Abstract
Epilepsy is a chronic neurological disorder marked by recurrent seizures, significantly affecting the population in Saudi Arabia across all age demographics. The global prevalence of active epilepsy is around 6.38/1,000 persons and in the Arabian region, the median prevalence of active epilepsy is 4.4/1,000 persons. However, over 75% of individuals are untreated. Consequently, the development of therapeutic strategies with increased efficacy and safety profiles is essential to improve the survival rate among epilepsy patients. The current study integrates network pharmacology along with Bioinformatics approaches to explore the potential molecular mechanisms of local flora of Saudi Arabia including Solanum incanum, Abrus precatorius, Withania somnifera, and Azadirachta indica in epilepsy treatment. In the preliminary phase, data related to the bioactive components of the local plants and the associated target genes of both these plants and epilepsy were gathered from scientific literature and open-source databases. This data was then analyzed to identify common targets between the plants and ovarian cancer. Based on these common targets, a protein–protein interaction (PPI) network was constructed utilizing the STRING database, which was subsequently incorporated into the Cytoscape software for identification of hub genes based on their degree of connectivity. Lastly, an interplay network depicting the associations between the compounds and the overlapping genes was formulated via Cytoscape, to study the potential network pharmacology implications of these active compounds in relation to ovarian cancer. Following that, a compound-target protein-pathway network was constructed which uncovered that namely abrectorin, genistin, (+)-catechin, precatorine, (+)-ascorbic acid, licoflavanone, skrofulein, stigmasterone, 5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone could potentially be used as antagonists for the therapeutic management of epilepsy by targeting TNF and TP53 proteins. Furthermore, the implementation of molecular docking reinforces the binding affinity of the compound, indicating a robust stability of the forecasted compounds at the docked site. This research lays both a theoretical and experimental groundwork for more profound investigations and establishes a practical method for the strategic employment of active compounds in the development of anti-epileptic therapeutics.
Keywords: Indigenous plants, Saudi Arabia, Multi-target approach, Network pharmacology, Epilepsy, Active compounds, Molecular docking
1. Introduction
Epilepsy is a prevalent neurological disorder in Saudi Arabia that holds a significant status among the most critical brain disorders worldwide, influencing the lives of>70 million people (Thijs et al., 2019). Epilepsy is not characterized by a singular manifestation or etiology; rather, it encompasses a myriad of symptoms linked to an assortment of risk factors and demonstrates a notable genetic inclination (Camfield and Camfield, 2015). Anxiety, depression, and cardiovascular disorders, migraines, and dementia are observed to be 8 times more prevalent in epilepsy patients. An estimated 80% of individuals with epilepsy reside in developing countries (Beghi, 2020). This leads to a significantly amplified comorbidity burden in those with this neurological disorder. Despite the significant advancements in the development of over 20 antiepileptic drugs for seizure management, it remains perplexing that a considerable proportion of individuals with epilepsy, approximately one-third, continue to endure seizures that are unresponsive to these medications (Löscher et al., 2020).
The Kingdom Saudi Arabia boasts a significant floral biodiversity, with an estimated 2250 species distributed across its varied ecosystems (Rahman et al., 2004). The region is distinguished by its rich assortment of both indigenous and cultivated plant species (Sher et al., 2010). Unfortunately, the progression of indigenous knowledge across generations faces obstacles due to limited dissemination of information, a dearth of data concerning their practical uses, and the complexities in discerning wild medicinal plants. These hurdles complicate the efforts to preserve and employ traditional knowledge, potentially putting both cultural heritage and potential therapeutic resources at risk. Previous studies has provided indications that native plants of Saudi Arabia, including Solanum incanum, Abrus precatorius, Withania somnifera, and Azadirachta indica, possess anti-epileptic properties (Worku, 2022, Attal et al., 2010, Anju et al., 2018, Birhan, 2022). Despite these promising indications, it is imperative to highlight the comparative scarcity of rigorous clinical trials that investigate the efficacy of these botanical entities in a clinical milieu for the amelioration of epilepsy. Consequently, the present study is designed to decipher the synergistic influence of these phytogenic extracts in epilepsy management, thereby establishing a foundation for prospective clinical exploration.
Recognizing the potential of phytoconstituents, Hopkins (Hopkins, 2007) formulated an integrative computational approach “network pharmacology”. Network pharmacology transitioned the paradigm that single gene disordered required single target drugs while complicated diseases where gene network is involved required more holistic multiple-targeted therapies (Noor et al., 2023, Noor et al., 2022). Thus, network pharmacology has now been emerged as an asset in the process of drug development, contributing significantly to the reinvigoration of traditional knowledge (Chandran et al., 2017). This strategy is employed as a foundation for the initial identification and screening of phytoconstituents, as well as the discovery of innovative therapeutic targets, thereby advancing our understanding of disease mechanisms (Noor et al., 2022). Therefore, it fundamentally aids in the internationalization and modernization of medicinal herbs, fostering a significant change in the drug discovery paradigm. In essence, the successful identification of potential drugs often necessitates a comprehensive understanding of the complex domain of network pharmacology. Numerous studies have highlighted the successful application of network pharmacology in identifying new therapeutic targets, repurposing existing drugs, and designing combination therapies. These endeavors have resulted in significant advancements in the treatment of various diseases, ranging from cancer to neurological disorders. However, it is crucial to critically evaluate and validate these approaches through well-designed clinical trials and observational studies that assess their true impact on patient outcomes. While clinical trials specifically focused on network pharmacology interventions are limited, Ongoing research endeavors are continuously exploring the clinical implications of network pharmacology approaches across various disease areas. Network pharmacology-based findings, although primarily based on computational predictions and experimental evidence, provide valuable insights into the potential clinical implications of network pharmacology.
This study endeavors to elucidate the active constituents, corresponding targets, and complex pharmacodynamic mechanisms driving the anti-epileptic potential of Solanum incanum, Abrus precatorius, Withania somnifera, and Azadirachta indica through the application of network pharmacology integrated with bioinformatics techniques. Network pharmacology deploys multi-target approaches to elucidate the complex interplay between bioactive compounds and their respective protein targets from a network-centric perspective. Later, the findings were further validated through molecular docking analysis. To the best of our understanding, this research marks the inaugural investigation into the therapeutic potential and mechanistic ac tion of native Saudi Arabian flora in the treatment of epilepsy, thereby offering conceptual reinforcement and trajectories for subsequent fundamental research.
2. Materials and methods
2.1. Data mining
2.1.1. Construction of phytochemical database
Information on the constituents of the four indigenous plants - Solanum incanum, Abrus precatorius, Withania somnifera, and Azadirachta indica - were collected from scholarly literature and open-source databases like KNApSAcK (Nakamura et al., 2013) and Indian Medicinal Plants, Phytochemistry And Therapeutics (IMPPAT) (Mohanraj et al., 2018) databases. Specific keywords relating to these plants were employed for searches within the KNApSAcK and IMPPAT databases. In parallel, an extensive review of existing studies was undertaken via online scholarly platforms, including PubMed and Google Scholar.
After retrieval of phytochemicals, these compounds underwent a selection process on the basis of Drug Likeness (DL) and Oral Bioavailability (OB) properties. OB, in pharmacological context, indicates the percentage of an orally ingested drug that becomes accessible in the circulatory system to exhibit its therapeutic influence (Sadaqat et al., 2023). An OB value equal to or>30% is typically utilized as a benchmark in compound screening, as it implies that a notable proportion of the drug administered orally can be effectively absorbed and distributed throughout the circulatory system (Yu et al., 2020). Consequently, a OB value > 0.30 augments the drug-like potential of active constituents, and is thus considered a crucial factor in the progression and optimization of drug development. Furthermore, a qualitative assessment was performed using DL analysis to evaluate the potential of the active constituents to be transformed into oral drugs based on their drug-like properties. We only considered active compounds having OB > 0.30 and DL > 0.18 as potential candidates with drug-like properties. Tools like MolSoft (DOĞAN et al., 2021) and SwissADME (Daina et al., 2017a) were later utilized for screening of active compounds based on their drug-like properties.
2.1.2. ADMET profiling
Though OB and DL are significant aspects in the evaluation of a compound's drug-like potential, they are not exclusive determinants of a compound's suitability for advanced development. Other critical characteristics, including absorption, distribution, metabolism, excretion, and toxicity (ADMET), are also necessitated to be assessed for determining a compound's potential to be used as drug. In relation to this, the SwissADME server (Daina et al., 2017b) and Protox II (Banerjee et al., 2018) tools were employed to examine the ADMET properties of active constituents. Those compounds demonstrating non-active toxicity, superior absorption, and commendable solubility characteristics were selected for further study.
2.1.3. Target prediction
The synergistic effect of compounds from the four indigenous plants was evaluated by scrutinizing interactions derived from two disparate platforms, STITCH (Gfeller et al., 2014) and Swiss Target Prediction (Kuhn et al., 2007) databases, confining the species to “Homo sapiens”. In both databases, those target proteins having 0.7 or above combined score were selected as the statistically significant targets. In the similar vein, identifying disease-associated genes is another essential initial step to investigate the molecular mechanisms of medicinal herbs used in the treatment of various diseases and disorders. A couple of databases, namely GeneCard (Stelzer et al., 2016), Online Mendelian Inheritance in Man (OMIM) (Amberger and Hamosh, 2017), DrugBank (Wishart et al., 2018), PharmGkb (Barbarino et al., 2018), DisGeNET (Piñero et al., 2020), and Therapeutic Target Database (TTD) (Li et al., 2018)were explored with the term 'epilepsy' to forecast genes correlated with the disease. Subsequently, the anticipated targets of the screened active ingredients and the epilepsy-associated targets were compared, and a Venn diagram was constructed to identify the shared targets among plants and epilepsy.
2.2. Compound-target network construction
The shared targets of Saudi Arabian plants and epilepsy were than considered as the putative targets of indigenous plants, having ability to halt the pathogenesis of epilepsy. Subsequently, the Cytoscape software (version 3.8) was used to construct a compound-target interaction network (Shannon et al., 2003) to analyze the interrelationship among these nodes. Within compound-target network, the bioactive constituents and target proteins were represented as the nodes, whereas the interaction among these target proteins and compounds were indicated with black lines (edges). Further, we used NetworkAnalyzer plugin of cytoHubba (Chin et al., 2014) for the identification of those nodes with highest connectivity in the network.
2.3. Functional annotation of overlapped genes
After successfully identifying the intersecting genes, we used Gene Ontology (GO) and pathway enrichment analyses to reveal their inherent cellular components (CC), biological processes (BP), molecular functions (MF), as well as the central signaling pathways. To identify statistically significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and GO terms, we employed the Database for Annotation, Visualization and Integrated Discovery (DAVID) database (Dennis et al., 2003). A rigorous screening process was implemented by setting a threshold of p-value less than 0.05. This ensured that only the most meaningful pathways and terms were selected for further analysis. To visualize the outcomes, we utilized the ggplot2 package in the R programming language. The top GO terms and KEGG pathways were chosen based on their p-value significance and counts, ensuring that only the most relevant and significant findings were presented in the visualization.
2.4. Protein-protein interaction (PPI) network construction
To explore the functional relationships among the identified common genes, we employed the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database, which is known for its comprehensive protein–protein interaction (PPI) information (Von Mering et al., 2005). PPI networks are highly valuable due to their versatility, specificity, and adaptability in uncovering biological interactions. The resulting PPI network was then subjected to network analysis using Cytoscape version 3.8 (Shannon et al., 2003) for retrieval of hub genes, which are key players within the network and have significant functional importance. Hub genes are nodes within PPI network that exhibit extensive connections and interact with multiple other proteins. These genes has a key role to play in the PPI network, and thus contributing significantly to the maintenance of network stability and integrity. In this study, the degree method provided by CytoHubba (Chin et al., 2014) was employed to select the hub genes from pool of overlapped target genes.
2.5. Compound-target-disease network construction
To gain a better understanding of how Saudi plants exert their therapeutic effects on epilepsy, we utilized Cytoscape version 3.8 [19] to construct two separate networks: the compound-target protein network and the target protein-pathways network. These networks were then merged to create a comprehensive compound-target-pathways network. Within this network, nodes represent proteins, targeted compounds, and the pathways in which these proteins are involved while the connections between them are represented by solid lines. By integrating these networks, we can gain useful insights into the synergistic effects of compounds when these native plants are used to treat epilepsy.
2.6. Molecular docking analysis
We performed molecular docking analysis to confirm the binding affinity among compounds and their target proteins. This analysis allows us to identify potential combinations of drugs that could have synergistic effects in treating the disease. In this study, we employed Autodock Vina 1.1.2 in PyRx 0.8 (Dallakyan and Olson, 2015) for the docking analysis. Initially, the 3D structure of the phytocompounds was obtained from the PubChem database for docking studies. Ligands optimization was carried out using an energy minimization parameter Universal Force Field (UFF) with a conjugate gradient optimization algorithm at 2000 steps. Energy minimization was carried out to attain the lowest free energy by open babel in PyRx and converted them into PDBQT formats for molecular docking analysis. Similarly, the 3D crystal structure of TNF accession number 1o8m and TP53 with accession number 1qkt as target proteins retrieved from Protein Data Bank (PDB). They are refined by preparing the protein for docking study, by adding hydrogen atoms, heteroatoms and non-essential water molecules are removed from the target protein structures Discovery Studio (Studio, 2008) and saved as PDB format. These processed protein structures are converted to the PDBQT file by selecting make macromolecule using the PyRx tool. To identify the binding pockets of the target proteins, an online CASTp tool was utilized (Tian et al., 2018). After identification of binding pockets, docking analysis was performed among proteins and active constituents. The best results were assessed based on their binding affinity, with a value below −5.00 kcal/mol indicating a good binding strength and a value below −7.00 kcal/mol indicating a very good affinity. The docked complexes were visualized using Discovery Studio (Studio, 2008), PyMOL (Yuan et al., 2017), and ChimeraX (Goddard et al., 2018) programs. The complete methodology used in current study was also presented in Fig. 1.
Fig. 1.
Graphical synopsis representing the overall methdology used in current study.
3. Results
3.1. Identification of bioactive components
After applying filtering criteria, we identified 6 compounds from Solanum incanum, 9 compounds from Abrus precatorius, 13 compounds from Withania somnifera, and 16 compounds from Azadirachta indica. Duplicate compounds were eliminated from the selection process, resulting in a total of 38 compounds considered as potential components derived from Saudi Arabian plants. These 38 compounds fulfilled specific criteria, including DL ≥ 0.18, OB ≥ 0.30, and a molecular weight below 500 g/mol, ensuring their suitability for further analysis (Table 1).
Table 1.
Selected active compound, their Oral Bioavailiability(OB), Drug-Likeness (DL), Molecular weight (MW), and PubChem IDs.
| Plant Source | Phytochemical Names | Drug Likeness (DL > 0.28) | OBOral (Bioavailability > 0.30) | MW(Molecular Weight > 500) |
|---|---|---|---|---|
| Solanum incanum | Ursolic acid | 0.66 | 0.85 | 456.70 |
| beta-Sitosterol | 0.78 | 0.55 | 414.7 | |
| Lanosterol | 0.55 | 0.55 | 413.64 | |
| Kaempferol | 0.5 | 0.55 | 286.24 | |
| Quercetin | 0.52 | 0.55 | 302.23 | |
| Protocatechuic acid | 0.23 | 0.56 | 154.12 | |
| Abrus precatorius | Abrisapogenol J | 0.74 | 0.55 | 456.78 |
| Precatorine | 0.19 | 0.56 | 289.26 | |
| Sophoradiol | 0.76 | 0.55 | 442.8 | |
| Abrectorin | 0.31 | 0.55 | 314.31 | |
| Isoorientin | 0.76 | 0.55 | 448.41 | |
| Cycloartenol | 0.78 | 0.55 | 426.8 | |
| Amyrin | 0.76 | 0.55 | 426.8 | |
| luteolin | 0.25 | 0.55 | 286.25 | |
| Skrofulein | 0.3 | 0.55 | 314.31 | |
| Withania somnifera | (+)-Catechin | 0.64 | 0.55 | 290.27 |
| 24-Methyldesmosterol | 0.76 | 0.55 | 398.66 | |
| Beta-Sitosterol | 0.78 | 0.55 | 414.7 | |
| Campesterol | 0.59 | 0.55 | 400.7 | |
| Fucosterol | 0.85 | 0.55 | 412.7 | |
| Kaempferol | 0.5 | 0.55 | 286.24 | |
| Oleanolic acid | 0.37 | 0.85 | 456.7 | |
| Quercetin | 0.52 | 0.55 | 302.23 | |
| Stigmasterol | 0.62 | 0.55 | 412.7 | |
| Stigmasterone | 0.5 | 0.55 | 410.7 | |
| Withaferin A | 0.37 | 0.55 | 470.6 | |
| Withanolide J | 0.46 | 0.55 | 470.6 | |
| Withanone | 0.45 | 0.55 | 470.6 | |
| Azadirachta indica | (+)-Catechin | 0.64 | 0.55 | 290.27 |
| (-)-Epicatechin | 0.29 | 0.55 | 290.27 | |
| (+)-Ascorbic acid | 0.29 | 0.56 | 176.12 | |
| Genistein 7-O-glucoside | 0.64 | 0.55 | 432.38 | |
| Ferulic acid | 0.29 | 0.85 | 194.18 | |
| alpha-Copaene | 0.29 | 0.55 | 204.35 | |
| Campesterol | 0.59 | 0.55 | 400.68 | |
| Stigmasterol | 0.62 | 0.55 | 412.69 | |
| Quercetin | 0.52 | 0.55 | 302.24 | |
| 5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone | 0.29 | 0.55 | 422.51 | |
| Licoflavanone | 1.15 | 0.55 | 340.37 | |
| 5,7-Dihydroxy-4′-methoxy-8-C-prenyl-3′-(3-hydroxy-3-methylbutyl)flavanone | 1.06 | 0.55 | 440.53 | |
| Flowerine | 1.33 | 0.55 | 368.42 | |
| alpha-Calacorene | 0.29 | 0.55 | 200.32 | |
| tau-Muurolol | 0.29 | 0.55 | 222.37 | |
| Palustrol | 0.29 | 0.55 | 222.37 |
Through ADMET analysis, 21 compounds namely Beta-Sitosterol, Lanosterol, Kaempferol, Protocatechuic acid, Precatorine, Sophoradiol, Abrectorin, Cycloartenol, Skrofulein, (+)-Catechin, 24-Methyldesmosterol, Campesterol, Fucosterol, Stigmasterol, Stigmasterone, (+)-Ascorbic acid, Genistein 7-O-glucoside, 5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone, Licoflavanone, 5,7-Dihydroxy-4′-methoxy-8-C-prenyl-3′-(3-hydroxy-3-methylbutyl)flavanone, and alpha-Calacorene were found to have non-toxic effect with minimal BBB permeant and high GI absorption (Table 2). ADMET analysis includes the assessment of various factors, such as hepatotoxicity, carcinogenicity, and mutagenicity, to evaluate the potential risks and safety profiles of active compounds. Hepatotoxicity refers to the capacity of compounds to induce liver damage, leading to impaired liver function or failure. In our study, it is important to highlight that all selected compounds demonstrated inactive hepatotoxicity, indicating their low potential for causing liver-related adverse effects. Additionally, our findings revealed that all selected compounds exhibited inactive carcinogenicity and mutagenicity, suggesting their favorable safety profiles in terms of these parameters. In conclusion, these results further support the findings of our study, emphasizing that Saudi Arabian plants possess active compounds with drug-like properties. These compounds hold significant potential in disease prevention and treatment, while exhibiting low risks of hepatotoxicity, mutagenicity, and carcinogenicity.
Table 2.
ADMET profiling of active compounds.
| Plant Source | Active compounds | GI absorption | BBB permeant | P-gp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | Log Kp (skin permeation) | Hepatotoxicity | Carcinogenicity | Mutagenicity | Cytotoxicity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Solanum incanum | Beta-Sitosterol | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −2.2 | ✗ | ✗ | ✗ | ✗ |
| Lanosterol | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −2.58 | ✗ | ✗ | ✗ | ✗ | |
| Kaempferol | High | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | −7.05 | ✗ | ✗ | ✗ | ✗ | |
| Protocatechuic acid | High | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ | −6.42 | ✗ | ✗ | ✗ | ✗ | |
| Abrus precatorius | Precatorine | High | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | −7.21 | ✗ | ✗ | ✗ | ✗ |
| Sophoradiol | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −3.59 | ✗ | ✗ | ✗ | ✗ | |
| Abrectorin | High | ✗ | ✗ | ✓ | ✗ | ✓ | ✓ | ✓ | −6.17 | ✗ | ✗ | ✗ | ✗ | |
| Cycloartenol | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −1.96 | ✗ | ✗ | ✗ | ✗ | |
| Skrofulein | High | ✗ | ✗ | ✓ | ✗ | ✓ | ✓ | ✓ | −5.86 | ✗ | ✗ | ✗ | ✗ | |
| Withania somnifera | (+)-Catechin | High | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | −7.82 | ✗ | ✗ | ✗ | ✗ |
| 24-Methyldesmosterol | Low | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ | −2.55 | ✗ | ✗ | ✗ | ✗ | |
| Beta-Sitosterol | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −2.2 | ✗ | ✗ | ✗ | ✗ | |
| Campesterol | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −2.5 | ✗ | ✗ | ✗ | ✗ | |
| Fucosterol | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −2.53 | ✗ | ✗ | ✗ | ✗ | |
| Kaempferol | High | ✗ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | −7.05 | ✗ | ✗ | ✗ | ✗ | |
| Stigmasterol | Low | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ | −2.74 | ✗ | ✗ | ✗ | ✗ | |
| Stigmasterone | Low | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ | −2.98 | ✗ | ✗ | ✗ | ✗ | |
| Azadirachta indica | (+)-Catechin | High | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | −7.82 | ✗ | ✗ | ✗ | ✗ |
| (+)-Ascorbic acid | High | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −8.54 | ✗ | ✗ | ✗ | ✗ | |
| Genistein 7-O-glucoside | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −8.33 | ✗ | ✗ | ✗ | ✗ | |
| Campesterol | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | −2.5 | ✗ | ✗ | ✗ | ✗ | |
| Stigmasterol | Low | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ | −2.74 | ✗ | ✗ | ✗ | ✗ | |
| 5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone | High | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✓ | −4.21 | ✗ | ✗ | ✗ | ✗ | |
| Licoflavanone | High | ✗ | ✗ | ✓ | ✗ | ✓ | ✓ | ✓ | −5.22 | ✗ | ✗ | ✗ | ✗ | |
| 5,7-Dihydroxy-4′-methoxy-8-C-prenyl-3′-(3-hydroxy-3-methylbutyl)flavanone | High | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ | −5.3 | ✗ | ✗ | ✗ | ✗ | |
| alpha-Calacorene | Low | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | −4.39 | ✗ | ✗ | ✗ | ✗ |
3.2. Compound-target network construction
In our study, we utilized the Swiss Target Prediction database to retrieve 907 potential target genes associated with 21 active constituents. To further investigate the potential targets of these compounds, we extracted a total of 8347 genes linked to epilepsy from databases such as GeneCards and OMIM. Next, we employed a Venn diagram analysis to identify the common targets shared between the target genes of the compounds and the epilepsy-related genes. This analysis yielded 368 potential anti-epileptic genes specific to the indigenous plants under investigation. These genes were subsequently considered as key targets, highlighting their significance in the context of epilepsy and the potential therapeutic effects of the compounds derived from these plants.
To analyze the interaction among compounds and their respective targets, we imported the compound-target data into Cytoscape. This allowed us to create a network representation where the hub compounds were highlighted. The analysis of the compound-target network unveiled the potential synergistic effects of the predicted targets, indicating that the utilization of these medicinal plants as anti-epileptic agents may result in a combined and enhanced therapeutic impact. This visualization provides valuable insights into the interactions among compounds and their targets, supporting the potential efficacy of these plants in combating epilepsy.
3.3. GO and KEGG enrichment analysis
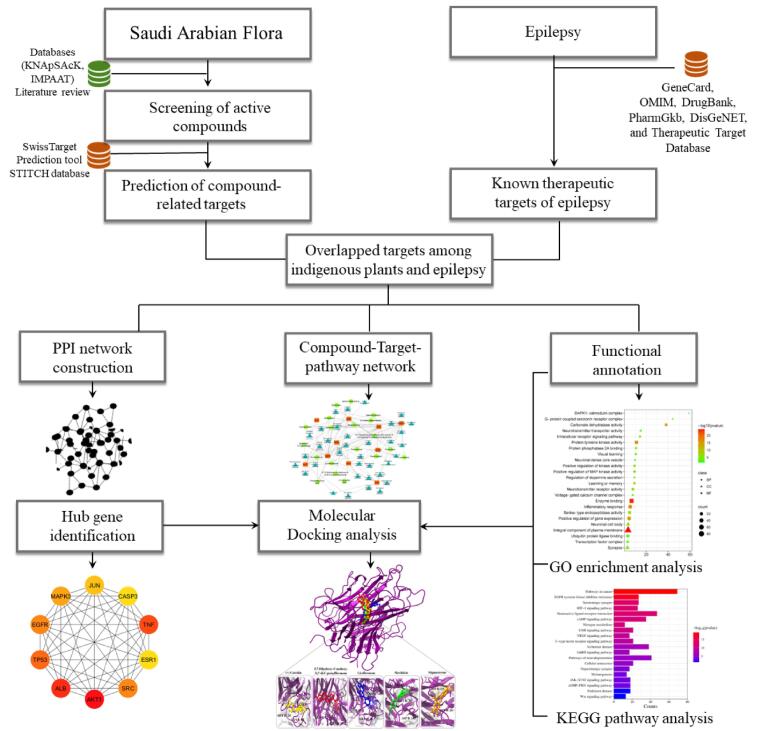
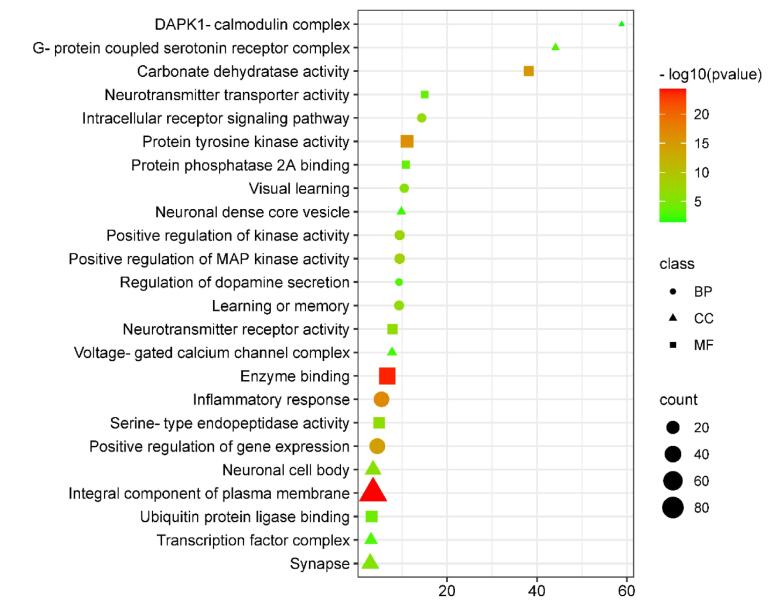
For the identification of the biological characteristics of the overlapped proteins, we performed functional annotation using the DAVID. Through this analysis, we obtained a total of 749 BP terms, 109 CC terms, and 201 MF terms. These terms were found to meet the significance criterion of a p-value less than 0.05. The GO and pathway enrichment analysis allowed us to gain insights into the functional roles and molecular functions associated with the overlapped proteins. The top GO terms indicated the overlapped gene mainly involved in inflammatory response, positive regulation of MAP kinase activity, learning or memory, positive regulation of kinase activity, intracellular receptor signaling pathway, visual learning, and regulation of dopamine secretion, protein tyrosine kinase activity, enzyme binding, carbonate dehydratase activity, neurotransmitter receptor activity, serine-type endopeptidase activity, ubiquitin protein ligase binding, and neurotransmitter transporter activity (Fig. 2). While a total of 170 KEGG pathways were obtained which demonstrated that the overlapped genes were mainly enriched in EGFR tyrosine kinase inhibitor resistance, serotonergic synapse, HIF-1 signaling pathway, neuroactive ligand-receptor interaction, cAMP signaling pathway, dopaminergic synapse, JAK-STAT signaling pathway, cGMP-PKG signaling pathway,Alzheimer disease,GnRH signaling pathway, Pathways of neurodegeneration, Cellular senescence, Parkinson disease, and Wnt signaling pathway (Fig. 3).
Fig. 2.
Gene Ontology (GO) enrichment analysis. The square represented molecular functions (MF), triangle represents cellular components (CC), while circle indicates the biological process (BP). Meanwhile, size and color of circle, square, and triangle within plot represents the count represents the count and p-value.
Fig. 3.
Bar plot representing the KEGG pathways in which the overlapped genes are mainly enriched.
3.4. Identification of hub genes
To construct the PPI network of the 368 targets, we utilized the STRING database. The resulting network consists of 4511 edges representing interaction among 349 nodes. Among these nodes, we identified the top 10 nodes based on their degree of connectivity as the hub genes. The hub genes, ranked by their degree of connectivity, are AKT1 (158 connections), ALB (149 connections), TNF (134 connections), TP53 (132 connections), EGFR (122 connections), SRC (122 connections), MAPK3 (114 connections), JUN (113 connections), CASP3 (110 connections), and ESR1 (110 connections) (Fig. 4). These hub genes are highly interconnected within the PPI network, suggesting their crucial roles in mediating the anti-epileptic effects. Their central positions in the network indicate their potential significance in modulating the biological processes associated with epilepsy treatment.
Fig. 4.
(A) The Venn plot exhibits the overlapping genes (368 anti-epileptic genes) between the plants and the disease (B) The compound-target network displays the interactions between compounds and targets, with node size indicating their connectivity (C) The top 10 genes are ranked using the degree algorithm, highlighting their significance in the network (D) A bar plot represents the degree of each hub gene, providing additional insights into their importance.
3.5. Compound-target-disease network
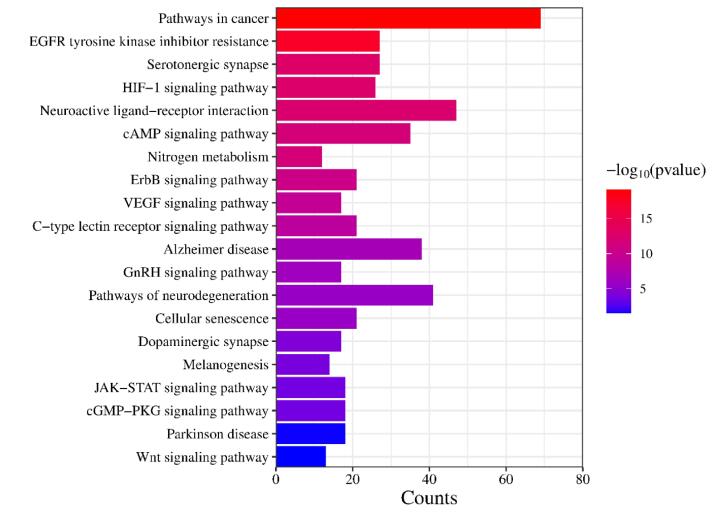
In order to develop a thorough understanding of how indigenous plants influence epilepsy, we generated an integrated network that encompasses compounds, targets, and the disease itself. This comprehensive “compound-target-disease” network assists in exploring the interconnected relationships between these elements and gain insights into the underlying mechanisms of the plants on epilepsy. This network was constructed based on the information obtained from GO and KEGG pathway analyses (Fig. 5).
Fig. 5.
Compound-Target-Disease network. The size of nodes indicates their degree of connectivity. The circle represents hub proteins, the square represents targeted pathways, while arrows represent the active compounds.
After network analysis, TNF and TP53 were found to be targeted by most of the active constituents as well as linked with various disease-relevant pathways. Additionally, KEGG pathway analysis highlighted the involvement of TNF and TP53 in various pathways, such as the cAMP signaling pathway, neurotrophin signaling pathway, Ras signaling pathway, calcium signaling pathway, HIF-1 signaling pathway, and TNF signaling pathway, among others. These findings offer valuable insights into the potential molecular mechanisms that underlie the impact of indigenous plants on epilepsy. By shedding light on these mechanisms, we gain a deeper understanding of how these plants may exert their therapeutic effects and contribute to the management of epilepsy. The identification of key proteins and pathways strengthens our understanding of the therapeutic effects of these plants in epilepsy treatment.
3.6. Molecular docking analysis
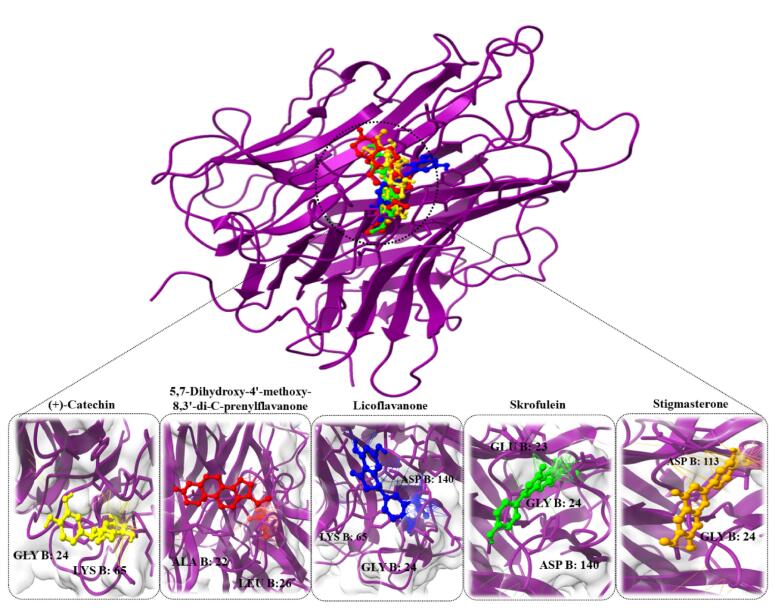
Molecular docking was performed to assess the binding affinity, stability, and free energy between the selected 21 compounds and TNF/TP53. Docking analysis enables us to predict the interaction between the compounds and the active site of the target proteins. In docking protocols, estimating the binding free energy is a key objective. These factors collectively provide insights into the strength and stability of the compound-protein interactions. Following the docking analysis, the top compounds exhibiting the highest binding energy (less than −5.00 kcal/mol) were selected for further analysis. These compounds are of particular interest as they are predicted to have strong and favorable interactions with TNF and TP53, suggesting their potential efficacy in modulating the activity of these proteins. Among the active compounds, licoflavanone (-12.4 kcal/mol), skrofulein (-11.2 kcal/mol), (+)-catechin (-10.2 kcal/mol), stigmasterone (-9.45 kcal/mol), and 5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone (-9.42 kcal/mol) exhibited the highest binding affinity with the TNF protein (Fig. 6). These compounds demonstrated strong interactions with the TNF protein, suggesting their potential as effective modulators. In the case of the TNF- licoflavanone complex, the binding affinity was attributed to hydrogen bonding interactions with specific residues. licoflavanone formed hydrogen bonds with GLY B: 24, LYS B: 65, ASP B: 140 residues, indicating their involvement in stabilizing the complex. Regarding skrofulein, it formed hydrogen bonds with GLY B: 24, GLU B: 23, ASP B: 140 residues of the TP53 protein. (+)-catechin exhibited hydrogen bond interactions with GLY B: 24, LYS B: 65 residues of TP53 protein. stigmasterone showed hydrogen bonding interactions with GLY B: 24, ASP B: 143 residues, while l5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone demonstrated hydrogen bonding interactions with LEU B:26, ALA B: 22 residues of TP53 protein. These findings indicate that these compounds have favorable binding affinities and specific hydrogen bond interactions with key residues of the TP53 protein. This information contributes to our understanding of the potential mechanisms of action and therapeutic relevance of these compounds in modulating TP53 protein activity (Table 3).
Fig. 6.
The docked complexes of TNF protein along with their strongest binding compounds. (A) (+)-Catechin, (B) 5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone, (C) Licoflavanone, (D) Skrofulein, (E) Stigmasterone.
Table 3.
Binding energy and interactions of active compounds with TNF protein.
| Protein-ligand complex | Binding Affinity (kcal/mol) | RMSD | Interacting residues |
|---|---|---|---|
| TNF_Licoflavanone | −12.4931 | 1.738838 | GLY B: 24, LYS B: 65, ASP B: 140 |
| TNF_Skrofulein | −11.2147 | 1.383699 | GLY B: 24, GLU B: 23, ASP B: 140 |
| TNF_(+)-Catechin | −10.2839 | 2.247661 | GLY B: 24, LYS B: 65 |
| TNF_Stigmasterone | −9.45824 | 2.158878 | GLY B: 24, ASP B: 143 |
| TNF_5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone | −9.4248 | 1.283021 | LEU B:26, ALA B: 22 |
Among the active compounds, abrectorin (-10.7 kcal/mol), genistin (-10.5 kcal/mol), (+)-catechin (-9.6 kcal/mol), precatorine (-9.56 kcal/mol), (+)-ascorbic acid (-8.18 kcal/mol) exhibited the highest binding affinity with the TP53 protein (Fig. 7). These compounds demonstrated strong interactions with TP53, indicating their potential as effective modulators. In the case of the TP53-abrectorin complex, the binding affinity was attributed to hydrogen bonding interactions with specific residues. Abrectorin formed hydrogen bonds with LYS A:249, LEU A:213, VAL A: 252 residues of TP53 protein, indicating their involvement in stabilizing the complex. For genistin, hydrogen bond interactions were observed with LYS A:249, ASP A:212, VAL A: 252, PRO A:250, GLN A: 253 residues of TP53 protein. (+)-Catechin demonstrated hydrogen bonding interactions with Ile LYS A:249, LEU A:213, VAL A: 252 residues of TP53 protein. Precatorine showed hydrogen bonding interactions with LYS A: 214 residue, while (+)-ascorbic acid exhibited hydrogen bonding interactions with LYS A: 214, VAL A: 252 residues of TP53 protein. These findings indicate that these compounds have strong binding affinities and specific hydrogen bond interactions with key residues of the TP53 protein. This information contributes to our understanding of the potential mechanisms of action and therapeutic relevance of these compounds in modulating TP53 protein activity (Table 4).
Fig. 7.
The docked complexes of TP53 protein along with their strongest binding compounds. (A) Abrectorin, (B) Genistin, (C) (+)-Catechin, (D) Precatorine, (E) (+)-Ascorbic Acid.
Table 4.
Binding energy and interactions of active compounds with TP53 protein.
| Protein-ligand complex | Binding Affinity (kcal/mol) | RMSD | Interacting residues |
|---|---|---|---|
| TP53_Abrectorin | −10.7851 | 1.641525 | LYS A:249, LEU A:213, VAL A: 252 |
| TP53_Genistin | −10.5223 | 2.115034 | LYS A:249, ASP A:212, VAL A: 252, PRO A:250, GLN A: 253 |
| TP53_(+)-Catechin | −9.68152 | 2.509125 | LYS A:249, LEU A:213, VAL A: 252 |
| TP53_Precatorine | −9.56072 | 4.877544 | LYS A: 214 |
| TP53_(+)-Ascorbic Acid | −8.18279 | 0.855859 | LYS A: 214, VAL A: 252 |
4. Discussion
The prevalence of epilepsy in Saudi Arabia is relatively high compared to other countries (Taha and Hussein, 2014). A recent study has provided compelling evidence indicating a considerably higher prevalence of epilepsy in Saudi Arabia compared to Iran and Egypt (Benamer and Grosset, 2009). These findings highlight the distinct epidemiological characteristics of epilepsy in Saudi Arabia and emphasize the need for further research and targeted interventions in the region to address this significant public health concern. To combat this significant health concern, the government has implemented various initiatives aimed at managing and mitigating the impact of epilepsy. These initiatives encompass improved diagnostic programs and easy availability of antiepileptic drugs. However, continuous efforts are still necessary to further advance the diagnosis, treatment, and public awareness surrounding epilepsy in Saudi Arabia. It is crucial to educate the public about the condition, its causes, symptoms, and available treatment options to dispel stigmas and misconceptions associated with epilepsy. By increasing awareness and understanding, individuals with epilepsy can receive better support and enhance their quality of life. In summary, this prevalence rate underscores the ongoing need for comprehensive approaches to address this health issue. By focusing on diagnosis, treatment, and public education, significant progress can be made in managing epilepsy and improving the well-being of affected individuals in the country.
This study employed a network pharmacology approach to gain a comprehensive understanding of the molecular interactions underlying epilepsy. Initially, the anti-epileptic properties of four plants, namely Solanum incanum, Abrus precatorius, Withania somnifera, and Azadirachta indica, were investigated based on their drug-like properties. The native plants of Saudi Arabia possess significant medicinal potential and offer promising benefits to individuals' well-being. To investigate their therapeutic applications, disease-related data on epilepsy were collected from public repositories. The overlapping genes obtained from comparing the plants with epilepsy data were then subjected to a network pharmacology approach, which allowed for the analysis of the multi-target effects of the selected active constituents against epilepsy. Additionally, pathway enrichment analysis revealed that the overlapping genes primarily participate in various signaling pathways, including the cAMP signaling pathway, neurotrophin signaling pathway, MAPK signaling pathway, PI3K-Akt signaling pathway, among others. These findings provide valuable insights into the potential mechanisms underlying the anti-epileptic effects of the identified compounds.
In the context of epilepsy, the role of cyclic adenosine monophosphate (cAMP) and its downstream effectors, such as protein kinase A (PKA), is significant. They can influence the excitability of neurons by modulating various processes, including ion channel function, neurotransmitter release, and gene transcription. For example, alterations in cAMP levels have been observed to impact the activity of specific potassium channels, which has key role to play in regulating neuronal excitability (Wasterlain and Csiszar, 1980). Dysregulation of cAMP signaling could therefore contribute to epileptogenesis by promoting abnormal neuronal activity. Neurotrophins, including nerve growth factor (NGF) and brain-derived neurotrophic factor (BDNF) are key players in the development, functioning, and survival of neurons. Increased levels of BDNF are often observed in the brain following seizures, and BDNF has been implicated in the process of epileptogenesis, which refers to the development of epilepsy, in various animal models. Understanding the role of neurotrophins, particularly BDNF, in epileptogenesis and neuronal excitability offers valuable insights into the underlying mechanisms of epilepsy and may pave the way for novel therapeutic approaches (Lin et al., 2020).
Further, dysregulation of PI3K-Akt signaling has been implicated in various neurological disorders, including epilepsy. For instance, mutations in genes that encode components of the PI3K-Akt pathway have been identified in some forms of genetic epilepsy. Furthermore, experimental evidence suggests that activation of the PI3K-Akt pathway could have neuroprotective effects in epilepsy, potentially by promoting neuron survival and inhibiting neuronal death (Xiao et al., 2017). Thus, understanding the intricate interplay between these interconnected pathways and their contributions to epilepsy pathogenesis is crucial for identifying potential targets for therapeutic interventions.
After functional annotation and network analysis, TNF and TP53 were selected as the main target proteins for epilepsy. In the context of epilepsy, studies have shown that TNF levels are elevated during seizures and may contribute to increased neuron excitability (Vezzani et al., 2008, Rana and Musto, 2018). Anti-TNF therapies are being studied as potential treatments for epilepsy. These work by blocking the action of TNF, therefore potentially reducing inflammation and neuronal excitability, and preventing seizure generation.TP53 Also known as p53, this protein plays a crucial role in controlling cell division and apoptosis. When DNA damage occurs, p53 halts the cell cycle to allow for repair or triggers programmed cell death if the damage is too severe. Mutations that disrupt p53 function can contribute to uncontrolled cell proliferation, leading to cancer. In the brain, p53 has been implicated in neurodegenerative diseases and acute neurological disorders (Huang et al., 2015, Teocchi and D’Souza-Li, 2016). Its role in epilepsy is less clear but it's believed that p53 might contribute to seizure-induced neuronal death. Therefore, therapies that modulate p53 activity could potentially protect neurons from damage following seizures. However, because p53 is also involved in suppressing tumor formation, such therapies would need to be carefully controlled to avoid promoting cancerous growth. In conclusion, targeting TNF and p53 could provide a novel therapeutic strategy for epilepsy. However, these proteins are involved in numerous biological processes and their manipulation could have widespread effects. Therefore, a detailed understanding of their roles in epilepsy and other physiological functions is required to exploit their potential as therapeutic targets effectively.
More importantly, various phytochemical compounds present in different plant species have been explored in current study for their potential anti-epileptic properties. Specifically, compounds such as abrectorin, genistin, (+)-catechin, precatorine, (+)-ascorbic acid, licoflavanone, skrofulein, stigmasterol, and 5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone have demonstrated promising effects. Flavonoids like abrectorin, genistin, (+)-catechin, and precatorine are well-documented for their neuroprotective effects, acting through a myriad of mechanisms including modulation of GABAergic and glutamatergic neurotransmission, attenuation of oxidative stress, and suppression of inflammation (Zhang et al., 2012). For instance, genistin has been shown to modulate neurotransmission and potentially suppress epileptic seizures (Rebas et al., 2020, Huang et al., 2021).
As an antioxidant, (+)-ascorbic acid, commonly known as Vitamin C, could play a pivotal role in preventing neuronal damage in epilepsy. In fact, Covarrubias-Pinto et al. (Covarrubias-Pinto et al., 2015) suggested that its antioxidant properties could mitigate the oxidative stress often associated with epileptic seizures. In the similar vein, Stigmasterol, a plant sterol, has been investigated for its potent anti-inflammatory properties. In a study by Kumar et al.(Kumar and Pandey, 2013), stigmasterol showed potential for alleviating neuroinflammatory conditions, which are often associated with epilepsy.
As for licoflavanone, skrofulein, and 5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone, the current body of literature is limited regarding their specific role in epilepsy. However, as members of the flavonoid and prenylflavonoid families, they could potentially share the neuroprotective and anti-epileptic effects generally associated with these classes of compounds. It is pertinent to underscore that while these findings are compelling, they are primarily derived from in vitro and animal studies. Rigorous clinical trials are necessary to translate these promising preclinical findings into viable therapeutic options for humans suffering from epilepsy.
In conclusion, our study provides a solid foundation for investigating the multi-target effects of indigenous plants from Saudi Arabia as potential treatment options for epilepsy. The combination of bioinformatics and network approaches allows for the identification of key molecular pathways and interactions involved in epilepsy, leading to the discovery of potential drug targets for intervention. However, it is important to acknowledge the limitations of our study. Firstly, further experimental validation through in vivo and in vitro studies is necessary to confirm the efficacy and safety of the identified compounds and their interactions with the target proteins. These studies will provide more concrete evidence for the therapeutic potential of the indigenous plants. Secondly, expanding the database of traditional medicines and target genes would enhance the accuracy of network pharmacology analysis results. A broader and more comprehensive dataset would provide a more robust foundation for the identification of potential targets and pathways involved in epilepsy. In summary, while our study provides valuable insights into the multi-target effects of indigenous plants for epilepsy treatment, further research is needed to validate our findings, expand the dataset, and gain a deeper understanding of the precise therapeutic mechanisms involved. These efforts will contribute to the development of effective and targeted therapeutic strategies for epilepsy.
5. Conclusion
The burden of epilepsy in Saudi Arabia emphasizes the critical need for effective strategies in preventing, diagnosing, and managing this neurological disorder and its associated complications. The high prevalence of epilepsy in the country underscores the urgency to develop new and improved treatments for epilepsy. In this context, our study proposed a novel scientific methodology for evaluating the multi-component, multi-target effects of active compounds derived from local plants, particularly those found in Saudi Arabia. Our study employed an integrated approach combining network pharmacology and bioinformatics techniques to identify and evaluate the potential therapeutic effects of active compounds. Among the identified compounds, abrectorin, genistin, (+)-catechin, precatorine, (+)-ascorbic acid, licoflavanone, skrofulein, stigmasterone, 5,7-Dihydroxy-4′-methoxy-8,3′-di-C-prenylflavanone were highlighted as potential candidates for treating epilepsy. Furthermore, our findings shed light on the potential therapeutic targets for reducing seizure activity and neuronal cell death associated with epilepsy, namely TNF and TP53. These targets offer promising avenues for intervention and could contribute to the development of more effective treatments for epilepsy. In summary, our study has enriched our understanding of the chemical composition of indigenous plants in Saudi Arabia and the synergistic mechanisms by which their active compounds may exert therapeutic effects against epilepsy. By integrating network pharmacology and bioinformatics approaches, we have identified potential compounds and therapeutic targets, providing valuable insights for further research and the development of improved epilepsy treatments.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The author would like to thank the Deanship of Scientific Research at Shaqra University for supporting this work.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Faez Falah Alshehri, Email: Falshehri@su.edu.sa.
Fuad M Alzahrani, Email: Fuadmubarak@tu.edu.sa.
Abdulaziz Alkhoshaiban, Email: khshiebana@qu.edu.sa.
Zafer Saad Al Shehri, Email: zaf@su.edu.sa.
References
- Amberger J.S., Hamosh A. Searching online mendelian inheritance in man (OMIM): a knowledgebase of human genes and genetic phenotypes. Curr. Protoc. Bioinformatics. 2017;58:1.2. 1–1.2. 12. doi: 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anju T., Smijin S., Jobin M., Paulose C. Altered muscarinic receptor expression in the cerebral cortex of epileptic rats: Restorative role of Withania somnifera. Biochem. Cell Biol. 2018;96:433–440. doi: 10.1139/bcb-2017-0198. [DOI] [PubMed] [Google Scholar]
- Attal A.R., Otari K.V., Shete R.V., Upasani C., Nandgude T. Abrus precatorius Linnaeus: a phytopharmacological review. J. Pharm. Res. 2010;3:2585–2587. [Google Scholar]
- Banerjee, P., Eckert, A. O., Schrey, A. K. & Preissner, R. J. N. a. r. 2018. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res, 46, W257-W263. [DOI] [PMC free article] [PubMed]
- Barbarino J.M., Whirl-Carrillo M., Altman R.B., Klein T.E. PharmGKB: a worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018;10:e1417. doi: 10.1002/wsbm.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghi E. The epidemiology of epilepsy. Neuroepidemiology. 2020;54:185–191. doi: 10.1159/000503831. [DOI] [PubMed] [Google Scholar]
- Benamer H.T., Grosset D.G. A systematic review of the epidemiology of epilepsy in Arab countries. Epilepsia. 2009;50:2301–2304. doi: 10.1111/j.1528-1167.2009.02058.x. [DOI] [PubMed] [Google Scholar]
- Birhan Y.S. Medicinal plants utilized in the management of epilepsy in Ethiopia: ethnobotany, pharmacology and phytochemistry. Chinese Med. 2022;17:1–37. doi: 10.1186/s13020-022-00686-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camfield P., Camfield C. Febrile seizures and genetic epilepsy with febrile seizures plus (GEFS+) Epileptic Disord. 2015;17:124–133. doi: 10.1684/epd.2015.0737. [DOI] [PubMed] [Google Scholar]
- Chandran U., Mehendale N., Patil S., Chaguturu R., Patwardhan B. Network pharmacology. Innov. Approaches Drug Discovery. 2017;127 [Google Scholar]
- Chin C.-H., Chen S.-H., Wu H.-H., Ho C.-W., Ko M.-T., Lin C.-Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8:1–7. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias-Pinto A., Acuña A.I., Beltrán F.A., Torres-Díaz L., Castro M.A. Old things new view: ascorbic acid protects the brain in neurodegenerative disorders. Int. J. Mol. Sci. 2015;16:28194–28217. doi: 10.3390/ijms161226095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. AJ.2015.;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R. DAVID: database for annotation, visualization, and integrated discovery. Nucleic Acids Res. 2003;4:1–11. [PubMed] [Google Scholar]
- DOĞAN, A., ÖZDEMİR, S., Yalcin, M., Sari, H. & Nural, Y. 2021. Naphthoquinone-thiazole hybrids bearing adamantane: Synthesis, antimicrobial, DNA cleavage, antioxidant activity, acid dissociation constant, and drug-likeness. J Pharm Res., 25.
- Gfeller, D., Grosdidier, A., Wirth, M., Daina, A., Michielin, O. & Zoete, V. J. N. a. r. 2014. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res., 42, W32-W38. [DOI] [PMC free article] [PubMed]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, A. L. J. N. b. 2007. Network pharmacology. Nat Biotechnol., 25, 1110-1111. [DOI] [PubMed]
- Huang J., He Y., Chen M., Du J., Li G., Li S., Liu W., Long X. Adenosine deaminase and adenosine kinase expression in human glioma and their correlation with glioma-associated epilepsy. Mol. Med. Rep. 2015;12:6509–6516. doi: 10.3892/mmr.2015.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Ke Y., Zhu J., Liu S., Cong J., Ye H., Guo Y., Wang K., Zhang Z., Meng W. TRESK channel contributes to depolarization-induced shunting inhibition and modulates epileptic seizures. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109404. [DOI] [PubMed] [Google Scholar]
- Kuhn M., von Mering C., Campillos M., Jensen L.J., Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2007;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. & Pandey, A. K. 2013. Chemistry and biological activities of flavonoids: an overview. Sci. World J., 2013. [DOI] [PMC free article] [PubMed]
- Li Y.H., Yu C.Y., Li X.X., Zhang P., Tang J., Yang Q., Fu T., Zhang X., Cui X., Tu G. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018;46:D1121–D1127. doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.W., Harward S.C., Huang Y.Z., McNamara J.O. Targeting BDNF/TrkB pathways for preventing or suppressing epilepsy. Neuropharmacology. 2020;167 doi: 10.1016/j.neuropharm.2019.107734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löscher W., Potschka H., Sisodiya S.M., Vezzani A. Drug resistance in epilepsy: clinical impact, potential mechanisms, and new innovative treatment options. Pharmacol. Rev. 2020;72:606–638. doi: 10.1124/pr.120.019539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraj K., Karthikeyan B.S., Vivek-Ananth R., Chand R., Aparna S., Mangalapandi P., Samal A. IMPPAT: a curated database of Indian medicinal plants, phytochemistry and therapeutics. Sci. Rep. 2018;8:1–17. doi: 10.1038/s41598-018-22631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Shimura N., Otabe Y., Hirai-Morita A., Nakamura Y., Ono N., Ul-Amin M.A., Kanaya S.J.P., Physiology C. KNApSAcK-3D: a three-dimensional structure database of plant metabolites. Plant Cell Physiol. 2013;54:e4. doi: 10.1093/pcp/pcs186. [DOI] [PubMed] [Google Scholar]
- Noor, F., Tahir ul Qamar, M., Ashfaq, U. A., Albutti, A., Alwashmi, A. S. & Aljasir, M. A. 2022. Network pharmacology approach for medicinal plants: review and assessment. Pharmaceuticals, 15, 572. [DOI] [PMC free article] [PubMed]
- Noor, F., Asif, M., Ashfaq, U. A., Qasim, M. & Tahir ul Qamar, M. 2023. Machine learning for synergistic network pharmacology: a comprehensive overview. Brief. Bioinformatics., bbad120. [DOI] [PubMed]
- Noor F., Rehman A., Ashfaq U.A., Saleem M.H., Okla M.K., Al-Hashimi A., AbdElgawad H., Aslam S. Integrating network pharmacology and molecular docking approaches to decipher the multi-target pharmacological mechanism of Abrus precatorius L. acting on diabetes. Pharmaceuticals. 2022;15:414. doi: 10.3390/ph15040414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.A., Mossa J.S., Al-Said M.S., Al-Yahya M.A. Medicinal plant diversity in the flora of Saudi Arabia 1: a report on seven plant families. Fitoterapia. 2004;75:149–161. doi: 10.1016/j.fitote.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Rana A., Musto A.E. The role of inflammation in the development of epilepsy. J. Neuroinflamm. 2018;15:1–12. doi: 10.1186/s12974-018-1192-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebas E., Rzajew J., Radzik T., Zylinska L. Neuroprotective polyphenols: a modulatory action on neurotransmitter pathways. Curr. Neuropharmacol. 2020;18:431–445. doi: 10.2174/1570159X18666200106155127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaqat, M., Qasim, M., ul Qamar, M. T., Masoud, M. S., Ashfaq, U. A., Noor, F., Fatima, K., Allemailem, K. S., Alrumaihi, F. & Almatroudi, A. 2023. Advanced network pharmacology study reveals multi-pathway and multi-gene regulatory molecular mechanism of Bacopa monnieri in liver cancer based on data mining, molecular modeling, and microarray data analysis. Comput. Biol. Med., 161, 107059. [DOI] [PubMed]
- Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B. & Ideker, T. J. G. r. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. , Genome Res., 13, 2498-2504. [DOI] [PMC free article] [PubMed]
- Sher H., Al-Yemeni M., Masrahi Y.S., Shah A.H. Ethnomedicinal and ethnoecological evaluation of Salvadora persica L.: a threatened medicinal plant in Arabian Peninsula. J. Med. Plants Res. 2010;4:1209–1215. [Google Scholar]
- Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016;54:1.30. 1–1.30. 33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- Studio, D. J. A. 2008. Discovery studio.
- Taha G.R., Hussein H. Autism spectrum disorders in developing countries: Lessons from the Arab world. Compreh. Guide Autism. 2014:2509–2531. [Google Scholar]
- Teocchi, M. A. & D’Souza-Li, L. 2016. Apoptosis through death receptors in temporal lobe epilepsy-associated hippocampal sclerosis. Mediators Inflamm., 2016. [DOI] [PMC free article] [PubMed]
- Thijs R.D., Surges R., O'Brien T.J., Sander J.W. Epilepsy in adults. Lancet. 2019;393:689–701. doi: 10.1016/S0140-6736(18)32596-0. [DOI] [PubMed] [Google Scholar]
- Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzani A., Balosso S., Ravizza T. The role of cytokines in the pathophysiology of epilepsy. Brain Behav. Immun. 2008;22:797–803. doi: 10.1016/j.bbi.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Von Mering, C., Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., Jouffre, N., Huynen, M. A. & Bork, P. J. N. a. r. 2005. STRING: known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res., 33, D433-D437. [DOI] [PMC free article] [PubMed]
- Wasterlain C.G., Csiszar E. Cyclic nucleotide metabolism in mouse brain during seizures induced by bicuculline or dibutyryl cyclic guanosine monophosphate. Exp. Neurol. 1980;70:260–268. doi: 10.1016/0014-4886(80)90025-4. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worku, M. 2022. Antibacterial Activity of Gizawa (Withania somnifera) and Embuay (Solanum incanum) Leaf Extracts and Their Synergistic Effect Against Standard and Clinical Pathogenic Bacterial Isolates.
- Xiao Z., Peng J., Gan N., Arafat A., Yin F. Interleukin-1β plays a pivotal role via the PI3K/Akt/mTOR signaling pathway in the chronicity of mesial temporal lobe epilepsy. Neuroimmunomodulation. 2017;23:332–344. doi: 10.1159/000460254. [DOI] [PubMed] [Google Scholar]
- Yu S., Wang J., Shen H. Network pharmacology-based analysis of the role of traditional Chinese herbal medicines in the treatment of COVID-19. Ann. Palliat. Med. 2020;9:437–446. doi: 10.21037/apm.2020.03.27. [DOI] [PubMed] [Google Scholar]
- Yuan S., Chan H.S., Hu Z. Using PyMOL as a platform for computational drug design. WIRES. 2017;7:e1298. [Google Scholar]
- Zhang Z.-J., Cheang L.C., Wang M.-W., Li G.-H., Chu I.K., Lin Z.-X., Lee S.M. Ethanolic extract of fructus Alpinia oxyphylla protects against 6-hydroxydopamine-induced damage of PC12 cells in vitro and dopaminergic neurons in zebrafish. Cell. Mol. Neurobiol. 2012;32:27–40. doi: 10.1007/s10571-011-9731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]