Abstract
Acinic cell carcinoma (AciCC) is a rare salivary gland cancer with excellent prognosis in most cases. However, a subset of patients will develop distant metastasis and die of disease. Recently, a two-tiered grading scheme in AciCC was proposed to recognize patients at risk for poor outcome. We performed a genetic analysis of AciCC to explore the underlying molecular correlates of the tumor grade and examined PD-L1 expression to identify potential candidates for immunotherapy. A retrospective cohort of 55 patients included 34 high-grade and 21 low-grade AciCCs. Forty-three cases were subjected to a targeted exome sequencing by MSK-IMPACT. PD-L1 immunohistochemistry was performed on 33 cases. Tumor mutation burden was low with a median 1 and 2 mutations in low-grade and high-grade AciCCs, respectively. CDKN2A/B was the most frequently altered gene and loss-of-function mutations were found only in high-grade but not in low-grade AciCCs (58.1%, 18/31 vs. 0/12, p<0.001). CDKN2A/B alterations were significantly associated with distant metastasis, which occurred in 88.9% (16/18) CDKN2A/B mutants vs. 44% (11/25) wild-type cases (p=0.004, Fisher’s exact test). Sequential profiling of multiple, temporally distant samples from the same patient demonstrated intratumor heterogeneity including detection of CDKN2A/B deletion in the second, high-grade metastasis only. ATM and PTEN mutations were detected in 19.4% (6/31) and 16.1% (5/31), ARID2, BIRC3 and FBXW7 mutations each in 12.9% (4/31), and TP53, MTAP and FAT1 each in 9.7% (3/31) HG AciCC. PD-L1 positive labeling was more common in high-grade AciCC (9/17, 52.9% vs 3/16, 18.9%, p=0.071). CDKN2A/B mutations in AciCC represent a molecular marker of high-grade histology and disease progression providing a rationale for further studies to determine their prognostic and therapeutic significance in this salivary gland cancer. AciCC with ATM mutations may be amenable to targeted therapy. Immunotherapy can be considered as a treatment option for a subset of AciCC patients.
Keywords: CDKN2A/B, ATM, PTEN, high-grade acinic cell carcinoma
Introduction
Acinic cell carcinoma (AciCC) is a rare malignant salivary gland neoplasm and vast majority of cases arise in the parotid and affects patients of a wide age range including children and young adults (1, 2). Although most cases show an indolent biological behavior, about 35%–45% cases recur and 15–20% have distant metastases, most commonly in lung (3). Overall 20-year survival is 90%, but is only 22 % in patients with distant metastasis (4). Most AciCCs display low-grade (LG) histology but high-grade (HG) histological features such as increased mitotic activity, tumor necrosis and cytologic pleomorphism occasionally referred to as “high-grade transformation” have been long recognized and linked to worse prognosis (5–7). Although earlier studies demonstrated associations between histologic grade and tumor biology (8), a systematic histological grading scheme to help better AciCC patient risk stratification is lacking at present. Recently, Xu et al. explored 3 histologic grades in a large retrospective AciCC cohort (9). The prognosis of LG and intermediate grade (IG) did not significantly differ and 5-year overall survival (OS), disease-specific survival (DSS), disease-free survival (DFS) and distant metastasis-free survival (DMFS) were 100%, 100%, 80%, 90% for LG or IG AciCC, respectively, but was only 50%, 50%, 37%, and 48% for HG AciCC. Ultimately, the authors proposed a two-tiered grading scheme (LG and HG) using a cut off of 5 or more mitoses per 10 high power fields (HPF) and/or presence of necrosis for HG AciCC (9). HG histology emerged as an independent adverse prognostic factor for OS, DFS and DMFS (9). At the genetic level, irrespective of the tumor histology or grade, the vast majority of AciCC harbor recurrent rearrangements [t(4;9)(q13;q31)]. Highly active chromatin regions (enhancers) from the secretory Ca-binding phosphoprotein gene cluster at 4q13 are translocated to 9p31 chromosomal region, upstream of transcription factor Nuclear Receptor Subfamily 4 Group A Member 3 (NR4A3) leading to the constitutive activation and overexpression of NR4A3 through “enhancer hijacking”. This event has been recognized as the initial oncogenic driver event in AciCC with [t(4;9)(q13;q31)] rearrangement (10). Consequently, nearly all AciCC are positive for NR4A3 by immunohistochemistry, which has been utilized as a highly specific and sensitive marker for AciCC in surgical pathology practice (11).
However, genetic markers associated with HG histology that could help separate cases with more aggressive biology have not been sufficiently explored. Here, we performed a genetic analysis of AciCC enriched for HG carcinomas aiming to identify the underlying molecular correlates of the tumor grade and to search for molecular biomarkers of prognostic and/or therapeutic value. In addition, given the limited treatment options for recurrent and/or metastatic AciCC and the potential treatment benefits of immunotherapy in various malignancies (12, 13), we explored the expression of programmed death-ligand 1(PD-L1) in AciCC.
Materials and methods
Cases
Upon approval of the Institutional Research Board of Memorial Sloan Kettering Cancer Center (MSKCC), a retrospective cohort of 55 AciCC patients diagnosed from 1985 to 2021. A total of 62 formalin-fixed paraffin embedded (FFPE) samples comprising 45 primaries and 17 metastatic tumors were collected. These included 23 LG and 39 HG tumors. An overview of basic demographic information and tests performed is provided in Table S1. Pathology slides on all cases were reviewed by head and neck pathologists (BX, NK, SD) and diagnoses were rendered following the WHO 4th edition criteria (2). A recently proposed grading scheme was used to define tumors as HG AciCC if they showed mitotic index of ≥ 5/10 HPFs (> 4/2 mm2) or tumor necrosis (9).
Molecular profiling by targeted exome sequencing
Molecular profiles on 50 samples were obtained from 43 patients. In 5 cases, both, primary and metastasis were sequenced, including a case with microdissected LG and HG component of the primary tumor (ACI_09). In one case, 2 temporally separate metastases were profiled (ACI_29). With the exception of 4 molecular profiles (ACI_39-ACI_42) performed in an outside laboratory using a similar method (14), the remaining 46 samples were sequenced in house as research (n=27) or clinical (n=19) samples. Minimum required tumor content was 20%. Genomic DNA was extracted from FFPE unstained tumor sections in all cases. Matched normal DNA from FFPE non-neoplastic tissue (n=21) or unmatched pool normal DNA (n =5) was used for all research samples, and matched normal blood was used for all clinical samples as previously described (15). A targeted hybridization capture-based massive parallel sequencing was performed by Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT), a clinically validated molecular assay to interrogate somatic variants in 341–505 cancer-related genes (15, 16). Oncogenic potential of somatic alterations and therapeutic levels of evidence were assessed by OncoKB (Table S2), as previously reported (17).
Immunohistochemistry
Immunohistochemistry for NR4A3 was performed using a monoclonal antibody (clone: NOR1 [H7], SC-393902, dilution: 1:100; Santa Cruz Biotechnology Inc., Heidelberg, Germany) as previously described and the was considered positive if >5% of tumor cells exhibited nuclear staining of any intensity (9). PD-L1 antibody E1L3N (Cell Signaling Technology, Danvers, MA, USA) was used according to manufacturer’s recommendations. The immunoexpression was scored as previously described; the percentage of positive tumor cells was calculated as the numbers of tumor cells with partial or complete membranous staining of any intensity divided by the number of tumor cells (18). The percentage of PD-L1-positive immune cells was calculated as the number of immune cells showing cytoplasmic and/or membranous staining of any intensity divided by the number of tumor cells. PD-L1 was interpreted as positive if a combined positive score (CPS) (calculated as the sum of positive tumor cells and positive immune cells divided by the total tumor cells multiplied by 100) was at least 1 (18).
Results
Patients
Of 55 patients, there were 32 women presented at a median age 59 years (range 21–88 years) and 23 men presented at a median age 57 years (range 31–77 years). Out of 43 cases subjected for molecular analysis in the current study, 62.8% (n=27) had history of distant metastasis. Outcome for 46 patients was previously reported by our group (9) and clinico-pathological features of 9 new cases are provided in Table S3.
Molecular analysis
a). Most HG AciCC harbor oncogenic mutations.
The tumors were sequenced at the median depth of coverage of 350x (range, 73–1927x). The median mutation count for all samples was 1 mutation per sample (range 0–11) and was higher in HG than in LG tumors (median 2 vs 1 mutation, p=0.024, Student t test). Eighty-five (likely) oncogenic genetic alterations included 51.8% (44/85) whole gene deletions, 27.1% (23/85) truncating mutations, 11.8% (10/85) missense mutations, 3.5% (3/85) structural variants, 3.5% (3/85) amplifications and 2.3% (2/85) promoter mutations (Figure 1A). Cell-cycle and TP53 pathways were most frequently affected pathways in AciCC. (Figure 1B). At least one (likely) oncogenic mutation was identified in 90.3% (28/31) of HG vs. 41.7% (5/12) in LG cases (p=0.002, Fisher’s exact test, Figure 2). A complete list of mutations is provided in Table S4.
Figure 1.
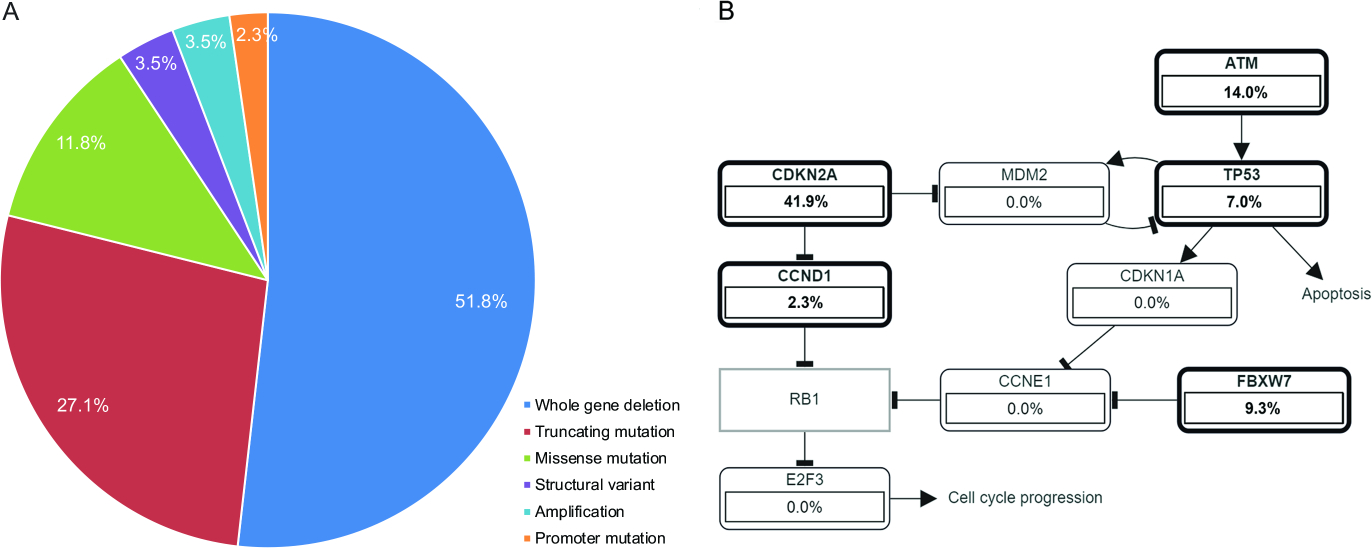
Distribution of (likely) oncogenic genetic alterations in acinic cell carcinoma (A). Signaling pathways affected in acinic cell carcinoma (B).
Figure 2.

Oncoprint of oncogenic and/or recurrent genetic alterations in acinic cell carcinoma. Top row represents patients. Frequencies of mutations are calculated per case/patient and represented as percentage occurrence in the entire cohort (left column), in low-grade acinic cell carcinomas (middle column) and in high-grade acinic cell carcinomas (right column).
b). Highly recurrent CDKN2A/B loss-of-function mutations are present in most HG AciCCs and are associated with distant metastasis.
Most frequent recurrent, mostly oncogenic loss-of-function alterations CDKN2A/B affected 41.9% (18/43) cases but were exclusively found in HG tumors (58.1%, 18/31 vs 0, p<0.001, Fisher’s exact test; Figure 3). These included CDKN2A/B whole gene deletion (n=15), three CDKN2A/B structural variants/rearrangements and one CDKN2A R80* mutation. The presence of oncogenic CDKN2A/B alteration was significantly associated with distant metastasis; 88.9% (16/18) CDKN2A/B mutants had distant metastasis vs. 44% (11/25) CDKN2A/B wild-type cases (p=0.004, Fisher’s exact test). A complete list of copy number alterations is provided in Table S5 and details on structural variants are in Table S6.
Figure 3.
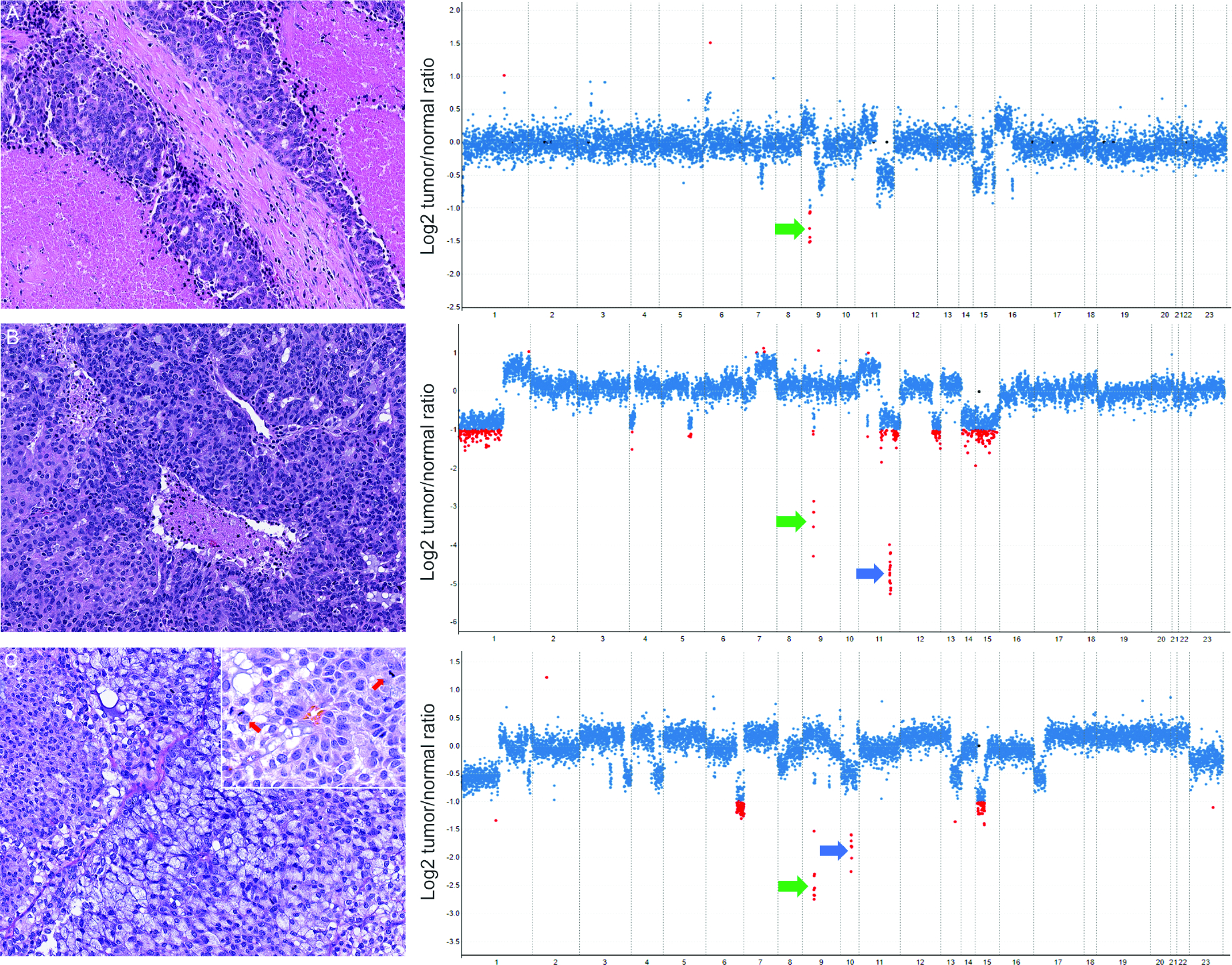
High-grade acinic cell carcinoma with CDKN2A/B deletion. Haemotoxylin and Eosin(H&E) microphotographs (left) and copy number plots (MSK-IMPACT; right); Y-axis depicts copy number changes expressed as the log2 transformed tumor/normal ratio according to their genomic positions indicated on the x-axis. Each dot represents one exon and ≥2-fold tumor/normal ratio is indicated with red dots. ACI_30 with extensive necrosis and CDKN2A/B deletion (green arrow; A). ACI_37 with focal necrosis CDKN2A (green arrow) and ATM deletion (blue arrow; B). ACI_38 showed high mitotic activity (yellow arrows, inset), CDKN2A (green arrow) and PTEN deletion (blue arrow; C).
c). HG AciCCs are enriched for mutations in ATM and PTEN.
ATM and PTEN mutations were found in 19.4% (6/31) and 16.1% (5/31) HG AciCCs. Other recurrent mutations detected only in HG tumors involved ARID2, BIRC3 and FBXW7, each in 12.9% (4/31), TP53, MTAP and FAT1, each in 9.7% (3/31), and MED12, MET, APC, PRKN, RAD50 and SETD2 each in 6.5% (2/31) HG AciCCs. In contrast to HG, mutations in LG AciCCs were overall less common and NF1, TERT, BAP1 and ARID1A oncogenic variants were each detected in a single case (Figure 2).
d). Most HG AciCCs harbor mutations associated with targeted molecular therapies.
Overall, 46.5% (20/43) AciCCs including the majority of HG AciCCs (61.3%, 19/31) showed mutations potentially targetable with standard or experimental therapies in clinical trials (Table 1). More specifically, in 25% (5/20) cases, these alterations qualified as a “standard care or investigational biomarker predictive of response to an FDA-approved or investigational drug in another indication” (OncoKB level 3B). In the remaining 75% (15/20) cases, potentially targetable mutations met the criteria for OncoKB level 4, “compelling biological evidence supports the biomarker as being predictive of response to a drug” (Table 1). Details on OncoKB therapeutic levels of evidence are provided in Table S2.
Table 1.
Genetic alterations in acinic cell carcinoma associated with molecular therapeutic agents.
| Case | Gene | Mutation | OnkoKB Level | Therapeutic Agent |
|---|---|---|---|---|
|
| ||||
| ACI_01 | NF1 | A1873fs | 4 | MEK1/2 inhibitor |
|
| ||||
| ACI_06 | CDKN2A | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_12 | CDKN2A/B | Deletion | 4 | CDK4/6 inhibitor |
| CHEK2 | Y445* | 3B | PARP inhibitor | |
| PTEN | H93L | 4 | PI3K inhibitor | |
|
| ||||
| ACI_16 | CDKN2A | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_17 | CDKN2A | R80* | 4 | CDK4/6 inhibitor |
| CDKN2B | Structural variant | |||
| NF1 | Deletion | 4 | MEK1/2 inhibitor | |
|
| ||||
| ACI_21 | ATM | X1313_splice | 3B | PARP inhibitor |
|
| ||||
| ACI_22 | CDKN2A | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_23 | CDKN2A/B | Deletion | 4 | CDK4/6 inhibitor |
| ATM | E2245* | 3B | PARP inhibitor | |
|
| ||||
| ACI_26 | PIK3CA | C420R | 3B | PI3K inhibitor |
| CDKN2A | Structural variant | |||
|
| ||||
| ACI_28 | CDKN2A2B | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_29 | CDKN2A/B | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_30 | CDKN2A/B | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_31 | CDKN2A/B | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_33 | CDKN2A/B | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_34 | CDKN2A | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_37 | ATM | Deletion | 3B | PARP inhibitor |
| CHEK1 | Deletion | 3B | PARP inhibitor | |
| CDKN2A | Deletion | 4 | CDK4/6 inhibitor | |
|
| ||||
| ACI 38 | CDKN2A | Deletion | 4 | CDK4/6 inhibitor |
| PTEN | Deletion | 4 | PI3K inhibitor | |
|
| ||||
| ACI_39 | CDKN2A | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_42 | CDKN2A | Deletion | 4 | CDK4/6 inhibitor |
|
| ||||
| ACI_55 | CDKN2A | Structural variant | 4 | CDK4/6 inhibitor |
| PTEN | E242* | 4 | PI3K inhibitor | |
e). AciCC exhibits intratumor heterogeneity.
Sequencing of more than one sample per patient demonstrated intratumor heterogeneity in AciCC. In ACI_09, PTEN deletion present in the LG component of the primary was absent in the HG component as well as in the metastasis. HG component of the primary and metastatic ACI_09 showed ATM mutation. In ACI_17, oncogenic FAT1 and NF1 deletions were detected in the primary but were both absent in the subsequent metastasis. In ACI_21, ATM X1313_splice oncogenic variant seen only in the primary, and TP53 and WT1 deletions, and CDK4 K22Q and CYLD G150* were found only in the subsequent metastasis. In ACI_29, FBXW7 and FAT1 deletions detected in the first, LG metastasis were absent in the second, HG metastasis, which acquired CDKN2A/B and MTAP deletions (Figure 2).
NR4A3 expression
Out of 55 cases, NR4A3 immunohistochemistry was performed on 31 cases with available material and all (100%) cases, 14 LG and 17 HG AciCC were positive (Table S1). These included 8 LG and 14 HG carcinomas were with available molecular profile. No difference in NR4A3 immunoexpression was observed between CDKN2A/B mutant (n=7) and CDKN2A/B wild-type (n=7) HG AciCC (Figure 2).
PD-L1 expression
Of 33 cases, 36.4% (n=12) were positive for PD-L1 and trended towards HG than in LG tumors (9/17, 52.9% vs 3/16, 18.9%, p=0.071). CPS ranged from 1–20 in HG, and 3–10 in LG AciCCs. Notably, in all positive cases, positive CPS was based only on PD-L1 expression in immune cells; tumor cells were PD-L1 negative in all cases (Figure 4).
Figure 4.
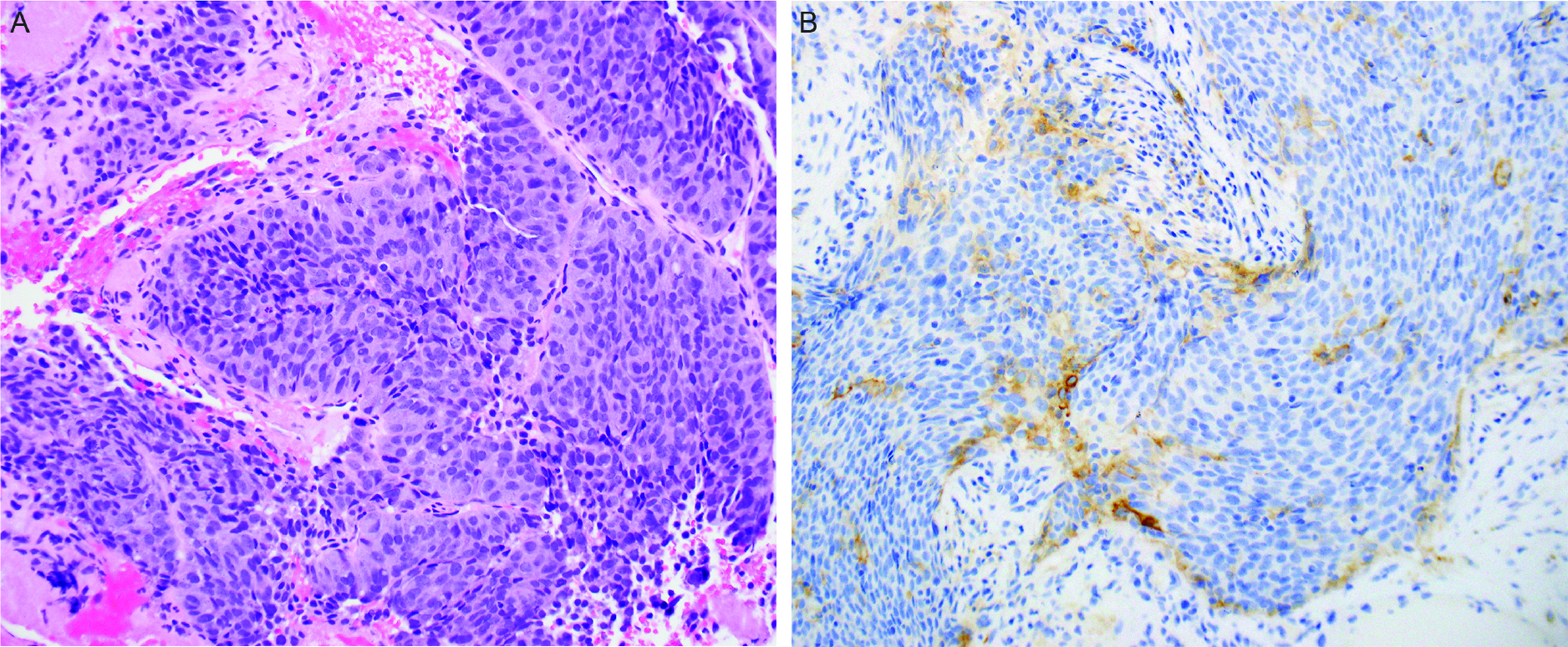
High-grade acinic cell carcinoma (A) demonstrates positive programmed cell death ligand-1(PD-L1) expression in the immune cells (B).
Discussion
Most AciCCs, stage I and II, are successfully treated with surgery alone and prognosis in such cases is excellent (1). These would include AciCCs with LG histology, whereas HG carcinomas have a higher propensity to recur/metastasize and to show poor outcome (9). The treatment options for recurrent/metastatic AciCC are limited and include radiation therapy and chemotherapy (1, 19). Experience with molecular therapies, mTOR inhibitors for instance, is limited and no specific targeted therapeutic agent has yet been identified (20). Herein, we studied a relatively large cohort of AciCCs enriched for HG tumors and identified recurrent somatic mutations of potential prognostic and therapeutic significance. We assessed the tumor immune microenvironment by PD-L1 and showed that AciCC patients can be candidates for immunotherapy.
Using a targeted exome sequencing assay, we found that most HG carcinomas harbored recurrent oncogenic mutations affecting mainly cell cycle and TP53 pathways. The most frequently altered gene CDKN2A is a tumor suppressor gene that inhibits CDK4/6 activity and is frequently deleted across cancers. Two reported AciCCs, one with CDKN2A/B deletion and nonsense ARID2 mutation (21), and the other with CDKN2A/B and MTAP deletion (22) both presented as locally aggressive cancers and developed a widely metastatic disease. This is in line with the present study showing CDKN2A/B mutations exclusively in HG carcinomas and being significantly associated with distant metastasis. Indeed, CDKN2A/B genetic alterations were indicators of poor prognosis in multiple cancer types, including salivary gland tumors such as mucoepidermoid carcinoma (23–26). More recently, loss of CDKN2A/B was reported in various salivary gland tumors with HG features, including AciCC (27). In meningiomas, they were associated with early recurrence (28). In intrahepatic cholangiocarcinoma, CDKN2A/B loss was associated with shorter overall survival (29). In oral squamous cell carcinomas, deleted CDKN2A/B was also an indicator of low survival rate and recurrent tumors showed a significant CDKN2A/B downregulation via whole gene deletion or truncating mutations in comparison to their primaries (30). In agreement with the latter, we found CDKN2A/B deletion only in the second, HG metastasis of ACI_29, further suggesting this could be a late event in carcinogenesis and associated with the progression of disease.
CDKN2A/B loss-of-function mutations have been associated with resistance to immunotherapy in multiple cancers (31, 32). A significant proportion of our cases were PD-L1 positive, including those with altered CDKN2A/B. Although the response to check-point blockade is partly determined by the tumor type (33), CDKN2A/B mutation status could be explored as a parameter of response if AciCC is considered for this treatment modality. Finally, CDKN2A/B mutations in AciCC may be an indication for targeted therapy agents such as CDK4/6 inhibitors. For instance, non-small cell lung carcinomas harboring loss of CDKN2A/B showed promising results after treatment with CDK4/6 inhibitors (34, 35). These agents are also currently explored for treatment of sarcomas and early clinical success has been documented in select cases with altered CDKN2A-CCND-CDK4/6-Rb pathway including CDKN2A/B loss-of-function somatic variants (36). A similar treatment approach could be investigated for patients with CDKN2A/B mutated AciCC.
Other recurrent genetic events of potential clinical significance in HG AciCCs included DNA repair genes mutations such as ATM, CHEK1 and CHEK2, which can be targeted with PARP inhibitors. Patients with metastatic castration-resistant prostate cancer with mutations in homologous recombination repair genes can benefit from Olaparib (37, 38) raising a possibility that the same treatment option might be considered for select AciCC patients. Furthermore, somatic mutations in ATM were associated with exceptional responses to radiotherapy in patients with head and neck squamous cell carcinoma (39) and these associations can be explored in AciCC cases undergoing radiation treatment. Lastly, carcinomas with activating PTEN or PIK3CA mutations may become amenable for targeted treatment with PI3K inhibitors (40).
Finally, sequential sequencing of multiple tumors from the same patient showed that AciCC can display significant intratumor heterogeneity. In addition to acquiring new alterations such as CDKN2A/B deletion (ACI_29) in certain instances, potentially targetable alterations, namely ATM mutation (ACI_21) detected in the primary tumor were later absent in the subsequent metastasis. While the underlying mechanisms of such phenomena would be a subject of further studies, this observation per se emphasizes the importance of obtaining the molecular profile on the most recent recurrent/metastatic tumor sample if targeted therapies come into consideration.
Our study has a few limitations. First, despite studying a relatively large cohort, the number of HG cases was still insufficient to perform a meaningful survival analysis in respect to the presence of CDKN2A/B mutations. Second, we were unable to obtain the status of NR4A3 gene and correlate the presence of rearrangement with other tumor features. Nevertheless, positive NR4A3 immunoexpression in all tested cases irrespective of the histologic grade or CDKN2A/B mutation status may suggest there are no particular associations between [t(4;9)(q13;q31)] rearrangement and the tumor grade or provided molecular features in this cohort. Several important conclusions emerged from this cohort which, to our knowledge, includes the largest number of genetically profiled HG AciCCs. Although characterized by a “quiet” genome, the large majority of HG AciCCs harbor biologically significant genetic alterations detectable by a targeted exome sequencing assay. CDKN2A/B mutations emerged as a genetic correlate of HG histology and a marker of disease progression in AciCC. Alternate treatment options such as targeted therapies and/or immunotherapy can be considered for qualifying AciCC patients. The data presented here provide a strong rationale for further studies to determine the prognostic significance and therapeutic relevance of recurrent oncogenic mutations in AciCC and particularly those involving CDKN2A/B and ATM genes.
Supplementary Material
Funding
Research reported in this publication was supported by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748.
Footnotes
Disclosure Statement:
No competing financial interests exist for all contributory authors. Research reported in this publication was supported by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748.
Ethics Approval / Consent to Participate
The study was approved by the Institutional Research Board of Memorial Sloan Kettering Cancer Center and consents to participate were obtained.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding or the senior author on reasonable request.
References
- 1.Vander Poorten V, Triantafyllou A, Thompson L. et al. Salivary acinic cell carcinoma: reappraisal and update. Eur Arch Otorhinolaryngol. 2016;273:3511–31. [DOI] [PubMed] [Google Scholar]
- 2.El-Naggar AK, Chan JKC, Grandis JR, Takata T, Slootweg PJ. World Health Organization Classification of Head and Neck Tumours (4th edition). Lyon: International Agency for Reseach on Cancer (IARC); 2017. p.166–167. [Google Scholar]
- 3.Neskey DM, Klein JD, Hicks S, et al. Prognostic factors associated with decreased survival in patients with acinic cell carcinoma. JAMA Otolaryngol Head Neck Surg. 2013;139:1195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel NR, Sanghvi S, Khan MN, Husain Q, Baredes S, Eloy JA. Demographic trends and disease-specific survival in salivary acinic cell carcinoma: an analysis of 1129 cases. Laryngoscope. 2014;124:172–8. [DOI] [PubMed] [Google Scholar]
- 5.Yue LE, Samankan S, Liu X. et al. Ten patients with high-grade transformation of acinic cell carcinomas: expression profiling of β-catenin and cyclin D1 is useful. Pathol Res Pract. 2020;216:152767. [DOI] [PubMed] [Google Scholar]
- 6.Thompson LD, Aslam MN, Stall JN, Udager AM, Chiosea S, McHugh JB. Clinicopathologic and immunophenotypic characterization of 25 cases of acinic cell carcinoma with high-grade transformation. Head Neck Pathol. 2016;10:152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanley RJ, Weiland LH, Olsen KD, Pearson BW. Dedifferentiated acinic cell (acinous) carcinoma of the parotid gland. Otolaryngol Head Neck Surg. 1988;98:155–61. [DOI] [PubMed] [Google Scholar]
- 8.Gomez DR, Katabi N, Zhung J. et al. Clinical and pathologic prognostic features in acinic cell carcinoma of the parotid gland. Cancer. 2009;115:2128–37. [DOI] [PubMed] [Google Scholar]
- 9.Xu B, Saliba M, Ho A, et al. Head and Neck Acinic Cell Carcinoma: A New Grading System Proposal and Diagnostic Utility of NR4A3 Immunohistochemistry. Am J Surg Pathol. 2022;46:933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haller F, Bieg M, Will R. et al. Enhancer hijacking activates oncogenic transcription factor NR4A3 in acinic cell carcinomas of the salivary glands. Nat Commun. 2019;10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haller F, Skálová A, Ihrler S, Märkl B, Bieg M, Moskalev EA, et al. Nuclear NR4A3 immunostaining is a specific and sensitive novel marker for acinic cell carcinoma of the salivary glands. Am J Surg Pathol 2019;43:1264–72. [DOI] [PubMed] [Google Scholar]
- 12.Negrao MV, Lam VK, Reuben A, Rubin ML, Landry LL, Roarty EB, et al. PD-L1 expression, tumor mutational burden, and cancer gene mutations are stronger predictors of benefit from immune checkpoint blockade than HLA class I genotype in non–small cell lung cancer. J Thorac Oncol. 2019;14:1021–31. [DOI] [PubMed] [Google Scholar]
- 13.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014:371, 2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frampton GM, Fichtenholtz A, Otto GA. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng DT, Mitchell TN, Zehir A. et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zehir A, Benayed R, Shah RH. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravarty D, Gao J, Phillips SM. et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu B, Jungbluth AA, Frosina D. et al. The immune microenvironment and expression of PD-L1, PD-1, PRAME and MHC I in salivary duct carcinoma. Histopathology. 2019;75:672–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller SK, Haderlein M, Lettmaier S. et al. Targeted Therapy, Chemotherapy, Immunotherapy and Novel Treatment Options for Different Subtypes of Salivary Gland Cancer. J Clin Med. 2022;11:720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ettl T, Schwarz-Furlan S, Haubner F. et al. The PI3K/AKT/mTOR signalling pathway is active in salivary gland cancer and implies different functions and prognoses depending on cell localisation. Oral Oncol. 2012;48:822–30. [DOI] [PubMed] [Google Scholar]
- 21.Warner WA, Wong DJ, Palma-Diaz F, Shibuya TY, Momand J. Clinicopathological and targeted exome gene features of a patient with metastatic acinic cell carcinoma of the parotid gland harboring an ARID2 nonsense mutation and CDKN2A/B deletion. Case Rep Oncol Med. 2015; 893694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols AC, Chan-Seng-Yue M, Yoo J. et al. A case report and genetic characterization of a massive acinic cell carcinoma of the parotid with delayed distant metastases. Case Rep Oncol Med. 2013; 270362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anzick SL, Chen WD, Park Y. et al. Unfavorable prognosis of CRTC1-MAML2 positive mucoepidermoid tumors with CDKN2A deletions. Genes Chromosomes Cancer. 2010;49:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Appay R, Dehais C, Maurage C-A. et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol. 2019;21:1519–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jardin F, Jais J-P, Molina T-J. et al. Diffuse large B-cell lymphomas with CDKN2A deletion have a distinct gene expression signature and a poor prognosis under R-CHOP treatment: a GELA study. Blood, 2010;116:1092–104. [DOI] [PubMed] [Google Scholar]
- 26.Shima K, Nosho K, Baba Y. et al. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review. Int J Cancer. 2011;128:1080–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakaguro M, Faquin W, Snuderl M. et al. CDKN2A/B Loss in High-Grade Transformation of Salivary Gland Carcinoma: High-Grade Malignant Potential in Histologically Low-Grade Component. United States & Canadian Academy of Pathology 111th Annual Meeting Abstracts: Head and Neck Pathology (800–850). Mod Pathol 2022; 35:960–1015.35302095 [Google Scholar]
- 28.Sievers P, Hielscher T, Schrimpf D. et al. CDKN2A/B homozygous deletion is associated with early recurrence in meningiomas. Acta Neuropathol. 2020;140:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takada K, Kubo T, Kikuchi J. et al. Effect of comprehensive cancer genomic profiling on therapeutic strategies and clinical outcomes in patients with advanced biliary tract cancer: A prospective multicenter study. Front Oncol. 2022;12:988527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padhi SS, Roy S, Kar M. et al. Role of CDKN2A/p16 expression in the prognostication of oral squamous cell carcinoma. Oral Oncol. 2017;73:27–35. [DOI] [PubMed] [Google Scholar]
- 31.Horn S, Leonardelli S, Sucker A, Schadendorf D, Griewank KG, Paschen A. Tumor CDKN2A-associated JAK2 loss and susceptibility to immunotherapy resistance. J Natl Cancer Inst. 2018;110:677–81. [DOI] [PubMed] [Google Scholar]
- 32.Gutiontov SI, Turchan WT, Spurr LF. et al. CDKN2A loss-of-function predicts immunotherapy resistance in non-small cell lung cancer. Sci Rep. 2021;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aldea M, Andre F, Marabelle A, Dogan S, Barlesi F, Soria J-C. Overcoming Resistance to Tumor-Targeted and Immune-Targeted Therapies. Cancer Discov. 2021;11:874–99. [DOI] [PubMed] [Google Scholar]
- 34.Ahn ER, Mangat PK, Garrett-Mayer E. et al. Palbociclib in patients with non–small-cell lung cancer with CDKN2A alterations: Results from the targeted agent and profiling utilization registry study. JCO Precis Oncol. 2020;4:757–66. [DOI] [PubMed] [Google Scholar]
- 35.Ke Y, Liao CG, Zhao ZQ. et al. Combining a CDK4/6 Inhibitor With Pemetrexed Inhibits Cell Proliferation and Metastasis in Human Lung Adenocarcinoma. Front Oncol. 2022;12:880153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu JY, Seligson ND, Hays JL, Miles WO, Chen JL. Clinical Utility of CDK4/6 Inhibitors in Sarcoma: Successes and Future Challenges. JCO Precis Oncol. 2022;6:e2100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Bono J, Mateo J, Fizazi K. et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–102. [DOI] [PubMed] [Google Scholar]
- 38.Neeb A, Herranz N, Arce-Gallego S. et al. Advanced prostate cancer with ATM loss: PARP and ATR inhibitors. Eur Urol. 2021;79:200–11. [DOI] [PubMed] [Google Scholar]
- 39.Ma J, Setton J, Morris L. et al. Genomic analysis of exceptional responders to radiotherapy reveals somatic mutations in ATM. Oncotarget. 2017;8:10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castel P, Toska E, Engelman JA, Scaltriti M. The present and future of PI3K inhibitors for cancer therapy. Nat Cancer. 2021;2:587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding or the senior author on reasonable request.


