiments are necessary to understand the mechanisms used by these domains to constrain the size of an emerging particle, these results indicate that this function of L domain is conserved among retroviruses.
ACKNOWLEDGMENTS
We thank Ronald Montelaro and Bridget Puffer for providing EIAV antibodies and EIAV infected equine dermal cells. The HIV immunoglobulin (from A. Prince) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
This work was supported by grants from the National Institutes of Health awarded to J.W.W. (CA47482) and L.J.P. (AI01148) and from the American Cancer Society awarded to J.W.W. (FRA-427). Support for L.G. was provided by Pasteur Merieux-Connaught Canada.
REFERENCES
- 1.Ball E, Karlik C C, Beall C J, Saville D L, Sparrow J C, Bullard B, Fyrberg E A. Arthrin, a myofibrillar protein of insect flight muscle, is an actin-ubiquitin conjugate. Cell. 1987;51:221–228. doi: 10.1016/0092-8674(87)90149-8. [DOI] [PubMed] [Google Scholar]
- 2.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chazal N, Carriere C, Gay B, Boulanger P. Phenotypic characterization of insertion mutants of the human immunodeficiency virus type 1 Gag precursor expressed in recombinant baculovirus-infected cells. J Virol. 1994;68:111–122. doi: 10.1128/jvi.68.1.111-122.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corsi D, Galluzzi L, Crinelli R R, Magnani M. Ubiquitin is conjugated to the cytoskeletal protein α-spectrin in mature erythrocytes. J Biol Chem. 1995;270:8928–8935. doi: 10.1074/jbc.270.15.8928. [DOI] [PubMed] [Google Scholar]
- 5.Craven R C, Bennett R P, Wills J W. Role of the avian retroviral protease in the activation of reverse transcriptase during virion assembly. J Virol. 1991;65:6205–6217. doi: 10.1128/jvi.65.11.6205-6217.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorfman T, Mammano F, Haseltine W A, Göttlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type I envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature (London) 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 8.Garnier L, Ratner L, Rovinski B, Cao S-X, Wills J W. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J Virol. 1998;72:4667–4677. doi: 10.1128/jvi.72.6.4667-4677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnier L, Bowzard J B, Wills J W. Recent advances and remaining problems in HIV assembly. AIDS. 1998;12:S5–S16. [PubMed] [Google Scholar]
- 10.Gheysen D, Yancey R, Petrovskis E, Timmins J, Post L. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 11.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processing, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 14.Huang M, Orenstein J M, Martin M A, Freed E O. p6gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter E. Biological techniques for avian sarcoma viruses. Methods Enzymol. 1994;58:379–392. doi: 10.1016/s0076-6879(79)58153-1. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karacostas V, Nagashima K, Gonda M A, Moss B. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc Natl Acad Sci USA. 1989;86:8964–8967. doi: 10.1073/pnas.86.22.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krausslich H G, Ochsenbauer C, Traenckner A M, Mergener K, Facke M, Gelderblom H R, Bosch V. Analysis of protein expression and virus-like particle formation in mammalian cell lines stably expressing HIV-1 gag and env gene products with or without active HIV proteinase. Virology. 1993;192:605–617. doi: 10.1006/viro.1993.1077. [DOI] [PubMed] [Google Scholar]
- 19.Krausslich H G, Facke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous Sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murti K G, Smith H T, Fried V A. Ubiquitin is a component of the microtubule network. Proc Natl Acad Sci USA. 1988;85:3019–3023. doi: 10.1073/pnas.85.9.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno H, Stewart J, Fournier M C, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino J S. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 23.Ott D E, Coren L V, Kane B P, Busch L K, Johnson D G, Sowder II R C, Chertova E N, Arthur L O, Henderson L E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J Virol. 1996;70:7734–7743. doi: 10.1128/jvi.70.11.7734-7743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ott D E, Coren L V, Copeland T D, Kane B P, Johnson D G, Sowder II R C, Yoshinaka Y, Oroszlan S, Arthur L O, Henderson L E. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of moloney murine leukemia virus. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearce-Pratt R, Malamud D, Phillips D M. Role of the cytoskeleton in cell-to-cell transmission of human immunodeficiency virus. J Virol. 1994;68:2898–2905. doi: 10.1128/jvi.68.5.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prince A M, Horowitz B, Baker L, Shulman R W, Ralph H, Valinsky J, Cudell A, Brotman B, Boehle W, Rey F, Barbosa L, Piet M, Reesink H, Lelie N, Tersmette M, Miedema F, Nemo G, Nastala C L, Allan J S, Lee D R, Eichberg J W. Failure of a human immunodeficiency virus (HIV) immune globulin to protect chimpanzees against experimental challenge with HIV. Proc Natl Acad Sci USA. 1988;85:6944–6948. doi: 10.1073/pnas.85.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puffer B A, Parent L J, Wills J W, Montelaro R C. Equine infectious anemia virus utilizes a YLLL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puffer B A, Watkins S C, Montelaro R C. Equine infectious anemia virus gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J Virol. 1998;72:10218–10221. doi: 10.1128/jvi.72.12.10218-10221.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putterman D, Pepinsky R B, Vogt V M. Ubiquitin in avian leukosis virus particles. Virology. 1990;176:633–637. doi: 10.1016/0042-6822(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 31.Reicin A S, Ohagen A, Yin L, Hoglund S, Goff S P. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J Virol. 1996;70:8645–8652. doi: 10.1128/jvi.70.12.8645-8652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rey O, Canon J, Krogstad P. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology. 1996;220:530–534. doi: 10.1006/viro.1996.0343. [DOI] [PubMed] [Google Scholar]
- 33.Royer M, Cerutti M, Gay B, Hong S-S, Devauchelle G, Boulanger P. Functional domains of HIV-1 gag-polyprotein expressed in baculovirus-infected cells. Virology. 1991;184:417–422. doi: 10.1016/0042-6822(91)90861-5. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sasaki H, Namakura M, Ohno T, Matsuda Y, Yuda Y, Nonomura Y. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc Natl Acad Sci USA. 1995;92:2026–2030. doi: 10.1073/pnas.92.6.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weldon R A, Jr, Erdie C R, Oliver M G, Wills J W. Incorporation of chimeric Gag protein into retroviral particles. J Virol. 1990;64:4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wills J W, Craven R C, Weldon R A, Jr, Nelle T D, Erdie C R. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J Virol. 1991;65:3804–3812. doi: 10.1128/jvi.65.7.3804-3812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]