Abstract
Intensive production can cause immunological stress in commercial broilers. Chlorogenic acid (CGA) regulates the intestinal microbiota, barrier function, and immune function in chickens. As complex interrelations regulate the dynamic interplay between gut microbiota, the host, and diverse health outcomes, the aim of this study was to elucidate the immunoregulatory mechanisms of CGA using multi-omics approaches. A total of 240 one-day-old male broilers were assigned to a 2 × 2 factorial design with 2 CGA levels (0 or 500 mg/kg) either with or without dexamethasone (DEX) injection for a 21-day experimental period. Therefore, there were 4 dietary treatments: control, DEX, CGA, and DEX + CGA, with 6 replicates per treatment. CGA supplementation improved (P < 0.05) growth performance, jejunal morphology, jejunal barrier function, and immune function in DEX-treated broilers. Moreover, in DEX + CGA-treated broilers, the increase in gut microbiome diversity (P < 0.05) was consistent with a change in taxonomic composition, especially in the Clostridiales vadin BB60_group. Additionally, the levels of short-chain fatty acids increased remarkably (P < 0.01) after CGA supplementation. This was consistent with the Kyoto Encyclopedia of Genes and Genomes analysis results that the “pyruvate fermentation to butanoate” pathway was more enriched (P < 0.01) in the DEX + CGA group than in the DEX group. Proteomics revealed that CGA treatment increased the expression of several health-promoting proteins, thymosin beta (TMSB4X) and legumain (LGMN), which were verified by multiple reaction monitoring. Metabolomics revealed that CGA treatment increased the expression of health-promoting metabolites (2,4-dihydroxy benzoic acid and homogentisic acid). Proteomic and metabolic analyses showed that CGA treatment regulated the peroxisome proliferator-activated receptor (PPAR) and mitogen-activated protein kinase (MAPK) pathways. Western blotting results support these findings. Pearson’s correlation analyses showed correlations (P < 0.01) between altered immune function, jejunal barrier function, different microbiota, proteins, and metabolites parameters. Overall, our data indicate that CGA treatment increased growth performance and improved the immunological functions of DEX-treated broilers by regulating gut microbiota and the PPAR and MAPK pathways. The results offer novel insights into a CGA-mediated improvement in immune function and intestinal health.
Keywords: Broiler, Immunological stress, Microbiome, Metabolomics, Proteomics
1. Introduction
Intensive farming practices have been popular in recent decades due to the increased demand for poultry products. However, high stocking densities have increased the vulnerability of commercial broiler chickens to various stress factors, including immunological stress (Li et al., 2015a, Li et al., 2015b). These stress factors can easily damage the intestine because the chickens are continuously exposed to multiple antigens from food, resident bacteria, and invading viruses (Söderholm and Perdue, 2001).
Intestinal health can be regulated by gut microbiota (Gao et al., 2018; Shi et al., 2019), which participates in the development and maintenance of the immune system and intestinal homeostasis by stimulating immune responses and maintaining epithelial barrier functions (Broom and Kogut, 2018). A decrease in gut microbiota biodiversity and disruption of microbe-host equilibrium has been observed in animals with intestinal inflammation (Zou et al., 2019; Yang et al., 2020). Additionally, immunological stress has been demonstrated to influence the gut microbiota of broilers; for example, resulting in lower abundances of Gammaproteobacteria and Enterobacteriales (Chen and Yu, 2021). Recently, the prevention or treatment of diseases through regulating gut microbiota has received considerable research attention. For example, phenolics, such as epicatechin, catechin, gallic acid, and caffeic acid have been implicated in inhibiting the growth of Clostridium perfringens, Clostridium difficile, and Bacteroides spp. (Selma et al., 2009). Chlorogenic acid (CGA) is a phenolic acid produced by several plants, including tea, coffee, and several Chinese herbs, such as the buds of Lonicera japonica Thunb and the leaves of Eucommia ulmodies (Upadhyay and Mohan Rao, 2013; Naveed et al., 2018). Lou et al. (2011) showed that CGA could kill pathogenic bacteria strains (Shigella dysenteria and Streptococcus pneumoniae) by provoking irreversible permeability changes in cell membranes. CGA also resists immune stress and regulates gut microbiota (Liang and Kitts, 2015; Chen et al., 2021). Furthermore, CGA has been reported to increase intestinal barrier function and the abundance of Lactobacillus spp. in the cecum of pigs (Chen et al., 2019) and reduce small intestine injury and inflammation in chickens challenged with Clostridium perfringens type A (Zhang et al., 2020). Thus, it is hypothesised that CGA may attenuate the immunological stress of chickens by regulating the gut microbiota.
Gut microbiota modulates signalling pathways involved in intestinal mucosa homeostasis by producing specific metabolites, indicating that metabolomics could be used to obtain detailed information on gut metabolic pathways (Vernocchi et al., 2016). Moreover, proteomic techniques can provide detailed information on the function and activity of identified metabolites (Xiong et al., 2015; Haange and Jehmlich, 2016). Studies have examined the effect of stress on the performance of domesticated animals using synthetic glucocorticoids, such as dexamethasone (DEX) (Gao et al., 2010; Njagi et al., 2012). In a previous study, we observed that DEX induced immunological stress and impaired the intestinal immune function of broilers (Liu et al., 2021). Although the beneficial effects of CGA are associated with the gut microbiota, little is known about how CGA intake influences the crosstalk between gut microbiota, host metabolism, and protein expression in the intestinal tract. Therefore, the aim of this study was to examine the effect and mechanism of CGA on immune function and intestinal health in DEX-treated broilers using multi-omics techniques.
2. Materials and methods
2.1. Animal ethics statement
All experimental protocols were approved by the Animal Care and Use Committee of Qingdao Agricultural University (protocol number 20200916115). We have followed the ARRIVE guidelines for reporting animal research (Kilkenny et al., 2010).
2.2. Chemicals and reagents
The DEX was obtained from Beian Feilong Animal Pharmaceutical Factory (Beian, China). The CGA (98% purity) was purchased from Changsha E.K Herb (Changsha, China).
2.3. Experimental design and sample collection
A total of 240 one-day-old male Cobb 500 broilers were assigned to a 2 × 2 factorial design with 2 CGA levels (0 or 500 mg/kg feed) and 2 DEX levels (0 or 3 mg/kg body weight), resulting in 4 dietary treatments: control, DEX, CGA, and DEX + CGA with 6 replicates per treatment and 10 broilers per replicate. The doses of CGA and DEX were according to Liu et al. (2022) and Wang (2012), respectively. The mixed feed was prepared according to the requirements of the National Research Council (NRC, 1994; Table S1), and the correct quantity of CGA was mixed with the basal diet to obtain the prefixed inclusion level. The DEX was injected intraperitoneally once a day from the 15th to the 21st day of the experiments. The experiments were performed for 21 d. On the 21st day, 6 broilers from each group were randomly selected for fasting treatments for 12 h. Thereafter, blood was sampled from the wing vein, centrifuged at 3,000 × g for 10 min at 4 °C, and stored at −20 °C for further analysis of d-lactate (d-LA), diamine oxidase (DAO), immunoglobulin (Ig) A, IgM, interleukin (IL)-1β, IL-4, IL-6, IL-10, IL-12, IL-18, IL-22, tumour necrosis factor-α (TNF-α), interferon-γ (IFN-γ), CXC chemokine ligand (CXCL) 1, CXCL2 and metabolites. Chickens were sacrificed by cervical dislocation, and the jejunal segments were fixed in 4% paraformaldehyde for analysing the intestinal morphology. Certain part of the jejunum was cut open, and the contents were rinsed with pre-cooled saline. After rinsing, filter paper was used to absorb water at the edge and the jejunal mucosa was collected by scraping the surface with a slide. The jejunal mucosa was used for the detection of levels of biochemical parameters (IgA, IgM, IL-1β, IL-4, IL-6, IL-10, IL-12, IL-18, IL-22, TNF-α, IFN-γ, CXCL1 and CXCL2), mRNA expression (IL-1β, IL-4, IL-6, IL-10, IL-12, IL-18, IL-22, TNF-α, IFN-γ, cysteinyl aspartate specific proteinase [caspase-3] and caspase-9), protein expression (β-actin, occludin, zonula occluden-1 [ZO-1], extracellular regulated protein kinases [ERK], p-ERK, c-Jun N-terminal kinase [JNK], p-JNK, P38, p-P38 and peroxisome proliferator-activated receptor [PPAR]), immunohistochemistry analysis (occludin and ZO-1), proteomics and multiple reaction monitoring (MRM). The cecal contents were collected for microbiome detection and short-chain fatty acid (SCFA) analysis. All jejunal samples, except for the jejunal samples for morphological analysis, and cecal contents were immediately placed in liquid nitrogen and then stored at −80 °C.
2.4. Growth performance measurement
The amounts of provided and refused feed were measured daily on a replicate basis to calculate the average daily feed intake (ADFI). Body weight was measured at d 14 and 21 to calculate average daily gain (ADG), and feed:gain ratio (F:G) on a per replicate basis. The ADFI, ADG and F:G formulae were calculated using the following equations:
ADG = (final body weight − initial body weight)/number of days of the rearing period;
ADFI = (feed offer weight − feed residue weight)/number of days of the rearing period;
F:G = ADFI/ADG.
2.5. Intestinal morphology
The intestinal morphology of the jejunal parts was evaluated as previously described (Livak and Schmittgen, 2001). Briefly, the jejunal segments were fixed using 4% paraformaldehyde, embedded with paraffin, sliced, placed on glass slides, and stained using hematoxylin and eosin (H&E) stain. Villi heights and crypt depths were observed using an HMIAS-2000 image analysis system and an Olympus microscope (Olympus, Tokyo, Japan).
2.6. Analysis of biochemical indices
The biochemical indices were detected using ELISA kits and a continuous wavelength microplate reader (Infinite 200 PRO, Tecan Life Sciences, Männedorf, Switzerland). The kits were acquired from Jiangsu Enzymatic Co., Ltd. (Yancheng, China), and the procedures were carried out according to the manufacturer’s instructions. d-LA and DAO levels in serum, and IgA, IgM, IL-1β, IL-4, IL-6, IL-10, IL-12, IL-18, IL-22, TNF-α, IFN-γ, CXCL1, and CXCL2 levels in the serum and jejunal mucosa were evaluated.
2.7. Determination of mRNA expression levels by RT-qPCR
RNA extraction, cDNA synthesis, qPCR analysis, and analysis of the relative levels of expression of mRNA were carried out as described previously (Wang et al., 2021). Total RNA was extracted from the jejunal mucosa using TRIzol (Tiangen Biochemical Technology, Beijing, China). The integrity and purity were analysed using agarose gel electrophoresis and a spectrophotometer. DNA was amplified using a BioRad CFX96 Real-Time PCR system (Bio-Rad Laboratories, Hercules, CA, USA), and expression levels of the target genes were determined using the 2−ΔΔCt method and normalized to glyceraldehyde-3-phosphate dehydrogenase (GADPH) expression. Reverse transcription and quantification kits were purchased from TAKARA (Takara Biotechnology, Dalian, China). The primers used in the present study are listed in Table S2.
2.8. Determination of protein expression levels by Western blotting
Proteins were isolated from the jejunal mucosa of the broilers using RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China), according to the manufacturer’s instructions. The protein expression levels of β-actin, occludin, ZO-1, ERK, p-ERK, JNK, p-JNK, P38 and p-P38 were detected using western blotting, as previously described (Hu et al., 2020). Equal quantities of protein were separated using sulfate-polyacrylamide gel electrophoresis (Beyotime Biotechnology). The protein was transferred onto polyvinylidene difluoride membranes (Merck Millipore, Darmstadt, Germany) and incubated with the primary antibodies. Subsequently, the membranes were incubated with HRP-labelled goat anti-rabbit IgG antibody (Beyotime Biotechnology). The proteins were detected using the iBright FL1000 electrochemiluminescence detection system (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions and quantitated using ImageJ (National Institutes of Health, USA). Anti-β-actin antibody was obtained from Beyotime Institute of Biotechnology (Shanghai, China); anti-occludin and anti-ZO-1 antibodies were obtained from Servicebio (Wuhan, China); anti-PPAR antibody was obtained from Novus Biologicals (Littleton, CO, USA); anti-ERK, anti-p-ERK, anti-JNK, anti-p-JNK, anti-P38 and anti-p-P38 antibodies were obtained from Beijing Bioss Biotechnology (Beijing, China).
2.9. Immunohistochemistry analysis
Immunohistochemical analyses were conducted as previously described (Chávez-Carbajal et al., 2019). The jejunal sections were paraffin-embedded, dewaxed and rehydrated, retried in antigen, blocked in endogenous peroxidase activity, sealed with rabbit serum, incubated with specific primary antibodies (occludin or ZO-1) and the corresponding HRP-conjugated secondary antibody, immunostaining with DAB chromogenic and counterstaining the nucleus, dehydrated, and mounted. An Olympus microscope was used to examine the stained sections.
2.10. DNA extraction and microbiome analysis
Bacterial genomes were isolated using a TIANamp stool DNA kit (Tiangen Biotech). The purity was examined using 0.8% agarose gel electrophoresis. It was quantified using a Qubit 2.0 Fluorometer (Invitrogen), and sequenced using a HiSeq 4000 system (Illumina Inc., San Diego, CA, US) at Lianchuan Biotech Co., Ltd. (Hangzhou, China).
High-quality clean tags were obtained by quality filtering of the raw tags as described in QIIME (Quantitative Insights Into Microbial Ecology v1.2.1) (http://qiime.org/). Uclust v1.2.22 (https://arc.umich.edu/software-item/uclust/) was used to prepare clusters of operational taxonomic units (OTU) at a 97% sequence identity level. Sequences with reference meanings per cluster were annotated for taxonomic classification against the SILVA database (https://www.arb-silva.de/). Additionally, the alpha diversity of the dataset was evaluated. R (Version 2.15.3) and PICRUSt (https://picrust.github.io/picrust/) were used independently for principal coordinate analysis (PCoA) of the OTU in different groups and functional prediction of the genes in the gut microbiota, using a closed-reference OTU table in BIOM-format from the pick_closed_reference_otus.py script generated in QIIME.
Taxonomic classification was achieved based on homology (>97% identity) between queried and reference sequences from the Greengenes database v13.8 (https://mothur.org/wiki/greengenes-formatted_databases). Subsequently, the OTU table was normalised using Langille Lab Online Galaxy Instance (http://galaxy.morganlangille.com), followed by metagenome functional prediction based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Significant differences in gene function among the groups were revealed, in addition to metagenome data prediction by PICRUSt, the Kruskal–Wallis test, and multiple test correction based on Storey’s false discovery rates (FDR) (Furuhashi et al., 2018). All sequence data have been submitted to the NCBI under BioProject ID PRJNA789475.
2.11. SCFA analysis
Short-chain fatty acids were quantified at MetWare Biotechnology Co., Ltd. (Wuhan, China) as previously described (Kuttappan et al., 2017). In brief, SCFA were extracted following homogenising 20 mg caecal content in isobutanol:water (1:9, vol/vol) solution. The supernatant was obtained by centrifugation at 12,000 × g for 10 min at 4 °C. Purification of SCFA from each 100 μL supernatant was carried out following extraction of the the-top-layer supernatant by centrifugation at 12,000 × g for 10 min at 4 °C, after mixing with chloroform and NaOH (20 mmol/L). Subsequently, the samples were mixed with isobutanol, pyridine, and isobutyl chloroformate. Purified SCFA (pretreated by hexane) were quantified using an Agilent 5977 B mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) with helium as the carrier gas. The injector and detector were set to 260 °C. The column temperature was set to 50 °C for 5 min, increased to 150 °C at a rate of 5 °C/min, increased to 325 °C at a rate of 40 °C/min, and finally held at 325 °C for 1 min.
2.12. Proteomic analysis
Proteomic analysis was performed by MetWare Biotechnology Co., Ltd. (Wuhan, China). The jejunal samples were homogenized in lysis buffer (2.5% SDS, 100 mmol/L Tris–HCl, pH 8.0), subjected to ultrasonication and centrifuged at 10,000 × g for 10 min at 4 °C. The proteins were precipitated by adding 4-fold volumes of pre-cooled acetone and lysed with 8 M urea and 100 mmol/L Tris-Cl. This was used in a reduction reaction with 10 mmol/L dithiothreitol, followed by an alkylation reaction with sulfhydryl and 40 mm iodoacetamide. Subsequently, 100 mM Tris–HCl was added, and the urea concentration was diluted to less than 2 M. Equal amounts of samples were used for TMT labelling, which was performed according to the manufacturer’s instructions. Each fraction was vacuum-dried and stored at −80 °C until mass spectrometry (MS) analysis.
Liquid chromatography-tandem (LC)-MS/MS analysis was carried out using a hybrid LTQ-OrbitrapXL mass spectrometer equipped with 40 cm C18 columns. The mobile phase (0 to 40% acetonitrile) was applied over a 2-h period (Lee et al., 2019). LC-MS/MS analysis was performed in triplicate. Data are available via ProteomeXchange with the following identifier: IPX0003857000.
2.13. MRM analysis
Peptide samples were desalted using a Monospin column. The dried mixed peptide samples were dissolved in trifluoroacetic acid (TFA, 0.1%), transferred to the desalting columns and centrifuged at 300 × g for 10 min at 4 °C. Thereafter, TFA (0.1%) solution was added to remove contaminants, and acetonitrile solution (50%) was used to elute the peptide. The elution solutions were collected and dried in tubes.
The peptide samples were analysed using a nanoACQUITY HPLC system with a nanoACQUITY ultra-performance liquid chromatography (UPLC) C18 column (100 m × 100 mm, 1.7 μm particle size; Waters, Milford, MA, USA) and a triple quadrupole mass spectrometer (QTRAP 5500; AB Sciex, Redwood, CA, US). The mobile phase contained acetonitrile (2%)/formic acid (0.1%) and acetonitrile (98%)/formic acid (0.1%) in HPLC-grade water as solvents A and B, respectively. Metabolites were eluted using the following step gradient at a flow rate of 5 L/min: 0–5 min: 95% A, 5% B; 5–50 min: 70% A, 30% B; 50.5–55 min: 20% A, 80% B; and 55.5–60 min: 98% A, 2% B. The mass spectrometry system conditions included ion source, electrospray ion source, positive ion detection, and scanning mode. The electrospray ionization source operation parameters were as follows: injection voltage, 5,500 eV; temperature, 150 °C; curtain gas (CUR, N2), 206.850 Pa; collision gas pressure (CAD, N2), high mode; auxiliary gas GAS1 pressure, 137,900 Pa; auxiliary gas GAS2 pressure, 103,425 Pa; scanning time, 5 ms. All analyses were performed in triplicate. Peptide peak areas were used to calculate percentage coefficient of variation (CV), and the average percentage CV for each target peptide in the sample was calculated (Wu et al., 2021).
2.14. Metabolomic analysis
The samples were thawed on ice, vortexed for 10 s and thoroughly mixed. Thereafter, 300 μL of pure methanol was added to 50 μL of serum. The mixture was rotated for 3 min and then centrifuged at 12,000 × g for 10 min at 4 °C. The resulting supernatant was centrifuged twice at 12,000 × g for 5 min at 4 °C. The samples were incubated at −20 °C for 30 min, centrifuged at 12,000 × g for 3 min at 4 °C, and 150 μL of the supernatant transferred into injection bottles for analysis.
The chromatography-mass spectrometry acquisition conditions of serum were as described previously (Zhang et al., 2022). The data acquisition instrument system mainly includes an ExionLCTM AD UPLC system (AB Sciex) and a QTRAP tandem mass spectrometry (MS) system (Applied Biosystems, Waltham, MA, USA).
2.15. Statistics
The effects of DEX and CGA and their interaction on inflammatory parameters, jejunal morphology and barrier function, mRNA and protein expression, and SCFA were assessed using analysis of variance (ANOVA) and the general linear model procedure using IBM SPSS Statistics 18.0 (IBM Corp., Armonk, NY, USA). Multiple mean comparisons were performed using univariate ANOVA and Duncan’s multiple range test. Cecum microbial domains, phyla, and genera were compared using the Wilcoxon rank-sum test, and cecal microbial species were compared using linear discriminant analysis effect size (LEfSe) analysis. Differentially expressed metabolites were screened based on a fold change (FC) > 2 or < 0.5 and variable importance in projection (VIP) > 1.2. Orthogonal partial least squares discriminant analysis (OPLS-DA) was used to obtain VIP values, including score and permutation plots. The MetaboAnalyzer package (Version 2.15.3) was used to generate the terms. TMT analysis was performed, and proteins with FC > 1.2 were identified as differentially expressed proteins (DEP). Spearman’s rank correlation analysis was performed to determine the relationships between serum and jejunal inflammatory parameters, jejunal morphology, mRNA expression of inflammatory cytokines, significantly different microbiome assemblages, SCFA contents, proteins, and metabolites. All means and comparison groups were considered statistically significant at P < 0.05.
3. Results
3.1. CGA improves the growth performance in DEX-induced broilers
Compared with the control group, DEX injection decreased the ADG (P < 0.01) and increased the F:G value (P < 0.01) during d 14 to 21 and d 1 to 21. In contrast, CGA supplementation increased the ADG (P < 0.01) and reduced the F:G value (P < 0.01). The interaction between CGA and DEX also had significant effects on ADG and the F:G value during d 14 to 21 and d 1 to 21 (P < 0.01) (Table 1).
Table 1.
Effects of dexamethasone, chlorogenic acid, or their interaction on the growth performance of broilers.
| Group | Day 1–14 |
Day 14–21 |
Day 1–21 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ADG, g | ADFI, g | F:G | ADG, g | ADFI, g | F:G | ADG, g | ADFI, g | F:G | |
| Control | 26.11 | 31.10 | 1.19 | 55.56a | 74.35 | 1.35b | 36.60a | 45.04 | 1.23c |
| DEX | 26.52 | 30.73 | 1.17 | 26.96c | 64.89 | 2.44a | 26.70c | 43.18 | 1.62a |
| CGA | 27.30 | 32.16 | 1.18 | 53.67a | 73.45 | 1.37b | 35.96a | 45.95 | 1.28bc |
| DEX + CGA | 26.49 | 30.76 | 1.16 | 46.00b | 69.21 | 1.51b | 32.42b | 43.49 | 1.34b |
| SEM | 1.13 | 0.67 | 0.04 | 2.25 | 2.14 | 0.10 | 1.16 | 1.30 | 0.05 |
| Main effect | |||||||||
| CGA | |||||||||
| − | – | – | – | 41.26 | 69.63 | 1.89 | 31.65 | 44.11 | 1.43 |
| + | – | – | – | 49.83 | 69.21 | 1.44 | 34.19 | 44.72 | 1.31 |
| DEX | |||||||||
| − | – | – | – | 54.61 | 73.90 | 1.36 | 36.28 | 45.50 | 1.26 |
| + | – | – | – | 36.48 | 67.06 | 1.97 | 29.56 | 43.34 | 1.48 |
| P-value | |||||||||
| CGA | 0.76 | 0.15 | 0.81 | <0.01 | 0.27 | <0.01 | <0.01 | 0.51 | <0.01 |
| DEX | – | – | – | <0.01 | <0.01 | <0.01 | <0.01 | 0.03 | <0.01 |
| Interaction | – | – | – | <0.01 | 0.10 | <0.01 | <0.01 | 0.75 | <0.01 |
DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid.
a,b,c Within a column, means without a common superscriptdiffer significantly (P < 0.05). n = 6 for each group.
3.2. Immunoregulation effects of CGA in DEX-induced broilers
Compared with the control group, DEX treatment increased the serum IL-1β (P = 0.02), IL-6 (P < 0.01), IL-18 (P < 0.01), IL-22 (P < 0.01), TNF-α (P = 0.01), CXCL1 (P = 0.02), and CXCL2 (P < 0.01) levels of the broilers and decreased the serum IL-4 and IFN-γ levels (P < 0.01). In contrast, CGA supplementation increased serum IgM (P = 0.02), IL-4 (P < 0.01), and IFN-γ (P = 0.01) levels and decreased serum IL-1β (P < 0.01), IL-12 (P = 0.01), IL-18 (P < 0.01), IL-22 (P < 0.01), CXCL1 (P < 0.01), and CXCL2 (P < 0.01) levels. Moreover, the interaction between CGA and DEX (CGA × DEX) reversed the DEX-induced changes in serum IL-1β (P = 0.03), IL-4 (P = 0.04), IL-6 (P < 0.01), IL-10 (P < 0.01), IL-12 (P < 0.01), IL-18 (P < 0.01), IL-22 (P = 0.03), IFN-γ (P < 0.01), CXCL1 (P < 0.01), and CXCL2 (P < 0.01) levels (Table 2).
Table 2.
Effects of dexamethasone, chlorogenic acid, or their interaction on the serum immune parameters of broilers.
| Item | IgA, ng/mL | IgM, ng/mL | IL-1β, pg/mL | IL-4, pg/mL | IL-6, pg/mL | IL-10, pg/mL | IL-12, pg/mL | IL-18, ng/L | IL-22, ng/L | TNF-α, pg/mL | IFN-γ, pg/mL | CXCL1, ng/L | CXCL2, ng/L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 333.77 | 1097.70 | 709.56b | 187.46ab | 86.61c | 56.45a | 534.33b | 105.19bc | 19.16b | 93.33 | 12.86a | 189.78b | 187.87b |
| DEX | 349.95 | 887.38 | 800.67a | 131.52c | 122.55a | 37.43b | 651.00a | 202.99a | 28.10a | 114.00 | 7.23b | 258.61a | 280.04a |
| CGA | 381.08 | 1241.75 | 693.44b | 195.78a | 98.64b | 47.01ab | 496.33c | 124.84bc | 14.88b | 89.76 | 11.34a | 164.78c | 145.83c |
| DEX + CGA | 341.13 | 1176.67 | 698.44b | 170.46b | 100.05b | 54.29a | 403.67c | 76.26c | 17.01b | 96.85 | 11.27a | 140.40c | 142.76c |
| SEM | 12.24 | 48.63 | 12.53 | 6.24 | 3.14 | 2.18 | 24.60 | 10.31 | 1.24 | 3.03 | 0.49 | 10.01 | 12.81 |
| Main effect | |||||||||||||
| CGA | |||||||||||||
| − | 341.86 | 992.54 | 755.11 | 160.49 | 104.58 | 46.94 | 592.67 | 154.09 | 23.63 | 103.67 | 10.04 | 224.20 | 232.46 |
| + | 361.10 | 1209.21 | 695.94 | 183.13 | 99.34 | 50.65 | 450.00 | 100.55 | 15.94 | 93.31 | 11.31 | 152.29 | 144.30 |
| DEX | |||||||||||||
| − | 357.43 | 1169.72 | 701.50 | 192.62 | 92.62 | 51.73 | 515.33 | 115.02 | 17.02 | 91.54 | 12.10 | 177.28 | 165.35 |
| + | 329.61 | 1032.02 | 749.56 | 151.00 | 111.30 | 45.86 | 527.33 | 139.62 | 22.55 | 105.43 | 9.25 | 199.51 | 211.40 |
| P-value | |||||||||||||
| CGA | 0.12 | 0.02 | <0.01 | <0.01 | 0.14 | 0.27 | 0.01 | <0.01 | <0.01 | 0.05 | 0.01 | <0.01 | <0.01 |
| DEX | 0.22 | 0.12 | 0.02 | <0.01 | <0.01 | 0.09 | 0.73 | <0.01 | <0.01 | 0.01 | <0.01 | 0.02 | <0.01 |
| Interaction | 0.59 | 0.41 | 0.03 | 0.04 | <0.01 | <0.01 | <0.01 | <0.01 | 0.03 | 0.19 | <0.01 | <0.01 | <0.01 |
DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid; Ig = immunoglobulin; IL = interleukin; TNF-α = tumor necrosis factor α; IFN-γ = interferon γ; CXCL= CXC chemokine ligand.
a,b,c Within a column, means without a common superscriptdiffer significantly (P < 0.05). n = 6 for each group.
Furthermore, DEX treatment increased (P < 0.01) the concentrations of IL-1β, IL-6, IL-18, IL-22, TNF-α, CXCL1, and CXCL2 in the jejunal mucosa of the broilers. In contrast, CGA supplementation increased (P < 0.01) the jejunal concentration of IgM and decreased (P < 0.01) the jejunal expression of IL-1β, IL-6, IL-12, IL-18, IL-22, and CXCL2. Additionally, CGA × DEX reversed the DEX-induced changes in the jejunal expression of IL-1β, IL-4, IL-6, IL-12, IL-18, IL-22, CXCL1, CXCL2 (P < 0.01) and TNF-α (P = 0.03) (Table 3). Moreover, gene expression analysis showed that DEX treatment increased jejunal expression of IL-1β (P < 0.01), IL-6 (P < 0.01), IL-12 (P < 0.01), IL-18 (P < 0.01), IL-22 (P = 0.01), TNF-α (P = 0.03), caspase-3 (P = 0.02), and caspase-9 (P = 0.04) genes compared with the control group. In contrast, CGA supplementation decreased the jejunal expression of IL-1β (P < 0.01), IL-6 (P < 0.01), IL-12 (P = 0.02), IL-18 (P < 0.01), IL-22 (P = 0.04), TNF-α (P = 0.01), caspase-3 (P = 0.02), and caspase-9 (P < 0.01). Additionally, CGA × DEX reversed the DEX-induced changes in the jejunal expression of IL-1β (P < 0.01), IL-4 (P < 0.01), IL-6 (P < 0.01), IL-10 (P < 0.01), IL-12 (P = 0.05), IL-18 (P < 0.01), IL-22 (P = 0.01), TNF-α (P = 0.04), caspase-3 (P < 0.01), and caspase-9 (P = 0.03) genes (Fig. 1).
Table 3.
Effects of dexamethasone, chlorogenic acid, or their interaction on the jejunal immune parameters of broilers.
| Item | IgA, ng/mg prot | IgM, ng/mg prot | IL-1β, pg/mg prot | IL-4, pg/mg prot | IL-6, pg/mg prot | IL-10, pg/mg prot | IL-12, pg/mg prot | IL-18, ng/mg prot | IL-22, ng/mg prot | TNF-α, pg/mg prot | IFN-γ, pg/mg prot | CXCL1, ng/mg prot | CXCL2, ng/mg prot |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 29.53 | 63.77 | 5.34b | 1.56a | 0.26b | 0.53 | 714.41b | 10.22b | 2.72b | 0.44c | 18.75 | 12.42c | 16.47b |
| DEX | 18.39 | 50.06 | 7.34a | 1.15c | 0.34a | 0.48 | 889.90a | 14.87a | 4.48a | 0.53a | 16.64 | 21.75a | 26.01a |
| CGA | 27.29 | 72.32 | 5.33b | 1.38b | 0.22c | 0.56 | 700.69b | 9.51b | 2.54b | 0.49b | 17.31 | 15.00bc | 15.63b |
| DEX + CGA | 22.26 | 68.54 | 5.41b | 1.33b | 0.23c | 0.54 | 635.98b | 10.38b | 2.68b | 0.51ab | 16.80 | 16.09b | 17.12b |
| SEM | 1.26 | 2.31 | 0.21 | 0.039 | 0.011 | 0.013 | 24.78 | 0.50 | 0.18 | 0.01 | 1.19 | 0.87 | 0.95 |
| Main effect | |||||||||||||
| CGA | |||||||||||||
| − | 23.96 | 56.91 | 6.34 | 1.35 | 0.30 | 0.50 | 802.16 | 12.55 | 3.60 | 0.49 | 17.70 | 17.08 | 21.24 |
| + | 24.78 | 70.43 | 5.37 | 1.35 | 0.22 | 0.55 | 668.33 | 9.95 | 2.61 | 0.50 | 17.06 | 15.54 | 16.38 |
| DEX | |||||||||||||
| − | 28.41 | 68.04 | 5.33 | 1.47 | 0.24 | 0.54 | 707.55 | 9.87 | 2.63 | 0.47 | 18.03 | 13.71 | 16.05 |
| + | 20.32 | 59.30 | 6.38 | 1.24 | 0.28 | 0.51 | 762.94 | 12.63 | 3.58 | 0.52 | 16.72 | 18.92 | 21.56 |
| P-value | |||||||||||||
| CGA | 0.67 | <0.01 | <0.01 | 0.98 | <0.01 | 0.08 | <0.01 | <0.01 | <0.01 | 0.43 | 0.80 | 0.16 | <0.01 |
| DEX | <0.01 | 0.01 | <0.01 | <0.01 | <0.01 | 0.16 | 0.10 | <0.01 | <0.01 | <0.01 | 0.61 | <0.01 | <0.01 |
| Interaction | 0.12 | 0.14 | <0.01 | <0.01 | <0.01 | 0.58 | <0.01 | <0.01 | <0.01 | 0.03 | 0.75 | <0.01 | <0.01 |
DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid; Ig = immunoglobulin; IL = interleukin; TNF-α = tumor necrosis factor α; IFN-γ = interferon γ; CXCL = CXC chemokine ligand.
a,b,c Within a column, means without a common superscriptdiffer significantly (P < 0.05). n = 6 for each group.
Fig. 1.
Effects of DEX, CGA, or their interaction on mRNA expressions of inflammation- and apoptosis-related genes. DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid; IL = interleukin; TNF-α = tumor necrosis factor α. n = 6 for each group.
3.3. CGA improves the jejunal morphology and barrier function of DEX-treated broilers
Compared with the control group, histological analysis showed that DEX treatment decreased (P < 0.01) villus height and villus height to crypt depth ratio (V:C ratio) and increased (P < 0.01) crypt depth. In contrast, CGA supplementation increased villus height (P = 0.02) and V:C ratio (P < 0.01) and decreased crypt depth (P = 0.04). Additionally, CGA × DEX reversed the DEX-induced decrease in villus height (P = 0.02) and V:C ratio (P < 0.01) (Fig. 2A, Table 4).
Fig. 2.
Effects of DEX, CGA, or their interaction on jejunal morphology and expressions of tight junction proteins. (A) H&E staining. (B) Western blotting for ZO-1 and occludin. (C) Immunohistochemistry for occludin. (D) Immunohistochemistry for ZO-1. DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid; ZO-1 = zonula occluden-1. n = 6 for each group.
Table 4.
Effects of dexamethasone, chlorogenic acid, or their interaction on the jejunal morphology of broilers.
| Item | Villus height, μm | Crypt depth, μm | V:C ratio |
|---|---|---|---|
| Control | 347.86a | 73.26 | 4.76a |
| DEX | 224.59b | 110.71 | 2.11b |
| CGA | 344.57a | 72.33 | 4.81a |
| DEX + CGA | 332.15a | 79.89 | 4.18a |
| SEM | 29.21 | 10.06 | 0.43 |
| Main effect | |||
| CGA | |||
| − | 286.23 | 91.98 | 3.43 |
| + | 338.36 | 76.11 | 4.50 |
| DEX | |||
| − | 346.22 | 72.79 | 4.78 |
| + | 278.37 | 95.30 | 3.15 |
| P-value | |||
| CGA | 0.02 | 0.04 | <0.01 |
| DEX | <0.01 | <0.01 | <0.01 |
| Interaction | 0.02 | 0.05 | <0.01 |
DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid; V:C = villus height to crypt depth.
a,b Within a column, means without a common superscriptdiffer significantly (P < 0.05). n = 6 for each group.
Moreover, DEX treatment increased (P < 0.01) the d-LA levels of the broilers, whereas CGA supplementation decreased (P < 0.01) the d-LA levels. Additionally, CGA × DEX reversed (P < 0.01) the DEX-induced increase in d-LA levels. However, the DAO level was not significantly affected by CGA, DEX, or their interaction (P > 0.05) (Table 5).
Table 5.
Effects of dexamethasone, chlorogenic acid, or their interaction on the intestinal permeability of broilers.
| Item | d-LA, μg/L | DAO, pg/mL |
|---|---|---|
| Control | 769.86b | 95.72 |
| DEX | 1,294.86a | 101.37 |
| CGA | 815.69b | 97.21 |
| DEX + CGA | 793.99b | 91.66 |
| SEM | 60.65 | 4.26 |
| Main effect | ||
| CGA | ||
| − | 1,032.36 | 98.54 |
| + | 804.84 | 94.43 |
| DEX | ||
| − | 792.77 | 96.46 |
| + | 1,044.42 | 96.52 |
| P-value | ||
| CGA | <0.01 | 0.19 |
| DEX | <0.01 | 0.99 |
| Interaction | <0.01 | 0.08 |
DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid; d-LA = d-lactate; DAO = diamine oxidase.
a,b Within a column, means without a common superscriptdiffer significantly (P < 0.05). n = 6 for each group.
Furthermore, the expression of tight junction proteins was examined using western blotting and immunohistochemical analysis. The results showed that DEX treatment downregulated (P < 0.01) occludin expression, whereas CGA supplementation upregulated (P < 0.01) occludin expression. Additionally, CGA × DEX reversed the DEX-induced decreases in occludin (P = 0.01) and ZO-1 (P = 0.04) expression (Fig. 2B), which was confirmed by immunohistochemical analysis (Fig. 2C and D).
3.4. The gut microbiota and SCFA were altered by CGA
High-throughput 16S rRNA sequencing was performed to determine the effect of DEX treatment and CGA supplementation on the gut microbiome of the broilers. The α-diversity indices, including chao1, goods_coverage, observed_otus, Shannon, and Simpson, were not significantly influenced (P > 0.05) by CGA or DEX treatments. However, DEX + CGA treatment increased chao1 (P = 0.02) and observed_otus (P = 0.01) indices compared with the DEX group (Fig. 3A). PCoA showed that different treatments induced distinct (P = 0.02) clustering of bacterial communities (Fig. 3B), with different gut microbiota compositions at the phylum, family, genus, and species levels. At the phylum level, CGA supplementation decreased (P = 0.04) the abundance of Actinobacteria. At the family level (top 20), CGA supplementation decreased the abundance of Firmicutes_unclassified (P = 0.04), Christensenellaceae (P < 0.01), and Mollicutes_RF39_unclassified (P = 0.02). DEX treatment decreased (P < 0.01) the abundance of Clostridiales vadin BB60_group. Compared with the DEX group, DEX + CGA treatment increased (P = 0.02) the abundance of Clostridiales vadin BB60_group. At the genus level (top 20), DEX treatment decreased (P < 0.01) the abundance of Clostridiales vadin BB60_group_unclassified and increased (P < 0.01) the abundance of Erysipelatoclostridium. Dietary supplementation with CGA increased (P = 0.01) the abundance of Intestinimonas and decreased the abundances of Ruminococcaceae_UCG-014 (P < 0.01), Firmicutes_unclassified (P = 0.02), and Ruminiclostridium_5 (P < 0.01). Additionally, DEX + CGA treatment increased (P < 0.01) the abundance of Clostridiales vadin BB60_group_unclassified compared with the DEX group. At the species level, DEX treatment decreased (P = 0.03) the abundance of Clostridiales vadin BB60_group unclassified. Moreover, CGA supplementation increased (P = 0.01) the abundance of Intestinimonas_unclassified and decreased the abundance of Ruminococcaceae_UCG-014_unclassified (P = 0.01), Firmicutes_unclassified (P = 0.03), and Ruminiclostridium_5_unclassified (P < 0.01). Moreover, compared with the DEX group, DEX + CGA treatment increased (P < 0.01) the abundance of Clostridiales vadin BB60_group_unclassified (Fig. 3C).
Fig. 3.
Effects of DEX, CGA, and DEX + CGA on the gut microbiota of broilers. (A) Changes in the α-diversity of gut microbiota communities, as indicated by chao1, goods_coverage, observed_otus, Shannon’s, and Simpson’s indices. (B) PCoA of gut microbiota. (C) The abundance of gut microbiota at phylum, family, genus, and species levels. (D) LDA score. DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid. n = 6 for each group.
LEfSe analysis was performed to identify taxonomic biomarkers in the gut microbiota. There was an increase in the relative abundance of bacteria, including Coprobacter (genus), Coprobacter_fastidiosus (species), Anaerotruncus_unclassified (species), DTU089 (genus), and DTU089_unclassified (species), in non-treated broilers. Additionally, CGA supplementation increased the relative abundance of Intestinimonas (genus), Intestinimonas_unclassified (species), UC5_1_2E3 (genus), UC5_1_2E3_unclassified (species), and Eubacterium_unclassified (species). DEX treatment increased the relative abundances of Shuttlewothia (genus) and Erysipelatoclostridium_unclassified (species). DEX + CGA treatment increased the relative abundance of Clostridiales vadin BB60_group (family), Clostridiales vadin BB60_group_unclassified (genus and species), Erysipelatoclostridium (genus), Shuttleworthia_unclassified (species), and Lactobacillus_hilgardii (species) (Fig. 3D).
PICRUSt analysis was conducted to determine the potential functional differences of the gut microbiota between the groups and predict their classification based on the KEGG pathways. Compared with the control group, there was a decrease in 4 terms, including “methanogenesis from acetate” (P = 0.03), “starch degradation V” (P = 0.03), and “galactose degradation I (Leloir pathway)” (P = 0.02), and an increase in 26 terms, including “myo-, chiro-, and scyllo-inositol degradation” (P = 0.03), “d-fructuronate degradation” (P = 0.03), and “superpathway of sulfur oxidation (Acidianus ambivalens)” (P = 0.02), in the CGA group. Compared with the control group, there was a decrease in 7 terms, including “l-glutamate degradation V (via hydroxyglutarate)” (P = 0.05), “pyrimidine deoxyribonucleotide biosynthesis from CTP” (P = 0.05), and “GDP-mannose biosynthesis” (P = 0.04), and an increase in 2 terms, i.e., “sucrose degradation IV (sucrose phosphorylase)” (P < 0.01) and “sucrose degradation III (sucrose invertase)” (P = 0.04), in the DEX treatment group. Compared with the DEX group, there was a decrease in “glycerol degradation to butanol” (P = 0.01) and “sucrose degradation IV (sucrose phosphorylase)” (P < 0.01) and an increase in 9 terms, including “superpathway of polyamine biosynthesis II” (P < 0.01), “d-fructuronate degradation” (P = 0.05), and “pyruvate fermentation to butanoate” (P = 0.02), in the DEX + CGA group (Fig. 4).
Fig. 4.
Predicted function of gut microbiota genes in the cecal contents of broilers. KEGG metabolic pathway enrichment analysis based on significant differential bacteria. DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid. n = 6 for each group.
Short-chain fatty acids are the main metabolites generated by gut microbiota. In the present study, DEX treatment had no significant effects on SCFA levels. In contrast, CGA supplementation increased the levels of acetic (P < 0.01), propanoic (P = 0.03), butyric (P < 0.01), isovaleric (P < 0.01), valeric (P < 0.01), and hexanoic acid (P < 0.01). Additionally, CGA × DEX reversed (P < 0.01) DEX-induced decreases in acetic, propanoic, butyric, isovaleric, valeric, and hexanoic acid levels (Table 6).
Table 6.
Dexamethasone and chlorogenic acid effects or interactions on short-chain fatty acid (SCFA) levels of broilers (μmol/g).
| Item | Acetic acid | Propanoic acid | Butyric acid | Isovaleric acid | Valeric acid | Hexanoic acid |
|---|---|---|---|---|---|---|
| Control | 115.56b | 21.38ab | 27.19b | 2.08b | 3.31b | 0.07b |
| DEX | 6.58c | 7.63c | 5.74c | 0.89c | 1.04c | 0.04b |
| CGA | 115.95b | 17.10b | 26.56b | 2.23ab | 2.64b | 0.08b |
| DEX + CGA | 169.00a | 27.38a | 47.45a | 3.23a | 5.40a | 0.13a |
| SEM | 22.20 | 4.54 | 7.75 | 0.5 | 0.72 | 0.02 |
| Main effect | ||||||
| CGA | ||||||
| − | 61.07 | 14.51 | 16.46 | 1.48 | 2.17 | 0.06 |
| + | 142.47 | 22.24 | 37.01 | 2.73 | 4.02 | 0.10 |
| DEX | ||||||
| − | 115.75 | 19.24 | 26.88 | 2.15 | 2.97 | 0.07 |
| + | 87.79 | 17.50 | 26.60 | 2.06 | 3.21 | 0.08 |
| P-value | ||||||
| CGA | <0.01 | 0.03 | <0.01 | <0.01 | <0.01 | <0.01 |
| DEX | 0.09 | 0.59 | 0.96 | 0.79 | 0.64 | 0.60 |
| Interaction | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
DEX = dexamethasone, CGA = chlorogenic acid, DEX + CGA = dexamethasone + chlorogenic acid.
a,b,c Within a row, means without a common superscript differ significantly (P < 0.05). n = 6 for each group.
3.5. CGA altered the jejunal protein profiles
Differentially expressed proteins (DEP) are represented using volcano plots (Fig. 5A). Compared with the control group, 25 DEP were upregulated and 33 were downregulated in the DEX group; 27 DEP were upregulated, and 10 were downregulated in the CGA group. Compared with the DEX group, 61 DEP were upregulated, and 48 were downregulated in the DEX + CGA group. The top 10 up- and downregulated DEP are presented based on fold change (Table 7, Table 8, Table 9).
Fig. 5.
Effects of DEX, CGA, and DEX + CGA on the jejunal proteomics. (A) Volcano map of jejunal proteins. (B) GO analysis of jejunal proteins. (C) KEGG metabolic pathway enrichment analysis based on significant differentially expressed proteins. (D) PPI network. DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid; BP = biological process; CC = cellular components; MF = molecular functions. n = 3 for each group.
Table 7.
Top 10 up- and downregulated DEP between control and DEX groups (fold-change ranked).
| Accession | Protein symbol | Protein name | Fold change | P-value |
|---|---|---|---|---|
| P04354 | CALB1 | Calbindin | 2.80 | 0.04 |
| A0A1D5P6N4 | MEP1A | Meprin A subunit | 1.87 | 0.01 |
| A0A1L1RS59 | MEP1B | Metalloprotease meprin beta gene | 1.82 | 0.05 |
| F1NN74 | ANO5 | Anoctamin 5 | 1.61 | 0.03 |
| R4GFW3 | CLCN2 | Chloride channel protein 2 | 1.58 | 0.01 |
| H9KYX6 | SELENBP1 | Selenium-binding protein 1 | 1.54 | 0.02 |
| R4GGG4 | CYP2U1 | Cytochrome P450 CYP2 subfamily U member 1 | 1.53 | 0.04 |
| F1NB67 | PLEKHO2 | Pleckstrin homology domain containing, family O member 2 | 1.50 | 0.01 |
| P11183 | GCSH | Glycine cleavage system H protein (lipoate-binding) | 1.43 | 0.04 |
| P21642 | PCK2 | Phosphoenolpyruvate carboxykinase 2 | 1.41 | 0.02 |
| A0A3Q2U8K0 | N/A | Uncharacterized protein | 0.57 | 0.01 |
| R4GF71 | TMSB4X | Thymosin beta | 0.56 | 0.01 |
| A0A1D5PXP9 | TGFBI | Transforming growth factor-beta-induced protein ig-h3 | 0.56 | 0.05 |
| P11602 | LPL | Lipoprotein lipase | 0.55 | <0.01 |
| F1NBD7 | CDK1 | Cyclin-dependent kinase 1 | 0.51 | 0.01 |
| P33145 | K-CAM | B-cadherin | 0.49 | <0.01 |
| A0A1D5PDE6 | MARCKS | Myristoylated alanine-rich C-kinase substrate | 0.49 | 0.05 |
| A0A3Q3AU25 | N/A | TED_complement domain-containing protein | 0.45 | 0.03 |
| A0A1D5NTE7 | N/A | Fibrinogen C-terminal domain-containing protein | 0.37 | 0.04 |
| F1P1P5 | FXN | Frataxin, mitochondrial | 0.37 | 0.03 |
DEX = dexamethasone; N/A = not applicable; DEP = differentially expressed proteins.
n = 3 for each group.
Table 8.
The top 10 up- and downregulated DEP between control and CGA groups (fold-change ranked).
| Accession | Protein symbol | Protein name | Fold change | P-value |
|---|---|---|---|---|
| F1NPA3 | ARID4A | ARID domain-containing protein | 2.26 | <0.01 |
| A0A3Q2TTI3 | FBRSL1 | Uncharacterized protein | 1.97 | 0.04 |
| A0A1D5PTI4 | ARID1A | ARID domain-containing protein | 1.94 | 0.04 |
| F1NI13 | SYAP1 | Synapse-associated protein 1 | 1.92 | 0.04 |
| A0A1D5PQJ7 | CYP1C1 | Cytochrome P450 CYP1 subfamily | 1.79 | 0.04 |
| A0A1D5PYB7 | LIMD1 | LIM domain-containing protein 1 | 1.73 | 0.02 |
| R4GF71 | TMSB4X | Thymosin beta | 1.64 | 0.04 |
| E1C2V9 | ARFGAP2 | Arf-GAP domain-containing protein | 1.61 | 0.01 |
| F1ND55 | ADD1 | Aldolase_II domain-containing protein | 1.61 | 0.01 |
| E1C667 | LAD1 | Uncharacterized protein | 1.59 | 0.01 |
| A0A3Q2UD05 | N/A | Aldolase_II domain-containing protein | 0.81 | 0.02 |
| F1NT33 | IKZF1 | DNA-binding protein Ikaros | 0.81 | 0.05 |
| F1P0B2 | APP | Amyloid-beta A4 protein | 0.78 | 0.05 |
| F1NBT0 | STK10 | Serine/threonine-protein kinase 10 | 0.76 | 0.03 |
| E1C1X1 | TMEM126A | Uncharacterized protein | 0.72 | 0.02 |
| E1BXC7 | MALL | MARVEL domain-containing protein | 0.71 | 0.02 |
| F1NPL9 | COX17 | Cytochrome c oxidase copper chaperone protein | 0.54 | 0.01 |
| A0A1D5P7X3 | CAPG | Macrophage-capping protein | 0.54 | 0.02 |
| A5HUL4 | BLB2 | MHC class II beta chain 2 | 0.50 | <0.01 |
| Q95601 | BFIV21 | MHC class II beta chain 2 | 0.47 | <0.01 |
CGA = chlorogenic acid; N/A = not applicable; DEP = differentially expressed proteins.
n = 3 for each group.
Table 9.
The top 10 up- and downregulated DEP between DEX and DEX + CGA groups (fold-change ranked).
| Accession | Protein symbol | Protein name | Fold change | P-value |
|---|---|---|---|---|
| R4GF71 | TMSB4X | Thymosin beta | 2.79 | <0.01 |
| A0A3Q2U3Y3 | CNN1 | Calponin | 2.65 | 0.04 |
| A0A3Q2U295 | C19orf43 | Uncharacterized protein | 2.53 | 0.05 |
| F1NVA3 | FHOD1 | Formin homology 2 domain containing 1 | 2.40 | 0.03 |
| A0A1D5PTI4 | ARID1A | ARID domain-containing protein | 2.28 | 0.05 |
| F1NPA3 | ARID4A | ARID domain-containing protein | 2.19 | <0.01 |
| A0A3Q2U530 | MAP7D3 | Uncharacterized protein | 2.18 | 0.04 |
| A0A3Q2UF99 | NASP | Cell cycle-regulated SHNi- Nuclear autoantigenic sperm protein | 2.05 | 0.02 |
| A0A1D5PYB7 | LIMD1 | LIM domain-containing protein 1 | 2.05 | 0.02 |
| A0A1D5PDE6 | MARCKS | Myristoylated alanine-rich C-kinase substrate | 2.02 | 0.05 |
| F1NHR4 | ACE2 | Angiotensin-converting enzyme | 0.64 | 0.03 |
| F1NYM0 | ACE | Angiotensin-converting enzyme | 0.60 | 0.04 |
| F1NPI1 | SLC6A19 | Neutral amino acid transporter B0AT1 | 0.60 | 0.02 |
| A0A1D5PHR1 | ABCD2 | Uncharacterized protein | 0.58 | 0.05 |
| F1NN74 | ANO5 | Anoctamin 5 | 0.56 | 0.02 |
| F1NAN4 | LCT | Uncharacterized protein | 0.56 | 0.03 |
| F1NY83 | LOC101747844 | Carbohydrate sulfotransferase | 0.55 | 0.02 |
| E1C958 | LGMN | Asparaginyl endopeptidase | 0.53 | <0.01 |
| A0A1L1RS59 | MEP1B | Metalloprotease meprin beta gene | 0.52 | 0.02 |
| A0A1D5P6N4 | MEP1A | Meprin A metalloprotease | 0.49 | 0.03 |
DEX = dexamethasone; CGA = chlorogenic acid; N/A = not applicable; DEP = differentially expressed proteins.
n = 3 for each group.
GO enrichment analysis showed that DEP between the DEX and control groups were enriched in biological process (BP) terms such as “oxidation-reduction process” (P < 0.01), “chemical homeostasis” (P < 0.01), and “drug metabolic process” (P < 0.01); cellular component (CC) terms such as “extracellular region” (P = 0.01), “cytoskeleton” (P = 0.01), “extracellular space” (P < 0.01); and molecular function (MF) terms such as “transition metal ion binding” (P < 0.01), “oxidoreductase activity” (P = 0.02), and “protein dimerization activity” (P < 0.01). DEP between the CGA and control groups were enriched in BP terms such as “cytoskeleton organization” (P < 0.01) and “cellular protein-containing complex assembly” (P = 0.01); CC terms such as “cytoskeleton” (P = 0.04) and “plasma membrane part” (P = 0.04); and MF terms such as “cytoskeletal protein binding” (P = 0.02) and “DNA binding” (P = 0.04). DEP between the DEX + CGA and DEX groups were enriched in BP terms such as “carbohydrate metabolic process” (P < 0.01), “myeloid cell differentiation” (P < 0.01), and “organic anion transport” (P = 0.02); CC terms such as “plasma membrane part” (P < 0.01), “cytoskeletal part” (P = 0.03), and “plasma membrane region” (P < 0.01); and MF terms such as “cytoskeletal protein binding” (P = 0.03), “protein dimerization activity” (P = 0.04), and “protein homodimerization activity” (P = 0.02) (Fig. 5B).
KEGG metabolic pathway enrichment analysis showed that DEP between the DEX and control groups were enriched in “protein digestion and absorption” (P < 0.01), “PPAR signalling pathway” (P = 0.01), and “proximal tubule bicarbonate reclamation” (P < 0.01). DEP between the CGA and control groups were enriched in “endocytosis” (P = 0.01), “viral myocarditis” (P = 0.03), and “type I diabetes mellitus” (P < 0.01). Additionally, DEP between the DEX + CGA and DEX groups were enriched in “protein digestion and absorption” (P < 0.01), “RNA transport” (P < 0.01), and “PPAR signalling pathway” (P < 0.01) (Fig. 5C). MRM analysis was performed to validate the presence and levels of relevant proteins identified by proteomics. The MRM results verified that eukaryotic translation initiation factor 3 subunit J (EIF3J, accession number: Q5ZKA4) was downregulated (P = 0.04), whereas pyridoxal phosphate homeostasis protein (PROSC, accession number: E1C516) (P = 0.03) and apolipoprotein A-I (APOA1, accession number: P08250) (P = 0.03) were upregulatedby DEX treatment. Additionally, DEX + CGA treatment downregulated (P < 0.01) APOA1 and calcineurin B homologous protein 1 (CHP1, accession number: Q5ZM44) (Fig. S1). According to the KEGG results of proteomic analysis, EIF3J is involved in the MAPK signalling pathway, PROSC is involved in butanoate metabolism, APOA1 is involved in the PPAR signalling pathway, and CHP1 is involved in the apoptosis signalling pathway.
Furthermore, a protein–protein interaction (PPI) network was generated using the STRING database (Fig. 5D). The network diagram illustrates the interactions between the DEP in the screened pathways. Among the PPIs, cyclin-dependent kinase 1 (CDK1, accession number: F1NBD7) was the core PPI node in the DEX vs control groups, with 8 interactions. DNA-binding protein Ikaros (IKZF1, accession number: FINT33) was the core PPI node in the CGA vs control groups, with 4 interactions. Moreover, CDK1 was the core PPI node in the DEX + CGA vs DEX groups, with 19 interactions.
3.6. CGA altered the serum metabolic profiles of the broilers
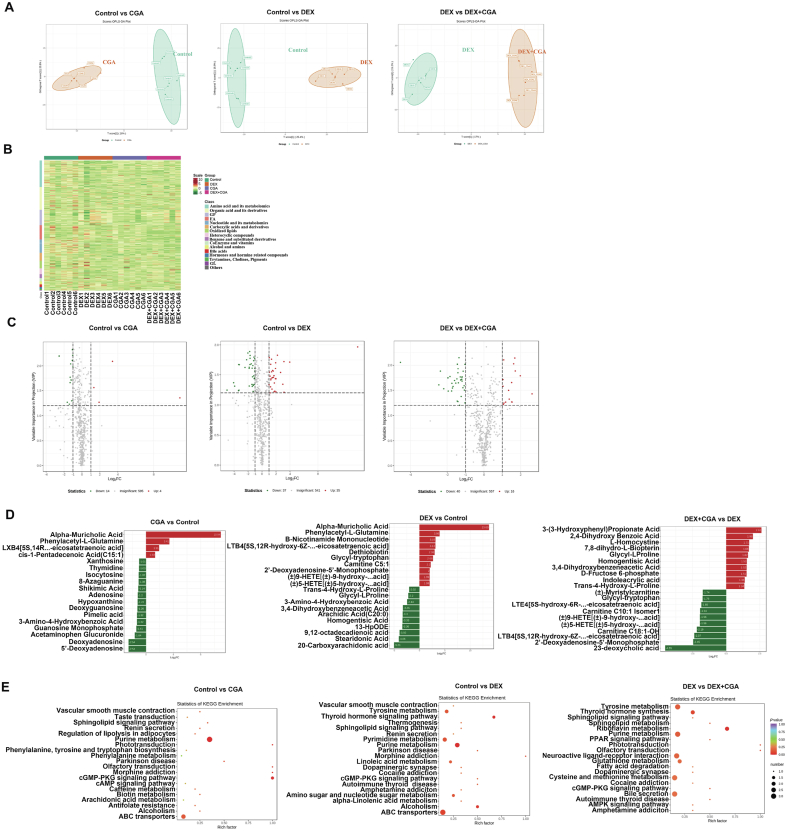
Broad-spectrum metabolomics was used to evaluate the serum profiles of the broilers. We observed a clear separation from the OPLS-DA score plots between the control vs CGA groups, control vs DEX groups, and DEX vs DEX + CGA groups (Fig. 6A). Differentially expressed metabolites between the groups were screened at a fold change ≥2 or ≤0.5, which was illustrated using a heatmap (Fig. 6B). Compared with the control group, CGA supplementation significantly increased the levels of 14 metabolites and decreased the levels of 4 metabolites, whereas DEX treatment significantly increased the levels of 37 metabolites and decreased the levels of 35 metabolites. Moreover, DEX + CGA treatment significantly increased the levels of 40 metabolites and decreased the levels of 16 metabolites compared with the DEX group (Fig. 6C). The top 20 metabolites with multiple differences between the groups are displayed in Fig. 6D. Compared to the control group, CGA supplementation increased the levels of α-muricholic acid, phenylacetyl-l-glutamine, and cis-pentadecenoic acid and decreased the levels of 5′-deoxyadenosine, deoxyadenosine, and acetaminophen glucuronide. Additionally, DEX treatment increased the levels of α-muricholic acid, phenylacetyl-l-glutamine, and B-nicotinamide mononucleotide and decreased the levels of 20-carboxyarachidonic acid, stearidonic acid, and 9,12-octadecadienoic acid compared with the control group. Moreover, DEX + CGA treatment increased the levels of 3-(3-hydroxyphenyl) propionate acid, 2,4-dihydroxy benzoic acid, and homogentisic acid, and decreased the levels of 23-deoxycholic acid, 2′-deoxyadenosine-5′-monophosphate, and carnitine C18:1-OH, compared with the DEX group. KEGG analysis showed that the differentially expressed metabolites in the control vs CGA groups were enriched in “purine metabolism” (P < 0.01), “ABC transporters” (P = 0.04), and the “cyclic guanosine monophosphate-protein kinase G signalling pathway” (P = 0.03). Differentially expressed metabolites in the control vs DEX groups were enriched in “tyrosine metabolism” (P = 0.03), “thyroid hormone signaling pathway” (P = 0.04), and “alpha-linolenic acid metabolism” (P = 0.03). Additionally, differentially expressed metabolites in the DEX vs DEX + CGA groups were enriched in “riboflavin metabolism” (P = 0.02), “tyrosine metabolism” (P = 0.04), “purine metabolism” (P = 0.03), “glutathione metabolism” (P = 0.02) and the “PPAR signalling pathway” (P = 0.01) (Fig. 6E).
Fig. 6.
Effects of DEX, CGA, and DEX + CGA on the metabolites of serum. (A) OPLS-DA analysis. (B) Heatmap analysis. (C) Volcano analysis. (D) The top 20 metabolites with multiple differences between groups. (E) KEGG metabolic pathway enrichment analysis based on significant differential metabolites. DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid. n = 3 for each group.
3.7. Effects of CGA on the PPAR and MAPK signalling pathways
Proteomic and metabolomic analyses revealed that CGA plays an important role in the PPAR signalling pathway. Additionally, MRM analysis showed that CGA participates in regulating the MAPK signalling pathway. Thus, western blotting was used to examine the effect of CGA on the activation of PPAR and MAPK signalling pathways. The results showed that DEX decreased p-JNK (P < 0.01), P-38 (P = 0.03), p-P38 (P < 0.01), and ERK (P < 0.01) expression. In contrast, CGA treatment increased JNK (P < 0.01), p-JNK (P < 0.01), P-38 (P = 0.01), and p-P38 (P < 0.01) expression. Additionally, CGA supplementation (CGA × DEX) reversed DEX-induced decreases in JNK (P = 0.02), p-JNK (P < 0.01), P38 (P = 0.02), and p-P38 (P < 0.01) (Fig. 7A). Regarding the PPAR signalling pathway, DEX treatment significantly downregulated (P < 0.01) PPAR expression, whereas CGA supplementation upregulated (P < 0.01) PPAR expression. Additionally, DEX × CGA interaction increased (P < 0.01) PPAR expression (Fig. 7B).
Fig. 7.
Effects of DEX, CGA or their interaction on the (A) MAPK and (B) PPAR signaling pathway in the jejunum. DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid. n = 6 for each group. Bars with different letters differ at P < 0.05.
3.8. Crosstalk between gut microbiota, SCFA, and biochemical parameters
Spearman’s correlation analysis was performed to identify the relationships between biochemical parameters and differential gut bacteria, proteins, and metabolites (Fig. 8). A total of 4 bacterial genera were common between the control vs DEX and DEX vs DEX + CGA groups (Fig. 8A). Based on this, parameters with a correlation coefficient (r) > 0.7 or < −0.7 and P < 0.01 were selected. Among the 4 genera, Mordavella was positively correlated (P < 0.01) with villus height (r = 0.866), and negatively correlated with jejunal CXCL1 level (r = −0.714) and serum IL-6 level (r = −0.710). Coprobacter was negatively correlated (P < 0.01) with jejunal IL-18 (r = −0.866) and IL-12 levels (r = −0.700) and positively correlated (P < 0.01) with serum IL-4 level (r = 0.797) and IL-10 transcription (r = 0.708). Clostridiales vadin BB60_group_unclassified was negatively correlated (P < 0.01) with serum IL-18 (r = −0.740), jejunal IL-18 (r = −0.740), serum CXCL2 (r = −0.707), jejunal CXCL2 (r = −0.733), serum CXCL1 (r = −0.730), and jejunal IL-12 levels (r = −0.712). Additionally, 7 bacterial genera were common between the control vs CGA and CGA vs DEX + CGA groups; however, there was no significant correlation between the different genera and the biochemical parameters under the screening condition (r < −0.7 or r > 0.7). Regarding the correlation between biochemical parameters and SCFA, results with r > 0.8 or < −0.8 and P < 0.01 were selected. A total of 4 SCFA were correlated with biochemical parameters in the control vs DEX and DEX vs DEX + CGA comparison groups, among which acetic acid was negatively correlated (P < 0.01) with d-LA level (r = −0.827), jejunal IL-6 level (r = −0.886), IL-18 transcription (r = −0.820), and serum CXCL1 (r = −0.802). Butyric acid was negatively correlated (P < 0.01) with jejunal IL-6 levels (r = −0.853), jejunal IL-22 levels (r = −0.813), and serum IL-18 levels (r = −0.808). Additionally, valeric acid was negatively correlated (P < 0.01) with jejunal IL-12 levels (r = −0.823), whereas isovaleric acid was negatively correlated (P < 0.01) with serum IL-1β levels (r = −0.819) and IL-1β transcription (r = −0.805). Regarding the control vs CGA and CGA vs DEX + CGA comparison group, there were no significant correlations between the parameters under the screening conditions (r < −0.8 or > 0.8) (Fig. 8B).
Fig. 8.
Spearman’s correlation analysis between biochemical parameters and omics parameters. (A) Spearman’s correlation analysis between biochemical parameters and gut bacteria at the genus level. (B) Spearman’s correlation analysis between biochemical parameters and SCFA. (C) Spearman’s correlation analysis between biochemical parameters and jejunal proteins. (D) Spearman’s correlation analysis between biochemical parameters and serum metabolites. DEX = dexamethasone; CGA = chlorogenic acid; DEX + CGA = dexamethasone + chlorogenic acid; G-IL = gene expression of interleukin; G-TNF-α = gene expression of tumor necrosis factor α; G-Caspase = gene expression of cysteinyl aspartate specific proteinase; J-IL = interleukin level in jejunal mucosa; J-TNF-α = tumor necrosis factor α level in jejunal mucosa; J-CXCL = CXC chemokine ligand level in jejunal mucosa; S-IL = interleukin level in serum; S-CXCL = CXC chemokine ligand level in serum; S-IFN-γ = interferon γ level in serum.
3.9. Crosstalk between proteomic and biochemical parameters
A total of 15 proteins were common between the control vs DEX and DEX vs DEX + CGA comparison groups. Based on this, parameters with r > 0.85 or < −0.85 and P < 0.01 were selected. Regarding the control vs DEX and DEX vs DEX + CGA comparison groups, legumain (LGMN, accession number: E1C958) was negatively correlated (P < 0.01) with d-LA (r = −0.867), and serum IL-12 (r = −0.983), CXCL2 (r = −0.917) and CXCL1 levels (r = −0.900). Meprin A subunit (MEP1A, accession number: A0A1D5P6N4) was positively correlated (P < 0.01) with IL-10 transcription (r = 0.983) and serum IL-10 levels (r = 0.867), and CDK1 was positively correlated (P < 0.01) with caspase-9 transcription (r = 0.933) and negatively correlated (P < 0.01) with serum IL-10 levels (r = −0.900). Nuclear autoantigenic sperm protein (NASP, accession number: A0A3Q2UF99) was positively correlated (P < 0.01) with caspase-9 transcription (r = 0.933) and negatively correlated with serum IL-10 levels (r = −0.867). Additionally, metalloprotease meprin beta gene (MEP1B, accession number: A0A1L1RS59) was positively correlated (P < 0.01) with serum IL-10 levels (r = 0.933), and anoctamin 5 (ANO5, accession number: F1NN74) was positively correlated (P < 0.01) with IL-10 transcription (r = 0.933). Thymosin beta (TMSB4X, accession number: R4GF71) was positively correlated (P < 0.01) with caspase-9 transcription (r = 0.933), serum CXCL1 levels (r = 0.917), and serum IL-18 levels (r = 0.867). Nucleolar complex protein 2 homolog (NOC2L, accession number: F1NV71) was positively correlated (P < 0.01) with serum IL-22 levels (r = 0.917), reelin adaptor protein (Dab1, accession number: Q6XBN7) was positively correlated (P < 0.01) with IL-10 transcription (r = 0.900), and myristoylated alanine-rich C-kinase substrate (MARCKS, accession number: A0A1D5PDE6) was negatively correlated (P < 0.01) with IL-10 transcription (r = −0.867). Additionally, mitochondrial genome maintenance exonuclease 1 (MGME1, accession number: A0A1L1RXX7) was positively correlated (P < 0.01) with IL-18 transcription (r = 0.867), PROSC was positively correlated (P < 0.01) with serum IL-10 levels (r = 0.867). Furthermore, 4 proteins were common between the control vs CGA and CGA vs DEX + CGA comparison groups, among which dynein regulatory complex subunit 4 (GAS8, accession number: F1NLA8) and Apolipoprotein C-III (APOC3, accession number: A0A1D5PK48) were negatively correlated (P < 0.01) with serum IL-18 levels (r = −0.933 and r = −0.900, respectively) (Fig. 8C).
3.10. Crosstalk between metabolomic and biochemical parameters
A total of 25 metabolites were common between the control vs DEX and DEX vs DEX + CGA groups, and parameters with r > 0.8 or < −0.8 and P < 0.01 were selected. Among the 25 metabolites, α-muricholic acid was negatively (P < 0.01) correlated with V:C ratio (r = −0.866), villus height (r = −0.815), and jejunal IL-4 level (r = −0.901), and positively correlated with jejunal CXCL1 levels (r = 0.843) and serum IL-6 level (r = 0.840). Additionally, 7,8-dihydro-L-biopterin was positively correlated (P < 0.01) with villus height (r = 0.829), whereas Asp–Phe was negatively correlated (P < 0.01) with villus height (r = −0.814) and positively correlated with IL-1β transcription (r = 0.874) and jejunal IL-12 (r = 0.862) and CXCL2 levels (r = 0.805). Glycyl-l-proline was negatively correlated (P < 0.01) with jejunal CXCL2 levels (r = −0.807), whereas 2,4-hexadienoic acid was negatively correlated (P < 0.01) with IL-4 transcription (r = −0.860). Moreover, (±)5-HETE (r = 0.846), (±)9-HETE (r = 0.846), and LTE4 (r = 0.836) levels were positively correlated (P < 0.01) with jejunal CXCL2 levels. Additionally, uracil was negatively correlated (P < 0.01) with IL-4 transcription (r = −0.840). Furthermore, only 1 metabolite was common between the control vs CGA and CGA vs DEX + CGA groups; however, there was no significant correlation between the metabolite and the biochemical parameters (Fig. 8D).
4. Discussion
In the present study, we evaluated the effect of CGA supplementation on the gut microbial composition, intestinal protein profiles, serum metabolites, intestinal barrier function, immune function and growth performance of broilers with DEX-induced immunological stress. The findings showed that CGA supplementation effectively improved the growth performance and reversed DEX-induced inflammation and jejunal permeability. Additionally, CGA × DEX improved jejunal morphology and expression of tight junction proteins in DEX-treated broilers, which was similar to the findings of previous studies on the growth-promoting effects and anti-inflammatory activities of CGA in chickens and pigs (Chen et al., 2018a,b; Xu et al., 2020; Zhang et al., 2020).
Although the gut microbial diversity of the broilers was not significantly altered by DEX treatment, CGA supplementation significantly increased the chao1 and observed_otus indices of broilers in the DEX + CGA group. Moreover, the gut microbiota of broilers in the DEX + CGA group was significantly affected at the family (Clostridiales vadin BB60_group), genus (Clostridiales vadin BB60_group_unclassified genus), and species (Clostridiales vadin BB60_group_unclassified) levels compared with those of broilers in the DEX group. Although a study indicated that the abundance of Clostridiales vadin BB60 was enriched in mice with enteritis (Liu et al., 2020), recent research has shown that Clostridium butyricum MIYAIRI 588 supplementations increased the abundance of Clostridiales vadin BB60 in mice under stress (Tian et al., 2019). Additionally, Kang et al. (2019) found that Clostridiales vadin BB60 was positively correlated with the expression of intestinal ZO-1 and negatively correlated with serum inflammatory parameters, such as TNF-α, IL-6, and LPS. Similarly, the results of the present study showed that Clostridiales vadin BB60_group_unclassified was negatively correlated with inflammatory parameters, including serum IL-18, CXCL1, and CXCL2 levels, and jejunal IL-12, IL-18, and CXCL2 levels.
Studies have shown that gut bacteria ferment non-digestible carbohydrates to produce SCFA, conferring several health benefits (Gibson et al., 2017; Ojo et al., 2021). Clostridiales vadin BB60_group are potential SCFA-producing bacteria (Cheng et al., 2021). SCFA affect gut epithelial integrity, which may regulate exposure of the mucosal immune system to bacteria or innate signals that affect immune tolerance (Macia et al., 2012). In the present study, CGA and CGA + DEX treatments significantly increased the levels of SCFA, such as acetic, propanoic, and butyric acids. These data are in accordance with the KEGG results of gut microbiota, which showed an upregulation in the pyruvate fermentation pathway, an SCFA-related pathway (Liang et al., 2020), in the DEX + CGA group compared with the DEX group. Additionally, the SCFA were negatively correlated with d-LA and pro-inflammatory cytokines. These findings indicated that CGA supplementation improved inflammatory responses and enhanced gut barrier function.
Furthermore, CGA intake may also play a role in the host proteome (Lin et al., 2017). The results obtained in the present study showed altered protein profiles in broilers treated with CGA and/or DEX. In particular, SELENBP1 and CLCN2 were decreased by DEX. SELENBP1 is a member of the selenium-binding protein family, which has been shown to bind covalently to selenium (Porat et al., 2000). The role of SELENBP1 in the intestine is to modulate the differentiation and function of immune cells, contributing to a reduction in excessive immune response (Speckmann and Steinbrenner, 2014). Moreover, CLCN2 can enhance the intestinal epithelial tight junction barrier function (Nighot et al., 2017). The expression of COX17 in the CGA group was increased compared to the control. It has been reported that in the gastrointestinal tract of weaned piglets, LPS significantly decreased the expression of COX17, while epidermal growth factor treatment significantly increased the expression of COX17 (Xue et al., 2020). In addition, compared with the DEX group, DEX + CGA significantly decreased the expression of FHOD1, which is upregulated in epithelial–mesenchymal transition, and participates in cancer cell migration and invasion (Gardberg et al., 2013). It is also worth noting that DEX increased the expression of the TMSB4X protein and decreased LGMN expression. However, DEX + CGA significantly reversed the above trends. TMSB4X is a naturally occurring peptide (Vasilopoulou et al., 2016) that exhibits several functions. Although exogenous TMSB4X has been shown to have beneficial effects on diverse pathologies, including myocardial infarction (Smart et al., 2011), stroke (Morris et al., 2014), and inflammatory lung disease (Conte et al., 2013), a recent study has revealed that the ethyl acetate extract of Cremastra appendiculata inhibits the growth of breast cancer tissues and reduces the expression of the TMSB4X gene in breast cancer cells in a tumour transplanted mouse model (Cao et al., 2021). Additionally, to participate in immune response, LGMN can process self-antigen peptides and foreign proteins, deliver them to T cells in the form of MHC II molecular complexes, and trigger the activation of toll-like receptors (TLRs) or other cathepsins via hydrolysis (Dall and Brandstetter, 2016), indicating the important role of LGMN in the immune system. Thus, decreased expression of TMSB4X and increased expression of LGMN after CGA supplementation confirmed the positive immunoregulatory activity of CGA in DEX-challenged broilers.
MRM analysis further confirmed that DEX treatment causes increased APOA1 expression. However, CGA supplementation caused a decrease in APOA1 expression. KEGG pathways analysis demonstrated that the DEP, including APOA1, were enriched in the PPAR signalling pathway. PPARs are involved in energy homeostasis. Moreover, a study showed that PPARs are expressed in immune cells and play an emerging critical role in immune cell differentiation and fate commitment (Christofides et al., 2021). Similar to our results, Ma et al. (2015) also reported that CGA caused a decrease in PPAR mRNA expression. Furthermore, MRM analysis also showed that DEX treatment caused increased PROSC expression, and CGA supplementation caused a decrease in CHP1 expression. According to the KEGG result, PROSC and CHP1 were enriched in the butanoate metabolism and the apoptosis signalling pathways. These findings confirmed that CGA plays positive roles in SCFA metabolism, PPAR signalling pathway, and apoptosis. Furthermore, EIF3J was downregulated in the DEX group. KEGG analysis indicated that EIF3J was involved in the MAPK signalling pathway. The MAPK signalling pathway is involved in the regulation of immune function. A previous study showed that Se-enriched Grifola frondosa polysaccharides improve immune function by activating the MAPK signalling pathway (Li et al., 2018). Additionally, sulfated modification enhanced the immunomodulatory effect of Cyclocarya paliurus polysaccharides in immunosuppressed mice through the MyD88-dependent MAPK signalling pathway (Yu et al., 2021). In the present study, western blotting confirmed that CGA supplementation reversed DEX-induced inactivation of the MAPK signalling pathway, confirming the immunoregulatory activity of CGA.
Normally, proteins interact with each other to perform various biological functions. Therefore, a PPI network was generated to visualize the interactions between the DEP identified in this study. CDK1 was the core PPI node in the DEX vs control groups in this network, with 8 interactions. IKZF1 was the core PPI node in the CGA vs control groups, with 4 interactions, and CDK1 was the core PPI node in the DEX + CGA vs DEX groups, with 19 interactions. The considerable overlap among the pathways indicated that a particular protein could exist in diverse signalling pathways and that various proteins could regulate a particular pathway. Moreover, Li et al., 2015a, Li et al., 2015b reported a significant increase in CDK1 expression in DF-1 cells after infection with subgroup J avian leukosis virus, indicating the immunoregulatory role of CDK1. Correlation analysis demonstrated that several upregulated DEP in the DEX + CGA group, such as ANO5, were positively correlated with the anti-inflammatory cytokine IL-10 level. In contrast, several downregulated DEP in the DEX + CGA group, such as TMSB4X, were positively correlated with the levels of pro-inflammatory cytokines and chemokines, such as CXCL1, IL-18, and IL-22. Although the functions of these proteins have rarely been reported, a study showed a decrease in ANO5 expression in DF-1 cells infected with subgroup J avian leukosis virus (Li et al., 2015a, Li et al., 2015b). Similarly, as mentioned above, a decrease in TMSB4X expression may be beneficial for animal health. The proteomic analysis indicated that CGA supplementation played important roles in regulating intestinal health or inflammation-related proteins and the PPAR, MAPK, butanoate metabolism and apoptosis signalling pathways.
Metabolomic analyses corroborated several key findings from microbiome and proteome analyses, providing valuable insights into the immunoregulatory effects of CGA. OPLS-DA analysis clustered the metabolites according to the treatments. Compared to the DEX group, there was an increase in the levels of 2,4-dihydroxy benzoic acid (a derivative of hydroxybenzoic acid), homogentisic acid, 7,8-dihydro-l-biopterin and a decrease in the levels of 23-deoxycholic acid, 2′-deoxyadenosine-5′-monophosphate, and carnitine C18:1-OH in the DEX + CGA treatment group. Reports have shown that hydroxybenzoic acids and their derivatives possess antioxidant properties (Hubková et al., 2014). Moreover, homogentisic acid exhibits antioxidant and antiradical activities (Rosa et al., 2011). Therefore, the increased concentrations of 2,4-dihydroxy benzoic acid and homogentisic acid may imply an increase in the antioxidant capacity of broilers in the DEX + CGA group. KEGG analysis demonstrated that the different metabolites between the DEX + CGA and DEX groups were enriched in glutathione metabolism and the PPAR signalling pathway. Glutathione possesses antioxidant capacity (Gaucher et al., 2018), indicating that CGA can exert antioxidant activity by promoting glutathione metabolism (Miao and Xiang, 2020) and reverse acetaminophen-induced decrease in liver glutathione levels and glutamate-cysteine ligase and glutathione reductase activities (Ji et al., 2013). Moreover, PPAR signalling pathway enrichment was downregulated in the DEX + CGA group compared with the DEX group, which was in accordance with the results of the KEGG analysis of the DEP. Western blotting further confirmed the effects of CGA on PPAR expression. Additionally, there were significant correlations between the altered metabolites and biochemical parameters. For instance, 7,8-dihydro-l-biopterin, which was increased by CGA treatment, was positively correlated with villus height. In contrast, glycyl-l-proline was negatively correlated with jejunal CXCL2 levels. This indicates that CGA regulated intestinal health and immune function of broilers through serum metabolites.
5. Conclusions
In conclusion, the multi-omics analysis showed that for the supplementation of CGA to immunologically stressed broilers, the gut microbes (Clostridiales vadin BB60), jejunal proteins (TMSB4X, LGMN, APOA1, PROSC, CHP1 and EIF3J), and serum metabolites, such as 2,4-dihydroxy benzoic acid and homogentisic acid, were the primary targets. Correlation analysis between biochemical and omics parameters indicated that CGA exerted beneficial effects by regulating gut microbiota, jejunal protein, and serum metabolites. Moreover, the increase in SCFA levels after CGA treatment verified the increased abundance of the SCFA-producing bacteria Clostridiales vadin BB60 in CGA-treated broilers. Proteomic and metabolomic analyses and western blotting corroborated the predicted PPAR and MAPK signalling pathway changes. However, further studies are necessary to identify additional mechanisms of CGA, including specific protein and metabolite targets.
Author contributions
Huawei Liu: Conceptualization, Funding acquisition. Xuemin Li: Investigation, Visualization. Kai Zhang: Methodology. Xiaoguo Lv: Investigation. Quanwei Zhang: Investigation. Peng Chen: Investigation. Yang Wang: Conceptualization, Writing-Original Draft. Jinshan Zhao: Conceptualization, Writing-Original Draft.
Availability of data and materials
Microbiomic and proteomic sequencing data have been deposited under BioProject PRJNA789475 and IPX0003857000, respectively.
Declaration of competing interest
We declare that we have no financial or personal relationships with other people or organizations that might inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be constructed as influencing the content of this paper.
Acknowledgements
This study was supported by the Qingdao Science and Technology Program (22-3-7-xdny-11-nsh) and Shandong Provincial Natural Science Foundation (ZR2021MC118).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2023.05.009.
Contributor Information
Yang Wang, Email: yangwang@qau.edu.cn.
Jinshan Zhao, Email: jszhaoqau@163.com.
Appendix. Supplementary data
The following is the Supplementary data to this article:
Multimedia component 1
References
- Broom L.J., Kogut M.H. The role of the gut microbiome in shaping the immune system of chickens. Vet Immunol Immunopathol. 2018;204:44–51. doi: 10.1016/j.vetimm.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Cao X.D., Nan Z., Yang D.D., Liu Y., Wang H., Zhang Z.Y. New revelation of TMT-based quantitative proteomic analysis on anti-4T1 breast cancer of ethyl acetate extract of Cremastra appendiculata. Nat Prod Res Dev. 2021;33:246. [Google Scholar]
- Chávez-Carbajal A., Nirmalkar K., Pérez-Lizaur A., Hernández-Quiroz F., Ramírez-del-Alto S., García-Mena J., et al. Gut microbiota and predicted metabolic pathways in a sample of Mexican women affected by obesity and obesity plus metabolic syndrome. Int J Mol Sci. 2019;20:438. doi: 10.3390/ijms20020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xie H., Chen D., Yu B., Mao X., Zheng P., et al. Chlorogenic acid improves intestinal development via suppressing mucosa inflammation and cell apoptosis in weaned pigs. ACS Omega. 2018;3:2211–2219. doi: 10.1021/acsomega.7b01971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Yu B., Chen D., Huang Z., Mao X., Zheng P., et al. Chlorogenic acid improves intestinal barrier functions by suppressing mucosa inflammation and improving antioxidant capacity in weaned pigs. J Nutr Biochem. 2018;59:84–92. doi: 10.1016/j.jnutbio.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Chen J., Yu B., Chen D., Zheng P., Luo Y., Huang Z., et al. Changes of porcine gut microbiota in response to dietary chlorogenic acid supplementation. Appl Microbiol Biotechnol. 2019;103:8157–8168. doi: 10.1007/s00253-019-10025-8. [DOI] [PubMed] [Google Scholar]
- Chen J.Y., Yu Y.H. Bacillus subtilis–fermented products ameliorate the growth performance and alter cecal microbiota community in broilers under lipopolysaccharide challenge. Poultry Sci. 2021;100(2):875–886. doi: 10.1016/j.psj.2020.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Zhang H., Zhao N., Yang X., Du E., Huang S., et al. Effect of chlorogenic acid on intestinal inflammation, antioxidant status, and microbial community of young hens challenged with acute heat stress. Anim Sci J. 2021;92 doi: 10.1111/asj.13619. [DOI] [PubMed] [Google Scholar]
- Cheng R., Cheng L., Zhao Y., Wang L., Wang S., Zhang J. Biosynthesis and prebiotic activity of a linear levan from a new Paenibacillus isolate. Appl Microbiol Biotechnol. 2021;105:769–787. doi: 10.1007/s00253-020-11088-8. [DOI] [PubMed] [Google Scholar]
- Christofides A., Konstantinidou E., Jani C., Boussiotis V.A. The role of peroxisome proliferator-activated receptors (PPAR) in immune responses. Metabolism. 2021;114 doi: 10.1016/j.metabol.2020.154338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte E., Genovese T., Gili E., Esposito E., Iemmolo M., Fruciano M., et al. Thymosin β4 protects C57BL/6 mice from bleomycin-induced damage in the lung. Eur J Clin Invest. 2013;43:309–315. doi: 10.1111/eci.12048. [DOI] [PubMed] [Google Scholar]
- Dall E., Brandstetter H. Structure and function of legumain in health and disease. Biochimie. 2016;122:126–150. doi: 10.1016/j.biochi.2015.09.022. [DOI] [PubMed] [Google Scholar]
- Furuhashi T., Sugitate K., Nakai T., Jikumaru Y., Ishihara G. Rapid profiling method for mammalian feces short chain fatty acids by GC-MS. Anal Biochem. 2018;543:51–54. doi: 10.1016/j.ab.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Gao J., Lin H., Wang X.J., Song Z.G., Jiao H.C. Vitamin E supplementation alleviates the oxidative stress induced by dexamethasone treatment and improves meat quality in broiler chickens. Poultry Sci. 2010;89:318–327. doi: 10.3382/ps.2009-00216. [DOI] [PubMed] [Google Scholar]
- Gao J., Xu K., Liu H., Liu G., Bai M., Peng C., et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher C., Boudier A., Bonetti J., Clarot I., Leroy P., Parent M. Glutathione: antioxidant properties dedicated to nanotechnologies. Antioxidants (Basel) 2018;7:62. doi: 10.3390/antiox7050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardberg M., Kaipio K., Lehtinen L., Mikkonen P., Heuser V.D., Talvinen K., et al. FHOD1, a formin upregulated in epithelial-mesenchymal transition, participates in cancer cell migration and invasion. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0074923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G.R., Hutkins R., Sanders M.E., Prescott S.L., Reimer R.A., Salminen S.J., et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- Haange S.B., Jehmlich N. Proteomic interrogation of the gut microbiota: potential clinical impact. Expert Rev Proteomics. 2016;13:535–537. doi: 10.1080/14789450.2016.1190652. [DOI] [PubMed] [Google Scholar]
- Hu R., He Z., Liu M., Tan J., Zhang H., Hou D.X., et al. Dietary protocatechuic acid ameliorates inflammation and up-regulates intestinal tight junction proteins by modulating gut microbiota in LPS-challenged piglets. J Anim Sci Biotechnol. 2020;11:92. doi: 10.1186/s40104-020-00492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubková B., Veliká B., Birková A., Guzy J., Mareková M. Hydroxybenzoic acids and their derivatives as peroxynitrite scavengers. Free Radic Biol Med. 2014;75(Suppl 1):S33–S34. doi: 10.1016/j.freeradbiomed.2014.10.770. [DOI] [PubMed] [Google Scholar]
- Ji L., Jiang P., Lu B., Sheng Y., Wang X., Wang Z. Chlorogenic acid, a dietary polyphenol, protects acetaminophen-induced liver injury and its mechanism. J Nutr Biochem. 2013;24:1911–1919. doi: 10.1016/j.jnutbio.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Kang Y., Li Y., Du Y., Guo L., Chen M., Huang X., et al. Konjaku flour reduces obesity in mice by modulating the composition of the gut microbiota. Int J Obes. 2019;43:1631–1643. doi: 10.1038/s41366-018-0187-x. [DOI] [PubMed] [Google Scholar]
- Kuttappan V.A., Bottje W., Ramnathan R., Hartson S.D., Coon C.N., Kong B.W., et al. Proteomic analysis reveals changes in carbohydrate and protein metabolism associated with broiler breast myopathy. Poultry Sci. 2017;96:2992–2999. doi: 10.3382/ps/pex069. [DOI] [PubMed] [Google Scholar]
- Kilkenny C., Browne W.J., Cuthill I.C., Emerson M., Altman D.G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. J Pharm Pharmacol. 2010;1(2):94–99. doi: 10.4103/0976-500X.72351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Mun S., Kim D., Lee Y.R., Sheen D.H., Ihm C., et al. Proteomics analysis for verification of rheumatoid arthritis biomarker candidates using multiple reaction monitoring. Proteonomics Clin Appl. 2019;13 doi: 10.1002/prca.201800011. [DOI] [PubMed] [Google Scholar]
- Li Q., Chen G., Chen H., Zhang W., Ding Y., Yu P., et al. Se-enriched G. frondosa polysaccharide protects against immunosuppression in cyclophosphamide-induced mice via MAPKs signal transduction pathway. Carbohydr Polym. 2018;196:445–456. doi: 10.1016/j.carbpol.2018.05.046. [DOI] [PubMed] [Google Scholar]
- Li X., Wang Q., Gao Y., Qi X., Wang Y., Gao H., et al. Quantitative iTRAQ LC-MS/MS proteomics reveals the proteome profiles of DF-1 cells after infection with subgroup J Avian leukosis virus. BioMed Res Int. 2015;2015 doi: 10.1155/2015/395307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhang H., Chen Y.P., Yang M.X., Zhang L.L., Lu Z.X., et al. Bacillus amyloliquefaciens supplementation alleviates immunological stress in lipopolysaccharide-challenged broilers at early age. Poultry Sci. 2015;94:1504–1511. doi: 10.3382/ps/pev124. [DOI] [PubMed] [Google Scholar]
- Liang S., Mao Y., Liao M., Xu Y., Chen Y., Huang X., et al. Gut microbiome associated with APC gene mutation in patients with intestinal adenomatous polyps. Int J Biol Sci. 2020;16:135–146. doi: 10.7150/ijbs.37399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang N., Kitts D.D. Role of chlorogenic acids in controlling oxidative and inflammatory stress conditions. Nutrients. 2015;8:16. doi: 10.3390/nu8010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Hu J., Zhou X., Cheung P.C.K. Inhibition of vascular endothelial growth factor-induced angiogenesis by chlorogenic acid via targeting the vascular endothelial growth factor receptor 2-mediated signalling pathway. J Funct Foods. 2017;32:285–295. [Google Scholar]
- Liu H., Zhao F., Zhang K., Zhao J., Wang Y. Investigating the growth performance, meat quality, immune function and proteomic profiles of plasmal exosomes in Lactobacillus plantarum-treated broilers with immunological stress. Food Funct. 2021;12:11790–11807. doi: 10.1039/d1fo01936h. [DOI] [PubMed] [Google Scholar]
- Liu H., Chen P., Lv X., Zhou Y., Li X., Ma S., et al. Effects of chlorogenic acid on performance, anticoccidial indicators, immunity, antioxidant status, and intestinal barrier function in coccidia-infected broilers. Animals. 2022;12(8):963. doi: 10.3390/ani12080963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Liu F., Wang W., Sun C., Gao D., Ma J., et al. Study of the alleviation effects of a combination of Lactobacillus rhamnosus and inulin on mice with colitis. Food Funct. 2020;11:3823–3837. doi: 10.1039/c9fo02992c. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lou Z., Wang H., Zhu S., Ma C., Wang Z. Antibacterial activity and mechanism of action of chlorogenic acid. J Food Sci. 2011;76:M398–M403. doi: 10.1111/j.1750-3841.2011.02213.x. [DOI] [PubMed] [Google Scholar]
- Ma Y., Gao M., Liu D. Chlorogenic acid improves high fat diet-induced hepatic steatosis and insulin resistance in mice. Pharm Res (N Y) 2015;32:1200–1209. doi: 10.1007/s11095-014-1526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia L., Thorburn A.N., Binge L.C., Marino E., Rogers K.E., Maslowski K.M., et al. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol Rev. 2012;245:164–176. doi: 10.1111/j.1600-065X.2011.01080.x. [DOI] [PubMed] [Google Scholar]
- Miao M., Xiang L. Pharmacological action and potential targets of chlorogenic acid. Adv Pharmacol. 2020;87:71–88. doi: 10.1016/bs.apha.2019.12.002. [DOI] [PubMed] [Google Scholar]
- Morris D.C., Cui Y., Cheung W.L., Lu M., Zhang L., Zhang Z.G., et al. A dose-response study of thymosin β4 for the treatment of acute stroke. J Neurol Sci. 2014;345:61–67. doi: 10.1016/j.jns.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nighot P.K., Leung L., Ma T.Y. Chloride channel ClC-2 enhances intestinal epithelial tight junction barrier function via regulation of caveolin-1 and caveolar trafficking of occludin. Exp Cell Res. 2017;352(1):113–122. doi: 10.1016/j.yexcr.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njagi L.W., Nyaga P.N., Bebora L.C., Mbuthia P.G., Minga U.M. Effect of immunosuppression on Newcastle disease virus persistence in ducks with different immune status. ISRN Vet Sci. 2012;2012 doi: 10.5402/2012/253809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveed M., Hejazi V., Abbas M., Kamboh A.A., Khan G.J., Shumzaid M., et al. Chlorogenic acid (CGA): a pharmacological review and call for further research. Biomed Pharmacother. 2018;97:67–74. doi: 10.1016/j.biopha.2017.10.064. [DOI] [PubMed] [Google Scholar]
- Ojo B.A., Lu P., Alake S.E., Keirns B., Anderson K., Gallucci G., et al. Pinto beans modulate the gut microbiome, augment MHC II protein, and antimicrobial peptide gene expression in mice fed a normal or western-style diet. J Nutr Biochem. 2021;88 doi: 10.1016/j.jnutbio.2020.108543. [DOI] [PubMed] [Google Scholar]
- Porat A., Sagiv Y., Elazar Z. A 56-kDa selenium-binding protein participates in intra-Golgi protein transport. J Biol Chem. 2000;275:14457–14465. doi: 10.1074/jbc.275.19.14457. [DOI] [PubMed] [Google Scholar]
- Rosa A., Tuberoso C.I., Atzeri A., Melis M.P., Bifulco E., Dessì M.A. Antioxidant profile of strawberry tree honey and its marker homogentisic acid in several models of oxidative stress. Food Chem. 2011;129:1045–1053. doi: 10.1016/j.foodchem.2011.05.072. [DOI] [PubMed] [Google Scholar]
- Shi D., Bai L., Qu Q., Zhou S., Yang M., Guo S., et al. Impact of gut microbiota structure in heat-stressed broilers. Poultry Sci. 2019;98:2405–2413. doi: 10.3382/ps/pez026. [DOI] [PubMed] [Google Scholar]
- Smart N., Bollini S., Dubé K.N., Vieira J.M., Zhou B., Davidson S., et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderholm J.D., Perdue M.H. Stress and gastrointestinal tract. II. Stress and intestinal barrier function. Am J Physiol Gastrointest Liver Physiol. 2001;280:G7–G13. doi: 10.1152/ajpgi.2001.280.1.G7. [DOI] [PubMed] [Google Scholar]
- Selma M.V., Espín J.C., Tomás-Barberán F.A. Interaction between phenolics and gut microbiota: role in human health. J Agric Food Chem. 2009;57(15):6485–6501. doi: 10.1021/jf902107d. [DOI] [PubMed] [Google Scholar]
- Speckmann B., Steinbrenner H. Selenium and selenoproteins in inflammatory bowel diseases and experimental colitis. Inflamm Bowel Dis. 2014;20:1110–1119. doi: 10.1097/MIB.0000000000000020. [DOI] [PubMed] [Google Scholar]
- Tian T., Xu B., Qin Y., Fan L., Chen J., Zheng P., et al. Clostridium butyricum miyairi 588 has preventive effects on chronic social defeat stress-induced depressive-like behaviour and modulates microglial activation in mice. Biochem Biophys Res Commun. 2019;516:430–436. doi: 10.1016/j.bbrc.2019.06.053. [DOI] [PubMed] [Google Scholar]
- Upadhyay R., Mohan Rao L.J. An outlook on chlorogenic acids—occurrence, chemistry, technology, and biological activities. Crit Rev Food Sci. 2013;53:968–984. doi: 10.1080/10408398.2011.576319. [DOI] [PubMed] [Google Scholar]
- Vasilopoulou E., Kolatsi-Joannou M., Lindenmeyer M.T., White K.E., Robson M.G., Cohen C.D., et al. Loss of endogenous thymosin β4 accelerates glomerular disease. Kidney Int. 2016;90:1056–1070. doi: 10.1016/j.kint.2016.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernocchi P., Del Chierico F., Putignani L. Gut microbiota Profiling: metabolomics based approach to unravel compounds affecting human health. Front Microbiol. 2016;7:1144. doi: 10.3389/fmicb.2016.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lv X., Li X., Zhao J., Zhang K., Hao X., et al. Protective effect of Lactobacillus plantarum P8 on growth performance, intestinal health, and microbiota in Eimeria-infected broilers. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.705758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Northwest A&F University; 2012. Dietary selenium and vitamin E alleviate DEX-induced oxidative stress in broilers. [Google Scholar]
- Wu Y., Zhang H., Zhang R., Cao G., Li Q., Zhang B., et al. Serum metabolome and gut microbiome alterations in broiler chickens supplemented with lauric acid. Poultry Sci. 2021;100 doi: 10.1016/j.psj.2021.101315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Abraham P.E., Li Z., Pan C., Hettich R.L. Microbial metaproteomics for characterizing the range of metabolic functions and activities of human gut microbiota. Proteomics. 2015;15:3424–3438. doi: 10.1002/pmic.201400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chang J., Wang P., Yin Q., Liu C., Li M., et al. Effect of chlorogenic acid on alleviating inflammation and apoptosis of IPEC-J2 cells induced by deoxyniyalenol. Ecotoxicol Environ Saf. 2020;205 doi: 10.1016/j.ecoenv.2020.111376. [DOI] [PubMed] [Google Scholar]
- Xue J., Xie L., Liu B., Zhou L., Hu Y., Ajuwon K.M., et al. Epidermal growth factor ameliorates essential trace element absorption in the gastrointestinal tract by regulating the expression of microelement transport-relative genes in lipopolysaccharide challenged early weaning piglets. Res Square. 2020 doi: 10.21203/rs.3.rs-102470/v1. [DOI] [Google Scholar]
- Yang X., Liang S., Guo F., Ren Z., Yang X., Long F., et al. Gut microbiota mediates the protective role of Lactobacillus plantarum in ameliorating deoxynivalenol-induced apoptosis and intestinal inflammation of broiler chickens. Poultry Sci. 2020;99(5):2395–2406. doi: 10.1016/j.psj.2019.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Mo S., Shen M., Chen Y., Yu Q., Li Z., et al. Sulfated modification enhances the immunomodulatory effect of Cyclocarya paliurus polysaccharide on cyclophosphamide-induced immunosuppressed mice through MyD88-dependent MAPK/NF-κB and PI3K-Akt signaling pathways. Food Res Int. 2021;150 doi: 10.1016/j.foodres.2021.110756. [DOI] [PubMed] [Google Scholar]
- Zhang K., Li X., Zhao J., Wang Y., Hao X., Liu K., et al. Protective effects of chlorogenic acid on the meat quality of oxidatively stressed broilers revealed by integrated metabolomics and antioxidant analysis. Food Funct. 2022;13:2238–2252. doi: 10.1039/d1fo03622j. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhao Q., Ci X., Chen S., Xie Z., Li H., et al. Evaluation of the efficacy of chlorogenic acid in reducing small intestine injury, oxidative stress, and inflammation in chickens challenged with Clostridium perfringens type A. Poultry Sci. 2020;99:6606–6618. doi: 10.1016/j.psj.2020.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Ji J., Qu H., Wang J., Shu D.M., Wang Y., et al. Effects of sodium butyrate on intestinal health and gut microbiota composition during intestinal inflammation progression in broilers. Poultry Sci. 2019;98(10):4449–4456. doi: 10.3382/ps/pez279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1
Data Availability Statement
Microbiomic and proteomic sequencing data have been deposited under BioProject PRJNA789475 and IPX0003857000, respectively.