Summary
Cytokine storms caused by viruses are associated with elevated cytokine levels and uncontrolled inflammatory responses that can lead to acute respiratory distress syndrome. Current antiviral therapies are not sufficient to prevent or treat these complications. Cyclophilin A (CypA) is a key factor that regulates the production of multiple cytokines and could be a potential therapeutic target for cytokine storms. Here, three proteolysis targeting chimeras (PROTACs) targeting CypA were designed. These PROTACs bind to CypA, enhance its ubiquitination, and promote its degradation in both cell lines and mouse organs. During influenza B virus (IBV) infection, PROTAC-mediated CypA depletion reduces P65 phosphorylation and NF-κB-mediated proinflammatory cytokine production in A549 cells. Moreover, Comp-K targeting CypA suppresses excessive secretion of proinflammatory cytokines in bronchoalveolar lavage fluid, reduces lung injury, and enhances survival rates of IBV-infected mice. Collectively, we provide PROTACs targeting CypA, which are potential candidates for the control of cytokine storms.
Subject areas: Immunology, Molecular biology, Virology
Graphical abstract

Highlights
-
•
PROTAC binds to CypA and promotes its ubiquitination-mediated degradation
-
•
PROTAC alleviates cytokine storm during virus infection
-
•
PROTAC relieves lung injury and enhances survival rates of IBV-infected mice
Immunology; Molecular biology; Virology
Introduction
Cytokine storm is a phenomenon characterized by exaggerated inflammation due to elevated circulating cytokines that most often occur after virus infection, such as those caused by influenza viruses,1 coronaviruses,2 and herpesviruses.3 For example, the H5N1 influenza A virus (IAV) can trigger cytokine storm, including but not limited to the upregulation of IL-1β, IL-6, IL-8, TNFα, CCL2 (MCP-1), CCL3 (MIP-1α), CCL5 (RANTES), and CXCL10 (IP-10).4,5 Influenza B virus (IBV) is more likely to infect children.6,7 The cytokine storm and clinical manifestations caused by IBV infection in children are similar to those caused by IAV.8 Several studies have found that patients with severe COVID-19 exhibit higher levels of IL-2, IL-6, IL-7, IL-10, CXCL10, CCL2, TNF-α, macrophage inflammatory protein 1 alpha (MIP-1α), and granulocyte-colony stimulating factor (G-CSF) than patients with mild and moderate infections. The disproportionate levels of chemokines and cytokines recruit excessive amounts of macrophages and neutrophils to the site of infection, culminating in a cytokine storm.9,10,11 Cytokine storm-induced pneumonia can lead to high morbidity and mortality.12 Currently, the therapeutic options to control cytokine storms include steroids, intravenous immunoglobulins, JAK inhibitors, and selective cytokine blockade treatments, such as anakinra, IL-1 receptor antagonist, and IL-6 receptor antagonist.13,14,15 However, the various treatments mentioned above have different side effects and risk profiles. Corticosteroids can lead to fractures, additional infections, Cushing’s syndrome, and psychiatric disorders, which may eventually lead to side effects that are more damaging than the original disease.16,17,18 As for cytokine-targeting therapy, the results of clinical trials targeting either the IL-1 or IL-6 pathway show no survival benefits in COVID-19 patients19,20 unless combined with corticosteroids.21,22 These results indicate that the inhibition of a single cytokine pathway may not be sufficient to control cytokine storms. Therefore, targeting key cellular and molecular determinants of multiple cytokine pathways should be investigated.
Cyclophilin A (CypA), an 18 kD protein encoded by PPIA and ubiquitously expressed in all types of cells. CypA has peptidyl prolyl cis-trans isomerase activity which can be inhibited by immunosuppressive drug cyclosporine A (CsA).23 As a multifunctional protein, CypA is mainly involved in protein folding and transport,24 signal transduction, and immunoregulation.25 Several lines of evidence indicate a pivotal role for CypA in cytokine production. CypA positively regulates IL-6-soluble IL-6R-gp130–JAK-STAT3 signaling pathway, increasing downstream iNOS and IL-6 by inhibiting the degradation of gp130.26 CypA promotes IL-1β maturation and secretion by enhancing K63-linked ubiquitination of pro-IL-1β in the early stages of LPS-mediated inflammation.27 CypA increases interferon (IFN) and proinflammation cytokines production by targeting RIG-I, MAVS, and P65, and CypA deficiency leads to reduced inflammatory responses in the lungs of SeV-infected mice.28,29 Moreover, CypA positively regulates the expression of integrin α5 and actin rearrangement via the FAK/Akt signaling pathway to facilitate group A Streptococcus adhesion and invasion, and enhances lung inflammatory infiltration.30 Furthermore, it is well documented that CypA is secreted in response to oxidative stress and virus infection, and promotes inflammation.31 Extracellular CypA plays an essential role in inflammation under different conditions, promoting leukocyte redistribution to inflamed tissues and local production of cytokines.32 Evaluated extracellular CypA levels were observed in the bronchoalveolar lavage fluids of patients with acute respiratory distress syndrome.33 Large amounts of CypA were found in the joint fluid of patients with rheumatoid arthritis and the extracellular CypA enhanced immune cell migration.34 Extracellular CypA induces activation of ERK1/2, JNK, and p38 MAPK pathways, leading to proinflammatory cytokine expression.35 Together, these findings suggest that CypA is a key player in multiple cytokine production, suggesting that CypA could be a potential therapeutic target for cytokine-mediated inflammatory diseases.
Proteolysis targeting chimeras (PROTACs) is a new technology developed in recent years for the degradation of intracellular target proteins based on the ubiquitin-proteasome system. PROTAC can connect E3 ubiquitin ligase and intracellular target protein into ternary complexes, thus allowing the target protein to be modified by ubiquitination and finally degraded by proteasome recognition.36 Currently, PROTAC technology is widely used in the field of drug development, especially for cancer treatment, such as breast cancer,37 intestinal cancer,38 lymphocytic leukemia,39 and prostate cancer.40 It is also applied in other fields, such as Huntington’s disease,41 Alzheimer’s disease, and Parkinson’s disease.42 Application of PROTAC in cytokine-mediated inflammatory diseases, however, has not been reported yet. Given that many functions of CypA are not dependent on its enzymatic activity,43,44 PROTACs that degrade CypA should control the production of multiple cytokines more effectively than CypA inhibitors.
In the present study, we design PROTACs to degrade CypA and determine the role of these compounds in the control of cytokine storm induced by IBV infection. Our data indicate that PROTACs can degrade CypA both in vitro and in vivo, and PROTACs-mediated CypA depletion reduces inflammatory responses and lung injury in IBV-infected mice.
Results
Design and identification of PROTACs targeting CypA
Three PROTACs (Comp-K, L, and M) targeting CypA were designed and synthesized based on two CypA ligands, 2d (CypA-L1) and 3c (CypA-L2)45 and the Hippel-Lindau (VHL) ligand VH032,46 which are conjugated with a linker (Figures 1A-1C, Data S1–S6). To investigate the binding characteristics of these compounds to CypA, the molecular docking studies were performed using Autodock Vina 4,47 which gives deep insight into the binding pattern between these PROTACs and CypA. Crystal structures of CypA were obtained from the Protein DataBank (PDB ID:3k0m). CypA has two main pockets. Pocket A is hydrophilic and pocket B is hydrophobic.48 We discovered that the cyclohexane of Comp-K and L are both binding to the amino acid PHE60 (Comp-K, 5.8 Å), MET61 (Comp-L, 4.7 Å), PHE113 (Comp-K, 5.0 Å; Comp-L, 4.6 Å), LEU122 (Comp-K, 5.4 Å; Comp-L, 4.6 Å), and HIS126 (Comp-K, 4.9 Å; Comp-L 5.5 Å), which together make up the hydrophobic pocket B, in Alkyl or Pi/Alkyl interaction. In addition, Comp-K also binds to ASN102 and GLN63 with hydrogen bond, and ALA103 with Alkyl interaction (Figure 1D). Comp-L binds to ARG55 through donor-acceptor interaction (Figure 1E). Comp-M has a fluorene group, which binds to PHE60 (5.9 Å), PHE113 (8.3 Å), LEU122 (5.2 Å), and HIS126 (5.9 Å). Comp-M also binds to ASN102 and ALA103 (Figure 1F). Meanwhile, there are many amino acids that bind to compounds through van der Waals force, such as PHE53, ALA101, GLN111, ASN71, HIS54, TRP121, and GLN63. Furthermore, surface plasmon resonance (SPR) was employed to assess the affinity of PROTACs to CypA. CypA was immobilized on the surface of Biacore Chip CM5. Then, various concentrations of compounds were prepared and injected to pass over the surface. The equilibrium dissociation constant (KD) values for CypA against Comp-K, L, and M were 11.74μM, 84.6μM, and 47.67μM (Figures 1G-1I), respectively. Collectively, all three PROTACs designed in this study could bind to CypA.
Figure 1.
Design strategy and identification of PROTACs targeting CypA
(A–C) PROTACs with CypA ligands (red), linkers (black), and E3 ligase ligands (blue) used in this study. A (Comp-K) and B (Comp-L): CypA-recruiting ligand with a cyclohexane. C (Comp-M): CypA-recruiting ligand with a fluorene.
(D–F) The main binding sites of Comp-K (D), L (E), and M (F) on CypA were described by PyMOL. The structure of compounds and CypA were imported to PyMOL, and the bond lengths between amino acids and compounds were measured. Crystal structure of CypA interacting with ligand in the hydrophobic pocket. The pocket is rendered as a white surface with the compounds visualized in stick representation and colored by atoms (gray carbon atoms, red oxygen atoms, blue nitrogen atoms, and yellow sulfur atoms). The bonds are colored by interaction (purple Alkyl hydrophobic bonds, green hydrogen bonds, light green carbon hydrogen bonds).
(G–I) The affinity and binding kinetics of PROTACs for CypA measured by SPR. 2-fold serial dilutions from 3.125 μM compounds in injected onto the captured CypA protein. Kinetic data from one representative experiment were fit to a 1:1 binding model. The profiles are shown for Comp-K (G), L (H), and M (I) (left). The KD values were calculated using a steady affinity state model by Biacore evaluation software 3.2 (right). Data are representative of three independent experiments. The data are presented as the mean ± SD. See also Data S1-S6.
PROTACs promote the degradation of CypA
To investigate the roles of these PROTACs to degrade endogenous CypA, A549 cells were treated with different concentrations of Comp-K, L, or M for 12 h. The western blot results showed that Comp-K, L, and M degraded CypA in a concentration-dependent manner, and the half maximal degraded concentration (DC50) are 44.66 nM, 57.86 nM and 32.9 nM, with the maximum degradation efficiency of over 90% at a concentration of 200 nM (Figures 2A–2C). Then the time dependence of degradation experiment was determined. The degradation efficiencies of these three PROTACs were gradually enhanced after 1 or 3 h treatment, with highest level of CypA degradation being observed at 12 h (Figures 2D–2F). In addition, the cytotoxicity of these PROTACs was evaluated using a CCK-8 assay. Serial dilutions of compound were added to A549 cells and incubated for 12 h. Comp-K and Comp-M did not affect the cell viability at concentrations 1–100 μM. At 1–10 μM, Comp-L did not affect cell viability, but at 100 μM, Comp-L was toxic and the cell survival rate decreased to about 30% (Figure 2G). In addition, CypA expression were not decreased in A549 cells treated with CypA-L1 (CypA ligand in Comp-K and Comp-L), CypA-L2 (CypA ligand in Comp-M), and VH032 (VHL ligand) (Figure 2H). Collectively, Comp-K, L, and M (200 nM) were able to effectively degrade CypA had no effect on A549 cell survival.
Figure 2.
PROTACs degrade endogenous CypA in A549 cells
(A–C) Immunoblot analysis of CypA in A549 cells treated with different concentrations of Comp-K (A), L (B), or M (C) for 12 h. As control, DMSO was treated to A549 cells for 12 h (top). The relative band intensity was quantified with ImageJ and normalized to β-tubulin (bottom).
(D–F) Immunoblot analysis of CypA in A549 cells treated with 200 nM Comp-K (D), L (E), or M (F) for the indicated time (top). The relative band intensity was quantified with ImageJ and normalized to β-tubulin (bottom).
(G) Cell viability of A549 cells treated with different doses of Comp-K, L, or M for 12 h. Cell survival was determined by the CCK-8 assay.
(H) Immunoblot analysis of CypA in A549 cells treated with DMSO, CypA-L1, CypA-L2, VH032, or Comp-K for 12 h (left). The relative band intensity was quantified with ImageJ and normalized to β-tubulin (right). Data are representative of three independent experiments. The data are presented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01 (unpaired, two-tailed Student’s t test). See also Figures S1 and S2.
We then examined the degradation of overexpressed CypA by these PROTACs in 293T/CypA-cells. We observed that these compounds degraded overexpressed CypA in a concentration-dependent manner with the highest degradation efficiency at 200 nM (Figures S1A–S1C). In the time-dependent assays, CypA expression levels were gradually increased in 293T/CypA-cells treated with DMSO, but not in 293T/CypA-cells treated with compounds (Figures S1D–S1F). Moreover, these PROTACs degraded CypA in THP-1 and Jurkat cells (Figures S2A and S2B). To further investigate the specificity of these PROTACs, the expression levels of CypA (Figure S2C), CypB (Figure S2D), CypE (Figure S2E), and CypF (Figure S2F) were detected in A549 cells treated with PROTAC. These compounds did not decrease the expression levels of CypB, CypE, and CypF, compared to CypA degradation, suggesting that these PROTACs specifically degraded CypA. Together, the results demonstrate that these three PROTACs can effectively and specifically degrade CypA.
PROTACs induce proteasomal degradation of CypA by enhancing its K48-linked ubiquitination
These three PROTACs include VHL ligand, which is supposed to recruit VHL to trigger ubiquitin-mediated proteasomal degradation of CypA. As expected, PROTACs enhanced the K48-linked ubiquitination of CypA in 293T cells co-transfected with CypA and K48-Ub (Figure 3A). To investigate whether PROTACs will recruit more VHL to CypA, immunoprecipitation experiments were performed. Anti-CypA antibodies-binding protein A beads were used to detect the interaction between endogenous CypA and VHL in 293T cells. PROTACs recruited more VHL to interact with CypA than DMSO (Figure 3B). We then investigated whether proteasome inhibitors could inhibit the degradation of endogenous CypA via PROTAC. Results showed that the proteasome inhibitor MG132 blocked PROTACs-mediated CypA degradation (Figure 3C). These data suggest that PROTACs promote CypA degradation through recruiting more VHL to CypA via the ubiquitin-proteasome pathway.
Figure 3.
PROTACs enhance proteasomal degradation of CypA by increasing ubiquitination of CypA
(A) Immunoblot analysis of lysates in 293T cells transfected with K48-HA and CypA-Flag plasmids for 18 h and subsequently treated with different concentrations of PROTACs or DMSO and 10 μM MG132 for 12 h, followed by immunoprecipitation with anti-Flag beads.
(B) Immunoblot analysis of lysates in 293T cells treated with 200 nM PROTACs or DMSO for 12 h, followed by immunoprecipitation with anti-CypA-binding protein A beads.
(C) Immunoblot analysis of the lysates in 293T cells treated with or without MG132 (10 μM) and PROTACs (200 nM) or DMSO for 12 h.
(D) Immunoblot analysis of the CypA in 293T cells transfected with CypA-Myc or mutants (K28R-Myc, K154R-Myc, K155R-Myc) for 18 h, then treated with 200 nM PROTACs or DMSO for 12 h.
(E) Immunoblot analysis of lysates in 293T cells transfected with K48-HA and CypA-Myc or mutant plasmids (K28R-Myc, K154R-Myc, K155R-Myc) for 18 h and subsequently treated with 200 nM PROTACs and 10 μΜ MG132 for 12 h, followed by immunoprecipitation with anti-Myc beads.
We then sought to identify the key ubiquitination sites of CypA for PROTAC-mediated degradation. Our previous studies have shown that K154 and K155 are the key ubiquitination sites for CypA degradation.26 We found that PROTACs induced the degradation of wildtype CypA and K28R mutants, but no longer promoted CypA degradation with of K154R or 155R mutant (Figure 3D). This suggests that K154 and K155 are the key sites for PROTAC-mediated CypA degradation. Similarly, K154R and K155R mutations led to a decrease in PROTACs-induced CypA ubiquitination, while the K28R mutation did not (Figure 3E). Given that K154 and K155 are adjacent, it is possible that a single amino acid mutation will affect the ubiquitination at both sites or confer a confirmational change to the target. Together, PROTACs enhance the K48-linked ubiquitination of CypA by recruiting more VHL to CypA, which facilitates the proteasomal degradation of CypA.
PROTAC-mediated CypA degradation reduces IBV-induced inflammatory responses
It is well documented that CypA is a key regulator of multiple cytokine production and may be a promising therapeutic target for treating cytokine storm. To investigate the effects of PROTAC on CypA-regulated proinflammatory cytokine production, an IBV infection model was established in A549 cells. Our previous studies have shown that influenza A virus is able to induce CypA expression.28,30 Immunoblot assays (Figures 4A–4C) showed that CypA expression was gradually up-regulated by IBV and reached a peak at 12 h post infection, and PROTACs significantly degraded CypA at 12, 24, and/or6 h post infection (Figures 4A–4C). As the controls of PROTACs, CypA-L1, CypA-L2, and VH032 did not degrade CypA (Figure 4D). It has been reported that CypA interacts with RIG-I, MAVS, and P65 to promote the expression of downstream inflammatory cytokines.49,50 As expected, PROTACs inhibited K63-linked RIG-I ubiquitination (Figure S2G) and promoted MAVS degradation (Figure S2H). We further observed an increase in phosphorylation levels of P65 in A549 (Figures 4E–4G) cells during IBV infection, but noted a significant decrease when cells were treated with PROTACs. CypA-L1 seemed to have a slight effect, but CypA-L2 and VH032 have no obvious effect on P65 phosphorylation (Figure 4H). Moreover, mRNA levels of downstream IL-1B, TNFA, and CXCL10 were all inhibited by PROTACs at 12 h post infection (Figure 4I). In addition, PROTACs inhibited P65 phosphorylation and proinflammatory cytokine production in THP-1 cells triggered by IBV or LPS (Figure S3). Interestingly, we observed that CypA-L1 and CypA-L2, especially Comp-K, decreased IBV nucleoprotein (BNP) expression in A549 cells (Figure 4J), suggesting that CypA might promote IBV replication. To explain this phenomenon, BNP expression was detected in the wild-type and CypA-knockdown A549 cells. We found that CypA knockdown reduced the mRNA levels of BNP (Figure 4K). These data demonstrate that PROTACs targeting CypA significantly reduce IBV-induced inflammatory responses in vitro.
Figure 4.
PROTACs decrease NF-κB-mediated proinflammatory cytokine production in IBV-infected A549 cells by degrading CypA
(A–C) Immunoblot analysis of the CypA in A549 cells treated with 200 nM Comp-K (A), L (B), or M (C) and IBV (MOI = 1) for 0, 6, 12, and 24 h (top). The CypA expression levels were quantified with ImageJ and normalized to β-tubulin (bottom).
(D) The immunoblot analysis of CypA in A549 cells treated with 200 nM Comp-K, CypA-L1, CypA-L2, or VH032 and IBV for 0, 12, and 24 h (left). The relative band intensity was quantified with ImageJ and normalized to β-tubulin (right).
(E–G) The immunoblot analysis of P65 phosphorylation in A549 cells treated with 200 nM Comp-K (E), L (F), or M (G) and IBV for 0, 1, 2, 4, and 8 h.
(H) The immunoblot analysis of P65 phosphorylation in A549 cells treated with 200 nM Comp-K, CypA-L1, CypA-L2, or VH032 and IBV for 0, 3, and 6 h.
(I) qPCR analysis of IL-1β, TNF-α, and CXCL10 mRNA in A549 cells treated with 200 nM Comp-K, L, or M and IBV for 12 h.
(J) qPCR analysis of BNP mRNA levels in A549 cells treated with 200 nM Comp-K, CypA-L1, CypA-L2 or VH032 and IBV for 8 h.
(K)qPCR analysis of BNP mRNA levels in A549/CypA+ and A549/CypA-cells infected with IBV (MOI = 1) at the indicated time points. Data are representative of three independent experiments. The data are presented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01 (unpaired, two-tailed Student’s t test). See also Figures S2 and S3.
Comp-K promotes CypA degradation in mice
Considering that Comp-K has the highest affinity and solubility among these three PROTACs, we next investigated the effect of Comp-K on CypA degradation in mice. Mice were infected intranasally with IBV, and then intravenously injected with Comp-K on day 1 post-infection. The results of immunoblotting assays showed that Comp-K can effectively degrade CypA in lung tissues of mice with or without IBV infection for 3 to 4 days (Figure 5A). Pneumonia is a common consequence of cytokine storms caused by pathogen infection, such as influenza virus, coronavirus, and respiratory syncytial virus.10 The anti-inflammatory effects of PROTACs were then evaluated in IBV-infected mice. Comp-K was used as a representative of the PROTACs for anti-inflammatory therapy and oseltamivir (OSE) was used for antiviral therapy.51 Mice infected with IBV were then treated with OSE, CypA-L1, Comp-K, combination of OSE and Comp-K, or vehicle (10% DMSO). Intraperitoneal injections of OSE were performed daily and intravenous injections of Comp-K or CypA-L1 were performed on days 1 and 3 post-infection. Samples of lung tissue, spleen tissue, and bronchoalveolar lavage fluid were collected on days 2, 4, and 6 post-infection (Figure 5B). The protein levels of CypA in the lungs and spleens of mice were tested. The results showed that the CypA levels in mice treated with Comp-K or the combination of OSE and Comp-K were significantly reduced, compared with those in mice treated with vehicle, OSE, or CypA-L1 (Figures 5C and 5D). Notably, the combination of OSE and Comp-K showed apparent effects on reducing CypA levels compared to Comp-K monotherapy. Considering the induction effect of IBV on CypA expression (Figures 4A–4C), the most likely reason is that OSE might reduce CypA expression by inhibiting IBV replication, but this effect was not significant and was enhanced when OSE was used in combination with Comp-K. Additionally, the results of hematoxylin and eosin staining (Figure S4A) and blood routine analysis (Figure S4B) show that Comp-K is safe for mice.
Figure 5.
Comp-K effectively degrades CypA in vivo
(A) Immunoblot analysis of CypA in the lungs of mice (n = 5) infected intranasally with IBV (3000 pfu) or PBS for 1 day then intravenously injected with Comp-K (10 mg/kg) or Vehicle at 1-day post infection. The lung tissue was collected at 0, 1, 2, 3, 4, 5, and 6 days after treated with IBV or PBS. The CypA levels were quantified with ImageJ and normalized to β-tubulin, the ratio was marked between CypA blot and β-tubulin blot.
(B) Schematic diagram of a therapy experiment of IBV-induced cytokine storm. Mice (n = 5) were infected with IBV (3000 pfu) or PBS, then treated with OSE (10 mg/kg), CypA-L1 (10 mg/kg), Comp-K (10 mg/kg), combination of OSE and Comp-K, or vehicle (10% DMSO). Intraperitoneal injections of OSE were performed daily and intravenous injections of CypA-L1 or Comp-K were performed on day 1 and 3 post-infection. The lung tissue, spleen tissue, and bronchoalveolar lavage fluid samples were collected at days 2, 4, and 6 post-IBV infection.
(C-D) Immunoblot analysis of CypA in the lungs (C) or spleens (D) of mice (n = 5) (left). The CypA expression levels were quantified with ImageJ and normalized to β-tubulin (right).
Data are representative of three independent experiments. The data are presented as the mean ± SD. ∗∗p < 0.01 (unpaired, two-tailed Student’s t test). See also Figure S4.
Inhibitory effect of Comp-K on cytokine storm in IBV-infected mice
Cytokine storm can lead to acute respiratory distress syndrome and multi-organ failure, as has been found in influenza virus-infected patients.52 Moreover, in A549 cells, the robust production of type I and III IFNs, IFN-stimulated genes, and proinflammatory factors can be induced by IBV rather than IAV infection.8,53 Therefore, the 36 major cytokines in bronchoalveolar lavage fluid from mice in Figure 5B was detected using ProcartaPlex Multiplex Immunoassays. Results showed that at all three time points, the overall cytokine levels in mice treated with Comp-K or OSE and Comp-K combination were significantly lower than those in mice treated with vehicle, OSE, or CypA-L1 (Figure 6A). Changes in the expression of key inflammatory cytokines are shown in Figure 6B. The cytokine levels in mice treated with Comp-K, or the combination of OSE and Comp-K were significantly reduced, compared with those in mice treated with vehicle, OSE, or CypA-L1. Downregulation of cytokines on day 2 after infection included chemokines, such as CXCL1 and CXCL2, as well as interleukins, such as IL-1β and IL-6. On day 4 post-infection, Comp-K inhibited the production of interferons, such as IFN-α and IFN-γ, and interleukins, such as IL-5 and IL-6. On day 6 post-infection, Comp-K inhibited the production of cytokines, such as IL-5, IFN-γ, CCL3, and M-CSF. Furthermore, we quantified the major pro-inflammatory cytokines in bronchoalveolar lavage fluid by ELISA. Results showed that cytokines IL-1β, IL-6, CCL2, and CXCL10 were reduced to varying degrees in OSE-, CypA-L1-, Comp-K-, or the combination of OSE and Comp-K-treated mice, among which OSE and Comp-K combination had the highest inhibition efficiency (Figure 6C). To further investigate the effects of Comp-K-mediated CypA knockdown on immune cell infiltration, mouse lung sections were subjected to immunofluorescence assays. There were fewer macrophages (Figure S5A) and T lymphocytes (Figure S5B) were found in the lungs of mice treated with Comp-K, OSE, CypA-L1, or the combination of OSE and Comp-K compared with those in the lungs of mice treated with vehicle, which is consistent with the results of lung injury and inflammatory responses. These data clearly demonstrate that Comp-K attenuates cytokine storm and immune cell infiltration in IBV-infected mice.
Figure 6.
Comp-K inhibits cytokine storm in IBV-infected mice
(A-B) ProcartaPlex Multiplex immunoassays of cytokines/chemokines in the bronchoalveolar lavage fluid of mice (n = 5). Mice were treated with OSE, CypA-L1, Comp-K, combination of OSE and Comp-K, or vehicle as described in Figure 5B and samples were collected at days 2, 4, and 6 post-IBV infection. (A) Ridge plot of the density distribution of log2 fold change (LFC). Dotted lines indicate mean LFC. (B) Heatmap of cytokines/chemokines panels altered in different groups at the indicated time points. Blue indicate LFC = 0 and orange indicate LFC = 1.2.
(C) ELISA analysis of the levels of IL-1β, IL-6, CCL2, and CXCL10 in bronchoalveolar lavage fluid of mice (n = 5) at day 2, 4, and 6 post IBV infection. Data are representative of three independent experiments. The data are presented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01 (unpaired, two-tailed Student’s t test). See also Figure S5.
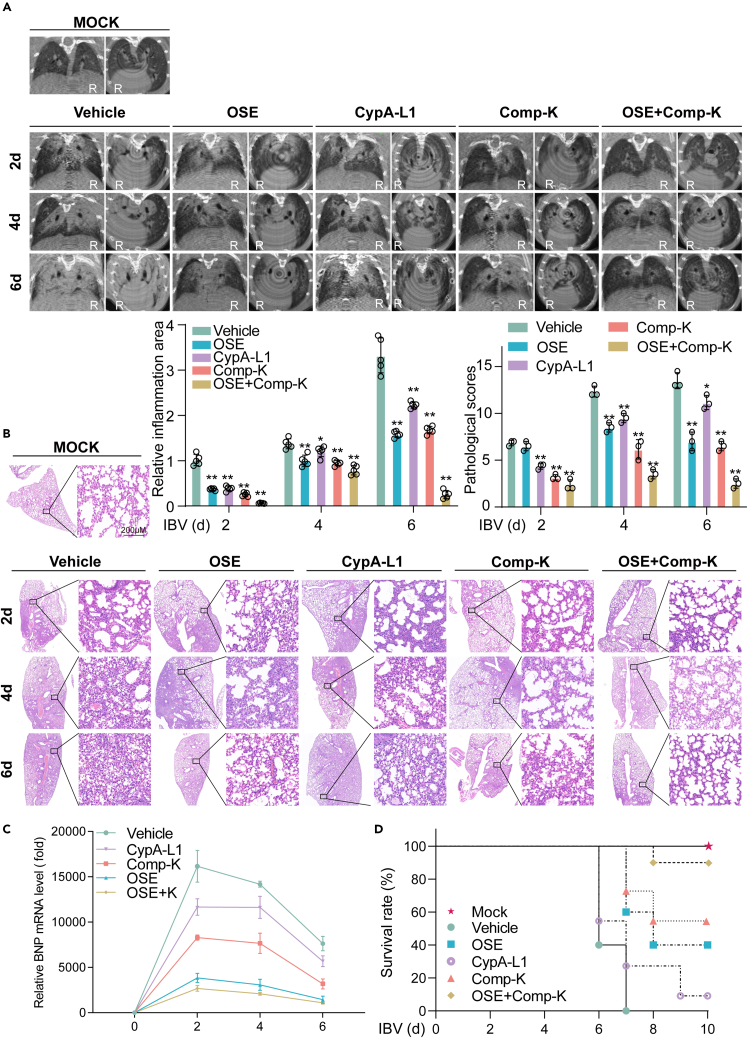
Comp-K alleviates lung damage and improves survival rate of IBV-infected mice
Micro-X-ray computed tomography (μCT) is one of the standard tools used to observe the anatomical basis and pathophysiology of lung injury in live animals.54 To determine the therapeutic effect of Comp-K on viral pneumonia caused by IBV, the mouse lungs were imaged with Micro-CT. Slight lesions were observed in all mice on day 2 post-infection. On day 4 post-infection, the area of high-density shadow in the lungs of mice treated with OSE, CypA-L1, Comp-K, or the combination of OSE and Comp-K was smaller than that of the vehicle-treated mice. On day 6 post-infection, the lungs of vehicle-treated mice were almost completely solidified. In contrast, the area of high-density shadow in the lungs of mice treated with OSE, CypA-L1, or Comp-K was smaller than that in the vehicle-treated mice. Notably, the mildest lung injury was observed in the lungs of mice treated with the combination of OSE and Comp-K (Figure 7A). The lung injury was then assessed by hematoxylin and eosin staining and pathological scores. Consist with the results of Micro-CT, mice treated with OSE or Comp-K showed less inflammatory cells infiltration, lung consolidation, and congested blood vessels. The combination of OSE and Comp-K had the best therapeutic effectiveness (Figure 7B). In addition, CypA-L1, Comp-K, especially OSE and combination of OSE and Comp-K, decreased BNP mRNA expression level in lungs from IBV-infected mice (Figure 7C). Meanwhile, the survival rates of IBV-infected mice treated with combination of OSE and Comp-K, Comp-K, or OSE were 90%, 60%, and 40% respectively, while those treated with CypA-L1 were just 10% and those treated with vehicle were all dead (Figure 7D). It is indicated that Comp-K treatment effectively enhanced the survival rates of IBV-infected mice. Collectively, we demonstrate that Comp-K, especially the combination of Comp-K and OSE, alleviates lung damage and increases survival rates of mice infected by IBV.
Figure 7.
Comp-K reduces lung damage and increases survival rates of mice infected by IBV
(A) Micro-X-ray computed tomography imaging of the lungs of IBV-infected mice (n = 5). Mice were treated with OSE, CypA-L1, Comp-K, combination of OSE and Comp-K, or vehicle as described in Figure 5B and examined at days 2, 4, and 6 post-IBV infection. Low density shadow (black) represents normal lung tissue. Moderate density shadow (gray) represents the inflammatory part or muscle-connective tissue. High density shadow (white) represents bone. Plain film and tomographic images were shown. Voltage 90 kV, Current 180 μA, FOV 24 mm, Scan Technique 4.5 min. The relative inflammation areas of lungs from five mice per group (moderate density shadow area/whole lung area) was quantified with ImageJ (bottom-left).
(B) Hematoxylin and eosin (H & E) staining of the lung tissue from mice (n = 5). Mice were treated with OSE, CypA-L1, Comp-K, combination of OSE and Comp-K, or vehicle as described in Figure 5B and samples were collected at days 2, 4, and 6 post-IBV infection. The lung injury (three sections from five mice per group) was analyzed in a blinded manner (top-right). Scale bars, 200 μm.
(C) qPCR analysis of BNP mRNA level in lungs of mice (n = 5) at days 2, 4, and 6 post IBV infection.
(D) Survival was evaluated in mice (n = 10). Mice were treated with OSE, CypA-L1, Comp-K, combination of OSE and Comp-K, or vehicle as described in Figure 5B and monitored daily for survival until day 10 after challenged with IBV. Mice that lost more than 25% of their initial body weight were humanely sacrificed. Data are representative of three independent experiments. The data are presented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01 (unpaired, two-tailed Student’s t test).
Discussion
Cytokine storm can be observed with virus infection. The common therapies include steroids and selective cytokine blockers. However, inhibiting a single cytokine pathway is usually not sufficient for controlling the entire cytokine storm. CypA is a key regulator of multiple cytokine production, indicating that it plays important roles in cytokine-mediated inflammatory diseases, such as viral pneumonia. In the present study, we constructed PROTACs to recruit E3 ubiquitin ligase to CypA to promote the ubiquitination of CypA and accelerate its degradation through the proteasome pathway instead of simply inhibiting the enzymatic activity of CypA. We discover that PROTACs specifically promote CypA degradation, and Comp-K significantly decreases the cytokine storm caused by IBV infection both in cell and mouse models. These data suggest that PROTACs targeting CypA are potential drugs for controlling virus-induced cytokine storms.
A known drug to inhibit the enzymatic activity of CypA is CsA, which prevents T cell activation via the formation of a tri-partite complex that includes CypA, CsA, and calcineurin.55 Our previous study showed that CypA could inhibit IAV56 and CsA inhibits IAV replication through CypA-dependent and -independent pathways.57 Recent evidence suggests that CsA impairs SARS-CoV-2 infection and dampens the virus-triggered synthesis of cytokines (including IL-6, IL-8, IL1α, and TNF-α) that are involved in cytokine storm in patients.58 However, CypA inhibitors, such as CsA, CypA-L1, and CypA-L2, are not ideal anti-inflammatory or antiviral drugs targeting CypA, because many functions of CypA are not dependent on its enzymatic activity and cannot be inhibited by CypA inhibitors.43,44 Notably, Comp-K not only reduced IBV-induced cytokine storm, but also inhibited IBV replication in cell and mouse models. Therefore, PROTACs targeting CypA are ideal drugs for the treatment of IBV-induced pneumonia. How CypA or its PROTAC regulate IBV replication is an exciting future area of investigation.
PROTACs consist of a ligand for recruiting a target protein of interest and a ligand for an E3 ubiquitin ligase, joined with an appropriate linker. The best and most widely used ligands are CRBN ligand and VHL ligand. In this study, we select VHL ligand for E3 ubiquitin ligase and two ligands of CypA. Comp-K contains the same CypA ligand as Comp-L but has a shorter linker than L, while Comp-M contains another CypA ligand and has the same linker as Comp-K. Many factors affect the affinity of PROTAC. For example, PROTAC linkers represent a delicate balance between affinity contributions and steric effects and that single atom changes to the linker can drastically shift that balance by a yet unresolved mechanism.59 Moreover, several studies have shown that degradation efficiency is not completely correlated with target affinity. Degradation was observed even with some weak-binding kinases, such as p38α, likely due to positive cooperativity via the interaction between p38α and VHL in the ternary complex.60 Similarly, a CRBN-recruiting PROTAC with a promiscuous kinase ligand exhibited different target degradation profiles unrelated to their binding affinities across different cell lines.61 In this study, we found that these three PROTACs have similar effects on the degradation of CypA, although they have different KD values. Additionally, Comp-K has the highest solubility relative to Comp-M and L, and Comp-M is more difficult to dissolve at high concentrations, probably because Comp-M has more hydrophobic groups of ligands. Therefore, Comp-K has a wide range of potential applications in the treatment of inflammatory disease.
The spread of IBV can be dominant during an influenza season and cause severe inflammatory response and disease, particularly in children and adolescents.62 Our previous studies show that the robust production of type I and III IFNs, IFN-stimulated genes, and proinflammatory factors can be induced by IBV rather than IAV infection in A549 cells.8,53 The damage to tissues is mainly due to both virus replication and virus-induced exaggerated immune responses. In many cases, the excessive immune response is even more harmful than the virus itself. Here, we used IBV-infected cell and mouse models to investigate the effect of PROTACs on cytokine production and cytokine-mediated inflammatory responses. We provide several potential PROTAC drugs targeting CypA for immunomodulatory therapy. We find that PROTACs can degrade CypA and inhibit P65 phosphorylation, leading to reduced multiple pro-inflammatory cytokine expression triggered by IBV infection, which might be the central mechanism of PROTACs in regulating inflammatory responses. Moreover, Comp-K, CypA-L1, OSE, or the combination of Comp-K and OSE all suppressed excessive inflammatory responses, alleviated lung injury, and increased the survival rate, but the combination of Comp-K and OSE, among which the combination of Comp-K and OSE had the best therapeutic effect, indicated that there are synergistic effects between Comp-K and OSE.
IL-6 is a non-redundant differentiation factor for Th17 cells and T follicular helper cells.63,64 CCL2 is a key factor for Th2 cells.65 The addition of MCP-1/CCL2 to naive T cells in the presence of antigen drives their differentiation in a Th2 direction.66 In addition, CXCL10 is a classical chemokine to induce migration of T cells.67,68 We observed that PROTAC treatment resulted in decreased IL-6, CCL2, and CXCL10 production both in vitro and in vivo, which might affect the immune cell populations and phenotypes in IBV-infected mice.
We demonstrate that PROTACs targeting CypA are potential drugs for the treatment of IBV-induced pneumonia. Due to the multiple functions of CypA, we believe that these PROTAC drugs might also play important roles in treating some autoimmune diseases, such as rheumatoid arthritis69 and systemic lupus erythematosus,70 and some oncological diseases, such as breast cancer and lung cancer,71 which is interesting and worthy of further study. In addition, since these PROTACs are able to degrade CypA quickly and effectively, they could be used for instantaneous knockdown of CypA in various cell lines or animals to explore more functions of CypA.
In conclusion, PROTACs targeting CypA in this study are potential anti-inflammatory drugs for controlling cytokine storm induced by IBV infection with good specificity and high degradation efficiency. Meanwhile, Comp-K, especially the combination of Comp-K and OSE, can significantly reduce lung damage and effectively enhanced the survival rates of IBV-infected mice. Our data provide an additional, promising option for drug development targeting cytokine storms and instantaneous knockdown of CypA.
Limitations of the study
Three PROTACs have a low affinity for CypA (μM), but degradation of CypA was observed, possibly due to positive cooperativity via the interaction between CypA and VHL in the ternary complex. Additionally, the in vivo data presented in the study is based on a mouse model of a single viral infection, which may not fully recapitulate the complex cytokine storm seen in actual patients.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-CypA | In this paper | N/A |
| Mouse monoclonal anti-β-tubulin | Abcam | Cat#ab6046; RRID: AB_2210370 |
| Mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat#F3165; RRID: AB_259529 |
| Mouse monoclonal anti-HA | Abcam | Cat#ab236632; RRID: AB_2864361 |
| Mouse monoclonal anti-c-Myc | Santa Cruz Biotechnology | Cat#sc-40; RRID: AB_2857941 |
| Rabbit monoclonal anti-p-P65 | Cell Signaling Technology | Cat#3033S; RRID: AB_331284 |
| Rabbit monoclonal anti-P65 | Cell Signaling Technology | Cat#8242S; RRID: AB_10859369 |
| Mouse monoclonal anti-GAPDH | Abcam | Cat#ab8245; RRID: AB_2107448 |
| Rabbit monoclonal anti-CypB | Abcam | Cat#ab178397; RRID:AB_2924975 |
| Rabbit monoclonal anti-CypE | Abcam | Cat#ab154865; RRID:AB_2942047 |
| Rabbit polyclonal anti-CypF | Abcam | Cat#ab126573; RRID:AB_2942048 |
| Rabbit monoclonal anti-VHL | Abcam | Cat#ab270968; RRID:AB_2942049 |
| Bacterial and virus strains | ||
| influenza virus B/Shanghai/PD114/2018 | Zheng, et al.72 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Compound-K | In this paper | N/A |
| Compound-L | In this paper | N/A |
| Compound-M | In this paper | N/A |
| CypA-L1 | In this paper | N/A |
| CypA-L1 | In this paper | N/A |
| VH032 | MedChemExpress | Cat# HY-120217 |
| Lipopolysaccharide | Sigma-Aldrich | Cat#L3024 |
| DMSO | Sigma-Aldrich | Cat#W387520 |
| ANTI-FLAG M2 affifinity gel | Sigma-Aldrich | Cat#A2220 |
| MG132 | Selleck Chemicals | Cat#S2619 |
| Cycloheximide (CHX) | Selleck Chemicals | Cat#S7418 |
| Anti-c-Myc agarose affifinity gel | Sigma-Aldrich | Cat#A7470 |
| TRIzol | Thermo Fisher Scientifific | Cat#15596026 |
| M-MLV reverse transcriptase | Promega | Cat#M1701 |
| SYBR Premix Ex Taq | TaKaRa | Cat#RR420L |
| Protease Inhibitor Cocktail | Roche | Cat#4693116001 |
| PhosSTOPTM phosphatase inhibitor tablets | Roche | Cat#4906837001 |
| ProteinA-Sepharose | Sigma-Aldrich | Cat#P9424 |
| TPCK-treated trypsin | Sigma-Aldrich | Cat#4370285 |
| Lipofectamine 2000 | Thermo Fisher Scientifific | Cat#11668-030 |
| Sulfobutylether-β-Cyclodextrin | MedChemExpress | Cat#HY-17031 |
| Oseltamivir phosphate | Sigma-Aldrich | Cat# PHR1781 |
| Critical commercial assays | ||
| Cell Counting Kit-8 | Beyotime | Cat# C0038 |
| Mouse IL-1b ELISA kit | Sigma-Aldrich | Cat#RAB0274 |
| Mouse IL-6 ELISA kit | Sigma-Aldrich | Cat#RAB0308 |
| Mouse CCL2 ELISA kit | Sigma-Aldrich | Cat# RAB0055 |
| Mouse CXCL10 ELISA kit | Sigma-Aldrich | Cat# RAB0120 |
| Experimental models: Cell lines | ||
| Human: A549 cells | ATCC | Cat#CCL-185 |
| Human: HEK293T cells | ATCC | Cat#CR-3216 |
| Human: HEK293T/CypA- cells | Liu et al.73 | N/A |
| Human: THP-1 cells | ATCC | Cat# TIB-202 |
| Human: jurkat cells | ATCC | Cat#CRL-2899 |
| Canine: MDCK (NBL-2) cells | ATCC | Cat#CCL-34 |
| Experimental models: Organisms/strains | ||
| Mouse: Balb/c | This paper | N/A |
| Oligonucleotides | ||
| IL-1B | CATGGGATAACGAGGCTTATGT | CATATGGACCAGACATCACCAA |
| TNFA | CCAGGGACCTCTCTCTAATCA | TCAGCTTGAGGGTTTGCTAC |
| CXCL10 | GATTTGCTGCCTTATCTTTCTGAC | TTGCAGGAATAATTTCAAGTTTTT |
| GAPDH | TTGTCTCCTGCGACTTCAACAG | GGTCTGGGATGGAAATTGTGAG |
| IL6 | GATTCAATGAGGAGACTTGCC | TGTTCTGGAGGTACTCTAGGT |
| BNP | GGAACCAGTGGGACAACCAG | CCGACATCAGCTTCACTGCT |
| Recombinant DNA | ||
| Plasmid: pCDNA3.0-Flag-CypA | Liu et al.28 | N/A |
| Plasmid: pCMV-K48-Ub-HA | Liu et al.28 | N/A |
| Plasmid: pCMV-Myc-CypA | Liu et al.28 | N/A |
| Plasmid: pCAGGS-Myc-CypA-K28mut | Luan et al.26 | N/A |
| Plasmid: pCAGGS-Myc-CypA-K154mut | Luan et al.26 | N/A |
| Plasmid: pCAGGS-Myc-CypA-K155mut | Luan et al.26 | N/A |
| Plasmid: pCMV-Flag-MAVS | Liu et al.28 | N/A |
| Plasmid: pcDNA3.0-Flag-RIG-I | Liu et al.28 | N/A |
| Plasmid: pCMV-HA-K63-Ub | Liu et al.28 | N/A |
| Software and algorithms | ||
| ImageJ | NIH software | https://imagej.nih.gov/ij/ |
| GraphPad 9.0 | OriginLab | https://www.graphpad.com/scientifific software/prism/ |
| AutoDock Tools | Molecular Graphics Lab | https://vina.scripps.edu/ |
| Pymol | Schrödinger | https://pymol.org/2/ |
| Other | ||
| RPMI 1640 | GIBCO | Cat# 11875085 |
| Fetal Bovine Serum (FBS) | GIBCO | Cat#10437-028 |
| Penicillin-streptomycin | GIBCO | Cat#15140-122 |
| DMEM | GIBCO | Cat#11995-065 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lei Sun (sunlei362@im.ac.cn).
Materials availability
The plasmids used in this study are available from the lead contact.
The compounds used in this study are available from the lead contact.
Experimental model and study participant details
Mice
All animal experiments were reviewed and approved by the Research Ethics Committee of the Chinese Academy of Sciences and complied with the Beijing Laboratory Animal Welfare and Ethical Guidelines of the Beijing Administration Committee of Laboratory Animals.
Mice were maintained under specific pathogen-free (SPF) conditions. Seven-week-old female BALB/c mice were used. All mice were maintained in a barrier facility with free access to food and water.
Cell lines and cell culture
The 293T, A549, THP-1, and Jurkat cells were obtained from ATCC, and shRNA-based knockdown of CypA in 293T (293T/CypA-) has been described previously.73 Cells were cultured at 37°C in a humidified CO2 (5%) incubator. The 293T, 293T/CypA-, MDCK, and A549 cells were cultured in Dulbecco’s Modified Eagle Media (DMEM) high glucose (Gibco) supplemented with 10% FBS (Gibco), 100 U/mL penicillin (Sigma), and 100 μg streptomycin (Sigma). The THP-1 and Jurkat cells were grown in RPMI 1640 supplemented with 10% FBS (Gibco), 100 U/ml penicillin (Sigma), and 100 μg streptomycin (Sigma). THP-1 cells were differentiated with 50 ng/mL phorbol-12-myristate-13-acetate (PMA) for 48 h.
Virus strains
The influenza virus B/Shanghai/PD114/2018 (B-SH, Yamagata lineage) was isolated from patients in Shanghai, China, and was provided by Prof. Dayan Wang of the Chinese Center for Disease Control and Prevention.72 The virus stock was propagated in the allantoic cavities of 9-day-old specific-pathogen-free embryonated chicken eggs at 33°C. The virus titer was determined by 10-fold serial titration in MDCK cells according to plaque assays. The virus was stored at −80°C before use.
IBV infection model
A549 cells and THP-1 cells were infected with IBV (MOI = 1) in DMEM or 1640 containing 0.5 μg/mL TPCK-treated trypsin for different time. THP-1 cells were stimulated by PMA for 48 h before infection. Cells were collected at indicated time for the subsequent real-time PCR and Western blot. Seven-week-old female BALB/c mice (female) were anesthetized with isoflurane by inhalation, then intranasally treated with 3,000 plaques forming unit (pfu) IBV (B-SH) in a volume of 50 μL per mice.
Method details
Molecular docking
AutoDock Vina is an open-source molecular docking program designed by the Scripps Research Institute for the computation of semiflexible molecular docking. The CypA crystal structure (3k0m) was obtained through the PDB database (https://www.rcsb.org/). The PDB files of the CypA and PROTACs were imported into AutoDock Tools (ADT, La Jolla, CA, USA) and were prepared before docking by fusing nonpolar hydrogen atoms, adding partial charges and atom types. After being computed by Autodock Vina, the lowest energy and best conformation was taken as the final result. The complexes were then observed and plotted using PyMOL.
Surface plasmon resonance assay
Evaluation of the binding affinity was performed by using Biacore T200 (Cytiva). Specifically, CypA was diluted with sodium acetate (pH=4.0, 10mM) to 20 μg/mL and immobilized on the channel two of CM5 chip with the channel one of CM5 as control. Comp-K/L/M were diluted by mobile phase buffer (10 mM PBS, pH7.4; 137 mM NaCl; 2.7 mM KCl; 0.05% P20) to the indicated concentrations. The compound was injected into the chip with a flow rate 30 μL/min for 120 s and allowed to dissociate for 120 s with running buffer as the blank control. The KD values were calculated by Biacore evaluation software 3.2.
Immunoblotting and co-immunoprecipitation
Cells or tissue homogenate were lysed on ice by lysis buffer74 containing PhosSTOP™ phosphatase inhibitor (Roche). Lysates were centrifuged at 4°C for 10 min. Add loading buffer to the supernatant and boiled sample for 10 min. Samples were then resolved on SDS-PAGE gel followed by immunoblotting with the indicated antibodies as described previously.30 For co-immunoprecipitation, the cells lysis was incubated with Pierce™ protein A magnetic beads (Thermo), anti-c-Myc agarose affinity gel (Sigma), or anti-Flag M2 affinity gel (Sigma) for 6 h. After incubation, the beads were washed with washing buffer (300 mM NaCl, 20 mM HEPES, 1 mM EDTA, 1% Triton X-100, and 10% glycerin) for 3-5 times, then the bound proteins were eluted by boiling for 10 min in SDS protein loading buffer and analyzed by immunoblotting assay.
Cell viability assay
Cell viability was determined by using the WST-8 cell counting kit-8 (CCK8) viability assay (Beyotime, Jiangsu, China). The A549 cells were cultured in 96-well plates in complete DMEM medium (DMEM medium with 10% FBS, 100 U/ml penicillin and 100 μg streptomycin) and cultured overnight. After being treated with PROTACs for 12 h, CCK-8 solution (10 μL) was added to each well, and the system was incubated for 120 min at 37°C. Absorbance at 450 nm was measured using a microplate reader.
RNA extraction, cDNA synthesis, and qPCR analysis
Total RNA was extracted from cells with TRIzol (Invitrogen), according to the manufacturer’s instructions. cDNA was synthesized from 1 mg of total RNA using an oligo (dT) primer and M-MLV reverse transcriptase (Promega), according to the manufacturer’s instructions. Relative gene expression was analyzed by qPCR using SYBR Premix Ex Taq (TaKaRa). The primers are listed in key resources table. The Ct values generated from an ABI 7500 were analyzed using the 2-ΔΔCT method. The expression of target genes was normalized to that of GAPDH.
Micro-X-ray computed tomography
At day 2, 4, and 6 post IBV infection, mice were anesthetized with isoflurane by inhalation and then transferred to Micro-computed tomography imaging system (PE Quantum FX, Perkin-Elmer). The parameters: Voltage 90 kV, Current 180 μA, FOV 24 mm, Scan Technique 4.5 min.
Histopathology staining
Lung tissues were fixed with 4% paraformaldehyde overnight at room temperature. Paraffin-embedded lung sections were stained with H & E or Masson’s trichrome. The lung pathology was scored by blinded experts. Alveolar congestion, alveolar edema, inflammatory cell infiltration and alveolar hemorrhage, and thickness of the alveolar wall were assessed. The scores of 0 to 4 represent normal lungs, lower than 25%, 25%–50%, 50%–75%, and higher than 75% lung involvement, respectively.
Immunofluorescence histochemistry
Frozen sections of lung tissue were fixed with cold acetone for 10 min and blocked with 5% normal serum in PBS from the same species. The sections were then incubated overnight at 4°C with anti-F4/80 and anti-CD3 mAbs (CST), followed by incubation with Alexa Fluor 488-labeled secondary Abs or Alexa Fluor 555-labeled streptavidin (Invitrogen). The sections were then mounted and counterstained using DAPI (Vector Laboratories) and analyzed using a BX51 fluorescence microscope (Olympus), BIOREVO BZ-9000 fluorescence microscope, BZ-H1C Dynamic Cell Count software (KEYENCE), and a DM-IRE2 confocal laser-scanning microscope (Leica Microsystems).
ProcartaPlex multiplex immunoassays and ELISA
Cytokines and chemokines in mice BALF were measured using the mice ProcartaPlex Multiplex Immunoassays kit (LAIZEE BIOTECH, EPX360-26092-901) and enzyme linked immunosorbent assay (ELISA, Sigma-Aldrich), according to the manufacturer’s protocol. BALF was collected from the IBV-infected mice at days 2, 4, and 6 post-infection.
Quantification and statistical analysis
Statistical analyzes were performed using GraphPad Prism 9 software. Data are presented as the mean values ±SD of at least three independent experiments. Comparisons between two groups were performed using the two-tailed Student’s t-test, with P value < 0.05 being considered significant (∗ p < 0.05, ∗∗ p < 0.01).
Acknowledgments
This work was supported by grants from the Strategic Priority Research Program of Chinese Academy of Sciences (XDB29010000) and the National Natural Science Foundation of China (31972657, 32070164, 32200711, and 32061123001).
Author contributions
L.S. and W.L. initiated and supervised the project. L.S., H.L., and W.Y. designed the experiments, analyzed the data, and wrote the paper; H.L. and W.Y. performed the experiments; X.B., H.L., H.Z., and W.F. helped with some experiments; W.L. helped analyze the data and revised the manuscript.
Declaration of interests
W.L., L.S., H.L., W.Y., X.B., and H.Z. have three patents related to this work. These applications claim the benefit of Chinese Patents (ZL202210958956.7, ZL202210957971.X, and 201710804078.2).
Published: August 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107535.
Contributor Information
Wenjun Liu, Email: liuwj@im.ac.cn.
Lei Sun, Email: sunlei362@im.ac.cn.
Supplemental information
Data and code availability
-
•
This paper does not report original code.
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Kash J.C., Tumpey T.M., Proll S.C., Carter V., Perwitasari O., Thomas M.J., Basler C.F., Palese P., Taubenberger J.K., García-Sastre A., et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443:578–581. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polizzotto M.N., Uldrick T.S., Wang V., Aleman K., Wyvill K.M., Marshall V., Pittaluga S., O'Mahony D., Whitby D., Tosato G., et al. Human and viral interleukin-6 and other cytokines in Kaposi sarcoma herpesvirus-associated multicentric Castleman disease. Blood. 2013;122:4189–4198. doi: 10.1182/blood-2013-08-519959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perrone L.A., Plowden J.K., García-Sastre A., Katz J.M., Tumpey T.M. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuyama S., Kawaoka Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr. Opin. Immunol. 2011;23:481–486. doi: 10.1016/j.coi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu Y., Zuo X., Zhang S., Ouyang Z., Jiang S., Wang F., Wang G. The Mechanism behind Influenza Virus Cytokine Storm. Viruses. 2021;13:1362. doi: 10.3390/v13071362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul Glezen W., Schmier J.K., Kuehn C.M., Ryan K.J., Oxford J. The burden of influenza B: a structured literature review. Am. J. Public Health. 2013;103:e43–e51. doi: 10.2105/AJPH.2012.301137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang J., Li J., Fan W., Zheng W., Yu M., Chen C., Sun L., Bi Y., Ding C., Gao G.F., Liu W. Robust Lys63-Linked Ubiquitination of RIG-I Promotes Cytokine Eruption in Early Influenza B Virus Infection. J. Virol. 2016;90:6263–6275. doi: 10.1128/JVI.00549-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., Li W., Tong Q., Yi J., Zhao L., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu B., Huang S., Yin L. The cytokine storm and COVID-19. J. Med. Virol. 2021;93:250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo X.H., Zhu Y., Mao J., Du R.C. T cell immunobiology and cytokine storm of COVID-19. Scand. J. Immunol. 2021;93 doi: 10.1111/sji.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee D.W., Gardner R., Porter D.L., Louis C.U., Ahmed N., Jensen M., Grupp S.A., Mackall C.L. Current concepts in the diagnosis and management of cytokine release syndrome. Blood. 2014;124:188–195. doi: 10.1182/blood-2014-05-552729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., HLH Across Speciality Collaboration, UK COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramiro S., Mostard R.L.M., Magro-Checa C., van Dongen C.M.P., Dormans T., Buijs J., Gronenschild M., de Kruif M.D., van Haren E.H.J., van Kraaij T., et al. Historically controlled comparison of glucocorticoids with or without tocilizumab versus supportive care only in patients with COVID-19-associated cytokine storm syndrome: results of the CHIC study. Ann. Rheum. Dis. 2020;79:1143–1151. doi: 10.1136/annrheumdis-2020-218479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Staa T.P., Leufkens H.G.M., Cooper C. The epidemiology of corticosteroid-induced osteoporosis: a meta-analysis. Osteoporos. Int. 2002;13:777–787. doi: 10.1007/s001980200108. [DOI] [PubMed] [Google Scholar]
- 17.Rice J.B., White A.G., Scarpati L.M., Wan G., Nelson W.W. Long-term Systemic Corticosteroid Exposure: A Systematic Literature Review. Clin. Ther. 2017;39:2216–2229. doi: 10.1016/j.clinthera.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Buchman A.L. Side effects of corticosteroid therapy. J. Clin. Gastroenterol. 2001;33:289–294. doi: 10.1097/00004836-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 19.Caricchio R., Abbate A., Gordeev I., Meng J., Hsue P.Y., Neogi T., Arduino R., Fomina D., Bogdanov R., Stepanenko T., et al. Effect of Canakinumab vs Placebo on Survival Without Invasive Mechanical Ventilation in Patients Hospitalized With Severe COVID-19: A Randomized Clinical Trial. JAMA. 2021;326:230–239. doi: 10.1001/jama.2021.9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosas I.O., Bräu N., Waters M., Go R.C., Hunter B.D., Bhagani S., Skiest D., Aziz M.S., Cooper N., Douglas I.S., et al. Tocilizumab in Hospitalized Patients with Severe Covid-19 Pneumonia. N. Engl. J. Med. 2021;384:1503–1516. doi: 10.1056/NEJMoa2028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyriazopoulou E., Poulakou G., Milionis H., Metallidis S., Adamis G., Tsiakos K., Fragkou A., Rapti A., Damoulari C., Fantoni M., et al. Early treatment of COVID-19 with anakinra guided by soluble urokinase plasminogen receptor plasma levels: a double-blind, randomized controlled phase 3 trial. Nat. Med. 2021;27:1752–1760. doi: 10.1038/s41591-021-01499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO Rapid Evidence Appraisal for COVID-19 Therapies REACT Working Group. Shankar-Hari M., Vale C.L., Godolphin P.J., Fisher D., Higgins J.P.T., Spiga F., Savovic J., Tierney J., Baron G., et al. Association Between Administration of IL-6 Antagonists and Mortality Among Patients Hospitalized for COVID-19: A Meta-analysis. JAMA. 2021;326:499–518. doi: 10.1001/jama.2021.11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handschumacher R.E., Harding M.W., Rice J., Drugge R.J., Speicher D.W. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 24.Göthel S.F., Marahiel M.A. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell. Mol. Life Sci. 1999;55:423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nigro P., Pompilio G., Capogrossi M.C. Cyclophilin A: a key player for human disease. Cell Death Dis. 2013;4:e888. doi: 10.1038/cddis.2013.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luan X., Yang W., Bai X., Li H., Li H., Fan W., Zhang H., Liu W., Sun L. Cyclophilin A is a key positive and negative feedback regulator within interleukin-6 trans-signaling pathway. FASEB J. 2021;35 doi: 10.1096/fj.202101044RRR. [DOI] [PubMed] [Google Scholar]
- 27.Yang W., Bai X., Luan X., Min J., Tian X., Li H., Li H., Sun W., Liu W., Fan W., et al. Delicate regulation of IL-1beta-mediated inflammation by cyclophilin A. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110513. [DOI] [PubMed] [Google Scholar]
- 28.Liu W., Li J., Zheng W., Shang Y., Zhao Z., Wang S., Bi Y., Zhang S., Xu C., Duan Z., et al. Cyclophilin A-regulated ubiquitination is critical for RIG-I-mediated antiviral immune responses. Elife. 2017;6 doi: 10.7554/eLife.24425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun S., Guo M., Zhang J.B., Ha A., Yokoyama K.K., Chiu R.H. Cyclophilin A (CypA) interacts with NF-κB subunit, p65/RelA, and contributes to NF-κB activation signaling. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai X., Yang W., Luan X., Li H., Li H., Tian D., Fan W., Li J., Wang B., Liu W., Sun L. Induction of cyclophilin A by influenza A virus infection facilitates group A Streptococcus coinfection. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109159. [DOI] [PubMed] [Google Scholar]
- 31.Geng J., Chen L., Yuan Y., Wang K., Wang Y., Qin C., Wu G., Chen R., Zhang Z., Wei D., et al. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Signal Transduct. Target. Ther. 2021;6:347. doi: 10.1038/s41392-021-00760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arora K., Gwinn W.M., Bower M.A., Watson A., Okwumabua I., MacDonald H.R., Bukrinsky M.I., Constant S.L. Extracellular cyclophilins contribute to the regulation of inflammatory responses. J. Immunol. 2005;175:517–522. doi: 10.4049/jimmunol.175.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koh M.W., Baldi R.F., Soni S., Handslip R., Tan Y.Y., O'Dea K.P., Malesevic M., McAuley D.F., O'Kane C.M., Patel B.V., et al. Secreted Extracellular Cyclophilin A Is a Novel Mediator of Ventilator-induced Lung Injury. Am. J. Respir. Crit. Care Med. 2021;204:421–430. doi: 10.1164/rccm.202009-3545OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Billich A., Winkler G., Aschauer H., Rot A., Peichl P. Presence of cyclophilin A in synovial fluids of patients with rheumatoid arthritis. J. Exp. Med. 1997;185:975–980. doi: 10.1084/jem.185.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin Z.G., Lungu A.O., Xie L., Wang M., Wong C., Berk B.C. Cyclophilin A is a proinflammatory cytokine that activates endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:1186–1191. doi: 10.1161/01.ATV.0000130664.51010.28. [DOI] [PubMed] [Google Scholar]
- 36.Lai A.C., Crews C.M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 2017;16:101–114. doi: 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He S., Gao F., Ma J., Ma H., Dong G., Sheng C. Aptamer-PROTAC Conjugates (APCs) for Tumor-Specific Targeting in Breast Cancer. Angew. Chem. Int. Ed. Engl. 2021;60:23299–23305. doi: 10.1002/anie.202107347. [DOI] [PubMed] [Google Scholar]
- 38.Liao H., Li X., Zhao L., Wang Y., Wang X., Wu Y., Zhou X., Fu W., Liu L., Hu H.G., Chen Y.G. A PROTAC peptide induces durable β-catenin degradation and suppresses Wnt-dependent intestinal cancer. Cell Discov. 2020;6:35. doi: 10.1038/s41421-020-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan S., Zhang X., Lv D., Zhang Q., He Y., Zhang P., Liu X., Thummuri D., Yuan Y., Wiegand J.S., et al. A selective BCL-X(L) PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 2019;25:1938–1947. doi: 10.1038/s41591-019-0668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang W., Zhao L., Han X., Qin C., Miao B., McEachern D., Wang Y., Metwally H., Kirchhoff P.D., Wang L., et al. Discovery of ARD-2585 as an Exceptionally Potent and Orally Active PROTAC Degrader of Androgen Receptor for the Treatment of Advanced Prostate Cancer. J. Med. Chem. 2021;64:13487–13509. doi: 10.1021/acs.jmedchem.1c00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barker R.A., Fujimaki M., Rogers P., Rubinsztein D.C. Huntingtin-lowering strategies for Huntington's disease. Expert Opin. Investig. Drugs. 2020;29:1125–1132. doi: 10.1080/13543784.2020.1804552. [DOI] [PubMed] [Google Scholar]
- 42.Kargbo R.B. PROTAC Compounds Targeting α-Synuclein Protein for Treating Neurogenerative Disorders: Alzheimer's and Parkinson's Diseases. ACS Med. Chem. Lett. 2020;11:1086–1087. doi: 10.1021/acsmedchemlett.0c00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song F., Zhang X., Ren X.B., Zhu P., Xu J., Wang L., Li Y.F., Zhong N., Ru Q., Zhang D.W., et al. Cyclophilin A (CyPA) induces chemotaxis independent of its peptidylprolyl cis-trans isomerase activity: direct binding between CyPA and the ectodomain of CD147. J. Biol. Chem. 2011;286:8197–8203. doi: 10.1074/jbc.C110.181347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis T.L., Walker J.R., Campagna-Slater V., Finerty P.J., Paramanathan R., Bernstein G., MacKenzie F., Tempel W., Ouyang H., Lee W.H., et al. Structural and biochemical characterization of the human cyclophilin family of peptidyl-prolyl isomerases. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ni S., Yuan Y., Huang J., Mao X., Lv M., Zhu J., Shen X., Pei J., Lai L., Jiang H., Li J. Discovering potent small molecule inhibitors of cyclophilin A using de novo drug design approach. J. Med. Chem. 2009;52:5295–5298. doi: 10.1021/jm9008295. [DOI] [PubMed] [Google Scholar]
- 46.Galdeano C., Gadd M.S., Soares P., Scaffidi S., Van Molle I., Birced I., Hewitt S., Dias D.M., Ciulli A. Structure-guided design and optimization of small molecules targeting the protein-protein interaction between the von Hippel-Lindau (VHL) E3 ubiquitin ligase and the hypoxia inducible factor (HIF) alpha subunit with in vitro nanomolar affinities. J. Med. Chem. 2014;57:8657–8663. doi: 10.1021/jm5011258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pflügl G., Kallen J., Schirmer T., Jansonius J.N., Zurini M.G., Walkinshaw M.D. X-ray structure of a decameric cyclophilin-cyclosporin crystal complex. Nature. 1993;361:91–94. doi: 10.1038/361091a0. [DOI] [PubMed] [Google Scholar]
- 49.Sun S., Guo M., Zhang J.B., Ha A., Yokoyama K.K., Chiu R.H. Cyclophilin A (CypA) interacts with NF-kappaB subunit, p65/RelA, and contributes to NF-kappaB activation signaling. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo M., Shen J., Kwak J.H., Choi B., Lee M., Hu S., Zhang X., Ting K., Soo C.B., Chiu R.H. Novel role for cyclophilin A in regulation of chondrogenic commitment and endochondral ossification. Mol. Cell Biol. 2015;35:2119–2130. doi: 10.1128/MCB.01414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee J.J., Smith M., Bankhead C., Perera Salazar R., Kousoulis A.A., Butler C.C., Wang K. Oseltamivir and influenza-related complications in children: a retrospective cohort in primary care. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.02246-2019. [DOI] [PubMed] [Google Scholar]
- 52.Liu Q., Zhou Y.H., Yang Z.Q. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell. Mol. Immunol. 2016;13:3–10. doi: 10.1038/cmi.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiao P., Fan W., Cao Y., Zhang H., Tian L., Sun L., Luo T., Liu W., Li J. Robust induction of interferon and interferon-stimulated gene expression by influenza B/Yamagata lineage virus infection of A549 cells. PLoS One. 2020;15 doi: 10.1371/journal.pone.0231039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Z., Kozlowski J., Schuster D.P. Physiologic, biochemical, and imaging characterization of acute lung injury in mice. Am. J. Respir. Crit. Care Med. 2005;172:344–351. doi: 10.1164/rccm.200503-343OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J., Farmer J.D., Jr., Lane W.S., Friedman J., Weissman I., Schreiber S.L. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 56.Liu X., Sun L., Yu M., Wang Z., Xu C., Xue Q., Zhang K., Ye X., Kitamura Y., Liu W. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell Microbiol. 2009;11:730–741. doi: 10.1111/j.1462-5822.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 57.Liu X., Zhao Z., Li Z., Xu C., Sun L., Chen J., Liu W. Cyclosporin A inhibits the influenza virus replication through cyclophilin A-dependent and -independent pathways. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fenizia C., Galbiati S., Vanetti C., Vago R., Clerici M., Tacchetti C., Daniele T. Cyclosporine A Inhibits Viral Infection and Release as Well as Cytokine Production in Lung Cells by Three SARS-CoV-2 Variants. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.01504-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith B.E., Wang S.L., Jaime-Figueroa S., Harbin A., Wang J., Hamman B.D., Crews C.M. Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 2019;10:131. doi: 10.1038/s41467-018-08027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bondeson D.P., Smith B.E., Burslem G.M., Buhimschi A.D., Hines J., Jaime-Figueroa S., Wang J., Hamman B.D., Ishchenko A., Crews C.M. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem. Biol. 2018;25:78–87.e5. doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang H.T., Dobrovolsky D., Paulk J., Yang G., Weisberg E.L., Doctor Z.M., Buckley D.L., Cho J.H., Ko E., Jang J., et al. A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell Chem. Biol. 2018;25:88–99.e6. doi: 10.1016/j.chembiol.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pascua P.N.Q., Mostafa H.H., Marathe B.M., Vogel P., Russell C.J., Webby R.J., Govorkova E.A. Pathogenicity and peramivir efficacy in immunocompromised murine models of influenza B virus infection. Sci. Rep. 2017;7:7345. doi: 10.1038/s41598-017-07433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korn T., Hiltensperger M. Role of IL-6 in the commitment of T cell subsets. Cytokine. 2021;146 doi: 10.1016/j.cyto.2021.155654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nurieva R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L., Wang Y.H., Watowich S.S., Jetten A.M., Tian Q., Dong C. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu L., Tseng S., Horner R.M., Tam C., Loda M., Rollins B.J. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407–411. doi: 10.1038/35006097. [DOI] [PubMed] [Google Scholar]
- 66.Karpus W.J., Lukacs N.W., Kennedy K.J., Smith W.S., Hurst S.D., Barrett T.A. Differential CC chemokine-induced enhancement of T helper cell cytokine production. J. Immunol. 1997;158:4129–4136. [PubMed] [Google Scholar]
- 67.Antonelli A., Ferrari S.M., Corrado A., Ferrannini E., Fallahi P. CXCR3, CXCL10 and type 1 diabetes. Cytokine Growth Factor Rev. 2014;25:57–65. doi: 10.1016/j.cytogfr.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 68.Hyun J.G., Lee G., Brown J.B., Grimm G.R., Tang Y., Mittal N., Dirisina R., Zhang Z., Fryer J.P., Weinstock J.V., et al. Anti-interferon-inducible chemokine, CXCL10, reduces colitis by impairing T helper-1 induction and recruitment in mice. Inflamm. Bowel Dis. 2005;11:799–805. doi: 10.1097/01.mib.0000178263.34099.89. [DOI] [PubMed] [Google Scholar]
- 69.Yang Y., Lu N., Zhou J., Chen Z.N., Zhu P. Cyclophilin A up-regulates MMP-9 expression and adhesion of monocytes/macrophages via CD147 signalling pathway in rheumatoid arthritis. Rheumatology. 2008;47:1299–1310. doi: 10.1093/rheumatology/ken225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kratz A., Harding M.W., Craft J., Mackworth-Young C.G., Handschumacher R.E. Autoantibodies against cyclophilin in systemic lupus erythematosus and Lyme disease. Clin. Exp. Immunol. 1992;90:422–427. doi: 10.1111/j.1365-2249.1992.tb05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J. Role of cyclophilin a during oncogenesis. Arch Pharm. Res. (Seoul) 2010;33:181–187. doi: 10.1007/s12272-010-0200-y. [DOI] [PubMed] [Google Scholar]
- 72.Zheng W., Fan W., Zhang S., Jiao P., Shang Y., Cui L., Mahesutihan M., Li J., Wang D., Gao G.F., et al. Naproxen Exhibits Broad Anti-influenza Virus Activity in Mice by Impeding Viral Nucleoprotein Nuclear Export. Cell Rep. 2019;27:1875–1885.e5. doi: 10.1016/j.celrep.2019.04.053. [DOI] [PubMed] [Google Scholar]
- 73.Liu X., Zhao Z., Xu C., Sun L., Chen J., Zhang L., Liu W. Cyclophilin A restricts influenza A virus replication through degradation of the M1 protein. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mahesutihan M., Zheng W., Cui L., Li Y., Jiao P., Yang W., Liu W., Li J., Fan W., Yang L., et al. CypA Regulates AIP4-Mediated M1 Ubiquitination of Influenza A Virus. Virol. Sin. 2018;33:440–448. doi: 10.1007/s12250-018-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report original code.
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.