Abstract
Coreceptor usage of primary human immunodeficiency virus type 1 (HIV-1) isolates varies according to biological phenotype. The chemokine receptors CCR5 and CXCR4 are the major coreceptors that, together with CD4, govern HIV-1 entry into cells. Since CXCR4 usage determines the biological phenotype for HIV-1 isolates and is more frequent in patients with immunodeficiency, it may serve as a marker for viral virulence. This possibility prompted us to study coreceptor usage by HIV-2, known to be less pathogenic than HIV-1. We tested 11 primary HIV-2 isolates for coreceptor usage in human cell lines: U87 glioma cells, stably expressing CD4 and the chemokine receptor CCR1, CCR2b, CCR3, CCR5, or CXCR4, and GHOST(3) osteosarcoma cells, coexpressing CD4 and CCR5, CXCR4, or the orphan receptor Bonzo or BOB. The indicator cells were infected by cocultivation with virus-producing peripheral blood mononuclear cells and by cell-free virus. Our results show that 10 of 11 HIV-2 isolates were able to efficiently use CCR5. In contrast, only two isolates, both from patients with advanced disease, used CXCR4 efficiently. These two isolates also promptly induced syncytia in MT-2 cells, a pattern described for HIV-1 isolates that use CXCR4. Unlike HIV-1, many of the HIV-2 isolates were promiscuous in their coreceptor usage in that they were able to use, apart from CCR5, one or more of the CCR1, CCR2b, CCR3, and BOB coreceptors. Another difference between HIV-1 and HIV-2 was that the ability to replicate in MT-2 cells appeared to be a general property of HIV-2 isolates. Based on BOB mRNA expression in MT-2 cells and the ability of our panel of HIV-2 isolates to use BOB, we suggest that HIV-2 can use BOB when entering MT-2 cells. The results indicate no obvious link between viral virulence and the ability to use a multitude of coreceptors.
The discovery that the replication of some human immunodeficiency virus type 1 (HIV-1) isolates can be suppressed in vitro by the CC chemokines MIP-1α, MIP-1β, and RANTES (15) and the cDNA cloning of a cofactor, CXCR4, that, along with CD4, allows the entry of T-cell-line-adapted (TCLA) HIV-1 strains into target cells (23) are two independent findings of great importance for research on HIV attachment, penetration, and cell tropism. It has been shown that HIV-1 isolates suppressed by the CC chemokines use the receptor for these ligands, CCR5, as a coreceptor (5, 19, 21), whereas isolates resistant to this suppressive effect use CXCR4. Infection of the latter group of viruses can be blocked by the CXC chemokine SDF-1, a ligand for CXCR4 (9, 38). CCR5 usage corresponds to a slow/low, non-syncytium-inducing (NSI) phenotype in previous classifications of primary HIV-1 isolates, whereas isolates with a rapid/high, syncytium-inducing (SI) phenotype use CXCR4 (8, 19, 23, 49). Some isolates are dualtropic, capable of using both CCR5 and CXCR4 and, in some instances, CCR3. A new, alternative HIV-1 phenotype classification system, based on coreceptor usage, was recently proposed; in this system, isolates using CCR5 are called R5 viruses, while isolates using CXCR4 are called X4 viruses. Isolates able to use both coreceptors are called R5X4 viruses (6).
The importance of the CCR5 molecule in HIV-1 transmission in vivo is supported by the observation that individuals homozygous for a 32-bp deletion in the CCR5 gene are almost completely resistant to HIV-1 infection (32, 42). This deletion leads to the production of defective CCR5 that is not transported to the cell surface, with the consequence that such cells are resistant to infection with HIV-1 using CCR5, the phenotype most frequently associated with sexual transmission (40, 47, 50).
HIV-2 was first isolated from West African patients with AIDS (2, 14). HIV-2 infection, which is prevalent in West African countries (reviewed in reference 34), is associated with a slower sexual spread (29) and a reduced rate of disease progression, compared to HIV-1 infection (35). The reason for the reduced virulence of HIV-2 remains unclear. HIV-2, like HIV-1, has been phenotypically divided into a rapid/high or slow/low phenotype (1, 3). The host range of HIV-2 in human cells is similar to that of HIV-1, yet differences exist in the ability to infect certain established cell lines. For example, human CD4+ U87 glioma cells have been shown to be susceptible to infection by HIV-2 and simian immunodeficiency virus (SIV) but not by HIV-1 (13).
While a considerable amount of knowledge concerning chemokine receptor usage by HIV-1 has accumulated during the last few years, less is known about the role of chemokine receptors in HIV-2 infection. SIV, which is genetically closer to HIV-2 than to HIV-1, has been shown to use CCR5 as a coreceptor for entry, regardless of other phenotypic properties (12). HIV-2 envelope glycoproteins, like those of HIV-1 and SIV, are able to use CCR5 as a coreceptor for viral entry and undergo CD4-dependent interactions with the same chemokine receptor (27). CXCR4, CCR3, and the orphan receptor V28 have been shown to mediate CD4-independent HIV-2 infection or infection in the presence of soluble CD4 of certain laboratory-adapted strains (22, 39). Bron and colleagues found that a laboratory-adapted HIV-2 strain showed considerable promiscuity in chemokine receptor usage (10). Other studies have indicated that promiscuity in chemokine receptor usage is common among primary HIV-2 isolates as well (26, 36, 44). However, there were differences in the patterns of HIV-2 coreceptor usage presented in these three studies, and only a limited number of primary isolates were tested in each study.
Recently, several additional HIV coreceptors were identified. Deng and coworkers reported on two novel coreceptors, Bonzo (also known as STRL33 [31]) and BOB (also known as GPR15 [25]), mainly used by SIV and HIV-2 isolates but also used by some HIV-1 isolates (20). Bonzo mRNA was detected in U87 cells, and antiserum raised against the N-terminal sequence of Bonzo blocked SIV Env-mediated infection. Moreover, the chemokine receptor CCR8 was shown to function as a coreceptor for several different HIV-1, HIV-2, and SIV Env proteins, whereas the orphan receptor V28 was used only by a few HIV-1 and HIV-2 isolates (41).
The tropism of HIV-1 for T-cell lines has been correlated with determinants within the loop structure in the third variable region (V3) of the HIV-1 envelope glycoprotein (27) and with coreceptor usage (8, 19, 21, 23). Moreover, the V3 loop has been identified as a critical determinant for susceptibility to the inhibitory effects of chemokines (16). It has also been reported that selective chemokine receptor usage by HIV-1 is predicted by determinants within the V3 region (45, 48). Less is known about determinants of cell tropism for HIV-2, even though the V3 loop has been suggested to be involved in viral fusion (24). Albert and colleagues found a correlation between viral biological phenotype and HIV-2 V3 genotype, as measured by net charge, sequence heterogeneity, and positively charged mutations at positions 313 and 314 (4).
To investigate which cellular receptors are involved when primary HIV-2 isolates infect cells, we used human U87 glioma cell lines stably expressing CD4 and the chemokine receptor CCR1, CCR2b, CCR3, CCR5, or CXCR4. GHOST(3) osteosarcoma cells coexpressing CD4 and CCR5, CXCR4, Bonzo, or BOB were also used. Eleven primary HIV-2 isolates previously characterized for phenotype (rapid/high or slow/low) and with known V3 sequences were tested for coreceptor usage and were characterized for MT-2 cell tropism. The expression of BOB mRNA in MT-2 cells was also investigated.
MATERIALS AND METHODS
Patients and virus isolates.
Eight primary HIV-2 isolates originating from Guinea-Bissau, two originating from the Ivory Coast (1653 and 1654), and one originating from an individual of Gambian origin (6669) were used in this study. Isolates 1816 and 2300 were obtained sequentially from one individual over a 6-month interval. The isolates were obtained by cocultivation of peripheral blood mononuclear cells (PBMC) from HIV-2-infected individuals with PBMC from healthy blood donors as previously described (3). The biological phenotype of each isolate was previously determined by cocultivation of infected PBMC with CEM, Jurkat-tat, and U937 clone 2 (U937-2) cell lines (3). Four isolates displayed a rapid/high phenotype, while seven isolates were classified as slow/low (Table 1). Isolate 6669 was included only in the cell-free infection experiments. HIV-1IIIB, known to use CXCR4 exclusively as a coreceptor, was included as a control in all experiments. The primary isolates had all been passaged in human PBMC.
TABLE 1.
Characteristics of the primary HIV-2 isolates used in this study
| Virus isolate | Subtypea | Clinical status | Biological phenotypeb | MT-2 tropismc
|
|
|---|---|---|---|---|---|
| Cell-free virus infection | Cocultivation | ||||
| 1010 | A | AIDS | Rapid/high | + | + |
| 1654 | A | AIDS | Rapid/high | * | * |
| 1682 | A | Asymptomatic | Slow/low | * | * |
| 1808 | A | Asthenia | Slow/low | * | * |
| 1812 | A | AIDS | Slow/low | − | *d |
| 1816 | A | AIDS | Slow/low | * | * |
| 2297 | A | Asthenia | Slow/low | + | + |
| 2298 | A | Asymptomatic | Slow/low | * | * |
| 2300 | A | AIDS | Slow/low | − | *d |
| 6669 | A | AIDS | Rapid/high | + | + |
| 1653 | B | AIDS | Rapid/high | * | * |
Determined by phylogenetic tree analysis of V3 sequences (4).
Four experiments were carried out, two by cocultivation with infected PBMC and two by cell-free virus infection. +, HIV-2 antigen production and syncytium formation; *, late HIV-2 antigen production and late single-cell death; −, no HIV-2 antigen production or syncytium formation.
Negative for both HIV-2 antigen production and cytopathic effects in one of two experiments.
Virus stocks were prepared by infecting 5 × 106 phytohemagglutinin P (Pharmacia, Uppsala, Sweden)-stimulated PBMC from two healthy blood donors with 2 ml of supernatant from infected PBMC. Supernatants were harvested on day 7 after infection and stored at −80°C. The 50% tissue culture infective doses (TCID50) of the supernatants were determined as previously described (7) with PBMC and an in-house HIV-2 capture enzyme-linked immunosorbent assay (ELISA) (46). The PBMC cultures were maintained in RPMI 1640 medium with 3 mM glutamine (Gibco BRL, Paisley, United Kingdom) and supplemented with 10% fetal calf serum (FCS) (Flow, Costa Mesa, Calif.), 5 U of recombinant interleukin 2 (Amersham, Buckinghamshire, United Kingdom) per ml, 2 μg of Polybrene (Sigma, St. Louis, Mo.) per ml, and antibiotics.
Cell lines.
Human glioma U87.CD4 cells, stably expressing CD4 and the chemokine receptor CCR1, CCR2b, CCR3, CCR5, or CXCR4, were previously described (8, 19). The GHOST(3) cell lines constitute an HIV-1 or HIV-2 indicator cell panel whose individual lines express a specific viral coreceptor molecule in conjunction with human CD4 (30). In brief, the GHOST(3) cell panel was derived from a clone of human osteosarcoma cells which stably express human CD4, HOS.T4neo. HOS.T4neo cells were stably transfected with a Tat-dependent reporter construct consisting of the HIV-2ROD long terminal repeat enhancer-promoter directing the expression of a humanized allele of the green fluorescent protein (GFP) (18). A clone (clone 3) which expressed a low basal level of GFP but which induced a high level of GFP expression in response to HIV-1LAI Tat was isolated. The GFP reporter-containing HOS.T4 neo cells were subsequently stably transduced with retroviral vectors containing different HIV coreceptor molecules, including CCR5, CXCR4, BOB/GPR15, and Bonzo/STRL33. Lines are designated by the coreceptor molecule expressed, except for the non-coreceptor-expressing progenitor, which is referred to as the GHOST(3) parental line. The GHOST(3) cell panel with the described coreceptors is now available through either the U.S. NIH AIDS Reagent Program or the British NIBSC AIDS Reagent Project. Note that the GHOST(3) cell panel is distinct from the GHOST(34) cell panel distributed previously (11, 33).
The U87.CD4 and GHOST(3) cells were grown in high-glucose Dulbecco’s modified Eagle’s medium (D-MEM) supplemented with 10% FCS and antibiotics. Cultures were kept in 25-cm2 tissue culture flasks (Costar) and detached and split 1:2 (U87.CD4) or 1:5 [GHOST(3)] every 2 to 3 days with trypsin-EDTA (Gibco BRL).
For infection experiments, cells were seeded in 24-well (U87.CD4) or 48-well [GHOST(3)] plates (Costar) 1 to 2 days prior to infection to obtain a subconfluent cell layer by the time of infection.
Cocultivation of infected PBMC with the U87.CD4 cell lines.
PBMC cultures were infected with virus as described above. Seven days postinfection, when cultures showed HIV-2 antigen production (optical density values ranging from 1.4 to 1.9 in our in-house HIV-2 capture ELISA), 150 × 103 PBMC from infected cultures were added to each well of a 24-well plate containing U87.CD4 cells (ratio, 1:2). After 24 h, the plate was washed with phosphate-buffered saline, and fresh D-MEM was added. The cultures were monitored for 10 days, and the medium was changed every second day. Inspection for syncytium formation was performed daily. Confluent cultures with no observable cytopathic effects were split 1:2 on day 5.
Cell-free infection of the U87.CD4 cell lines.
We used 1,000 to 2,000 TCID50 of each isolate in a final volume of 1 ml of D-MEM to infect U87.CD4 cells expressing the different chemokine receptors. Twenty-four hours later, 1 ml of fresh medium was added. Cultures were washed twice on day 2 and then were monitored for 10 days as described above.
Cell-free infection of the GHOST(3) cell lines.
GHOST(3) cells expressing CD4 and the different HIV coreceptors were infected with 1,000 to 2,000 TCID50 of each isolate diluted to a final volume of approximately 1 ml in D-MEM supplemented with 10% FCS, 2 μg of Polybrene per ml, and antibiotics. Twelve hours postinfection, the cultures were washed, and fresh medium was added. The cultures were trypsinized and split 1:5 every second day after supernatants were harvested and were visually inspected every day in a light microscope for cytopathic effects and in a fluorescence microscope for fluorescence induced by HIV-2 infection. Cultures were terminated 8 to 10 days postinfection.
MT-2 assay.
MT-2 cells cultured in 25-cm2 flasks with RPMI 1640 medium supplemented with 10% FCS and antibiotics were tested both by cell-free infection and by cocultivation with infected PBMC (ratio, 2:1). Cultures were maintained for at least 21 days, split and monitored for HIV-2 antigen production twice a week, and visually inspected daily for cytopathic effects.
Capture enzyme immunoassay for detection of HIV-2 antigen.
The in-house HIV-2–SIV capture ELISA has been described elsewhere (46). In brief, cell culture supernatants were added to 96-well microtiter plates previously coated with purified immunoglobulin G from an asymptomatic HIV-2-positive blood donor and blocked with 1% bovine serum albumin. Captured HIV-2 antigen was detected with rabbit anti-SIVmac serum (cross-reactive with HIV-2) and then with horseradish peroxidase-conjugated swine anti-rabbit immunoglobulin. o-Phenylenediamine substrate with H2O2 was added, and optical density was measured at 490 nm after the reaction was stopped with 2.5 M H2SO4.
Northern blot analysis.
Total RNA (15 μg) isolated from the different cell lines with Ultraspec RNA (Biotecx Laboratories, Inc., Houston, Tex.) was electrophoresed in a 1.2% agarose–formaldehyde gel and transferred to a Hybond-N+ nylon membrane (Amersham). Full-length BOB cDNA labelled with 32P by use of Rediprime II (Amersham) was used for probing the membrane. Hybridization was performed at 42°C with hybridization solution (50% formamide, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt’s solution, 0.5% sodium dodecyl sulfate [SDS], 100 μg of denatured salmon sperm DNA per ml). The membrane was washed twice in 0.1× SSC–0.1% SDS at room temperature. Equivalent levels of mRNA among the samples were verified by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probing. Dehybridization of the membrane was confirmed before repeat or control hybridizations were carried out.
RT-PCR.
Two micrograms of total RNA isolated from the different cell lines with Ultraspec RNA was used for cDNA synthesis with a 1st Strand cDNA Synthesis Kit for reverse transcriptase (RT) PCR (RT-PCR) (Boehringer Mannheim Biochemicals), random hexamer primers, and avian myeloblastosis virus RT. Four microliters of the 40-μl reaction mixture was used as a template for PCR amplification with AmpliTaq DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.) at 94°C for 1 min, followed by 35 cycles of 94°C for 1 min, 58°C (55°C for GAPDH) for 1 min, and 72°C for 1 min, and finally a 10-min extension at 72°C. The primers used were as follows: for BOB, 5′-CATCTGCTCTTTGGTGATG-3′ and 5′-GTATGGCTTATCATCAATCAGC-3′; for GAPDH, 5′-GGTGAAGGTCGGAGTCAACG-3′ and 5′-CAAAGTTGTCATGGATGACC-3′. Parallel cDNA synthesis reactions in which the RT enzyme was left out to exclude the possibility of contaminating genomic DNA were uniformly negative.
RESULTS
Infection of MT-2 cells.
All isolates were tested for their ability to replicate and induce syncytia in MT-2 cells, both by cocultivation with infected PBMC and by cell-free virus infection (Table 1). Typical syncytia and HIV-2 antigen production a few days after infection were induced only by three HIV-2 isolates. The majority of the remaining isolates showed an atypical cytopathic effect, without evident syncytia but with single-cell killing accompanied by late virus production 10 to 25 days postinfection. Two of the SI isolates (1010 and 6669) were recovered from AIDS patients, while the third (2297) originated from a patient with asthenia. Moreover, of the four rapid/high isolates, classified by their ability to infect and replicate in established CEM and U937-2 cell lines, only two (1010 and 6669) induced typical syncytia in MT-2 cells. Conversely, isolate 2297 did not replicate in CEM or U937-2 cells but replicated and induced syncytia in MT-2 cells. With isolates 1812 and 2300, cell-free infection of MT-2 cells was unsuccessful. These results indicate that HIV-2, unlike HIV-1, may enter the MT-2, CEM, and U937-2 cell lines through more than one pathway.
Chemokine receptor usage by primary HIV-2 isolates. (i) Infection of U87.CD4 cells stably expressing chemokine receptors.
Infection was carried out in two ways: by cocultivation with PBMC cultures infected with each of 11 primary HIV-2 isolates 7 days earlier and with cell-free virus. Syncytium formation was recorded at day 7 (cocultivation) and at day 10 (cell-free infection) postinfection. HIV-2 antigen production was recorded at day 10 postinfection in the cell-free infection experiment.
All HIV-2 isolates, except for 6669, efficiently induced syncytia in U87.CD4.CCR5 cells, whereas only two isolates, 1010 and 6669, both recovered from AIDS patients, formed syncytia in U87.CD4.CXCR4 cells (Table 2). HIV-2 antigen production was detected in all syncytium-positive cultures. Low levels of HIV-2 antigen, in the absence of syncytia, were detected in cultures of CXCR4-expressing cells upon infection with isolates 1682 and 1808. In addition, the majority of the isolates induced syncytia in CCR1- and CCR3-expressing cells, particularly upon cocultivation with infected PBMC, whereas syncytium formation in CCR2b-expressing cells was seen only with some of the isolates. None of the isolates replicated in parental U87.CD4 cells, but 7 of 11 isolates induced syncytia after cocultivation with infected PBMC. Interestingly, this effect was seen with viruses able to use several coreceptors. Syncytium formation upon inoculation with the TCLA HIV-1IIIB isolate was seen only in CXCR4-expressing cells.
TABLE 2.
Chemokine receptor usage by primary HIV-2 isolates tested in U87.CD4 cells expressing chemokine receptors
| Virus isolate | HIV-2 antigen production (Ag) and syncytium induction (S) in U87.CD4 cells expressing the following chemokine receptora:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| None
|
CCR1
|
CCR2bb
|
CCR3
|
CCR5
|
CXCR4
|
|||||||
| Ag | S | Ag | S | Ag | S | Ag | S | Ag | S | Ag | S | |
| 1010 | 0 | − | 1.3 | +++ | 1.1 | + | 1.4 | ++++ | 1.1 | +++ | 1.2 | ++++ |
| 1654 | 0 | − | 0.8 | + | 0 | − | 1.1 | + | 1.2 | ++++ | 0 | − |
| 1682 | 0 | − | 1.3 | ++++ | 1.0 | + | 1.3 | +++ | 1.2 | ++++ | 0.4 | − |
| 1808 | 0.2 | − | 1.2 | +++ | 0.6 | − | 1.2 | ++ | 1.2 | ++++ | 0.4 | − |
| 1812 | 0 | − | ND | − | 0.2 | − | 1.0 | − | 1.3 | ++++ | 0 | − |
| 1816 | 0 | − | 0.4 | + | 0.7 | + | 0.9 | − | 1.3 | ++++ | 0 | − |
| 2297 | 0 | − | 0.9 | + | 0 | − | 0.5 | − | 1.1 | +++ | 0 | − |
| 2298 | 0 | − | ND | ∗ | 0.3 | + | 1.3 | − | 1.2 | ++++ | 0 | − |
| 2300 | 0 | − | 1.2 | ++ | 0 | − | 1.0 | ++ | 1.2 | ++++ | 0 | − |
| 6669c | 0 | − | 0 | − | 0 | − | 1.6 | +++ | 0 | − | 1.2 | ++ |
| 1653 | 0 | − | 0 | − | 0 | − | 0 | − | 1.1 | ++++ | 0 | − |
| IIIBd | ND | − | ND | − | ND | − | ND | − | 0e | − | >2.7e | ++++ |
Infection assays were performed both by cocultivation of U87.CD4 cells with infected PBMC and by cell-free virus infection with 1,000 to 2,000 TCID50 of each isolate. The data shown indicate results obtained with both methods. Differences, whenever apparent, are indicated. Experiments were repeated two to four times. Ag, cell culture supernatants were harvested at day 10 postinfection and monitored for HIV-2 antigen with an in-house capture ELISA; values represent the mean optical density at 490 nm from independent experiments. ND, not determined. S, syncytium formation was visually quantitated at day 10 postinfection: ++++, syncytia covering the well; +++, large syncytia covering >50% of the well; ++, large syncytia covering <50% of the well; +, occasional small syncytia; −, no syncytia; ∗, syncytia formed in cocultivation experiment, cell-free virus infection not performed. Boldfacing indicates that syncytium formation occurred only after cocultivation with infected PBMC.
Results were obtained at day 8 postinfection.
6669 was not included in the cocultivation experiment.
Prototype TCLA laboratory HIV-1 strain.
p24 production measured with an in-house HIV-1 p24 capture ELISA.
Comparison between the modes of infection generally showed a lower efficiency of syncytium induction after cell-free virus infection than after cocultivation with infected PBMC (Tables 2 and 3). However, this variation in efficiency was isolate and coreceptor dependent, exemplified by isolate 1682, which induced syncytia with equal efficiencies after cocultivation and cell-free infection in CCR1- and CCR5-expressing cells but not in CCR2b- or CCR3-expressing cells, and by isolate 1010, which used CCR3 and CXCR4 with equal efficiencies but which showed differences when tested for the usage of all other receptors.
TABLE 3.
Comparison of chemokine receptor usage patterns of primary HIV-2 isolates tested by cocultivation versus cell-free virus infection and in U87.CD4 cells versus GHOST(3) cells
| Chemokine receptor | Cell line | Mode of infectiona | Results for isolateb
|
|||||
|---|---|---|---|---|---|---|---|---|
| 1653
|
1682
|
1010
|
||||||
| Ag | s or f | Ag | s or f | Ag | s or f | |||
| None | U87.CD4 | Cocultivation | NA | − | NA | SSS | NA | SSS |
| U87.CD4 | Cell-free infection | 0 | − | 0 | − | 0 | − | |
| GHOST(3) | Cell-free infection | 0 | − | 0 | − | 0 | F | |
| CCR1 | U87.CD4 | Cocultivation | NA | − | NA | SSSS | NA | SSSS |
| U87.CD4 | Cell-free infection | 0 | − | 1.3 | SSSS | 1.3 | SSS | |
| CCR2b | U87.CD4 | Cocultivation | NA | − | NA | SSSS | NA | SSSS |
| U87.CD4 | Cell-free infection | 0 | − | 1.0 | S | 1.1 | S | |
| CCR3 | U87.CD4 | Cocultivation | NA | − | NA | SSSS | NA | SSSS |
| U87.CD4 | Cell-free infection | 0 | − | 1.3 | SSS | 1.4 | SSSS | |
| CCR5 | U87.CD4 | Cocultivation | NA | SSSS | NA | SSSS | NA | SSS |
| U87.CD4 | Cell-free infection | 1.1 | SSSS | 1.2 | SSSS | 1.1 | SS | |
| GHOST(3) | Cell-free infection | 0.2 | FFF | 0 | FFF | 0.5 | FF | |
| CXCR4 | U87.CD4 | Cocultivation | NA | − | NA | − | NA | SSSS |
| U87.CD4 | Cell-free infection | 0 | − | 0.4 | − | 1.2 | SSSS | |
| GHOST(3) | Cell-free infection | 0 | − | 0 | − | 2.2 | FFF | |
Infection assays were performed both by cocultivation of U87.CD4 cells with infected PBMC and by cell-free virus infection with 1,000 to 2,000 TCID50 of each isolate.
Cell culture supernatants were harvested at day 8 [GHOST(3)] or day 10 (U87.CD4) postinfection and monitored for HIV-2 antigen production (Ag) with an in-house capture ELISA; values indicate the optical density at 490 nm. NA, not applicable. Syncytium formation (s) in U87.CD4 cell cultures was visually quantitated at day 10 postinfection: SSSS, syncytia covering the well; SSS, large syncytia covering >50% of the well; SS, large syncytia covering <50% of the well; S, occasional small syncytia; −, no syncytia. HIV-induced GFP activation (f) in GHOST(3) cell cultures was detected by observing the cultures with a fluorescence microscope. The percentage of cells showing fluorescence was estimated at day 8 postinfection; FFF, >30%; FF, 5 to 30%; F, 1 to 5%; −, 0%.
(ii) Infection of GHOST(3) cells expressing chemokine receptors and the orphan receptors Bonzo and BOB.
Since 7 of 11 HIV-2 isolates formed syncytia in parental U87.CD4 cells, known to express low levels of Bonzo mRNA (20), it was of interest to test the coreceptor usage of HIV-2 isolates in another cell line, devoid of endogenous Bonzo mRNA expression. GHOST(3) cells expressing CD4 and the CCR5, CXCR4, Bonzo, or BOB receptor and engineered to express GFP upon HIV infection were infected with 1,000 to 2,000 TCID50 of the HIV-2 isolates. The cultures were microscopically inspected for fluorescence and monitored for HIV-2 antigen production. The pattern of CCR5 and CXCR4 usage was confirmed; all HIV-2 isolates, except for 6669, used CCR5, whereas isolates 1010 and 6669 used CXCR4 (Table 4). Isolates 1682 and 1808, which showed a low level of HIV-2 antigen production in U87.CD4.CXCR4 cells, showed no production in GHOST(3).CXCR4 cells.
TABLE 4.
Coreceptor usage by primary HIV-2 isolates tested in GHOST(3) cells coexpressing CD4 and CCR5, CXCR4, or the orphan receptor Bonzo or BOB
| Virus isolate | HIV-2 antigen production (Ag) and GFP activation (F) in GHOST(3) cells coexpressing CD4 and the following HIV coreceptora:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| None
|
CCR5
|
CXCR4
|
Bonzo
|
BOB
|
||||||
| Ag | F | Ag | F | Ag | F | Ag | F | Ag | F | |
| 1010 | 0 | + | 0.5 | ++ | 2.2 | +++ | 0.2 | ++ | 0.4 | ++ |
| 1654 | 0 | − | 1.3 | +++ | 0 | − | 0 | + | 0.2 | ++ |
| 1682 | 0 | − | 0 | +++ | 0 | − | 0 | + | 0 | + |
| 1808 | 0 | − | 1.2 | +++ | 0 | − | 0 | + | 0 | + |
| 1812 | 0 | − | 1.3 | +++ | 0 | − | 0 | + | 0.2 | ++ |
| 2297 | 0 | + | 1.4 | +++ | 0 | + | 0 | + | 2.2 | +++ |
| 2298 | 0 | − | 1.2 | +++ | 0 | − | 0 | + | 0 | + |
| 2300 | 0 | − | 1.2 | +++ | 0 | − | 0 | + | 0 | ++ |
| 6669 | 0 | + | 0 | + | 1.2 | +++ | 0 | + | 0 | + |
| 1653 | 0 | − | 0.2 | +++ | 0 | − | 0 | − | 0 | + |
| IIIBb | 0c | + | 0c | + | >2.7c | +++ | 0 | + | 0 | + |
Infection assays were performed by cell-free virus infection with 1,000 to 2,000 TCID50 of each HIV-2 isolate. Experiments were carried out twice. Ag, cell culture supernatants were harvested at day 8 postinfection and monitored for HIV-2 antigen with an in-house capture ELISA; values represent the mean optical density at 490 nm from two independent experiments (0 indicates an optical density of <0.2). F, HIV-induced GFP activation in GHOST(3) cell cultures was detected by observing the cultures with a fluorescence microscope. The percentage of cells showing fluorescence was estimated at day 8 postinfection: +++, >30%; ++, 5 to 30%; +, 1 to 5%; −, 0%.
Prototype TCLA laboratory HIV-1 strain.
p24 production measured with an in-house HIV-1 p24 capture ELISA.
Bonzo-expressing cells showed no or weak fluorescence and remained antigen negative after infection with all HIV-2 isolates tested, except for 1010. In BOB-expressing cells, five of the isolates induced fluorescence in more than 5% of the cells at day 8 postinfection, with four isolates also inducing the production of HIV-2 antigen (Table 4). Even when the level of replication was barely detectable at day 8, HIV-2 antigen production increased for all four isolates 2 days later (data not shown). The same result was observed with isolate 1010 in Bonzo-expressing cultures. GFP activation paralleled antigen production in BOB-expressing cells. The results showed that 5 of 10 HIV-2 isolates efficiently used BOB in addition to CCR5, whereas only one isolate used Bonzo efficiently.
HIV-2 infection detected by GFP activation correlated with HIV-2 antigen production, even when fluorescence was observed earlier than antigen production and, in some cases, also in antigen-negative cultures. These findings indicate that long terminal repeat-driven GFP expression in GHOST(3) cells is a more sensitive assay for HIV-2 infection than our in-house HIV-2 capture ELISA. The weak fluorescence seen in some of the HIV-2 antigen-negative cultures may indicate a low-efficiency function of a particular coreceptor or may, for CXCR4-using isolates, be due to low levels of endogenously expressed CXCR4. The latter possibility is suggested by the fact that HIV-26669 and HIV-1IIIB infections induced fluorescence across the panel of cells, although productive infection could be detected only in cells engineered to express CXCR4.
Detection of BOB mRNA in the MT-2, CEM, and U937-2 cell lines.
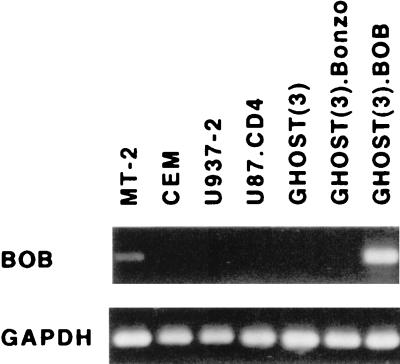
Since infection and replication in MT-2 cells for our panel of primary HIV-2 isolates were not restricted to CXCR4 usage, we investigated the expression of alternative HIV-2 coreceptors in MT-2 cells. The observation that the isolate with the most prominent ability to use BOB, isolate 2297, was also the only non-CXCR4-using isolate able to induce syncytia in MT-2 cells prompted us to investigate BOB mRNA expression in MT-2 cells. With GHOST(3).BOB cells as a positive control, low levels of BOB mRNA could be detected in MT-2 cells by RT-PCR (Fig. 1) but not by Northern blot analysis. In contrast to MT-2 cells, neither CEM nor U937-2 cells expressed BOB mRNA at levels detectable by RT-PCR or Northern blot analysis. Moreover, no BOB mRNA could be detected in parental U87.CD4 cells or GHOST(3) cells.
FIG. 1.
Expression of BOB mRNA in various cell lines analyzed by RT-PCR. Four microliters from a total of 40 μl of cDNA synthesized from 2 μg of total RNA was subjected to PCR amplification with BOB primers or GAPDH primers as an internal control. Primer specificity was verified with GHOST(3).BOB cells. The PCR-amplified products are shown (∼600 bp for BOB and 496 bp for GAPDH).
V3 sequence alignment.
The V3 sequences of the isolates were previously determined and aligned to a consensus of 10 published HIV-2 sequences (4, 37). In the present study, CXCR4 usage was associated with substitutions to positively charged residues at positions 314 and/or 313. For isolate 1010, there was a V314R substitution, while isolate 6669 exhibited two positively charged mutations, L313R and V314R. Only isolate 6669 had a V3 net charge (+9) significantly higher than the mean (+5.82). None of the other isolates had positively charged mutations at these positions. All isolates contained one N-linked glycosylation site within V3, at positions 302 to 304 (N-K-T).
DISCUSSION
In the present work, we studied coreceptor usage by 11 primary HIV-2 isolates in U87.CD4 cells expressing chemokine receptors previously shown to function as coreceptors for HIV-1 (CCR1, CCR2b, CCR3, CCR5, and CXCR4) and in GHOST(3) cells expressing CCR5, CXCR4, or orphan receptors implicated in SIV infection (Bonzo and BOB) together with CD4. We found that 10 of 11 primary HIV-2 isolates were able to use CCR5. In contrast, only two isolates, both from patients with advanced disease, were able to efficiently use CXCR4. These two isolates also promptly induced syncytia in MT-2 cells, a pattern described for HIV-1 isolates using CXCR4. However, in contrast to HIV-1 isolates, many of the HIV-2 isolates were promiscuous in their coreceptor usage in that they were able to use, in addition to CCR5, one or more of the CCR1, CCR2b, CCR3, and BOB receptors. This result is in line with findings obtained by other groups (10, 20, 26, 36, 44). Interestingly, the least promiscuous isolate, 1653, efficiently using only CCR5 but still able to replicate in MT-2 cells, was the only HIV-2 isolate of subtype B. Due to its promiscuous use of coreceptors, HIV-2 does not easily fit into the classification system based on coreceptor usage that has been proposed for HIV-1 (6).
Another difference between HIV-1 and HIV-2 was that the ability to replicate in MT-2 cells appeared to be a general property of HIV-2 isolates. Nevertheless, although all HIV-2 isolates replicated in MT-2 cells, they differed in replication kinetics and syncytium-inducing capacity; syncytium induction was correlated with CXCR4 usage (except for isolate 2297), while the majority of the remaining isolates, not using CXCR4, replicated with a 1- or 2-week delay and did not induce syncytia. These findings suggest a CXCR4-independent pathway for HIV-2 infection of MT-2 cells. Observations in accordance with these have been made for HIV-2 (44) and for SIV (12). We now show that MT-2 cells express BOB mRNA. Considering the frequent ability of our HIV-2 isolates to use BOB, we suggest that HIV-2 can use BOB when entering MT-2 cells.
This suggestion explains why, unlike those of HIV-1, phenotypically distinct groups of HIV-2 could not be distinguished clearly in MT-2 cells. For HIV-1, replication in established cell lines, such as CEM and U937-2, with few exceptions correlates with replication and syncytium induction in MT-2 cells and can be explained by the use of CXCR4 as a coreceptor. While such a correlation can also be found among HIV-2 isolates, exceptions are frequently encountered. Based on replicative capacity in CEM and U937-2 cells, four HIV-2 isolates (all from patients with advanced disease) were previously classified as having a rapid/high phenotype. However, only two of these isolates showed the typical SI phenotype in MT-2 cells which was coincident with CXCR4 usage. Conversely, HIV-2 isolate 2297 was SI in MT-2 cells but was unable to replicate in CEM, U937-2, U87.CD4.CXCR4, and GHOST(3).CXCR4 cells. Since isolate 2297 was the isolate which used BOB most efficiently, we suggest that BOB could be the coreceptor mediating syncytium induction in MT-2 cells for this isolate. The ability to detect low levels of BOB mRNA by RT-PCR but not by Northern blotting indicates a low level of BOB expression in MT-2 cells and is in line with the slow replication kinetics of the non-CXCR4-using HIV-2 isolates in these cells. For the exceptional isolate 2297, efficient BOB usage and syncytium induction in MT-2 cells were coincident.
Cocultivation appeared to be a more effective mode of infection than cell-free virus (Table 3). In fact, after cocultivation, several isolates induced syncytia even in parental U87.CD4 cells. These cells are devoid of transfected chemokine receptors but spontaneously express Bonzo (20). The difference in infection efficiency could simply reflect virus dose dependency, assuming that the infected PBMC cultures used for cocultivation produce amounts of virus larger than the amounts used for cell-free infection. In addition to prolonged virus production, cell-to-cell contact between gp125-expressing PBMC and U87.CD4 cells may intensify the fusion reaction. In such a situation, syncytia may be produced in the absence of productive infection. Comparison of the results obtained with U87.CD4 and GHOST(3).Bonzo cells suggests that spontaneous Bonzo expression may contribute to the low level of syncytium formation in parental U87.CD4 cells. In addition, other cell surface molecules present on PBMC may also assist in cell-to-cell fusion.
The frequent usage of CCR5 by HIV-2 was confirmed with GHOST(3) osteosarcoma cells, as was rare CXCR4 usage. This CXCR4 usage pattern is in agreement with that found in two previous studies (26, 44) but is at variance with that found in another (36), in which all HIV-2 isolates were found to use CXCR4. Moreover, the frequent CCR5 usage reported here is supported by two of these studies (26, 36), whereas Sol and coworkers found a relative inability of HIV-2 to use CCR5 (44). Whether these differences are due to the collections of HIV-2 isolates used in the different studies or to differences in indicator cell systems remains to be clarified. In the previous studies, the orphan receptors Bonzo and BOB were not included. In our experiments, BOB was used by all HIV-2 isolates, except for 6669, while Bonzo was used less efficiently. It is an open question whether cooperation between different coreceptors on the cell surface may promote HIV entry. If so, the promiscuity of HIV-2 isolates may result in different patterns of permissiveness in different cell types.
What in vivo relevance has the broad coreceptor usage of HIV-2? The lower pathogenicity of HIV-2 than of HIV-1 is difficult to explain by a more broad coreceptor usage. Hence, other viral properties are strongly suggested as being responsible for the difference in virulence between these two viruses. Moreover, the observation that coreceptor usage by HIV-1 often broadens with disease progression (8, 17, 43) does not seem to apply to HIV-2, since in our experiments all isolates, whether from AIDS patients or asymptomatic persons, were equally promiscuous. However, as for HIV-1, CXCR4 usage by HIV-2 appears to be most frequent in late-stage infection.
The observation that mutations to positively charged amino acid residues at positions 314 and/or 313 for isolates 1010 and 6669 were associated with CXCR4 usage is concordant with the reported correlation between these mutations and a rapid/high phenotype (4). The role of the V3 region as a determinant for coreceptor usage and susceptibility to chemokine inhibition for HIV-1 implies the possibility of a similar function for the HIV-2 V3 region. Our results indicate that determinants within the V3 loop appear to predict CXCR4 usage. Infection inhibition experiments with peptides corresponding to the HIV-2 V3 region and the cells used in this study may answer the question of whether V3 is indeed the determinant for HIV-2 coreceptor usage.
In the present study, we found that 10 of 11 primary HIV-2 isolates recovered from patients at different stages of disease use CCR5 and that all of the isolates can use at least one additional coreceptor. CXCR4 usage was observed for only two isolates, both recovered from AIDS patients, suggesting that a switch in coreceptor usage from CCR5 to CXCR4, which is associated with disease progression in HIV-1 infection, may also occur in HIV-2-infected patients. The in vivo role of the promiscuous use of coreceptors by HIV-2 remains to be clarified.
ACKNOWLEDGMENTS
We thank Kajsa Aperia and Kerstin Andréasson for technical assistance.
This work was supported by grants from the Tobias Foundation and the Swedish Medical Research Council. V.N.K. is a postdoctoral fellow of the Damon Runyon-Walter Winchell Foundation, and D.R.L. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Albert J, Böttiger B, Biberfeld G, Fenyö E M. Replicative and cytopathic characteristics of HIV-2 and severity of infection. Lancet. 1989;1:852–853. doi: 10.1016/s0140-6736(89)92321-0. [DOI] [PubMed] [Google Scholar]
- 2.Albert J, Bredberg U, Chiodi F, Böttiger B, Fenyö E M, Norrby E, Biberfeld G. A new human retrovirus isolate of West African origin (SBL-6669) and its relationship to HTLV-IV, LAV-II, and HTLV-IIIB. AIDS Res Hum Retroviruses. 1987;3:3–10. doi: 10.1089/aid.1987.3.3. [DOI] [PubMed] [Google Scholar]
- 3.Albert J, Naucler A, Böttiger B, Broliden P A, Albino P, Ouattara S A, Björkegren C, Valentin A, Biberfeld G, Fenyö E M. Replicative capacity of HIV-2, like HIV-1, correlates with severity of immunodeficiency. AIDS. 1990;4:291–295. doi: 10.1097/00002030-199004000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Albert J, Stålhandske P, Marquina S, Karis J, Fouchier R A, Norrby E, Chiodi F. Biological phenotype of HIV type 2 isolates correlates with V3 genotype. AIDS Res Hum Retroviruses. 1996;12:821–828. doi: 10.1089/aid.1996.12.821. [DOI] [PubMed] [Google Scholar]
- 5.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 6.Berger E A, Doms R W, Fenyö E M, Korber B T M, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 7.Björling E, Scarlatti G, von Gegerfelt A, Albert J, Biberfeld G, Chiodi F, Norrby E, Fenyö E M. Autologous neutralizing antibodies prevail in HIV-2 but not in HIV-1 infection. Virology. 1993;193:528–530. doi: 10.1006/viro.1993.1160. [DOI] [PubMed] [Google Scholar]
- 8.Björndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyö E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 10.Bron R, Klasse P J, Wilkinson D, Clapham P R, Pelchen-Matthews A, Power C, Wells T N, Kim J, Peiper S C, Hoxie J A, Marsh M. Promiscuous use of CC and CXC chemokine receptors in cell-to-cell fusion mediated by a human immunodeficiency virus type 2 envelope protein. J Virol. 1997;71:8405–8415. doi: 10.1128/jvi.71.11.8405-8415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cecilia D, KewalRamani V N, O’Leary J, Volsky B, Nyambi P, Burda S, Xu S, Littman D R, Zolla-Pazner S. Neutralization profiles of human immunodeficiency virus type 1 primary isolates in the context of coreceptor usage. J Virol. 1998;72:6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clapham P R, Blanc D, Weiss R A. Specific cell surface requirements for the infection of CD4-positive cells by human immunodeficiency virus types 1 and 2 and by simian immunodeficiency virus. Virology. 1991;181:703–715. doi: 10.1016/0042-6822(91)90904-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey M A, Santos-Ferreira M O, Laurent A G, Dauguet C, Katlama C, Rouzioux C, et al. Isolation of a new human retrovirus from West African patients with AIDS. Science. 1986;233:343–346. doi: 10.1126/science.2425430. [DOI] [PubMed] [Google Scholar]
- 15.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 16.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 17.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 19.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 20.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 21.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 22.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 24.Freed E O, Myers D J. Identification and characterization of fusion and processing domains of the human immunodeficiency virus type 2 envelope glycoprotein. J Virol. 1992;66:5472–5478. doi: 10.1128/jvi.66.9.5472-5478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heiber M, Marchese A, Nguyen T, Heng H H, George S R, O’Dowd B F. A novel human gene encoding a G-protein-coupled receptor (GPR15) is located on chromosome 3. Genomics. 1996;32:462–465. doi: 10.1006/geno.1996.0143. [DOI] [PubMed] [Google Scholar]
- 26.Heredia A, Vallejo A, Soriano V, Epstein J S, Hewlett I K. Chemokine receptors and HIV-2. AIDS. 1997;11:1198–1199. doi: 10.1097/00002030-199709000-00025. [DOI] [PubMed] [Google Scholar]
- 27.Hill C M, Deng H, Unutmaz D, KewalRamani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 29.Kanki P J, Travers K U, Mboup S, Hsieh C C, Marlink R G, Gueye-Ndiaye A, Siby T, Thior I, Hernandez-Avila M, Sankale J L, Ndoye I, Essex M E. Slower heterosexual spread of HIV-2 than HIV-1. Lancet. 1994;343:943–946. doi: 10.1016/s0140-6736(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 30.KewalRamani, V. N., B. Volsky, D. S. Kwon, W.-K. Xiang, J. Gao, D. Unutmaz, C. M. Hill, R. E. Sutton, and D. R. Littman. 1998. Unpublished data.
- 31.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 33.Mack M, Luckow B, Nelson P J, Cihak J, Simmons G, Clapham P R, Signoret N, Marsh M, Stangassinger M, Borlat F, Wells T N C, Schlöndorff D, Proudfoot A E I. Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling: a novel inhibitory mechanism of HIV infectivity. J Exp Med. 1998;187:1215–1224. doi: 10.1084/jem.187.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marlink R. Lessons from the second AIDS virus, HIV-2. AIDS. 1996;10:689–699. doi: 10.1097/00002030-199606001-00002. [DOI] [PubMed] [Google Scholar]
- 35.Marlink R, Kanki P, Thior I, Travers K, Eisen G, Siby T, Traore I, Hsieh C C, Dia M C, Gueye E H, Hellinger J, Gueye-Ndiaye A, Sankale J L, Ndoye I, Mboup S, Essex M. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265:1587–1590. doi: 10.1126/science.7915856. [DOI] [PubMed] [Google Scholar]
- 36.McKnight Á, Dittmar M T, Moniz-Periera J, Ariyoshi K, Reeves J D, Hibbitts S, Whitby D, Aarons E, Proudfoot A E I, Whittle H, Clapham P R. A broad range of chemokine receptors are used by primary isolates of human immunodeficiency virus type 2 as coreceptors with CD4. J Virol. 1998;72:4065–4071. doi: 10.1128/jvi.72.5.4065-4071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers G, Korber B, Wain-Hobson S, Smith R F, Pavlakis G N, editors. Human retroviruses and AIDS 1993. Theoretical Biology and Biophysics Group T-10. Los Alamos, N.Mex: Los Alamos National Laboratory; 1993. [Google Scholar]
- 38.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 39.Reeves J D, McKnight A, Potempa S, Simmons G, Gray P W, Power C A, Wells T, Weiss R A, Talbot S J. CD4-independent infection by HIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 40.Roos M T, Lange J M, de Goede R E, Coutinho R A, Schellekens P T, Miedema F, Tersmette M. Viral phenotype and immune response in primary human immunodeficiency virus type 1 infection. J Infect Dis. 1992;165:427–432. doi: 10.1093/infdis/165.3.427. [DOI] [PubMed] [Google Scholar]
- 41.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 43.Scarlatti G, Tresoldi E, Björndal Å, Fredriksson F, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyö E M, Lusso P. In vivo evolution of HIV-1 coreceptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 44.Sol N, Ferchal F, Braun J, Pleskoff O, Treboute C, Ansart I, Alizon M. Usage of the coreceptors CCR-5, CCR-3, and CXCR-4 by primary and cell line-adapted human immunodeficiency virus type 2. J Virol. 1997;71:8237–8244. doi: 10.1128/jvi.71.11.8237-8244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorstensson R, Walther L, Putkonen P, Albert J, Biberfeld G. A capture enzyme immunoassay for detection of HIV-2/SIV antigen. J Acquired Immune Defic Syndr. 1991;4:374–379. [PubMed] [Google Scholar]
- 47.van’t Wout A B, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Investig. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao L, Owen S M, Goldman I, Lal A A, deJong J J, Goudsmit J, Lal R B. CCR5 coreceptor usage of non-syncytium-inducing primary HIV-1 is independent of phylogenetically distinct global HIV-1 isolates: delineation of consensus motif in the V3 domain that predicts CCR-5 usage. Virology. 1998;240:83–92. doi: 10.1006/viro.1997.8924. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L, Huang Y, He T, Cao Y, Ho D D. HIV-1 subtype and second-receptor use. Nature. 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 50.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]