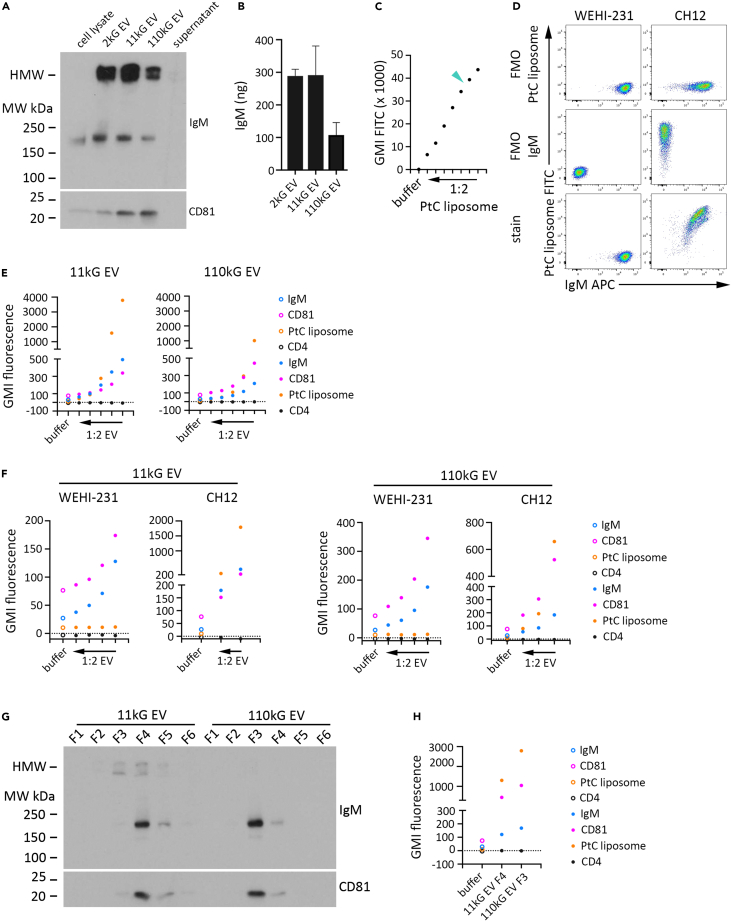
Figure 6.
IgM contained in B cell-derived EV can bind antigen
CH12 cells were cultured in media alone for two days, after which the conditioned media was subjected to differential ultracentrifugation (DUC) to isolate EV. The EV samples (2kG, 11kG, and 110kG) were resuspended in an equivalent volume of buffer for subsequent analysis by immunoblot and ELISA.
(A) An equivalent proportion of each sample was resolved by SDS/PAGE under unreduced conditions, and immunoblotted for IgM and CD81. CH12 cell lysate (lane 1; 7.5 μg) and 110kG supernatant (lane 5), were run as controls.
(B) The amount of IgM in each EV sample under buffer +0.25% TX100 conditions was determined by anti-mouse IgM ELISA. For (A) and (B), data are representative of two independent experiments.
(C) CH12 cells were incubated with PtC liposomes for 30 min on ice at 1:2000 stock and at sequential 1:2 dilutions thereof, after which FITC fluorescence was determined by flow cytometry and GMI determined. The extreme left data point is the GMI FITC for CH12 cells incubated in buffer only. The turquoise arrowhead indicates the reagent concentration (1:5000 stock) utilized for subsequent experiments.
(D) WEHI 231 and CH12 cells were stained with IgM APC and PtC liposome FITC reagent for flow cytometry analysis. Dot plots indicate FMO PtC liposome (first row), FMO IgM APC (second row), and complete stain (third row) fluorescence profiles of single, viable WEHI-231 (left column) and CH12 (right column) cells.
(E) EV samples (11kG and 110kG) were isolated by DUC from CH12 conditioned media, resuspended in an equivalent volume of buffer, sequentially titrated 1:2, incubated with an identical volume of aldehyde sulfate microbeads, and subsequently stained with a fluorescent panel of IgM APC, CD81 BV241, CD4 PE/Cy5 and PtC liposome FITC reagent for flow cytometry analysis. GMI for each fluorophore is shown for the most concentrated EV preparation (extreme right data point) and each titrated sample (progressing left). GMI fluorescence of beads incubated in buffer only is indicated by the extreme left data point. Data are representative of two independent experiments.
(F) EV (11kG and 110kG) were isolated from WEHI-231 and CH12 conditioned media by differential ultracentrifugation and then titrated 1:2, adsorbed onto an equivalent volume of aldehyde sulfate microbeads, and then stained with the reagent panel described above for flow cytometry analysis (11 kG EV, left graphs; 110kG right graphs). GMI for each fluorophore is shown for the most concentrated EV preparation (extreme right data point) and each titrated sample (progressing left). GMI fluorescence of beads incubated in buffer only is indicated by the extreme left data point.
(G) EV (11kG and 110kG) were isolated from CH12 conditioned media by DUC and subjected to sucrose gradient fractionation. Equivalent volumes of resultant fractions were subjected to SDS/PAGE under non-reducing conditions and immunoblotted for IgM and CD81. Data are representative of two independent experiments.
(H) Fraction 4 from 11 kG EV and fraction 3 from 110 kG EV were analyzed by flow cytometry following adsorption to aldehyde sulfate microbeads and staining with the reagent panel described above. GMI fluorescence for each fluorophore is shown. GMI fluorescence of beads incubated in buffer only is indicated by the extreme left data point. Data are represented as mean ± SEM.