Summary
Prions are deadly infectious agents made of PrPSc, a misfolded variant of the cellular prion protein (PrPC) which self-propagates by inducing misfolding of native PrPC. PrPSc can adopt different pathogenic conformations (prion strains), which can be resistant to potential drugs, or acquire drug resistance, hampering the development of effective therapies. We identified Zn(II)-BnPyP, a tetracationic porphyrin that binds to distinct domains of native PrPC, eliciting a dual anti-prion effect. Zn(II)-BnPyP binding to a C-terminal pocket destabilizes the native PrPC fold, hindering conversion to PrPSc; Zn(II)-BnPyP binding to the flexible N-terminal tail disrupts N- to C-terminal interactions, triggering PrPC endocytosis and lysosomal degradation, thus reducing the substrate for PrPSc generation. Zn(II)-BnPyP inhibits propagation of different prion strains in vitro, in neuronal cells and organotypic brain cultures. These results identify a PrPC-targeting compound with an unprecedented dual mechanism of action which might be exploited to achieve anti-prion effects without engendering drug resistance.
Subject areas: Pharmacology, Molecular neuroscience, Cell biology
Graphical abstract

Highlights
-
•
Zn(II)-BnPyP binds to distinct domains of PrPC
-
•
Binding to the C-terminal domain is sufficient to inhibit conversion to PrPSc
-
•
Binding to the N-terminal tail triggers PrPC endocytosis and degradation
-
•
Zn(II)-BnPyP is active against different prion strains
Pharmacology; Molecular neuroscience; Cell biology
Introduction
Prion diseases, including scrapie of sheep, chronic wasting disease of cervids, and human Creutzfeldt-Jakob disease (CJD), are deadly brain disorders for which there is not yet any cure.1 They are caused by the conformational conversion of PrPC, encoded by the PRNP gene, into PrPSc, which self-propagates by converting native PrPC. Inhibiting this conversion is, therefore, the primary target for therapy.
PrPC is a cell surface glycoprotein with an elusive function, expressed in neurons and many other body cells.2 It is synthesized in the endoplasmic reticulum (ER), where it undergoes oxidative folding, N-linked glycosylation and addition of a glycosyl-phosphatidyl-inositol (GPI) anchor that attaches the protein C terminus to the lipid bilayer. After transit in the Golgi, PrPC is delivered to lipid rafts, cholesterol-rich microdomains of the plasma membrane. PrPC molecules are constitutively endocytosed through clathrin- and/or caveolae/raft-dependent pathways, and either recycled to the plasma membrane or delivered to lysosomes for degradation.3
PrPC has a flexible N-terminal tail [residues 23–128, human (hu) PrP numbering] containing an octapeptide repeat (OR) region, which can interact with Cu(II) and Zn(II) ions, and a C-terminal globular domain (residues 129–231) comprising three α-helices (α1, α2, and α3) and two short anti-parallel β strands (β1 and β2).4 Cu(II) and Zn(II) promote tertiary contacts between the N- and C-terminal PrPC domains.5,6,7,8
PrPSc is rich in β-sheet, aggregated and protease-resistant.1 It can acquire different structures with distinct pathogenic properties (prion strains), which respond differently to potential drugs, and acquire drug resistance through strain selection and adaptation.9,10,11,12 PrPSc is not toxic per se, with neurodegeneration stemming from PrPC-mediated signaling upon contact with extracellular PrPSc and/or from intracellular PrPC misfolding.2 Thus, therapeutic strategies that target PrPC may be more effective than those targeting PrPSc.
There is ample evidence that downregulation of PrPC expression is a safe and valuable therapeutic approach. Genetic ablation of Prnp in mice, cows and goats has little phenotypic effect,13,14,15 and the most consistent phenotype – a mild sensorimotor defect in aged PrP knockout (KO) mice – is not seen in heterozygotes.16 Humans with only one functional PRNP allele are in fact healthy.17 However, PrP KO mice are resistant to prions,18 and heterozygotes show a significant delay in disease onset, surviving more than twice as long as wild-type (wt) mice.19 Conditional PrP KO in neurons of mice in the early stage of prion disease rescues clinical signs and prevents neurodegeneration.20 Finally, RNA interference and antisense oligonucleotides targeting PrPC significantly increase the survival of prion-diseased mice.21,22,23,24 Thus, lowering PrPC is well tolerated and beneficial in prion disease.
Cyclic tetrapyrroles, including porphyrins and phthalocyanines, are potent inhibitors of PrPSc replication in prion-infected cells, with some of them prolonging the survival of prion-infected animals, especially when given early for peripheral infection.25,26,27,28 Their activity is influenced by the type of metal coordinated at the center of the molecule and generally correlates with their tendency to self-aggregate.29 Among a series of porphyrins first studied as photosensitizers for the treatment of cancer or bacterial infections,30,31 we identified a zinc-tetracationic porphyrin [Zn(II)5,10,15,20-tetra(N-benzyl-4-pyridyl)porphyrin tetrachloride; hereafter Zn(II)-BnPyP] that binds to distinct domains of native PrPC inducing its degradation while inhibiting conversion to PrPSc. By simultaneously reducing the PrPC substrate for prion propagation and blocking the conversion of PrPC to PrPSc, Zn(II)-BnPyP acts upstream of PrPSc formation, thus evading the problem of drug resistance. Here, we describe the cellular and molecular mechanisms responsible for its dual activity and provide proof-of-principle of efficacy against different prion strains.
Results
Zn(II)-BnPyP induces clathrin-dependent endocytosis and lysosomal degradation of cell surface PrPC
Zn(II)-BnPyP markedly lowered PrPC levels in mouse (mo) primary neurons (Figures 1 and S1A) and different cell lines, including hu glioblastoma H4 (Figures S1B–S1D), mo neuroblastoma N2a (Figure S1G) and hu HEK293 cells expressing transgenic mo or bank vole (bv) PrP (Figure S1H). The effect was dose- and time-dependent, with EC50 0.75 μM in hippocampal neurons (Figures 1C–1F), and was reversible, with PrPC returning to normal levels within 24h once treatment stopped (Figure S1I). Fe(III)-TMPyP [Fe(III)-5,10,15, 20-tetra(N-methyl-4-pyridyl)porphyrin tetratosylate], a porphyrin that binds to the PrPC globular domain and inhibits PrPSc replication,26,27 did not lower PrPC (Figures 1B, 1G, and 1H). Neither did the well-known anti-prion compound pentosan polysulfate (PPS) which interacts with both the globular and N-terminal domains of PrPC32,33,34,35,36 (Figures S1D and S1E).
Figure 1.
Zn(II)-BnPyP down-regulates PrPC
(A) Chemical structure of Zn(II)-BnPyP.
(B) Chemical structure of Fe(III)-TMPyP.
(C) Primary hippocampal neurons were treated with 0–10 μM Zn(II)-BnPyP for 24h and analyzed by Western blot (WB) with anti-PrP and anti-actin antibodies.
(D) PrP signals were analyzed by densitometry of blots like the one shown in (C), normalized on the levels of actin, and expressed as percentages of untreated controls; mean ± SEM of 4–5 replicates from 5 independent experiments.
(E) Neurons were treated with 2.5 μM Zn(II)-BnPyP for the times indicated and analyzed by WB.
(F) Actin-normalized PrP levels; mean ± SEM of four experiments like the one shown in (E).
(G) Confocal immunofluorescence analysis of PrP (green) and MAP2 (red) of neurons treated with 2.5 μM Zn(II)-BnP or Fe(III)-TMPyP for 6h. Cells were reacted with Hoechst 33258 (blue) to stain the nuclei. Scale bar 50 μm.
(H) Neurons were exposed to 2.5 μM Zn(II)-BnPyP or Fe(III)-TMPyP for 24h, and analyzed by WB with anti-PrP and anti-actin antibodies.
(I) Neurons treated with 2.5 μM Zn(II)-BnPyP for 24h were analyzed by WB to detect PrP, amyloid precursor protein (APP), tau, α-synuclein (α-syn) and actin.
(J) H4 cells were incubated without (−) or with (+) 20 μM chloroquine and 5 μM Zn(II)-BnPyP, and analyzed by WB after 6h.
(K) Actin-normalized PrP levels in H4 cells treated as in (J), relative to untreated controls, set as 100%; mean ± SEM. F3,16 = 7.443, p = 0.0024 by one-way ANOVA; ∗p < 0.05 vs. untreated, §§p < 0.01 vs. CQ, #p < 0.05 vs. Zn(II)-BnPyP by Tukey’s post-hoc test; CQ, chloroquine.
We exploited the intrinsic Zn(II)-BnPyP fluorescence to investigate the porphyrin distribution in HEK293 cells stably expressing mo PrPC. Cells were exposed to 10 μM Zn(II)-BnPyP and immunostained with an anti-PrP antibody to visualize also PrPC distribution at different times (from 5 min to 24h). At short time points Zn(II)-BnPyP fluorescence was not visible. After 1h Zn(II)-BnPyP fluorescence started becoming visible around and within cells, and progressively accumulated intracellularly where it colocalized in part with PrPC which concomitantly disappeared (Figure S2).
Zn(II)-BnPyP was not toxic at the active concentrations (Figure S1J). The effect of Zn(II)-BnPyP appeared to be selective for PrPC since it did not affect the levels of other proteins, including the amyloid precursor protein, tau, α-synuclein, synaptophysin, β3-tubulin, actin and vinculin, or cause any evident alteration in total cellular proteins visualized by SDS-PAGE (Figures 1I, S1A, S1P, and S3G). Zn(II)-BnPyP did not induce PrPC release from the cell surface (Figure S1K), nor did it down-regulate PrPC mRNA (Figure S1L), or inhibit global protein synthesis (Figure S1M).
Cycloheximide chase experiments to assess steady-state protein stability indicated that Zn(II)-BnPyP increased the rate of PrPC degradation (Figures S1N and S1O). The PrPC lowering effect was reduced by chloroquine and bafilomycin A1, which inhibit lysosome activity (Figures 1J, 1K, S1P, and S1Q). Immunofluorescence analysis of cells expressing EGFP-tagged PrPC treated with Zn(II)-BnPyP and bafilomycin A1 showed increased co-localization of the protein and the lysosome marker LAMP1, supporting lysosomal PrPC destruction (Figures 2A and 2B).
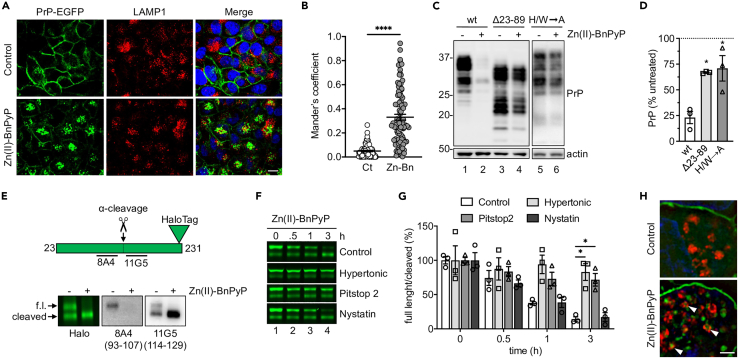
Figure 2.
Zn(II)-BnPyP induces clathrin-dependent PrPC endocytosis and lysosomal degradation
(A) Immunofluorescence analysis of LAMP1 in PrP-EGFP-expressing HeLa cells treated with the vehicle (control) or 5 μM Zn(II)-BnPyP, with 0.1 μg/μL bafilomycin A1 for 30 min. Scale bar 10 μm.
(B) Mander’s coefficient M1: PrP-EGFP fraction that co-localizes with LAMP1. ∗∗∗∗p < 0.0001 by Student’s t test. Zn-Bn: Zn(II)-BnPyP.
(C) Western blot of PrP and actin in HEK293 cells expressing moPrP wild-type (wt), Δ23-89, or with histidine- and tryptophan-to-alanine substitutions in the OR region (H/W→A), after 24h incubation without (−) or with (+) 5 μM Zn(II)-BnPyP.
(D) Actin-normalized PrP levels in HEK293 cells relative to untreated controls, set as 100%. Mean ± SEM of three experiments like the one shown in C. ∗p < 0.05 by one-way ANOVA, Tukey’s post-hoc test.
(E) Scheme of the PrP-Halo protein, indicating the site of α-cleavage and 8A4 and 11G5 antibody epitopes, and analysis of its expression in HEK293 cells visualized by cell impermeant Alexa Fluor 488 ligand fluorescence (Halo) or WB using 8A4 and 11G5 antibodies (epitopes in parentheses), before (−) and after (+) treatment with 5 μM Zn(II)-BnPyP for 6h. Arrows point to full-length (f.l.) and N-terminally cleaved PrP-Halo.
(F) PrP-Halo visualized by cell impermeant Alexa Fluor 488 ligand in HEK293 cells treated with 5 μM Zn(II)-BnPyP for the times indicated, in standard (control) or hypertonic medium or with Pitstop 2 or nystatin.
(G) Ratio of full-length to N-terminally cleaved PrP-Halo in cells treated as in (F). Mean ± SEM of three experiments. ∗p < 0.05 by two-way ANOVA, Dunnett’s post-hoc test.
(H) SIM microscopy of PrP-Halo (green) and LAMP1 (red) in HEK293 cells before and after treatment with 5 μM Zn(II)-BnPyP for 6h with 0.1 μg/μL bafilomycin A1. Scale bar 2 μm.
Immature (endoglycosidase H-sensitive) PrPC accumulating in cells treated with brefeldin A to block ER to Golgi transport was not lowered by Zn(II)-BnPyP, whereas mature (endoglycosidase H-resistant) PrPC was (Figure S3A; the ratio of mature to immature PrP was 1.34 ± 052 in control cells and 0.55 ± 0.43 in Zn(II)-BnPyP-treated cells; mean ± SD, n = 5; p = 0.0298, unpaired t-test). Zn(II)-BnPyP did not reduce the levels of PrPC molecules artificially targeted to the ER (Figure S3B), suggesting that the porphyrin preferentially degrades PrPC that has reached post-ER compartments.
During ER stress misfolded PrPC may be rapidly exported from the ER to downstream compartments of the secretory pathway for subsequent lysosomal degradation (RESET).37 Zn(II)-BnPyP did not cause PrPC misfolding, assessed in a detergent insolubility assay (Figure S3C) and structural investigations (see below), or activation of the unfolded protein response (Figure S3D), suggesting that the RESET degradation pathway was not involved.
Deletions of the PrPC N-terminal region, including Δ23-89 and Δ23-111, significantly reduced the PrPC-lowering effect of Zn(II)-BnPyP (Figure 2C, lanes 1–4, and 2D; Figures S3E and S3F). Since the N-terminal tail is essential for PrPC endocytosis,38 these results suggested that Zn(II)-BnPyP might stimulate internalization and lysosomal degradation of cell surface PrPC. To test this, we checked the effect of Zn(II)-BnPyP on PrPC levels in HEK293 cells exposed to a hypertonic medium which blocks clathrin-dependent endocytosis by disrupting clathrin lattices,39 or nystatin which lowers cholesterol levels and inhibits caveolae/raft-dependent endocytosis. The hypertonic medium, but not nystatin, prevented Zn(II)-BnPyP-induced PrPC lowering, indicating that the porphyrin triggered PrPC endocytosis through clathrin-coated pits (Figures S3G and S3H).
To assess the effect of Zn(II)-BnPyP specifically on cell surface PrPC, we generated a moPrP-HaloTag fusion construct (PrP-Halo) which can be labeled using a cell impermeant fluorescent reagent. PrP-Halo was efficiently expressed and glycosylated in HEK293 cells (Figure S3I). A substantial amount of cell surface-labeled PrP-Halo was N-terminally cleaved between amino acid 107 and 114, consistent with physiological PrPC α-cleavage at site 110/111 during endocytic recycling3 (Figure 2E). Full-length but not N-terminally cleaved PrP-Halo was down-regulated by Zn(II)-BnPyP, confirming that the N-terminal tail was necessary for the PrPC lowering effect (Figure 2E). Zn(II)-BnPyP-induced downregulation of cell surface PrP-Halo was prevented by exposing cells to the hypertonic milieu or Pitstop 2, which inhibits clathrin-mediated endocytosis by binding to the terminal domain of clathrin, but not nystatin (Figures 2F and 2G). Super-resolution microscopy of cells treated with Zn(II)-BnPyP with bafilomycin A1 showed PrP-Halo accumulation in LAMP1-positive structures (Figure 2H), consistent with lysosomal delivery of the internalized protein.
Zn(II)-BnPyP has strain-independent anti-prion activity
To test the effect of Zn(II)-BnPyP on PrPSc replication, in a first set of experiments we used protein misfolding cyclic amplification (PMCA), an in vitro reaction which allows ultra-efficient amplification of tiny amounts of PrPSc from prion-diseased brains using healthy brain homogenates as a source of PrPC. Zn(II)-BnPyP inhibited the amplification of proteinase K (PK)-resistant PrP (a surrogate biochemical marker of PrPSc) from different prion strains, including variant and sporadic CJD, scrapie and bank vole prion strain, more efficiently than Fe(III)-TMPyP (Figures 3A, 3B, S4A, and S4B). Zn(II)-BnPyP did not induce PrPC degradation during the PMCA (Figure S4C), indicating that its inhibitory effect was not due to depletion of the PrPC substrate. Thus, Zn(II)-BnPyP, besides lowering PrPC levels in live cells, inhibited PrPSc propagation in a cell-free environment, potentially resulting in a strong anti-prion effect.
Figure 3.
Zn(II)-BnPyP inhibits replication of different prion strains in PMCA, N2a cells and COCS
(A and B) Western blot analysis of PK-resistant PrP after a single 24h round of protein misfolding cyclic amplification (PMCA), with different dilutions of vCJD (A) or sCJD VV2 subtype (B) PrPSc seeds, with the vehicle or 5 μM Zn(II)-BnPyP or Fe(III)-TMPyP. Results are representative of two independent experiments.
(C and D) WB analysis of total (-PK) and protease-resistant (+PK) PrP and actin in ScN2a-22L cells treated with 0, 0.5, 1, 2.5 and 5 μM Zn(II)-BnPyP or Fe(III)-TMPyP for 72h.
(E) Quantification of actin-normalized PK-resistant PrP in ScN2a-22L cells treated with increasing concentrations of Zn(II)-BnPyP or Fe(III)-TMPyP. Data are expressed as percentages of untreated cells; mean ± SEM of three experiments.
(F) WB of total (-PK) and protease-resistant (+PK) PrP in ScN2a-22L cells cultured without (−) or with (+) 5 μM Zn(II)-BnPyP for 16 days, then without porphyrin (wash-out) for the days indicated.
(G) WB of PrP and actin in RML- and 22L-infected COCS treated with Zn(II)-BnPyP at the concentrations indicated. Blots were overexposed to show the presence of full-length PrPC and the absence of PK-resistant PrP in the treated COCS (shorter exposure of a replicate experiment is shown in Figure S4F).
Next, we analyzed the effect of Zn(II)-BnPyP in prion-infected cells. Neuroblastoma N2a cells chronically infected with the 22L (ScN2a-22L) or RML (ScN2a-RML) scrapie strains were treated with 0.25–5 μM Zn(II)-BnPyP or Fe(III)-TMPyP, and the amount of PK-resistant PrP was measured after 72h by Western blot. Both porphyrins reduced PK-resistant PrP, indicating inhibition of prion replication, but Zn(II)-BnPyP was stronger, with IC50 0.7 μM, compared to 1.5 μM for Fe(III)-TMPyP (Figures 3C–3E, S4D, and S4E). Consistent with its PrPC-lowering activity, Zn(II)-BnPyP reduced not only PK-resistant PrPSc but also the higher molecular weight PrP bands between 37 and 25 kDa seen in the undigested cell lysates, whereas Fe(III)-TMPyP did not (compare the -PK lanes in Figures 3C, 3D, S4D, and S4E).
To see whether Zn(II)-BnPyP cured prion infection, ScN2a-22L cells were treated with 5 μM Zn(II)-BnPyP for 9 or 16 days, and lysed for analysis of PK-resistant PrP at the end of the treatment, or after 2, 7 or 21 more days in culture without the porphyrin (wash-out). No PK-resistant PrP was detectable at the end of the treatment or after wash-out (Figure 3F and data not shown), indicating that the cells had been cleared of prions.
Zn(II)-BnPyP was also tested in prion-infected cultured organotypic cerebellar slices (COCS). COCS from C57BL/6J mice were infected with RML or 22L prions. Five weeks after infection they were treated with 1–5 μM Zn(II)-BnPyP or Fe(III)-TMPyP every second day for one week, then lysed for analysis of PK-resistant PrP. In untreated COCS most PrPC had converted to PrPSc and was N-terminally cleaved, as shown by the typical protease-resistant PrP band pattern and the virtual absence of full-length PrP even before PK digestion (Figure 3G, lanes 1 and 2, and 5 and 6; Figure S4F, lanes 1 and 2). Treatment with as little as 1 or 2.5 μM Zn(II)-BnPyP completely eliminated protease-resistant PrP and promoted the appearance of full-length PrP, albeit at low levels due to the porphyrin’s PrPC-lowering activity (Figure 3G, lanes 3 and 4, and 7 and 8; see also Figure S4F, lanes 3 and 4, showing shorter exposure in a similar experiment). At the same concentrations Fe(III)-TMPyP had no effect (Figure S4F, lanes 5 and 6), and at higher concentrations it was toxic to COCS. PPS was not toxic, but still less efficient than Zn(II)-BnPyP even at much higher concentrations (Figure S4G).
To quantitatively compare the activity of the Zn(II)-BnPyP and Fe(III)-TMPyP we treated 22L-infected COCS acutely with 1–10 μM and measured PrPSc levels after 48h, when there was no evidence of Fe(III)-TMPyP toxicity. Zn(II)-BnPyP markedly reduced PrPSc replication, more efficiently than Fe(III)-TMPyP (Figures S4H–S4J). For example, 5 μM Zn(II)-BnPyP reduced PK-resistant PrP >85%, compared to ∼50% reduction with the same concentration of Fe(III)-TMPyP (Figures S4H–S4J).
Zn(II)-BnPyP binds to distinct PrPC regions
To assess whether Zn(II)-BnPyP interacted directly with PrPC and identify the binding site, we conducted NMR titrations, monitoring chemical shift perturbations (CSPs) and peak intensity ratio (I/I0) differences in 1H-15N heteronuclear single quantum coherence (HSQC) spectra on addition of increasing concentrations of Zn(II)-BnPyP to 15N huPrP23-231. Significant spectral perturbations were evident in two distinct PrPC regions: i) the α2 helix and the C-terminus of the α3 helix of the globular domain, ii) the OR and the non-OR region of the N-terminal tail (Figure 4A). This bipartite interaction mode suggested that more than one Zn(II)-BnPyP molecule interacted with huPrP23-231; this is in line with isothermal titration calorimetry (ITC) showing binding saturation at high molar ratios of porphyrin (estimated apparent Kd 28 ± 2.4 μM) (Figure S5B). Dynamic mass redistribution (DMR) confirmed the low micromolar apparent Kd (3.1 ± 0.1 μM, Figure S5C), in agreement with the fast-intermediate exchange regime of the interaction on the NMR timescale (Figure S5D).
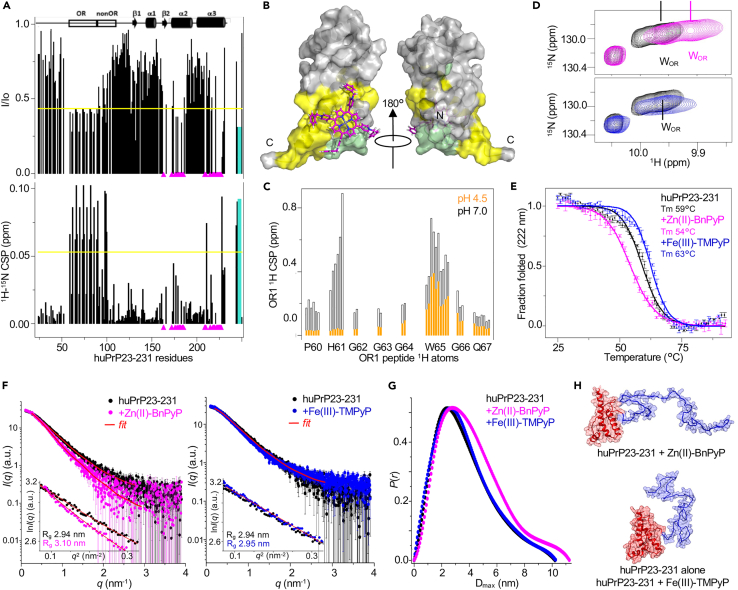
Figure 4.
Structural characterization of the Zn(II)-BnPyP interaction with huPrP23-231
(A) Intensity ratio (I/Io) and 1H15N chemical shift perturbations (CSPs) of 15N huPrP23-231 amide resonances on addition of Zn(II)-BnPyP (1:2). Magenta triangles: peaks that disappeared; yellow line: avg - sd (I/Io) or avg + sd (CSP); cyan bars: data refer to OR W indole NH.
(B) Docking model of huPrP23-231 globular domain in complex with Zn(II)-BnPyP. Yellow: residues whose resonances are significantly affected by Zn(II)-BnPyP; pale green: unassigned resonances because of overlap or solvent exchange.
(C) CSPs of OR1 peptide 1H resonances on addition of Zn(II)-BnPyP (1:1) at pH 7 (black) or pH 4.5 (orange).
(D) Superposition of 1H-15N HSQC spectra of 15N huPrP23-231 without (black) and with (1:1) Zn(II)-BnPyP (magenta) or Fe(III)-TMPyP (blue), zoom into tryptophan (W) indole NH region.
(E) Circular dichroism melting curves of huPrP23-231 alone (black), on addition (1:15) of Zn(II)-BnPyP (magenta) or Fe(III)-TMPyP (blue). Mean of two replicates ±SEM.
(F) SAXS curves of huPrP23-231 alone (black) and in complex (1:5) with Zn(II)-BnPyP (magenta) or Fe(III)-TMPyP (blue); red: GNOM fitting curve. The insets show the Guinier fits.
(G) Pair distance distributions of huPrP23-231 without (black) and with (1:5) Zn(II)-BnPyP (magenta) or Fe(III)-TMPyP (blue).
(H) Illustrative protein structures after EOM modeling of SAXS data, describing huPrP23-231 in the presence of Zn(II)-BnPyP (upper panel) and alone or with Fe(III)-TMPyP (lower panel). The C-terminal globular domain (red) was used as a rigid body and the N-terminal tail (blue) was modeled by EOM.
Dissection of the interaction through NMR titrations with different PrPC fragments did in fact indicate that Zn(II)-BnPyP could independently bind to the N-terminal region (residues 23–91, Figures S6A–S6C), to double (residues 60–75; OR2) and single (residues 60–67; OR1) octapeptide repeats (Figures S6D–S6L) and to huPrP90-231 (Figures S7A–S7D). Each OR motif was potentially able to bind one Zn(II)-BnPyP molecule, the affinity increasing with the number of OR motifs (Figures S6C, S6G, and S6L). The interaction of Zn(II)-BnPyP with the isolated non-OR region (residues 90–112) was negligible (Figures S7E–S7H).
Based on the well-documented ability of PrPC to bind Zn(II) ions through the OR histidines (H),7,8 we hypothesized that Zn(II)-BnPyP bound to the N-terminal region through coordination of the histidine imidazole rings and displacement of the water molecule, completing the tetrahedral coordination of Zn(II) inside the porphyrin (Figure S8A).40 Titration of 15N huPrP23-231 with Zn(II)-BnPyP at pH 4.5, where histidines are mainly protonated, did in fact cause only small changes in both peak intensities and CSPs in the N-terminal region (Figures S8B–S8D). Accordingly, the CSPs induced by Zn(II)-BnPyP on the histidine resonances of OR1 at pH 4.5 were negligible compared to those at pH 7 (Figures 4C, S8E, and S8F). Moreover, titration of 15N huPrP23-231 with Fe(III)-TMPyP (Figures S9A–S9D) and Fe(III)-BnPyP (Figures S9E–S9H) showed significant spectral perturbations only in the globular domain, conceivably because of the lower affinity of Fe(III) porphyrins to nitrogen-containing axial ligands compared to Zn(II) porphyrins.41,42 Therefore, Zn(II) in the porphyrin scaffold is crucial for binding to the PrPC N-terminal tail through histidine coordination.
We next questioned whether the phenyl rings of the BnPyP scaffold played a role in the binding. A three-dimensional model of the PrPC globular domain complexed with Zn(II)-BnPyP indicated that BnPyP docks on the surface formed by the α2 and α3 helices, establishing favorable interactions with the aromatic side chains of tyrosines (Y) (Figures 4B and S5E), while Fe(III)-TMPyP docks on the opposite face, on the surface made by the β-sheet and α3 (Figures S9C and S9D). In addition, the significant CSPs of the resonances of the OR tryptophan (W) side-chains induced by Zn(II)-BnPyP binding (Figures 4D and S6), together with the high percentage of saturation of the porphyrin phenyl rings in saturation transfer difference (STD) experiments (Figures S5F–S5H), suggest that the BnPyP scaffold contributes to π-π interactions with the indole moieties of the OR tryptophans. Titration of Zn(II)-TMPyP into 15N huPrP23-231 only negligibly perturbed the W indole amine resonances (Figure S10). Attempts to investigate huPrP23-231 interaction with non-metalated BnPyP were inconclusive because traces of Zn(II) in the protein solutions were readily incorporated into BnPyP (Figure S11).
These data indicate that Zn(II)-BnPyP binds independently to both the globular domain and the N-terminal tail of PrPC. Engagement of the N-terminal tail is a peculiarity of Zn(II)-BnPyP, which can interact with both the histidine and tryptophan residues of the ORs.
Zn(II)-BnPyP destabilizes and opens the PrPC conformation
We examined the effects of Zn(II)-BnPyP and Fe(III)-TMPyP binding on PrPC stability by circular dichroism (CD) thermal denaturation. In agreement with previous studies,27 Fe(III)-TMPyP increased the Tm of huPrP90-231 by 2°C, while Zn(II)-BnPyP reduced it by 12°C (Figure S12A), suggesting destabilization of the globular domain. On huPrP23-231, Fe(III)-TMPyP raised the Tm 3°C while Zn(II)-BnPyP lowered it 5°C (Figure 4E). Thus, in stark contrast with Fe(III)-TMPyP, Zn(II)-BnPyP markedly reduced the thermal stability of PrPC.
Next we questioned whether the Zn(II)-BnPyP interaction with the ORs interfered with the transient intramolecular interactions between the N-terminal tail and the globular domain.5,6 We used Small-Angle X-ray Scattering (SAXS) to analyze the conformational equilibrium on huPrP23-231 alone and in the presence of Zn(II)-BnPyP or Fe(III)-TMPyP. Ensemble Optimization Method (EOM) analysis of the SAXS curves in the 0.01 to 3.5 nm−1 q-range indicated that Zn(II)-BnPyP shifted the structural parameters to larger Rg (gyration radius) and Dmax (maximum diameter) compared to huPrP23-231 alone, while Fe(III)-TMPyP, which binds only to the globular domain, did not alter these structural parameters (Figures 4F–4H and S12B–S12F). The EOM structural models that contribute most to the final assembly of huPrP23-231 alone or in complex with Fe(III)-TMPyP predict more compact structures (Rg ∼2.9 nm and Dmax ∼9.4 nm) with the N-terminal tail close to the globular domain (Figure S12G). Conversely, the most frequent huPrP23-231 conformational state in the presence of Zn(II)-BnPyP had larger structural parameters (Rg 3.4 nm and Dmax ∼11.1 nm), indicating that binding of this porphyrin reduces the interdomain contacts (Figure S12H). In all experimental conditions, the I(0)-based mass values were in agreement with the expected monomeric huPrP23-231 molecular weight (i.e., ∼25 kDa, Table S1).
Taken together, these CD and SAXS analyses indicate that Zn(II)-BnPyP destabilizes the PrPC globular domain and shifts PrPC conformational equilibrium toward more open structural states.
Zn(II)-BnPyP binding to distinct PrPC domains accounts for its ability to downregulate PrPC and inhibit conversion to PrPSc
We questioned whether Zn(II)-BnPyP’s ability to bind to N- and C-terminal PrPC domains was responsible for its dual anti-prion activity. First we tested the effect of substituting Fe(III) for Zn(II) on the porphyrin’s ability to downregulate PrPC. Treatment of H4 cells with Zn(II)-BnPyP caused dose-dependent reduction in PrPC, whereas Fe(III)-BnPyP, which does not bind to the N-terminal tail, had no effect (Figures S1B, S1F, and S1D).
Next we tested the roles of histidine and tryptophan residues in the OR which, on the basis of NMR analysis, are essential for Zn(II)-BnPyP binding to the N-terminal tail. HEK293 cells expressing PrP wt or a PrP mutant with histidine/tryptophan-to-alanine substitutions in the OR region (PrP H/W→A) were treated with Zn(II)-BnPyP, and PrP levels were measured by WB after 24h. Zn(II)-BnPyP downregulated PrP H/W→A significantly less than PrP wt (Figure 2C, compare lanes 1 and 2 with 5 and 6; Figure 2D). This indicates that Zn(II)-BnPyP binding to the N-terminal tail is required for lowering PrPC.
Finally, we asked whether Zn(II)-BnPyP had to interact with the OR region to inhibit conformational conversion to PrPSc. We compared the inhibitory activity of Zn(II)-BnPyP in a modified PMCA reaction which uses recombinant PrP full-length (bvPrP23-231) or lacking the OR region (bvPrPΔ52-92), as substrates for PrPSc amplification. Zn(II)-BnPyP efficiently inhibited conversion of both substrates (Figure S4K), indicating that the porphyrin binding to the C-terminal pocket was sufficient to hinder PrPC to PrPSc conversion.
Zn(II)-BnPyP does not penetrate the brain efficiently enough to prolong the survival of prion-infected mice
As a first step toward assessing the therapeutic potential of Zn(II)-BnPyP, we characterized its pharmacokinetic parameters and blood-brain barrier (BBB) passage. C57BL/6 mice were given a single intraperitoneal (ip) injection of 10 mg/kg Zn(II)-BnPyP, and the plasma and brain levels were analyzed at different time points. The porphyrin plasma levels peaked 1h after the dose and then rapidly declined, with ∼80% of the compound removed from circulation in the first 24h (Figure 5A). Porphyrin levels in the CNS peaked 2h after the dose (0.249 ± 0.039 μg/g) and declined slowly with time, still being high (0.157 ± 0.137 μg/g) after 7 days (Figure 5B). Considering the Zn(II)-BnPyP maximal concentrations in plasma and brain, the brain-to-plasma ratio was 0.035, indicating very poor brain penetration; however, its slow clearance from the CNS suggested that therapeutically useful concentrations could be achieved with chronic treatment. To investigate this, mice were treated with 10 mg/kg Zn(II)-BnPyP ip every second day and culled after 1, 2, 4, 8 and 12 weeks of treatment for analysis of the porphyrin brain levels. There was progressive accumulation of Zn(II)-BnPyP in the CNS, reaching 1855 ± 160 ng/g (∼1.5 μM) after 12 weeks (Figure 5C).
Figure 5.
Zn(II)-BnPyP pharmacokinetics and effect on brain PrPC levels
(A and B) C57BL/6 mice were injected ip with 10 mg/kg Zn(II)-BnPyP and euthanized at different times after the dose (5 min, 30 min, 1h, 2h, 6h, 24h, 3 days and 7 days). Porphyrin in the plasma (A) and the brain (B) was quantified by HPLC coupled with a fluorescence detector. Data are the mean ± SD of three mice.
(C) C57BL/6 mice were injected ip with 10 mg/kg Zn(II)-BnPyP every second day for the weeks indicated, and euthanized 6h after the last dose for quantification of brain Zn(II)-BnPyP. Data are the mean ± SD of 3–5 mice.
(D–F) Quantification (mean ± SD) of actin-normalized PrPC levels in the brains of the mice treated for 4, 8 or 12 weeks shown in C.
To test target engagement, we quantified brain PrPC levels in mice that had been treated for ≥4 weeks, in which Zn(II)-BnPyP had reached brain concentrations equal to or higher than its PrPC-lowering EC50 in cultured hippocampal neurons (0.75 μM). There were no differences in brain PrPC levels between mice treated with Zn(II)-BnPyP or the vehicle for 4 or 8 weeks (Figures 5D and 5E). A significant, albeit modest, reduction was seen in the brain of mice treated with Zn(II)Bn-PyP for 12 weeks (Figure 5F), indicating that the threshold concentration for activity in the CNS was ∼1.5 μM.
To test whether this was sufficient to protect against CNS prion infection, groups of C57BL/6 mice were intracerebrally inoculated with RML and given the vehicle (saline) or 10 mg/kg Zn(II)-BnPyP ip every second day starting from 40 days post-inoculation (dpi) in order to reach a bioactive concentration at approximately 120 dpi, which is ∼80% of the time to terminal disease endpoint. There was no difference between vehicle- and Zn(II)-BnPyP-treated mice in time to disease onset (median 141.5 versus 135 dpi; Figure 6A) or terminal disease (median 153 versus 152 dpi; Figure 6B). There was also no difference in the amount of PK-resistant PrPSc, or its degree of protease resistance, between vehicle- and Zn(II)-BnPyP-treated mice culled at the terminal stage of disease (Figure 6C).
Figure 6.
Zn(II)-BnPyP does not affect RML-induced prion disease
(A and B) Kaplan-Meier plots showing time to onset (A) and to terminal disease (B) of C57BL/6 mice intracerebrally inoculated with RML and treated ip with saline (vehicle; n = 6) or 10 mg/kg Zn(II)-BnPyP (n = 9) every second day from 40 dpi.
(C) Representative WB analysis of PK-resistant PrP in the brains of RML-inoculated mice, treated with vehicle or Zn(II)-BnPyP and euthanized at the terminal stage of the disease. Brain extracts were digested with 0, 5, 25, and 50 μg/mL of PK and analyzed by WB with anti-PrP 12B2 monoclonal antibody. The undigested samples (0 μg/mL PK) amount to 10 μg of protein, and the other samples 20 μg.
Discussion
Zn(II)-BnPyP binds to distinct domains of native PrPC, producing biological effects through a bimodal mechanism of action: it makes PrPC less prone to PrPSc conversion and it triggers endocytosis and lysosomal destruction of cell surface PrPC, reducing the substrate for PrPSc formation. Because it targets the prion precursor, Zn(II)-BnPyP is active against different prion strains. To our knowledge, this is the first evidence of a compound that can bind different PrPC regions, eliciting a dual anti-prion effect. The challenge for future studies will be to exploit this mechanism of action to obtain therapeutic effects in vivo.
Zn(II)-BnPyP binding to the C-terminal globular domain reduces its thermal stability, as shown in CD melting experiments, while Fe(III)-TMPyP increases it. This difference may be attributable to the different binding modalities of the two porphyrins: Fe(III)-TMPyP fills a shallow cleft made by the C terminus of the α3 helix and the first β-strand, while Zn(II)-BnPyP docks onto the opposite face, on the surface comprising the α2 and α3 helices, and establishes hydrophobic interactions with the aromatic side chains of Y226 and Y225, most likely destabilizing the hydrophobic core of the globular domain. Moreover, in contrast to Fe(III)-TMPyP, which binds only to the PrPC globular domain, Zn(II)-BnPyP also interacts with the flexible N-terminal tail through Zn(II)-mediated coordination of the OR histidine side chains (H61, H69, H77 and H85), and π-π interactions between the BnPyP scaffold and the OR tryptophan side chains (W65, W73, W81, W89).
There is evidence that the PrPC N-terminal tail establishes tertiary contacts with the C-terminal globular domain.4 SAXS data analysis indicates that, differently from Fe(III)-TMPyP, which had no effect on the overall PrPC conformation, Zn(II)-BnPyP shifts PrPC conformational equilibrium toward more open structures. Conceivably Zn(II)-BnPyP, through its ability to engage the OR region, counteracts the intramolecular interactions between the two domains of the protein, favoring a less compact fold.
Zn(II)-BnPyP’s ability to interact with both the PrPC N-terminal tail and the globular domain may explain its dual anti-prion activity. Zn(II)-BnPyP inhibits cell-free PrPC to PrPSc conversion irrespective of whether the reaction employs PrPC full-length or lacking the OR domain, indicating that binding to the C-terminal globular domain is sufficient for this effect. Fe(III)-TMPyP too inhibits PrPSc replication in PMCA by binding to the globular domain, but it stabilizes the fold whereas Zn(II)-BnPyP destabilizes it. Therefore, Zn(II)-BnPyP must act through a different mechanism from Fe(III)-TMPyP and other pharmacological chaperones.27,43
PrPC to PrPSc conversion is thought to occur through a refolding process involving a partially structured PrP intermediate. Biophysical studies identified several partially folded PrP conformers, in addition to the native and fully unfolded states, which may represent the monomeric precursor on the pathway to PrPSc conversion.44 These generally present rearrangements of the α2 and α3 helices, the region of PrPC globular domain engaged by Zn(II)-BnPyP. The porphyrin may conceivably boost the local dynamics of this region, shifting the conformational equilibrium toward an off-pathway state. Alternatively, Zn(II)-BnPyP may block the α2 and α3 rearrangements leading to the PrPSc state. Regardless of the exact molecular mechanism, these results indicate that prion propagation can be inhibited not only by pharmacological chaperones that stabilize the native PrPC fold, but also by ligands that weaken the protein stability, and identify the α2 and α3 pocket recognized by Zn(II)-BnPyP as a new target region for in silico screening of anti-prion drugs.
Our observations indicate that Zn(II)-BnPyP binding to the flexible N-terminal tail is necessary to trigger clathrin-mediated internalization and lysosomal destruction of cell surface PrPC. Fe(III)-BnPyP, which cannot coordinate the OR histidines and binds only to the globular domain, does not lower PrPC levels. Moreover, PrP H/W→A molecules, in which the OR histidines and tryptophans mediating Zn(II)-BnPyP binding to the N-terminal tail are replaced with alanines, are not downregulated by the porphyrin.
Cu(II) binds the PrPC OR with high affinity45 and promotes tertiary contacts between the N- and C-terminal domains like Zn(II),6 raising the question of whether Cu(II)-BnPyP had anti-prion properties like Zn(II)-BnPyP. Preliminary data indicate that Cu(II)-BnPyP binds the N-terminal tail, and down-regulates PrPC and inhibits PrPSc replication like Zn(II)-BnPyP (Figure S13).
The natural porphyrin hemin was shown to interact with the N-terminal region of PrPC and cause aggregation, endocytosis and lysosomal degradation of cell surface PrPC.46 Likewise, it has been recently found that the anti-PrP antibody POM2, which recognizes repetitive epitopes in the OR region, stimulates multimeric clustering of cell surface PrPC, followed by endocytic uptake and lysosomal degradation of the antibody-PrPC complex.47 This depends on POM2’s ability to crosslink PrPC on the cell surface, as it is not seen when cells are treated with the single-chain form of POM2. Thus, ligands that induce clustering of cell surface PrPC by interacting with the N-terminal tail may eventually promote the protein internalization and turnover. Several cyclic tetrapyrroles have an intrinsic tendency to assemble into supramolecular aggregates in aqueous solutions, a property that correlates with their anti-prion activity and may mediate PrPC multimerization.29,48
Our findings indicate that Zn(II)-BnPyP does not self-aggregate (Figure S14), and stimulates PrPC endocytosis and degradation through a mechanism not involving the formation of PrPC multimers. First, we did not observe any PrPC clustering on the surface of Zn(II)-BnPyP-treated cells; second, Zn(II)-BnPyP did not induce PrPC oligomerization, as assessed by a detergent insolubility assay; finally, NMR spectroscopy gave no evidence of huPrP23-231 aggregation in the presence of Zn(II)-BnPyP.
Cu(II) and Zn(II) ions induce the internalization and recycling of PrPC back to the cell surface, stimulating a PrPC endocytic pathway that normally operates constitutively in the absence of metal.3 Zn(II)-BnPyP instead stimulates lysosomal destruction of endocytosed PrPC. This, and the fact that Zn(II)-BnPyP destabilizes and opens the protein conformation – unlike Cu(II) and Zn(II) ions which promote a more compact PrPC fold5,6,7,8 – suggest that Zn(II)-BnPyP may activate a plasma membrane quality control pathway for selective internalization and lysosomal destruction of PrPC. We do not yet know the mechanisms that enables the cell to distinguish folded from misfolded GPI-anchored proteins at the cell surface. Zn(II)-BnPyP may be useful for investigating how the cell detects structurally challenged GPI-anchored PrPC.
Limitations of the study
Zn(II)-BnPyP has strain-independent anti-prion activity in PMCA, prion-infected cells and COCS, outcompeting Fe(III)-TMPyP and PPS, two of the most potent anti-prion compounds described so far. However, like other porphyrins, Zn(II)-BnPyP does not cross the BBB efficiently, and a 12-week chronic treatment is required to reach a brain concentration of ∼1.5 μM, which engages the target, as shown by the ∼20% reduction in PrPC, but is not sufficient to prolong the survival of mice in the advanced stage of prion infection close to symptomatic disease. Future studies may test whether starting the treatment just after infection achieves bioactive brain concentrations earlier and delays disease. Alternatively, to bypass the BBB, Zn(II)-BnPyP could be administered directly into the CNS via a head pump. However, preliminary attempts to deliver Zn(II)-BnPyP into the mo brain ventricle by either bolus injection or chronic infusion with osmotic minipumps have been unrewarding due to the poor porphyrin diffusion into the brain parenchyma. Therefore, Zn(II)-BnPyP pharmacokinetic properties will have to be improved through chemical modification, or its BBB penetration boosted using brain-targeted nanocarriers. However, a more drug-like molecule would be desirable. Zn(II)-BnPyP presents a starting point for the design of small molecules with a similar mode of action. Given the discovery that PrPC acts as receptor for a variety of neurotoxic protein oligomers, including amyloid-β, α-synuclein and tau,49 compounds downregulating cell surface PrPC like Zn(II)-BnPyP may be able to prevent the toxic signaling mediated by these interactions, with broad therapeutic benefits in neurodegenerative diseases of protein aggregation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-actin clone C4 | Millipore | Cat# MAB1501 |
| anti-APP | Invitrogen | Cat# PA14648 |
| anti-LAMP1 | Abcam | Cat# ab24170 |
| anti-MAP2 | Abcam | Cat# ab5392 |
| anti-PrP 3F4 | Provided by Dr R.J. Kascsak, New York State Institute for Basic Research, Staten Island, NY, U.S.A. / Millipore | Cat#: MAB1562 (Millipore) |
| anti-PrP 6D11 | Provided by Dr R.J. Kascsak, New York State Institute for Basic Research, Staten Island, NY, U.S.A. | RRID:AB_1027165 |
| anti-PrP 8A4 | Provided by S.J. Collins, University of Melbourne, Parkville, Melbourne, VIC, Australia | N/A |
| anti-PrP 11G5 | Provided by Man Sun-Sy, Case Western Reserve University, Cleveland, OH, USA | N/A |
| anti-PrP 12B2 | Wageningen University and Research | Cat# 12B2/500 |
| anti-PrP 94B4 | Wageningen University and Research | Cat# 94B4/500 |
| anti-PrP D18 | Provided by E. Biasini, University of Trento, Italy | N/A |
| anti-PrP SAF83 | Vitro S.A. | Cat# SPI-A03207.200 UG |
| anti-puromycin | Zymed | Cat# 51-1600 |
| anti-α-synuclein | Thermo Fisher | Cat# 35-8300 |
| anti-total tau | Dako | Cat# A0024 |
| anti-vinculin | Sigma-Aldrich | Cat# V9264 |
| Bacterial and virus strains | ||
| E. coli BL21(DE3) | EMD Millipore | Cat# 70954 |
| Max efficiency DH5α competent cells | Invitrogen | Cat# 18258012 |
| Biological samples | ||
| sCJD subtype VV2 brain homogenate | From the Basque Biobank, Spain | N/A |
| vCJD brain homogenate | From Biobanco Hospital Universitario, Fundación Alcorcón, Spain |
N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 5,10,15,20-Tetrakis(4-trimethylammoniophenyl)porphyrin tetra(p-toluenesulfonate) | Merck Life Science | CAS No.: 69458-20-4 |
| Ammonium chloride (15N, 99%) | Cambridge Isotope Laboratories, Inc. | Cat# NLM-467-10 |
| Ampicillin sodium salt | Sigma-Aldrich | Cat# A9518 |
| Bafilomycin A1 | Sigma-Aldrich | Cat# B1793 |
| Biotin | Sigma-Aldrich | Cat# B4501 |
| Brefeldin A | Sigma-Aldrich | Cat# B7651 |
| Calcium chloride dihydrate | Sigma-Aldrich | Cat# 31307 |
| Chloroquine | Sigma-Aldrich | Cat# C6628 |
| cOmplete, EDTA-free | Roche | Cat# 05056489001 |
| Cycloheximide | Sigma-Aldrich | Cat# C4859 |
| Deoxyribonuclease I from bovine pancreas | Sigma-Aldrich | Cat# DN25 |
| Deuterium oxide (D, 99.9%) | Cambridge Isotope Laboratories, Inc. | Cat# DLM-4-10X0.7 |
| Diethyl ether | Carlo Erba Reagents | Cat# 340751 |
| Diisopropylcarbodiimide | Sigma-Aldrich | Cat# 38370 |
| Dimethylformamide | Carlo Erba Reagents | Cat# 528221 |
| DMEM High glucose | Sigma-Aldrich | Cat# D5671 |
| Fe(III) 5,10,15,20-tetra(N-methyl-4-pyridyl)porphyrin pentachloride | Frontiers scientific | CAS No.: 133314-07-5 |
| Fetal bovine serum | Sigma-Aldrich | Cat# F7524 |
| Fetal bovine serum, defined | HyClone | Cat# SH30070.02 |
| Fetal bovine serum, dyalized | Life technologies | Cat# 26400-044 |
| Fluorenylmethyloxycarbonyl chloride | Sigma-Aldrich | Cat# 23186 |
| D-glucose (U13C6, 99%) | Cambridge Isotope Laboratories, Inc. | Cat# CLM-1396-10 |
| L-Glutamine | Sigma-Aldrich | Cat# G7513 |
| Guanidine hydrochloride | Sigma-Aldrich | Cat# G3272 |
| HaloTag Alexa Fluor 488 cell-impermeant ligand | Promega | Cat# G1001 |
| Hank's balanced salt solution | Gibco | Cat# 14170-088 |
| HEPES | Sigma-Aldrich | Cat# H3375 |
| Hygromycin | Invitrogen | Cat# 10687010 |
| Imidazole | Sigma-Aldrich | Cat# 792527 |
| Isopropyl b-D-1-thiogalactopyranoside | Giotto Biotech | Cat# 367-93-1 |
| LB broth | Sigma-Aldrich | Cat# L3522 |
| Lysozyme from chicken egg white | Sigma-Aldrich | Cat# L6876 |
| Magnesium sulfate heptahydrate | Sigma-Aldrich | Cat# 230391 |
| Ni-NTA agarose | Qiagen | Cat# 30230 |
| Nystatin | Sigma-Aldrich | Cat# N1638 |
| Oxyma | Sigma-Aldrich | Cat# 8510860250 |
| Papain | Sigma-Aldrich | Cat# P3125 |
| Penicillin/Streptomycin | Sigma-Aldrich | Cat# P4333 |
| Phenylmethylsulphonyl fluoride | Merck | Cat# 52332 |
| Phosphate buffer saline | Life technologies | Cat# 10010056 |
| Phosphatidylinositol-specific phospholipase C (PIPLC) | Invitrogen | Cat# P6466 |
| Pitstop 2 | Sigma-Aldrich | Cat# SML1169 |
| Poly-L-lysine | Sigma-Aldrich | Cat# P2636 |
| Proteinase-K | Roche | Cat# 03115801001 |
| Puromycin | Sigma-Aldrich | Cat# P8833 |
| Sodium acetate | Sigma-Aldrich | Cat# S2889 |
| Sodium chloride | Sigma-Aldrich | Cat# S3014 |
| Sodium phosphate monobasic | Sigma-Aldrich | Cat# 71496 |
| Sodium phosphate dibasic | Carlo Erba Reagents | Cat# 480141 |
| Thiamine | Sigma-Aldrich | Cat# T-4625 |
| Trifluoroacetic acid | Carlo Erba Reagents | Cat# 411543 |
| Triisopropylsilane | Sigma-Aldrich | Cat# 233781 |
| Trypsin-EDTA | Sigma-Aldrich | Cat# T4174 |
| Trypsin inhibitors | Sigma-Aldrich | Cat# T9003 |
| Trizma base | Sigma-Aldrich | Cat# T1503 |
| Zn(II) 5,10,15,20-tetra(N-methyl-4-pyridyl)porphyrin tetrachloride | Frontiers scientific | CAS No.: 28850-44-4 |
| Critical commercial assays | ||
| Clarity Western ECL Substrate | Bio-rad | Cat# 170-5061 |
| Fugene HD | Promega | Cat# E2311 |
| GoTaq qPCR Master Mix | Promega | Cat# A6002 |
| High Capacity cDNA Reverse Transcription Kit | Applied biosystem | Cat# 4368814 |
| Pierce™ BCA protein assay kit | Thermo Fisher | Cat# 23227 |
| RNeasy maxi kit | Qiagen | Cat# 75162 |
| RNeasy mini kit | Qiagen | Cat# 74104 |
| Deposited data | ||
| Raw data | This paper: Mendelay data | https://doi.org/10.17632/r6s3krtzt4.1 |
| SAXS data | Small Angle Scattering Biological Data Bank (SASBDB) | SASDNB8, SASDNC8, SASDND8 |
| Experimental models: Cell lines | ||
| H4 | ATCC | Cat# HTB-148 |
| HEK293 | ATCC | Cat# CRL-1573 |
| HeLa | ATCC | Cat# CCL-2 |
| N2a | ATCC | Cat# CCL-131 |
| ScN2a-RML | Provided by E. Biasini (Biggi et al.50https://doi.org/10.1111/jnc.14805) | N/A |
| ScN2a-22L | Provided by S. Lehmann (Nishida et al.51https://doi.org/10.1128/jvi.74.1.320-325.2000) | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | Charles River | RRID:IMSR_JAX:000664 |
| Mouse: Tg(CJD-66) | Generated by the Chiesa lab (Dossena et al.52https://doi.org/10.1016/j.neuron.2008.09.008; Bouybayoune et al.53 https://doi.org/10.1371/journal.ppat.1004796) |
RRID:IMSR_EM:12879 |
| Oligonucleotides | ||
| Table S2 | This paper | N/A |
| Recombinant DNA | ||
| pCDNA3.1(+) bvPrPI109 | Provided by J.C. Watts (Watts et al.54https://doi.org/10.1007/s00401-016-1585-6) | N/A |
| pCDNA3.1(+) bvPrPM109 | Provided by J.C. Watts (Watts et al.54https://doi.org/10.1007/s00401-016-1585-6.) | N/A |
| pCDNA3.1(+) moPrPΔ23-89 | Provided by D.A. Harris, Boston University, Boston, MA, USA | N/A |
| pCDNA3.1(+) moPrPΔ23-111 | Provided by D.A. Harris (Turnbaugh et al.55https://doi.org/10.1371/journal.pone.0025675) | N/A |
| pCDNA3.1(+) moPrP-EGFP | Massignan et al.56https://doi.org/10.1074/mcp.M900271-MCP200 | N/A |
| pCDNA3.1(+) moPrPKDEL | Senatore et.57https://doi.org/10.1016/j.neuron.2012.02.027 | N/A |
| pCDNA3.1(+) moPrP H/W -> A | This paper | N/A |
| pCDNA3.1(+) moPrP-Halo | This paper | N/A |
| pCDNA3.1(+) moPrPwt | Provided by D.A. Harris (Turnbaugh et al.55https://doi.org/10.1371/journal.pone.0025675) | N/A |
| pOPINE bvPrPΔ52-92 (I109) | This paper | N/A |
| pOPINE bvPrP23-231 (I109) | Generated in the Castilla lab (Fernández-Borges et al.58https://doi.org/10.1007/s00401-017-1782-y) | N/A |
| pOPINE huPrP23-231 (M129) | Generated in the Castilla lab (Elezgarai et al.59https://doi.org/10.1038/s41598-017-09489-3) | N/A |
| pET-41 huPrP90-231 (M129) | Provided by B. Caughey, Rocky Mountain Laboratories, Hamilton, MT, USA | N/A |
| Software and algorithms | ||
| ATSAS 3.2.1 | EMBL | https://www.embl-hamburg.de/biosaxs/software.html |
| BioRender | biorender.com | N/A |
| CCPN Analysis 2.4.2 | https://www.ccpn.ac.uk/v2-software/software/analysis | RRID:SCR_016984 |
| Empower 2.0 | Waters | https://www.waters.com |
| Fiji | NIH | RRID:SCR_002285 |
| Glide | Schrodinger, LLC | RRID:SCR_000187 |
| Graphpad Prism 9 | Graphpad | RRID:SCR_002798 |
| ImageLab 6.0 | Bio-rad | RRID:SCR_014210 |
| JaCoP | Fabrice P. Cordelières, Bordeaux Imaging Center (France) - Susanne Bolte, IFR 83, Paris (France) - https://imagej.net/plugins/jacop | N/A |
| Kaleido software | Perkin Elmer | N/A |
| PyMOL 1.8.4.0 | Schrodinger, LLC | RRID:SCR_000305 |
| TopSpin 4.0.8 | Bruker BioSpin GmbH | RRID:SCR_014227 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Roberto Chiesa (roberto.chiesa@marionegri.it).
Materials availability
Plasmids and other reagents generated in this study will be made available upon request to the lead contact, Roberto Chiesa (roberto.chiesa@marionegri.it).
Experimental model and study participant details
In this study, we utilized newborn mice to prepare primary neurons and cultured organotypic cerebellar slices, and eight weeks old male and female mice for pharmacokinetics and inoculation studies. Procedures involving animals and their care were conducted in conformity with the institutional guidelines at the Istituto di Ricerche Farmacologiche Mario Negri IRCCS in compliance with national (D.lgs 26/2014; Authorization no. 19/2008-A issued March 6, 2008 by Ministry of Health) and international laws and policies (EEC Council Directive 2010/63/UE; the NIH Guide for the Care and Use of Laboratory Animals, 2011 edition). They were reviewed and approved by the Mario Negri Institute Animal Care and Use Committee, which includes ad hoc members for ethical issues, and by the Italian Ministry of Health (Decreto no. 212-2016-PR, 363-2020-PR, 656-2021-PR and 9F5F5.N.ULW). Animal facilities meet international standards and are regularly checked by a certified veterinarian who is responsible for health monitoring, animal welfare supervision, experimental protocols and review of procedures.
Method details
Porphyrin synthesis
Zn(II) 5,10,15,20-tetra(N-benzyl-4-pyridyl)porphyrin tetrachloride was synthesized as previously reported.31 500 mg (0.73 mmol) of Zn(II) 5,10,15,20-tetra(4-pyridyl)porphyrin and 3.5 mL of benzylchloride (30 mmol) were reacted in 60 mL of dimethylformamide to obtain 737 mg of the desired product (85% yield). C68H52Cl4N8Zn Mw 1188.46. UV-vis(H2O): ε426 nm 165,000 M-1 cm-1. 1H NMR (D2O) δ: 6.08 (s, 8H); 7.60 (m, 12H); 7.73 (m, 8H); 8.89 (d, 16H); 9.34 (d, 8H). HPLC: retention time 16′54″ (100%).
Fe(III) 5,10,15,20-tetra(N-benzyl-4-pyridyl)porphyrin pentachloride was produced by reacting 50 mg (0.07 mmol) of Fe(III) 5,10,15,20-tetra(4-pyridyl)porphyrin chloride and 330 μL benzylchloride (2.8 mmol) as above; 28 mg of product were obtained (0.023 mmol; yield 32.8%). C68H52Cl5N8Fe Mw 1214.38. UV-vis(H2O): ε422 nm 145,700 M-1 cm-1. HPLC: retention time 15′41″ (100%).
Cu(II) 5,10,15,20-tetra(N-benzyl-4-pyridyl)porphyrin tetrachloride was produced by reacting 25 mg (0.037 mmol) of Cu(II) 5,10,15,20-tetra(4-pyridyl)porphyrin and 160 µL benzylchloride (1.4 mmol) as above; 19.9 mg of product were obtained (0.017 mmol; yield 45.9%). C68H52N8CuCl4 Mw 1186.63. UV-vis(H2O): ε429 nm 132,500 M−1 cm-1. HPLC: retention time 17′03″ (100%).5,10,15,20-tetra(N-benzyl-4-pyridyl)porphyrin tetrachloride was synthetized as previously reported.31
UV-vis absorption spectra were measured with a Jasco V-560 spectrophotometer. 1H NMR spectra were recorded on a Bruker 500 MHz spectrometer in CDCl3; chemical shifts, expressed in ppm, are reported as s (singlet), d (doublet), and m (multiplet). HPLC analyses were done with an Agilent 1100 Series instrument coupled with a diode array detector. The instrument was fitted with a 250×4.6 mm column (Supelco, Ascentis) packed with C-18 reversed-phase particles (5 μm), operating with 1.5 mL/min flux and the following elution gradient: A - water 20 nM trifluoroacetic acid + 20 mM triethylamine (pH 2.70); B – acetonitrile. Initial A/B 90/10 (5 min); then to A/B 10/90 in 30 min.
Fe(III) 5,10,15,20-tetra(N-methyl-4-pyridyl)porphyrin pentachloride, Cu(II) 5,10,15,20-tetra(4-pyridyl)porphyrin and Zn(II) 5,10,15,20-tetra(N-methyl-4-pyridyl)porphyrin tetrachloride were from Frontier Scientific.
Plasmids and transfection
pCDNA3.1 plasmids encoding mouse PrP wt, PrP-EGFP, PrPKDEL, PrPΔ23-89, PrPΔ23-111, bvPrPI109 and bvPrPM109 have been described.54,55,56,57,60,61 Mouse PrP-Halo, containing the HaloTag sequence with a GS linker (GGGGS, repeated four times) at its 5’ end, inserted after codon 229 of mouse PrP, and PrP H/W→A in which the OR histidines and tryptophans were substituted with alanines, were synthesized by GeneArt (Thermo Fisher) and inserted between the HindIII and BamHI restriction sites of pCDNA3.1(+). Cells were transfected using FuGENE HD (Promega).
Cell lines and primary neurons
N2a, ScN2a-RML,50 ScN2a-22L,51 H4, HEK293 and HeLa cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Sigma-Aldrich) supplemented with 1% L-glutamine (Sigma-Aldrich), 10% fetal bovine serum Δ56 (FBS, Sigma-Aldrich), 100 U/ml penicillin/streptomycin (PenStrep, Sigma-Aldrich), at 37°C in 5% CO2/95% air. Stably transfected clones of HeLa expressing PrP-EGFP and HEK293 expressing PrP-Halo were selected by limiting dilution with 400 μg/mL Hygromycin (Invitrogen) and were then maintained in 200 μg/mL Hygromycin. ScN2a-22L51 were cultured with 200 μg/mL G418 (Life technologies). Murine primary hippocampal and cerebellar granule neurons were prepared from C57BL6/J (RRID:IMSR_JAX:000664) or Tg(CJD-66+/-)/Prnp0/0 (RRID:IMSR_EM:12879) mice.52,53,62 Hippocampal neurons were prepared from 1–2-day-old mice. Animals were euthanized and hippocampi were isolated under a surgical stereomicroscope. Tissues were digested for 30 min at 34°C with papain (200 U/mL) in CNDM medium (5.8 mM MgCl2, 0.5 mM CaCl2, 3.2 mM HEPES, 0.2 mM NaOH, 30 mM K2SO4 and 90 mM Na2SO4 pH 7.4, 292 mOsm) supplemented with 0.4% glucose. Enzymatic activity was blocked with trypsin inhibitors (10 μg/mL, Sigma) in CNDM plus 0.4% glucose for 45 min at room temperature (RT). Mechanical dissociation was done in MEM (Invitrogen) supplemented with 10% FBS (HyClone) and 0.4% glucose. 0.5 × 105 cells per cm2 were plated on polylysine-coated plates in the same medium with PenStrep (Sigma, 100 U/mL). After attachment, culture medium was switched to Neurobasal-A (Invitrogen) supplemented with 2% B27 (Invitrogen), 200 mM glutamine, and 100 U/mL PenStrep. Medium was changed after 7 days in culture.
Cerebella were dissected from 6–8-day-old mice, sliced into ∼1-mm pieces and incubated in Hank’s balanced salt solution (HBSS, Sigma) containing 0.3 mg/ml trypsin (Sigma) at 37°C for 15 min. Trypsin inhibitor (Sigma) was added to a final concentration of 0.5 mg/mL and the tissue was mechanically dissociated by passing through a flame-polished Pasteur pipette. Cells were plated at 2.5-3 × 105 cells/cm2 on poly-L-lysine (0.1 mg/mL)-coated plates. Cells were maintained in DMEM High Glucose (Sigma) supplemented with 10% dialyzed fetal bovine serum (FBS, Life Technologies), penicillin/streptomycin and KCl 25 mM, at 37°C in an atmosphere of 5% CO2, 95% air.
Cell treatments
Porphyrins were dissolved in sterile water at the concentration of 1 mM in LoBind tubes (Eppendorf) using a Vortex mixer and sonication (30 sec at maximum power in a Bandelin HD 2200 cup sonicator). The stock solutions were kept at 4°C in the dark and sonicated before each use. To block clathrin-mediated endocytosis cells were incubated in hypertonic medium containing 120 mM NaCl, 12 mM MgSO4, 1 mM EDTA, 1% bovine serum albumin (BSA, Sigma-Aldrich), 100 mM HEPES pH 7.0, 5 mM KCl, 0.4 M sucrose (Sigma-Aldrich).39 Pharmacological inhibitors were used at the following final concentrations: 50 nM Bafilomycin A1 (Sigma-Aldrich), 50 μg/mL cycloheximide (Sigma-Aldrich), 20 μM chloroquine (Sigma-Aldrich) and 1 μM Brefeldin A (Sigma-Aldrich), 100 U/mL Nystatin (Sigma-Aldrich), 50 μM Pitstop 2 (Sigma-Aldrich). For all treatments half the culture medium was replaced with fresh medium containing the compounds at double their final concentrations; control cells were treated by replacing half the culture medium with fresh medium containing the vehicle.
To rule out any confounding effects due to the porphyrin photodynamic activity, we avoided exposing treated cells and cell extracts to direct light. We worked in laboratories with shaded natural light and during cell culture experiments, we turned the tissue culture cabinet lights off and checked the treated cells by phase contrast microscopy at low light intensity for the shortest possible time. Preliminary analysis indicated that these precautions were sufficient to avoid reactive oxygen species (ROS)-mediated cell toxicity or protein degradation.
Cultured organotypic cerebellar slices
Prion-infected cultured organotypic cerebellar slices were prepared from 9-10 day-old C57BL/6J (RRID:IMSR_JAX:000664) mice and cultured on 30-mm diameter Millicell Cell Culture Inserts (Millipore) (4 slices each).63 COCS were treated after 5 weeks in culture by replacing the culture medium with fresh medium containing the porphyrin; for chronic treatment this was repeated every second day. At the end of the treatment COCS were washed three times with ice-cold phosphate buffered saline (PBS, Life Technologies), detached from the cell culture insert by gentle pipetting in ice-cold PBS and collected in 1.5 mL Eppendorf tube. After 1-min centrifugation at 10,600xg, the PBS was discarded and the pelleted slices were lysed in 120 μL of 0.5% sodium deoxycholate, 0.5% Nonidet P-40 (Igepal) in PBS by Vortex mixing, three freeze/thaw cycles using ethanol/dry ice and a water bath at 37°C, and homogenization using polypropylene Pellet Pestles (Merck) attached to a cordless motor. After centrifugation at 10,600xg for 1 min, the supernatant was collected and PK-resistant PrP was analyzed by WB.63
RT-qPCR
Total RNA was extracted with RNeasy kit (Qiagen). One μg RNA was reverse-transcribed with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) using random primers. Quantitative PCR was done in Optical 96- or 384-well plates (Applied Biosystems) using 7900 Real-Time PCR System (Applied Biosystems) and GoTaq qPCR Master Mix (Promega). Reaction conditions were 50°C for 2 min, 95°C for 10 min, then 95°C for 15 s alternating with 60°C for 1 min for 41 cycles, followed by 95°C for 15 s and 60°C for 15 s. The amplifications were always run in triplicate. The primer pair sequences are reported in Table S2.
Biochemical analysis
Global protein synthesis was analyzed using the SUnSET method.64 N2a cells were treated with 5 μM Zn(II)-BnPyP or the vehicle, and after 16h puromycin (10 μg/mL) was added to the culture medium. After 60 min cells were lysed 50 mM Tris-HCl pH 7.4, 1% SDS and analyzed by WB with anti-puromycin and anti-actin antibodies. As control for inhibition of protein synthesis cyclohexamide (CHX, 50 μg/mL) was added 10 minutes before puromycin.
PrP molecules released in the culture medium were analyzed as described.65 Stably transfected HEK293 cells expressing moPrP were left untreated, or treated with 5 μM Zn(II)-BnPyP or 1 U/mL phosphatidylinositol-specific phospholipase C (PIPLC) from Bacillus thuringiensis (Invitrogen) for 6h in OptiMEM. Conditioned media were collected and cleared of cell debris by centrifugation at 10,000xg for 1 min, and proteins were precipitated by the addition of 5 volumes of methanol. Cells were lysed in 50 mM Tris-HCl pH 7.4, 1% SDS. Cell lysates corresponding to 50 μg of proteins and all proteins from the media were analyzed by WB with anti-PrP antibody.
Detergent insolubility of PrP was measured as described.66 Cells were lysed in LB (lysis buffer: 0.5% Triton X-100, 0.5% Na deoxycholate in 5 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA) containing a protease inhibitor cocktail (Roche) and cleared by centrifugation at 10,000xg for 1 min. Lysates corresponding to 100 μg of protein were diluted in 300 μL of lysis buffer and centrifuged at 100,000xg for 45 min in a Beckman Optima Max-E ultracentrifuge using a TLA-55 rotor. Proteins in the pellet and supernatant were analyzed by WB.
To assay PK-resistant PrP, prion-infected N2a cells were lysed in LB. After centrifugation at 10,000xg for 1 min, cell extracts corresponding to 100 μg of total proteins were incubated with 10 μg/mL PK in a final volume of 400 μL for 30 min at 37°C. PK digestion was terminated by adding 5 mM phenylmethylsulphonyl fluoride (PMSF, Merck) with 5 min incubation at 4°C. Proteins were methanol-precipitated overnight at -20°C. The precipitated proteins were dissolved in 20 μL Laemmli sample buffer (LMSB) 2X and analyzed by WB. The undigested samples represent 50 μg of protein; the PK-digested samples 100 μg.
Western blot
Cells were lysed in 50 mM Tris-HCl pH 7.4, 1% SDS, then incubated at 95°C for 5 min. Proteins were separated by 12% SDS-PAGE and electro-transferred onto polyvinylidene fluoride or nitrocellulose membranes (Bio-Rad) by Trans-blot turbo (Bio-Rad). Protein transfer was checked by Ponceau S staining (Sigma-Aldrich) followed by washings in 100 mM Tris pH 7.5, 150 mM NaCl and 0.1% Tween 20 (TTBS) and blocking in 5% non-fat dry milk (NFDM, Sigma-Aldrich) in TTBS for 30 min. Membranes were incubated overnight at 4°C or 1h at room temperature with the following antibodies: mouse monoclonal antibodies (mAb) anti-PrP 3F4 and 6D1167,68,69 (provided by Dr R.J. Kascsak, New York State Institute for Basic Research, Staten Island, NY, U.S.A.; 1:5000), 12B2 and 94B4 (WUR; 1:5000), 11G570 (provided by Man Sun-Sy; 1:5000), and 8A471 (provided by S.J. Collins, University of Melbourne, Parkville, Melbourne, VIC, Australia; 1:5000); rabbit polyclonal anti-APP (Invitrogen PA14648; 1:1000); rabbit polyclonal anti-total tau (Dako A0024; 1:1000); mAb anti-α-synuclein (Thermo Fisher 35-8300; 1:1000); mAb anti-actin (Millipore MAB1501 clone C4; 1:10000); mAb anti-vinculin (Sigma-Aldrich V9264; 1:5000); and rabbit polyclonal anti-puromycin (Zymed 51-1600; 1:1000). All antibodies were diluted in 5% non-fat dry milk (Sigma-Aldrich) in TTBS, then rinsed three times with TTBS and incubated for 45 min with secondary antibody goat anti-mouse diluted 1:5000 conjugated with horseradish peroxidase (Bio-Rad). The chemiluminescent signal was revealed using Clarity Western ECL Substrate (Bio-Rad) and acquired with a Chemidoc Imager MP (Bio-Rad). Quantitative densitometry of protein bands was done using ImageLab 6.0 software (Bio-Rad) by selecting regions of interested (ROI) including all PrP glycoforms and the internal standard (actin or vinculin). An example of quantitative analysis of western blot is given in Figure S15.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde (Sigma-Aldrich) for 10 min at room temperature (RT) and incubated in blocking solution (0.05% saponin, 0.5% BSA, 50 mM NH4Cl and 10% FBS in PBS) for 30 min at RT. The cells were then incubated with anti-PrP mAb 12B2 (WUR; 1:500), rabbit polyclonal anti-LAMP1 (Abcam ab24170; 1:1000), chicken polyclonal anti-MAP2 (Abcam ab5392; 1:50000) primary antibodies diluted in blocking solution, for 1h at RT, or overnight at 4°C, then rinsed three times in PBS, and incubated with a fluorescent conjugated anti-IgG secondary antibody (Invitrogen) for 1h at RT.
Images were acquired using an Olympus FV-500 or Olympus IX81 microscope with a 60X oil-immersion objective. Super-resolution images were acquired using a Structured Illumination Microscope Nikon N-SIM with a 100X oil-immersion objective. Images were acquired under nonsaturating conditions and analyzed with NIH ImageJ software. Mander’s correlation coefficient was measured using ImageJ and JaCoP plugin.72
PrP-Halo labeling
To label cell surface PrP-Halo, cells were incubated for 15 min at 37°C with the HaloTag Alexa Fluor 488 cell-impermeant ligand (Promega G1001) diluted 1:1000 in the culture medium, then rinsed three times with PBS. PrP-Halo fluorescence was analyzed by structured illumination microscopy, or in-gel after SDS-PAGE, using a Chemidoc Imager MP (Bio-Rad), and quantitative densitometry of protein bands with ImageLab 6.0 software (Bio-Rad).
In vitro prion propagation
Brain PMCA was done as described,73 using serial dilutions of the following PrPSc seeds, consisting of 10% brain homogenates (w/v) in PBS containing protease inhibitors (Roche): vCJD (from Biobanco Hospital Universitario, Fundación Alcorcón, Spain), sCJD subtype VV2 (from the Basque Biobank, Spain), PMCA-adapted classical scrapie (Dawson strain, provided by Olivier Andreoletti, INRAE, Toulouse) in Tg338,59 and a de novo PMCA-generated bank vole prion. The PrPC substrates were 10% (w/v) brain homogenates from cattle, TgZWV,74 Tg338 and TgVole mice. After PMCA, samples were diluted 1:4 in digestion buffer [1% Tween-20 (Sigma-Aldrich), 1% NP-40 (Sigma-Aldrich) and 5% Sarkosyl (Sigma-Aldrich) in PBS (Fisher BioReagents)] and incubated with 85 μg/mL of PK (Roche) for 1h at 56°C with agitation at 450 rpm, and PrP was immunodetected by WB59 using mAb SAF83 (Vitro S.A.; 1:400) for TgVole, D1875 (a kind gift from Emiliano Biasini, Univ. of Trento; 1:5000) for cattle and Tg338, 3F4 (Millipore; 1:10000) for TgZWV.
Recombinant prion propagation was done by protein misfolding shaking amplification (PMSA).73 Recombinant bvPrP23-231 and bvPrPΔ52-92 substrates were seeded respectively with serial dilutions of L-seeded-PMSA recombinant prion,58 or the same strain adapted by serial rounds of PMSA to bvPrPΔ52-92. Propagation reactions were run with dextran sulphate sodium salt, from Leuconostoc spp. with molecular weights from 6500 to 10000 (Sigma-Aldrich), and 1.0 mm zirconium silicate beads (BioSpec products) at 39°C, with continuous 700 rpm shaking for 18h (Monoshake, Thermo Scientific). PMSA reaction products were PK-digested, and analyzed by WB using mAb SAF83.73 PMCA and PMSA reactions were carried out in light-proof conditions to avoid the photodynamic production of ROS by Zn(II)-BnPyP.
Expression and purification of recombinant PrP proteins
HuPrP23-231, huPrP90-231, bvPrP23-231 and bvPrPΔ52-92 were expressed in E. coli BL21(DE3) cells and purified from the inclusion bodies on a Ni-NTA column.73 Briefly, protein expression was induced by culturing bacteria in M9 minimal medium containing 1 mM isopropil-β-D-1-tiogalattopiranoside (IPTG, Giotto Biotech) overnight at 37°C. For production of uniformly 15N and 13C labeled recombinant hPrPs bacteria were cultured with 15NH4Cl (1 g/liter, Cambridge Isotope Laboratories) and 13C D-glucose (3 g/liter, Cambridge Isotope Laboratories) as unique nitrogen and carbon sources. Bacteria were lysed by sonication, and the inclusion bodies containing the recombinant protein were collected by centrifugation at 18,000xg for 45 min at 4°C, and washed twice with milli-Q water. The recombinant protein was solubilized from the inclusion bodies by incubation in 6M guanidine-HCl pH 8 overnight at 37°C and sonication. The solubilized protein was purified using a histidine affinity column (Ni-NTA agarose, Quiagen), taking advantage of the natural histidines present in the PrP protein, and buffers containing 2M guanidine-HCl pH 8. The purified protein was refolded through three dialysis steps (4h, overnight, 4h) at 4°C against 20 mM sodium acetate pH 5. Correct folding was assessed by circular dichroism and NMR spectra. The protein concentration was determined from A280 values and molar extinction coefficients.
Synthesis of huPrP peptides
huPrP N-terminal (23-91), OR1 (60-67), OR2 (60-75), non-OR (90-112) peptides were synthesized by solid-phase chemistry using fluorenylmethyloxycarbonyl (Fmoc) chloride group protected amino acid with Initiator+Alstra peptide synthesizer (Biotage) at 0.1 mM scale on PEG-PS-copolimer, functionalized with 4-hydroxy-methylphenoxyacetic acid (TGA). Fmoc deprotection was performed automatically at room temperature by treating the peptide-resin with 20% piperidine in dimethylformamide (DMF) for 3 min, followed by another cycle of 10 min and 4 times DMF washing. Amino acids were activated using N, N′-diisopropylcarbodiimide and Oxyma pure, both at 0.5 mM in DMF. Peptides were cleaved from the resin with trifluoroacetic acid (TFA): triisopropylsilane solution (95:5 vol/vol), precipitated, and washed with cold diethyl ether. Purification of crude peptides was carried out in reverse phase HPLC using a semi-preparative C4 column (Symmetry 300, Waters), with mobile phases of 0.1% TFA in water (eluent A) / 0.08% TFA in acetonitrile (eluent B) and a linear gradient from 5 up to 100% of eluent B in 60 min. The peaks were collected and characterized by matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry using an ABI 4800 mass spectrometer, operating in reflector mode. Solutions containing the peptides with a purity greater than 95% were frozen, dried, and the powders stored at -20°C until use.
All Fmoc-amino acid and reagents used for peptides synthesis were from Sigma-Aldrich.
NMR spectroscopy experiments
NMR experiments were run at 298 K on a Bruker Avance 600 MHz spectrometer equipped with a triple-resonance TCI cryoprobe and x, y, z-shielded pulsed-field gradient coil, with samples containing 0.15 mM DSS and 10% D2O. Data were processed with TopSpin® 4.0.8 (Bruker, Karlsruhe, Germany) and analyzed using CcpNmr Analysis 2.4.2.76 Proteins, peptides and porphyrins were in the same buffer: 10 mM CH3COONa pH 4.5 or 10 mM Na2HPO4/NaH2PO4 pH 7. 1H, 15N and 13C backbone resonances of huPrP23-231 and huPrP90-231 (0.2 mM) were assigned by standard triple-resonance experiments77 (HNCA, HNCO, CBCA(CO)NH, CBCANH). Backbone assignments were nearly complete, except for M22, K23, G30, W31, G35, G46, G53-G58, G63/71/79/87 (in OR), G64/72/80/88 (in OR), R164, D167, E168, Y169, S170, N171, H187 in huPrP23-231 and M90, Q91, G93, H96, S97, R164, D167, E168, Y169, S170, N171, H187 in huPrP90-231, because of overlapping peaks or solvent exchange. For NMR titrations of 15N-labeled huPrP23-231 and huPrP90-231 (80 μM), water-flip-back 1H-15N HSQC spectra were acquired for each titration point (3-6 mM ligand stock solution). Resonances were assigned following the individual peaks along the titration.1H, 15N and 13C resonances of N-terminal, OR2, OR1 and non-OR peptides (2-6 mM) and of Zn(II)-BnPyP (1 mM were assigned through 1H-1H-TOCSY (mixing time tmix 60 ms), 1H-1H-NOESY (tmix 150 ms), 1H-1H-ROESY (tmix 300 ms), 1H-15N and 1H-13C HSQC spectra (natural abundance).
For peptide binding curves, 1D 1H spectra at each titration point were acquired (0.3 mM peptide samples, 3-6 mM Zn(II)-BnPyP stock solution). For analysis of the chemical shift perturbations (CSPs) of the peptides we acquired 1H resonances induced by Zn(II)-BnPyP binding, 1H-1H-TOCSY (tmix 60 ms), 1H-1H-NOESY (tmix 150 ms), 1H-1H-ROESY (tmix 300 ms), 1H-15N and 1H-13C HSQC (natural abundance) spectra on 2-3 mM peptide samples containing 1 molar equivalent of Zn(II)-BnPyP, except for the N-terminal peptide, for which we used 0.25 molar equivalent of porphyrin because of the strong line-broadening effects on binding.
Saturation Transfer Difference (STD) experiments (stddiffesgp.3 Bruker pulse sequence) were done as in78 (on- and off-resonance irradiations at respectively 1 ppm and 107 ppm) on 2.8 μM huPrP23-231 with 200 μM Zn(II)-BnPyP, 10 mM Na2HPO4/NaH2PO4 pH 7. To avoid T1 relaxation time bias, we considered the slopes of the STD build-up curves (0.5, 1, 2, 3, 5 s) at tsat=0, normalized to the highest slope value and shown as a percentage.79
NMR spectroscopy data
For NMR titrations of 15N-labeled huPrP23-231 and huPrP90-231, the weighted average of the 1HN and 15N chemical shifts perturbation (CSP) was calculated for each protein residue as:80
| (Equation 1) |
where ΔδHN and ΔδN are respectively the differences of 1HN and 15N chemical shifts between free and bound protein. Because of extensive line-broadening due to ligand binding in the intermediate exchange regime on the NMR time scale or due to the paramagnetic effect of Fe(III)-containing ligands, we also monitored changes in the intensity ratio (I/I0) of the 1HN-15N amide resonances, where I0 and I are the peak intensities in the free and bound protein.
For peptides complexed with Zn(II)-BnPyP, we analyzed the CSPs of all the peptide 1H resonances (ΔδH). We calculated the KD of Zn(II)-BnPyP for N-terminal, OR1, OR2, non-OR peptides, fitting the 1H peptide CSPs as a function of the total ligand concentration at each titration point. For non-OR and OR1 peptides we used the equation describing a one-binding site model:
| (Equation 2) |
where CSPi is the change in 1H chemical shift at each titration point, CSPmax is the maximum change in 1H chemical shift, Li is the ligand concentration at each titration point and P is the peptide concentration. CSPmax and KD were used as fitting parameters in Xmgrace software (https://plasma-gate.weizmann.ac.il/Grace). For OR2 and N-term peptides containing respectively two and four OR motifs, the Hill–Langmuir equation, reflecting the occupancy of macromolecules, was applied:
| (Equation 3) |
where n is the Hill coefficient, describing the cooperativity of ligand binding. CSPmax, KD and n were used as fitting parameters in Xmgrace software. The plots report the average plus sample standard deviation of the KD and n values obtained fitting the well-resolved 1H resonances during the titration.
Circular dichroism experiments and data analysis
CD melting experiments were done in duplicate using a Jasco J-815CD spectrometer (0.1 cm path length). CD melting curves (25°C-95°C, 1°C/min) of huPrP90-231 (8 μM) and huPrP23-231 (5 μM) in 10 mM Na2HPO4/NaH2PO4 pH 7, with and without 15 molar equivalent of Zn(II)-BnPyP or Fe(III)-TMPyP, were collected at 222 nm. Duplicates were merged point to point, calculating the average CD signal and the standard error of the mean (SEM). The protein melting temperature (Tm) was obtained as in.81
Isothermal titration calorimetry
Experiments were done at 25°C using a PEAQ-ITC MicroCalorimeter (MicroCal, Malvern Instruments Ltd., Malvern, UK) following the general procedure.82 A 24 μM huPrP23-231 solution was titrated with Zn(II)-BnPyP (1 mM) using injections of 2 μL. Protein and ligand were prepared and diluted with the same buffer (10 mM Na2HPO4/NaH2PO4 pH 7). Calorimetric data were analyzed with the instrument software. Heat change peaks were integrated and normalized per mole of Zn(II)-BnPyP injected. The ITC binding curve was fitted to the one binding site model equation.83
Dynamic mass redistribution
The EnSight Multimode Plate Reader (Perkin Elmer, Waltham, MA) was used to carry out DMR analyses, as described.84 Immobilization of recombinant huPrP23-231 (15 μL/well of a 2.5 μM huPrP23-231 solution in 10 mM sodium acetate buffer, pH 5) on label-free microplates (EnSpire-LFB high sensitivity microplates, Perkin Elmer) was obtained by amine-coupling chemistry. The interaction between Zn(II)-BnPyP, diluted to different concentrations in assay buffer (10 mM PO4, pH 7.5, 2.4 mM KCl, 138 mM NaCl, 0.05% Tween-20) and huPrP23-231 was monitored after 30 min incubation at RT. A Zephyr Compact Liquid Handling Workstation (Perkin Elmer) was used for all the steps. The Kaleido software (Perkin Elmer) was used to acquire and process the data.
Docking models
Molecular docking of Zn(II)-BnPyP and Fe(III)-TMPyP bound to the huPrP globular domain was done with the Glide docking program (Schrödinger, LLC, New York, NY, 2019). Porphyrin structure and geometry were constructed starting from the porphyrin scaffold available in the Maestro Builder Tool. The ligands were prepared with the LigPrep module available in the Schrödinger Suite, adding hydrogens and optimizing the molecules with OPLS_2005 force field.
For huPrP we used the NMR structure 5YJ5 (PDB id) comprising part of the N-terminal tail (90-128) and the globular domain (129-231). We selected structure number 12 of the NMR bundle where the globular domain is not occluded or hampered by the N-terminal tail. The huPrP90-231 structure was pre-processed, minimized and refined with the Protein Preparation Wizard tool of Maestro, setting the histidines as singly protonated. The docking settings (grid centered on H177 and R220; docking ligands allowed 25 Å; inner box length 20 Å in each x,y,z direction; outer box 46 Å edges) were selected to enable Zn(II)-BnPyP and Fe(III)-TMPyP to explore the whole surface of the globular domain. The hydroxyl and thiol groups of residues S132, T183, Y218, S222, Y225, Y226 were free to rotate during docking. We used the “Standard Precision” (SP) docking and “expanded sampling” for the initial pose selection and considered ten docking poses per ligand, selecting the best poses in terms of energy ranking (Emodel and GScore combination). The Gscores were respectively -5.466 and -2.710 kcal/mol for Zn(II)-BnPyP and Fe(III)-TMPyP.
The best Zn(II)-BnPyP docking pose shows the porphyrin bound to the globular domain in a pocket made of α2 and the C-terminus of α3; this agrees with the spectral perturbation induced in the globular domain by Zn(II)-BnPyP binding, in terms of CSPs (E211, R228), residues disappeared (S132, Y163, Q172, V176, D178, V180, N181, I182, I184, K185, V209, V110, I215, Y218, E219, R220, S222, Q223, A224, Y225, Y226, Q227) and I/I0 changes (H177, C178, T183, T188, T216, R228, G229, S230). In the best Fe(III)-TMPyP model the ligand docks into a pocket made of the β1 strand and the C-terminus of the α3 helix, in agreement with I/I0 changes (M129, S132, A133, M134, Y157, M166, N173, H177, D178, V180. Q186, C214, Q227, R228, S230, S231) and residues disappeared (L130, G131, V161, Y163, Q172, N174, V176, D178, I182, I184, V210, Q217, R220, S222, Q223, A224, Y226, G229) in the NMR titration.
Small Angle X-ray Scattering (SAXS) data collection and analysis
All the experiments were done at the ESRF bioSAXS beamline BM29 in Grenoble, France.85 Fifty μL of huPrP23-231 samples with and without a 5 molar equivalent of Zn(II)-BnPyP or Fe(III)-TMPyP were prepared in 100 mM NaCl, 10 mM HEPES pH 7.0 immediately before SAXS measurements, incubated 15 min at 20°C, centrifuged 30 min at 12,000 g to remove precipitates and measured in batch mode at 20°C. We measured three concentrations (4, 2, 1 mg/mL) of huPrP23-231 alone and one concentration (2 mg/mL) for huPrP23-231 complexed with Zn(II)-BnPyP and Fe(III)-TMPyP, because the complex precipitated at other concentrations. Therefore only the 2 mg/mL SAXS data were analyzed for both huPrP23-231 alone and in complex.
Analysis of the overall parameters was as previously described.86 For low-resolution structural models, an Ensemble Optimization Method (EOM) analysis was conducted, treating the globular domain (125-228) as a rigid body derived from the huPrP90-230 NMR structure 1QM0 (PDB id) and modeling the N-terminal tail (23-124) by EOM.87 Ten independent EOM internal replica were done for each SAXS curve and the frequency distributions of selected radius of gyration (Rg) and maximum diameter (Dmax) ensembles were averaged. Plots and huPrP23-231 models were generated using OrginPro 9.0 and UCSF Chimera software.
Plasma and brain levels of Zn(II)-BnPyP
Blood samples of treated mice were collected in Eppendorf tubes PCR clean (to prevent drug adsorption) containing heparin, gently shaken by inversion and kept on ice. Plasma was obtained after blood centrifugation at 10,000xg for 15 min at 4°C, then immediately frozen on dry ice and kept at -80°C until analysis. Brain tissues were removed, weighed and homogenized, 1 g in 6 mL of PBS 10 mM (pH 7.4). For Zn(II)-BnPyP extraction from the biological matrices, brain homogenate (50 μL) or plasma (50 μL) samples were spiked with 5,10,15,20-Tetrakis(4-trimethylammoniophenyl)porphyrin tetra(p-toluenesulfonate) (Merck), used as internal standard (IS), diluted with an equal volume of 0.5 M ammonium acetate (pH 5.5) and mixed 1:4 (v/v) with acetonitrile for protein precipitation. After centrifugation at 9,390xg for 5 min at room temperature the supernatant was mixed with equal volume of 0.5 M ammonium acetate (pH 5.5) before injection into the HPLC-FLU system.
Plasma and brain samples of treated mice were run and analyzed in parallel with a 7-point calibration curve prepared by spiking 2 μL of 0.25 mg/mL solution of Zn(II)-BnPyP in 198 μL of blank homogenate or blank plasma, to obtain the highest concentration of calibration standard; the following standard points were then obtained by serial dilutions 1:1 (v/v) with blank homogenate or blank plasma. Plasma and brain calibration curve were linear in the range 0.039-2.5 μg/mL and 0.234-15 μg/g, respectively.
Chromatographic separation was achieved on a 2695 Alliance HPLC system (Waters Corp.) using a Kinetex EVO C18 column (150 x 2.1 mm 5 μm 100Å, Phenomenex) at 30°C, with SecurityGuard™ ULTRA cartridges EVO C18 (Phenomenex Inc.). The elution solvents were 0.5 M ammonium acetate (pH 5.5) (mobile phase A, MP-A) and CH3CH/CH3ON (90/10, v/v (mobile phase B, MP-B). The injection volume was 40 μL and the flow rate 400 μL/min. The auto-sampler temperature was maintained at 20°C. Elution started with 90% of MP-A and 10% MP-B for 1 min, followed by a 11-min linear gradient to 100% of MP-B which was maintained for 1 min; then a 1-minute linear gradient to 90% of MP-A was maintained for 5 min to equilibrate the column. The total run time was 18 min; retention times for IS and Zn(II)-BnPyP were 6.5 min and 7.5 min, respectively.
Detection was achieved with a 2475 fluorescence detector (Waters) operating at excitation and emission wavelengths of 435 and 630 nm, respectively. The HPLC-FLU system was controlled by Empower 2.0 (Waters) and data were collected with the same software.
Prion inoculation
Ten-percent (w/v) of RML scrapie-infected brain homogenate was prepared in PBS; cleared by centrifugation at 900xg for 5 min and diluted to 1% in PBS. 25 μL was injected intracerebrally into the right parietal lobe of recipient mice using a 25-gauge needle. Mice were observed weekly for signs of neurological dysfunction according to a set of objective criteria.53 Terminal disease was scored when mice became unable to right themselves from a supine position in < 60 s, to walk on a horizontal metal grid, and/or had cumulative weight loss equivalent to 20% of their body mass at the onset of disease.
Quantification and statistical analysis
Results were analyzed using Prism 9 (GraphPad, Software Inc.). After testing the data for normal distribution with the Kolmogorov-Smirnov test, the appropriate statistical tests were used (see Figure legends). Data are presented as mean ± standard error of the mean (SEM) of the number of elements analyzed. P < 0.05 was considered statistically significant.
Acknowledgments
We thank Sylvain Lehmann for the ScN2a-22L cells, Emiliano Biasini for the ScN2a-RML cells and the D18 antibody; David A. Harris for plasmids encoding PrPΔ23-89 and PrPΔ23-111; Joel Watts for plasmids encoding bvPrP; Byron Caughey for plasmid encoding huPrP90-231; Ester Zito for the anti-puromycin antibody; Richard J. Kascsak for the 3F4 and 6D11 antibodies; Man Sun-Sy for the 11G5 antibody; Steven J. Collins for the 8A4 antibody; Olivier Andreoletti for the Dawson scrapie strain; Wenquan Zou, Vincent Béringue and Glenn Telling for the TgZWV, Tg338 and TgVole brains. We are grateful to Rosanna Piccirillo for participating in the initial phase of this project, and to Stefano Fumagalli for advice on super-resolution microscopy. We also thank the BM29 BIOSAXS, ESRF for provision of the beamtime and assistance during data collection, and Massimo Degano for access to the Italian block allocation group (BAG) for structural biology (proposal number MX-2291). This study was supported by Telethon Italy award GGP15225 (R.C., G.M, S.B.), Italian Ministry of Health award RF-2016-02362950 (R.C., C.Z., M.A.D.B.), the CJD Foundation USA (R.C.), the Associazione Italiana Encefalopatie da Prioni (AIEnP; R.C.), AIRC award IG 2018 - ID. 21440 (G.M.), Spanish Ministry of Science award PID2021-122201OB-C21 co-funded by ERDF (J.C). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The Graphical abstract was created with BioRender.com (Agreement number: XB25KL15E2).
Author contributions
A.M., L.T., L.C., E.R., I.R., L.Taiarol, and M.O. conducted experiments in cells. C.Z. and G.Q. conducted and analyzed NMR and CD experiments. E.C. synthesized the porphyrins. G.L. ran the experiments in cultured cerebellar organotypic slices. F.D.L. did the molecular docking. G.G. and M.D.T. acquired and analyzed SAXS data. A.C. and S.M. synthesized and purified PrP peptides. J.L. and M.G. ran pharmacokinetics studies. H.E., S.G-M., I.V., R.N., M.A.D.B., and J.C. did the in vitro prion amplification experiments. V.C., G.C., and S.P. ran and analyzed the ITC experiments. S.R.E. ran and analyzed the DMR experiments. E.V.A. gave advice and reagents for PrP-Halo analysis. H.E., N.L.L., J.C. and J.R.R. produced the recombinant PrPs. R.C., G.M., and S.B. conceptualized and supervised the project. A.M., C.Z., G.G., G.M. and R.C. wrote the manuscript with input from all authors.
Declaration of interests
J.C. and H.E., as part of the company ATLAS Molecular Pharma S.L., declare that they have no conflicts of interest, as the company had no role in study design or funding, nor will they, or their immediate family members, benefit financially from the findings reported. All the other authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. We support inclusive, diverse, and equitable conduct of research.
Published: July 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107480.
Supplemental information
Data and code availability
-
•
The underlying numerical data for Figure panels 1D, 1F, 1K, 2B, 2D, 2G, 3E, 4A, 4B, 4C, 4E, 5A, 5B, 5C, 5D, 5E, 5F, 6A, 6B, S1D, S1J, S1L, S1O, S1Q, S3D, S3F, S3H, S4J, S5B, S5E, S5G, S6C, S6F, S6G, S6K, S6L, S7C, S7F, S7G, S8D, S8E, S9B, S9C, S9D, S9G, S10C, S11A, S11B, S12A, S13B, S13E, S14 were deposited on Mendeley Data, and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
SAXS data were deposited in the SASBDB data bank. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Colby D.W., Prusiner S.B. Prions. Cold Spring Harb. Perspect. Biol. 2011;3:a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiesa R. The elusive role of the prion protein and the mechanism of toxicity in prion disease. PLoS Pathog. 2015;11:e1004745. doi: 10.1371/journal.ppat.1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris D.A. Trafficking, turnover and membrane topology of PrP. Br. Med. Bull. 2003;66:71–85. doi: 10.1093/bmb/66.1.71. [DOI] [PubMed] [Google Scholar]
- 4.Zahn R., Liu A., Lührs T., Riek R., von Schroetter C., López García F., Billeter M., Calzolai L., Wider G., Wüthrich K. NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spevacek A.R., Evans E.G.B., Miller J.L., Meyer H.C., Pelton J.G., Millhauser G.L. Zinc drives a tertiary fold in the prion protein with familial disease mutation sites at the interface. Structure. 2013;21:236–246. doi: 10.1016/j.str.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thakur A.K., Srivastava A.K., Srinivas V., Chary K.V.R., Rao C.M. Copper alters aggregation behavior of prion protein and induces novel interactions between its N- and C-terminal regions. J. Biol. Chem. 2011;286:38533–38545. doi: 10.1074/jbc.M111.265645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gielnik M., Taube M., Zhukova L., Zhukov I., Wärmländer S.K.T.S., Svedružić Ž., Kwiatek W.M., Gräslund A., Kozak M. Zn(II) binding causes interdomain changes in the structure and flexibility of the human prion protein. Sci. Rep. 2021;11:21703. doi: 10.1038/s41598-021-00495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markham K.A., Roseman G.P., Linsley R.B., Lee H.-W., Millhauser G.L. Molecular features of the Zn2+ binding site in the prion protein probed by 113Cd NMR. Biophys. J. 2019;116:610–620. doi: 10.1016/j.bpj.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J., Browning S., Mahal S.P., Oelschlegel A.M., Weissmann C. Darwinian evolution of prions in cell culture. Science. 2010;327:869–872. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berry D.B., Lu D., Geva M., Watts J.C., Bhardwaj S., Oehler A., Renslo A.R., Dearmond S.J., Prusiner S.B., Giles K. Drug resistance confounding prion therapeutics. Proc. Natl. Acad. Sci. USA. 2013;110:E4160–E4169. doi: 10.1073/pnas.1317164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghaemmaghami S., Ahn M., Lessard P., Giles K., Legname G., DeArmond S.J., Prusiner S.B. Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 2009;5:e1000673. doi: 10.1371/journal.ppat.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burke C.M., Mark K.M.K., Kun J., Beauchemin K.S., Supattapone S. Emergence of prions selectively resistant to combination drug therapy. PLoS Pathog. 2020;16:e1008581. doi: 10.1371/journal.ppat.1008581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benestad S.L., Austbø L., Tranulis M.A., Espenes A., Olsaker I. Healthy goats naturally devoid of prion protein. Vet. Res. 2012;43:87. doi: 10.1186/1297-9716-43-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Büeler H., Fischer M., Lang Y., Bluethmann H., Lipp H.P., DeArmond S.J., Prusiner S.B., Aguet M., Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 15.Richt J.A., Kasinathan P., Hamir A.N., Castilla J., Sathiyaseelan T., Vargas F., Sathiyaseelan J., Wu H., Matsushita H., Koster J., et al. Production of cattle lacking prion protein. Nat. Biotechnol. 2007;25:132–138. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bremer J., Baumann F., Tiberi C., Wessig C., Fischer H., Schwarz P., Steele A.D., Toyka K.V., Nave K.A., Weis J., Aguzzi A. Axonal prion protein is required for peripheral myelin maintenance. Nat. Neurosci. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- 17.Minikel E.V., Vallabh S.M., Lek M., Estrada K., Samocha K.E., Sathirapongsasuti J.F., McLean C.Y., Tung J.Y., Yu L.P.C., Gambetti P., et al. Quantifying prion disease penetrance using large population control cohorts. Sci. Transl. Med. 2016;8:322ra9. doi: 10.1126/scitranslmed.aad5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Büeler H., Aguzzi A., Sailer A., Greiner R.A., Autenried P., Aguet M., Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 19.Büeler H., Raeber A., Sailer A., Fischer M., Aguzzi A., Weissmann C. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol. Med. 1994;1:19–30. [PMC free article] [PubMed] [Google Scholar]
- 20.Mallucci G., Dickinson A., Linehan J., Klöhn P.C., Brandner S., Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 21.Nazor Friberg K., Hung G., Wancewicz E., Giles K., Black C., Freier S., Bennett F., DeArmond S.J., Freyman Y., Lessard P., et al. Intracerebral infusion of antisense oligonucleotides into prion-infected mice. Mol. Ther. Nucleic Acids. 2012;1:e9. doi: 10.1038/mtna.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raymond G.J., Zhao H.T., Race B., Raymond L.D., Williams K., Swayze E.E., Graffam S., Le J., Caron T., Stathopoulos J., et al. Antisense oligonucleotides extend survival of prion-infected mice. JCI Insight. 2019;5:e131175. doi: 10.1172/jci.insight.131175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White M.D., Farmer M., Mirabile I., Brandner S., Collinge J., Mallucci G.R. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc. Natl. Acad. Sci. USA. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minikel E.V., Zhao H.T., Le J., O’Moore J., Pitstick R., Graffam S., Carlson G.A., Kavanaugh M.P., Kriz J., Kim J.B., et al. Prion protein lowering is a disease-modifying therapy across prion disease stages, strains and endpoints. Nucleic Acids Res. 2020;48:10615–10631. doi: 10.1093/nar/gkaa616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priola S.A., Raines A., Caughey W.S. Porphyrin and phthalocyanine antiscrapie compounds. Science. 2000;287:1503–1506. doi: 10.1126/science.287.5457.1503. 8296 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Caughey W.S., Raymond L.D., Horiuchi M., Caughey B. Inhibition of protease-resistant prion protein formation by porphyrins and phthalocyanines. Proc. Natl. Acad. Sci. USA. 1998;95:12117–12122. doi: 10.1073/pnas.95.21.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicoll A.J., Trevitt C.R., Tattum M.H., Risse E., Quarterman E., Ibarra A.A., Wright C., Jackson G.S., Sessions R.B., Farrow M., et al. Pharmacological chaperone for the structured domain of human prion protein. Proc. Natl. Acad. Sci. USA. 2010;107:17610–17615. doi: 10.1073/pnas.1009062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kocisko D.A., Caughey W.S., Race R.E., Roper G., Caughey B., Morrey J.D. A porphyrin increases survival time of mice after intracerebral prion infection. Antimicrob. Agents Chemother. 2006;50:759–761. doi: 10.1128/AAC.50.2.759-761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caughey W.S., Priola S.A., Kocisko D.A., Raymond L.D., Ward A., Caughey B. Cyclic tetrapyrrole sulfonation, metals, and oligomerization in antiprion activity. Antimicrob. Agents Chemother. 2007;51:3887–3894. doi: 10.1128/AAC.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banfi S., Caruso E., Buccafurni L., Murano R., Monti E., Gariboldi M., Papa E., Gramatica P. Comparison between 5,10,15,20-tetraaryl- and 5,15-diarylporphyrins as photosensitizers: synthesis, photodynamic activity, and quantitative structure-activity relationship modeling. J. Med. Chem. 2006;49:3293–3304. doi: 10.1021/jm050997m. [DOI] [PubMed] [Google Scholar]
- 31.Banfi S., Caruso E., Buccafurni L., Battini V., Zazzaron S., Barbieri P., Orlandi V. Antibacterial activity of tetraaryl-porphyrin photosensitizers: an in vitro study on Gram negative and Gram positive bacteria. J. Photochem. Photobiol., B. 2006;85:28–38. doi: 10.1016/j.jphotobiol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Brimacombe D.B., Bennett A.D., Wusteman F.S., Gill A.C., Dann J.C., Bostock C.J. Characterization and polyanion-binding properties of purified recombinant prion protein. Biochem. J. 1999;342:605–613. [PMC free article] [PubMed] [Google Scholar]
- 33.Petrosyan R., Patra S., Rezajooei N., Garen C.R., Woodside M.T. Unfolded and intermediate states of PrP play a key role in the mechanism of action of an antiprion chaperone. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2010213118. e2010213118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diringer H., Ehlers B. Chemoprophylaxis of scrapie in mice. J. Gen. Virol. 1991;72:457–460. doi: 10.1099/0022-1317-72-2-457. [DOI] [PubMed] [Google Scholar]
- 35.Caughey B., Raymond G.J. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J. Virol. 1993;67:643–650. doi: 10.1128/jvi.67.2.643-650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taubner L.M., Bienkiewicz E.A., Copié V., Caughey B. Structure of the flexible amino-terminal domain of prion protein bound to a sulfated glycan. J. Mol. Biol. 2010;395:475–490. doi: 10.1016/j.jmb.2009.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satpute-Krishnan P., Ajinkya M., Bhat S., Itakura E., Hegde R.S., Lippincott-Schwartz J. ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell. 2014;158:522–533. doi: 10.1016/j.cell.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shyng S.L., Moulder K.L., Lesko A., Harris D.A. The N-terminal domain of a glycolipid-anchored prion protein is essential for its endocytosis via clathrin-coated pits. J. Biol. Chem. 1995;270:14793–14800. doi: 10.1074/jbc.270.24.14793. [DOI] [PubMed] [Google Scholar]
- 39.Heuser J.E., Anderson R.G. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J. Cell Biol. 1989;108:389–400. doi: 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cremers J., Richert S., Kondratuk D.V., Claridge T.D.W., Timmel C.R., Anderson H.L. Nanorings with copper(II) and zinc(II) centers: forcing copper porphyrins to bind axial ligands in heterometallated oligomers. Chem. Sci. 2016;7:6961–6968. doi: 10.1039/C6SC01809B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin C.l., Fang M.-Y., Cheng S.-H. Substituent and axial ligand effects on the electrochemistry of zinc porphyrins. J. Electroanal. Chem. 2002;531:155–162. doi: 10.1016/S0022-0728(02)01056-2. [DOI] [Google Scholar]
- 42.Coyle C.L., Rafson P.A., Abbott E.H. Equilibria of imidazole with iron(III) tetraphenylporphine. Inorg. Chem. 1973;12:2007–2010. [Google Scholar]
- 43.Yamaguchi K., Kamatari Y.O., Ono F., Shibata H., Fuse T., Elhelaly A.E., Fukuoka M., Kimura T., Hosokawa-Muto J., Ishikawa T., et al. A designer molecular chaperone against transmissible spongiform encephalopathy slows disease progression in mice and macaques. Nat. Biomed. Eng. 2019;3:206–219. doi: 10.1038/s41551-019-0349-8. [DOI] [PubMed] [Google Scholar]
- 44.Surewicz W.K., Apostol M.I. Prion protein and its conformational conversion: a structural perspective. Top. Curr. Chem. 2011;305:135–167. doi: 10.1007/128_2011_165. [DOI] [PubMed] [Google Scholar]
- 45.Brown D.R., Qin K., Herms J.W., Madlung A., Manson J., Strome R., Fraser P.E., Kruck T., von Bohlen A., Schulz-Schaeffer W., et al. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 46.Lee K.S., Raymond L.D., Schoen B., Raymond G.J., Kett L., Moore R.A., Johnson L.M., Taubner L., Speare J.O., Onwubiko H.A., et al. Hemin interactions and alterations of the subcellular localization of prion protein. J. Biol. Chem. 2007;282:36525–36533. doi: 10.1074/jbc.M705620200. [DOI] [PubMed] [Google Scholar]
- 47.Linsenmeier L., Mohammadi B., Shafiq M., Frontzek K., Bär J., Shrivastava A.N., Damme M., Song F., Schwarz A., Da Vela S., et al. Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies. Sci. Adv. 2021;7:eabj1826. doi: 10.1126/sciadv.abj1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caughey B., Caughey W.S., Kocisko D.A., Lee K.S., Silveira J.R., Morrey J.D. Prions and transmissible spongiform encephalopathy (TSE) chemotherapeutics: a common mechanism for anti-TSE compounds? Acc. Chem. Res. 2006;39:646–653. doi: 10.1021/ar050068p. [DOI] [PubMed] [Google Scholar]
- 49.Corbett G.T., Wang Z., Hong W., Colom-Cadena M., Rose J., Liao M., Asfaw A., Hall T.C., Ding L., DeSousa A., et al. PrP is a central player in toxicity mediated by soluble aggregates of neurodegeneration-causing proteins. Acta Neuropathol. 2020;139:503–526. doi: 10.1007/s00401-019-02114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biggi S., Pancher M., Stincardini C., Luotti S., Massignan T., Dalle Vedove A., Astolfi A., Gatto P., Lolli G., Barreca M.L., et al. Identification of compounds inhibiting prion replication and toxicity by removing PrPC from the cell surface. J. Neurochem. 2020;152:136–150. doi: 10.1111/jnc.14805. [DOI] [PubMed] [Google Scholar]
- 51.Nishida N., Harris D.A., Vilette D., Laude H., Frobert Y., Grassi J., Casanova D., Milhavet O., Lehmann S. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 2000;74:320–325. doi: 10.1128/jvi.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dossena S., Imeri L., Mangieri M., Garofoli A., Ferrari L., Senatore A., Restelli E., Balducci C., Fiordaliso F., Salio M., et al. Mutant prion protein expression causes motor and memory deficits and abnormal sleep patterns in a transgenic mouse model. Neuron. 2008;60:598–609. doi: 10.1016/j.neuron.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 53.Bouybayoune I., Mantovani S., Del Gallo F., Bertani I., Restelli E., Comerio L., Tapella L., Baracchi F., Fernández-Borges N., Mangieri M., et al. Transgenic fatal familial insomnia mice indicate prion infectivity-independent mechanisms of pathogenesis and phenotypic expression of disease. PLoS Pathog. 2015;11:e1004796. doi: 10.1371/journal.ppat.1004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watts J.C., Giles K., Bourkas M.E.C., Patel S., Oehler A., Gavidia M., Bhardwaj S., Lee J., Prusiner S.B. Towards authentic transgenic mouse models of heritable PrP prion diseases. Acta Neuropathol. 2016;132:593–610. doi: 10.1007/s00401-016-1585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turnbaugh J.A., Westergard L., Unterberger U., Biasini E., Harris D.A. The N-terminal, polybasic region is critical for prion protein neuroprotective activity. PLoS One. 2011;6:e25675. doi: 10.1371/journal.pone.0025675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Massignan T., Biasini E., Lauranzano E., Veglianese P., Pignataro M., Fioriti L., Harris D.A., Salmona M., Chiesa R., Bonetto V. Mutant prion protein expression is associated with an alteration of the Rab GDP dissociation inhibitor alpha (GDI)/Rab11 pathway. Mol. Cell. Proteomics. 2010;9:611–622. doi: 10.1074/mcp.M900271-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senatore A., Colleoni S., Verderio C., Restelli E., Morini R., Condliffe S.B., Bertani I., Mantovani S., Canovi M., Micotti E., et al. Mutant PrP suppresses glutamatergic neurotransmission in cerebellar granule neurons by impairing membrane delivery of VGCC alpha(2)delta-1 subunit. Neuron. 2012;74:300–313. doi: 10.1016/j.neuron.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernández-Borges N., Di Bari M.A., Eraña H., Sánchez-Martín M., Pirisinu L., Parra B., Elezgarai S.R., Vanni I., López-Moreno R., Vaccari G., et al. Cofactors influence the biological properties of infectious recombinant prions. Acta Neuropathol. 2018;135:179–199. doi: 10.1007/s00401-017-1782-y. [DOI] [PubMed] [Google Scholar]
- 59.Elezgarai S.R., Fernández-Borges N., Eraña H., Sevillano A.M., Charco J.M., Harrathi C., Saá P., Gil D., Kong Q., Requena J.R., et al. Generation of a new infectious recombinant prion: a model to understand Gerstmann-Sträussler-Scheinker syndrome. Sci. Rep. 2017;7:9584. doi: 10.1038/s41598-017-09489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fioriti L., Dossena S., Stewart L.R., Stewart R.S., Harris D.A., Forloni G., Chiesa R. Cytosolic prion protein (PrP) is not toxic in N2a cells and primary neurons expressing pathogenic PrP mutations. J. Biol. Chem. 2005;280:11320–11328. doi: 10.1074/jbc.M412441200. [DOI] [PubMed] [Google Scholar]
- 61.Restelli E., Capone V., Pozzoli M., Ortolan D., Quaglio E., Corbelli A., Fiordaliso F., Beznoussenko G.V., Artuso V., Roiter I., et al. Activation of Src family kinase ameliorates secretory trafficking in mutant prion protein cells. J. Biol. Chem. 2021;296:100490. doi: 10.1016/j.jbc.2021.100490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Restelli E., Fioriti L., Mantovani S., Airaghi S., Forloni G., Chiesa R. Cell type-specific neuroprotective activity of untranslocated prion protein. PLoS One. 2010;5:e13725. doi: 10.1371/journal.pone.0013725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Falsig J., Aguzzi A. The prion organotypic slice culture assay--POSCA. Nat. Protoc. 2008;3:555–562. doi: 10.1038/nprot.2008.13. [DOI] [PubMed] [Google Scholar]
- 64.Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 65.Chiesa R., Harris D.A. Nerve growth factor-induced differentiation does not alter the biochemical properties of a mutant prion protein expressed in PC12 cells. J. Neurochem. 2000;75:72–80. doi: 10.1046/j.1471-4159.2000.0750072.x. [DOI] [PubMed] [Google Scholar]
- 66.Biasini E., Tapella L., Restelli E., Pozzoli M., Massignan T., Chiesa R. The hydrophobic core region governs mutant prion protein aggregation and intracellular retention. Biochem. J. 2010;430:477–486. doi: 10.1042/BJ20100615. [DOI] [PubMed] [Google Scholar]
- 67.Kascsak R. Reiterating the epitope specificity of prion-specific mAb 3F4. J. Biol. Chem. 2010;285:le5. doi: 10.1074/jbc.L109.le5. author reply le6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kascsak R.J., Rubenstein R., Merz P.A., Tonna-DeMasi M., Fersko R., Carp R.I., Wisniewski H.M., Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spinner D.S., Kascsak R.B., Lafauci G., Meeker H.C., Ye X., Flory M.J., Kim J.I., Schuller-Levis G.B., Levis W.R., Wisniewski T., et al. CpG oligodeoxynucleotide-enhanced humoral immune response and production of antibodies to prion protein PrPSc in mice immunized with 139A scrapie-associated fibrils. J. Leukoc. Biol. 2007;81:1374–1385. doi: 10.1189/jlb.1106665. [DOI] [PubMed] [Google Scholar]
- 70.Pan T., Chang B., Wong P., Li C., Li R., Kang S.C., Robinson J.D., Thompsett A.R., Tein P., Yin S., et al. An aggregation-specific enzyme-linked immunosorbent assay: detection of conformational differences between recombinant PrP protein dimers and PrPSc aggregates. J. Virol. 2005;79:12355–12364. doi: 10.1128/JVI.79.19.12355-12364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan F.F., Biffin S., Brazier M.W., Suarez M., Cappai R., Hill A.F., Collins S.J., Sullivan J.S., Middleton D., Multhaup G., et al. Detection of prion epitopes on PrP and PrP of transmissible spongiform encephalopathies using specific monoclonal antibodies to PrP. Immunol. Cell Biol. 2005;83:632–637. doi: 10.1111/j.1440-1711.2005.01384.x. [DOI] [PubMed] [Google Scholar]
- 72.Bolte S., Cordelières F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 73.Eraña H., Charco J.M., Di Bari M.A., Díaz-Domínguez C.M., López-Moreno R., Vidal E., González-Miranda E., Pérez-Castro M.A., García-Martínez S., Bravo S., et al. Development of a new largely scalable in vitro prion propagation method for the production of infectious recombinant prions for high resolution structural studies. PLoS Pathog. 2019;15:e1008117. doi: 10.1371/journal.ppat.1008117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan J., Zhan Y.-A., Abskharon R., Xiao X., Martinez M.C., Zhou X., Kneale G., Mikol J., Lehmann S., Surewicz W.K., et al. Recombinant human prion protein inhibits prion propagation in vitro. Sci. Rep. 2013;3:2911. doi: 10.1038/srep02911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Williamson R.A., Peretz D., Pinilla C., Ball H., Bastidas R.B., Rozenshteyn R., Houghten R.A., Prusiner S.B., Burton D.R. Mapping the prion protein using recombinant antibodies. J. Virol. 1998;72:9413–9418. doi: 10.1128/jvi.72.11.9413-9418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 77.Bax A., Grzesiek S. Methodological advances in protein NMR. Acc. Chem. Res. 1993;26:131–138. doi: 10.1021/ar00028a001. [DOI] [Google Scholar]
- 78.Nardelli F., Paissoni C., Quilici G., Gori A., Traversari C., Valentinis B., Sacchi A., Corti A., Curnis F., Ghitti M., Musco G. Succinimide-based conjugates improve isoDGR cyclopeptide affinity to αv β3 without promoting integrin allosteric activation. J. Med. Chem. 2018;61:7474–7485. doi: 10.1021/acs.jmedchem.8b00745. [DOI] [PubMed] [Google Scholar]
- 79.Mayer M., James T.L. NMR-based characterization of phenothiazines as a RNA binding scaffold. J. Am. Chem. Soc. 2004;126:4453–4460. doi: 10.1021/ja0398870. [DOI] [PubMed] [Google Scholar]
- 80.Grzesiek S., Stahl S.J., Wingfield P.T., Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 81.Greenfield N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saponaro A. Isothermal titration calorimetry: a biophysical method to characterize the interaction between label-free biomolecules in solution. Bio. Protoc. 2018;8:e2957. doi: 10.21769/BioProtoc.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brautigam C.A., Zhao H., Vargas C., Keller S., Schuck P. Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions. Nat. Protoc. 2016;11:882–894. doi: 10.1038/nprot.2016.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Massignan T., Cimini S., Stincardini C., Cerovic M., Vanni I., Elezgarai S.R., Moreno J., Stravalaci M., Negro A., Sangiovanni V., et al. A cationic tetrapyrrole inhibits toxic activities of the cellular prion protein. Sci. Rep. 2016;6:23180. doi: 10.1038/srep23180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pernot P., Round A., Barrett R., De Maria Antolinos A., Gobbo A., Gordon E., Huet J., Kieffer J., Lentini M., Mattenet M., et al. Upgraded ESRF BM29 beamline for SAXS on macromolecules in solution. J. Synchrotron Radiat. 2013;20:660–664. doi: 10.1107/S0909049513010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salzano G., Brennich M., Mancini G., Tran T.H., Legname G., D’Angelo P., Giachin G. Deciphering copper coordination in the mammalian prion protein amyloidogenic domain. Biophys. J. 2020;118:676–687. doi: 10.1016/j.bpj.2019.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernadó P., Mylonas E., Petoukhov M.V., Blackledge M., Svergun D.I. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The underlying numerical data for Figure panels 1D, 1F, 1K, 2B, 2D, 2G, 3E, 4A, 4B, 4C, 4E, 5A, 5B, 5C, 5D, 5E, 5F, 6A, 6B, S1D, S1J, S1L, S1O, S1Q, S3D, S3F, S3H, S4J, S5B, S5E, S5G, S6C, S6F, S6G, S6K, S6L, S7C, S7F, S7G, S8D, S8E, S9B, S9C, S9D, S9G, S10C, S11A, S11B, S12A, S13B, S13E, S14 were deposited on Mendeley Data, and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
SAXS data were deposited in the SASBDB data bank. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.