Abstract
The members of the family Filoviridae, Marburg virus (MBGV) and Ebola virus (EBOV), are very similar in terms of morphology, genome organization, and protein composition. To compare the replication and transcription strategies of both viruses, an artificial replication system based on the vaccinia virus T7 expression system was established for EBOV. Specific transcription and replication of an artificial monocistronic minireplicon was demonstrated by reporter gene expression and detection of the transcribed and replicated RNA species. As it was shown previously for MBGV, three of the four EBOV nucleocapsid proteins, NP, VP35, and L, were essential and sufficient for replication. In contrast to MBGV, EBOV-specific transcription was dependent on the presence of the fourth nucleocapsid protein, VP30. When EBOV VP30 was replaced by MBGV VP30, EBOV-specific transcription was observed but with lower efficiency. Exchange of NP, VP35, and L between the two replication systems did not lead to detectable reporter gene expression. It was further observed that neither MBGV nor EBOV were able to replicate the heterologous minigenomes. A chimeric minigenome, however, containing the EBOV leader and the MBGV trailer was encapsidated, replicated, transcribed, and packaged by both viruses.
Marburg virus and Ebola virus (MBGV and EBOV) are the only members of the family Filoviridae. Both originated from Africa and both cause a severe hemorrhagic fever in humans and nonhuman primates (5, 25). The symptoms of the disease, caused by MBGV and EBOV, are very similar and include high fever, severe hemorrhages, and shock in the final state of the illness (24, 30). Both viruses cause systemic infections and display a widespread organ distribution (11, 18, 33). Also the genomic organization of the viruses is similar (17). Both possess a nonsegmented negative-strand RNA genome ca. 19 kb in length with a coding capacity for seven structural proteins. The genes are flanked by highly conserved transcriptional start and stop signals (6, 17, 29, 34). Noncoding sequences containing the signals for replication and encapsidation are located at the 3′ and 5′ ends of the genomes (28).
In contrast to most of the other members of the order Mononegavirales, MBGV and EBOV contain four nucleocapsid proteins instead of three (3, 13). The surface proteins of MBGV and EBOV are inserted in the viral membrane as homotrimers and are both cleaved by the cellular prohormone convertase furin (16, 36, 39). Phosphorylation can be observed with the first and the fifth gene products of MBGV and EBOV. The second gene product, which for MBGV is shown to be the P equivalent of paramyxo- and rhabdoviruses (28) is only very weakly phosphorylated, if at all (2, 13). This is a feature which clearly separates Filoviridae from the other nonsegmented negative-strand (NNS) RNA viruses, since the P protein of the other NNS RNA viruses, as far as it has been investigated, is strongly phosphorylated. Beside these similarities among the two members of the Filoviridae, differences can be found. The noncoding sequences located at the 3′ and 5′ ends of the viral genomes are of significantly different lengths (34, 41). The EBOV genome reveals three gene overlaps, whereas the MBGV genome contains only one (34). The translation of EBOV glycoprotein (GP) needs the editing activity of the viral polymerase (35, 40), whereas the glycoprotein of MBGV (MBGV GP) is transcribed from a single open reading frame (ORF) (4, 6, 42).
Recently, we were able to establish an artificial replication system for MBGV showing that three of the four nucleocapsid proteins, NP, VP35, and L, were sufficient to support replication and transcription of an MBGV-specific monocistronic minigenome (28). To investigate the relationship between MBGV and EBOV, we have compared the functions of the nucleocapsid proteins of both viruses in transcription and replication. We also examined whether the signals for replication and encapsidation were functionally conserved between the two members of the family Filoviridae. Therefore, an artificial replication system for EBOV was established based on the vaccinia virus T7 expression system (9). Further, we constructed chimeric minigenomes containing the leader sequence of the MBGV genome and the trailer sequence of the EBOV genome and vice versa. These minigenomes were employed to transfect MBGV- or EBOV-infected cells or were tested for their suitability in the respective artificial replication system.
In contrast to the previously established MBGV-specific replication system, it was demonstrated for the EBOV replication system that the fourth nucleocapsid protein, VP30, was essential for transcription. Although the nucleocapsid proteins NP, VP35, and L and the EBOV- and MBGV-specific minigenomes could not be exchanged between MBGV and EBOV, it was possible to construct a chimeric minigenome which was accepted by both viruses.
MATERIALS AND METHODS
Viruses and cell lines.
The Musoke strain of MBGV, isolated in 1980 in Kenya (37), was grown in E6 cells, a Vero cell line clone (ATCC CRL1586) as described by Mühlberger et al. (27). For cloning of the EBOV NP, VP35, and VP30 genes, EBOV strain Mayinga, subtype Zaire, guinea pig adapted (EBOV 8mc [38a]), was used. For cloning of the L gene and the minigenomes, EBOV strain Mayinga, subtype Zaire, was used (34). This EBOV strain was also used for infection of E6 cells as described by Feldmann et al. (16). All experiments with MBGV and EBOV were performed in a high-containment laboratory. For T7 RNA polymerase expression, the recombinant vaccinia virus MVA-T7 (38), which was grown in chicken embryo fibroblasts, was used. Expression of recombinant proteins by using the vaccinia virus T7 expression system was performed in HeLa cells. All cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum.
Molecular cloning. (i) Cloning of the EBOV nucleocapsid protein genes.
cDNAs containing the ORFs of NP, VP35, VP30, and L were cloned into the T7 expression vector pTM1 (kindly provided by Bernhard Moss, National Institutes of Health). The ORF and parts of the nontranslated regions of all genes were amplified by reverse transcription-PCR (RT-PCR) with specific primers containing appropriate restriction sites. The amplified products were subsequently inserted into the pTM1 vector. The NP gene (nucleotides 456 to 3085; nucleotide numbers refer to the EBOV Zaire genome sequence, GenBank accession number AF086833) was cloned into pTM1, which was cut with BamHI and HindIII. The VP35 gene (nucleotides 3043 to 4456) was cloned into pTM1 by using BamHI and SalI; the VP30 gene (nucleotides 8415 to 10193) was cloned into pTM1 by using BamHI and PstI. The resulting plasmids were designated as pT/NPEBO, pT/VP35EBO, and pT/VP30EBO. Cloning of the L gene (nucleotides 11577 to 18403) involved the synthesis of four PCR fragments. The fragments were inserted into pTM1 to receive the vector pT/LEBO containing the complete ORF of the EBOV L gene. The sequences of the recombinant nucleocapsid protein genes were verified by sequencing.
(ii) Cloning of artificial minigenomes.
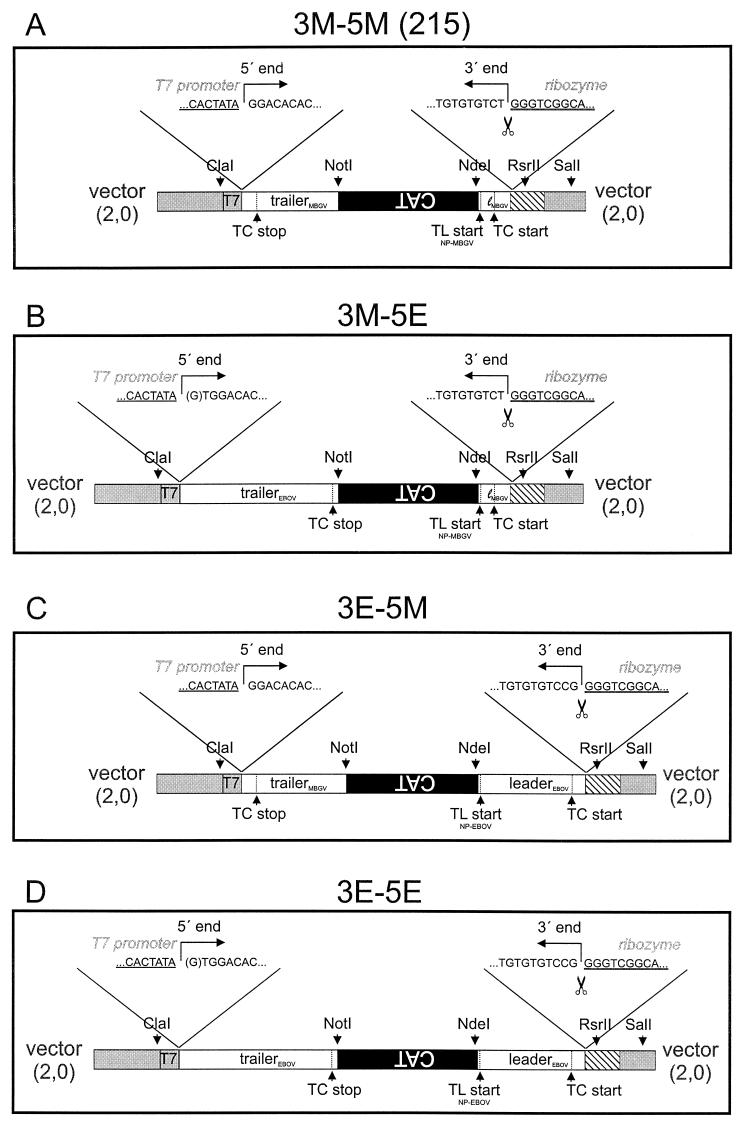
Plasmid 215 containing the CAT gene flanked by the leader and trailer regions of the MBGV genome was used for construction of the artificial minigenomes. Cloning of plasmid 215 is described elsewhere (28). This vector possesses a ClaI restriction site which had been inserted by site-directed mutagenesis and is positioned 15 nucleotides upstream of the T7 RNA polymerase promoter. Plasmid 215 is designated 3M-5M in the present study (Fig. 1A).
FIG. 1.
Construction of minigenomes. The minigenomes were inserted in the transcription vector 2,0 (gray segments) between the T7 RNA polymerase promoter (T7) and the hepatitis delta virus ribozyme (striped segments). (A) Diagram of MBGV-specific minigenome 3M-5M (previously designated 215), consisting of 106 nucleotides of the MBGV leader (white; lMBGV), 668 nucleotides of the CAT gene (black), and 439 nucleotides of the MBGV trailer (white). (B) Diagram of the chimeric minigenome 3M-5E consisting of 106 nucleotides of the MBGV leader (white; lMBGV), the CAT gene (black), and 731 nucleotides of the EBOV trailer (white). (C) Diagram of the chimeric minigenome 3E-5M consisting of 472 nucleotides of the EBOV leader (white), the CAT gene (black), and 439 nucleotides of the MBGV trailer (white). (D) Diagram of the EBOV-specific minigenome 3E-5E, consisting of 472 nucleotides of the EBOV leader (white), the CAT gene (black), and 731 nucleotides of the EBOV trailer (white). Above each scheme are indicated the boundary between the T7 RNA polymerase promoter sequence (underlined) and the 5′ ends of the minigenome (negative-sense orientation) (left side) and the boundary between the hepatitis delta virus ribozyme sequence (underlined) and the 3′ end of the minireplicon (negative-sense orientation) (right side). ClaI, NotI, NdeI, and RsrII, restriction enzymes used for cloning; SalI, restriction site used for linearization of the plasmids for in vitro transcription; TL start, translation start codon of the NP gene. Below, in smaller fonts, it is indicated whether the start site originates from NP of MBGV or EBOV. TC start, transcription start site of the NP gene of MBGV or EBOV; TC stop, transcription stop site of the L gene of MBGV or EBOV. The cleavage site of the ribozyme is symbolized by a pair of scissors.
To construct plasmid 3M-5E (Fig. 1B), the last 731 nucleotides of the 5′ end of the EBOV genome were amplified by RT-PCR. cDNA synthesis was performed with viral RNA (vRNA) of EBOV and a primer complementary to nucleotides 18227 to 18256 which contained a NotI restriction site. For PCR, a second primer was added spanning the last 26 nucleotides of the EBOV genome and the T7 RNA polymerase promoter sequence of plasmid 3M-5M, including the ClaI restriction site. To enhance T7 RNA polymerase-dependent RNA synthesis, an additional G residue was introduced between the T7 RNA polymerase promoter and the EBOV-specific sequence (Fig. 1B and D, in brackets). The resulting PCR fragment was digested with ClaI and NotI and inserted into the ClaI and NotI sites of vector 3M-5M. Thus, the MBGV trailer sequence was replaced by the EBOV trailer (Fig. 1B).
For generating plasmid 3E-5M, the first 472 nucleotides of the 3′ end of the EBOV genome, including the translational start codon of the NP gene, were amplified by RT-PCR. The primer used for cDNA synthesis was complementary to the first 26 nucleotides of the EBOV genome and contained parts of the ribozyme sequence, including an RsrII restriction site. The second primer spanning nucleotides 443 to 472 of the vRNA was flanked by an NdeI restriction site. The amplified fragment was cloned into the NdeI-RsrII sites of vector 3M-5M, thus replacing the MBGV leader (Fig. 1C).
The EBOV-specific minigenome 3E-5E was constructed by replacing the MBGV leader sequence of plasmid 3M-5E by the PCR fragment comprising the EBOV leader region (see above) by using the NdeI and RsrII restriction sites (Fig. 1D).
RNA transfection of EBOV- and MBGV-infected cells and passaging of CAT activity.
Subconfluent E6 cells (approximately 106) were infected with EBOV or MBGV at a multiplicity of infection (MOI) of 1 PFU per cell. At 1 h postinfection (p.i.), cells were washed once with DMEM and transfected with 5 to 10 μl of the respective in vitro-transcribed minigenomic RNA by using the transfection reagent DOTAP (Boehringer Mannheim). At 1 day p.i., the medium was replaced by DMEM supplemented with 2% fetal calf serum, and 3 days later the cells were assayed for chloramphenicol acetyltransferase (CAT) activity. For passaging, supernatants of MBGV- or EBOV-infected and transfected cells were clarified at 5 days p.i. and serially passaged to fresh E6 cells. The cells were harvested and processed for CAT assay.
Combined DNA-RNA transfection of MVA-T7-infected cells.
HeLa cells (106 in a 7-cm2 well) were infected with MVA-T7 at an MOI of 5 PFU per cell. At 1 h p.i., the cells were transfected with various amounts of plasmids encoding the nucleocapsid proteins of EBOV and MBGV, as indicated in the text as well as in the figures, by using the Lipofectin (Gibco-BRL) precipitation technique. At 3.5 h after transfection, the cells were washed three times with DMEM without fetal calf serum and subjected to the RNA transfection procedure, which was carried out as described previously (28). After RNA transfection, the plates were further incubated for 48 to 72 h at 33°C.
CAT assay.
CAT activity was determined according to a standard protocol (19). Quantification of the radioactivity was done with a Fuji BAS-1000 BioImaging analyzer (Fujifilm) and by using the TINA software (Raytest).
RNA isolation and MCN treatment.
HeLa cells grown in 7-cm2 wells were infected with MVA-T7 and transfected with DNA as described above. At 2 days p.i., cells were washed two times with phosphate-buffered saline, scraped into the washing buffer, and pooled (three wells). After centrifugation for 10 min at 3,000 rpm in a microcentrifuge at 4°C, the pellets were resuspended in 200 μl of micrococcal nuclease (MCN) buffer (10 mM NaCl, 10 mM Tris [pH 7.5], 1.5 mM MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 10 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride [15]), sheared, and sonicated for 1 min. Then, 3 μl of MCN (15 U/μl; Boehringer Mannheim) was added, and the samples were incubated for 75 min at 30°C. Finally, RNA was isolated by using an RNeasy kit (Qiagen) and subjected to Northern blot analysis. For oligo(dT) purification, total cellular RNA corresponding to three 7-cm2 wells of MVA-T7-infected and transfected HeLa cells was isolated at 2 days p.i. by using an RNeasy kit. Polyadenylated RNA was purified from total RNA by oligo(dT) cellulose binding (New England Biolabs). Unbound and eluted bound RNA were ethanol precipitated and subjected to Northern blot analysis, which was performed as described previously (20, 28).
RESULTS
Ebola virus replication system.
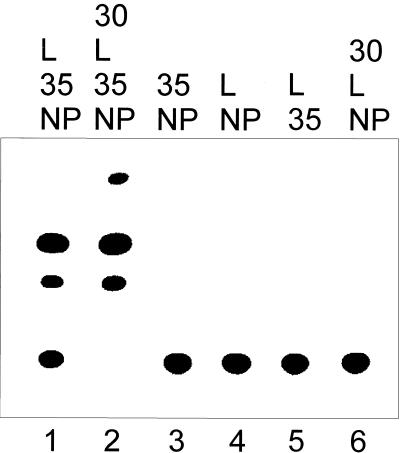
To establish an artificial EBOV replication-transcription system, an EBOV-specific minigenome was constructed containing the leader and trailer sequence of the EBOV genome and a CAT reporter gene in antisense orientation (Fig. 1D, 3E-5E). These genetic elements were cloned into the vector 2,0 (kindly provided by A. Ball, University of Alabama Medical School [31]) between the T7 RNA polymerase promoter and the hepatitis delta virus ribozyme. Thus, transcription of the resulting vector by the phage T7 RNA polymerase led to an RNA construct with exactly defined 5′ ends and, by the self-cleavage activity of the ribozyme, exact 3′ ends corresponding to the very ends of the EBOV genome (34, 41). The function of the minigenome was tested by transfection of EBOV-infected cells with the in vitro-transcribed minigenomic RNA. At 4 to 5 days p.i., CAT activity was detected in the cell lysates which was caused by EBOV-specific transcription of the minigenome (data not shown). Cells which were only transfected without prior EBOV infection did not show CAT activity. In a next step, the four nucleocapsid protein genes of EBOV were cloned into the vector pTM1 under the control of the T7 RNA polymerase promoter. These plasmids were used for transfection of HeLa cells previously infected with the recombinant vaccinia virus MVA-T7. After DNA transfection, in vitro-transcribed EBOV-specific minigenomic RNA was transfected into the same cells to serve as a template for the artificial EBOV replication complex formed by the recombinant nucleocapsid proteins. To examine the activity of the artificial replication system, lysates of infected and transfected cells were assayed for CAT activity at 3 days p.i. It is shown in Fig. 2 that CAT activity was detected in the presence of EBOV NP, VP35, and L (lane 1). VP30 was not essential for reporter gene expression but led to a considerable increase in CAT activity (lane 2). As it was previously observed with MBGV, titration of the nucleocapsid protein genes exhibited an optimum for the amount of NP and VP35 input DNA (Fig. 3A). In contrast to MBGV (28), the EBOV replication system was more tolerant towards the amount of NP input DNA. Increase of the amount of pT/LEBO led to increased CAT activity, which reached a plateau at 500 ng of input DNA (Fig. 3A).
FIG. 2.
Reporter gene expression mediated by recombinant EBOV nucleocapsid proteins. HeLa cells were infected with MVA-T7; transfected with 500 ng of pT/NPEBO, 500 ng of pT/VP35EBO, 100 ng of pT/VP30EBO, and/or 1 μg of pT/LEBO; and subsequently transfected with RNA 3E-5E. At 3 days p.i., cells were lysed. CAT activity was determined, and acetylated products were separated by thin-layer chromatography. DNA transfection was performed with different combinations of nucleocapsid protein genes as indicated. NP, pT/NPEBO; 35, pT/VP35EBO; 30, pT/VP30EBO; L, pT/LEBO.
FIG. 3.
Characterization of the artificial EBOV replication system. HeLa cells were infected with MVA-T7 and subsequently transfected with plasmids encoding the nucleocapsid proteins. After DNA transfection, the cells were transfected with the in vitro-transcribed EBOV-specific minigenome RNA 3E-5E. At 3 days p.i. cells were probed for CAT activity and the amount of acetylated chloramphenicol was quantified with a BioImaging Analyzer (Fuji BAS-1000) with the Raytest TINA software. (A) Titration of plasmids encoding the different nucleocapsid proteins. Titration experiments were performed with fixed amounts of two of the plasmids encoding nucleocapsid proteins (500 ng of pT/NPEBO, 500 ng of pT/VP35EBO, and 1 μg of pT/LEBO) and various amounts of one of the plasmids as indicated in the legend. (B) Functional complex between L and VP35. HeLa cells were infected with MVA-T7 and subsequently transfected with 200 ng of pT/NPEBO and various amounts of pT/VP35EBO and pT/LEBO. The ratio between pT/LEBO and pT/VP35EBO was either held constant (4:1) and increased simultaneously, or only the amount of pT/LEBO was increased. In this case the amount of transfected pT/VP35EBO was 20 ng.
Since it is known for several NNS RNA viruses that L and P form the functional polymerase complex (1, 22), it was of interest to determine whether EBOV polymerase activity was dependent on the stoichiometrical relation between L and VP35, which is presumed to represent the P analogue. The amount of L input DNA was therefore titrated in the presence of fixed amounts of pT/VP35EBO and pT/NPEBO, or else pT/LEBO and pT/VP35EBO were titrated with constant amounts of pT/NPEBO (Fig. 3B). The amount of VP35 in the experiment with fixed concentration was previously optimized to give maximal CAT activity with the start concentration of L (data not shown). It is shown that only a simultaneous increase of L and VP35 led to an enhanced reporter gene expression, suggesting that indeed the complex between L and VP35 represents the active polymerase.
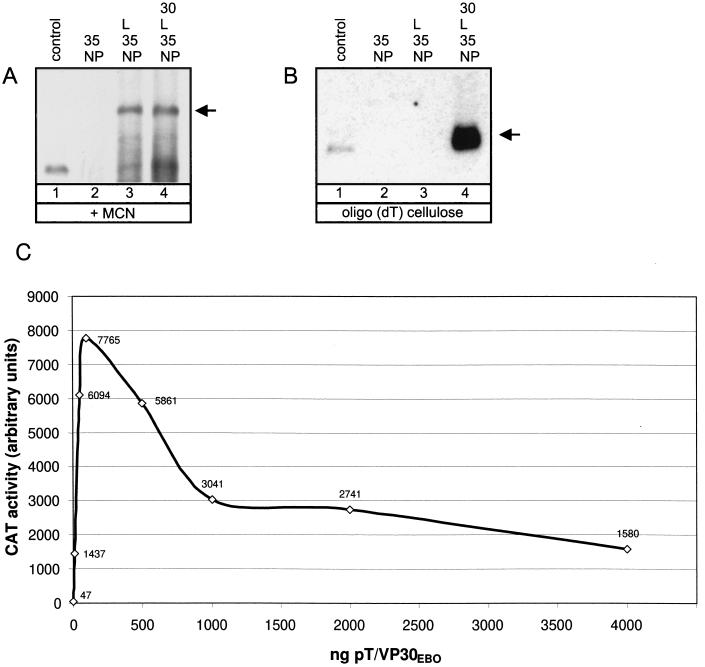
To prove that the artificial replication system supported transcription and replication, the EBOV-specific RNA species synthesized were analyzed. These are the replicated full-length minigenomic RNA and the transcribed polyadenylated mRNA. Discrimination of the RNA species was performed by nuclease treatment or oligo(dT) binding. Only replicated RNA is encapsidated and thus nuclease resistant, whereas the polyadenylated mRNA is nuclease sensitive. HeLa cells were infected with MVA-T7 and subsequently transfected with plasmids encoding the nucleocapsid proteins of EBOV and the EBOV-specific minigenome 3E-5E. At 2 days p.i., cells were lysed and total RNA was purified either with or without previous MCN treatment of the cell lysates. To collect polyadenylated mRNA, total RNA extracted from nontreated cell lysates was further purified by oligo(dT) cellulose binding. The RNA fractions were subjected to Northern blot analysis and finally hybridized with a negative-sense-orientated probe to detect replicated positive-sense RNA and mRNA. As can be seen in Fig. 4A, the encapsidated positive-sense minigenome, representing a replicative intermediate, was detected in the presence of NP, VP35, and L (lane 3). Surprisingly, no EBOV-specific polyadenylated RNA could be detected under these conditions (Fig. 4B, lane 3). Only in the presence of VP30 was the EBOV transcription complex able to synthesize detectable levels of mRNA (Fig. 4B, lane 4). Replication, however, was not influenced by VP30 (Fig. 4A, lane 4). Titration experiments performed with pT/VP30EBO revealed that even at small amounts of VP30 input DNA (10 ng) reporter gene expression was drastically enhanced (1,437 arbitrary units; Fig. 4C). Under the chosen experimental conditions, only 47 arbitrary units could be measured in the absence of VP30. Since it is assumed that CAT activity reflected transcription and not replication (28), this result was in line with the results of the Northern blot analysis. The optimal concentration of VP30 input DNA was shown to be about 100 ng, which is considerably lower than the optimal concentration of the other nucleocapsid proteins (ca. 500 ng). It is important to note that residual CAT activity was still detectable in the absence of VP30 (Fig. 2 and 4C). From these data, it is concluded that EBOV VP30 acts as a transcriptional cofactor.
FIG. 4.
Characterization of RNA species synthesized by the artificial EBOV nucleocapsid complex. HeLa cells were infected with MVA-T7 and subsequently transfected with 500 ng of pT/NPEBO, 500 ng of pT/VP35EBO, and 2 μg of plasmid 3E-5E. One hundred nanograms of pT/VP30EBO and 1 μg of pT/LEBO were added to the transfection mixture as indicated. At 2 days p.i., cells were lysed. Lysates were either subjected to MCN digestion and subsequent RNA purification (A) or RNA was directly isolated from cell lysates and further purified by treatment with oligo(dT) cellulose (B). Purified RNA species were transferred onto nylon membranes and finally probed by using the negative-sense digoxigenin-labeled riboprobe DIG-BS/CAT (28). As a control, in vitro-transcribed positive-sense MBGV-specific minigenome 2.1-CAT was used (28) which is identical in size to 3M-5M. The arrows indicate the positions of positive-sense full-length RNA (A) or polyadenylated mRNA (B). NP, pT/NPEBO; 35, pT/VP35EBO; 30, pT/VP30EBO; L, pT/LEBO. (C) Influence of VP30 on CAT activity. HeLa cells were infected and transfected as described for Fig. 2. DNA transfection was performed with various amounts of pT/VP30EBO and fixed amounts of the other nucleocapsid protein genes (500 ng of pT/NPEBO, 500 ng of pT/VP35EBO, and 1 μg of pT/LEBO).
Chimeric minigenomes as templates for MBGV and EBOV polymerases.
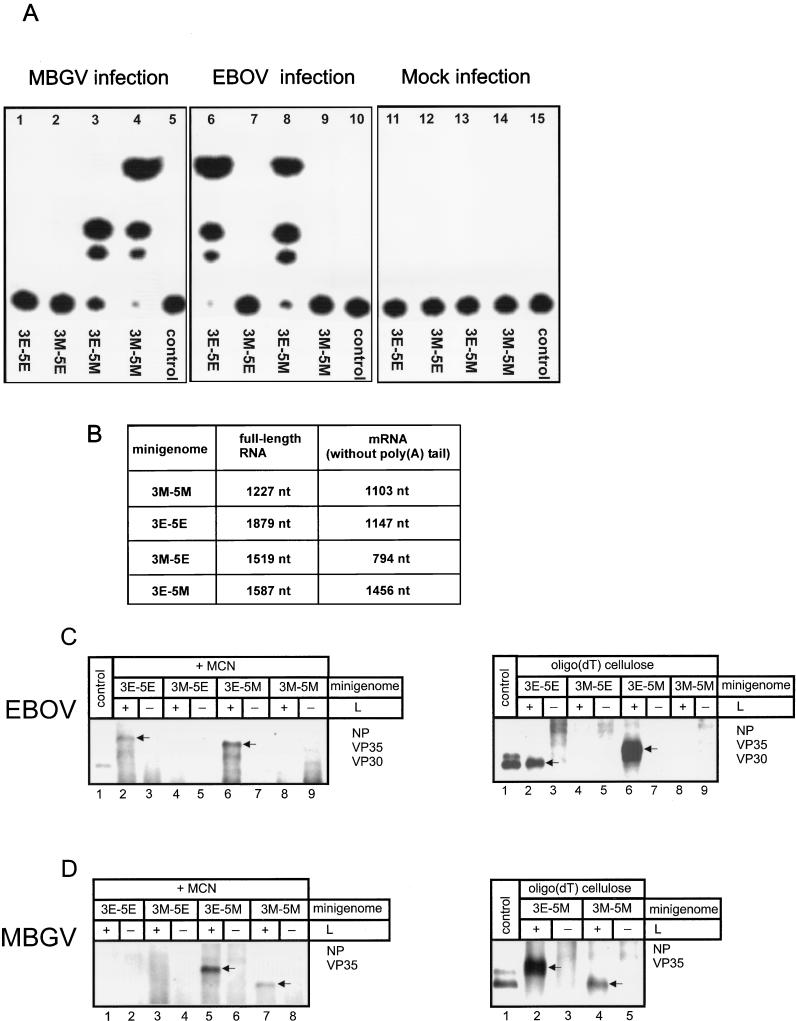
It was now of interest to determine whether the polymerase of MBGV or EBOV can recognize the heterologous signals for encapsidation, replication, and transcription. To this end, chimeric minigenomes were constructed whose 3′ or 5′ ends were replaced by the respective leader or trailer region of the heterologous virus (Fig. 1B and C). These constructs and the authentic minigenomes were in vitro transcribed, and the resulting RNA was employed to transfect MBGV- or EBOV-infected cells. At 4 to 5 days p.i., lysates of these cells were subjected to CAT assay. As expected, MBGV accepted the MBGV-specific minigenome and EBOV the EBOV-specific minigenome as a template for replication and transcription (Fig. 5A, lanes 4 and 6). Neither MBGV nor EBOV was able to recognize the heterologous minigenome (lanes 1 and 9). In addition, both viruses did not recognize the chimeric minigenome containing the 3′ end of the MBGV genome and the 5′ end of the EBOV genome (3M-5E, lanes 2 and 7). However, the minigenome with the leader sequence of EBOV and the trailer sequence of MBGV (3E-5M) was replicated by both viruses (lanes 3 and 8). Whenever CAT activity was detected, it could be passaged to fresh cells showing that the minigenomes were also packaged into virions (data not shown). These data indicate that the signals for transcription, replication, and encapsidation localized in the leader and trailer regions of minigenome 3E-5M are recognized by the replication complexes of both viruses, MBGV and EBOV.
FIG. 5.
Replication and transcription of chimeric minigenomes. (A) E6 cells were infected with either MBGV (lanes 1 to 5) or EBOV (lanes 6 to 10) at an MOI of 0.1 PFU per cell or were not infected (Mock, lanes 11 to 15). Subsequently, cells were transfected with one of the in vitro-transcribed minigenomic RNAs. At 5 days p.i., cells were assayed for CAT activity. Transfected minigenomes are indicated below the panels. Control, no RNA transfection. (B) Table of the theoretical nucleotide numbers of the expected replicated or transcribed RNA species derived from the employed minigenomes. The mRNA nucleotide number is given without the poly(A) tail. nt, nucleotides. (C and D) Northern blot analysis of the different RNA species. (C) HeLa cells were infected with MVA-T7 and subsequently transfected with 500 ng of pT/NPEBO, 500 ng of pT/VP35EBO, 100 ng of pT/VP30EBO, and 2 μg of the respective minigenome DNA as indicated, with (+) or without (−) 1 μg of pT/LEBO. At 3 days p.i., cells were lysed. Total RNA was either purified after MCN digestion (left side), or RNA was purified without MCN digestion and further treated with oligo(dT) cellulose (right side). Purified RNA was analyzed by Northern hybridization with the negative-sense digoxigenin-labeled riboprobe DIG-BS/CAT (28). (D) HeLa cells were infected with MVA-T7 and subsequently transfected with 100 ng of pT/NPMBG, 500 ng of pT/VP35MBG, and 2 μg of the respective minigenome DNA as indicated, with (+) or without (−) 1 μg of pT/LMBG. RNA purification was performed as described for Fig. 5C. Control, in vitro-transcribed positive-sense MBGV-specific minigenome 2.1-CAT (see Fig. 4). The arrows indicate the specific RNA species.
The chimeric minigenomes were then checked for their function in the established MBGV- and EBOV-specific artificial replication systems. HeLa cells were infected with MVA-T7 and transfected with plasmids encoding MBGV or EBOV nucleocapsid proteins and the above-described minigenomes. At 3 days p.i., cells were lysed and CAT activity was measured. The artificial replication systems showed the same specificity for replication of the chimeric minigenomes as had the authentic EBOV and MBGV replication complexes (data not shown).
Since CAT activity is caused by transcription and not by replication (28), it was possible that in the cases where the employed minireplicons did not lead to CAT activity the minigenomes were only replicated but not transcribed. Therefore, RNA was purified from lysates of MVA-T7-infected HeLa cells transfected with plasmids encoding the minigenomes and the respective nucleocapsid proteins. Figure 5B indicates the expected length of the replicated and transcribed RNA species. Differences in length between replicated RNA and mRNA (Fig. 1 and 5B) resulted from the different length of the 3′ and 5′ noncoding sequences of the MBGV and EBOV genomes. While the transcriptional stop signal of MBGV L gene is localized only 87 nucleotides upstream of the genome end, the respective signal of EBOV is located 688 nucleotides upstream of the genome end (41). Furthermore, the translational start AUG of the EBOV NP gene spans nucleotides 470 to 472, and the start AUG of the MBGV NP gene spans nucleotides 104 to 106. Thus, the differences in length between replicated RNA and mRNA are considerable with EBOV minigenomes. Replicated RNA and mRNA of the MBGV replication system, however, are of similar size, since the only slightly smaller mRNA is elongated by the poly(A) tail. Northern blot analysis revealed that those minigenomes generating CAT activity in the EBOV replication system were indeed replicated and transcribed by the EBOV nucleocapsid proteins. The detected RNA species showed the expected size for positive-sense minireplicons and mRNA (Fig. 5C). The analogous result was received with the MBGV replication system (Fig. 5D). All minigenomes leading to CAT activity served as a template for replication and transcription. The chimeric minigenome 3M-5E was neither replicated nor transcribed. The prerequisite for replication of 3E-5M by EBOV is that the replication intermediate containing the positive-sense trailer of MBGV as 3′ end is recognized by the EBOV replication complex. It was therefore expected that an MBGV-specific copy-back minigenome containing 105 nucleotides complementary to the 5′ end of the MBGV genome as leader and the 5′ end of the MBGV genome as trailer (28) also could serve as a template for the EBOV polymerase. Surprisingly, this copy-back minigenome was not replicated by EBOV (data not shown), indicating that the leader and the trailer primary sequences were not sufficient to serve as replication signal.
Exchange of nucleocapsid proteins.
We then investigated whether the MBGV nucleocapsid proteins NP, VP35, and L could support replication and transcription of the EBOV-specific minigenome and vice versa. MVA-T7-infected HeLa cells were transfected with all possible combinations of EBOV and MBGV genes coding for NP, VP35, and L. For DNA transfection, 100 ng or 1 μg of pT/NP (MBGV or EBOV), respectively, 500 ng of pT/VP35 (MBGV or EBOV), and 1 μg of pT/L (MBGV or EBOV) were used. The cells were subsequently transfected with the respective minigenome RNA (3M-5M or 3E-5E) and finally tested for CAT activity at 2 days p.i. Using this protocol, CAT activity could only be observed with the homologous proteins (data not shown). None of the tested nucleocapsid proteins could be replaced by its respective analogue, indicating that functional complexes between heterologous proteins were not formed.
It was further investigated whether VP30 of MBGV could replace VP30 of EBOV. Cells were infected with MVA-T7 and transfected with pT/NPEBO, pT/VP35EBO, and pT/LEBO and either with pT/VP30EBO or with pT/VP30MBG. As a control, the vector pTM1 containing the green fluorescence protein gene (pT/GFP) was used. Thereafter, the cells were transfected with 3E-5E RNA and checked for CAT activity. As demonstrated in Fig. 6, the addition of MBGV VP30 led to a significant increase in reporter gene expression in a dose-dependent manner (lanes 4 and 5), indicating that MBGV VP30 could act as an enhancer for EBOV-specific transcription. However, the effect of MBGV VP30 was less pronounced compared to the effect of EBOV VP30 (lane 3).
FIG. 6.
Exchange of EBOV VP30 by MBGV VP30. HeLa cells were infected with MVA-T7 and transfected with DNA and RNA as described under Fig. 2. At 2 days p.i., the cell lysates were analyzed for CAT activity. DNA transfection was performed with the following plasmids: 500 ng of pT/NPEBO, 500 ng of pT/VP35EBO, and 1 μg of pT/LEBO; pT/VP30MBG and pT/GFP were added as indicated in the figure. As a negative control, pT/LEBO was omitted (negative control). For RNA transfection, minigenome 3E-5E was used. For the CAT assay, 1.5 μl of each cell lysate (except lane 3) was used. For the sample shown in lane 3, only a fifth of this amount was subjected to CAT assay. After quantification, the obtained value for the sample shown in lane 3 was multiplied by five and set as the maximal CAT activity (100%). −, DNA transfection with pT/NPEBO, pT/VP35EBO, and pT/LEBO.
DISCUSSION
In this study we describe an artificial replication system for EBOV which is based on the vaccinia virus T7 expression system. The established EBOV replication system extends the number of reverse genetic systems for NNS RNA viruses and allowed us to compare both members of the family of Filoviridae regarding their mode of replication and transcription.
Titration experiments performed with the plasmids encoding the EBOV nucleocapsid proteins revealed optimal amounts for NP and VP35 input DNA, which led to maximal CAT activity. When the amount of input DNA was further increased, CAT activity declined. It is not likely that simply the overexpression of one component (NP or VP35) resulted in an inhibition of the expression of other proteins since large amounts of pT/LEBO were tolerated without suppressing CAT activity. Similar results were obtained concerning titration of NP and VP35 DNA in the artificial replication system established for MBGV (28). Further, it has been shown for the Sendai and measles virus replication complexes that the amount of input DNA encoding the nucleoprotein could not be increased above a certain level without a decrease in reporter gene activity (7, 22). The observed decrease of reporter gene activity might be caused by an imbalance between the different components of the replication complex. This presumption is supported by titration experiments performed at optimal start concentrations of the L and VP35 input DNA. In these experiments, titration of the L input DNA led to an increase of reporter gene activity only when VP35, the second gene product, was increased simultaneously, indicating that L and VP35 form a functional complex, presumably the polymerase. This result is in line with the observation that the functional polymerase of other Mononegavirales types (Sendai virus, Newcastle disease virus, and vesicular stomatitis virus [VSV]) is composed of complexes between the second gene product (P) and L (14, 21–23). There is accumulating evidence that the filovirus VP35 is homologous to the P proteins of other Mononegavirales. First, it was observed previously that EBOV and MBGV VP35 are components of the nucleocapsid complex (3, 13); second, VP35 is essential for replication and transcription of both viruses; third, interaction between NP and VP35 (first and second gene product) parallels interaction between N (NP) and P of paramyxo-, rhabdo-, and borna viruses (3); and finally, EBOV VP35 is functionally linked to L. It is therefore proposed to further designate VP35 of the Filoviridae as P.
Comparison of the artificial replication systems for MBGV and EBOV revealed one striking difference. While replication and transcription of monocistronic MBGV minireplicons is not regulated by VP30, reporter gene expression mediated by EBOV nucleocapsid proteins is strongly increased by VP30. Moreover, EBOV-specific polyadenylated mRNA could only be detected in the presence of VP30. Replication and encapsidation of the EBOV-specific minigenomes, however, were not dependent on VP30. A similar effect has been described for the M2 protein of respiratory syncytial virus (RSV [8]). M2 represents the fourth nucleocapsid protein of RSV besides NP, P, and L. Bona fide transcripts of RSV genes could only be observed in the presence of M2. Otherwise, transcription was prematurely terminated. Since the EBOV-specific minigenomic DNA served as a template for vaccinia virus RNA polymerases, high-background signals were observed in the Northern hybridization experiments when the isolated RNA was neither treated with nuclease nor purified with oligo(dT) cellulose. Thus, under the chosen experimental conditions, prematurely terminated nonpolyadenylated mRNA species could not be detected. However, a residual transcriptional activity of the EBOV replication complex was found in the absence of VP30 as shown by the detected CAT activity. Thus, transcription was initiated, but possibly with a very low efficiency. The fact that no mRNA was detected in the absence of VP30 might be due to a lower sensitivity of the Northern hybridization compared to the CAT assay or to missing polyadenylation of the nascent RNA chains. Taken together, these results lead to the following hypotheses concerning the function of VP30: (i) VP30 acts as a positive regulator for transcription initiation, (ii) VP30 acts as an elongation factor during mRNA synthesis or polyadenylation as shown for RSV M2, and (iii) VP30 stabilizes the mRNA. Studies are currently under way to clarify the function of VP30.
Interestingly, MBGV VP30 was able to replace EBOV VP30, although with reduced efficiency. This indicates that VP30 is well conserved on a functional level even though it does not seem to be involved in transcription of MBGV-specific monocistronic minigenomes. Whether MBGV VP30 interacts with the EBOV-specific transcription complex or acts independently remains unclear.
In contrast to VP30, exchange of NP, VP35, and L between MBGV and EBOV was impossible. It is presumed that functional complexes of heterologous nucleocapsid proteins have not been formed. So far, exchange of single nucleocapsid proteins of heterologous NNS RNA viruses was only successful with a cell-free system. Chandrika et al. (7) reported that it was possible to replace the Sendai virus nucleoprotein by the measles virus nucleoprotein.
Several attempts have been made to investigate in vivo whether NNS RNA virus replication complexes were able to recognize heterologous RNA templates. Curran and Kolakofsky (10) reported that human parainfluenza virus (hPIV) type 1 and type 3 accepted a Sendai virus minireplicon as a template for replication, whereas measles virus did not. Using a plasmid-based artificial replication system, Pelet et al. (32) obtained comparable results. On the other hand, Dimock and Collins (12) demonstrated that rescue of hPIV type 3 minigenomes was not supported by RSV and, unexpectedly, not supported by bovine PIV type 3. For VSV, it was shown that replacement of defective interfering particles between the very closely related serotypes New Jersey and Indiana was only possible when the replication complex was supplied by VSV New Jersey and the minigenome by VSV Indiana (26). Thus, it appears that in most cases the replication complexes are highly specific for the homologous RNA. This is underscored by the result that the MBGV-specific minigenome was not accepted as a template by the EBOV replication complex and vice versa, regardless of whether the replication complex was supplied by infection with a helper virus or by expression of recombinant nucleocapsid proteins. Against the background of these findings, it was unexpected that a chimeric minigenome containing the EBOV leader and the MBGV trailer (3E-5M) was encapsidated, replicated, transcribed, and packaged by both viruses. Transcription of 3E-5M by both viruses indicates that the transcription start and stop signals are not highly specific for MBGV or EBOV but can be recognized by both filoviruses. Sequence comparison of these regulatory elements revealed a high degree of homology between MBGV and EBOV (29, 34).
The protein requirements for transcription of the 3E-5M chimera by the EBOV- or MBGV-specific replication complexes were the same as for the homologous minigenomes, namely, NP, VP35, and L for MBGV transcription and NP, VP35, L, and VP30 for EBOV transcription. When EBOV VP30 was omitted, polyadenylated 3E-5M mRNA could not be detected (data not shown). Since both viruses were able to transcribe the same RNA, it is assumed that signals located on the minigenomic RNAs were not responsible for the different transcription strategies of EBOV and MBGV. It is more likely that the structure or the composition of the transcription complexes are different.
ACKNOWLEDGMENTS
We thank Angelika Lander for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 286).
REFERENCES
- 1.Banerjee A K, Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188:417–428. doi: 10.1016/0042-6822(92)90495-b. [DOI] [PubMed] [Google Scholar]
- 2.Becker S, Mühlberger E. Co- and posttranslational modifications and functions of Marburg virus proteins. In: Klenk H-D, editor. Marburg and Ebola viruses. Berlin, Germany: Springer-Verlag; 1998. [DOI] [PubMed] [Google Scholar]
- 3.Becker S, Rinne C, Hofsäss U, Klenk H-D, Mühlberger E. Interactions of Marburg virus nucleocapsid proteins. Virology. 1998;249:406–417. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- 4.Becker S, Klenk H-D, Mühlberger E. Intracellular transport and processing of the Marburg virus surface protein in vertebrate and insect cells. Virology. 1996;225:145–155. doi: 10.1006/viro.1996.0582. [DOI] [PubMed] [Google Scholar]
- 5.Bowen E T, Lloyd G, Harris W J, Platt G S, Baskerville A, Vella E E. Viral haemorrhagic fever in southern Sudan and northern Zaire. Preliminary studies on the aetiological agent. Lancet. 1977;i:571–573. doi: 10.1016/s0140-6736(77)92001-3. [DOI] [PubMed] [Google Scholar]
- 6.Bukreyev A A, Volchkov V E, Blinov V M, Dryga S A, Netesov S V. The complete nucleotide sequence of the Popp (1967) strain of Marburg virus: a comparison with the Musoke (1980) strain. Arch Virol. 1995;140:1589–1600. doi: 10.1007/BF01322532. [DOI] [PubMed] [Google Scholar]
- 7.Chandrika R, Myers T, Moyer S A. Measles virus nucleocapsid protein can function in Sendai virus defective interfering particle genome synthesis in vitro. Virology. 1995;206:777–782. doi: 10.1016/s0042-6822(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 8.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a non-segmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conzelmann K-K. Genetic manipulation of nonsegmented negative strand RNA viruses. J Gen Virol. 1996;77:381–389. doi: 10.1099/0022-1317-77-3-381. [DOI] [PubMed] [Google Scholar]
- 10.Curran J A, Kolakofsky D. Rescue of a Sendai virus DI genome by other parainfluenza viruses: implications for genome replication. Virology. 1991;182:168–176. doi: 10.1016/0042-6822(91)90660-4. [DOI] [PubMed] [Google Scholar]
- 11.Davis K J, Anderson A O, Geisbert T W, Steele K E, Geisbert J B, Vogel P, Connolly B M, Huggins J W, Jahrling P B, Jaax N K. Pathology of experimental Ebola virus infection in African green monkeys. Involvement of fibroblastic reticular cells. Arch Pathol Lab Med. 1997;121:805–819. [PubMed] [Google Scholar]
- 12.Dimock K, Collins P L. Rescue of synthetic analogs of genomic RNA and replicative-intermediate RNA of human parainfluenza virus type 3. J Virol. 1993;67:2772–2778. doi: 10.1128/jvi.67.5.2772-2778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott L, Kiley M P, McCormick J B. Descriptive analysis of Ebola virus proteins. Virology. 1985;147:169–176. doi: 10.1016/0042-6822(85)90236-3. [DOI] [PubMed] [Google Scholar]
- 14.Emerson S U, Yu Y. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol. 1975;15:1348–1356. doi: 10.1128/jvi.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearns R, Peeples M E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 16.Feldmann H, Will C, Schikore M, Slenczka W, Klenk H-D. Glycosylation and oligomerization of the spike protein of Marburg virus. Virology. 1991;182:353–356. doi: 10.1016/0042-6822(91)90680-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldmann H, Mühlberger E, Randolf A, Will C, Kiley M P, Sanchez A, Klenk H-D. Marburg virus, a filovirus: messenger RNAs, gene order, and regulatory elements of the replication cycle. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]
- 18.Geisbert T W, Jaax N K. Marburg hemorrhagic fever: report of a case studied by immunohistochemistry and electron microscopy. Ultrastruct Pathol. 1998;22:3–17. doi: 10.3109/01913129809032253. [DOI] [PubMed] [Google Scholar]
- 19.Gorman M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosfeld H, Hill M G, Collins P L. RNA replication by respiratory syncytial virus RSV is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol. 1995;69:5677–5686. doi: 10.1128/jvi.69.9.5677-5686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamaguchi M, Yoshida T, Nishikawa K, Naruse H, Nagai Y. Transcriptive complex of Newcastle disease virus. I. Both L and P proteins are required to constitute an active complex. Virology. 1983;128:105–117. doi: 10.1016/0042-6822(83)90322-7. [DOI] [PubMed] [Google Scholar]
- 22.Horikami S M, Curran J, Kolakofsky D, Moyer S A. Complexes of Sendai virus NP-P and P-L proteins are required for defective-interfering genome replication in vitro. J Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Ferla F M, Peluso R W. The 1:1 N-NS protein complex of vesicular stomatitis virus is essential for efficient genome replication. J Virol. 1989;63:3852–3857. doi: 10.1128/jvi.63.9.3852-3857.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martini G A. Marburg agent disease in man. Trans R Soc Trop Med Hyg. 1969;63:295–302. doi: 10.1016/0035-9203(69)90001-7. [DOI] [PubMed] [Google Scholar]
- 25.Martini G A, Siegert R. Marburg virus disease. Berlin, Germany: Springer Verlag; 1971. [Google Scholar]
- 26.Moyer S A. Replication of the genome RNAs of defective interfering particles of vesicular stomatitis and Sendai viruses using heterologous viral proteins. Virology. 1989;172:341–345. doi: 10.1016/0042-6822(89)90136-0. [DOI] [PubMed] [Google Scholar]
- 27.Mühlberger E, Sanchez A, Randolf A, Will C, Klenk H-D, Feldmann H. The nucleotide sequence of the L gene of Marburg virus, a filovirus: homologies with paramyxoviruses and rhabdoviruses. Virology. 1992;187:534–547. doi: 10.1016/0042-6822(92)90456-y. [DOI] [PubMed] [Google Scholar]
- 28.Mühlberger E, Lötfering B, Klenk H-D, Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J Virol. 1998;72:8756–8764. doi: 10.1128/jvi.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mühlberger E, Trommer S, Funke C, Volchkov V, Klenk H-D, Becker S. Termini of all mRNA species of Marburg virus: sequence and secondary structure. Virology. 1996;223:376–380. doi: 10.1006/viro.1996.0490. [DOI] [PubMed] [Google Scholar]
- 30.Murphy F A. Pathology of Ebola virus infection. In: Pattyn S R, editor. Ebola virus hemorrhagic fever. Amsterdam, The Netherlands: Elsevier-North Holland; 1978. pp. 61–82. [Google Scholar]
- 31.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 32.Pelet T, Marq J B, Sakai Y, Wakao S, Gotoh H, Curran J. Rescue of Sendai virus cDNA templates with cDNA clones expressing parainfluenza virus type 3 N, P, and L proteins. J Gen Virol. 1996;77:2465–2469. doi: 10.1099/0022-1317-77-10-2465. [DOI] [PubMed] [Google Scholar]
- 33.Ryabchikova E, Kolesnikova L, Smolina M, Tkachev V, Pereboeva L, Baranova S, Grazhdantseva A, Rassadkin Y. Ebola virus infection in guinea pigs: presumable role of granulomatous inflammation in pathogenesis. Arch Virol. 1996;141:909–921. doi: 10.1007/BF01718165. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez A, Kiley M P, Holloway B P, Auperin D D. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez A, Trappier S G, Mahy B W, Peters C J, Nichol S T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez A, Yang Z Y, Xu L, Nabel G J, Crews T, Peters C J. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol. 1998;72:6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith D H, Johnson B K, Isaäcson M, Swanapoel R, Johnson K M, Kiley M P, Bagshawe A, Siongok T, Keruga W K. Marburg-virus disease in Kenya. Lancet. 1982;i:816–820. doi: 10.1016/s0140-6736(82)91871-2. [DOI] [PubMed] [Google Scholar]
- 38.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 38a.Volchkov, V. E. Unpublished data.
- 39.Volchkov V E, Feldmann H, Volchkova V A, Klenk H-D. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volchkov V E, Becker S, Volchkova V A, Ternovoj V A, Kotov A N, Netesov S V, Klenk H D. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 41.Volchkov V E, Volchkova V A, Chepurnov A A, Blinov V M, Dolnik O, Netesov S, Feldmann H. Characterization of the L gene and 5′ trailer region of Ebola virus. J Gen Virol. 1999;80:355–362. doi: 10.1099/0022-1317-80-2-355. [DOI] [PubMed] [Google Scholar]
- 42.Will C, Mühlberger E, Linder D, Slenczka W, Klenk H-D, Feldmann H. Marburg virus gene four encodes the virion membrane protein, a type I transmembrane glycoprotein. J Virol. 1993;67:1203–1210. doi: 10.1128/jvi.67.3.1203-1210.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]