Abstract
Ex vivo gene correction with CRISPR-Cas9 and a recombinant adeno-associated virus serotype 6 (rAAV6) in autologous hematopoietic stem/progenitor cells (HSPCs) to treat sickle cell disease (SCD) has now entered early-phase clinical investigation. To facilitate the progress of CRISPR-Cas9/rAAV6 genome editing technology, we analyzed the molecular changes in key reagents and cellular responses during and after the genome editing procedure in human HSPCs. We demonstrated the high stability of rAAV6 to serve as the donor DNA template. We assessed the benefit of longer HSPC pre-stimulation in terms of increased numbers of edited cells. We observed that the p53 pathway was transiently activated, peaking at 6 h, and resolved over time. Notably, we revealed a strong correlation between p21 mRNA level and rAAV6 genome number in cells and beneficial effects of transient inhibition of p53 with siRNA on genome editing, cell proliferation, and cell survival. In terms of potential immunogenicity, we found that rAAV6 capsid protein was not detectable, while a trace amount of residual Cas9 protein was still detected at 48 h post-genome editing. We believe this information will provide important insights for future improvements of gene correction protocols in HSPCs.
Keywords: CRISPR, Cas9, rAAV6, genome editing, hematopoietic stem and progenitor cell, p53 activation, SCD, HBB gene
Graphical abstract

Xu and colleagues highlight dynamic molecular changes in key genome editing components and cellular responses in human hematopoietic stem and progenitor cells during and after β-globin gene correction with CRISPR-Cas9 and rAAV6 virus ex vivo to treat sickle cell disease. These findings will facilitate the development and application of this technology.
Introduction
Since the discovery of the CRISPR-Cas9 system in 2012 as a tool to make DNA double-strand breaks,1 genome editing has exploded as a research tool. Moreover, gene therapy with CRISPR-Cas9 has risen as a promising and powerful therapeutic approach for incurable diseases, particularly, genetic disorders with unmet medical needs.2,3 Currently, an increasing body of clinical trials has been opened worldwide with the CRISPR-Cas9 system operating a variety of genomic modifications,4,5,6 and promising results of safety and efficacy have been achieved in these clinical trials.4,5,7 CRISPR-Cas9-mediated genome editing comprises Cas9 nuclease and a single-guide RNA (sgRNA) that recognizes target DNA and guides Cas9 protein to the target loci adjacent to a protospacer adjacent motif (PAM) and generates site-specific double-strand breaks (DSBs), which are subsequently repaired by homologous recombination (HR) or non-homologous end joining (NHEJ) or microhomology-mediated end joining (MMEJ). A better understanding of the molecular changes and cellular responses during and after CRISPR-Cas9 genome editing ex vivo in clinically relevant cells is vital to ensure the progress and success of this technology.
Sickle cell disease (SCD) is the most common serious monogenic disease, with 300,000 births annually worldwide. It is an autosomal-recessive disease resulting from a single nucleotide variant in codon six of the β-globin (HBB) gene. The current standard treatments focus on symptomatic relief, which does not cure SCD. These treatments include periodic blood transfusions, administration of hydroxyurea or other small molecules, and pain control as needed (https://www.nhlbi.nih.gov/health-topics/sickle-cell-disease).8,9 The only curative treatment of SCD is allogeneic hematopoietic stem cell transplantation (allo-HSCT).10,11 However, HLA-matched donors are limited, and graft-vs-host disease (GVHD) may occur after allo-HSCT. Although several gene therapy clinical trials using lentiviral vectors have shown promise for SCD, safety concerns and transduction efficiency remain challenging.10,12,13,14,15,16 In addition, genome editing using nuclease-based insertions/deletions (INDELs) to reactivate the fetal hemoglobin in autologous hematopoietic stem/progenitor cells (HSPCs) has also shown very promising results.7 A third approach is to do direct HBB gene correction in HSPCs with CRISPR-Cas9 and recombinant adeno-associated virus serotype 6 (rAAV6) as the DNA donor template.5,17,18,19 Based on these results, the first in-human FDA-approved clinical trial for SCD gene correction has been opened (NCT04819841). To better understand the molecular mechanism of genome editing with CRISPR-Cas9 and rAAV6, and to ensure the safety of genome-edited drug products, we developed sensitive assays for detecting changes in manufactured reagents and for cellular responses during and after genome editing. In this study, all experiments were performed with commercial lots of rAAV6 (Viralgen VC, San Sebastián, Spain) produced with processes identical to the final good manufacturing practice (GMP)-grade clinical lot. Hence, we expected this vector to be highly concentrated and to have higher purity and potency in comparison with research-grade lots. We demonstrated consistently high frequencies of gene correction along with (1) stability and activity preservation of rAAV6 donor template virus when not stored at −80°C; (2) improved genome editing outcomes when frozen mobilized peripheral blood HSPCs (mPB-HSPCs) were pre-stimulated for 72 h compared with 48 h; (3) transient activation of the p53 pathway in HSPCs during and after HBB gene editing, which was strongly correlated with rAAV6 genome number in cells, and the beneficial effects of transiently inhibiting p53 activation via a p53-siRNA during the genome editing procedure; and (4) a rapid decrease in rAAV6 capsid and Cas9 proteins with no detectable residual rAAV6 capsid proteins and a trace amount of residual Cas9 protein in the cells at the completion of manufacture.
Results
Stability of rAAV6 donor template activity
Multiple different rAAV serotypes have been successfully used in gene therapy investigations.4 We previously optimized a gene correction protocol using CRISPR-Cas9 and rAAV6 vector as the DNA donor template for HR5,17,20 (Figure S1). rAAV6 stability is critical for ensuring reproducibility and safety of the ex vivo genome editing protocol. However, prior to this study, very few reports on AAV vector stability were available, leading most IND (Investigational New Drug Application) sponsors to repeat all necessary studies. To date, there are only two studies on rAAV1 stability reported based on its transduction efficiency.21,22 Temperature effects on genome editing efficiency of rAAV donor virus have not been reported. We investigated how several conditions (freeze-and-thaw cycles, 4°C storage, and room temperature storage) affected the capability of the rAAV6 virus (commercial lot) to serve as a DNA sequence donor by measuring functional readouts, including HR% at HBB gene and viability (live cells%) in edited HSPCs. Because cord blood CD34+ (CB-CD34+) HSPCs have features similar to mobilized peripheral blood CD34+ (mPB-CD34+) HSPCs and were easily accessed from our internal resource (the Binns Program for Cord Blood Research at Stanford University), we used CB-CD34+ HSPCs to test rAAV6 stability.
In our experience in this system, HR% from donor to donor ranges from 40% to 80% in HSPCs. Therefore, genome editing in HSPCs from each donor has its own 0 cycle or 0 week as its own control. We exposed aliquots of purified rAAV6 virus to multiple freeze-and-thaw cycles, as shown in the workflow (Figure 1A), and used these aliquots of virus to perform genome editing in CB-CD34+ HSPCs. Interestingly, we found that HR% (Figure 1A) and cell viability (Figure S2A) were equivalent in the cells edited with the virus that went through up to 16 cycles of freeze-and-thaw, indicating that rAAV6 was stable and did not change its genome editing potency or toxicity. These results are consistent with the finding that rAAV1 vector virus did not decrease its transduction efficiency after 10 cycles of freeze-and-thaw.22
Figure 1.
Detecting stability of HBB donor rAAV6
Each bar graph is an independent genome editing experiment in CB-CD34+ HSPCs from a different donor with its own 0-cycle control or 0-week control. UNT, untreated; GE, genome editing; RT, room temperature. Stability was assessed throughout: (A) rAAV6 repeated freeze-and-thaw cycles, (B) rAAV6 at 4°C storage conditions, (C) rAAV6 at room temperature storage conditions.
We then tested the stability of rAAV6 under non-recommended temperature storage conditions. We stored aliquots of rAAV6 at 4°C (Figure 1B) or at room temperature (Figure 1C) for different lengths of time, and we then used these aliquots of virus to perform genome editing in CB-CD34+ HSPCs.
We observed that HR% (Figure 1B) and cell viability (Figure S2B) were equivalent in the cells edited with the virus stored at 4°C for up to 29 weeks. Surprisingly, the virus retained 85% of HR efficiency after being stored at 4°C for up to 31 weeks. We also found that rAAV6 was tolerant of storage at room temperature for up to 18 weeks (Figures 1C and S2C).
These results demonstrate that there can be flexibility in handling rAAV6 in ex vivo genome editing, which is particularly relevant in GMP manufacturing protocols and may provide handling flexibility as well, if the process is applied globally.
A 72-h pre-stimulation of CD34+ HSPCs is more favorable for genome editing outcomes compared with 48-h pre-stimulation
A critical factor in ex vivo autologous HSPC gene therapy protocols is the yield of modified HSPCs, which affects the final dose of infusion in patients. CD34+ HSPCs are generally pre-stimulated in culture for 48 h before the genome editing procedure. To seek a higher yield of gene-corrected cells that underwent HR, we compared 48- and 72-h pre-stimulation times and monitored the percentage of HR alleles and total cell numbers in mPB-CD34+ HSPCs after genome editing.
As shown in Figure 2A, cells expanded on average 1.6- and 3.4-fold after 48- and 72-h pre-stimulation, respectively, resulting in significantly more cells for genome editing after 72-h pre-stimulation compared with 48-h pre-stimulation. Furthermore, cell viability was higher and more consistent among different donors when cells were pre-stimulated for 72 h compared with 48 h (Figure 2B). Importantly, HR% at harvest was also higher and more consistent among different donors when cells were pre-stimulated for 72 h compared with 48 h (Figures 2C and S3A). Seventy-two-hour pre-stimulation not only yielded higher HR frequencies but also generated a higher total number of HBB-corrected cells with >3-fold more HBB-corrected cells compared with 48-h pre-stimulation (Figure 2D) (3.4-fold HBB-corrected cells relative to the cells at thaw using 72-h pre-stimulation vs. 0.8-fold HBB-corrected cells relative to the cells at thaw for 48-h pre-stimulation). These data indicate a benefit to HBB-corrected cell dose in the final drug product by using a longer pre-stimulation time before editing.
Figure 2.
Improved outcomes with 72-h pre-stimulation compared with 48-h pre-stimulation in mPB-CD34+ HSPCs
Cell viability and cell number were measured with trypan blue. UNT, untreated; 48hr-pre, 48-h pre-stimulation; 72hr-pre, 72-h pre-stimulation; GE, genome editing. (A) Cell expansion in pre-stimulation expressed as the fold change in cell number at thaw. Bars represent the mean values of the results from four different donors. One-way ANOVA Tukey’s multiple comparison test was conducted for statistics analysis. ∗∗∗p < 0.001, ∗∗p < 0.01; NS, no significant difference. (B) Cell viability during the genome editing process. Dots are the means with standard deviation (SD) of experiments from four different donors. (C) Genome editing efficiency. Bars represent the mean values with SD of the results from four different donors. (D) HBB correction at 48 h post-genome editing expressed as the fold change in HBB-corrected cell number relative to the cell number at thaw. Bars are the mean values of the results from four different donors.
We evaluated the colony-forming unit (CFU) potential of edited HSPCs that underwent 72-h vs. 48-h pre-stimulation by plating cells in methylcellulose medium using a CFU assay. We observed no significant differences in colony numbers or colony types formed in the CFU assay (Figure S3B), indicating that 72-h pre-stimulation conditions did not change the multipotency and differentiation capacity of HSPCs compared with 48-h pre-stimulation.
Activation of p53 pathway in HBB-gene-edited HSPCs
The function of p53 as a tumor suppressor is well known, and it plays pivotal roles in the DNA damage response (DDR).23,24,25 Genome editing with CRISPR-Cas9 generates on-target and off-target DSBs, which are potentially toxic and trigger a p53-mediated DDR.26,27,28,29,30 It is also reported that transduction of AAV6 in hematopoietic cells activates the p53 pathway, which affects genome editing, HSPC proliferation, and engraftment of edited HSPCs.31,32 Ferrari et al. reported that the AAV genome triggered sustained p53 activation and p53-mediated DDR by increasing the mRNA level of a p53 downstream target, the p21 gene.33 In addition, several groups have demonstrated that transient inhibition of the p53 pathway during CRISPR-Cas9 genome editing can overcome these functional constraints and can also enhance HR.34,35,36,37,38,39,40
To better understand the molecular mechanism of CRISPR-Cas9/rAAV6 genome editing, we investigated the activation status of the p53 pathway by monitoring the dynamic changes in p21 (p21Waf1 or p21Cip1) mRNA and protein, the downstream target of p53 protein41,42,43 in HSPCs during and after the HBB gene correction procedure.
At 48 h post-genome-editing treatment, cells electroporated with Cas9 protein alone or with Cas9-ribonucleoprotein (RNP) complex showed p53 activation (Figures S4A, S4B, and S4E). Adding rAAV6 alone after electroporation (rAAV6) also induced p53 activation, and this activation was MOI dependent (Figure S4B). Finally, RNP electroporation plus rAAV6 treatment (RNP/rAAV6) activated p53 to a higher magnitude compared with electroporation plus rAAV6-alone treatment (rAAV6) (Figure S4B). The activation of the p53 pathway with both RNP and rAAV6 was also MOI dependent (Figure S4C).
Elevated levels of p21 protein were detectable at 3 h post-genome-editing treatment, reached the peak at 6 h post-treatment, and then started gradually decreasing (Figures 3A and S4D–S4H). This phenomenon was consistent across several technical and biological replicates (Figures 3A, S4G, and S4H), which had similar genome editing frequencies (Figure S5). The cells with electroporation only also activated p53 due to the electroporation procedure, but the magnitude was significantly lower compared with RNP/rAAV6 treatment, and it was completely resolved at 24 h post-genome editing (Figure S4G). There was a variability in the level of p53 activation across three different donors (biological replicates), but the general trend of a peak at 6 h with a slow decline in, but persistent, p53 activation at 48 h post-editing was observed (Figures 3A and S4H). p21 mRNA level was highly elevated at day 2 post-genome editing compared with its basal level in untreated cells. p21 levels gradually decreased over time and returned to the basal level at day 10 post-genome editing (Figure 3B). We noted that p21 levels increased over time in culture in untreated cells without genome editing, highlighting the importance of time in culture as another factor in regulating overall cell health (Figure S4I). These results were consistent with the findings by Ferrari et al., which showed a high level of p21 mRNA over 7 days post-genome editing.33 Interestingly, we observed that rAAV6 genome copies per cell also gradually decreased over time post-genome editing along with the decline in p21 mRNA level (Figure 3C). At day 10 post-genome editing, we detected only trace numbers of rAAV6 genomes inside cells, with an average of 8 vector genomes (vg/cell) (Figure 3C). Importantly, we revealed a strong correlation between p21 mRNA level and rAAV6 genome number per cell (Figure 3D), suggesting that rAAV6 is a main trigger of p53 activation.
Figure 3.
Activation of p53 pathway in CD34+ HSPCs during and after genome editing with CRISPR-Cas9/rAAV6
Cells at the 0-h time point were collected prior to electroporation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; NS, no significant difference. (A) Changes in p21 protein level up to 48 h post-genome editing in mPB-CD34+ HSPCs from three different donors. Bars represent the mean values of three donors. One-way ANOVA Tukey’s multiple comparison test was conducted for statistics analysis. (B) Changes in p21 mRNA level up to 10 days post-genome editing in CB-CD34+ HSPCs from four different donors. Bars represent the mean values of four different donors. p21 mRNA level is expressed as the fold change in p21 mRNA amount in genome-edited cells relative to untreated cells. p21 mRNA amount was normalized with TBP mRNA amount in the same ddPCRs. One-way ANOVA Tukey’s multiple comparison test was conducted for statistics analysis. (C) Changes in rAAV6 genome copies per cell up to 10 days post-genome editing in CB-CD34+ HSPCs from four different donors. Bars represent the mean values of four different donors. rAAV6 genome number per cell was normalized with Zeb2 gene in the same ddPCR. One-way ANOVA Tukey’s multiple comparison test was conducted for statistics analysis. (D) Correlation between p21 mRNA level and rAAV6 genome copies in cells. XY correlation analysis was performed for statistics analysis.
In 2021, Lehnertz et al. reported that transient downregulation of p53 with p53-specific siRNA increased HSPC survival after genome editing with Cas9-RNP electroporation and rAAV6 transduction.43 We observed that when p53 activation was transiently inhibited with a p53-specific siRNA during HBB gene editing, the p21 mRNA level was decreased (Figure 4A; Table 1). This transient downregulation of p53 helped cell survival and cell proliferation while maintaining equivalent numbers of rAAV6 virus genome copies per cell (Figures 4B and S6; Table 1), suggesting that the improved cell survival and cell proliferation in p53-siRNA-treated cells did not result from exposure to a different number of rAAV6 genomes in cells. These results were consistent in CB-CD34+ HSPCs from three different donors (Figures 4A, 4B, and S6; Table 1). Importantly, the transient downregulation of p53 with p53-siRNA during genome editing slightly increased the HR percentage, decreased the NHEJ percentage, and increased the CFU colony number without changing colony type distribution (Figures 4C and 4D; Table 1). These results are consistent with previous reports and confirm that transient inhibition of p53 activation during genome editing is beneficial for HR DNA repair and HSPC proliferation and preservation.36,38,39,40,44
Figure 4.
Beneficial effect of transient inhibition of p53 activation via p53-siRNA in HBB-edited CB-CD34+ HSPCs
UNT, untreated cells; RNP, RNP/electroporation without p53-siRNA; GE, genome-edited cells that were HBB gene-edited with CRISPR-Cas9/rAAV6 at day 0; no siRNA, genome editing without p53-siRNA; siRNA, genome editing with p53-siRNA. (A) Decrease in p21 mRNA level in p53-siRNA-treated HSPCs. (B) Improvement in cell proliferation post-genome editing in p53-siRNA-treated HSPCs. (C) Higher HR% and lower NHEJ% in p53-siRNA-treated HSPCs. (D) Slight increase in CFU colony formation without changing colony type distribution in p53-siRNA-treated HSPCs. BFU-E, primitive erythroid progenitors; CFU-E, mature erythroid progenitors; CFU-G/GM, granulocyte and macrophage progenitors; CFU-GEMM, granulocyte, erythrocyte, macrophage, and megakaryocyte.
Table 1.
Fold changes in different parameters at day 2 post-genome editing in CB-HSPCs edited with p53-siRNA and without p53-siRNA
| No. live cells | Cell viability | HR | NHEJ | p21 mRNA | Total no. CFU colonies | rAAV6 vg/cell | |
|---|---|---|---|---|---|---|---|
| Donor 1 | 1.15 ↑ | 1.18 ↑ | 1.16 ↑ | 0.68 ↓ | 0.79 ↓ | 1.04 ↑ | 0.99 |
| Donor 2 | 1.31 ↑ | 1.07 ↑ | 1.27 ↑ | 0.82 ↓ | 0.74 ↓ | 1.14 ↑ | 1.01 |
| Donor 3 | 1.03 ↑ | 1.08 ↑ | 1.03 ↑ | 0.80 ↓ | 0.79 ↓ | 1.25 ↑ | 1.03 |
| Average | 1.16 ↑ | 1.11 ↑ | 1.15 ↑ | 0.77 ↓ | 0.77 ↓ | 1.14 ↑ | 1.01 |
Genome editing was performed with and without p53-siRNA at day 0. Each parameter is presented as the fold change in p53-siRNA treatment relative to no p53-siRNA treatment.
Monitoring rAAV6 capsid protein level in HBB-gene-edited HSPCs
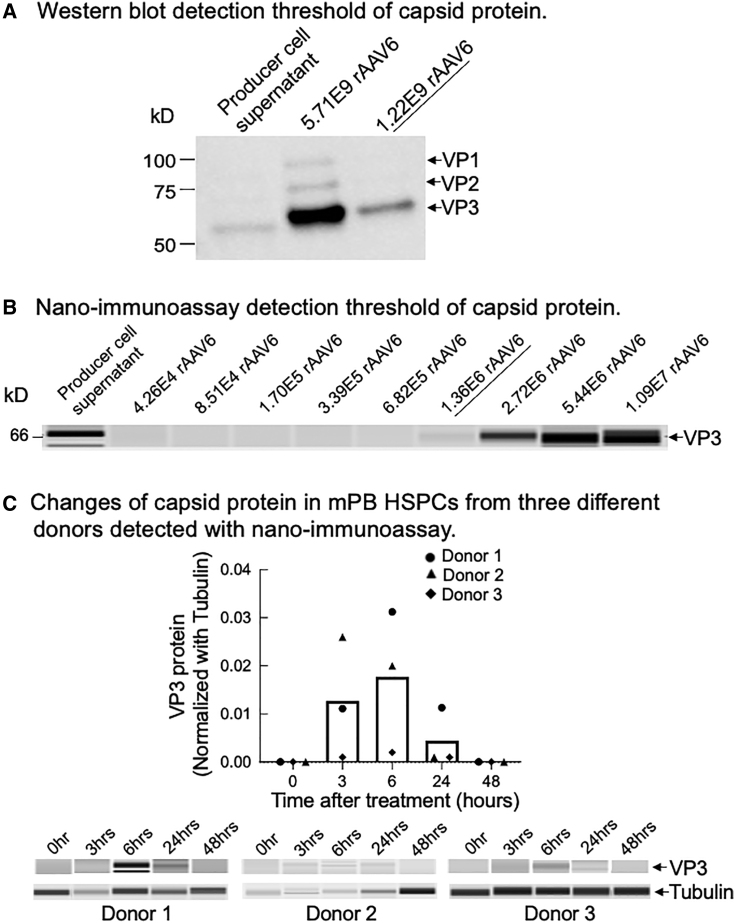
AAV is a single-stranded DNA packaged into an 18- to 25-nm capsid protein containing a 60-mer of VP1, VP2, and VP3 repeating monomers in a ratio of 1:1:10.45 Transduction of a cell by AAV requires interaction of the viral capsid protein with surface receptors of the cell followed by endocytosis and then capsid protein-mediated endosomal escape and nuclear import for uncoating and converting the single-stranded genome to a double-stranded DNA.46 Therefore, AAV capsid protein integrity is an important factor for transduction efficacy. On the other hand, it is known that AAV and its capsid protein can trigger innate and adaptive immune responses and cause inflammation.4,46,47,48,49,50,51 Therefore, the presence of residual rAAV6 capsid protein in genome-edited cell products is a safety concern. To better assess rAAV6 protein residuals, we developed a sensitive protein-based assay to detect rAAV6 capsid protein and monitored its changes during and after HBB gene correction in mPB-CD34+ HSPCs.
First, we attempted traditional western blot with the diluted purified rAAV6 virus (Viralgen VC) and with the rAAV6 producer cell supernatant from Stanford’s Laboratory of Cell and Gene Medicine (LCGM). We found that the detection threshold of the western blot was at approximately 1.22E9 vg (Figure 5A).
Figure 5.
Detection of rAAV6 capsid protein in HBB-gene-edited mPB-CD34+ HSPCs
Cells at the 0-h time point were collected prior to genome editing treatment. (A) Western blot detection threshold of rAAV6 capsid protein. (B) Nano-immunoassay detection threshold of rAAV6 capsid protein. (C) Changes in rAAV6 capsid protein post-genome-editing treatment in mPB-CD34+ HSPCs from three different donors detected with nano-immunoassay. Bars represent mean values of three different donors.
We then developed and optimized a capillary-based and automated nano-immunoassay52,53,54 with a serial dilution of purified rAAV6 virus (Viralgen VC). As shown in Figure 5B, the detection sensitivity of the nano-immunoassay was 3 logs higher than that of the western blot, with a detection threshold of 1.36E6 vg.
We monitored changes in the capsid protein in cell pellets from the genome editing procedure with the nano-immunoassay. We observed that capsid protein gradually increased and peaked at 6 h post-genome-editing treatment and then decreased (Figure 5C). These results were consistent in three technical replicates from one donor (Figure S7) and mPB-CD34+ HSPCs from three different donors (Figure 5C). The timing of rAAV6 capsid protein peak was aligned to that of p21 protein (Figures 3A and S4E–S4H), further suggesting that rAAV6 is a major trigger of p53 activation. Importantly, we did not detect residual rAAV6 capsid protein at 48 h after genome editing in any of the CB-CD34+ and mPB-CD34+ HSPCs from different biological donors when using different lots of rAAV6 (Tables 2 and 3), implying that the cell products manufactured for clinical application were safe at this time point, without any detectable amount of residual rAAV6 capsid protein.
Table 2.
Detection of rAAV6 capsid protein in CB-CD34+ HSPCs at 24 and 48 h post-genome-editing treatment
| Sample | HR% | NHEJ% | VP3 |
|---|---|---|---|
| Producer cell supernatant | N/A | N/A | detected |
| Untreated | N/A | N/A | not detecteda |
| RNP 24 h | N/A | 90.57 | not detecteda |
| RNP/rAAV6 24 h | 72.01 | 22.75 | not detecteda |
| RNP 48 h | N/A | 90.57 | not detecteda |
| RNP/rAAV6 48 h | 73.49 | 21.27 | not detecteda |
RNP, RNP/electroporation; RNP/rAAV6, RNP/electroporation/rAAV6.
Levels were lower than the reliable detection threshold, which was equivalent to 2.00E4 vg/cell.
Table 3.
Detection of rAAV6 capsid protein in mPB-CD34+ HSPCs at 48 h post-genome-editing treatment
| Sample | HR% | NHEJ% | VP3 |
|---|---|---|---|
| Untreated | N/A | N/A | not detecteda |
| rAAV6 (lot #3) MOI 2,500 rep1 (donor 3) | 56.67 | 33.92 | not detecteda |
| rAAV6 (lot #3) MOI 2,500 rep2 (donor 3) | 58.76 | 33.62 | not detecteda |
| rAAV6 (log #1) MOI 1,250 rep1 (donor 5) | 41.10 | 28.63 | not detecteda |
| rAAV6 (log #1) MOI 1,250 rep2 (donor 5) | 40.90 | 30.79 | not detecteda |
| rAAV6 (log #1) MOI 2,500 rep1 (donor 5) | 41.07 | 29.28 | not detecteda |
| rAAV6 (log #1) MOI 2,500 rep2 (donor 5) | 45.09 | 22.94 | not detecteda |
| rAAV6 (log #1) MOI 5,000 rep1 (donor 5) | 41.39 | 29.46 | not detecteda |
| rAAV6 (log #1) MOI 5,000 rep2 (donor 5) | 45.25 | 22.81 | not detecteda |
| rAAV6 (lot #2) MOI 2,500 rep1 (donor 5) | 43.52 | 26.22 | not detecteda |
| rAAV6 (lot #2) MOI 2,500 rep2 (donor 5) | 44.86 | 30.11 | not detecteda |
| rAAV6 (lot #1) MOI 2,500 (donor 7) | 52.77 | 29.93 | not detecteda |
| rAAV6 (lot #1) MOI 2,500 (donor 8) | 46.94 | 34.94 | not detecteda |
rAAV6, RNP/electroporation/rAAV6; rep, replicate.
Levels were lower than the reliable detection threshold, which was equivalent to 2.00E4 vg/cell.
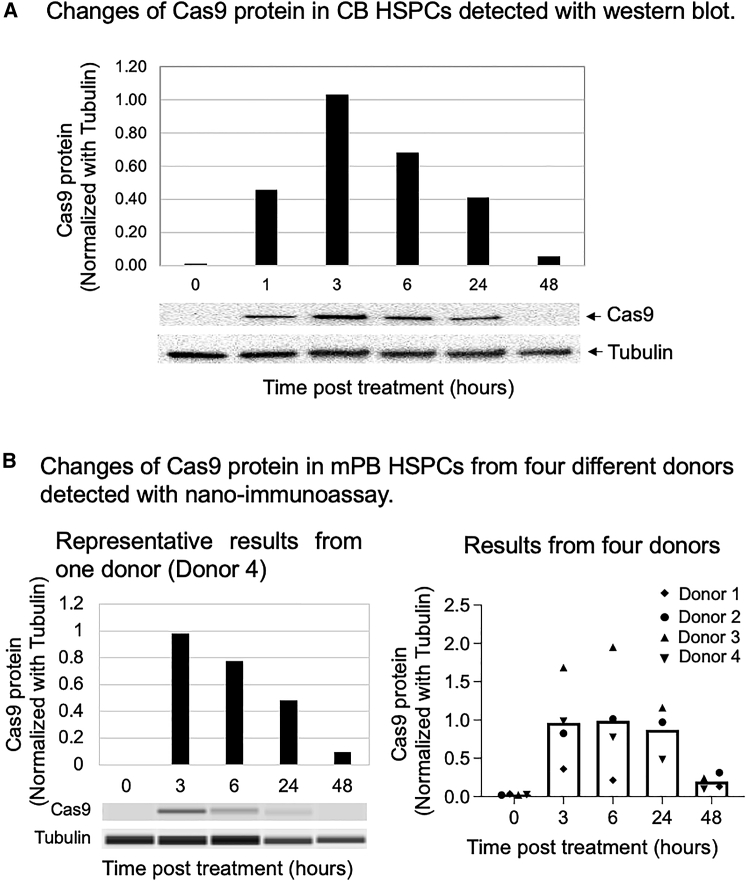
Monitoring Cas9 protein in HBB-gene-edited HSPCs
Residual Cas9 protein in ex vivo-edited human cells could serve as an antigenic factor to cause an adaptive immune response in patients if still present in the cell products,47,55,56 thereby raising concerns regarding the safety and efficacy of this technology and the need to develop sensitive assays for detection of residual Cas9 protein. In 2020, Stadtmauer et al. reported that they developed an immunoassay for Cas9 protein detection and quantified Cas9 in their manufacturing process, showing <0.75 fg/cell of Cas9 protein in the harvested engineered T cell products.57 We built on this work to measure Cas9 protein in edited HSPCs.
First, we electroporated RNP into CB-CD34+ HSPCs and collected cells at various time points after electroporation. We ran SDS-PAGE and performed western blots under denaturing conditions. We observed that Cas9 protein level peaked at 3 h and then gradually decreased. At 48 h post-RNP electroporation, Cas9 protein was significantly reduced but still detectable (Figure 6A).
Figure 6.
Detection of Cas9 protein in HBB-gene-edited HSPCs
Cells at the 0-h time point were collected prior to genome editing treatment. (A) Changes in Cas9 protein in CB HSPCs detected with western blot. (B) Changes in Cas9 protein in mPB-CD34+ HSPCs from four different donors detected with nano-immunoassay. Bars represent the mean values of the four donors.
To measure Cas9 protein more sensitively, we developed and optimized a capillary-based and automated nano-immunoassay.52,53,54
The pattern of Cas9 protein changes detected with the nano-immunoassay was consistent with the western blot analysis. Cas9 protein level peaked at 3 h post-genome-editing treatment and then gradually decreased. At 48 h post-genome editing, a trace amount of Cas9 protein still existed in the cells (Figure 6B).
Discussion
In previous studies, we demonstrated that a protocol of HBB gene correction in HSPCs derived from both healthy donors and SCD patients using RNP electroporation with rAAV6 transduction is highly efficient, reproducible, and safe in terms of low off-target activity, low translocations, no tumorigenesis in mice, and low transcriptional perturbation.5,17,58 As safety is always the most important factor for all investigational new drugs, in this study we deepened the knowledge of the key reagents deployed in the manufacturing protocol. Specifically, we investigated their stability, their residuals in the final drug products, and the related responses at the cellular level.
In the HBB gene correction protocol, rAAV6 provides the correct HBB DNA template for HR. Therefore, its stability is one of the main determinants for genome editing efficacy. Previously, the Flotte and Harvey laboratories21,22 reported rAAV1 stability at 4°C and after multiple cycles of freeze-and-thaw based on transduction efficiency. Our results confirmed rAAV6 stability upon multiple cycles of freeze-and-thaw, like rAAV1. We also observed that rAAV6 retained its genome editing potency under all temperature conditions we have tested (Figure 1), suggesting excellent stability. Of note, in this study we used a virus manufactured (Viralgen VC) identical to the lot being used in the phase I/II clinical trial.
In this study, we demonstrated that, compared with 48-h pre-stimulation, 72-h pre-stimulation provided significantly higher numbers of CD34+ HSPCs for genome editing, better cell viability, and overall higher numbers of gene-corrected HSPCs at harvest (Figure 2). This evidence suggests that 72-h HSPC pre-stimulation might be beneficial to cell manufacturing yield without compromising stem cell properties as indicated with the CFU assay (Figure S3B).
It is now well documented that CRISPR-Cas9-induced DNA DSBs activate the p53 pathway, which hampers genome editing efficacy, cell survival, and engraftment potential.23,24,25,33,34,35,36,37 Therefore, it is important to assess dynamic changes in p53 pathway activation during and after genome editing and its status in the final drug products. By monitoring changes in p21 mRNA and protein levels, we discovered that p53 was transiently activated during and after genome editing under all conditions combined with electroporation (Figures 3A and S4). We also found that there was a strong correlation between p21 mRNA level and rAAV6 genome copies per cell (Figures 3B–3D). In addition, we noticed that the pattern of capsid protein changes was aligned with that of p21 protein changes in genome-edited cells during and after genome editing (Figures 3A and 5C). These results suggest that rAAV6 virus is a main trigger to activate the p53 pathway during and after genome editing.
Since 2018, several groups have reported that transient inhibition of p53 activation in CRISPR-Cas9 genome editing can enhance HR and inhibit NHEJ but does not change HSC functions.34,36,38,39,40,44 Here, we also demonstrated that transient downregulation of p53 activation with p53-siRNA during genome editing increased HR% and decreased NHEJ% of HBB gene editing (Figure 4C; Table 1). Moreover, this transient inhibition improved cell survival and proliferation after genome editing (Figures 4B and S6A; Table 1). It also facilitated CFU colony formation without changing the distribution of different colony types (Figure 4D; Table 1). Our results are consistent with previous findings reported by other groups.34,36,38,39,40,44 However, when the DNA repair pathway does not work properly, translocation and other genome rearrangements will diminish cell viability and increase the chance of tumorigenesis.43,59,60 While p53 activation may be harmful to HR, HSPC proliferation, and HSPC engraftment, it may also serve as a protective mechanism to ensure that only healthy cells with a minimal stress response are able to be engrafted. For the sake of safety, investigations of the long-term effects of transient p53 inhibition during genome editing on edited HSPCs are needed before it is moved to clinical applications. As the p53 pathway was still partially activated at 48 h post-genome-editing treatment when drug products were harvested (Figures 3A, 3B, and S4), the edited HSPCs might not proliferate and engraft as well as untreated HSPCs. We previously reported engraftment of a human population in xenotransplant models with an average of 50% for RNP-only control and rAAV6-only control vs. 20% for genome-edited HSPCs, these genome-edited HSPCs bearing 20%–30% HBB-gene-corrected alleles.5 We acknowledge the drop in engraftment potential of gene-corrected cells and want to emphasize the importance of improving genome editing protocols and using high doses of autologous manipulated HSPCs in clinical settings for SCD patients as a strategy to mitigate the engraftment risk. This is particularly relevant in view of the recent press release about a phase I/II trial (NCT04819841) voluntary pause following an unexpected adverse event of prolonged low blood cell counts (pancytopenia) requiring transfusion and growth factor support in the first patient dosed with the drug.
Cas9 protein and rAAV6 capsid protein are immunogenic and can trigger immune responses,4,46,47,48,49,50,51,55,56,57 and therefore residual Cas9 and rAAV6 capsid proteins in drug products may lead to clearance of transplanted HSPCs from a patient immune response even when using autologous cells. They may also impair cellular functions of genome-edited cells, thereby raising concerns regarding the safety and efficacy of this technology and the need to develop sensitive assays for detection of these residual proteins in genome-edited cell products. Here, we showed a good profile of the cell drug products with no detectable residual rAAV6 capsid protein (Figure 5C; Tables 2 and 3) and only a trace amount of Cas9 protein (Figure 6) at the end of the manufacturing process.
In conclusion, we have developed sensitive assays to detect residual Cas9 protein, rAAV6 capsid protein, and p53 activation, and we have monitored dynamic changes in these molecules during and after genome editing. We believe that our findings are beneficial to better understand the molecular mechanism of ex vivo genome editing in HSPCs and they will also provide new knowledge on the further development and application of genome editing with CRISPR-Cas9/rAAV6.
Materials and methods
HBB-SCD genome editing with CRISPR-Cas9 and rAAV6 virus
Cloning of the rAAV6 donor plasmid
Single-stranded AAV6 vector DNA was cloned into the pAAV-MCS plasmid (Agilent Technologies, Santa Clara, CA, USA) using Gibson Assembly Mastermix (New England Biolabs, Ipswich, MA, USA). The ampicillin resistance was replaced with kanamycin resistance for a plasmid with a total length of 5,097 bp. The HBB rAAV6 donor contained arms of homology to the β-globin locus of 540 bp on the left side and 420 bp on the right side (GRCh38/hg38: ch11:5,224,356–5,229,388). The HR donor sequence for correction of the HBB mutation is 5′-ACCATGGTGCACCTGACTCCTGAGGAAAAATCCGCAGTCACTGCCCTGTGGGGCAAG-3′. Letters in bold are mutated nucleotides, letter in bold plus italic is the corrected adenosine at the place of the pathologic thymidine. No stuffer DNA was used, and the overall cassette enclosed between ITRs of AAV6 vector is 2,550 bp long.
Production of rAAV6 donor virus and virus producer cell supernatant
All experiments were performed with industrial lots of rAAV6 purchased from Viralgen VC (Donostia, Gipuzkoa, Spain) and produced with a process very similar to the final GMP-grade clinical lot. Briefly, a proprietary HEK293T cell bank was used for triple plasmid transfection in a 200-L bioreactor under suspension cell conditions. Three days after transfection, cells were harvested, and a downstream purification process was performed, including passages of affinity chromatography and ion-exchange chromatography. During the final ultrafiltration step, the product was exchanged into the final formulation buffer, DPBS with 0.001% Pluronic F68, and was aliquoted and stored at −80°C until use.
A crude rAAV6 producer cell supernatant was generated at Stanford’s LCGM.5 Briefly, cells were thawed at 37°C and expanded in Freestyle F17 medium supplemented with 5 mM GlutaMAX and 0.2% (w/v) Pluronic F68. The cells were expanded up to a final volume of 3 L in spinner flasks and co-transfected with the same plasmids described above, using a polyethylenimine (PEI)-based transfection method. Two days after transfection, HEK293T cells producing rAAV6 were harvested by centrifugation. Supernatant was collected and kept at −80°C until use.
Detection of rAAV6 donor virus titer
rAAV6 genomic DNA was extracted by using QuickExtract DNA Extraction Solution (Epicentre, Madison, WI, USA) following the manufacturer’s instruction. A series dilution of the extracted rAAV6 genomic DNA was made. The diluted rAAV6 genomic DNA was used to measure virus titer with droplet digital PCR (ddPCR) in the QX200 Droplet Digital PCR System (Bio-Rad, Hercules, CA, USA) with ddPCR Supermix for Probes (no dUTP) (Bio-Rad, Hercules, CA, USA). Droplets were generated and analyzed according to the manufacturer’s instructions by using the QX200 system (Bio-Rad, Hercules, CA, USA). ddPCR cycling conditions were as follows: 95°C (10 min); followed by 45 cycles (2°C ramp rate) of 94°C (30 s), 60°C (1 min); and then 98°C (10 min). rAAV6 ITR(Inverted Terminal Repeat) primer and probe sequences for ddPCR are described in Table S1.
Detection of rAAV6 donor virus genome copies in HBB-gene-edited HSPCs
HBB-edited HSPCs were harvested at different days post-genome-editing treatment, together with untreated cells as a control. Genomic DNA was extracted by using QuickExtract DNA Extraction Solution (Epicentre, Madison, WI, USA) following the manufacturer’s instructions. The extracted genomic DNA was used to measure rAAV6 donor virus genome copies with ddPCR in the QX200 Droplet Digital PCR System (Bio-Rad, Hercules, CA, USA) with ddPCR Supermix for Probes (no dUTP) (Bio-Rad, Hercules, CA, USA). Droplets were generated and analyzed according to the manufacturer’s instructions by using the QX200 system (Bio-Rad, Hercules, CA, USA). ddPCR cycling conditions were as follows: 95°C (10 min); followed by 45 cycles (2°C ramp rate) of 94°C (30 s), 60°C (1 min); and then 98°C (10 min). rAAV6 ITR primers and probe were used to detect rAAV6 genome copies, and the Zeb2 (zinc finger E-box binding homeobox 2) primers and probe were used as an internal control for detecting rAAV6 genome copies per cell in the same ddPCRs. rAAV6 ITR and Zeb2 primer/probe sequences are described in Table S1.
CD34+ HSPC culture conditions
Human CD34+ HSPCs were cultured as previously described.5,20 Human CB-CD34+ HSPCs were generously provided by the Binns Program for Cord Blood Research at Stanford University. Plerixafor mPB-CD34+ HSPCs were purchased from AllCells (Alameda, CA, USA) or received from the Stanford LCGM Facility. The cells were thawed per the manufacturer’s instructions and were cultured at 37°C, 5% CO2, and 5% O2 for 48 h (48-h pre-stimulation) or 72 h (72-h pre-stimulation) prior to genome editing. Cell pre-stimulation medium and culturing medium were the same, which was GMP SCGM medium from CellGenix (Portsmouth, NH, USA) supplemented with human cytokine cocktail (PeproTech, Rocky Hill, NJ, USA) containing stem cell factor (SCF) (100 ng/mL), thrombopoietin (100 ng/mL), Fms-like tyrosine kinase 3 ligand (100 ng/mL), and interleukin 6 (100 ng/mL). UM171 (35 nM) (StemCell Technologies, Vancouver, BC, Canada), streptomycin (20 mg/mL), and penicillin (20 U/mL) were added into the cell culture medium.
HBB gene editing procedure
HBB gene editing was performed as previously described with slight modification.5,20 Briefly, CD34+ HSPCs were cultured and pre-stimulated for 72 h after thaw and before genome editing except when specifically indicated. Chemically modified HBB sgRNA was synthesized by Agilent Technologies (Santa Clara, CA, USA). SpyFi Cas9 was purchased from Aldevron (Fargo, ND, USA). HBB donor rAAV6 virus was purchased from Viralgen VC (Donostia, Gipuzkoa, Spain). Electroporation of Cas9-RNP complex was performed using Lonza 4D-Nucleofector (Lonza Group, Alpharetta, GA, USA) in P3 Primary Cell Solution with program DZ-100 as described previously. Electroporated cells were then plated at 2.5 × 105 cells/mL in HSPC culture medium supplemented with cytokines. For RNP/rAAV6 or rAAV6 treatment, HBB donor rAAV6 was immediately dispensed onto cells at an MOI of 2.5 × 103 vg/cell based on the titer determined by ddPCR except for special indications. After incubation for 24 h, a medium addition or medium exchange was performed to dilute or remove residual HBB donor rAAV6. The CD34+ HSPCs were cultured for an additional 24 h for quantification of genome editing events. At different time points as indicated in the figures and tables, aliquots of cells were collected, washed once with PBS buffer, and pelleted, and the cell pellets were stored at −80°C until use.
For HBB gene editing with siRNA against p53 (p53-siRNA), 4 pmol of p53-siRNA (ID s605, Thermo Fisher Scientific, Waltham, MA, USA) was added into cells in 20 μL of P3 solution containing RNP before electroporation. At 24 h post-genome editing, a medium exchange was performed to remove p53-siRNA and rAAV6. The other steps of the genome editing procedure were the same as described above.
Quantification of HBB gene editing events
HBB-edited HSPCs were harvested at day 2 post-electroporation, together with untreated cells as a control. Genomic DNA was extracted by using QuickExtract DNA Extraction Solution (Epicentre, Madison, WI, USA) following the manufacturer’s instruction. To evaluate HR and NHEJ events, an in-out PCR approach was performed to amplify the HBB locus followed by a nested ddPCR.5,17,19 Briefly, to exclude episomal rAAV6, an HBB-specific 1.4-kb product was amplified with Phusion Green Hot Start II HF PCR 2× Master Mix (Thermo Fisher Scientific, Waltham, MA, USA) in a PCR program of 98°C (30 s); followed by 35 cycles of 98°C (10 s), 60°C (30 s), 72°C (1 min); and then 72°C (10 min). Primer sequences for the in-out PCR are provided in Table S1.
The PCR product was purified with the QIAquick Gel Extraction Kit (Qiagen, Redwood City, CA, USA). The purified PCR product was diluted to 10 fg/μL in nuclease-free water for ddPCR in the QX200 Droplet Digital PCR System (Bio-Rad, Hercules, CA, USA) with ddPCR Supermix for Probes (no dUTP) (Bio-Rad, Hercules, CA, USA). To quantify genome editing events in the HBB locus, two primers and three probes (REF [reference], WT [wild type], and HR) were used in one ddPCR. Droplets were generated and analyzed according to the manufacturer’s instructions by using the QX200 system (Bio-Rad, Hercules, CA, USA). ddPCR cycling conditions were as follows: 95°C (10 min); followed by 50 cycles (2°C ramp rate) of 94°C (30 s), 60.8°C (30 s), and 72°C (2 min); and then 98°C (10 min). Primer and probe sequences for ddPCR are provided in Table S1.
Methylcellulose colony-forming unit assay
CFU assay was started at 48 h post-genome editing. For each condition, 1.1 mL of semi-solid methylcellulose medium (StemCell Technologies, Seattle, WA, USA) containing 300 cells was plated in a well of a SmartDish (StemCell Technologies, Seattle, WA, USA) in duplicates and then the cells were incubated at 37°C, 5% O2, and 5% CO2 for 14 days. The resulting progenitor colonies were counted and scored with STEMVision analysis (StemCell Technologies, Seattle, WA, USA) following the manufacturer’s instructions.
Detection of HBB donor rAAV6 stability
HBB donor rAAV6 was divided into small aliquots. Some aliquots were kept at −80°C all the time until genome editing as 0-cycle control or 0-week control. The other aliquots were gone through certain cycles of freeze-and-thaw as indicated in Figure 1A or were kept at 4°C (Figure 1B) or at room temperature (Figure 1C) until genome editing.
These aliquots of virus were used to do genome editing in CB-CD34+ HSPCs. HBB gene editing in CB-CD34+ HSPCs from different donors was done as independent experiments with their own 0-cycle controls or 0-week controls.
Cell lysates for detection of Cas9, rAAV6 capsid, and p21 proteins
Cell pellets were lysed in RIPA lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) containing protease inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, cells were lysed in a ratio of 2 × 105 cells per 10 μL of cell lysis buffer containing protease inhibitor cocktail on ice for 45 min with brief vortexing at high speed every 10 min. After lysis, cell lysates were centrifuged at 13,000g at 4°C for 20 min. Each supernatant was transferred to a new tube and aliquoted in a small volume. All aliquots were stored at −80°C until use.
Detection of p53 pathway activation in HBB-gene-edited CD34+ HSPCs
Detection of p21 mRNA with RT-ddPCR assay
Total RNA was extracted from cell pellets with the Rneasy Micro Kit following the manufacturer’s instructions (Qiagen, Redwood City, CA, USA). cDNA was synthesized from total RNA with iScript RT Supermix following the manufacturer’s instructions (Bio-Rad, Hercules, CA, USA). Synthesized cDNA was used for ddPCR with human p21CIP1 and TBP (TATA box binding protein) PrimeTime qPCR primers (Integrated DNA Technologies, Coralville, IA, USA) in the QX200 Droplet Digital PCR System (Bio-Rad, Hercules, CA, USA) with ddPCR Supermix for Probes (no dUTP) (Bio-Rad, Hercules, CA, USA). Droplets were generated and analyzed according to the manufacturer’s instructions by using the QX200 system (Bio-Rad, Hercules, CA, USA). ddPCR cycling conditions were as follows: 95°C (10 min); followed by 45 cycles (2°C ramp rate) of 94°C (30 s), 60°C (1 min); and then 98°C (10 min). TBP was used as the internal housekeeping gene in the same ddPCRs. The p21 mRNA amount was normalized to the TBP mRNA amount. The p21 mRNA level is presented as the fold change in p21 mRNA relative to p21 mRNA in untreated cells.
Detection of p21 protein with nano-immunoassay
Cell lysates were subjected to the automated and capillary-based nano-immunoassay52,53,54 executed in the PeggySue machine (ProteinSimple, San Jose, CA, USA) according to the manufacturer’s protocol. All reagents except for primary antibodies were purchased from ProteinSimple (San Jose, CA, USA). Briefly, 1 μL of cell lysate was mixed with 11.5 μL of a master mix to a final concentration of 1× sample buffer, 1× fluorescent molecular weight markers, and 40 mM dithiothreitol (DTT) and then heated at 95°C for 5 min. The samples, blocking reagent, primary antibodies, HRP-conjugated secondary antibodies, chemiluminescent substrate, and separation and stacking matrices were dispensed to designated wells in a 384-well plate. After plate loading, the separation, electrophoresis, and immunodetection steps took place in a capillary microfluidic system and were fully automated. Nano-immunoassay was carried out at room temperature with instrument default settings. Quantification of α-tubulin and p21 proteins was performed with Compass software (ProteinSimple, San Jose, CA, USA) following the manufacturer’s instructions.
Anti-p21Waf1/Cip1 rabbit monoclonal antibody (12D1) (Cell Signaling, Danvers, MA, USA) and anti-α-tubulin antibody (DM1A) (AbCam, Cambridge, MA, USA) as an internal control were used in the nano-immunoassay to detect p53 activation.
Detection of rAAV6 capsid protein in HBB-gene-edited CD34+ HSPCs
Detection of rAAV6 capsid protein with western blot
HBB donor rAAV6 producer cell supernatant was provided by the Stanford LCGM Facility. Aliquots of rAAV6 producer cell supernatant were stored at −80°C until use.
Purified rAAV6 from Viralgen VC (Donostia, Gipuzkoa, Spain) was diluted in RIPA lysis buffer containing protease inhibitor cocktail. rAAV6 producer cell supernatant and diluted rAAV6 were loaded into 4%–15% Mini-Protean TGX pre-cast gel (Bio-Rad, Hercules, CA, USA) to run denaturing SDS-PAGE. Protein was transferred to PVDF membrane with a Mini Trans-Blot electrophoretic transfer kit (Bio-Rad, Hercules, CA, USA). The membrane was pre-blotted with 5% non-fat milk in TBS buffer containing 0.5% Tween 20. The membrane was then incubated with 1:500 diluted adeno-associated virus VP1, VP2, and VP3 antibody (B1) (GenTex, Irvine, CA, USA) at 4°C with shaking overnight. ECL anti-mouse IgG-HRP-linked whole antibody (Thermo Fisher Scientific, Waltham, MA, USA) was used as the secondary antibody and the membrane was developed with Clarity Max Western ECL Substrate (Bio-Rad, Hercules, CA, USA) and was imaged in a ChemiDoc MP machine (Bio-Rad, Hercules, CA, USA).
Development for detection of rAAV6 capsid protein with nano-immunoassay
rAAV6 producer cell supernatant and dilutions of purified rAAV6 were prepared as above.
Detection of rAAV6 capsid protein was performed by using the nano-immunoassay executed in the PeggySue machine (ProteinSimple, San Jose, CA, USA) according to the manufacturer’s protocol as described above. Adeno-associated virus VP1, VP2, and VP3 antibody (B1) (GenTex, Irvine, CA, USA) was used.
Detection of rAAV6 capsid protein in HBB-gene-edited CD34+ HSPCs with nano-immunoassay
Cell lysates of HBB-gene-edited CD34+ HSPCs were prepared and rAAV6 capsid protein was detected with nano-immunoassay as described above. Qualification of capsid and α-tubulin proteins was performed with Compass software (ProteinSimple, San Jose, CA, USA) following the manufacturer’s instructions.
Adeno-associated virus VP1, VP2, and VP3 antibody (B1) (GenTex, Irvine, CA, USA) and mouse anti-α-tubulin antibody (DM1A) (AbCam, Cambridge, MA, USA) were used.
Detection of Cas9 protein in HBB-gene-edited CD34+ HSPCs
Detection of Cas9 protein with western blot
Cell lysates of RNP-electroporated CB-CD34+ HSPCs were prepared as above. Cell lysates containing 210,000 cells per lane were loaded into 4%–15% Mini-Protean TGX pre-cast gel (Bio-Rad, Hercules, CA, USA) to run denaturing SDS-PAGE. Protein was transferred to PVDF membrane with the Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, CA, USA). The membrane was pre-blotted with 5% non-fat milk in TBS buffer containing 0.5% Tween 20. The membrane was then cut in half. The top half was incubated with 1:2,000 diluted mouse monoclonal anti-CRISPR-Cas9 antibody 7A9 (Diagenode, Denville, NJ, USA) and the bottom half was incubated with 1:5,000 diluted mouse anti-α-tubulin (DM1A) antibody (AbCam, Cambridge, MA, USA) in 5% non-fat milk in TBS buffer containing 0.5% Tween 20 at 4°C with shaking overnight. A 1:10,000 diluted ECL peroxidase-labeled anti-mouse antibody (Thermo Fisher Scientific, Waltham, MA, USA) was used as the secondary antibody, and the membrane was developed with Clarity Max Western ECL Substrate (Bio-Rad, Hercules, CA, USA) and was imaged in a ChemiDoc MP machine (Bio-Rad, Hercules, CA, USA). Quantification of band intensity was conducted with the Image Lab program.
Detection of Cas9 protein in HBB-gene-edited CD34+ HSPCs with nano-immunoassay
Cell lysates of HBB-gene-edited CD34+ HSPCs were prepared as described above. Cas9 protein detection was performed by using nano-immunoassay executed in a PeggySue machine (ProteinSimple, San Jose, CA, USA) according to the manufacturer’s protocol as described above. Qualification of Cas9 and α-tubulin proteins was performed with Compass software (ProteinSimple, San Jose, CA, USA) following the manufacturer’s instructions.
The same mouse monoclonal anti-CRISPR-Cas9 antibody 7A9 (Diagenode, Denville, NJ, USA) and mouse anti-α-tubulin antibody (DM1A) (AbCam, Cambridge, MA, USA) used for western blot were used in the nano-immunoassay.
Statistical analysis
All statistical analyses on experimental groups were conducted using Prism7 GraphPad software. The exact statistical tests used for each comparison are noted in the figure legends. One-way ANOVA Tukey’s multiple comparison test with one variable and two-way ANOVA Tukey’s multiple comparison test in the case of two variables were used. XY correlation analysis was performed for correlation analysis.
Data and code availability
Source data are provided with this paper.
Acknowledgments
We thank the Binns Program for Cord Blood Research at Stanford University for providing CB-CD34+ HSPCs. We thank the Stanford Laboratory for Cell and Gene Medicine Facility for providing some mPB-CD34+ HSPCs, some HBB-gene-edited mPB-CD34+ HSPC samples, and some reagents. We thank Dr. Fabian Suchy in Dr. Hiromitsu Nakauchi’s laboratory at Stanford University for providing the sequence information of Zeb2 primers and probe. We further thank members of the Porteus laboratory and Stanford Laboratory for Cell and Gene Medicine Facility for input, comments, and discussions. A.L. was an employee of Stanford University when she conducted this work. She currently is an employee of Genentech. P.L. and J.S. were employees of the Stanford Laboratory for Cell and Gene Medicine Facility when they worked on this study. They currently are employees of Graphite Bio. N.B. passed away after this work. We dedicate this paper to her.
This work was supported by CIRM CLIN1-10084, NIH grant R01HL135607, the Debbie's Dream Foundation, the Laurie Kraus Lacob Faculty Scholar Fund in Pediatric Translational Medicine, and the Sutardja Chuk Professorship.
Author contributions
L.X. conducted most experiments, analyzed and interpretated results, and wrote the manuscript. P.L. and J.S. did some genome editing experiments. M.H.P., A.L., and N.B. provided guidance to this study. M.H.P. and A.L. made significant contributions to data interpretation and manuscript editing.
Declaration of interests
M.H.P. holds equity in CRISPR Tx, holds equity and serves on the SAB of Allogene Tx and Ziopharm Therapeutics, on the board of directors of Graphite Bio, and as an advisor to Versant Ventures.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2023.07.009.
Contributor Information
Annalisa Lattanzi, Email: lattanzi.annalisa@gene.com.
Matthew H. Porteus, Email: mporteus@stanford.edu.
Supplemental information
References
- 1.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porteus M.H. A new class of medicines through DNA editing. N. Engl. J. Med. 2019;380:947–959. doi: 10.1056/NEJMra1800729. [DOI] [PubMed] [Google Scholar]
- 3.Doudna J.A. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verdera H.C., Kuranda K., Mingozzi F. AAV vector immunogenicity in humans: A long journey to successful gene transfer. Mol. Ther. 2020;28:723–746. doi: 10.1016/j.ymthe.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lattanzi A., Camarena J., Lahiri P., Segal H., Srifa W., Vakulskas C.A., Frock R.L., Kenrick J., Lee C., Talbott N., et al. Development of β-globin gene correction in human hematopoietic stem cells as a potential durable treatment for sickle cell disease. Sci. Transl. Med. 2021;13:eabf2444. doi: 10.1126/scitranslmed.abf2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical trials with CRISPR/Cas9 in ClinicalTrials.gov website. https://clinicaltrials.gov/ct2/results?cond=CRISPR%2FCas9&term=&cntry=&state=&city=&dist=
- 7.Frangoul H., Altshuler D., Cappellini M.D., Chen Y.S., Domm J., Eustace B.K., Foell J., de la Fuente J., Grupp S., Handgretinger R., et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 8.Ataga K.I., Kutlar A., Kanter J., Liles D., Cancado R., Friedrisch J., Guthrie T.H., Knight-Madden J., Alvarez O.A., Gordeuk V.R., et al. Crizanlizumab for the prevention of pain crises in sickle cell disease. N. Engl. J. Med. 2017;376:429–439. doi: 10.1056/NEJMoa1611770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vichinsky E., Hoppe C.C., Ataga K.I., Ware R.E., Nduba V., El-Beshlawy A., Hassab H., Achebe M.M., Alkindi S., Brown R.C., et al. HOPE Trial Investigators, A phase 3 randomized trial of voxelotor in sickle cell disease. N. Engl. J. Med. 2019;381:509–519. doi: 10.1056/NEJMoa1903212. [DOI] [PubMed] [Google Scholar]
- 10.Gluckman E., Cappelli B., Bernaudin F., Labopin M., Volt F., Carreras J., Pinto Simões B., Ferster A., Dupont S., de la Fuente J., et al. Sickle cell disease: An international survey of results of HLA-identical sibling hematopoietic stem cell transplantation. Blood. 2017;129:1548–1556. doi: 10.1182/blood-2016-10-745711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eapen M., Brazauskas R., Walters M.C., Bernaudin F., Bo-Subait K., Fitzhugh C.D., Hankins J.S., Kanter J., Meerpohl J.J., Bolaños-Meade J., et al. Effect of donor type and conditioning regimen intensity on allogeneic transplantation outcomes in patients with sickle cell disease: A retrospective multicenter cohort study. Lancet. Haematol. 2019;6:e585–e596. doi: 10.1016/S2352-3026(19)30154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeil J.A., Hacein-Bey-Abina S., Payen E., Magnani A., Semeraro M., Magrin E., Caccavelli L., Neven B., Bourget P., El Nemer W., et al. Gene therapy in a patient with sickle cell disease. N. Engl. J. Med. 2017;376:848–855. doi: 10.1056/NEJMoa1609677. [DOI] [PubMed] [Google Scholar]
- 13.Urbinati F., Campo Fernandez B., Masiuk K.E., Poletti V., Hollis R.P., Koziol C., Kaufman M.L., Brown D., Mavilio F., Kohn D.B. Gene therapy for sickle cell disease: A lentiviral vector comparison study. Hum. Gene Ther. 2018;29:1153–1166. doi: 10.1089/hum.2018.061. [DOI] [PubMed] [Google Scholar]
- 14.Ikawa Y., Miccio A., Magrin E., Kwiatkowski J.L., Rivella S., Cavazzana M. Gene therapy of hemoglobinopathies: Progress and future challenges. Hum. Mol. Genet. 2019;28:R24–R30. doi: 10.1093/hmg/ddz172. [DOI] [PubMed] [Google Scholar]
- 15.Orkin S.H., Bauer D.E. Emerging genetic therapy for sickle cell disease. Annu. Rev. Med. 2019;70:257–271. doi: 10.1146/annurev-med-041817-125507. [DOI] [PubMed] [Google Scholar]
- 16.Magrin E., Miccio A., Cavazzana M. Lentiviral and genome-editing strategies for the treatment of β-hemoglobinopathies. Blood. 2019;134:1203–1213. doi: 10.1182/blood.2019000949. [DOI] [PubMed] [Google Scholar]
- 17.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S., et al. CRISPR/Cas9 β-globin gene targeting in human hematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bak R.O., Dever D.P., Reinisch A., Cruz Hernandez D., Majeti R., Porteus M.H. Multiplexed genetic engineering of human hematopoietic stem and progenitor cells using CRISPR/Cas9 and AAV6. Elife. 2017;6:e27873. doi: 10.7554/eLife.27873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin R.M., Ikeda K., Cromer M.K., Uchida N., Nishimura T., Romano R., Tong A.J., Lemgart V.T., Camarena J., Pavel-Dinu M., et al. Highly efficient and marker-free genome editing of human pluripotent stem cells by CRISPR-Cas9RNP and AAV6 donor-mediated homologous recombination. Cell Stem Cell. 2019;24:821–828.e5. doi: 10.1016/j.stem.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Charlesworth C.T., Camarena J., Cromer M.K., Vaidyanathan S., Bak R.O., Carte J.M., Potter J., Dever D.P., Porteus M.H. Priming human repopulating hematopoietic stem and progenitor cells for Cas9/sgRNA gene targeting. Mol. Ther. Nucleic Acids. 2018;12:89–104. doi: 10.1016/j.omtn.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruntman A.M., Su L., Su Q., Gao G., Mueller C., Flotte T.R. Stability and compatibility of recombinant adeno-associated virus under conditions commonly encountered in human gene therapy trials. Hum. Gene Ther. Methods. 2015;26:71–76. doi: 10.1089/hgtb.2015.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard D.B., Harvey B.K. Assaying the stability and inactivation of AAV serotype 1 vectors. Hum. Gene Ther. Methods. 2017;28:39–48. doi: 10.1089/hgtb.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane D.P., Midgley C.A., Hupp T.R., Lu X., Vojtesek B., Picksley S.M. On the regulation of the p53 tumour suppressor, and its role in the cellular response to DNA damage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1995;347:83–87. doi: 10.1098/rstb.1995.0013. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Kulesz-Martin M. p53 protein at the hub of cellular DNA damage response pathways through sequence-specific and non-sequence-specific DNA binding. Carcinogenesis. 2001;22:851–860. doi: 10.1093/carcin/22.6.851. [DOI] [PubMed] [Google Scholar]
- 25.Cuella-Martin R., Oliveira C., Lockstone H.E., Snellenberg S., Grolmusova N., Chapman J.R. 53BP1 integrates DNA repair and p53-dependent. cell fate decisions via distinct mechanisms. Mol. Cell. 2016;64:51–64. doi: 10.1016/j.molcel.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 30.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., Scadden D.T., Crumpacker C.S. Primitive hematopoietic cells resist HIV-1 infection via p21Waf1/Cip1/Sdi1. J. Clin. Invest. 2007;117:473–481. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piras F., Riba M., Petrillo C., Lazarevic D., Cuccovillo I., Bartolaccini S., Stupka E., Gentner B., Cittaro D., Naldini L., Kajaste-Rudnitski A. Lentiviral vectors escape innate sensing but trigger p53 in human hematopoietic stem and progenitor cells. EMBO Mol. Med. 2017;9:1198–1211. doi: 10.15252/emmm.201707922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari S., Jacob A., Cesana D., Laugel M., Beretta S., Varesi A., Unali G., Conti A., Canarutto D., Albano L., et al. Choice of template delivery mitigates the genotoxic risk and adverse impact of editing in human hematopoietic stem cells. Cell Stem Cell. 2022;29:1428–1444.e9. doi: 10.1016/j.stem.2022.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canny M.D., Moatti N., Wan L.C.K., Fradet-Turcotte A., Krasner D., Mateos-Gomez P.A., Zimmermann M., Orthwein A., Juang Y.C., Zhang W., et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat. Biotechnol. 2018;36:95–102. doi: 10.1038/nbt.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schiroli G., Conti A., Ferrari S., della Volpe L., Jacob A., Albano L., Beretta S., Calabria A., Vavassori V., Gasparini P., et al. Precise Gene Editing Preserves Hematopoietic Stem Cell Function following Transient p53-Mediated DNA Damage Response. Cell Stem Cell. 2019;24:551–565.e8. doi: 10.1016/j.stem.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jayavaradhan R., Pillis D.M., Goodman M., Zhang F., Zhang Y., Andreassen P.R., Malik P. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat. Commun. 2019;10:2866. doi: 10.1038/s41467-019-10735-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurup S.P., Moioffer S.J., Pewe L.L., Harty J.T. p53 hinders CRISPR/Cas9-mediated targeted gene disruption in memory CD8 T cells in vivo. J. Immunol. 2020;205:2222–2230. doi: 10.4049/jimmunol.2000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferrari S., Jacob A., Beretta S., Unali G., Albano L., Vavassori V., Cittaro D., Lazarevic D., Brombin C., Cugnata F., et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat. Biotechnol. 2020;38:1298–1308. doi: 10.1038/s41587-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweeney C.L., Pavel-Dinu M., Choi U., Brault J., Liu T., Koontz S., Li L., Theobald N., Lee J., Bello E.A., et al. Correction of X-CGD patient HSPCs by targeted CYBB cDNA insertion using CRISPR/Cas9 with 53BP1 inhibition for enhanced homology-directed repair. Gene Ther. 2021;28:373–390. doi: 10.1038/s41434-021-00251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiroli G., Conti A., Ferrari S., Della Volpe L., Jacob A., Albano L., Beretta S., Calabria A., Vavassori V., Gasparini P., et al. Precise gene editing preserves hematopoietic stem cell function following transient p53-mediated DNA damage response. Cell Stem Cell. 2019;24:551–565.e8. doi: 10.1016/j.stem.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karimian A., Ahmadi Y., Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair. 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Georgakilas A.G., Martin O.A., Bonner W.M. p21: A Two-Faced Genome Guardian. Trends Mol. Med. 2017;23:310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Li T., Kon N., Jiang L., Tan M., Ludwig T., Zhao Y., Baer R., Gu W. Tumor suppression in the absence of p53-mediated cell cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehnertz B., Chagraoui J., MacRae T., Tomellini E., Corneau S., Mayotte N., Boivin I., Durand A., Gracias D., Sauvageau G. HLF expression defines the human hematopoietic stem cell state. Blood. 2021;138:2642–2654. doi: 10.1182/blood.2021010745. [DOI] [PubMed] [Google Scholar]
- 45.Wörner T.P., Bennett A., Habka S., Snijder J., Olga Friese O., Powers T., Agbandje-McKenna M., Heck A.J.R. Adeno-associated virus capsid assembly is divergent and stochastic. Nat. Commun. 2021;12:1642–1651. doi: 10.1038/s41467-021-21935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronzitti G., Gross D.A., Mingozzi F. Human immune responses to adeno-associated virus (AAV) vectors. Front. Immunol. 2020;11:1–13. doi: 10.3389/fimmu.2020.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chew W.L., Tabebordbar M., Cheng J.K.W., Mali P., Wu E.Y., Ng A.H.M., Zhu K., Wagers A.J., Church G.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaiss A.K., Liu Q., Bowen G.P., Wong N.C.W., Bartlett J.S., Muruve D.A. Differential activation of innate immune responses by adenovirus and adeno-associated virus vectors. J. Virol. 2002;76:4580–4590. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun J.Y., Anand-Jawa V., Chatterjee S., Wong K.K. Immune responses to adeno-associated virus and its recombinant vectors. Gene Ther. 2003;10:964–976. doi: 10.1038/sj.gt.3302039. [DOI] [PubMed] [Google Scholar]
- 50.Mingozzi F., High K.A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang Z., Kurupati R.K., Li Y., Kuranda K., Zhou X., Mingozzi F., High K.A., Ertl H.C.J. The effect of CpG sequences on capsid-specific CD8+ T Cell responses to AAV vector gene transfer. Mol. Ther. 2020;28:771–783. doi: 10.1016/j.ymthe.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu J., Allred C.C., Jensen M.D. Human adipose tissue protein analyses using capillary western blot technology. Nutr. Diabetes. 2018;8:26–35. doi: 10.1038/s41387-018-0030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eemirel E., Arnold C., Garg J., Jäger M.A., Sticht C., Li R., Kuk H., Wettschureck N., Hecker M., Korff T. RGS5 attenuates baseline activity of ERK1/2 and promotes growth arrest of vascular smooth muscle cells. Cells. 2021;10:1748–1768. doi: 10.3390/cells10071748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ban T., Kikuchi M., Sato G.R., Manabe A., Tagata N., Harita K., Nishiyama A., Nishimura K., Yoshimi R., Kirino Y., et al. Genetic and chemical inhibition of IRF5 suppresses pre-existing mouse lupus-like disease. Nat. Commun. 2021;12:4379–4393. doi: 10.1038/s41467-021-24609-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., Vakulskas C.A., Collingwood M.A., Zhang L., Bode N.M., et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mehta A., Merkel O.M. Immunogenicity of Cas9 protein. J. Pharmaceut. Sci. 2020;109:62–67. doi: 10.1016/j.xphs.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E., Mangan P.A., Kulikovskaya I., Gupta M., Chen F., et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367 doi: 10.1126/science.aba7365. eaba7365–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cromer M.K., Vaidyanathan S., Ryan D.E., Curry B., Lucas A.B., Camarena J., Kaushik M., Hay S.R., Martin R.M., Steinfeld I., et al. Global transcriptional response to CRISPR/Cas9-AAV6-based genome editing in CD34+ hematopoietic stem and progenitor cells. Mol. Ther. 2018;26:2431–2442. doi: 10.1016/j.ymthe.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ward I.M., Minn K., van Deursen J., Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X., Jiang W., Dubois R.L., Yamamoto K., Wolner Z., Zha S. Overlapping functions between XLF repair protein and 53BP1 DNA damage response factor in end joining and lymphocyte development. Proc. Natl. Acad. Sci. USA. 2012;109:3903–3908. doi: 10.1073/pnas.1120160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this paper.