Abstract
The program(s) of gene expression operating during murine gammaherpesvirus 68 (γHV68) latency is undefined, as is the relationship between γHV68 latency and latency of primate gammaherpesviruses. We used a nested reverse transcriptase PCR strategy (sensitive to approximately one copy of γHV68 genome for each genomic region tested) to screen for the presence of viral transcripts in latently infected mice. Based on the positions of known latency-associated genes in other gammaherpesviruses, we screened for the presence of transcripts corresponding to 11 open reading frames (ORFs) in the γHV68 genome in RNA from spleens and peritoneal cells of latently infected B-cell-deficient (MuMT) mice which have been shown contain high levels of reactivable latent γHV68 (K. E. Weck, M. L. Barkon, L. I. Yoo, S. H. Speck, and H. W. Virgin, J. Virol. 70:6775–6780, 1996). To control for the possible presence of viral lytic activity, we determined that RNA from latently infected peritoneal and spleen cells contained few or no detectable transcripts corresponding to seven ORFs known to encode viral gene products associated with lytic replication. However, we did detect low-level expression of transcripts arising from the region of gene 50 (encoding the putative homolog of the Epstein-Barr virus BRLF1 transactivator) in peritoneal but not spleen cells. Latently infected peritoneal cells consistently scored for expression of RNA derived from 4 of the 11 candidate latency-associated ORFs examined, including the regions of ORF M2, ORF M11 (encoding v-bcl-2), gene 73 (a homolog of the Kaposi’s sarcoma-associated herpesvirus [human herpesvirus 8] gene encoding latency-associated nuclear antigen), and gene 74 (encoding a G-protein coupled receptor homolog, v-GCR). Latently infected spleen cells consistently scored positive for RNA derived from 3 of the 11 candidate latency-associated ORFs examined, including ORF M2, ORF M3, and ORF M9. To further characterize transcription of these candidate latency-associated ORFs, we examined their transcription in lytically infected fibroblasts by Northern analysis. We detected abundant transcription from regions of the genome containing ORF M3 and ORF M9, as well as the known lytic-cycle genes. However, transcription of ORF M2, ORF M11, gene 73, and gene 74 was barely detectable in lytically infected fibroblasts, consistent with a role of these viral genes during latent infection. We conclude that (i) we have identified several candidate latency genes of murine γHV68, (ii) expression of genes during latency may be different in different organs, consistent with multiple latency programs and/or multiple cellular sites of latency, and (iii) regions of the viral genome (v-bcl-2 gene, v-GCR gene, and gene 73) are transcribed during latency with both γHV68 and primate gammaherpesviruses. The implications of these findings for replacing previous operational definitions of γHV68 latency with a molecular definition are discussed.
Gammaherpesviruses are characterized biologically by their association with tumors in immunosuppressed hosts. The prototypic gammaherpesvirus 2, herpesvirus saimiri (HVS), causes lymphomas in primates and can transform T lymphocytes (25, 31, 42, 48). Epstein-Barr virus (EBV) is associated with lymphomas and nasopharyngeal carcinoma in humans (33, 58). Kaposi’s sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) is associated with Kaposi’s sarcoma, body cavity-based lymphomas, and Castleman’s disease in humans (8, 11, 46, 65). Analysis of transcripts expressed by these primate viruses in tumors and latently infected cells has provided important information on both the mechanisms of pathogenesis for these viruses and the cellular machinery involved in host immune responses, cell cycle regulation, and cytokine signaling. The species specificity of primate viruses such as EBV and KSHV has, however, limited analysis of the role of these transcripts in vivo.
The availability of gammaherpesvirus 68 (γHV68 or MHV68), a murine virus well suited to pathogenesis studies, presents the opportunity to evaluate the role of individual gammaherpesvirus genes in a model amenable to both genetic and pathogenetic studies (76–78). Analysis of the γHV68 genome demonstrates that this virus is closely related to primate gammaherpesviruses, including EBV, KSHV, and HVS (21, 22, 76), but regions of the γHV68 genome transcribed during latency have not been defined. γHV68 is a natural pathogen of wild rodents (4, 44), capable of infecting both outbred and inbred mice (5, 44, 56, 71). In one study, a significant portion of mice infected with γHV68 developed lymphoproliferative disorders. Treatment with cyclosporine increased the frequency of lymphoproliferative disease (70). γHV68 infects multiple organs of inbred mice and can establish a latent infection in the spleen (5, 56, 71, 72, 77). Pending development of a molecular definition of γHV68 latency, we operationally define latency as the absence of preformed infectious virus, as measured by an assay of defined sensitivity, and the capacity to reactivate virus (77).
Two studies have suggested that B lymphocytes are the sole reservoir within the hematopoietic compartment for γHV68 (72, 75). In addition, a B-lymphoma cell line chronically infected with γHV68 has been isolated from an infected mouse (74). However, the issue of the cellular reservoir for latent virus within the lymphoid organs remains unclear since subsequent analyses have demonstrated efficient establishment of splenic latency in mice lacking mature B cells (77) and persistence of γHV68 DNA in lungs of B-cell-deficient mice (69, 75). The presence of latent virus in cells other than B cells was supported by a recent study suggesting that epithelial cells are a site of γHV68 persistence in lungs of B-cell-deficient mice (69). Data on latency sites in vivo therefore suggest that multiple cell types harbor latent virus. Consistent with this, members of our group have found that latent γHV68 can be found in spleen, bone marrow, and peritoneal exudate cells (PECs) (77, 78a) and identified macrophages as well as B cells as sites of γHV68 latency (78a). To further understand γHV68 latency and to generate a molecular, rather than functional, definition of latency for γHV68, we have performed experiments to identify regions of the γHV68 genome that are transcribed in spleens and peritoneal cells from latently infected mice.
MATERIALS AND METHODS
Viruses, tissue culture, and mice.
γHV68 was passaged and assayed as described previously (77). Experiments were performed with the γHV68 WUMS clone (American Type Culture Collection), which was doubly cloned by limiting dilution from a stock originally supplied by Peter Doherty and Tony Nash. This isolate has been sequenced in full (76). Plaque assays were performed with NIH 3T12 cells (CCL 164; American Type Culture Collection). γHV68 was handled with strict biosafety level 2 precautions, as γHV68 infects human cells (reference 4 and unpublished observations). NIH 3T12 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 100 U of penicillin/ml, 100 mg of streptomycin/ml, 10 mM HEPES, and 2 mM l-glutamine (complete DMEM). Murine embryonic fibroblasts (MEFs) were isolated and used as described previously (77). Mice were housed and bred at the Washington University School of Medicine at biosafety level 2 in accordance with all federal and university regulations. Sentinel mice screened negative every 2 to 4 months for adventitious mouse pathogens. B-cell-deficient MuMT mice on the C57BL/6 background (C57BL/6J-Igh-6tm1Cgn) were obtained from the Jackson Laboratory (Bar Harbor, Maine). B-cell-deficient mice were derived by interruption of the exon encoding the transmembrane domain of immunoglobulin M and are deficient in mature B cells (34).
Limiting-dilution analysis.
Assays for detection of latency and preformed infectious γHV68 were performed as previously reported (77). Briefly, serial twofold dilutions (24 wells/dilution) of stock γHV68 or of infected mouse splenocytes were plated on monolayers of 1 × 104 to 1.5 × 104 MEFs/well in 96-well tissue culture plates. MEFs were observed microscopically for viral cytopathic effect (CPE) for up to 4 weeks. To differentiate between latently infected cells and preformed infectious virus in cell samples, serial twofold dilutions of spleen cells or PECs were plated before or after mechanical disruption of viable cells. For mechanical disruption, cells were resuspended at a concentration of 1 × 106 to 5 × 106 cells/ml in hypotonic 1/3× DMEM in the presence of 100 μl of 0.5-mm-diameter zirconia-silica beads/ml and shaken twice for 1 min at high speed in a Mini-Beadbeater-8 (Biospec Products, Bartlesville, Okla.). Published studies have shown that mechanical disruption has at most a twofold effect on the infectivity of preformed infectious γHV68, while >99% of cells are killed (77). Thus, mechanical disruption has minimal effects on detection of preformed infectious virus in tissues but efficiently eliminates latent cells. The limiting-dilution assay is 5- to 10-fold more sensitive than plaque assay on NIH 3T12 cells for detecting preformed infectious γHV68 (77, 78a).
RNA samples for RT-PCR.
Adult B-cell-deficient mice were inoculated intraperitoneally (i.p.) with 106 PFU of γHV68 in 0.5 ml of complete DMEM, and peritoneal cells and spleens were harvested as previously described (28, 77). RNA used as a negative control for reverse transcriptase (RT)-PCR assays was isolated from uninfected thioglycolate-elicited PECs (52). Latent peritoneal cell RNA was derived from nonelicited peritoneal cells harvested from the following four groups of mice: (i) 7 mice 45 days after infection, (ii) 9 mice 46 days after infection, (iii) 28 mice 42 days after infection, and (iv) 19 mice 42 days after infection. Spleen RNA was harvested from mice 46 days after infection. Two separate pools of spleen RNA were made and analyzed, each containing RNA from three individual mouse spleens. RT-PCR was performed for every set of primers on at least two different pools of peritoneal cell RNA and on both pools of spleen RNA. RT-PCR data obtained from different pools of RNA were consistent.
RNA preparation and RT-PCR assays.
RNA was prepared by the method of Chomczynski and Sacchi (12, 54). Briefly, spleen or PEC samples were homogenized in 10 ml of GITC-phenol (mixed 1:0.1:1 with one of the following: [i] 4 M GITC–0.1 M β-mercaptoethanol–0.5% sarcosyl, [ii] 2 M sodium acetate [pH 4], and [iii] water-saturated phenol) and RNA was isolated. Spleens were homogenized with a Tissue Tearor (Biospec Products), while PECs were homogenized by repeated pipetting. Following initial RNA preparation, samples were resuspended in 25 μl of DMPC-treated water and treated for 15 min at 25°C with 2 μl of DNase I (2 U; Gibco BRL, Grand Island, N.Y.) per the manufacturer’s instructions. DNase-treated RNA was resuspended in 1 ml of GITC-phenol and reextracted by the same methods as described above. RNA was resuspended in 50 to 100 μl of DMPC-treated water and quantitated, and the DNase treatment was repeated. RNA (2 μg/experiment/primer set) was reverse transcribed by using the Superscript preamplification system for first-strand cDNA synthesis in a total volume of 20 μl (Gibco BRL), and a no-RT control was carried out in parallel. Individual nested PCRs were performed on 2 μl of the RT mixture in 1× reaction buffer (Promega, Madison, Wis.) containing 2.5 mM MgCl2, a 0.2 mM concentration of each nucleotide, a 0.15 μM concentration of each primer (see Table 1 for primer sequences), and 1.25 U of Taq DNA polymerase (Promega) in a final volume of 50 μl per PCR. Controls for each PCR included water controls, no-RT controls, and multiple samples containing 1 or 10 copies of the γHV68 genome diluted in 0.1-mg/ml tRNA (Sigma, St. Louis, Mo.) combined with cDNA made by RT treatment of RNA from uninfected mice. PCR was performed in a GeneAmp PCR System 9600 (Perkin-Elmer, Norwalk, Conn.). The initial round of PCR was performed with 25 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min. Two microliters of the initial reaction mixture was entered into the second round of each nested PCR. The second round of PCR was performed under the same conditions as described above for 35 cycles. Both PCR programs began with an initial denaturation at 94°C for 2 min and concluded with a final hold at 72°C for 5 min. PCR products were stored at 4°C until separation by electrophoresis on a 2% agarose gel. PCR products were visualized by staining with ethidium bromide.
TABLE 1.
Primers used in this study
| Genome region | Sequence and genome coordinate
|
Size (bp) of outer product | Sequence and genome coordinate
|
Size (bp) of inner product | ||
|---|---|---|---|---|---|---|
| Outer primer A | Outer primer B | Inner primer A | Inner primer B | |||
| K3 | GTCGCGATCGCCTCATCAATG, 24848 | GAGAGTTCTGTTGGATCTGC, 25322 | 475 | CGAGGACAACTTGCATTCTTGG, 24987 | AAAGATTCTGTGGCTGCAAGGG, 25280 | 294 |
| Gene 50 | GATGGAAACAGAAAACGAGCCC, 68351 | TCGCTTGTTTCTGGGGAGGTTT, 68775 | 425 | AAAAGTTCTGCATCCCAGACCC, 68409 | AGGGCTAATGGGTGAAAATGGC, 68701 | 293 |
| M8 | TGAGCTCTCAGATTCTCCAC, 76025 | CGCAGTTTACCAGTTGTAGG, 76450 | 426 | TTCTCCAAAGGCTAGAAAGCGC, 76145 | GGTTCCATCTGTTTGGTCTTCG, 76338 | 194 |
| ssDNABP gene | CAAGGCACCGTGTCAGAAAATC, 11400 | ATCATAGAAGGGCAGTAGCTGG, 11808 | 409 | CCATGCCATCGTCTTTCACAAC, 11463 | TGGTCACAGTTTTCATGGGTGC, 11728 | 266 |
| DNApol gene | AGTCAATTCAAGGGCAAGCG, 19727 | CAGGGAAAACAACAGCTTGGAG, 20221 | 495 | CAATTGCTGTATCCCATCTGCG, 19844 | GGAAACCCACATTCACCCAAAC, 20127 | 284 |
| M7 | GTCCGATCCCACCACCTCAG, 69753 | CGGACTGTCAGCGGTTTCAGG, 70239 | 487 | CCACCTCCAATGCAGATGTTTC, 69941 | ATTTTGGGAAGGGGTGGTTG, 70068 | 128 |
| gBa | CTGTTCGAACCACCGTTAAC, 17155 | TGTTTTCCAGTGCACCAGGTC, 17476 | 323 | ATTGTAGACATGGTGGCACGC, 17180 | TCTGGTGGCTGTTTTCCAGG, 17440 | 261 |
| MCP | AGATTTGAGGCGCTCCTGGG, 40407 | TTTACCAAACCCTCGCTGCG, 40883 | 477 | TATAGCGCACGGTGATGGAAGAAG, 40559 | GATTGGAGAATAAAGGTGGGCG, 40843 | 285 |
| M1 | ATGCAGCTGGCCACCTTATG, 2023 | CCATGGTGGGTAGTGGGAGTC, 2350 | 328 | AAAGCATAGCTCACTGGCCATC, 2066 | TCCAGCTTCTCGAAGGAATCAG, 2228 | 163 |
| M2 | TCGAGCCAGAGTCCAACATCAT, 4171 | CAAAATCCAGTGCTATGGGGTG, 4549 | 379 | TAAGGACCTCGTAGAGATTGGC, 4193 | ACGTTAAAGTCCCCATGGAAGC, 4336 | 144 |
| M3 | TGGCACTCAAACTTGGTTGTGG, 6566 | TAACAGGCAGATTGCCATTCCC, 6946 | 381 | ATTCTTCAGAGATGGCGCCAAG, 6593 | TCTTATGTGGATGTGGGCATGC, 6725 | 133 |
| M4 | AAAATCCATCGGCTGACCTTCG, 8643 | GCGACTTCCCACTGACTGGATC, 9025 | 383 | CACCCGCTGGAAAGATTTATCC, 8747 | GATGCAGGGAGATGAAAGTTGG, 8946 | 200 |
| M5 | GCATACAGCGTGGGAATGGG, 26184 | AAACAGGGGGAATATGTGGAGG, 26619 | 436 | CCCAGAGCAAACAACCAATCAG, 26308 | ATATTGGGGAAGCATCCGACAG, 26423 | 116 |
| M6 | ATGGCACACACCAACGGGACCCTCCA, 26554 | TCGGGAGGGGAACCCAAAAC, 26787 | 234 | TCCATTCTATAAACCCACGCCC, 26576 | CCAAAATGACCCAGACAAGCTC, 26742 | 167 |
| M9 | TAACCCTAAAGCTGTTCCCG, 94048 | AAAGACAGGGTCAAAGCTCCAG, 94513 | 466 | AGACAGGGTCCATCATTTTGGC, 94267 | TTGGCAAAGACCCAGAAGAAGC, 94425 | 159 |
| M11 | ATGAGTCATAAGAAAAGCGG, 103418 | TATATCCTGGGCAACACCTTCG, 103908 | 491 | AGAAAAGCGGGACTTATTGGGC, 103428 | TTTTCCAGTTCTTGAGGGCAGG, 103622 | 195 |
| M12 | TATGCGTGATACCAAACCTG, 118003 | CCTGGACTCTGGCTCTGGGG, 118455 | 453 | TATGCGTGATACCAAACCTGGC, 118003 | ATTTTCGTCTCCCGCGTCTT, 118188 | 186 |
| Gene 72 | TGTGATTAGCACTGGGCGTTTC, 102594 | TATCGCAGCGAAAGAGAACACG, 103109 | 516 | TTAGCACTGGGCGTTTCATG, 102599 | CACGTGGATGTTTTGTGTGTGC, 103023 | 425 |
| Gene 73 | TAGATCCAGGTGATCCTGTGGC, 103959 | CCGCATAATCCATCTGATCCAT, 104475 | 517 | TGTTGTGTGCCAGAAGCTTGTG, 104021 | TACCAGGCACAACACAACAGT, 104255 | 235 |
| Gene 74 | TCTGTGCTTGGAAATCTGACGG, 105234 | TTCTTACCCATTCTCATGCGCC, 105790 | 557 | GTGTTTTGTGCATATAGGGCGG, 105264 | TGGTCCCTCCTAGCTCCATAGC, 105672 | 409 |
Through a clerical error outer primer A for the gB region started with a C rather than a G. The genome number given is the first base in the primer that matches the γHV68 genome sequence (the T in position 2 of the primer). The predicted product is therefore 1 bp longer than predicted from the genome location of the primers.
Primers for RT-PCR and determination of PCR sensitivity.
Primers were designed (Table 1) by using vector NTI (Informax, Gaithersburg, Md.). In general we designed primers that had a G+C content of approximately 50% so that a single PCR program could be used for all primers. To determine the sensitivity of our nested PCR, we prepared the γHV68 genome as previously described (76). The genome copy number was calculated assuming a genome size of 150 kb and a molecular mass of 1.7 × 10−10 μg/molecule. Note that we assumed that the average γHV68 genome has approximately 150 kb, based on the unique sequence of 118,237 bp plus about 30,000 bp (or ca. 26 copies) of the 1,213-bp terminal repeats (76). While this is an arbitrary assignment, the KSHV genome has about 30,000 bp of terminal repeat sequence (ca. 35 to 45 copies of an 803-bp repeat) (37, 57, 59), and the γHV68 genome carries on average at least 10 repeats (22). Our estimate of the genome size, and therefore PCR assay sensitivity, is unlikely to be more than twofold off based on the assumptions used to calculate the genome copy number. We prepared stocks allowing us to add approximately 1 or approximately 10 copies of the γHV68 genome to samples containing cDNA made from naive mouse tissue RNA, and we then ran these samples in parallel with cDNA samples from latently infected tissues. We selected nested primer pairs that allowed consistent detection of one copy of the γHV68 genome in a background of cDNA (see Results and Table 2).
TABLE 2.
Detection of genome regions by PCR and RT-PCRa
| Genome region | No. of positives/no. of reactions with:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Naive tissue
|
PECs
|
Spleen cells
|
||||||||
| −RT | +RT | −RT | +RT | Sensitivity, 1 copy | Sensitivity, 10 copies | −RT | +RT | Sensitivity, 1 copy | Sensitivity, 10 copies | |
| K3 | 0/2 | 0/2 | 0/10 | 0/10 | 7/12 | 6/6 | 0/16 | 0/16 | 7/16 | 8/8 |
| Gene 50 | 0/2 | 0/2 | 1/28 | 6/32 | 12/36 | 17/18 | 0/16 | 0/16 | 8/16 | 8/8 |
| M8 | ND | ND | 0/8 | 1/8 | 5/16 | 8/8 | 0/16 | 1/16 | 13/24 | 8/8 |
| ssDNABP gene | 0/2 | 0/2 | 0/10 | 0/10 | 4/12 | 6/6 | 0/16 | 0/16 | 11/16 | 8/8 |
| DNApol gene | 0/6 | 0/6 | 0/22 | 0/22 | 11/20 | 12/12 | 0/16 | 2/16 | 8/16 | 8/8 |
| M7 | ND | ND | 0/8 | 0/8 | 6/16 | 8/8 | 0/16 | 0/16 | 9/16 | 8/8 |
| gB gene | 0/14 | 0/14 | 0/34 | 1/34 | 10/28 | 15/15 | 0/16 | 1/16 | 12/16 | 8/8 |
| MCP gene | 0/6 | 0/6 | 0/14 | 0/14 | 5/16 | 7/8 | 0/16 | 0/16 | 8/16 | 8/8 |
| M1 | 0/4 | 0/4 | 0/16 | 1/16 | 13/24 | 11/11 | 0/16 | 3/16 | 6/8 | 8/8 |
| M2 | 0/8 | 0/8 | 0/34 | 32/35 | 9/40 | 20/20 | 0/16 | 15/16 | 4/16 | 7/8 |
| M3 | 0/12 | 0/12 | 0/20 | 4/20 | 11/28 | 14/14 | 0/20 | 13/20 | 6/16 | 9/10 |
| M4 | 0/8 | 0/8 | 1/16 | 1/16 | 9/24 | 12/12 | 0/16 | 0/16 | 5/16 | 8/8 |
| M5 | 0/4 | 0/4 | 0/12 | 0/12 | 8/21 | 8/10 | 0/16 | 0/16 | 9/16 | 8/8 |
| M6 | 0/4 | 0/4 | 0/12 | 0/12 | 6/20 | 9/10 | 0/16 | 0/16 | 8/16 | 8/8 |
| M9 | 0/8 | 0/8 | 0/20 | 2/20 | 14/32 | 16/16 | 0/16 | 10/16 | 9/16 | 6/7 |
| M11 | 0/4 | 1/4 | 0/20 | 11/20 | 14/28 | 14/14 | 0/16 | 2/16 | 8/16 | 8/8 |
| Gene 72 | ND | ND | 0/16 | 4/16 | 14/24 | 12/12 | 0/16 | 1/16 | 7/16 | 8/8 |
| Gene 73 | 0/26 | 0/26 | 1/38 | 24/38 | 17/44 | 18/20 | 0/16 | 0/16 | 8/16 | 7/8 |
| Gene 74 | 0/8 | 0/8 | 0/18 | 16/23 | 19/40 | 20/20 | 0/16 | 2/16 | 7/16 | 7/8 |
| Total | 0/118 | 1b/118 | 3b/356 | ND | 194/481 | 233/240 | 0/308 | ND | 153/304 | 148/153 |
All positive RT-PCR results for experimental samples are presented in bold. RNA preparations are described in Materials and Methods. −RT, without transcriptase; +RT, with transcriptase. ND, not determined.
False positives.
Isolation of RNA from lytically infected fibroblasts and Northern blotting.
NIH 3T12 fibroblasts (2 × 107 cells/T175 flask) were pretreated with drugs as outlined below for 1 h and then infected at a multiplicity of infection of 7 in a volume of 25 ml containing drugs, and flasks were incubated at 37°C in a 5% CO2 atmosphere for 12 or 18 h prior to harvest of RNA. Flasks were either left untreated or treated with drugs as follows: (i) γHV68 DNA synthesis was inhibited by adding phosphonoacetic acid (PAA) at a final concentration of 200 μg/ml, and (ii) protein synthesis was inhibited by adding a combination of cycloheximide at a final concentration of 40 μM and anisomycin at a final concentration of 10 μM. Total cellular RNA was harvested as above and analyzed by Northern blot hybridization (60). Probes were generated by cloning the PCR product of the outer sets of primers for each genome region (Table 1). All probes were sequenced to confirm their identities. In this way Northern probes encompassing the region of the γHV68 genome evaluated for transcription in latently infected tissues were obtained. Blots were probed for rat cyclophilin (13) to assess loading and RNA quality. All probes were radiolabeled with the Megaprime DNA labeling system (Amersham, Arlington Heights, Ill.) per the manufacturer’s protocol. To provide an internal control on each blot for differences in probe-specific activity, we dotted 40 ng of γHV68 genomic DNA prepared as described previously (76) onto membranes prior to hybridization with labeled probe.
RESULTS
Selection of candidate regions of the genome to evaluate for transcription during latent infection.
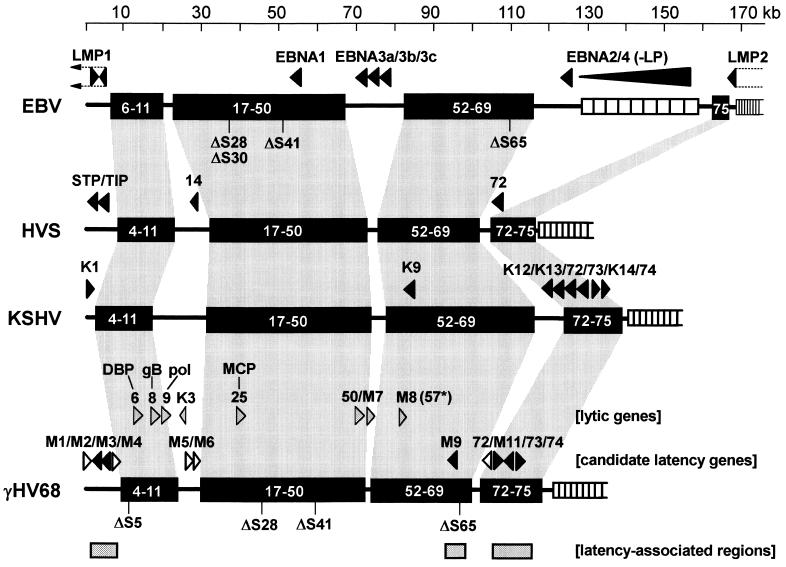
As there are currently no tightly latent γHV68-infected cell lines available, we evaluated viral transcription in spleen and peritoneal cells isolated from latently infected mice. Viral gene expression during gammaherpesvirus latency has been thoroughly characterized for EBV, and tumor- and latency-associated genes of KSHV and HVS have also been identified (Fig. 1). Although there are no clear homologs for many of these genes in γHV68, most of the tumor- or latency-associated genes of EBV, HVS, and KSHV map either between the conserved regions of the viral genomes or between KSHV and HVS genes 72 to 74 (Fig. 1), suggesting that the equivalent regions of γHV68 may also contain genes associated with latent infection or tumor induction.
FIG. 1.
Gammaherpesvirus latency-associated and transforming genes. The map positions of viral genes implicated in latency and/or cellular transformation by the human and primate gammaherpesviruses EBV, KSHV, and HVS are shown. The blocks of conserved viral genes present in the genomes of all, or most, gammaherpesviruses are indicated by the black rectangles. Genes missing from these blocks are designated as ΔS below- the black rectangles. The identification and characterization of EBV gene transcription in latently infected B cells and EBV-associated tumors are reviewed in references 33 and 58. Transcription of the HVS genome in latently infected T cells and T lymphomas has only partially been characterized but has revealed a role for the STP and Tip genes (25, 31, 42, 48) and gene 14 (18) and expression of gene 72 (the v-cyclin gene) (30). Transcription analyses of latently KSHV-infected primary effusion lymphoma cell lines has led to the identification of a latency locus containing KSHV K12 (47, 62, 79, 80), K13 (17, 62), gene 72 (14, 17, 62), gene 73 (17, 32, 55, 62), and K14 (62). In addition, gene K1 (37, 38), gene K9 (24), and gene 74 (3) have been shown to have transforming activity and may be expressed during viral latency. The indicated candidate latency-associated γHV68 genes were chosen based on either their location in the viral genome or homology to KSHV latency- or tumor-associated genes. The genome regions containing candidate γHV68 latency-associated genes shown by filled arrowheads scored strongly positive in the RT-PCR analysis of latently infected cells (Table 2). The locations of several known γHV68 lytic-cycle-associated genes, which were used as controls for the presence of lytic gene transcription, are shown (Table 2; see also Discussion). Transcription of γHV68 gene 8 (encoding gB) (68) and gene M7 (encoding gp150) (40) has been characterized. In addition, ORF M8 is contained within an early transcript (40).
Of the 80 ATG-initiated open reading frames (ORFs) (with products (of ≥100 amino acids in length) identified in the primary sequence of the γHV68 genome (76), 40 (including M7 and M8) have products that are homologous to tegument proteins, structural proteins, or other proteins known or suspected to function during the lytic cycle. Of these, we screened 8 for expression in latently infected tissue (Tables 2 and 3). This afforded a direct assessment of lytic gene transcription in latently infected tissue. Of the remaining 40 ORFs in γHV68, 23 are homologous to ORFs in other gammaherpesviruses but are of unknown function. We did not examine these ORFs in this study, although they may encode proteins expressed during latency (62). Of the remaining 17 ORFs, we selected 11 to screen for expression in latently infected tissue (Tables 2 and 3). Four of these are in a cluster of genes {M11 (encoding a protein containing a BH1 domain similar to bcl-2 family members, v-bcl-2), gene 72 (encoding a D-type cyclin, v-cyclin), gene 73 (homolog of the KSHV latency-associated nuclear antigen [LANA] gene [32, 55]), and gene 74 (encoding a G-protein coupled receptor homolog, v-GCR)} which are known to be, or suspected of being, involved in KSHV or HVS latency and/or tumorigenesis (Fig. 1 and Discussion). The other seven putative γHV68 ORFs that we targeted (ORFs M1, M2, M3, M4, M5, M6, and M9) do not have homologs in the known gammaherpesviruses but are located in regions of the γHV68 genome positionally homologous to regions in other gammaherpesviruses that contain genes expressed either in latently infected cell lines or in tumors (Fig. 1) (76). Six other ORFs unique to γHV68 (ORFs M10a, M10b, M10c, M12, M13, and M14) span either the G+C-rich 100-bp repeat region or the G+C-rich terminal repeats, making design of PCR primers difficult, and thus we did not screen for the presence of transcripts arising from these regions of the viral genome.
TABLE 3.
Characteristics of genome regions studieda
| Genome region | Lytic transcription | Transcribed in latent peritoneal cells | Transcribed in latent spleen cells | Latency candidate | Comment(s) and references |
|---|---|---|---|---|---|
| K3 | ND | No | No | No | BHV IE gene and KSHV K3 and K5 homolog (59, 76) |
| Gene 50 | IEb | No | No | No | EBV R transactivator homolog (27, 76) |
| M8 | E | No | No | No | Region transcribed with early kinetics (40) |
| ssDNABP gene | E | No | No | No | DNA replication (76) |
| DNApol gene | E | No | No | No | DNA replication (76) |
| M7 | ND | No | No | No | Envelope glycoprotein gene (67) |
| gB gene | L (Fig. 3) | No | No | No | Glycoprotein gene (68) |
| MCP gene | L (Fig. 3) | No | No | No | Capsid gene (76) |
| M1 | Yesc | No | No | No | Poxvirus serpin gene homolog, M3 homolog (6, 64, 76) |
| M2 | Low (Fig. 3) | Yes | Yes | Yes | No known homologs |
| M3 | Yes (Fig. 3) | No | Yes | Yes | No known homologs, secreted protein (75a), M1 homolog (6) |
| M4 | ND | No | No | No | No known homologs |
| M5 | ND | No | No | No | No known homologs |
| M6 | ND | No | No | No | No known homologs |
| M9 | Yes (Fig. 3) | No | Yes | Yes | No known homologs |
| M11 | Low (Fig. 3) | Yes | No | Yes | v-bcl-2 gene (EBV, KSHV, HVS) (76) |
| Gene 72 | Yesd | No | No | No | v-Cyclin gene (HVS, KSHV) (76) |
| Gene 73 | Low (Fig. 3) | Yes | No | Yes | KSHV LANA gene homolog (32, 55, 76) |
| Gene 74 | Low (Fig. 3) | Yes | No | Yes | Interleukin-8 receptor gene homolog (KSHV, HVS) (76) |
Development of sensitive nested RT-PCR assays for detection of γHV68 transcripts in tissue RNA.
To assess transcription of candidate γHV68 latency-associated genes, we developed sensitive RT-PCR assays for detection of γHV68 transcripts in RNA samples from infected tissue. We designed primers (Table 1) meeting the following criteria: (i) they are approximately 50% G+C, (ii) the outer primers amplify a region 300 to 500 bp in length, (iii) independent inner primers amplify a region 100 to 400 bp in length, and (iv) amplified sequences are completely within ATG-initiated ORFs whose products are at least 100 amino acids in length (76). These criteria were selected to optimize RT-PCR sensitivity by amplifying only short regions of RNA transcripts derived from regions containing candidate genes. This approach, which does not provide information on the direction of transcription or the structure of the RNA detected, serves to identify regions of the genome that are transcriptionally active.
To define the sensitivity of each nested set of primers, we prepared dilutions of the γHV68 genome calculated to contain either 1 or 10 genome copies (see Materials and Methods). To measure sensitivity under RT-PCR conditions, we performed all sensitivity assays in the presence of cDNA derived by reverse transcription of RNA isolated from uninfected mouse tissues (see Materials and Methods). Using this approach we generated 19 nested PCR primer sets that reproducibly detected 1 copy and 10 copies of the γHV68 genome (Table 2). Note that our false-positive rate (PCR products present after PCR of RNA samples without added RT or using cDNA generated from uninfected mice) was 4/900 PCRs, or 0.4% (Table 2). Three of four false-positives were seen in RNA derived from peritoneal cells (no RT), a site of latency that contains the highest frequency of γHV68 genome-positive cells of any tissue evaluated in our laboratory (78a). This raises the possibility that these false positives may have been due to very low levels of γHV68 DNA in RNA preparations from peritoneal cells, despite treatment with DNase prior to reverse transcription. To provide meaningful RT-PCR results we (i) performed multiple independent RT-PCRs (usually four to eight per condition) within each experiment, (ii) included data only from assays in which the sensitivity was 1 to 10 copies of genome, (iii) used cDNA derived from at least two independent RT reactions from each RNA preparation, and (iv) analyzed RNA isolated from multiple different groups of animals (Table 2; see also Materials and Methods).
Minimal expression of lytic transcripts in tissues from latently infected mice.
Previous studies have shown that γHV68 efficiently establishes latency in the spleens of B-cell-deficient mice after i.p. inoculation of γHV68 (77). Peritoneal cells were also selected for analysis in this study since it has been shown that they are also tightly latent and regularly carry approximately about 50- to 100-fold more cells that reactivate γHV68 than spleen cells (77, 78a). The definition of latency that members of our group have used in earlier studies is based on the use of a sensitive cocultivation assay that is approximately 5- to 10-fold more sensitive than the standard plaque assay for detecting preformed infectious γHV68 in tissue samples (77, 78a). Latency is defined as the presence of cells that can reactivate γHV68 in the absence of preformed infectious γHV68. The presence of preformed infectious γHV68 is determined by the presence of virus after mechanical disruption of live cells by the previously described cocultivation assay (77). Mechanical disruption of cells removes any contribution of reactivation of latent virus, since the latter requires live cells. Importantly, this is an operational definition of latency and has not been independently validated by evaluating γHV68 lytic gene transcription. For this reason we first examined transcription from regions of the genome predicted or known to encode lytic gene products in RNA from tissues operationally defined as latently infected (i.e., no detectable preformed virus).
RNA was prepared from latently infected peritoneal cells and spleens isolated 42 to 46 days after i.p. infection with γHV68. At this time both peritoneal cells and spleen cells did not contain any detectable preformed virus but did contain cells that reactivated γHV68 (Fig. 2) (77, 78a). To determine whether regions of the genome that encode lytic transcripts are transcriptionally active in latent tissues, we used RT-PCR targeted to genome regions (Tables 2 and 3) containing (i) two putative immediate-early genes (K3 and gene 50; the latter is predicted to encode a homolog of the EBV BRLF1 transactivator [27, 76]), (ii) three early genes (gene 6, encoding the ssDNA binding protein [ssDNABP]; gene 8, encoding the DNA polymerase [DNApol]; and M8, which is contained within the region encoding two early mRNAs, including a spliced transcript [40]), and (iii) three late genes (gene 25, encoding the major capsid protein [MCP]; gene 7, encoding glycoprotein B [gB] [68]); and M7, encoding glycoprotein 150 [gp150], a known virion protein [67]). We did not detect significant expression of seven of these lytic genes (K3, the ssDNABP gene, the DNApol gene, M8, the MCP gene, the gB gene, or M7) in either spleen or peritoneal cell RNA (Table 2). However, we did detect at a low frequency transcripts derived from the gene 50 region in peritoneal cells but not spleen cells (Table 2). The frequency of detection did not meet our conservative criterion for selecting a gene as a candidate latency gene (see below). Expression of a homolog of the EBV BRLF1 transactivator might reflect abortive reactivation or the presence of a very low level of RNA transcribed from the region of gene 50 during latency. Overall, the lack of significant transcription from multiple regions of the genome containing lytic genes substantiates our operational definition of latency and sets the stage for evaluation of transcription from other regions of the genome in latently infected tissue.
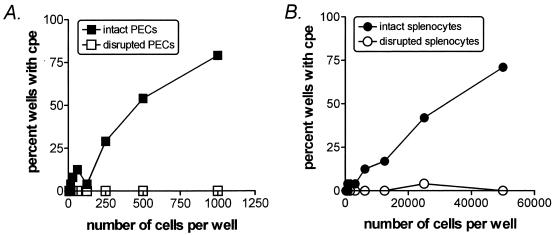
FIG. 2.
Limiting-dilution reactivation analysis of γHV68 from spleen and peritoneal cells. Shown are representative examples of limiting-dilution analysis performed on intact or mechanically disrupted PECs or spleen cells. B-cell-deficient mice were infected with 106 PFU of γHV68 i.p. At 42 or 45 days postinfection, animals were sacrificed and PECs and spleens were harvested. A portion of pooled PECs or spleen cells from infected animals was subjected to limiting-dilution reactivation analysis. RNA was prepared from the remaining cells and subjected to RT-PCR (Table 2). For the limiting-dilution assay, intact or mechanically disrupted cells were plated in twofold dilutions at the numbers indicated; replicates of 24 wells were plated onto MEF monolayers and scored for viral CPE at 2 and 3 weeks.
Identification of genome regions transcribed in latent tissues.
Because we were successful in developing nested PCR assays which exhibit single-copy sensitivity, and given the very low false-positive rate of 0.4%, we elected the conservative criterion that at least 50% of RT-PCR assays for transcription from a region of the γHV68 genome must be positive in order for us to make the determination that a specific genome region contains a candidate latency gene (Table 2). This conservative criterion will likely result in us failing to call attention to bona fide candidate latent transcripts, but we feel that this is preferable to falsely selecting candidate latency genes for further analysis.
Analysis of RNA prepared from spleen and peritoneal cells harvested from uninfected mice did not result in significant positive reactions for any of the PCR primers tested (Table 2). In contrast, more than 50% of PCRs were positive when peritoneal cell RNA was tested for transcription arising from the regions of ORF M2, ORF M11, gene 73, and gene 74. Transcription from the region of gene M2 was also detected in RNA from spleens of latently infected mice. Spleen RNA also consistently scored positive for transcription from the region of ORF M3 and ORF M9 but not ORF M11, gene 73, or gene 74. We detected transcripts from other areas of the genome in either spleen or peritoneal cell RNA at frequencies lower than 50% (Table 2). There was a general correlation between genes that met our criteria of >50% positive reactions in one tissue and low levels of positives in the other latent tissue. For example, the M3 region scored positive in 13 of 20 reactions with RNA prepared from latently infected spleen and 4 of 20 reactions with RNA prepared from latently infected peritoneal cells, and gene M11 scored positive in 10 of 16 reactions in latent spleen and 2 of 20 reactions in latent peritoneal cells. These data may suggest that the M11 and M3 genome regions are transcribed in latent peritoneal cells in addition to latent spleen cells. However, we do not believe that the data are conclusive given the small number of positive reactions in the setting of RT-PCRs with a sensitivity of one copy of the genome and the fact that we have no way of assessing the efficiency of reverse transcription of these transcripts.
Analysis of γHV68 gene expression in lytically infected fibroblasts by Northern blotting.
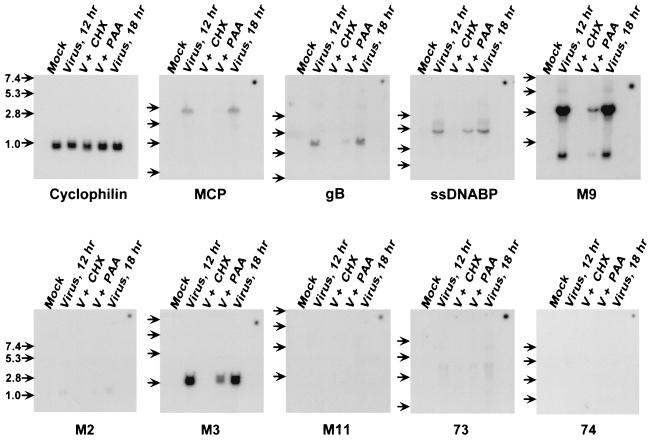
The above-described analysis identified ORFs M2, M3, M9, and M11 and genes 73 and 74 as regions of the viral genome that are actively transcribed during latency and thus as candidate latency genes. To further evaluate these candidates we analyzed transcription of these putative viral genes in lytically infected fibroblasts (Fig. 3 and Table 3). Our working assumption is that most viral genes expressed during latent infection will either not be transcribed or be transcribed only at a low level in lytically infected cells. To assess transcription of the candidate latency genes in lytically infected fibroblasts, Northern probes were derived by PCR amplification with the outer primers listed in Table 1. Thus, in all cases, the probe used for Northern analysis fully encompassed the region analyzed by RT-PCR (Tables 1 and 2). To allow comparison of relative transcript levels of Northern blots hybridized with different probes, we spotted 40 ng of denatured viral genome onto each blot to provide an internal hybridization standard (see Materials and Methods and the legend to Fig. 3). RNA for Northern analysis was prepared from mock-infected cells and cells infected at a multiplicity of infection of 7 and harvested at 12 h in the presence or absence of either cycloheximide and anisomycin (will detect transcription only of immediate-early genes) or PAA (will detect transcription of immediate-early and early genes) and at 18 h without drug treatment. These times were based on preliminary Northern analyses demonstrating that γHV68 late genes are expressed by 12 to 18 h in lytically infected fibroblasts (data not shown).
FIG. 3.
Northern blot analysis of γHV68 transcription. Permissive murine fibroblasts were mock or γHV68 infected for the times indicated and total cellular RNA was harvested for northern blot analysis. Cells were left untreated (12 or 18 h) or treated with a combination of cycloheximide and anisomycin (V + CHX) or PAA (V + PAA) as described in Materials and Methods. Multiple blots were stripped and reprobed with probes derived from the outer primers listed in Table 1 for the genes indicated beneath each panel. A probe for cyclophilin was used to ensure equal loading and integrity of RNA in each lane, and a representative cyclophilin blot is shown. The dot in the upper right corners of panels probed for various γHV68 genes is 40 ng of purified γHV68 DNA and provides an internal control for the exposure. Migration of 7.4-, 5.3-, 2.8-, and 1.0-kb molecular size standards is indicated by the arrows to the left of each panel.
Northern analyses assessing lytic gene expression demonstrated that the ssDNABP gene is an early lytic gene (transcript detected in the presence of PAA), while both the MCP and gB genes are transcribed as late genes (inhibited by PAA) (Fig. 3). Northern analysis showed that the DNApol gene is an early gene (data not shown). Notably, abundant lytic transcripts were detected when probes within two of the candidate latency-associated ORFs, M3 and M9, were used (Fig. 3). Both of these ORFs were transcribed with early kinetics, although the level of M9-hybridizing transcripts was significantly inhibited by PAA, suggesting that this is an early-late gene. In contrast to the results obtained with the M3 and M9 probes, low or undetectable levels of transcripts were obtained with the M2, M11, gene 73, and gene 74 probes (Fig. 3). Comparison of Northern analysis data (Fig. 3) with RT-PCR data from latent tissues (Table 2) revealed that (i) regions of genes M2, M11, 73, and 74 are transcribed in latent tissues but are not transcribed to a high level during lytic replication in fibroblasts, and (ii) the genes for MCP, gB, ssDNABP, and DNApol are all transcribed to detectable levels in lytically infected fibroblasts but are not transcribed to a detectable level in latent tissues.
DISCUSSION
In this study we made the following important observations (summarized in Table 3). First, we validated our operational definition of in vivo γHV68 latency by demonstrating that a different program of gene expression operates in latent tissues than is seen in lytically infected fibroblasts. Second, we have identified three distinct regions of the γHV68 genome that are transcriptionally active during latency (Fig. 1). The location of these regions correlates well with regions in KSHV, EBV, and HVS that contain known latency and/or tumor-associated genes, and some of the ORFs in these regions are homologous to known latency- or tumor-associated genes of EBV, HVS, or KSHV. This provides support for the use of γHV68 in a small-animal model for evaluating the molecular mechanisms underlying the latency of primate gammaherpesviruses. Third, we present data consistent with the idea that multiple programs of latency may operate in vivo, with different sites showing different patterns of transcriptional activity. Fourth, we have shown that several of the candidate genes that appear to be transcribed during latency are only weakly transcribed in lytically infected fibroblasts.
Operational definitions of γHV68 latency.
Analysis of transcription during in vivo latency depends on a quantitative and consistent operational definition of latency. Latency is the presence of the γHV68 viral genome in a reactivable form in the absence of ongoing production of preformed infectious virus. A critical aspect of this operational definition of latency is that the absence of preformed virus (a negative result) is of necessity interpretable only when information about the sensitivity of the assay used, and about the survival of preformed virus in samples processed for analysis, is provided. Several groups have operationally defined γHV68 latency in tissues by using different assays to detect preformed infectious virus. Initial studies used a plaque assay to test for preformed infectious virus, and investigators argued that virus detected as plaques by 5 to 7 days in an infectious-center assay reflected latently infected cells reactivating γHV68 since plaque assays failed to show infectious virus (71). With this assay, infectious centers were detected well after preformed infectious γHV68 was cleared from organs as measured by plaque assay, and several useful studies were performed using this criterion (7, 20, 61, 71, 72, 75). However, studies in the murine cytomegalovirus (MCMV) system from multiple groups have shown that typical plaque assays for MCMV lack sufficient sensitivity for preformed infectious virus in disrupted organs to be a criterion for latency (36, 43, 53). Similarly, two findings in the γHV68 system invalidate plaque assay results as a criterion for defining latency. First, using a limiting-dilution assay, infectious γHV68 has been detected in tissues after clearance of infectious virus as measured by plaque assay (77). Second, a recent study detected linear γHV68 genomes (demonstrated to be from lytic replication based on studies using a viral DNA synthesis inhibitor in vivo) in the lung (69) when standard assays for infectious virus fail to detect infectious virus (71, 75, 77). These studies show that plaque assays for γHV68, as has been shown for MCMV, are not sufficiently sensitive to provide a criterion for latency.
An alternative definition of latency has been the presence of cells staining by in situ hybridization with a probe specific for the γHV68 tRNA-like transcripts (6). While γHV68 tRNA-positive cells were observed in this study on day 21, the lack of preformed infectious virus was addressed by the failure to demonstrate viral DNA by in situ hybridization with a probe within the terminal repeats (a negative result without quantitative data on sensitivity) and reliance on previous studies showing by plaque assay that virus was cleared (6). In another study, in situ hybridization for transcripts from the glycoprotein H and thymidine kinase genes failed to demonstrate lytically infected cells despite the fact that sensitive PCR analysis demonstrated the presence of linear genomes and lytic replication (69). While the sensitivity of in situ hybridization for the viral terminal repeats versus transcripts from lytic genes has not been addressed, these data provide a clear caution against using the absence of a signal in in situ hybridization as a criterion for the lack of lytic replication. In addition, the viral tRNA-like genes of γHV68 are abundantly transcribed during lytic infection, and thus expression of the viral tRNA-like genes is not specific for latency (6). Given the limitations of the plaque assay (see above), the fact that in situ hybridization for viral DNA is of unknown sensitivity in tissue sections, and the lack of specificity of viral tRNA-like gene expression for latently infected cells, expression of the viral tRNAs as a marker for latent cells will require further substantiation.
Members of our group have used a different approach to assess organs for preformed infectious virus (77). In this assay cells from tissue are serially diluted onto permissive monolayers, and the presence of infectious virus is detected by CPE that occurs over 2 to 4 weeks in culture (a significantly longer time than required for detection of plaques formed by infectious centers [see above]). To distinguish preformed infectious virus from latent virus, cells are mechanically disrupted and diluted in parallel with live cells. This assay is 5- to 10-fold more sensitive than the plaque assay for detecting preformed infectious γHV68 in tissue, and controls have been performed to ensure that virus is not inactivated during tissue preparation (77, 78a). The major advantage of this assay is that it affords a direct comparison of the presence of preformed infectious virus and virus reactivation from latency, since the same assay with a defined sensitivity is used for both analyses. Thus, the detection of viral CPE upon plating live cells in the absence of viral CPE upon plating mechanically disrupted cells unambiguously demonstrates the presence of virus reactivation from latency. Using this assay, we have shown that γHV68 establishes latency in the absence of mature B cells (Fig. 2) (77), a finding confirmed and extended in subsequent studies from other groups (69, 75). Notwithstanding the sensitivity of this assay and that of others, such as PCR detection of linear genomes (69), all of the criteria for latency used to date depend on the sensitivity of assays to detect a negative result (no preformed virus, no lytic transcripts by in situ hybridization, and no linear genomes), and a positive molecular definition of γHV68 latency has been lacking.
Molecular definition of γHV68 latency.
Based on the limitations of the above-described assays, we embarked on generating a molecular definition of γHV68 latency. This study represents the first step in this direction. We felt that the strictest criteria that could apply to defining γHV68 latency were (i) the absence of preformed infectious virus as determined by the most sensitive assay currently available for detecting infectious γHV68 in tissue (limiting-dilution analysis [77]), (ii) the absence of lytic gene transcription from multiple regions of the viral genome in sensitive nested RT-PCR assays, and (iii) the identification of genome regions preferentially expressed in latent tissues as opposed to lytically infected cells.
Using our nested RT-PCR assays we detected three distinct regions of the genome that are transcribed in latent spleen and/or peritoneal cells (Tables 2 and 3 and Fig. 1). It is important to note that the RT-PCR screen does not unambiguously identify transcribed genes since (i) the RT-PCR assay does not distinguish which strand of the viral genome is transcribed, and (ii) we have assessed only the presence of short, unspliced regions of transcript which may or may not correspond to the ORF for which the PCR primers were designed. Thus, extension of our results will require that transcripts be identified. Furthermore, transcription in latent tissues does not prove that a specific transcript or genome region is important for latency. Mutagenesis studies with quantitative analysis of the latent genome and reactivation from latency will be required to prove relevance to latency, as opposed to mere association of transcription of a region with latency. Finally, this analysis does not provide any insight into what fraction of cells harboring the latent genome express the transcripts detected or how gene transcription relates to establishment of or reactivation from latency.
Notwithstanding these limitations, the data presented here provide support for our definition of latency, validate the use of the limiting-dilution assay for quantitating latently infected cells, and provide an initial molecular definition of γHV68 latency. We believe that we have detected true latency in vivo for several reasons. First, minimal transcription from multiple regions of the genome containing known lytic genes was detected in latent tissues by nested PCR assays with one-copy sensitivity (Tables 2 and 3 and Fig. 1). While the absence of transcription of a single lytic gene might be ignored based on the idea that there could be a problem with the assay, we believe that lack of transcription of seven different lytic genes strongly argues against the presence of significant lytic gene expression in the latently infected PEC and splenocyte populations. In contrast to results obtained by RT-PCR with RNA isolated from latent tissues, transcripts from these lytic genes was readily detected by Northern blot analysis of RNA isolated from lytically infected fibroblasts (Fig. 3). Notably, transcripts from several genome regions (gene 73, gene 74, M11, M2) were detected in latent tissues (Table 2), despite the fact that these regions were inefficiently transcribed in lytically infected fibroblasts (Fig. 3). In addition, we identified transcription in latent tissue from two regions of the γHV68 genome that contain genes (gene 73 and M11) homologous to genes transcribed during either EBV or KSHV latency (see below). Together, these data strongly argue that γHV68 has at least one latent program of gene expression that is distinct from the lytic program, and they provide for the first time a positive criterion (the presence of transcription from genome regions containing M2, M11, gene 73, and gene 74) for detecting latency.
We also detected transcription from regions of the genome encoding the M3 and M9 ORFs in latently infected tissues (Tables 2 and 3). However, in contrast to the situation with M2, M11, gene 73, and gene 74, the M3 and M9 genome regions are abundantly transcribed during lytic infection. This clouds selection of M3 and M9 as candidate latent genes. Notably, in EBV latent transcripts span multiple lytic genes, with the latency-associated antigens translated from mRNAs in which the regions containing the lytic-cycle genes have been spliced out. Thus, it is possible that latent transcripts exist for γHV68 that span lytic genes contained in the M3 and M9 regions. This possibility is supported by preliminary data showing that the M3 gene encodes an abundant protein that is secreted from lytically infected cells (75a). Further analysis of possible latent genes in the regions of the M3 and M9 ORFs will therefore require definition of latent transcript structure.
We were interested to see that the pattern of gene regions transcribed to detectable levels in latent tissue differed for peritoneal cells and spleens (Table 2). These apparent differences may be related to different loads of latent cells or to differences in efficiency of RT for specific mRNAs in RNA derived from different organs. An alternate possibility is that γHV68 has multiple latent gene programs (as has been demonstrated for EBV) or that multiple different cell types, each having a different gene program, carry γHV68 during latency. Interestingly, data for latency in both B cells and an additional cell type (not B cells) in the lung have been presented (69, 72, 77), and members of our group have obtained data consistent with γHV68 latency in both B cells and macrophages (78a). These studies support the idea that there are multiple sites of γHV68 latency, and they support the need for evaluation of γHV68 latent gene transcription in different cell types in vivo.
ORFs in regions of the viral genome actively transcribed in latent tissue.
Several of the candidate latency-associated genes (M2, M3, and M9) encode putative products with no clear homologs in the database (as determined by advanced Blast search of the nonredundant database at www.ncbi.nlm.nih.gov/BLAST/ in September 1998). These regions are particularly interesting because they may encode proteins with novel functions that are important for persistence of viral infection in the face of active immunity. However, three of the candidate latency-associated genes are predicted to encode proteins homologous to viral antigens encoded by EBV, KSHV, and/or HVS that have been studied in some detail.
v-bcl-2.
Data presented here demonstrate that the region of the γHV68 genome containing M11, predicted to encode a protein homologous to proteins of the bcl-2 family that regulate cell death, is transcriptionally active during latency. The putative γHV68 ORF M11-encoded v-bcl-2 protein is homologous to the v-bcl-2 proteins of EBV (BHRF1), KSHV (encoded by gene 16), and HVS (encoded by gene 16) (76). There has been consistent agreement from a number of studies that the gammaherpesvirus v-bcl-2 proteins inhibit apoptosis (10, 15, 23, 29, 49, 63, 73). However, the function of the v-bcl-2 proteins during viral infection is not clear, since the BHRF1 protein is not required for either viral replication or B-cell transformation in vitro (39, 41). There are four recognized domains that are conserved between different bcl-2 family members (BH1 to BH4). The γHV68 v-bcl-2 protein has a recognizable BH1-like domain but differs significantly from host and v-bcl-2 family members in regions corresponding to BH2 to BH4 (10, 76). In addition to the BH1 domain, γHV68 v-bcl-2 shares with the EBV, KSHV, and HVS proteins a hydrophobic C-terminal domain potentially involved in membrane localization. The preservation of these structural motifs across gammaherpesvirus v-bcl-2 molecules argues that these regions are functionally important.
Expression of v-bcl-2 genes during the lytic cycle has been documented for EBV, KSHV, and HVS. For KSHV, transcripts derived from the region of gene 16 (encoding v-bcl-2) have been detected in latently infected cell lines and tumors in one study (63). However, there is general agreement that KSHV v-bcl-2 is expressed as part of the lytic cycle (10, 62, 63). Similarly, HVS v-bcl-2 is expressed in lytically infected cells (35). Similarly for EBV, the BHRF1 gene is expressed as an early gene in lytically infected cells or lymphoblastoid cell lines induced into the lytic cycle (29, 39, 41, 51). African swine fever virus v-bcl-2 is expressed in lytically infected cells (50). While expression during the lytic cycle is clearly present for multiple viruses, BHRF1 may also be expressed in latently infected cells and tumors (2, 23), and transcriptional analyses shows that the structure of the mRNA encoding BHRF1 differs for lytic and latent infection (2). For γHV68, minimal transcription of the M11 ORF, encoding v-bcl-2, was detected in lytically infected fibroblasts, while RT-PCR detected transcription of this region in latent tissue. While low levels of expression of γHV68 v-bcl-2 may be critical to lytic replication, our data suggest that γHV68 v-bcl-2 may play an important role during latency.
Gene 73 (encoding LANA).
Gene 73 is shared among γHV68, KSHV, and HVS. The KSHV gene 73 protein (LANA is expressed in spindle cells of Kaposi’s sarcoma [9, 32, 55]). It is important to note that the amino acid homology among the products of gene 73 of γHV68, HVS, and KSHV is weaker than the homology seen between the majority of lytic genes conserved in these viruses (76). In particular, γHV68 gene 73 lacks a long complex repeat present (albeit with different structures and differing lengths) in both KSHV and HVS. The function of this repeat is not known. KSHV gene 73 and the KSHV v-cyclin gene are transcribed from a single promoter region as a bicistronic message and a shorter spliced form encoding v-cyclin (17). Thus, there is coregulation of the transcription of the KSHV v-cyclin gene and gene 73, suggesting that these genes play important roles in the same part of the viral life cycle. There is some upregulation of the KSHV v-cyclin-specific mRNA by treatment with phorbol esters (17) (perhaps consistent with a role for KSHV v-cyclin in lytic infection), but data for expression of both the KSHV v-cyclin gene and KSHV gene 73 in tumor cells is strong (9, 14, 32, 55). Thus, KSHV gene 73 and the KSHV v-cyclin gene, which are derived from the same promoter, are clear latent genes. The situation for γHV68 appears to be different in two respects. The γHV68 v-cyclin gene is abundantly expressed in lytically infected fibroblasts (75b), while we detected little or no transcription from the region of gene 73 in lytically infected fibroblasts (Fig. 3). Thus, there is independent regulation of γHV68 gene 73 and the γHV68 v-cyclin gene in lytically infected cells. The genome structure of γHV68, which has the v-bcl-2-encoding M11 gene interposed in the opposite orientation between gene 73 and the v-cyclin gene, provides a further basis for believing that the γHV68 v-cyclin gene and gene 73 may be independently regulated (76). In addition, the RT-PCR analysis of transcripts corresponding to the γHV68 v-cyclin gene region did not meet our conservative criteria for designating the v-cyclin gene a candidate latency gene (Table 2), while gene 73 did meet these criteria. This may reflect a preferential role of the γHV68 v-cyclin gene in lytic infection, although inefficient detection of γHV68 v-cyclin mRNA in latent tissues could be due to poor efficiency of reverse transcription of this mRNA. Further studies evaluating the role of the γHV68 v-cyclin gene and γHV68 gene 73 during latency and lytic infection will be needed to determine the contribution of the viral gene products to latency and virus replication.
Gene 74 (v-GCR).
Previous studies have shown that the KSHV interleukin-8 receptor-like antigen encoded by gene 74 (v-GCR [59]) is a constitutively signaling receptor linked to cell proliferation (1, 3) and is expressed in tumors (9, 26). Transcriptional mapping in a KSHV-infected B-cell line is consistent with the expression of the v-GCR gene as both a latent and lytic transcript (62), although in another study abundant expression of an mRNA encoding the KSHV-v-GCR in phorbol ester-treated cells was noted (17). These data are in contrast to the situation with γHV68. We found minimal transcription from the v-GCR gene region in lytically infected cells, but we did detect expression from this region in latently infected peritoneal cells. Thus, in contrast to the situation in KSHV, it seems likely that the γHV68 v-GCR plays an important role during latent but not lytic infection.
γHV68 as a model for analysis of in vivo gammaherpesvirus latency.
One of the fundamentally important issues confronted by those using animal models of viral infection is how well the model relates to human disease processes. The γHV68 model presents certain very compelling advantages for the study of the basic biology of gammaherpesvirus latency. These include the close genetic relatedness in certain regions of the γHV68 genome to other primate and human gammaherpesviruses, the availability of a manipulable animal model, and the ease with which mutants of γHV68 can be made (reference 64 and unpublished data). However, it is still unclear whether γHV68 will serve as a useful model at the level of molecular mechanisms. This study presents the first molecular data which afford an initial assessment of the issue of whether γHV68 serves as a model for latency of EBV and KSHV.
It is clear that γHV68 is distinct in some respects from other gammaherpesviruses, such as EBV and KSHV, insofar as latency- and tumor-associated gene expression is concerned. For example, none of the EBNA genes of EBV are clearly represented in the γHV68 genome (76). In addition, known or candidate latency- or tumor-associated KSHV genes (such as the viral IRF gene [24, 45]; the K12 gene, encoding kaposin protein [47, 66, 80]; and K1 [38]) are not easily recognized in the γHV68 genomic sequence (76). γHV68 contains no obvious homologs of the HVS transforming STP and Tip genes (16, 19, 25, 31, 76) or ORF 14 (18). This argues either that γHV68 uses distinct pathways for tumor generation and/or latency or that γHV68 has arrived at different strategies for addressing the problems addressed by the EBNA genes or certain KSHV and HVS genes. Similarly, there are no obvious homologs in KSHV, EBV, or HVS of the γHV68 M2, M3, and M9 genes. In addition, preliminary analysis suggests that even genes that may be held in common between the latency programs of KSHV and γHV68 (gene 73 and the v-GCR gene) or EBV and γHV68 (the v-bcl-2 gene) may be regulated differently (see above).
Despite these differences, the data presented here provide a compelling argument for the use of γHV68 to assess the role of certain gammaherpesvirus genes in latency and infection in vivo. These include the v-bcl-2 gene (M11), gene 73, and gene 74. The regions of the viral genome containing these genes are transcribed in latent tissues, tumors, and tumor cell lines infected with γHV68 and either both EBV and KSHV or KSHV alone. We believe that this argues strongly for the validity of γHV68 for assessing the function of certain genes in gammaherpesvirus biology.
ACKNOWLEDGMENTS
This work was supported by a grant to H.W.V. from the National Cancer Institute (RO1 CA74730). S.H.S. was supported by NIH RO1 grants CA43143, CA52004, CA58524, and CA74730 from the National Cancer Institute. R.M.P. was supported by NIH grants GM07200 and AI07163.
We acknowledge helpful discussions from members of the Speck and Virgin labs, as well as discussions that occurred during lab meetings shared with David Leib.
REFERENCES
- 1.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 2.Austin P J, Flemington E, Yandava C N, Strominger J L, Speck S H. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. Proc Natl Acad Sci USA. 1988;85:3678–3682. doi: 10.1073/pnas.85.11.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A. G-protein-coupled receptor of Kaposi’s sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. . (Erratum, 392:210.) [DOI] [PubMed] [Google Scholar]
- 4.Blaskovic D, Stancekova M, Svobodova J, Mistrikova J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980;24:468. [PubMed] [Google Scholar]
- 5.Blaskovic D, Stanekova D, Rajcani J. Experimental pathogenesis of murine herpesvirus in newborn mice. Acta Virol. 1984;28:225–231. [PubMed] [Google Scholar]
- 6.Bowden R J, Simas J P, Davis A J, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol. 1997;78:1675–1687. doi: 10.1099/0022-1317-78-7-1675. [DOI] [PubMed] [Google Scholar]
- 7.Cardin R D, Brooks J W, Sarawar S R, Doherty P C. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J Exp Med. 1996;184:863–871. doi: 10.1084/jem.184.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman E, Nador R G, Bai F, Bohenzky R A, Russo J J, Moore P S, Chang Y, Knowles D M. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang E H Y, Nicholas J, Bellows D S, Hayward G S, Guo H-G, Reitz M S, Hardwick J M. A bcl-2 homolog encoded by Kaposi’s sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc Natl Acad Sci USA. 1997;94:690–694. doi: 10.1073/pnas.94.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12a.Clambey, E. C., H. W. Virgin, and S. H. Speck. Unpublished observation.
- 13.Danielson P E, Forss-Petter S, Brow M A, Calavetta L, Douglass J, Milner R J, Sutcliffe J G. p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA. 1988;7:261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 14.Davis M A, Sturzl M A, Blasig C, Schreier A, Guo H G, Reitz M, Opalenik S R, Browning P J. Expression of human herpesvirus 8-encoded cyclin D in Kaposi’s sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1868–1874. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- 15.Dawson C W, Eliopoulos A G, Dawson J, Young L S. BHRF1, a viral homologue of the Bcl-2 oncogene, disturbs epithelial cell differentiation. Oncogene. 1995;10:69–77. [PubMed] [Google Scholar]
- 16.Desrosiers R C, Bakker A, Kamine J, Falk L A, Hunt R D, King N W. A region of the herpesvirus saimiri genome required for oncogenicity. Science. 1985;228:184–187. doi: 10.1126/science.2983431. [DOI] [PubMed] [Google Scholar]
- 17.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duboise M, Guo J, Czajak S, Lee H, Veazey R, Desrosiers R C, Jung J U. A role for herpesvirus saimiri orf14 in transformation and persistent infection. J Virol. 1998;72:6770–6776. doi: 10.1128/jvi.72.8.6770-6776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duboise S M, Guo J, Czajak S, Desrosiers R C, Jung J U. STP and Tip are essential for herpesvirus saimiri oncogenicity. J Virol. 1998;72:1308–1313. doi: 10.1128/jvi.72.2.1308-1313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutia B M, Clarke C J, Allen D J, Nash A A. Pathological changes in the spleens of gamma interferon receptor-deficient mice infected with murine gammaherpesvirus: a role for CD8 T cells. J Virol. 1997;71:4278–4283. doi: 10.1128/jvi.71.6.4278-4283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Efstathiou S, Ho Y M, Hall S, Styles C J, Scott S D, Gompels U A. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J Gen Virol. 1990;71:1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- 22.Efstathiou S, Ho Y M, Minson A C. Cloning and molecular characterization of the murine herpesvirus 68 genome. J Gen Virol. 1990;71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- 23.Foghsgaard L, Jaattela M. The ability of BHRF1 to inhibit apoptosis is dependent on stimulus and cell type. J Virol. 1997;71:7509–7517. doi: 10.1128/jvi.71.10.7509-7517.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao S J, Boshoff C, Jayachandra S, Weiss R A, Chang Y, Moore P S. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 25.Geck P, Whitaker S A, Medveczky M M, Medveczky P G. Expression of collagenlike sequences by a tumor virus, herpesvirus saimiri. J Virol. 1990;64:3509–3515. doi: 10.1128/jvi.64.7.3509-3515.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo H G, Browning P, Nicholas J, Hayward G S, Tschachler E, Jiang Y W, Sadowska M, Raffeld M, Colombini S, Gallo R C, Reitz M S J. Characterization of a chemokine receptor-related gene in human herpesvirus 8 and its expression in Kaposi’s sarcoma. Virology. 1997;228:371–378. doi: 10.1006/viro.1996.8386. [DOI] [PubMed] [Google Scholar]
- 27.Hardwick J M, Lieberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heise M T, Virgin H W., IV The T-cell-independent role of gamma interferon and tumor necrosis factor alpha in macrophage activation during murine cytomegalovirus and herpes simplex virus infections. J Virol. 1995;69:904–909. doi: 10.1128/jvi.69.2.904-909.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson S, Huen D, Rowe M, Dawson C, Johnson G, Rickinson A. Epstein-Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc Natl Acad Sci USA. 1993;90:8479–8483. doi: 10.1073/pnas.90.18.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung J U, Stäger M, Desrosiers R C. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung J U, Trimble J J, King N W, Biesinger B, Fleckenstein B W, Desrosiers R C. Identification of transforming genes of subgroup A and C strains of herpesvirus saimiri. Proc Natl Acad Sci USA. 1991;88:7051–7055. doi: 10.1073/pnas.88.16.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 34.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 35.Kraft M S, Henning G, Fickenscher H, Lengenfelder D, Tschopp J, Fleckenstein B, Meinl E. Herpesvirus saimiri transforms human T-cell clones to stable growth without inducing resistance to apoptosis. J Virol. 1998;72:3138–3145. doi: 10.1128/jvi.72.4.3138-3145.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurz S, Steffens H-P, Mayer A, Harris J R, Reddehase M J. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71:2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus. Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 38.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A, Desrosiers R C, Jung J U. Deregulation of cell growth by the K1 gene of Kaposi’s sarcoma-associated herpesvirus. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 39.Lee M-A, Yates J L. BHRF1 of Epstein-Barr virus, which is homologous to human proto-oncogene bcl2, is not essential for transformation of B cells or for virus replication in vitro. J Virol. 1992;66:1899–1906. doi: 10.1128/jvi.66.4.1899-1906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Liu, C., H. W. Virgin IV, and S. H. Speck. Unpublished observation.
- 40.Mackett M, Stewart J P, Pepper S D V, Chee M, Efstathiou S, Nash A, Arrand J R. Genetic content and preliminary transcriptional analysis of a representative region of murine gammaherpesvirus 68. J Gen Virol. 1997;78:1425–1433. doi: 10.1099/0022-1317-78-6-1425. [DOI] [PubMed] [Google Scholar]
- 41.Marchini A, Tomkinson B, Cohen J I, Kieff E. BHRF1, the Epstein-Barr virus gene with homology to Bcl2, is dispensable for B-lymphocyte transformation and virus replication. J Virol. 1991;65:5991–6000. doi: 10.1128/jvi.65.11.5991-6000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medveczky M, Geck P, Sullivan J L, Serbousek D, Djeu J Y, Medveczky P G. IL-2 independent growth and cytotoxicity of herpesvirus saimiri-infected human CD8 cells and involvement of two open reading frame sequences of the virus. Virology. 1993;196:402–412. doi: 10.1006/viro.1993.1495. [DOI] [PubMed] [Google Scholar]
- 43.Mercer J A, Wiley C A, Spector D H. Pathogenesis of murine cytomegalovirus infection: identification of infected cells in the spleen during acute and latent infections. J Virol. 1988;62:987–997. doi: 10.1128/jvi.62.3.987-997.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mistrikova J, Blaskovic D. Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol. 1985;29:312–317. [PubMed] [Google Scholar]
- 45.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 46.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 47.Muralidhar S, Pumfery A M, Hassani M, Sadaie M R, Azumi N, Kishishita M, Brady J N, Doniger J, Medveczky P, Rosenthal L J. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) transforming gene. J Virol. 1998;72:4980–4988. doi: 10.1128/jvi.72.6.4980-4988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murthy S C S, Trimble J J, Desrosiers R C. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J Virol. 1989;63:3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nava V E, Cheng E H-Y, Veliuona M, Zou S, Clem R J, Mayer M L, Hardwick J M. Herpesvirus saimiri encodes a functional homolog of the human bcl-2 oncogene. J Virol. 1997;71:4118–4122. doi: 10.1128/jvi.71.5.4118-4122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neilan J G, Lu Z, Afonso C L, Kutish G F, Sussman M D, Rock D L. An African swine fever virus gene with similarity to the proto-oncogene bcl-2 and the Epstein-Barr virus gene BHRF1. J Virol. 1993;67:4391–4394. doi: 10.1128/jvi.67.7.4391-4394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearson G R, Luka J, Petti L, Sample J, Birkenbach M, Braun D, Kieff E. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology. 1987;160:151–161. doi: 10.1016/0042-6822(87)90055-9. [DOI] [PubMed] [Google Scholar]
- 52.Pollock J L, Presti R M, Paetzold S, Virgin H W. Latent murine cytomegalovirus infection in macrophages. Virology. 1997;227:168–179. doi: 10.1006/viro.1996.8303. [DOI] [PubMed] [Google Scholar]
- 53.Pollock J L, Virgin H W., IV Latency, without persistence, of murine cytomegalovirus in spleen and kidney. J Virol. 1995;69:1762–1768. doi: 10.1128/jvi.69.3.1762-1768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puglielli M T, Woisetschlaeger M, Speck S H. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol. 1996;70:5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajcani J, Blaskovic D, Svobodova J, Ciampor F, Huckova D, Stanekova D. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 1985;29:51–60. [PubMed] [Google Scholar]
- 57.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2397–2446. [Google Scholar]
- 59.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 61.Sarawar S R, Cardin R D, Brooks J W, Mehrpooya M, Hamilton-Easton A-M, Mo X Y, Doherty P C. Gamma interferon is not essential for recovery from acute infection with murine gammaherpesvirus 68. J Virol. 1997;71:3916–3921. doi: 10.1128/jvi.71.5.3916-3921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi’s sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 64.Simas J P, Bowden R J, Paige V, Efstathiou S. Four tRNA-like sequences and a serpin homologue encoded by murine gammaherpesvirus 68 are dispensable for lytic replication in vitro and latency in vivo. J Gen Virol. 1998;79:149–153. doi: 10.1099/0022-1317-79-1-149. [DOI] [PubMed] [Google Scholar]
- 65.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M-F, Clauvel J-P, Raphael M, Degos L, Signaux F. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 66.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart J P, Janjua N J, Pepper S D V, Bennion G, Mackett M, Allen T, Nash A A, Arrand J R. Identification and characterization of murine gammaherpesvirus 68 gp150: a virion membrane glycoprotein. J Virol. 1996;70:3528–3535. doi: 10.1093/benz/9780199773787.article.b00034574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stewart J P, Janjua N J, Sunil-Chandra N P, Nash A A, Arrand J R. Characterization of murine gammaherpesvirus 68 glycoprotein B (gB) homolog: similarity to Epstein-Barr virus gB (gp110) J Virol. 1994;68:6496–6504. doi: 10.1128/jvi.68.10.6496-6504.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stewart J P, Usherwood E J, Ross A, Dyson H, Nash T. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J Exp Med. 1998;187:1941–1951. doi: 10.1084/jem.187.12.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sunil-Chandra N P, Arno J, Fazakerley J, Nash A A. Lymphoproliferative disease in mice infected with murine gammaherpesvirus 68. Am J Pathol. 1994;145:818–826. [PMC free article] [PubMed] [Google Scholar]
- 71.Sunil-Chandra N P, Efstathiou S, Arno J, Nash A A. Virological and pathological features of mice infected with murine gammaherpesvirus 68. J Gen Virol. 1992;73:2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 72.Sunil-Chandra N P, Efstathiou S, Nash A A. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol. 1992;73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 73.Tarodi B, Subramanian T, Chinnadurai G. Epstein-Barr virus BHRF1 protein protects against cell death induced by DNA-damaging agents and heterologous viral infection. Virology. 1994;201:404–407. doi: 10.1006/viro.1994.1309. [DOI] [PubMed] [Google Scholar]
- 74.Usherwood E J, Stewart J P, Nash A A. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J Virol. 1996;70:6516–6518. doi: 10.1128/jvi.70.9.6516-6518.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Usherwood E J, Stewart J P, Robertson K, Allen D J, Nash A A. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J Gen Virol. 1996;77:2819–2825. doi: 10.1099/0022-1317-77-11-2819. [DOI] [PubMed] [Google Scholar]
- 75a.Van Berkel, V., K. Preiter, H. W. Virgin, and S. H. Speck. Identification of a murine gammaherpes virus 68 gene encoding an abundantly secreted protein. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 75b.Van Dyk, L. F., J. D. Katz, M. Jacoby, S. H. Speck, and H. W. Virgin. Submitted for publication.
- 76.Virgin H W, IV, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weck K E, Barkon M L, Yoo L I, Speck S H, Virgin H W., IV Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol. 1996;70:6775–6780. doi: 10.1128/jvi.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weck K E, Dal Canto A J, Gould J D, O’Guin A K, Roth K A, Saffitz J E, Speck S H, Virgin H W. Murine gammaherpesvirus 68 causes large vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus induced vascular disease. Nat Med. 1997;3:1346–1353. doi: 10.1038/nm1297-1346. [DOI] [PubMed] [Google Scholar]
- 78a.Weck, K. E., S. S. Kim, H. W. Virgin, and S. H. Speck. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 79.Zhong W, Ganem D. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:1207–1212. doi: 10.1128/jvi.71.2.1207-1212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi’s sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]