Abstract
Cap-independent translation of the hepatitis C virus (HCV) genomic RNA is mediated by an internal ribosome entry site (IRES) within the 5′ untranslated region (5′UTR) of the virus RNA. To investigate the effects of alterations to the primary sequence of the 5′UTR on IRES activity, a series of HCV genotype 1b (HCV-1b) variant IRES elements was generated and cloned into a bicistronic reporter construct. Changes from the prototypic HCV-1b 5′UTR sequence were identified at various locations throughout the 5′UTR. The translation efficiencies of these IRES elements were examined by an in vivo transient expression assay in transfected BHK-21 cells and were found to range from 0.4 to 95.8% of the activity of the prototype HCV-1b IRES. Further mutational analysis of the three single-point mutants most severely defective in activity, whose mutations were all located in or near stem-loop IIIc, demonstrated that both the primary sequence and the maintenance of base pairing within this stem structure were critical for HCV IRES function. Complementation studies indicated that defective mutants containing either point mutations or major deletions within the IRES elements could not be complemented in trans by a wild-type IRES.
Hepatitis C virus (HCV) is a single-stranded, positive-sense RNA virus that is the main causative agent of posttransfusion non-A, non-B viral hepatitis worldwide. Infection with HCV frequently leads to chronic hepatitis and cirrhosis and is associated with the development of hepatocellular carcinoma. The genome of HCV contains approximately 9,500 nucleotides (nt), including highly conserved untranslated regions (UTRs) at both the 5′ and 3′ termini. The single large open reading frame encodes a polyprotein of 3,010 to 3,037 amino acids which is processed by cellular and viral proteases to produce the structural and nonstructural proteins (10). Based on phylogenetic analysis, it has been proposed that HCV be classified into six major genotypes, designated 1 to 6, and subdivided into subtypes a, b, c, etc. (21, 22). HCV has recently been placed into a separate genus, Hepacivirus, within the family Flaviviridae.
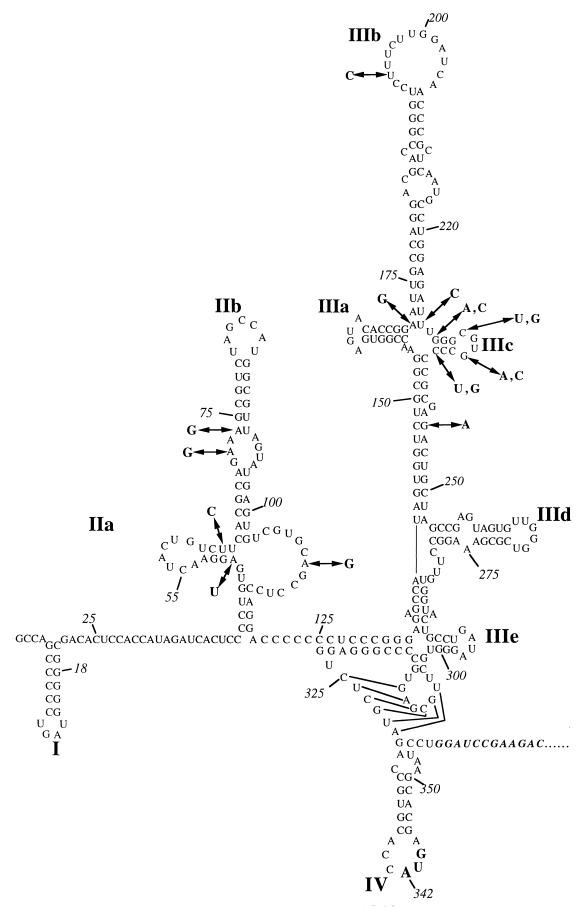
The HCV 5′UTR ranges in length from 332 to 343 nt and contains up to five AUG codons, depending on the HCV genotype or subtype (2). The sequence is highly conserved (25) and is predicted to fold into a complex secondary structure encompassing multiple stem-loops and an RNA pseudoknot (Fig. 1) (for a review, see reference 13). It is now clear that translation of the HCV open reading frame is initiated by a cap-independent internal ribosome entry mechanism mediated by an internal ribosome entry site (IRES) situated within the 5′UTR (28, 30).
FIG. 1.
Predicted secondary structure of the HCV 5′UTR based on the structures of Brown et al. (1) and Honda et al. (9). Stem-loop structures are labelled for reference. The sequence shown is that of the prototypic genotype 1b 5′UTR (14). Mutations described in the text are shown in boldface.
Comparison of different 5′UTR sequences from natural isolates of HCV serves to confirm the predicted secondary structure proposed by Brown et al. (1) and Honda et al. (9) and indicates that in addition to the primary sequence, the secondary and tertiary structures are also highly conserved in order to maintain the correct functioning of the IRES (3, 4, 12, 28). However, the role of the primary sequence and the sensitivity of different regions of the IRES to mutagenesis have yet to be fully elucidated (13). This information may well be important in predicting the mechanisms by which translation is initiated.
In this study, a series of clones containing point mutations and large deletions within the 5′UTR of HCV type 1b (HCV-1b) was generated and sequenced. By using a bicistronic luciferase reporter system (4), the abilities of different isolates of HCV to initiate translation of the downstream reporter were assessed. Our results show that mutations in the IRES of HCV cause a range of effects, depending on their location, with the region around stem-loop IIIc (Fig. 1) being particularly sensitive to alterations in the predicted secondary structure. None of these mutants could be complemented in trans by the wild-type (wt) IRES of HCV-1b.
MATERIALS AND METHODS
Plasmids.
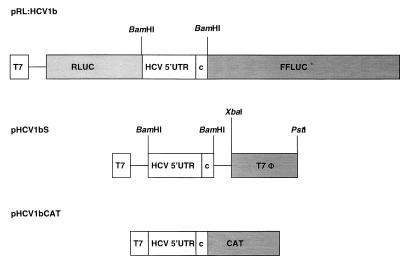
DNA manipulations were performed by standard methods (20). Plasmid DNA was purified with a plasmid miniprep kit (Qiagen). The bicistronic vector pRL was described previously (4) and encodes renilla luciferase (RLUC) and firefly luciferase (FFLUC) under the control of a T7 promoter. HCV-1b IRES fragments were inserted into the unique BamHI site of the pRL vector (Fig. 2). A T7 terminator sequence was amplified by PCR with primer pair T7F (5′CCGTCTAGAAGCTGAGTTGGCTGCTG3′) and T7B (5′GAGCTGCAGCATCCGGATATAGTTCCTC3′), using plasmid pT7ribo (6) as a template. The PCR product was digested with XbaI-PstI and inserted into similarly digested pHCV1b to produce pHCV1bS (Fig. 2). The monocistronic plasmid pHCV1bCAT was derived from plasmid pSGNTR (a gift from G. Lounsbach) and contained the chloramphenicol acetyltransferase (CAT) gene fused to the HCV-1b IRES (Fig. 2).
FIG. 2.
Structures of the plasmids transfected into vTF7-3-infected cells. The two luciferase reporter genes are indicated. The HCV RNA sequences are shown as open boxes. Restriction enzyme cleavage sites used for DNA manipulation are indicated over the cDNAs. T7, T7 promoter sequence; T7φ, T7 terminator sequence; c, core sequence.
Isolation of HCV cDNAs.
HCV RNA was isolated from sera of Chinese blood donors who had previously been confirmed to be infected with HCV-1b (27) by the use of a QIAamp viral RNA kit (Qiagen) as directed by the manufacturer. The extracted RNA was reverse transcribed, using antisense primer AC8 (5′CCGACGCTGCAGATGTACCCCATGAG3′) and avian myeloblastosis virus reverse transcriptase (Promega) as previously described (4). The entire 5′UTR plus the first 15 nt of the coding region (nt 1 to 356) was amplified by PCR with sense primer AC5 (5′TTGCTGGATCCGGCGACACTCCACCAT3′) and antisense primer AC6 (5′AGCAAGGATCCAGGATTCGTGCTCATGGTGC3′), using Taq DNA polymerase (Gibco-BRL). The PCR involved 30 cycles of heating at 94°C for 45 s, 58°C for 30 s, and 72°C for 1 min. The resulting PCR products were digested with BamHI and subsequently cloned into pTZ18R by standard procedures. Twenty positive recombinant clones were sequenced by the dideoxy-mediated chain termination method and by automated sequencing on an ABI Prism 377 DNA sequencer (Perkin-Elmer).
Mutagenesis of the HCV IRES.
Random mutations were introduced into the HCV 5′UTR in plasmid pHCV1b by high-cycle PCR as described by van der Velden et al. (29), using primer pair AC5-AC6. After a total of 55 cycles of amplification, PCR products were digested with BamHI, cloned into similarly digested pTZ18R, and sequenced as described above. For PCR-based site-directed mutagenesis, two sets of reactions were performed, the first with primer pairs AC5-primer 1 and AC6-primer 2 (Table 1), using the appropriate construct as a template, and the second with the two gel-purified PCR products from the first reaction and primers AC5 and AC6. The sequences of these cDNAs were confirmed and then subcloned into the pRL vector. Deletion mutant Δ5–20, which lacks domain I of the 5′UTR, was constructed by PCR amplification with primer pairs AC6 and AC5–20 (5′TTGCTGGATCCGCCAGACACTCCACCATAGATCACTC3′), using pHCV1b as a template. Again, the amplified fragments were cloned, sequenced, and subcloned into the pRL vector. Deletion mutant Δ1–62 was generated by PCR amplification with 10 ng of pHCV1b as the template and 50 pmol each of primers AC5B (5′TTCACGCAGAAAGCGTCTAG) and AC6. Deletion mutant Δ57–83 was constructed by deletion of the sequence between the BbsI and NcoI sites followed by blunt-end repair of the vector and religation of the two ends. Likewise, mutants Δ165–287 and Δ132–317 were created by deletion of the sequence between the RsaI or SmaI restriction sites, respectively.
TABLE 1.
Primer sequences used for directed mutagenesis of the HCV-1b 5′UTRa
| Mutant | Primer binding site location (nt) | Primer sequence |
|---|---|---|
| A172G-U227C | 240–218, 213–235 | 5′-GGGGGCACGCCCAGATCTCCAGG (primer 1), 5′-CAATGCCTGGAGATCTGGGCGTG (primer 2) |
| G229A-C238U | 252–227, 222–247 | 5′-TAGCAGTCTCGCGGAGGCACGCCTAA (primer 1), 5′-GAGATTTAGGCGTGCCTCCGCGAGAC (primer 2) |
| C232G-G235C | 245–224, 219–241 | 5′-CTCGCGGGGGGACCCCCAAATC (primer 1), 5′-CTGGAGATTTGGGGGTCCCCCCG (primer 2) |
| G235A-C232U | 245–216, 219–241 | 5′-CTCGCGGGGGTACACCCAAATC (primer 1), 5′-CTGGAGATTTGGGTGTACCCCCG (primer 2) |
| G229C-C238G | 250–222, 217–245 | 5′-GCAGTCTCGCGGCGGCACGCCGAAATCTC (primer 1), 5′-GCCTGGAGATTTCGGCGTGCCGCCGCGAG (primer 2) |
Positions of mutations are underlined in the mutant name and shown in boldface in the primer sequence. Template plasmid DNAs are shown in boldface in the mutant name.
Transient expression assay.
The transient expression assay used in this study was described previously (4). Briefly, subconfluent monolayers of BHK-21 cells in 6-well plates were infected with vTF7-3, a vaccinia virus expressing T7 RNA polymerase (7) (obtained from B. Moss, National Institutes of Health), at 5 PFU/cell in 300 μl of serum-free medium (OptiMEM; Gibco-BRL) for 30 min at 37°C. The inoculum was removed, and the cells were washed once with OptiMEM. The cells were then transfected with plasmid DNA in 500 μl of OptiMEM containing 15 μl of liposomes prepared as described by Rose et al. (19). Following a 2-h incubation at 37°C, 1 ml of growth medium was added to each of the wells. The cells were harvested 16 h later and assayed as described below. For each construct, four replicate wells were transfected, and standard deviations were calculated from the data obtained for these wells (4).
The amount of luciferase activity was established by using a dual luciferase reporter assay system (Promega) and a model M3 benchtop luminometer (Biotrace). For determination of CAT activity, 50 μl of cell lysate (diluted 1:1,000) was incubated with 50 μl of CAT assay mixture (consisting of 0.2 μCi of d-threo-[dichloroacetyl-1-14C]chloramphenicol, 25 μg of n-butyryl coenzyme A, and 10 μM Tris-HCl [pH 8.0]) for 30 min at 37°C. CAT activity was determined by liquid scintillation counting following tetramethylpentadecane-xylene phase extraction. The results presented are the mean values of data from four independent determinations (4).
RESULTS
In vivo activity of HCV IRES elements with point mutations.
PCR-amplified products of HCV cDNA from three HCV-infected sera corresponding to the entire 5′UTR and the first 15 nt of the core coding region were cloned into pTZ18R. A number of recombinant plasmids from each serum preparation were then sequenced. The sequence of one such isolate, pHCV1b, was identical to that of the prototype HCV-1b 5′UTR isolate (14) deposited in GenBank (accession no. D00832). Mutations arising from the presence of HCV quasispecies or from Taq DNA polymerase misincorporation were identified at various locations throughout the 5′UTR and were named according to the nucleotide positions of each change from the prototype sequence. These single-point mutants, together with the deletion mutants described in Materials and Methods, were tested for their ability to direct the translation of the downstream FFLUC reporter in a dual luciferase-bicistronic reporter assay system. This value was normalized against the activity of the upstream, control RLUC reporter (4). The FFLUC/RLUC ratio was then used as an index of IRES activity, with the translation efficiency of the prototype HCV-1b IRES being arbitrarily set at 100%. For convenience, BHK cells, rather than a hepatocyte-derived line, were used for these experiments; we previously reported that the level of activity of the HCV IRES in BHK cells was similar to that in the liver-derived cell line HepG2 and was about twofold lower than that in another liver derived line, HuH7 (4).
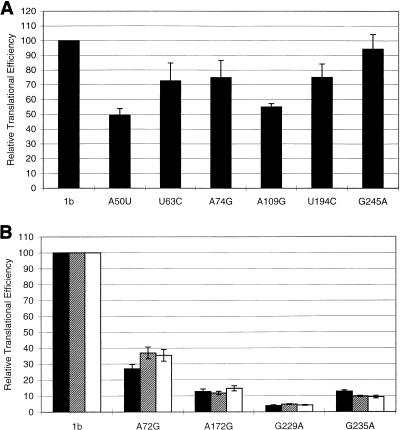
The relative translation initiation efficiencies (RTEs) of the different point mutants studied differed considerably (Fig. 3). IRES domain II (Fig. 1) mutations U63C and A74G showed a marginal effect on translation, resulting in RTEs of 70.7 and 74.8%, respectively, while domain II mutants A72G (28.6% RTE) and A50U (49.2% RTE) were intermediate in their activity. The RTEs of domain III mutants U194C (75.0%) and G245A (95.8%) also were only marginally affected. However, the three mutations at positions A172G, G229A, and G235A, which are all located within or near stem-loop IIIc (Fig. 1), had a more drastic effect on IRES activity (3.9 to 12.7% RTE).
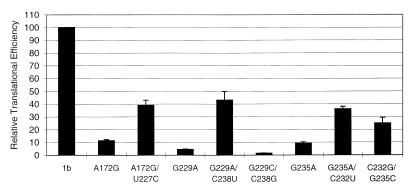
FIG. 3.
Effect of mutations in the 5′UTR of HCV-1b on IRES activity. IRES activity was assessed by measuring the ratio of the levels of RLUC and FFLUC produced from transfected bicistronic plasmid constructs in vTF7-3-infected cells. The activity of the prototype HCV-1b sequence (1b) was taken as 100%. (A) The activities of six 5′UTRs containing single point mutations expressed relative to that of the prototypic HCV-1b sequence are shown. (B) Complementation of mutated 5′UTRs with the prototypic genotype 1b 5′UTR was attempted. Biscistronic constructs expressing mutated 5′UTRs were transfected alone (black bars) or cotransfected with pHCV1bS (hatched bars) or with pHCV1bCAT (white bars) DNA. IRES activity was assessed as described above. Mutant 5′UTRs are designated according to the position of the changed nucleotide.
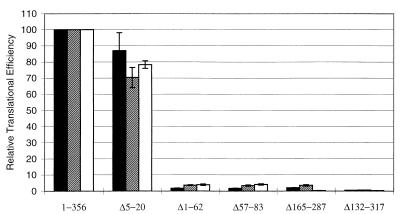
With regard to the five deletion mutations studied (Fig. 4), the loss of domain I (Δ5–20) caused a marginal effect on the translational efficiency, reducing the RTE to about 85%. A low RTE (less than 5% of the prototype level) was observed when the IRES was truncated from the 5′ end (Δ1–62) and when domains II and III were partially deleted (Δ57–83 and Δ165–287, respectively). Deletion of all of domain III (Δ132–317) completely abolished the function of the HCV IRES, resulting in background levels of FFLUC expression.
FIG. 4.
Attempted complementation of mutants with deletions in the HCV-1b 5′UTR. Different HCV-1b 5′UTR deletion mutants either were transfected into vTF7-3-infected cells alone (black bars) or were cotransfected with pHCV1bS (hatched bars) or pHCV1bCAT (white bars) DNA. Relative IRES activity was calculated as described in the legend to Fig. 3.
Site-specific mutagenesis of stem-loop IIIc.
In the secondary-structure model of the 5′UTR of HCV (9), base pairing is predicted to occur between nt A172 and U227 and between nt G229 and C238 while nt C232 and G235 form part of a tetranucleotide loop, all within stem-loop IIIc (Fig. 1). According to this model, point mutations at nt 172 or 229 will disrupt the base pairing required to form the structures in and around stem-loop IIIc, while a mutation at nt 235 alters only the primary sequence. It was stated above that all three mutants (A172G, G229A, and G235A) displayed a dramatic reduction in translation initiation. The large decrease in the ability of these mutants to function efficiently suggests that the stem structures around domain IIIc are particularly important to IRES activity. To investigate further the relevance of the base interactions in these mutants, compensatory base substitutions were introduced into mutants A172G and G229A in order to restore the predicted secondary structure. In addition, a C232U change was introduced into mutant G235A to confirm the single-stranded nature of the proposed IIIc tetraloop. Results from in vivo translation studies of these compensatory mutants are shown in Fig. 5. When predicted base pairings were restored by the introduction of compensatory mutations (A172G/U227C and G229A/C238U), the translation initiation efficiency was only partially recovered (40% RTE), suggesting that maintenance of the secondary structure alone is insufficient for IRES function and that the primary sequence also has an important role to play. Indeed, the computer-generated secondary structures (M-fold [32]) for the wt and the A172G and A172G/U227C mutants were identical, despite their wide differences in IRES activity, although, of course, the possibility that subtle changes to the secondary structure were not detected by the computer program cannot be discounted. Interestingly, mutant G235A/C232U showed increased activity compared to that of mutant G235A; this could be interpreted as indicating that in this system, nt 232 to 235 may not represent a single-stranded loop but may be involved in base pair interactions in a context other than the one suggested in the predicted secondary structure.
FIG. 5.
Comparison of the IRES activities of severely defective point mutants of HCV-1b 5′UTRs and constructs containing compensatory mutations which restore the predicted base pairing. Relative IRES efficiencies were calculated as described in the legend to Fig. 3.
To substantiate further the importance of the primary sequence in this region, two other mutants, G229C/C238G and C232G/G235C, were analyzed. The data showed that sequence alterations in mutant C232G/G235C resulted in a significant decrease in translation efficiency (25% RTE), while translation was essentially abolished in mutant G229C/C238G (1.5% RTE). Taken together, these results suggest a functional importance for both the primary sequence and the maintenance of a secondary structure within stem-loop IIIc.
Complementation of defective IRES elements in vivo.
In 1992, Percy et al. (15) showed that a defective poliovirus IRES could be complemented in trans by a complete infectious poliovirus cDNA. It was presumed that poliovirus infection, resulting in the down-regulation of cap-dependent translation, allowed the otherwise-defective poliovirus IRES to function efficiently. It was further suggested that this occurred by the increased availability of the canonical cellular translational machinery. Subsequently it has been shown that this is not the case and that complementation of a defective poliovirus IRES requires neither poliovirus-encoded proteins nor the inhibition of cap-dependent translation (26). These studies have since been extended to other members of the family Picornaviridae, such as foot-and-mouth disease virus (5) and encephalomyocarditis virus (EMCV) (29).
In the present study, using the mutants described above, we investigated whether these findings might also apply to the HCV IRES. Bicistronic constructs containing HCV-1b IRES point and deletion mutants were transfected into BHK-21 cells together with plasmid pHCV1bS, which contains the prototype (wt) HCV-1b IRES under the control of a T7 promoter, followed by a T7 terminator sequence. All of the plasmids tested expressed RLUC, but only the pRL:HCV1b bicistronic construct, containing the prototype HCV-1b IRES, produced FFLUC efficiently, albeit at a slightly reduced level. No enhancement of expression of FFLUC was shown by any of the constructs containing a defective HCV-1b IRES under these conditions (Fig. 3b and 4).
To confirm that the prototypic HCV-1b IRES sequence was functional under the conditions described above, the experiments were repeated, using a monocistronic plasmid, pHCV1bCAT, in place of pHCV1bS. pHCV1bCAT contains the prototype HCV-1b IRES element with a downstream CAT reporter gene. Hence, CAT, RLUC, and FFLUC could be assayed simultaneously. As expected, significant CAT activity was detected in all lysates of cells transfected with pHCV1bCAT (data not shown). As with the previous set of experiments, however, no enhancement of the translation efficiency of the defective IRES elements in the bicistronic luciferase construct was achieved (Fig. 3b and 4).
To ensure that the failure of the prototype HCV-1b IRES to complement the activity of various defective mutants was not an artifact of the experimental system employed, an attempt was made to complement the activity of a defective EMCV IRES with a wt EMCV sequence, using constructs generously supplied by G. Belsham (18, 29). BHK-21 cells were transfected with bicistronic reporter constructs pGEM-CAT/EMC/LUC, containing a wt EMCV IRES sequence, or pGEM-CAT/DAvr/LUC, containing a defective EMCV IRES sequence. Cells transfected with pGEM-CAT/DAvr/LUC were cotransfected with pD1+D2+D3, which represents the entire EMCV IRES sequence without any reporter sequences present, or a mock control. In the absence of pD1+D2+D3, pGEM-CAT/DAvr/LUC showed an activity of 0.35% compared to the activity of pGEM-CAT/EMC/LUC (100%). However, when cells were cotransfected with pD1+D2+D3, the activity of pGEM-CAT/DAvr/LUC increased to 10.5% of the wt activity (data not shown).
DISCUSSION
Domain III of the HCV 5′UTR represents a relatively large secondary structure with multiple stem-loops (Fig. 1), and deletion analysis (31) has demonstrated that the integrity of this domain is important for translation initiation. We have found that single point mutations in one of these stem-loops, domain IIIc, cause a dramatic loss of IRES function. However, a point mutation in the apical loop of domain III (domain IIIb, U194C) had only a marginal effect on translation, similar to results reported by Wang et al. (31) and Buratti et al. (3). The available data (this work and reference 17) suggest that the structure encompassing stem-loops IIIa and IIIc is critical for IRES function, and it is interesting that sequences in this region are also highly conserved among different genotypes of HCV, GB virus B (23), and the animal pestiviruses responsible for bovine viral diarrhea and hog cholera (1). This high degree of conservation also implies that this region is important for IRES function. One possibility is that these sequences are involved in the maintenance of the secondary and/or tertiary structure of the HCV IRES. Another possibility is that domain IIIc is a binding site for cellular or viral proteins. This is particularly relevant in light of the recent publications of Pestova et al. (16) and Sizova et al. (24), whose work suggests that the apical region of domain III, including stem-loops IIIa and IIIc, represents a binding site for the canonical eukaryotic initiation factor eIF-3 and that this structure may influence the binding of the ribosome at the initiation codon.
Complementation of defective picornavirus IRES elements has been described for poliovirus (15, 26), foot-and-mouth disease virus (5), and EMCV (18, 29). From these studies, it can be concluded that complementation takes place in trans in a highly sequence-specific manner and possibly involves RNA-RNA interactions. This might occur either by complementation of the defective IRES structure with regions of the wt IRES or by transfer of translational machinery between the two. However, when we used the prototypic IRES of HCV-1b (pHCV1bS and pHCV1bCAT) to attempt complementation of defective single-point and large deletion mutants, no complementation was observed. Two particular controls were employed to ensure that the failure of the wt HCV IRES to complement a defective IRES was not an artifact of the experimental system. First, the complementation of a defective EMCV IRES (29) was repeated and confirmed by using our expression system. Second, as described above, the inclusion of a third reporter, CAT, in the complementation experiments confirmed that the protypic HCV-1b IRES was active in this setting. It therefore seems unlikely that the failure to complement defective HCV IRES elements was due to our experimental conditions. It is also unlikely that it is a function of the mutants used in the study, since several mutants with mutations in different regions of the HCV IRES were randomly chosen for attempted complementation. Thus, these data suggest that unlike picornavirus IRES elements, HCV IRES elements can act only in cis.
To date, there is little understanding of the mechanism of IRES function. Based on the observation that mutations in different areas of the picornavirus IRES require distinct regions of the wt IRES for complementation, Roberts and Belsham (18) speculate that a discontinuous transfer of an initiation complex may be performed by the IRES and that this may occur in trans as well as in cis. According to our results, such a mechanism is not operative in translation of HCV RNA. Kamoshita et al. (12) recently utilized allele replacement experiments to identify the structures that influence the efficiency of HCV 5′UTR-directed translation initiation and found that IRES activity is determined by multiple regions within the IRES. Their results suggest that HCV IRES function is supported by a highly ordered structure formed by the entire IRES segments. The strong requirement for both the complete 5′UTR and some coding sequence is in sharp contrast to IRES elements of most picornaviruses (although recently Graff and Ehrenfeld [8] reported that viral coding sequences enhanced hepatitis A virus IRES activity in vitro and Kaminiski and Jackson (11) reported that the presence of EMCV coding sequences could have an influence on the cognate IRES, depending on the nature of the downstream reporter). This model is also supported by the results of studies by other groups (reviewed in references 13 and 16) which provide strong evidence that ribosomes enter the HCV IRES directly on the initiation codon, prior to translation initiation. This would allow little scope for transfer of translational machinery from one IRES to another. These previous observations, combined with the results of our complementation study, suggest another fundamental difference between the IRES elements of HCV and picornaviruses.
ACKNOWLEDGMENTS
This work was supported by a Medical Research Council (ROPA) project grant to R.M.E., a Wellcome Trust travelling fellowship (047027/2/96/2) to S.T., and a Wellcome Trust equipment grant (046745/Z/96).
We thank G. Belsham for reagents and helpful discussion and L. Taylor for automated sequencing.
REFERENCES
- 1.Brown E A, Zhang H, Ping L H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bukh J, Purcell R H, Miller R H. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buratti E, Gerotto M, Pontisso P, Alberti A, Tisminetzky S G, Baralle F E. In vivo translational efficiency of different hepatitis C virus 5′-UTRs. FEBS Lett. 1997;411:275–280. doi: 10.1016/s0014-5793(97)00715-1. [DOI] [PubMed] [Google Scholar]
- 4.Collier A J, Tang S X, Elliott R M. Comparison of the translation efficiencies of the six major genotypes of hepatitis C virus using a new novel bicistronic reporter assay system. J Gen Virol. 1998;79:2359–2366. doi: 10.1099/0022-1317-79-10-2359. [DOI] [PubMed] [Google Scholar]
- 5.Drew J, Belsham G J. trans complementation by RNA of defective foot-and-mouth disease virus internal ribosome entry site elements. J Virol. 1994;68:697–703. doi: 10.1128/jvi.68.2.697-703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn E F, Pritlove D C, Jin H, Elliott R M. Transcription of a recombinant bunyavirus RNA template by transiently expressed bunyavirus proteins. Virology. 1995;211:133–143. doi: 10.1006/viro.1995.1386. [DOI] [PubMed] [Google Scholar]
- 7.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graff J, Ehrenfeld E. Coding sequences enhance internal initiation of translation by hepatitis A virus in vitro. J Virol. 1998;72:3571–3577. doi: 10.1128/jvi.72.5.3571-3577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda M, Brown E A, Lemon S M. Stability of a stem loop involving the initiator AUG controls the efficiency of internal initiation of translation of hepatitis C virus. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 10.Houghton M, Selby M, Weiner A, Choo Q-L. Hepatitis C virus. Curr Stud Hematol Blood Transfus. 1994;61:1–11. doi: 10.1159/000423264. [DOI] [PubMed] [Google Scholar]
- 11.Kaminski A, Jackson R J. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamoshita N, Tsukiyama-Kohara K, Kohara M, Nomoto A. Genetic analysis of internal ribosome entry site on hepatitis C virus RNA: implication for involvement of the highly ordered structure and cell type-specific transacting factors. Virology. 1997;233:9–19. doi: 10.1006/viro.1997.8600. [DOI] [PubMed] [Google Scholar]
- 13.Lemon S M, Honda M. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin Virol. 1997;8:274–288. [Google Scholar]
- 14.Okamoto H, Okada S, Sugiyama Y, Hurai K, Iizuka H, Machida A, Miyakawa Y, Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991;72:2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- 15.Percy N, Belsham G J, Brangwyn J K, Sullivan M, Stone D M, Almond J W. Intracellular modifications induced by poliovirus reduce the requirement for structural motifs in the 5′ noncoding region of the genome involved in internal initiation of protein synthesis. J Virol. 1992;66:1695–1701. doi: 10.1128/jvi.66.3.1695-1701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pestova T V, Shatsky I N, Fletcher S P, Jackson R T, Hellen C U T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNA. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspé G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 18.Roberts L, Belsham G J. Complementation of defective picornavirus internal ribosome entry site (IRES) elements by the coexpression of fragments of the IRES. Virology. 1997;227:53–62. doi: 10.1006/viro.1996.8312. [DOI] [PubMed] [Google Scholar]
- 19.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Simmonds P, Holmes E C, Cha T A, Chan S W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into 6 major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 22.Simmonds P, Alberti A, Alter H J, Bonino F, Bradley D W, Brechot C, Brouwer J T, Chan S W, Chayama K, Chen D S, Choo Q L, Colombo M, Cuypers H T M, Date T, Dusheiko G M, Esteban J L, Fay O, Hadziyannis S J, Han J, Hatzakis A, Holmes E C, Hotta H, Houghton M, Irvine B, Kohara M, Kolberg J A, Kuo G, Lau J Y N, Lelie P N, Maertens G, McOmish F, Miyamura T, Mizokami M, Nomoto A, Prince A M, Reesink H W, Rice C, Roggendorf M, Schalm S W, Shikata T, Shimotohno K, Stuyver L, Trepo C, Weiner A, Yap P L, Urdea M S. A proposed nomenclature of hepatitis C genotypes. Hepatology. 1994;19:1321–1324. [PubMed] [Google Scholar]
- 23.Simons J N, Pilot-Matias T J, Leary T P, Dawson G J, Desai S M, Schlauder G G, Muerhoff A S, Erker J C, Buijk S L, Chalmers M L, Yansant C L, Mushahwar I K. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sizova D V, Kolupaeva V G, Pestova T V, Shatsky I N, Hellen C U T. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith D B, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P-L, Simmonds P the International HCV Collaborative Study Group. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J Gen Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 26.Stone D M, Almond J W, Brangwyn J K, Belsham G J. trans complementation of cap-independent translation directed by poliovirus 5′ noncoding region deletion mutants: evidence for RNA-RNA interactions. J Virol. 1993;67:6215–6223. doi: 10.1128/jvi.67.10.6215-6223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang S X, Meng Q H, Ma X K, Zhang X T, Jiang Y T. HCV RNA detection and genotyping by polymerase chain reaction in anti-HCV positive professional blood donors in China. Chin J Hepatol. 1994;2:33–35. [Google Scholar]
- 28.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site with hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Velden A, Kaminski A, Jackson R J, Belsham G J. Defective point mutants of the encephalomyocarditis virus internal ribosome entry site can be complemented in trans. Virology. 1995;214:82–90. doi: 10.1006/viro.1995.9952. [DOI] [PubMed] [Google Scholar]
- 30.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]