Abstract
Chronic obstructive pulmonary disease (COPD), which includes emphysema and chronic bronchitis, is now the third cause of death worldwide, and COVID-19 infection has been reported as an exacerbation factor of them. In this study, we report that the intratracheal administration of the keratan sulfate-based disaccharide L4 mitigates the symptoms of elastase-induced emphysema in a mouse model. To know the molecular mechanisms, we performed a functional analysis of a C-type lectin receptor, langerin, a molecule that binds L4. Using mouse BMDCs (bone marrow-derived dendritic cells) as langerin-expressing cells, we observed the downregulation of IL-6 and TNFa and the upregulation of IL-10 after incubation with L4. We also identified CapG (a macrophage-capping protein) as a possible molecule that binds langerin by immunoprecipitation combined with a mass spectrometry analysis. We identified a portion of the CapG that was localized in the nucleus and binds to the promoter region of IL-6 and the TNFa gene in BMDCs, suggesting that CapG suppresses the gene expression of IL-6 and TNFa as an inhibitory transcriptional factor. To examine the effects of L4 in vivo, we also generated langerin-knockout mice by means of genome editing technology. In an emphysema mouse model, the administration of L4 did not mitigate the symptoms of emphysema as well as the inflammatory state of the lung in the langerin-knockout mice. These data suggest that the anti-inflammatory effect of L4 through the langerin-CapG axis represents a potential therapeutic target for the treatment of emphysema and COPD.
Keywords: glycobiology, carbohydrate function, chronic obstructive pulmonary disease (COPD), dendritic cell, inflammation, CapG, langerin, keratan sulfate
Chronic obstructive pulmonary disease (COPD), which includes emphysema and chronic bronchitis, is the third most common cause of death in the world. The development of chronic bronchitis in the lung induced by long-term exposure to tobacco leads to bronchial obstruction and respiratory failure, conditions that are not fully reversible and which eventually cause emphysema and COPD in humans (1). Tobacco smoke can activate a variety of immune cells including macrophages, neutrophils, T-lymphocytes, B-lymphocytes, and dendritic cells that are present in the lung (2). The activated immune cells secrete various inflammatory mediators including chemokines, cytokines, growth factors, and proteolytic enzymes, and these in turn, stimulate each other cells (3, 4). These inflammatory mediators exacerbate bronchitis and induce bronchial obstruction. In such cases, corticosteroids and related, anti-inflammatory drugs are administrated to the patients to suppress the progression of COPD (5). Concerning the suppression of the inflammatory state, dendritic cells would be potential targets because they play a central role in the initiation and acceleration of immune responses (6). By sensing the harmful substances in tobacco smoke, dendritic cells activate the immune cell network to protect the body from them, but its responses often damage the bronchi, which leads to the onset and progression of emphysema and COPD (7). The infections such as COVID-19 can also exacerbate the symptoms by activating immune cells (8).
Langerin, a C-type lectin receptor that is expressed in Langerhans cells, is a type II membrane protein that contains a C-terminal carbohydrate recognition domain in its extracellular region (8). Langerhans cells are a subtype of dendritic cells that are located in the epidermis and it has been recently shown that a part of the resident dendritic cells in other organs also expresses langerin (9, 10). Importantly, it was demonstrated that langerin functions as a natural barrier against the HIV-1 (human immunodeficiency virus 1), the influenza virus, and related viruses by recognizing the glycans that are attached to their envelope proteins (11, 12, 13, 14). After being captured by langerin, they are incorporated by endocytosis, then degraded by lysosomes, and the antigen presentation then proceeds. It is possible that langerin as well as other lectin receptors could capture other pathogens by recognition of glycans, but to verify this a more detailed structural analysis would be needed to permit their potential glycan ligands to be identified.
Tateno et al. identified some potential glycan structures that are recognized by langerin (15). Using a langerin-Fc chimera and a glycan array, they found that langerin-Fc binds strongly to the 6′-sulfo-LacNAc (galactose-6-sulfate linked by a β1,4 linkage to N-acetylglucosamine). Importantly, in in vitro assays, we also demonstrated that 6′-sulfo- and 6-sulfo-LacNAc (galactose-6-sulfate β1,4-linked to N-acetylglucosamine-6-sulfate) binds to langerin more strongly than 6′-sulfo-LacNAc (16). These disaccharides are components of the glycosaminoglycan of the keratan sulfate proteoglycan (Fig. S1), therefore, these results indicate that the keratan sulfate side chain would be a natural ligand for langerin.
We refer to these, 6′-sulfo- and 6-sulfo-LacNAc disaccharides, as L4, and, in an independent study, our research group previously reported on the potential function of L4 against an emphysema mouse model. It was demonstrated that the intratracheal administration of L4 modulated the inflammatory state in the lung and mitigated the pathology of emphysema (17). While these data suggest therapeutic potentials of L4 against emphysema, we had no knowledge regarding the target of L4 and no information concerning the mechanism responsible for the action of L4. Taking these background findings into consideration we report on our attempts to address these questions in this study by using langerin-expressing cells and an emphysema mouse model in langerin-knockout mice. Our findings indicate that the protective function of L4 against emphysema represents one of the mechanisms in in this process.
Results
L4 suppresses the gene expression of inflammatory cytokines in BMDCs
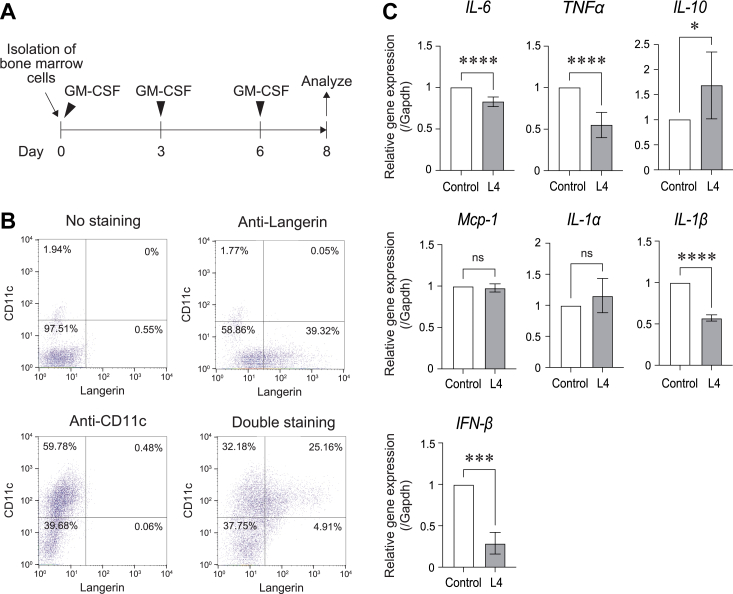
It is known that the expression of langerin is restricted in dendritic cells. To examine the function of langerin, we first analyzed the expression of langerin in a well-established mouse follicular dendritic cell line, FL-YB mice. However, no langerin expression was observed in that cell line. For this, we next isolated murine bone marrow cells and differentiated them into dendritic cells by incubation with granulocyte-macrophage colony-stimulating factor (GM-CSF) (Fig. 1A) (18, 19). These bone marrow-derived dendritic cells (BMDCs) expressed langerin as well as CD11c, a dendritic cell marker, in 39.32 % and 59.78 % of the total cells, respectively (Fig. 1B) and 25.16 % of the total cells were double positive (Fig. 1B). Using this information, we incubated the cells with L4 for 2 days and then, analyzed gene expression by quantitative RT-PCR. As shown in Figure 1C, L4 downregulated the inflammatory cytokines IL-6, TNFα, IL-1β, and IFN-β, while upregulated an anti-inflammatory cytokine IL-10. The expression of Mcp-1 and IL-1α did not change. These data suggest that L4 suppressed the inflammatory status of langerin-expressing BMDCs.
Figure 1.
L4 downregulated inflammatory cytokines in BMDCs. A, a time course for the isolation and differentiation of mouse bone-marrow cells into BMDCs (bone-marrow-derived dendritic cells). B, the expression of langerin and CD11c in BMDCs was examined by flow cytometry. The X-axis indicates langerin expression and the Y-axis indicates CD11c expression. The numbers indicate the percentages of the cells existing each square area. C, the gene expression of IL-6, TNFα, IL-10, Mcp-1, IL-1α, IL-1β, and IFN-β in BMDCs after incubation with L4 (100 μg/ml) for 2 days examined by qRT-PCR analysis. The expression levels after incubation with L4 were normalized to the levels without L4. The expression levels indicated as mean ± SD of three independent experiments. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns: not significant.
CapG (macrophage-capping protein) is immunoprecipitated with langerin in BMDCs
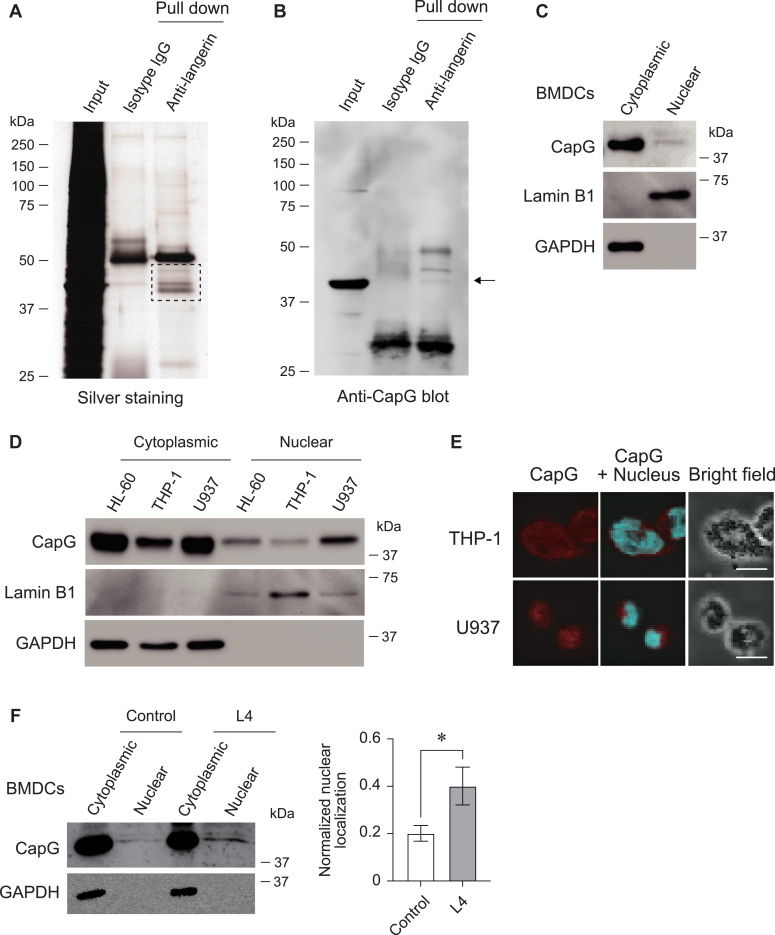
The molecules that functioned with langerin in a cooperative manner have not been identified yet. For this, we next attempted to identify the molecules that bind with langerin. In BMDCs, the cell lysate was immunoprecipitated with an anti-langerin antibody and SDS-PAGE was performed with the resulting immunoprecipitates. As shown in Figure 2A, we found unique bands in the molecular weight range of 37 to 50 kDa after visualizing the proteins by silver staining in molecules that were immunoprecipitated with an anti-langerin antibody. To identify these proteins, we cut the bands out and analyzed them by mass spectrometry. A total of 11 proteins were identified as langerin-binding molecules and are listed in Table 1. Among them, we confirmed the binding between langerin and CapG (a macrophage-capping protein) in the immunoprecipitate with an anti-CapG antibody by western blotting (Fig. 2B). In addition, we also analyzed the binding between them in HEK293 cells. We overexpressed mouse langerin and mouse CapG together and immunoprecipitation with an anti-langerin antibody was then performed. The immunoprecipitate was blotted with an anti-CapG antibody. As shown in Fig. S2, CapG was indeed detected as a molecule that binds to langerin. Moreover, we also found that langerin and CapG are endogenously expressed in a human monocytic leukemia cell line, THP-1 cells. In THP-1 cells, the binding between langerin and CapG and the association between them in the plasma membrane was also confirmed (Fig. S3). These data indicate that CapG would function with langerin cooperatively in BMDCs.
Figure 2.
CapG was identified as a langerin-binding molecule and localized in the nucleus.A, silver staining of the SDS-PAGE gel. The immunoprecipitates prepared from BMDCs with an anti-langerin antibody and a non-relevant IgG separated in SDS-PAGE were then subjected to silver staining. The dashed square area was cut out and the contents analyzed by mass spectrometry. B, Western blotting of the immunoprecipitates prepared from BMDCs with an anti-langerin antibody and a non-relevant IgG. The membrane was blotted with an anti-CapG antibody. Arrow indicates CapG. C, subcellular localization of CapG in BMDCs examined by western blotting. Nuclear-rich fraction and cytoplasm-rich fraction were prepared and analyzed. Lamin B1 was used as a maker for nuclear fraction and GAPDH was used as a marker for cytoplasmic fraction. D, subcellular localization of CapG in HL-60, THP-1, and U937 cells examined by western blotting. E, immunocytochemical staining of THP-1 and U937 cells with an anti-CapG antibody and DAPI. Red indicates CapG and blue indicates nuclear. Bars indicate 10 μm. Images were pictured under confocal microscopy. F, subcellular localization of CapG in BMDCs after incubation with L4 (100 μg/ml) for 2 days examined by western blotting. GAPDH was used as a marker for the cytoplasmic fraction. Independent experiments were performed and the normalized nuclear localization of CapG after incubation with or without L4 is shown in the right panel. ∗p < 0.05.
Table 1.
Identified molecules as langerin-associated proteins
| Name | Gene Name | Identified peptide sequence | Coverage (%) |
|---|---|---|---|
| Langerin | Cd207 | SQILSLETSMK | 16 |
| SDAQMLK | |||
| RPYVQTTE | |||
| KAHLTSVSSESEQK | |||
| AHLTSVSSESEQK | |||
| VQGLQNSLENVNK | |||
| IgG2a secreted form | N/A | VNNRALPSPIEK | 6 |
| VDKKIEPR | |||
| ALPSPIEK | |||
| 60S ribosomal protein L4 | Rpl4 | MMNTDLSR | 2 |
| Zinc finger protein 609 | Anf609 | CKKPSSLK | 1 |
| Transmembrane protease serine 13 | Tmprss13 | NKPGVYTK | 1 |
| WD repeat-containing protein 81 | Wdr81 | EAGLLAAVTLTQK | 1 |
| Macrophage-capping protein | Capg | VSDATGQMNLTK | 3 |
| UPF0711 protein C18orf21 homolog | N/A | SNAATAANKASPK | 6 |
| FK506-binding protein-like | Fkbpl | VLAVDPKNR | 3 |
| RCC1 and BTB domain-containing protein 2 | Rcbtb2 | REFDNPDTADLK | 2 |
| Renalase | Rnls | QREQLK | 2 |
L4 enhanced nuclear localization of CapG in BMDCs
Human and mouse CapG harbor a putative nuclear localization signal and their amino acid sequence lack the nuclear export signal (20). To further examine the function of CapG, we analyzed the subcellular localization of CapG in murine BMDCs and in a human promyelocytic leukemia cell line: HL-60, a human monocytic leukemia cell line: THP-1, and in U937 cells: a human diffuse histiocytic lymphoma cell line. We prepared cytoplasmic and nuclear fractions from these cells and performed western blotting. As shown in Figure 2, C and D, a portion of the CapG was detected in the nuclear fractions. The larger amount of GAPDH (glyceraldehyde-3-phosphate dehydrogenase), however, was detected in cytoplasmic fractions and Lamin B1 was enriched in nuclear fractions in these samples. We also stained the cells with an anti-CapG antibody and DAPI in THP-1 and U937 cells. As shown in Figure 2E, CapG was clearly localized in the nucleus in these cells. In addition, to determine if the CapG localization is regulated by an L4 treatment, we again analyzed the subcellular localization of CapG in BMDCs after incubation with L4. Interestingly, as shown in Figure 2F, the amount of CapG present in the nuclear fraction was increased by the L4 treatment. These data indicate that L4 promoted the nuclear localization of CapG in langerin-expressing BMDCs.
CapG functions as a transcription factor to suppress TNFα gene expression in BMDCs
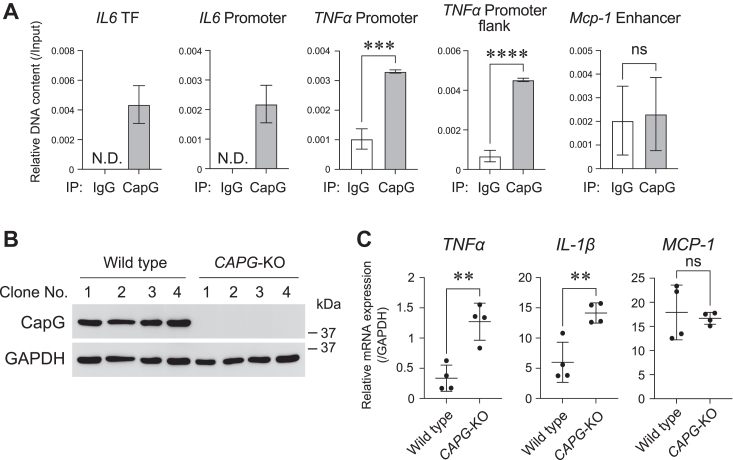
Concerning the nuclear localization of CapG, we hypothesized that CapG was involved in the regulation of gene expression. To address this, we performed ChIP (chromatin immunoprecipitation) analyses of BMDCs. After fixation of the cells, DNA fragmentation and immunoprecipitation with an anti-CapG antibody were carried out. DNA fragments bound to CapG were purified and pPCR (quantitative polymerase chain reaction) targeting the IL-6 promoter or transcription binding region, TNFα promoter region, and Mcp-1 enhancer region were also performed (Fig. S4). Interestingly, as shown in Figure 3A, qPCR targeting IL-6 was successful only in the immunoprecipitate prepared with an anti-CapG antibody, while it was not observed in the case of the immunoprecipitate prepared with non-relevant IgG. We also observed that the DNA fragments of the TNFα promoter region were enriched in the immunoprecipitate prepared with an anti-CapG antibody, while this enrichment was not observed in the sample prepared with non-relevant IgG. The DNA fragments of the Mcp-1 enhancer region were not enriched significantly. In addition, to analyze the roles of CapG in the regulation of TNFα gene expression, we knocked out the CAPG gene in THP-1 cells by the genome editing technology, CRISPR/Cas9. The knocking out of the CAPG gene was confirmed by western blotting as shown in Figure 3B. Using these cells, we analyzed the expression level of the TNFα, IL-1β, and MCP-1 genes by qRT-PCR and the results showed that the CAPG-knockout cells expressed the TNFα and IL-1β genes more strongly than the control cells (Fig. 3C). These data suggest that CapG binds to the TNFα promoter region and suppresses their gene expression as an inhibitory transcription factor.
Figure 3.
CapG functioned as an inhibitory transcription factor to suppress TNFa gene expression. A, ChIP-qPCR analysis targeting the promoter or enhancer region for the IL-6, TNFα, and Mcp-1 genes. Genomic DNA of BMDCs was fragmentated and immunoprecipitated with an anti-CapG antibody or a non-relevant IgG, then, immunoprecipitated DNAs were purified and analyzed by qPCR analysis. Relative DNA content relative to input samples were indicated as the mean±SD. N.D.: not detected. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns: not significant. TF: transcription factor binding site. B, Western blot analysis of CapG in clonal cell lines derived from THP-1 cells. After transfection of a vector to knockout CAPG gene by genome editing in THP-1 cells, four clonal cell lines were established and analyzed. GAPDH was used as a loading control. C, the gene expression level of TNFα, IL-1β, and MCP-1 in CAPG knockout cells (CAPG-KO) and control cells were examined by qRT-PCR analysis. Before the analysis, the cells were differentiated by incubation with PMA (phorbol 12-myristate 13-acetate) (20 nM) for 2 days. The expression levels were normalized to GAPDH genes. ∗∗p < 0.01, ns: not significant.
L4 did not suppress the expression of genes that encode for inflammatory cytokines in langerin-knockout BMDCs
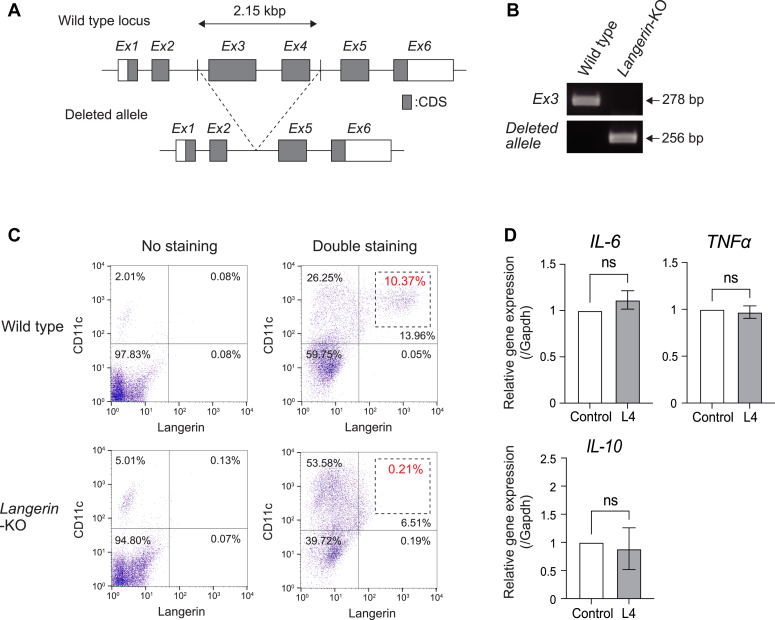
Our group previously reported that L4 binds to langerin as described above, however, the functional meaning of this event has not yet been demonstrated in vivo. For this, we generated a langerin-knockout mouse with genome editing technology, CRISPR/Cas9. The langerin-knockout mouse lacks exon three and exon four of langerin as illustrated in Figure 4A. The genomic PCR analysis confirmed exon three of langerin had been knocked out and that the generation of the deleted allele occurred in a knockout mouse (Fig. 4B). The sequencing analysis of genomic DNA also confirmed the deletion (Fig. S5). In BMDCs prepared from langerin-knockout mice, the expression of langerin was not observed by flow cytometric analysis (Fig. 4C). Additionally, to determine whether L4 functions through langerin, we examined the gene expression of IL-6, TNFα, and IL-10 in BMDCs prepared from the langerin-knockout mice. Very interestingly, incubation with L4 did not result in the downregulation of those genes, as evidenced by a quantitative RT-PCR analysis in BMDCs prepared from langerin-knockout mice (Fig. 4D). These data suggest that the anti-inflammatory function of L4 proceeded via langerin.
Figure 4.
L4 did not downregulate inflammatory cytokines in BMDCs prepared from langerin-knockout mice.A, the locus of the langerin gene in wild type mice and langerin-knockout mice. The exon No.3 (E x 3) and exon No.4 (E x 4) of langerin gene were deleted by genome editing in langerin-knockout mice. B, PCR analysis targeting exon No.3 (E x 3) of langerin gene and deleted allele. Genomic DNAs were prepared from wild type mouse and langerin-knockout mouse (langerin-KO). C, the expression of langerin and CD11c in BMDCs prepared from wild type mice and langerin-KO mice examined by flow cytometry. The X-axis indicates langerin expression and Y-axis indicates CD11c expression. The numbers shown by dushed squares indicates the percentage of the langerin-expressing cells. D, the gene expression of IL-6, TNFα, and IL-10 in BMDCs prepared from langerin-KO mice after incubation with L4 (100 μg/ml) for 2 days was examined by qRT-PCR analysis. The expression levels after incubation with L4 were normalized to the levels without L4. The expression levels were indicated as mean ± SD of three independent experiments. ns: not significant.
L4 suppressed the bronchial obstruction through langerin in an emphysema mouse model
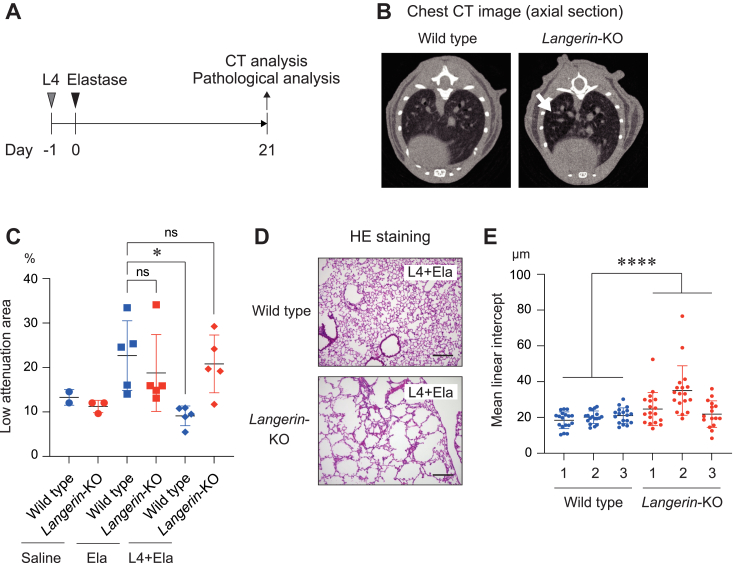
An elastase-induced emphysema mouse model is a well-established animal model in the respiratory research field (21). Elastase is an enzyme that catalyzes the degradation of elastin which is a major component of the extracellular matrix (22). The administration of elastase into the tracheae induces bronchial obstruction and bronchitis which are similar to the symptoms of emphysema in humans. To analyze the function of L4 against emphysema, we administrated L4 and elastase into the tracheae of langerin-knockout mice and wild-type mice (Fig. 5A). At 21 days after the administration, we assessed the state of emphysema with CT (computed tomography) analysis and found that bronchial obstruction had been induced as indicated by low attenuation of X-rays observed in langerin-knockout mice (Fig. 5B). To quantify this, we also assessed the LAA (low attenuation area) of the lungs in langerin-knockout mice and wild type mice after the administration of elastase with and without L4. The administration of elastase induced an equal level of bronchial obstruction between langerin-knockout mice and wild type mice because the LAA was similar between these two groups (Fig. 5C, central two groups). On the other hand, and very interestingly, the administration of L4 before the elastase treatment significantly decreased LAA in wild type mice, but not in the langerin-knockout mice (Fig. 5C, right two groups). Moreover, we performed pathological analyses in these mice and observed severe bronchial obstructions in the langerin-knockout mice, not in the wild0type mice after the administration of L4 and elastase (Fig. 5D). The mean linear intercept of the lung was increased in langerin-knockout mice compared to wild-type mice (Fig. 5E). Additionally, pulmonary fibrosis is sometimes combined with emphysema, which induces more severe respiratory failure (23). To analyze the extent of fibrosis, we performed Masson’s trichrome staining. As shown in Fig. S6, lung sections of the langerin-knockout mouse showed evidence of advanced fibrosis, but this was not observed in the wild type mouse. These data strongly suggest that L4 has a protective function against emphysema and that it is exhibited through langerin.
Figure 5.
L4 suppresses bronchial obstruction through langerin in an emphysema mouse model.A, the schedule for the administration of L4 (1 mg/mouse) and elastase (4 U/mouse). At 21 days after the administration of elastase, CT analysis, and pathological analyses were performed. B, representative images of the CT analysis in the lung of wild type and langerin-KO mouse after administration of L4 and elastase. Arrow indicates low attenuation of X-ray in the lung of langerin-KO mouse. C, the percentages of low attenuation area in the lung of wild type and langerin-KO mice after administration of elastase with or without L4 examined by CT analysis. Each five mice in each group were analyzed after elastase treatment. ∗p < 0.05. ns: not significant. D, representative images of HE staining of the lung sections prepared from wild type and langerin-KO mice after administration of L4 and elastase. Bars indicate 200 μm. E, the mean linear intercept for the lung prepared from wild type (n = 3) and langerin-KO mice (n = 3) after administration of L4 and elastase. Random 16−18 lines in each mouse were measured. ∗∗∗∗p < 0.0001.
L4 mitigated the inflammatory state through langerin in the emphysema mouse model
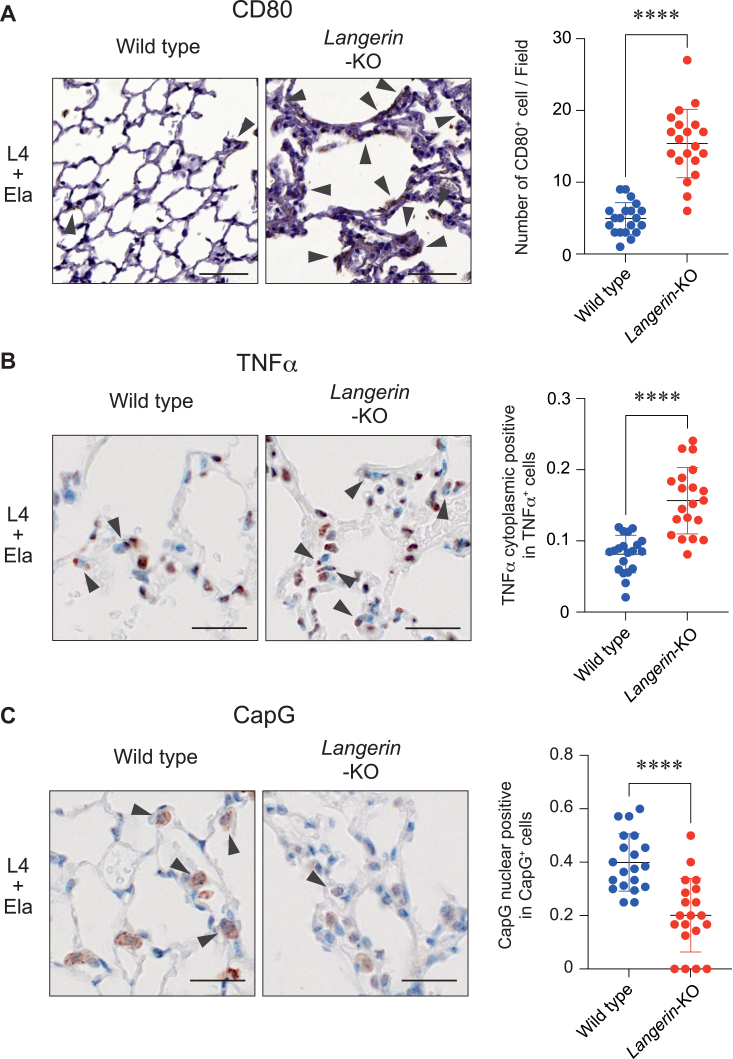
In BMDCs, L4 suppressed the gene expression of TNFα through the langerin-CapG axis, as demonstrated above (Figs. 3 and 4). We hypothesized that the downregulation mechanism for TNFα by L4 would also function in the emphysema mouse model. To address this, we analyzed the inflammatory status and the dynamic state of CapG in the lungs in the emphysema mouse model. CD80 is often used as a marker for the activation of antigen-presenting cells including dendritic cells (24). As shown in Figure 6A, we stained the lung sections of langerin-knockout mice and wild type mice with an anti-CD80 antibody after the administration of L4 and elastase. Interestingly, the number of CD80 positive cells was dramatically increased in the lungs of langerin-knockout mice compared to that of wild type mice. We also analyzed the expression of TNFα by immunohistochemical staining of lung sections and found that the lungs of langerin-knockout mice expressed TNFα more strongly than the wild type mice (Fig. 6B). Moreover, in the analysis of the subcellular localization of CapG in the lung, reduced nuclear expression of CapG was observed in the langerin-knockout mice compared to that in wild type mice (Fig. 6C). These data demonstrate the anti-inflammatory function of L4 through the langerin-CapG axis in in vivo and strongly suggest that the langerin-CapG axis represents a therapeutic target for emphysema (Fig. 7).
Figure 6.
L4 mitigates the inflamed state through the langerin-CapG axis in the lung of the emphysema mouse model.A, representative images of the staining for CD80 in the lung sections of wild type and langerin-KO mouse after administration of L4 and elastase. Arrowheads indicate CD80-positive cells. The number of CD80-positive cells were counted in random 20 fields in each mouse. ∗∗∗∗p < 0.0001. Bars indicate 50 μm. B, representative images of the staining for TNFα in the lung sections of wild type and langerin-KO mouse after administration of L4 and elastase. The number of cells expressing TNFα in cytoplasmic region were counted in 20 randomly selected fields in each mouse and the ratio against the total cell number was shown. ∗∗∗∗p < 0.0001. Bars indicate 20 μm. C, representative images of the staining for CapG in the lung sections of wild type and langerin-KO mouse after administration of L4 and elastase. The number of cells expressing CapG in nuclear were counted in 20 randomly selected fields in each mouse and the ratio against total CapG positive cells was shown. ∗∗∗∗p < 0.0001. Bars indicate 20 μm.
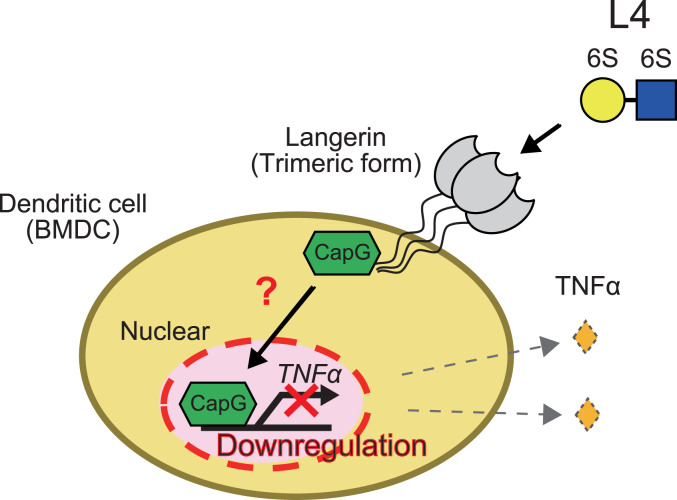
Figure 7.
Graphical summary of this study. In dendritic cells (BMDCs), L4 promotes the nuclear localization of CapG through langerin. In the nucleus, CapG functions as an inhibitory transcription factor for TNFα gene. The L4-langerin-CapG axis results in the downregulation of TNFα, which contributes to the suppression of the inflammatory state of the lung and mitigation of symptoms associated with emphysema.
Discussion
In this study, CapG was identified as a molecule that cooperates with langerin. It was also demonstrated that L4 enhanced the function of CapG to suppress the TNFα gene in BMDCs. Notably, both human and murine langerin harbor a proline-rich domain (PRD) inside their cytoplasmic region. The PRD is generally known to be a unique and functional amino acid domain for docking with a multitude of signaling molecules (25, 26). To determine whether the PRD of langerin is involved in the binding with CapG, we again performed immunoprecipitation with an anti-langerin antibody after the overexpression of human CapG and the PRD-deleted mutant of human langerin (ΔPRD-langerin) in HEK293 cells. However, contrary to our expectations, we observed an association between CapG and ΔPRD-langerin in this analysis (Fig. S7), which indicates that PRD of langerin is not required for langerin to bind with CapG. CapG was originally identified as playing a role in the organization of actin filament in macrophages (27), which suggests that CapG mediates the internalization and recruitment of langerin to lysosomes. Also, CapG could be phosphorylated by PKC (protein kinase C) (28). The dynamics of now CapG is regulated by phosphorylation has not been uncovered, although the phosphorylation would be one of the molecular mechanisms responsible for changing the subcellular localization of CapG. The possibility that phosphorylation of CapG is involved in the transport into the nuclear and function for translational regulation cannot be excluded, but further experiments would be needed to confirm this possibility. Notably, Huang S. and colleagues reported that CapG functions as a transcription factor in breast cancer (29). Upregulation of the STC-1 (stanniocalcin-1) gene by CapG in cancer cells enhances breast cancer metastasis. In this report, our ChIP-qPCR data (Fig. 3A), and that the CAPG gene potentially contains a homology sequence with basis helix-loop-helix family DNA-binding proteins (bHLH) (30) constitute strong pieces of evidence to suggest that CapG plays a role as a transcription factor in immune cells and cancer cells.
In patients with emphysema and/or COPD, a subset of dendritic cells expressing langerin selectively accumulate in the small airways (31). This langerin-expressing subset uniquely expresses IgE receptors, which might be involved in the cross-presentation of antigens to CD8+ T cells (32). Other lectin receptors in emphysema and/or COPD have also been studied. The production of sialic acid-binding immunoglobulin-type lectin-9 (Siglec-9) is also increased in alveolar neutrophils in the lungs of COPD patients. Inflammatory cytokines, for example, IL-6, IL-8, and TNFα induce the expression of Siglec-9 and its soluble form in neutrophils, which contributes to the negative feedback loop by suppressing neutrophil activation (33). C-type lectin domain family five-member A (CLEC5A) is also involved in the development of COPD. Using Clec5a-deficient mice, it was demonstrated that Clec5a was required for the expression of inflammatory cytokines that are linked to the enlargement of airspaces in the lungs of a COPD model (34). An increase in the levels of these lectin receptors, especially langerin, in the lung of patients with emphysema and/or COPD would be expected to mitigate the inflammatory state of the lung. However, there have not been any constructive research that clearly demonstrates the critical function of lectin receptors against emphysema and COPD.
Tobacco smoke can strongly induce the TNFα gene, which is involved in the onset and progression of emphysema and COPD (35, 36). In the lung, a variety of immune cells including macrophages, monocytes, and dendritic cells primarily produce TNFα as well as other types of cells, for example, alveolar epithelial cells also have the potential to express TNFα. Otherwise, the bronchioles mainly express the receptors for TNFα (37). In the analysis using a transgenic mouse that expresses TNFα in alveolar type II cells under the control of the human surfactant protein C promoter, the overexpression of TNFα resulted in the development of pulmonary emphysema by the activation of the elastase (38). In addition, it was reported that an increased expression of TNFα contributed to the progressive disease state in COPD patients (39). For these studies, the blockade of TNFα is considered to be a therapeutic strategy against emphysema and COPD (40). In this study, we demonstrated that L4 suppressed the expression of TNFα through a langerin-CapG axis in dendritic cells. This is strong evidence to suggest that L4 is a potential candidate as a novel TNFα suppressor. In the clinical setting, corticosteroids are often used to modulate the inflammatory state of emphysema and COPD by the suppression of TNFα. The treatment with corticosteroids inhibits the accumulation and activation of neutrophils and macrophages in the lung of these patients. Intriguingly, in a previous study, we reported that L4 also inhibited the accumulation of neutrophils and macrophages in the bronchoalveolar lavage fluid of an elastase-induced emphysema model mouse. The level of this inhibitory effect of L4 was similar to that of the corticosteroid, dexamethasone (17). In this study, we also demonstrated that the molecular mechanism of L4 involves the suppression of inflammation through langerin. As also reported in this study, uncovering the molecular mechanism of the target should be very important in terms of understanding the biological processes involved and in estimating the undesirable effects associated with its administration.
Glycomimetic is a term indicating a type of scientific innovation in which glycan analogs are used to improve biological functions (41). Some types of glycans that target C-type lectin receptors are being developed as novel specific delivery systems for this purpose (42, 43). Wamhoff and co-workers identified heparin-derived monosaccharides as glycomimetic langerin ligands and reported that it is possible to deliver liposomes modified with its glycomimetic ligand specifically to Langerhans cells (44, 45). As described above, L4 is also a glycan analog that mimics the glycosaminoglycan of keratan sulfate proteoglycan. Keratan sulfate is normally expressed in the human body, especially in joints, corneas, and brain. Hence, the administration of L4 would not be expected to show any severe side effects (46). L4 would be stable because there is no naturally produced enzyme to degrade it, and additionally, the molecular weight of L4 is small, therefore making tissue permeability of L4 high. These properties would be strong advantages for the development of L4-based glycomimetics as anti-inflammatory drugs. There are many inflammatory diseases that are exacerbated by the over-activation of dendritic cells in addition to emphysema. Also, it is known that chronic inflammation in the tissues increases the risk of cancer. Considering these issues, we conclude that L4 would be an ideal candidate for use in drug discovery. Our research group plans to continue our research into L4 for subjects who are suffering from abnormal inflammation including emphysema and COPD.
Experimental prodecures
Cells and cell culture
Human promyelocytic leukemia cell line: HL-60, human monocytic leukemia cell line: THP-1, and human embryonic kidney cell line: HEK293 were obtained from the RIKEN BioResource Research Center (Tsukuba, Japan). Human diffuse histiocytic lymphoma cell line: U937 was obtained from JCRB Cell Bank. HL-60, THP-1, and U937 cells were cultured in RPMI-1640 (FUJIFILM Wako Pure Chemical Corporation) supplemented with 10 % (v/v) fetal bovine serum (FBS) and 1 % (v/v) penicillin-streptomycin (Thermo Fisher Scientific) at 37 °C under 5 % CO2. HEK293 cells were cultured in DMEM (FUJIFILM Wako) supplemented with 10 % (v/v) of fetal bovine serum (FBS) and 1 % (v/v) penicillin-streptomycin.
Isolation and differentiation of bone marrow cells
Bone marrow cells were isolated from 10 to 20 weeks old male mice (C57BL/6J). The isolated cells were cultured in RPMI-1640 (FUJIFILM Wako) supplemented with 10 % (v/v) fetal bovine serum (FBS), 1 % (v/v) penicillin-streptomycin (Thermo Fisher Scientific), 50 μM 2-mercaptoethanol (FUJIFILM Wako), and a 20 ng/ml suspension of mouse granulocyte macrophage colony-stimulating factor (Abcam). The culture medium was replaced every 3 days. After incubation for totally 8 days, floating cells were analyzed as BMDCs (bone marrow-derived dendritic cells).
Flow cytometry
BMDCs were washed twice with ice-cold PBS and incubated with an anti-CD16/CD32 antibody (Thermo Fisher Scientific, catalog number 14–0161–85) to block the non-specific binding of antibodies in further procedures. After washing with ice-cold PBS, the cells were incubated with an anti-langerin antibody labeled with phycoerythrin (BioLegend Inc, catalog number 144203) and an anti-CD11c antibody labeled with allophycocyanin (Thermo Fisher Scientific, catalog number 17–0114–82) for 1 h on ice. After washing twice with ice-cold PBS, cells were resuspended in 500 μl of ice-cold PBS and analyzed with BD FACSCalibur (BD Biosciences).
Quantitative reverse transcription PCR
RNAs were extracted from cells by using RNeasy Mini Kit (QIAGEN, Venlo, Netherlands). Reverse transcription was performed with oligo dT primer and SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific) following manufacturer’s protocol. Real-time PCR analysis was also carried out with THUNDERBIRD Next SYBR qPCR Mix (TOYOBO) and analyzed with 7500 Real-Time PCR System (Applied Biosystems). Primers for mouse IL-6 were 5′-CTCATTCTGCTCTGGAGCCC-3′ and 5′-TGCCATTGCACAACTCTTTTCT-3′, for mouse TNFα were 5′-GTAGCCCACGTCGTAGCAAA-3′ and 5′-TAGCAAATCGGCTGACGGTG-3′, for mouse IL-10 were 5′-GGCGCTGTCATCGATTTCTC-3′ and 5′-CGGAGAGAGGTACAAACGAGG-3′, for human IL-6 were 5′- CCTTCGGTCCAGTTGCCTT-3′ and 5′- GTTGTTTTCTGCCAGTGCCT-3′, for human TNFα were 5′- CCTCTCTCCCCTGGAAAGGA-3′ and 5′- CTACAGGCTTGTCACTCGGG-3′, for human IL-10 were 5′- CAAAAGAAGGCATGCACAGCTC-3′ and 5′- ACTGGATCATCTCAGACAAGGC-3′, for mouse Mcp-1 were 5′- CACTCACCTGCTGCTACTCA-3′ and 5′- AGACCTTAGGGCAGATGCAG-3′, for mouse IL-1α were 5′-CCCGTGTTGCTGAAGGAGTTG-3′ and 5′-TGACTCAGGGTGAGGAAGGTT-3′, for mouse IL-1β were 5′-TGCCACCTTTTGACAGTGATG-3′ and 5′-AAGGTCCACGGGAAAGACAC-3′, for mouse IFN-β were 5′-TGGGAGATGTCCTCAACTGC-3′ and 5′-ACTACCAGTCCCAGAGTCCG-3′.
PCR for mouse genotyping
A portion of the murine tail was incubated with 200 μl of DNA extraction buffer (0.1 M Tris-HCl (pH 8.0), 1 mM EDTA (pH 8.0), 0.5 % (v/v) Tween20) and 100 μg of Proteinase K (FUJIFILM Wako) at 55 °C overnight. The tails were then heated at 95 °C for 10 min to inactivate Proteinase K. Using the resulting solution, PCR was performed with MightyAmp DNA Polymerase (Takara Bio Inc). Primers targeting exon No.3 of langerin gene were 5′- GAGGGTGCGTTCTCAGATCC-3′ and 5′- TGCTCAAAACAGGTAGGGGC-3′, and targeting deleted allele of langerin gene were 5′- TGTGTGGTAGCCCCTAATGC-3′ and 5′-CCTGGAGACGTGAGCAGAAA-3′. PCR products were separated by agarose gel electrophoresis at 100 V for 20 min and visualized with Midori Green Direct (NIPPON Genetics Co, Ltd).
Preparation of cell lysates and immunoprecipitation
Cells were washed 3 times with ice-cold PBS and lysed with cell lysis buffer (1% NP-40 in TBS) that included protease inhibitors (Roche, Basel, Switzerland) and PhosSTOP phosphatase inhibitor (Roche). The lysates were sonicated briefly and centrifuged at 15,000g at 4 °C for 5 min to remove the insoluble materials. The protein concentration of the lysate was determined with a BCA protein assay kit (BioRad Laboratories). For immunoprecipitation, a part of lysate was incubated with an anti-langerin antibody (BioLegend Inc, catalog number 144202) or non-relevant isotype IgG (Santa Cruz Biotechnology, Inc) at 4 °C overnight under rotation. Next day, 20 to 50 μl of Protein G Sepharose four Fast Flow (GE Healthcare) was added into the lysate and incubated for more 2 h at 4 °C under rotation. After washing 3 times with cell lysis buffer, Protein G Sepharose was added to the sample buffer (50 mM Tris (pH6.8), 2 % (w/v) SDS, 2.5 % (v/v) 2-mercaptoethanol, 9 % (v/v) glycerol) and heated at 95 °C for 5 min.
Preparation of cytoplasmic and nuclear fractions
Cells were washed 3 times with ice-cold PBS. After washing, 500 μl of hypotonic buffer (20 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2) including protease inhibitors (Roche) was added and then incubated on ice for 15 min. Twenty-5 μl of 10 % NP-40 was added and the resulting suspension was vortexed briefly. After incubation on ice for more 3 min, the resulting solution was centrifuged at 3000 rpm (TOMY TMP-1 rotor) at 4 °C for 10 min. The supernatant was transferred into new tube as cytoplasmic fraction. Additionally, 100 μl of RIPA buffer (50 mM Tris-HCl (pH 8.0), 150 mM sodium chloride, 0.5 % (w/v) sodium deoxycholate, 0.1 % (w/v) sodium dodecyl sulfate, 1 % (v/v) NP-40) including protease inhibitor (Roche) was added into the pellet. After incubation on ice for 30 min, the resulting solution was centrifuged at 14,000g at 4 °C for 30 min. The supernatant was transferred into new tube as nuclear fraction.
Mass spectrometry
The immunoprecipitates prepared from BMDCs with an anti-langerin antibody were separated by SDS-PAGE using 10 % (w/v) acrylamide gels. Silver staining was then performed with Pierce Silver Stain for Mass Spectrometry (Thermo Fisher Scientific) following manufacturer’s protocol. The bands were cut out and destaining was also performed by using regents in the Pierce Silver Stain for Mass Spectrometry. The resulting piece of the gel was taken to dryness with a KPI Rotary Vacuum Miro-concentrator RMC-24 (KOIKE PRECISION INSTRUMENTS) and EYELA Cold Trap UT-1000 (TOKYO RIKAKIKAI CO LTD). Proteins in the gel pieces were digested with trypsin for Mass Spectrometry (Thermo Fisher Scientific) after reduction with dithiothreitol and alkylation with iodoacetamide. The tryptic peptides were trapped using a short ODS column (PepMap 100; 5 μm C18, 5 mm × 300 μm ID; Thermo Fisher Scientific) and then separated using an ODS column (Nano HPLC Capillary Column; 3 μm C18, 120 mm × 75 μm ID; Nikkyo Technos) using nano-liquid chromatography (nanoLC) (Ultimate 3000; Thermo Fisher Scientific). The mobile phases for separation were solvent A (0.1% formic acid) and solvent B (0.1% formic acid in acetonitrile). After loading tryptic peptides to trap column with 0.1% TFA for 3 min at a flow rate of 30 μl/min, the concentrated tryptic peptides were eluted from trap column and separated on separation column using a sequence of isocratic and a linear gradient: 0 to 3 min, solvent A; 3 to 35 min, 0 to 35% (v/v) solvent B; and increasing to 90% solvent B for 10 min and re-equilibrated with solvent A for 15 min. The eluate from separation column was introduced continuously into a nanoESI source and analyzed by MS and MS/MS (LTQ Orbitrap XL; Thermo Fisher Scientific). MS and MS/MS spectra were obtained in the positive ion mode using Orbitrap (mass range: m/z 300–1500) and Iontrap (data dependent scan of top five peaks using CID), respectively. The voltage of the capillary source was set at 1.6 kV, and the temperature of the transfer capillary maintained at 200 °C. The capillary voltage and tube lens voltage were set at 30 V and 80 V, respectively. Assignment of MS/MS data to tryptic peptides was performed by Proteome Discoverer 2.4 (Thermo Fisher Scientific) using Mascot search engine 2.5.1 (Matrix Science) on UniProt data base. Doubly and triply charged peptide ions were subjected to the database search with precursor and fragment ion mass tolerance of ±10 ppm and ±0.8 Da, respectively, with static modification (carbamidomethylation of cysteine) and dynamic modification (oxidation of methionine and deamidation of asparagine and glutamine). The significance threshold on Proteome Discoverer for Mascot search was set at p < 0.05 and one and two missed trypsin cleavage was allowed.
Western blotting
Cell lysates were separated by SDS-PAGE using 10 % (w/v) acrylamide gels. After transferring to a nitrocellulose membrane (Merck Millipore, Burlington, MA), the membrane was blocked with 2 % (w/v) bovine serum albumin (Nakalai tesque, Inc) or 5 % (w/v) skim milk (Nakalai tesque, Inc) in TBST (0.1 % (v/v) Tween20 in TBS). The membrane was then incubated with an anti-CapG antibody (GeneTex, Irvine, CA, catalog number GTX114301), an anti-Lamin B1 antibody (BioLegend Inc, catalog number 8969801), or an anti-GAPDH antibody (Merck Millipore, catalog number MAB374). After washing 4 times with TBST, the membrane was then incubated with an anti-rabbit or -mouse IgG labeled with horseradish peroxidase (GE Healthcare). To visualize the bands, the membrane was analyzed with Amersham ECL Prime Western Blotting Detection Reagents (GE Healthcare) and ChemiDoc MP Imaging System (BioRad Laboratories).
Immunocytochemistry
Cells were washed 3 times with ice-cold PBS and fixed with 4 % (w/v) paraformaldehyde in PBS. After washing 3 times with PBS, cells were incubated for 10 min with 0.1 % (v/v) TritonX-100 in PBS at room temperature, The resulting cells were then washed with PBS and blocked with 2.5 % w/v bovine serum albumin (Nakalai tesque, Inc) in PBS. After blocking, cells were incubated with an anti-CapG antibody (Santa Cruz Biotechnology, Inc, catalog number sc-166428) and an anti-mouse IgG labeled with Alexa 594 (Thermo Fisher Scientific, catalog number A21044). To stain the nucleus, cells were also incubated with DAPI (DOJINDO LABORATORIES). The stained cells were analyzed with confocal microscopy, FV10i (Olympus Corporation) or LSM 900 (ZEISS).
Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR)
Cells were washed twice with PBS and fixed with 1 % (v/v) formaldehyde in PBS. After incubation at room temperature for 10 min, quenching was performed by incubation with 0.125 M glycine in PBS. The cells were then lysed with RIPA buffer including protease inhibitors (Roche) and the DNAs were fragmented by sonication. The size of fragmented DNAs was confirmed as approximately 600 to 1000 bp. The resulting solution was centrifuged at 15,000 g at 4 °C for 10 min to remove debris. The protein concentration was measured by using BCA protein assay kit (BioRad Laboratories) and a part of lysate including 2 mg of total protein was incubated with 4 μg of anti-CapG antibody (Santa Cruz Biotechnology, Inc, catalog number sc-166428) or non-relevant isotype IgG (Santa Cruz Biotechnology, Inc) overnight at 4 °C under rotation. On the next day, 30 μl of Protein G Sepharose four Fast Flow (GE Healthcare) was added and the resulting solution incubated at 4 °C for more 2 h under rotation. The resulting solution was centrifuged at 4 °C for 30 s and the supernatant was removed. Five hundred μl of TE was added into the Protein G Sepharose, then, RNaseA (50 μg/ml) (Merck Sigma-Aldrich, St Louis, MO) treatment at 37 °C for 30 min and proteinase K (1.68 unit/ml) (FUJIFILM Wako) treatment at 55 °C for 1 h were carried out. After heating at 95 °C for 10 min for inactivation proteinase K, DNA fragments were purified and concentrated by using phenol-chloroform. The pellet of DNA was resuspended in 50 μl of TE and a 2 μl aliquot was analyzed by qPCR. Primers targeting the promoter region of mouse IL-6 gene were 5′-TGCTCATGCTTCTTAGGGCT-3′ and 5′-TGGGGCTGATTGGAAACCTT-3′, targeting the transcription factor binding site of mouse IL-6 gene were 5′-GACTGAGCCTAAGGGTGCAT-3′ and 5′-ACCACTAGAGGGCCAAGTCA-3′, targeting promoter region of mouse TNFα gene were 5′-ACTGATGAGAGGGAGGCCAT-3′ and 5′-AACTGTAAGCGGGGCAATCA-3′, targeting promoter with the flank region of mouse TNFα gene were 5′-TGAGTCCTTGATGGTGGTGC-3′ and 5′-AGGGGATTATGGCTCAGGGT-3′, and targeting enhancer region of mouse Mcp-1 gene were 5′- GGACTCAGCCGACTTACTGG-3′ and 5′- AAGGAAGTGGCCAAGGAACC-3′. The genome regions were identified by using the Ensembl genome browser.
Generation of CapG knockout THP-1 cells
THP-1 cells were transfected with a pSpCas9(BB)-2A-GFP vector (Addgene, plasmid code 48138) harboring target sequences against the human CAPG gene by using jetPRIME (PolyPlus-transfection, Illkirch, France). After culturing for 2 days, GFP positive cells were enriched with a S3 Cell Sorter (BioRad Laboratories). Clonal cells then grown from single cells were prepared by limiting the dilution. Target sequences were 5′-GATCTGAAGACAGCATGTAC-3′ and 5′-GTGCCGTGCTGGCTGTGCAC-3′.
Generation of langerin-knockout mice
The pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (Addgene, plasmid code 42230) harboring target sequences against mouse langerin gene was implanted by micro-injected into embryonic stem cells. In the newborn mice, the mutation of langerin gene was confirmed by PCR and DNA sequencing analysis. Dr Satoru Takahashi (Tsukuba University) and Dr Yasunori Chiba (AIST: National Institute of Advanced Industrial Science and Technology) kindly contributed for generation of langerin-knockout mice. The genetic background of mice is C57BL/6J.
Intratracheal administration and computed tomography analysis
Under general anesthesia, a 100-μl solution of L4 in saline (10 mg/ml) and 100 μl of elastase from the porcine pancreas in saline (40 U/ml) (FUJIFILM Wako) were administrated into the tracheae by spray (17). After 21 days, CT analyses were performed with ALOKA Latheta Laboratory CT LCT-200 (Hitachi, Ltd). The area indicated by the intensity value between −900 to −610 was calculated as low attenuation area. The male mice (10–20 weeks old) were analyzed.
Preparation of paraffin sections and immunohistochemistry
Mice were euthanized by inhaling carbon dioxide gas. For the fixation of lung tissues, 10 % (v/v) formalin natural buffer solution (FUJIFILM Wako) was injected into the lung at a constant rate through the respiratory tract. After removing the lung tissues, paraffin blocks were prepared using an automatic fixation and embedding machine RH-12 (Sakura Finetek Japan Co, Ltd) and HistoCore Arcadia H (Leica Biosystems). Four μm-thick paraffin sections were prepared with Microm HM 360 Automated Microtome (Marshall Scientific LLC.). For immunohistochemistry, the sections were microwaved for epitope retrieval and deparaffinization was then performed. After washing with water, the sections were saturated in an Antigen Unmasking Solution (Vector Laboratories) and microwaved again. The sections were then incubated with 1 % (v/v) H2O2 in MeOH for 5 min at room temperature. After washing with PBST (0.05 % (v/v) Tween 20 in PBS), the sections were blocked with 1.5 % (w/v) bovine serum albumin (Nacalai tesque) in PBST. The resulting sections were incubated with an anti-CD80 antibody (Santa Cruz Biotechnology, Inc, catalog number sc-376012), an anti-TNFα antibody (Novus Biologicals, Littleton, CO, catalog number NBP1-19532SS), or an anti-CapG antibody (Santa Cruz Biotechnology, Inc, catalog number sc-166428). After incubation at 4 °C overnight, the sections were washed 4 times with PBST and incubated with Dako EnVision + Dual Link System-HRP (Agilent Technologies, Inc) at room temperature for 30 min. After washing with PBST, the sections were visualized by means of a Liquid DAB+ Substrate Chromogen System (Agilent Technologies, Inc). Counter staining with Mayer’s hematoxylin solution (ScyTec Laboratories, Inc) was also performed. The sections were then dehydrated and encapsulated with Multi Mount 220 (Matsunami Glass Ind., Ltd) and the resulting sections were observed by microscopy, BZ-X710 (KEYENCE CORPORATION).
Statistical analysis
All data are presented as the mean ± standard deviation. For comparison between multiple groups, the one-way ANOVA test was performed. For comparison between two groups, the student’s t test analysis was performed.
Study approval
All studies using mice were approved by the local committee at Osaka International Cancer Institute (Approval number, 22,030,316).
Data availability
All data are contained with the manuscript or associated supplementary files.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare no conflicts of interest associated with this study.
Acknowledgments
We wish to thank Dr Shigeki Higashiyama and Dr Yusuke Imagawa at Department of Oncogenesis and Growth Regulation in Osaka International Cancer Institute for their experimental supports. We also thank Dr Milton S Feather for examining the English use in this submission.
Author contributions
Y. O., Y. K., and methodology; Y. O., N. K., R. F., and E. L. J. investigation; M. N. formal analysis; Y. O. writing–original draft; Y. K., Y. H., J. T. and N. T. writing–review and editing.
Funding and additional information
This study was supported by Japan Agency for Medical Research and Development (AMED) (20ae0101070h0005 to N. T.), Grants-in-Aid for Scientific Research (C) (20K07629 to Y. O.) and Takeda Science Foundation.
Reviewed by members of the JBC Editorial Board. Edited by Robert Haltiwanger
Supporting information
References
- 1.Barnes P.J., Burney P.G.J., Silverman E.K., Celli B.R., Vestbo J., Wedzicha J.A., et al. Chronic obstructive pulmonary disease. Nat. Rev. Dis. Primers. 2015;1 doi: 10.1038/nrdp.2015.76. [DOI] [PubMed] [Google Scholar]
- 2.Strzelak A., Ratajczak A., Adamiec A., Feleszko W. Tobacco smoke induces and alters immune responses in the lung triggering inflammation, allergy, asthma and other lung diseases: a mechanistic review. Int. J. Environ. Res. Public Health. 2018;15:E1033. doi: 10.3390/ijerph15051033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu F., Liang C.-L., Liu H., Zeng Y.-Q., Hou S., Huang S., et al. Impacts of cigarette smoking on immune responsiveness: up and down or upside down? Oncotarget. 2017;8:268–284. doi: 10.18632/oncotarget.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang D.C., Chen C.-H. Cigarette smoking-mediated macrophage reprogramming: mechanistic insights and therapeutic implications. J. Nat. Sci. 2018;4:e539. [PMC free article] [PubMed] [Google Scholar]
- 5.Falk J.A., Minai O.A., Mosenifar Z. Inhaled and systemic corticosteroids in chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2008;5:506–512. doi: 10.1513/pats.200707-096ET. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granucci F., Zanoni I., Ricciardi-Castagnoli P. Central role of dendritic cells in the regulation and deregulation of immune responses. Cell Mol. Life Sci. 2008;65:1683–1697. doi: 10.1007/s00018-008-8009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung K.F., Adcock I.M. Multifaceted mechanisms in COPD: inflammation, immunity, and tissue repair and destruction. Eur. Respir. J. 2008;31:1334–1356. doi: 10.1183/09031936.00018908. [DOI] [PubMed] [Google Scholar]
- 8.Gerayeli F.V., Milne S., Cheung C., Li X., Yang C.W.T., Tam A., et al. COPD and the risk of poor outcomes in COVID-19: a systematic review and meta-analysis. EClinicalMedicine. 2021;33 doi: 10.1016/j.eclinm.2021.100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valladeau J., Clair-Moninot V., Dezutter-Dambuyant C., Pin J.-J., Kissenpfennig A., Mattéi M.-G., et al. Identification of mouse langerin/CD207 in langerhans cells and some dendritic cells of lymphoid tissues. J. Immunol. 2002;168:782–792. doi: 10.4049/jimmunol.168.2.782. [DOI] [PubMed] [Google Scholar]
- 10.Bigley V., McGovern N., Milne P., Dickinson R., Pagan S., Cookson S., et al. Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J. Leukoc. Biol. 2015;97:627–634. doi: 10.1189/jlb.1HI0714-351R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turville S.G., Arthos J., Donald K.M., Lynch G., Naif H., Clark G., et al. HIV gp120 receptors on human dendritic cells. Blood. 2001;98:2482–2488. doi: 10.1182/blood.v98.8.2482. [DOI] [PubMed] [Google Scholar]
- 12.Turville S.G., Cameron P.U., Handley A., Lin G., Pöhlmann S., Doms R.W., et al. Diversity of receptors binding HIV on dendritic cell subsets. Nat. Immunol. 2002;3:975–983. doi: 10.1038/ni841. [DOI] [PubMed] [Google Scholar]
- 13.Nasr N., Lai J., Botting R.A., Mercier S.K., Harman A.N., Kim M., et al. Inhibition of two temporal phases of HIV-1 transfer from primary langerhans cells to T cells: the role of langerin. J. Immunol. 2014;193:2554–2564. doi: 10.4049/jimmunol.1400630. [DOI] [PubMed] [Google Scholar]
- 14.GeurtsvanKessel C.H., Willart M.A.M., van Rijt L.S., Muskens F., Kool M., Baas C., et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b− but not plasmacytoid dendritic cells. J. Exp. Med. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tateno H., Ohnishi K., Yabe R., Hayatsu N., Sato T., Takeya M., et al. Dual specificity of Langerin to sulfated and mannosylated glycans via a single C-type carbohydrate recognition domain. J. Biol. Chem. 2010;285:6390–6400. doi: 10.1074/jbc.M109.041863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ota F., Hirayama T., Kizuka Y., Yamaguchi Y., Fujinawa R., Nagata M., et al. High affinity sugar ligands of C-type lectin receptor langerin. Biochim. Biophys. Acta Gen. Subj. 2018;1862:1592–1601. doi: 10.1016/j.bbagen.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Gao C., Fujinawa R., Yoshida T., Ueno M., Ota F., Kizuka Y., et al. A keratan sulfate disaccharide prevents inflammation and the progression of emphysema in murine models. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;312:L268–L276. doi: 10.1152/ajplung.00151.2016. [DOI] [PubMed] [Google Scholar]
- 18.Bowers W.E., Berkowitz M.R. Differentiation of dendritic cells in cultures of rat bone marrow cells. J. Exp. Med. 1986;163:872–883. doi: 10.1084/jem.163.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutz M.B., Kukutsch N., Ogilvie A.L.J., Rößner S., Koch F., Romani N., et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 20.Van Impe K., De Corte V., Eichinger L., Bruyneel E., Mareel M., Vandekerckhove J., et al. The nucleo-cytoplasmic actin-binding protein CapG lacks a nuclear export sequence present in structurally related proteins. J. Biol. Chem. 2003;278:17945–17952. doi: 10.1074/jbc.M209946200. [DOI] [PubMed] [Google Scholar]
- 21.Suki B., Bartolák-Suki E., Rocco P.R.M. Elastase-induced lung emphysema models in mice. Methods Mol. Biol. 2017;1639:67–75. doi: 10.1007/978-1-4939-7163-3_7. [DOI] [PubMed] [Google Scholar]
- 22.Werb Z., Banda M.J., McKerrow J.H., Sandhaus R.A. Elastases and elastin degradation. J. Invest. Dermatol. 1982;79:154s–159s. doi: 10.1111/1523-1747.ep12546056. [DOI] [PubMed] [Google Scholar]
- 23.Mejía M., Carrillo G., Rojas-Serrano J., Estrada A., Suárez T., Alonso D., et al. Idiopathic pulmonary fibrosis and emphysema: decreased survival associated with severe pulmonary arterial hypertension. Chest. 2009;136:10–15. doi: 10.1378/chest.08-2306. [DOI] [PubMed] [Google Scholar]
- 24.Tseng S.-Y., Otsuji M., Gorski K., Huang X., Slansky J.E., Pai S.I., et al. B7-Dc, a new dendritic cell molecule with potent costimulatory properties for T cells. J. Exp. Med. 2001;193:839–846. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williamson M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarrinpar A., Bhattacharyya R.P., Lim W.A. The structure and function of proline recognition domains. Sci. STKE. 2003;2003:RE8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 27.Dabiri G.A., Young C.L., Rosenbloom J., Southwick F.S. Molecular cloning of human macrophage capping protein cDNA. A unique member of the gelsolin/villin family expressed primarily in macrophages. J. Biol. Chem. 1992;267:16545–16552. [PubMed] [Google Scholar]
- 28.Murphy L.A., Sarge K.D. Phosphorylation of CAP-G is required for its chromosomal DNA localization during mitosis. Biochem. Biophys. Res. Commun. 2008;377:1007–1011. doi: 10.1016/j.bbrc.2008.10.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang S., Chi Y., Qin Y., Wang Z., Xiu B., Su Y., et al. CAPG enhances breast cancer metastasis by competing with PRMT5 to modulate STC-1 transcription. Theranostics. 2018;8:2549–2564. doi: 10.7150/thno.22523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prendergast G.C., Ziff E.B. Mbh 1: a novel gelsolin/severin-related protein which binds actin in vitro and exhibits nuclear localization in vivo. EMBO J. 1991;10:757–766. doi: 10.1002/j.1460-2075.1991.tb08007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Pottelberge G.R., Bracke K.R., Demedts I.K., De Rijck K., Reinartz S.M., van Drunen C.M., et al. Selective accumulation of langerhans-type dendritic cells in small airways of patients with COPD. Respir. Res. 2010;11:35. doi: 10.1186/1465-9921-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori M., Siddhuraj P., Roos A.B., Stämpfli M.R., Löfdahl C.-G., Erjefält J.S. New aspects of langerin dendritic cells in patients with COPD. Eur. Respir. J. 2017;50:PA983. [Google Scholar]
- 33.Zeng Z., Li M., Wang M., Wu X., Li Q., Ning Q., et al. Increased expression of Siglec-9 in chronic obstructive pulmonary disease. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-09120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wortham B.W., Eppert B.L., Flury J.L., Garcia S.M., Donica W.R., Osterburg A., et al. Cutting edge: CLEC5A mediates macrophage function and chronic obstructive pulmonary disease pathologies. J. Immunol. 2016;196:3227–3231. doi: 10.4049/jimmunol.1500978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Churg A., Dai J., Tai H., Xie C., Wright J.L. Tumor necrosis factor-alpha is central to acute cigarette smoke-induced inflammation and connective tissue breakdown. Am. J. Respir. Crit. Care Med. 2002;166:849–854. doi: 10.1164/rccm.200202-097OC. [DOI] [PubMed] [Google Scholar]
- 36.Lundblad L.K.A., Thompson-Figueroa J., Leclair T., Sullivan M.J., Poynter M.E., Irvin C.G., et al. Tumor necrosis factor–α overexpression in lung disease. Am. J. Respir. Crit. Care Med. 2005;171:1363–1370. doi: 10.1164/rccm.200410-1349OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimomoto H., Hasegawa Y., Nozaki Y., Takagi N., Shibagaki T., Nakao A., et al. Expression of tumor necrosis factor receptors in human lung cancer cells and normal lung tissues. Am. J. Respir. Cell Mol. Biol. 1995;13:271–278. doi: 10.1165/ajrcmb.13.3.7654383. [DOI] [PubMed] [Google Scholar]
- 38.Fujita M., Shannon J.M., Irvin C.G., Fagan K.A., Cool C., Augustin A., et al. Overexpression of tumor necrosis factor-α produces an increase in lung volumes and pulmonary hypertension. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;280:L39–L49. doi: 10.1152/ajplung.2001.280.1.L39. [DOI] [PubMed] [Google Scholar]
- 39.El-Shimy W.S., El-Dib A.S., Nagy H.M., Sabry W. A study of IL-6, IL-8, and TNF-α as inflammatory markers in COPD patients. Egypt J. Bronchol. 2014;8:91–99. [Google Scholar]
- 40.Matera M.G., Calzetta L., Cazzola M. TNF-alpha inhibitors in asthma and COPD: we must not throw the baby out with the bath water. Pulm. Pharmacol. Ther. 2010;23:121–128. doi: 10.1016/j.pupt.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Compain P. Glycomimetics: design, synthesis, and therapeutic applications. Molecules. 2018;23:1658. doi: 10.3390/molecules23071658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medve L., Achilli S., Guzman-Caldentey J., Thépaut M., Senaldi L., Le Roy A., et al. Enhancing potency and selectivity of a DC-SIGN glycomimetic ligand by fragment-based design: structural basis. Chemistry. 2019;25:14659–14668. doi: 10.1002/chem.201903391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Pietro S., Bordoni V., Iacopini D., Achilli S., Pineschi M., Thépaut M., et al. New lipophilic glycomimetic DC-SIGN ligands: stereoselective synthesis and SPR-based binding inhibition assays. Bioorg. Chem. 2021;107 doi: 10.1016/j.bioorg.2020.104566. [DOI] [PubMed] [Google Scholar]
- 44.Wamhoff E.-C., Schulze J., Bellmann L., Rentzsch M., Bachem G., Fuchsberger F.F., et al. A specific, glycomimetic langerin ligand for human langerhans cell targeting. ACS Cent. Sci. 2019;5:808–820. doi: 10.1021/acscentsci.9b00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rentzsch M., Wawrzinek R., Zelle-Rieser C., Strandt H., Bellmann L., Fuchsberger F.F., et al. Specific protein antigen delivery to human langerhans cells in intact skin. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.732298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caterson B., Melrose J. Keratan sulfate, a complex glycosaminoglycan with unique functional capability. Glycobiology. 2018;28:182–206. doi: 10.1093/glycob/cwy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained with the manuscript or associated supplementary files.