Abstract
Of the numerous endogenous retroviral elements that are present in the human genome, the abundant HERV-K family is distinct because several members are transcriptionally active and coding for biologically active proteins. A detailed phylogeny of the HERV-K family based on the partial sequence of the reverse transcriptase (RT) gene revealed a high incidence of an intact RT open reading frame within the HML-2 subgroup of HERV-K elements. In this study, we report the cloning of six full-length HML-2 RT genes, of which five contain an uninterrupted open reading frame. The RT enzymes were expressed as glutathione S-transferase fusion proteins in Escherichia coli, and several HERV-K RT enzymes demonstrated polymerase as well as RNase H activity. Several biochemical properties of the RT polymerase were analyzed, including the template requirements and optimal reaction conditions (temperature, type of divalent cation). Inspection of the nucleotide sequence of the HERV-K RT genes demonstrated a mosaic structure, suggesting that a high level of genetic recombination has occurred in this virus family, which is a hallmark of replication by means of reverse transcription. The selective pressure to maintain the RT coding potential is illustrated by the sequence of a particular HERV-K isolate that contains three 1-nucleotide deletions within a small RT segment, thus maintaining the open reading frame. These combined results may suggest that these endogenous RT enzymes still have a biological function. It is possible that the RT activity was involved in the spread of this major class of retroelements by retrotransposition, and in fact it cannot be excluded that this retrovirus group is still mobile. The endogenous RT activity may also have been involved in the shaping of the human genome, e.g., by formation of pseudogenes.
An intrinsic property of the retroviral replication cycle is the insertion of the reverse-transcribed viral genome into a chromosome of the host cell. Infections of germ line cells by exogenous retroviruses will lead to the stable introduction of new genetic information that is transmitted vertically to the offspring. Such endogenous retroviruses inhabit the genomes of all eukaryotes, indicating that such infections have occurred multiple times. Alternatively, retroviral elements that became endogenous may have remained biologically active, causing intracellular spread by retrotransposition after the initial infection. Over time, multiple infections and/or reiterative rounds of intracellular transposition can lead to significant expansion of these viral elements in the host genome. The human genome contains a variety of ancient endogenous retroviruses (52). Sequence studies imply that most of these elements are of considerable antiquity, as exemplified by their coding sequences, which are usually riddled with mutations, including deletions and in-frame stop codons (5). Nevertheless, the presence of such proviruses might have a variety of effects on the host. Detrimental effects can be caused by expression of viral transcripts or by insertional inactivation of important host genes. On the other hand, endogenous retroviruses may be beneficial for the host, e.g., by providing resistance to infection by related exogenous viruses (3). These mechanisms include using the Env protein of an endogenous virus to block the receptor for an exogenous virus (the mouse Fv4 gene), using the Gag protein to inhibit at the postentry level (the Fv1 gene), and using a virus-encoded protein with superantigen activity to specifically deplete cells that are the target for an exogenous virus (the sag genes) (44). Furthermore, these potentially mobile genetic elements may play a role in the evolution of the host genome by integration, recombination, duplication, and transposition events. In this study, we report the cloning and identification of an enzymatically active reverse transcriptase (RT) enzyme encoded by a human endogenous retrovirus of the HERV-K family.
The human endogenous retroviruses of the so-called HERV-K family (36) come closest to being biologically active. This element is relatively abundant, with at least 59 copies per haploid genome (33, 36, 53, 55). Cross-hybridization studies initially revealed nine subgroups (14). More recently, six subgroups (HML-1 to HML-6) were described with a nucleotide sequence dissimilarity of approximately 25% for a 244-bp fragment of the RT gene (33), and this phylogeny could be confirmed for a larger RT fragment (55). Although the HERV-K element was first introduced into the human germ line more than 25 million years ago, as judged from its distribution in different primates (28, 32), several members contain a complete genome with long terminal repeats and open reading frames encoding Gag, Pol, and Env proteins (25) and three chromosomes are candidates for harboring a completely intact HERV-K locus (30, 31). The idea that this virus group may still be biologically active is further supported by the finding that several HERV-K elements are transcriptionally active (1, 34). Perhaps more important, specific enzymatic activities have been reported for the HERV-K dUTPase, protease, and endonuclease (15, 20, 45), and we now report data for the two remaining enzymatic activities, that of the RT polymerase and its RNase H domain. RT enzyme activity has been found in a variety of HERV-K-containing biological samples. Viral particles containing an active RT enzyme have been detected in normal placenta, platelets from patients with thrombocythemia, culture supernatant of pancreatic cells of diabetic patients, and teratocarcinoma and breast cancer cell lines, and these particles were reported to contain nucleic acid sequences belonging to the HERV-K family (6–8, 40, 46, 47). Furthermore, weak RT activity was reported with an HERV-K genome expressed in a recombinant baculovirus system (51). We set out to clone a functional HERV-K RT enzyme for further biochemical analysis. To do so, we designed PCR primers to amplify the full-length RT gene of members of the HML-2 subgroup, which was previously suggested to contain a significant number of isolates with an intact RT open reading frame based on the sequence of a partial 600-bp RT fragment (53). Furthermore, we observed a strong preference for synonymous substitutions within the HERV-K RT open reading frames (54). In this study, we cloned and sequenced six RT gene fragments of approximately 1,800 bp, and all but one contained a continuous open reading frame for RT protein. The HERV-K RT enzymes were expressed as recombinant protein in Escherichia coli, and we measured both RT and RNase H activity for some of the clones. The biological pressure to maintain the HERV-K coding capacity is further illustrated by the sequence of individual RT clones, in which multiple insertions and deletions were found within short RT segments, apparently to restore the RT reading frame. Inspection of the RT gene sequences revealed that the HERV-K elements have a mosaic genome structure. This result indicates that these elements were formed by a process that is recombination prone, which is fully consistent with amplification by means of reverse transcription. The putative role of the HERV-K RT enzyme in the shaping of the human genome is discussed.
MATERIALS AND METHODS
Cloning and sequencing of full-length HERV-K RT genes.
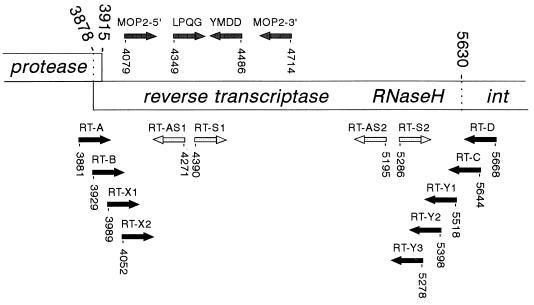
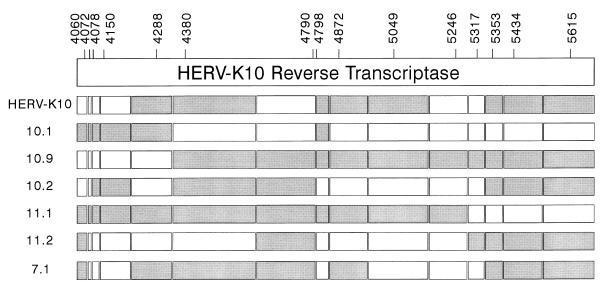
Two primer sets were used to amplify HERV-K RT genes (Fig. 1). These primers were designed on the basis of the prototype HERV-K10 sequence (36), which is a member of the extensive HML-2 subgroup (53). The borders of the RT gene were estimated by sequence alignment of the HERV-K10 sequence with those of RT genes of a variety of exogenous retroviruses (results not shown; see also reference 12). A large (1.8-kb) RT segment was amplified by primers RT-A (TCAGGATCCAAATCAAGAAAGAGAAGG) and RT-D (TCAGCGGCCGCTAAGCATGAAGTTCTTGTGC). The second primer set amplifies a shorter RT segment of approximately 1.7 kb: RT-B (TCAGGATCCGTAGAGCCTCCTAAACCC) and RT-C (TCAGCGGCCGCTAAGCATGAAGTTCTTGTGC). The sense and antisense primers contain a BamHI and NotI restriction enzyme site, respectively (underlined in the sequences). These sites are not present within the HERV-K10 RT gene, and they allow the in-frame fusion of the RT open reading frame with that of the glutathione S-transferase (GST) protein in the pGEX-4T-1 vector (Pharmacia Biotech). The antisense primers provide a UAG stop codon (marked in bold in the antisense sequences).
FIG. 1.
Schematic of part of the HERV-K genome encompassing the RT and RNase H domains. This genome structure was originally proposed by Ono et al. for the prototype HERV-K10 element (36), and all nucleotide numbers refer to positions in this genome. The protease gene is in the +1 reading frame compared with RT and ends at stop codon 3915. Thus, a −1 frameshift is expected to occur in the upstream region to fuse the protease and RT reading frames. The integrase enzyme (int) is in frame with the RT gene. The protease cleavage sites, which, by analogy to exogenous retroviruses, will determine the ends of the mature RT enzyme, are not known. The RT primer sets RT-A plus RT-D and RT-B plus RT-C (black arrows) were used in this study to amplify full-length RT genes, and the RT-X1 plus RT-X2 and RT-Y1 through RT-Y3 primers (black arrows) were used to construct N-terminal and C-terminal deletion variants of the RT enzyme (Fig. 7). The LPQG plus YIDD and MOP2-5′ plus MOP2-3′ primer sets shown above the RT gene (grey arrows) have been used previously to amplify internal RT fragments (24, 33). The sense primers S1 and S2 and the antisense primers AS1 and AS2 (open arrows) were used in this study for sequencing of the complete RT gene.
Cellular RNA was isolated from bone marrow mononuclear cells and converted into cDNA by reverse transcription with random hexamer primers and the avian myeloblastosis virus (AMV) RT enzyme as described previously (53). The bone marrow sample used in this study was from H6, a patient with common acute lymphoblastic leukemia, and was taken for standard diagnostic tests. We suggested previously that there is no gross difference in the number and types of HERV-K elements expressed in normal and leukemic bone marrow (53). The cDNA was subsequently PCR amplified with the RT-A plus RT-D and the RT-B plus RT-C primer sets. The PCR was performed with 100 μl of PCR buffer (20 mM Tris-HCl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2, 0.1 mg of bovine serum albumin per ml) with 0.5 μl of AmpliTaq (5 U/μl; Perkin-Elmer), 200 ng of each DNA primer, 1 μl of deoxynucleoside triphosphate mixture mM (100), and 1 μl of input cDNA for 35 cycles (1 min at 95°C, 1 min at 55°C, 2 min at 72°C). The PCR products were analyzed on Tris-borate-EDTA 1.5% agarose gels and revealed fragments of the appropriate length of approximately 1.7 to 1.8 kb. The remainder of the PCR products were concentrated by ethanol precipitation and subsequently digested with BamHI and NotI. The fragments were purified over an agarose gel, eluted, and ligated into the BamHI-NotI-digested pGEX-4T-1 vector. The ligation mixture was transformed into E. coli DH5α. Positive clones were identified by restriction enzyme digestion and subsequent sequence analysis.
The complete sequence of six HERV-K RT genes was analyzed on both strands with multiple primers on a 373 automated sequencer (ABI) by using the dye terminator cycle sequencing protocol. In addition to the RT-A through RT-D primers, we used several internal sequencing primers. New primers include the sense DNA oligonucleotides RT-S1 (TTGTCAGACTTTTGTAGG) and RT-S2 (GTTCCAGCAATGGAAAAG) and the antisense primers RT-AS-1 (GCTTTTTTACCATCCCTC) and RT-AS-2 (CATTACCCACAAAACAAAG). Figure 1 gives an overview of all primers used in this study and their nucleotide positions on the HERV-K10 genome. HERV-K RT clone 10.1 was used to introduce N- and C-terminal truncations. We used three 5′ primers and four 3′ primers to PCR amplify part of the RT open reading frame. The 5′ primers are RT-B (described above), RT-X1 (TCAGGATCCCAGTGGCCGCTACCAAAA), and RT-X2 (TCAGGATCCATTGAGCCTTCATTCTCG), and the 3′ primers are RT-C (described above), RT-Y1 (TCAGCGGCCGCTATAAATTGAATAGCTGGTT), RT-Y2 (TCAGCGGCCGCTAATCTTGTAACACTGTAAT), and RT-Y3 (TCAGCGGCCGCTAAAATACTGTTAGAGCATT). Restriction enzyme sites and stop codons are marked as described above. Because the new primers are based on the 10.1 sequence, they may differ from the HERV-K10 sequence (e.g., at two positions in RT-X2). The resulting DNA fragments were digested with BamHI and NotI, whose sites were encoded by the 5′- and 3′-primer sequence, respectively. The DNA fragments were cloned in pGEX-4T1 as described above. The RT domain encoded by the different constructs is schematically depicted in Fig. 7A.
FIG. 7.
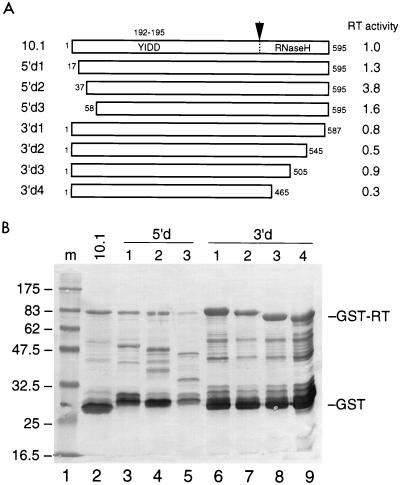
Deletion mapping of the HERV-K RT domain. (A) The HERV-K RT gene of clone 10.1, which expresses a relatively active RT enzyme, was used as the starting material for the generation of a nested set of 5′- and 3′-truncated RT proteins. Indicated are the amino acid positions, with those of clone 10.1 being arbitrarily set at 1 to 595. The position of the YIDD motif within the catalytic core (positions 192 to 195) and the putative position of the RT-RNase H border are indicated. (B) The different RT enzymes were expressed in E. coli as GST fusion proteins and visualized by Western blotting with the GST-specific antiserum. See Fig. 4 for further details. Note that a different protein marker was used in lane 1. This set of RT proteins was tested in the poly(rA)-oligo(dT) assay for enzyme activity. The relative RT activity is listed in panel A, with the activity of the “full-length” RT clone 10.1 being arbitrarily set at 1.0.
RT protein expression in E. coli and Western blot analysis.
An overnight culture of E. coli DH5α harboring one of the pGEX-4T-1 plasmids was diluted 1:10, and 100 ml was cultured for 2 h at 37°C in brain heart infusion broth. GST-RT protein expression was induced with 100 μl of 0.1 M isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were collected by centrifugation after a 4-h IPTG induction and resuspended in 1.5 ml of NET-N buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 8.0], 0.5% Nonidet-P40). The lysate was cleared by extensive sonication on ice (45-s pulse setting, 50% output microtip). The cellular debris was removed by centrifugation. The GST-RT fusion protein was allowed to bind to 25 μl of glutathione-agarose beads (Sigma; 1:1 suspension in phosphate-buffered saline) at room temperature for 1 h. The beads were collected by low-speed centrifugation, washed three times in phosphate-buffered saline, and incubated for 10 min at room temperature in 25 μl of freshly made elution buffer (10 mM glutathione in 50 mM Tris-HCl [pH 8.0]). This elution step was repeated once, and the combined eluate was used either directly in RT or RNase H assays or brought to 50% glycerol and stored at −70°C. We used 3 μl of this enzyme preparation in the RT assay and 0.1, 0.2, and 1.0 μl in the RNase H assay.
Western blotting was performed with the purified GST-RT proteins. The samples (5 μl of a HERV-K GST-RT preparation) were boiled in sodium dodecyl sulfate (SDS) sample buffer, separated on an SDS–12.5% polyacrylamide gel, and electrophoretically transferred to Immobilon nitrocellulose. The immunoblot was stained with a GST-specific monoclonal antibody (anti-GST; Pierce) and developed by using the 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium protocol (Sigma).
RT and RNase H assays.
The purified GST-RT proteins were tested for RT activity in a poly(rA)-oligo(dT) assay as described previously (2). In brief, reaction mixtures contained 60 mM Tris (pH 7.8), 75 mM KCl, 5 mM MgCl2, 0.1% Nonidet P-40, 1 mM EDTA, 5 μg of poly(rA)7000 per ml, 0.16 μg of oligo(dT)15 per ml, 4 mM dithiothreitol, and 50 μCi of [32P]dTTP per ml (3,000 Ci/mmol). In a standard assay, we used 3 μl of the HERV-K GST-RT preparations and 1 μl of 1:30-diluted human immunodeficiency virus type 1 (HIV-1) GST-RT. Samples were incubated at 37°C for 2 h. Duplicate 7.5-μl aliquots were taken after 1 and 2 h and spotted onto DEAE ion-exchange paper (DE81; Whatman). The filter paper was washed three times in 5% Na2HPO4 to remove unincorporated [32P]dTTP and dried after two 96% ethanol washes. The spots were visualized by autoradiography and quantitated on a PhosphorImager (Molecular Dynamics). The length distribution of the cDNA products was analyzed on a 6% polyacrylamide–7.1 M urea sequencing gel.
The RNase H assay was performed with an internally labeled RNA molecule that was made in vitro with the T7 RNA polymerase in the presence of [α-32P]UTP. The transcript consists of the HIV-1 leader sequence and is fully complementary to the CN1 DNA oligonucleotide. We mixed 2 μl of RNA transcript (approximately 10 ng) and 0.5 μl of DNA oligonucleotide (50 ng) in 9 μl of RT buffer (50 mM Tris-HCl [pH 8.5], 8 mM MgCl2, 30 mM KCl, 1 mM dithiothreitol) with 0.25 μl of RNasin (40 U/ml; Boehringer). The samples were incubated for 1 h at 37°C upon addition of the GST-RT protein (0.1, 0.2, and 1.0 μl). The reaction was stopped by addition of 4 μl of formamide sample buffer. The samples were heated at 100°C for 3 min and analyzed on a denaturing 6% polyacrylamide–7.1 M urea gel.
Phylogenetic analyses of nucleic acid and amino acid sequences.
Nucleotide sequences were either analyzed with the Clustal program (PC gene software; IntelliGenetics) or the PileUp program (GCG package). Both programs are based on the method of Higgins and Sharp (16). Clustal permits the alignment of multiple nucleic acid sequences in three steps: computation of pairwise similarity scores, construction of a dendrogram, and subsequent alignment. Similarly, PileUp creates a multiple sequence alignment of a group of related sequences by using progressive pairwise alignments. We used standard settings (gap weight, 3.00; gap length weight, 0.10). The reference strains used in the phylogenetic analysis have the following accession numbers: STPLU4 = HML-1, AF030038; N8.4 = HML-2, U87590; P1.3 = HML-3, AF030043; M3.10 = HML-4, AF030046; N8.4 = HML-2A, U87590; D1.2 = HML-2B, U87595; M3.5 = HML-2C, U87592; P1.4 = HML-2D (which is identical in the small RT segment to HML2.5 of reference 33); M3.8 = HML-2E, U87587; Stmin2 = HML-2F (which is identical in the small RT segment to clone HML2.2 of reference 33), and the prototype HERV-K10, M14123.
Nucleotide sequence accession numbers.
The nucleotide sequence of six full-length HERV-K RT genes presented in this study have been deposited in the Genbank database under accession no. AF080229 (clone 10.1), AF080230 (clone 10.2), AF080231 (clone 10.9), AF080232 (clone 11.1), AF080233 (clone 11.2), and AF080234 (clone 7.1).
RESULTS
Cloning of full-length HERV-K RT genes.
Primers were designed to amplify a complete RT gene on the basis of the nucleotide sequence of the prototype HERV-K10 isolate, a member of the extensive HML-2 subgroup. Because the exact borders of the protease-RT and RT-integrase proteins are not known (Fig. 1), we estimated the 5′ and 3′ ends of the HERV-K10 RT coding information by alignment with the homologous regions of several exogenous retroviruses (data not shown; see also reference 12). The complete RT region is expected to be amplified by the primer set RT-B plus RT-C, but we also amplified a slightly larger RT fragment with the primer set RT-A plus RT-D (Fig. 1). We restricted this search to the HERV-K elements that are transcriptionally active, because cellular RNA was used as starting material. Obviously, transcriptionally inactive genomes may in principle also be candidates for intact RT genes, but we focused on expressed HERV-K copies because they are more likely to exhibit a biological function. Total RNA was isolated from human bone marrow cells that were demonstrated previously to express numerous members of the HML-2 subgroup of HERV-K elements (53). The RNA was converted into cDNA with random primers. This cDNA was subsequently used for PCR amplification with the two HERV-K10-specific primer sets. Fragments of the expected length were produced by both primer sets and were cloned into the E. coli expression vector pGEX-4T-1. The upstream primers RT-A and RT-B were designed to fuse the RT gene in-frame with the GST gene, thus allowing the production of GST-RT fusion proteins. The downstream primers provided a stop codon to terminate translation.
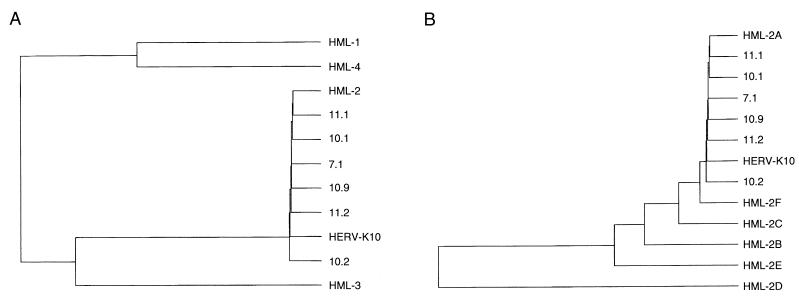
We first analyzed the complete nucleotide sequence of three RT genes obtained with the outer primers (clones 10.1, 10.2, and 10.9), and three inserts obtained with the inner primer set (clones 7.1, 11.1, and 11.2). All six HERV-K isolates were different from one another. We previously estimated that on average one mutation is generated by the RT-PCR protocol for a 1,700-bp RT fragment (53). Thus, clones differing by at least two nucleotides are thought to represent unique isolates. Strikingly, the new sequences were also different from any of the previously reported HERV-K members, indicating that this family may be more extended than was previously estimated. All new sequences showed the highest similarity score to the HERV-K10 sequence, which may not be surprising, because the PCR primers were based on this isolate. The similarity ranged from 93% (clone 10.2), to 95% (clone 10.9) and up to 98% for all other clones. It is likely that even the most similar sequences represent unique HERV-K clones. For instance, clone 10.1 differs from HERV-K10 at 31 nucleotide positions within the RT gene. The sequences were analyzed to determine the positions of the new isolates in the current HERV-K phylogeny. All six RT clones belong to the HML-2 subgroup (Fig. 2A). A more detailed comparison with members of the different clusters within the HML-2 subgroup (53) revealed that all new RT sequences belong to the HML-2A cluster (Fig. 2B). Thus, the six HERV-K RT genes represent unique but closely related members of the HML-2A cluster, of which HERV-K10 is the prototype member. The result of this specific PCR suggests that the HML-2A cluster contains many more members than was previously anticipated and that many sequences are closely related to the prototype HERV-K10 isolate.
FIG. 2.
Phylogenetic analysis of the HERV-K RT genes. For this comparison with known HML representatives, we used the 600-bp internal RT fragment amplified by the MOP-2 primer set (Fig. 1). Sequences were analyzed with the Clustal program (PC gene package). (A) The novel isolates described in this study are compared with the prototype HERV-K10 and members of the HML-1 through HML-4 subgroups (33). We used the following reference strains: STPLU4 (HML-1), N8.4 (HML-2), P1.3 (HML-3), and M3.10 (HML-4). (B) The new sequences are compared with HERV-K10 and representative members of the HML-2 clusters A through F (53). The following representative clones were used: N8.4 (HML-2A), D1.2 (HML-2B), M3.5 (HML-2C), P1.4 (HML-2D), M3.8 (HML-2E), and Stmin2 (HML-2F). This analysis unequivocally places all new HERV-K sequences and the HERV-K10 isolate in the HML-2A subgroup.
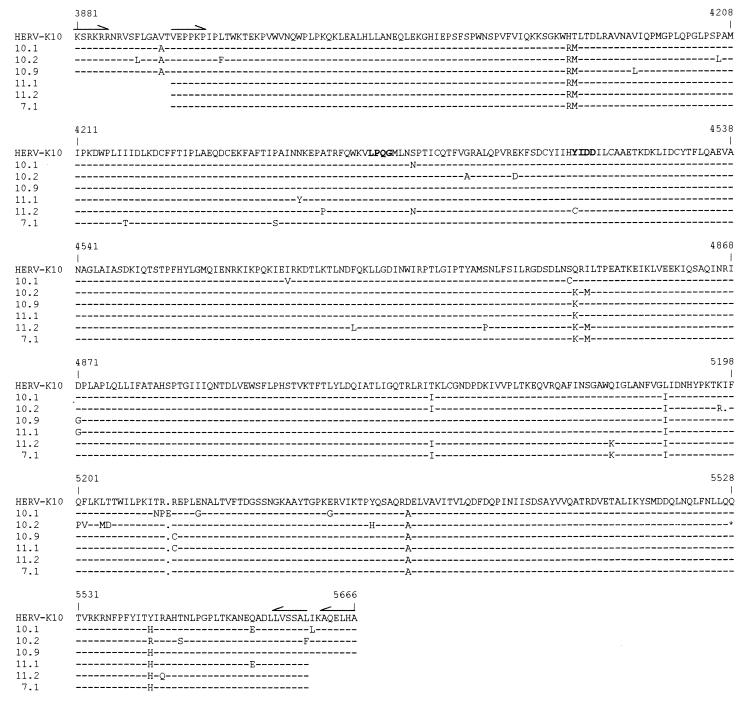
The translated RT amino acid sequences are shown in Fig. 3. Some of the conserved RT motifs are marked, for instance the well-conserved LPQG motif and the catalytically important YIDD motif that were previously used to design RT-specific primers (see, e.g., Fig. 1). Five of the six HERV-K RT genes were uninterrupted, testifying to the apparent conservation of the RT open reading frame in this HERV-K subgroup. A premature stop codon was present in clone 10.2 near the C terminus of the RT protein (Fig. 3). Obviously, it cannot be excluded that the single nucleotide change that creates this in-frame stop codon was generated during the RT-PCR procedure. With five potentially complete HERV-K RT genes in hand, we set out to express these enzymes as recombinant protein in E. coli to test for their activity in polymerase assays.
FIG. 3.
Alignment of the HERV-K RT amino acid sequences. The prototype HERV-K10 is shown at the top. Dashes indicate amino acids that are identical to this prototype. Gaps are indicated by dots. The in-frame stop codon at position 5528 in clone 10.2 is marked by an asterisk. The conserved LPQG and YIDD motifs are in boldface type. Numbers refer to the first nucleotide of the respective codons in the HERV-K10 nucleotide sequence. The nucleotide sequences of the hypermutated segments of clone 10.2 (starting at position 5192) and clone 10.1 (position 5240) are shown in detail in Fig. 11. The prototype HERV-K10 sequence is unique at certain positions, which may explain the inactivity of the corresponding RT enzyme (see the text for further details).
Several HERV-K RT enzymes exhibit RT activity.
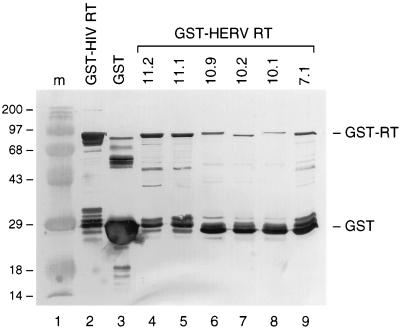
The HERV-K RT enzymes were expressed as GST fusion proteins in E. coli and purified in a single step with glutathione-agarose beads by a standard procedure (48). As controls, we expressed and purified the 30-kDa GST domain and an enzymatically active GST-RT fusion of HIV-1 (37). Although reasonable amounts of the GST-RT fusion proteins were expressed in this system (data not shown), the bulk of the HERV-K proteins could not be extracted in soluble form, indicating that this fusion protein is prone to aggregation or formation of inclusion bodies. Similar but less severe insolubility problems were encountered for the HIV-1 GST-RT fusion protein. We failed to significantly optimize the yield of the HERV-K fusion proteins by adaptation of the culture and/or extraction protocol (e.g., shorter IPTG induction period, reduced culture temperature, addition of detergents during extraction). The results of a typical experiment are presented in Fig. 4. E. coli cultures (100 ml) were used to prepare a 50-μl stock solution of purified GST-RT protein, of which 5 μl was analyzed by Western blotting and stained with an anti-GST monoclonal antibody. Whereas the control GST protein could be isolated in bulk amounts (Fig. 4, lane 3) and a reasonable yield was obtained for the HIV-1 GST-RT fusion (lane 2), dramatically low yields were apparent for the HERV-K GST-RT fusion proteins (lanes 4 to 9). The minor differences in migration of the individual HERV-K proteins on the SDS-gel are consistent with the length of the cloned RT fragments (primer set RT-A plus RT-D versus primer set RT-B plus RT-C) and the presence of a premature stop codon in clone 10.2. Besides a poor yield, we also observed significant degradation of the GST-RT fusion proteins. In particular, these proteins seem vulnerable to proteolytic cleavage near the junction of the GST and RT domains (near the thrombin site encoded by the pGEX-4T-1 vector). This proteolytic activity generates an approximately 30-kDa GST domain, and it is likely that a separate RT domain is also produced, but only the former protein could be visualized with the GST-specific antiserum.
FIG. 4.
Western blot analysis of the HERV-K GST-RT proteins. The type of HERV-K GST-RT protein that is expressed is indicated at the top (lanes 4 to 9). As a positive control, we expressed the HIV-1 GST-RT protein (lane 2). The control pGEX-4T-1 vector produces a large amount of soluble GST protein (lane 3). Lane 1 contains marker proteins (the molecular mass in kilodaltons is indicated on the left). The positions of the 30-kDa GST protein and the approximately 95-kDa GST-RT protein are indicated on the right. The SDS–12.5% polyacrylamide gel was electroblotted and stained with an anti-GST monoclonal antibody.
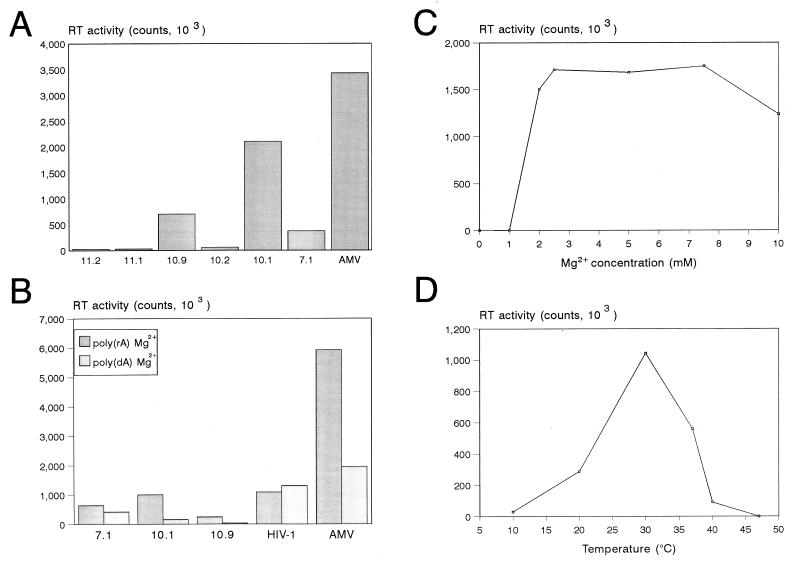
The purified GST-RT proteins were assayed for RT activity by measuring dTTP incorporation on a poly(rA)-oligo(dT) template-primer duplex. The reaction mixture was incubated at 37°C, and duplicate samples were taken after 1 and 2 h. The background activity obtained with the GST control sample was subtracted from the incorporation measured for the HERV-K GST-RT enzymes. The mean value of the 2-h samples is plotted in Fig. 5A, but similar results were measured for the 1-h samples. Two clones with a large (1.8-kb) RT insert (clones 10.1 and 10.9) and one clone with a short (1.7-kb) RT insert (clone 7.1) demonstrated significant polymerase activity (Fig. 5A). Three HERV-K RT clones were inactive (clones 10.2, 11.1, and 11.2) and thus provide additional negative controls. As a positive control, we included the RT enzyme of AMV. Inspection of the amino acid sequences (Fig. 3) provides some putative explanations for the apparent inactivity of some HERV-K RT proteins. For instance, the RT enzyme of clone 11.2 is likely to be inactive due to mutation of the catalytically important YIDD motif into CIDD, and clone 10.2 may be inactive due to the presence of the premature stop codon. The prototype HERV-K10 RT protein is enzymatically inactive (25), which may be related to the presence of a unique RM-to-HT amino acid substitution in the N-terminal domain of this RT enzyme (Fig. 3).
FIG. 5.
Enzyme characteristics of the HERV-K RT polymerase. (A) Activity of the different HERV-K RT enzymes in a regular poly(rA)-oligo(dT) assay. Duplicate samples were taken after 1 and 2 h at 37°C. The activity was quantitated with a PhosphorImager, and background values obtained with the control GST protein were subtracted. In these RT assays, less than 20% variation was measured between the duplicate samples. The results of the 2-h incubation are shown, but similar results were obtained for the 1-h samples. The AMV RT enzyme (Boehringer; 1 μl of a 1-U/μl stock) was used as a positive control. (B) dTTP incorporation onto an oligo(dT) primer annealed to either an RNA or DNA template; poly(rA) versus poly(dA). The HERV-K enzymes 7.1, 10.1, and 10.9 were compared with HIV-1 GST-RT and AMV RT. (C and D) Magnesium cation and temperature dependency of the HERV-K RT enzyme of clone 10.1. This assay was performed with the poly(rA)-oligo(dT) template primer at 37°C (C) and 5 mM MgCl2 (D).
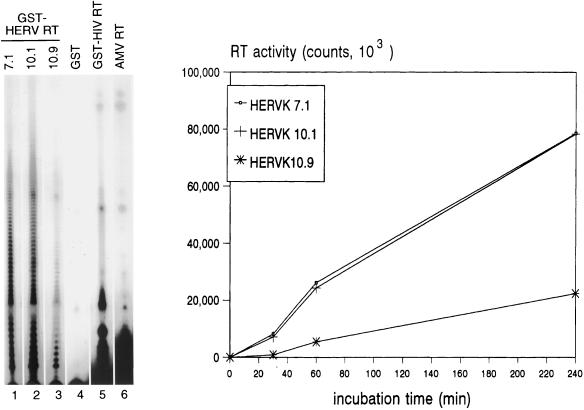
We next tested several characteristics of the HERV-K RT enzyme. First, we compared dTTP incorporation on RNA and DNA templates [Fig. 5B, poly(rA) and poly(dA)]. The HERV-K enzymes were two- to threefold more active on the RNA template. This pattern is consistent with the behavior of the AMV RT enzyme but differs from that of the HIV-1 RT enzyme, which is equally active on these two templates. A standard RT assay was performed in the presence of 5 mM MgCl2, but some retroviral RT enzymes are known to be active in the presence of Mn2+ as the divalent cation. However, none of the HERV-K enzymes demonstrated polymerase activity in buffers containing Mn2+ (not shown). The dose-response curve for Mg2+ is illustrated in Fig. 5C and demonstrates an absolute requirement for this cation. The temperature optimum of the HERV-K RT enzyme was 30 to 37°C (Fig. 5D). A kinetic analysis of this reverse transcription reaction is presented in Fig. 6 (right), indicating that the HERV-K RT enzyme is relatively stable for up to 4 h at 37°C. A characteristic of retroviral RT enzymes is their poor processivity, which means that short cDNAs are produced in a single cycle of polymerization. To test this, we analyzed some of the cDNA samples on a denaturing gel (Fig. 6, left). Indeed, only short cDNAs (less than 20 to 30 nucleotides [nt]) were synthesized by the RT enzymes of HERV-K (lanes 1 to 3), HIV-1 (lane 5), and AMV (lane 6). The initial biochemical characterization of this enzyme indicates that it is very similar to that of contemporary exogenous retroviruses. The specific activity of the HERV-K enzyme appears to be rather low, which is consistent with a previous report on the baculovirus-produced enzyme (51). On the basis of GST fusion protein concentrations as determined by Western blotting (Fig. 4), we estimate that the HERV-K enzyme of clone 10.1 exhibits only 5% of the polymerase activity of the HIV-1 enzyme.
FIG. 6.
Kinetics of reverse transcription and processivity the HERV-K RT enzyme. (Left) HERV-K RT samples of clones 7.1, 10.1, and 10.9 were used in the standard poly(rA)-oligo(dT) assay, and the length of the cDNA products of the HERV-K RT enzymes was analyzed on a denaturing sequencing gel (lanes 1 to 3) and compared with the products of the HIV-1 and AMV RT enzymes (lanes 5 and 6). The GST protein was used as negative control (lane 4). (Right) Standard RT assay with clones 7.1, 10.1, and 10.9. Samples were analyzed for dTTP incorporation at several times up to 4 h. The background values obtained for the GST control protein were subtracted.
It is possible that the naturally processed HERV-K RT enzyme, whose N and C termini are not known, is a more active RT enzyme than is the GST-RT protein used in this study. We performed several additional experiments to test this. It is possible that the RT domain is relatively inactive due to the N-terminal GST extension. For instance, such a detrimental effect has been reported in one study with the HIV-1 RT enzyme (17), although it could not be confirmed in another study (37). Another RT enzyme of the human T-cell leukemia virus type 1 retrovirus was also demonstrated to be active in the presence of a N-terminal extension (39). We measured no increase in HERV-K RT activity upon removal of the N-terminal GST domain in clone 10.1 by thrombin digestion (data not shown). To test RT forms with different N and C termini, we constructed a nested set of deletion mutants of RT clone 10.1 (illustrated in Fig. 7A). All truncated RT variants were expressed in E. coli at an extremely low level (Fig. 7B). A somewhat improved recovery was apparent for the C-terminally truncated RT forms (Fig. 7B, lanes 6 to 9), which may indicate increased protein stability and/or solubility. However, no increased RT activity was measured for these mutants. A modest increase in RT activity was measured upon deletion of 36 N-terminal amino acids, but no further increase in RT activity was measured upon removal of additional amino acids.
The HERV-K RT enzyme has a functional RNase H domain.
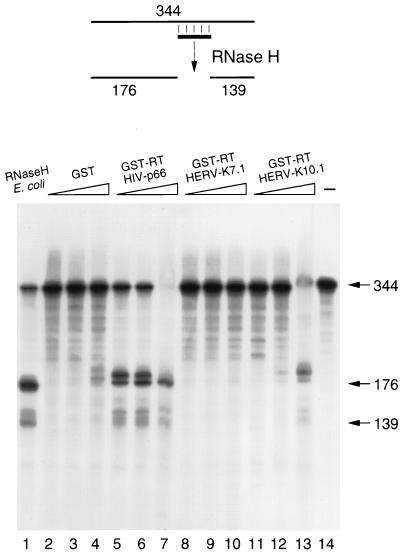
Retroviral RT enzymes contain an RNase H domain that degrades the template RNA after it is copied by the polymerase domain. This activity is thought to be essential in the intricate process of reverse transcription (50). The HERV-K10 genome has the potential to encode a C-terminal RT domain with similarity to RNase H (12, 36). Although the exact borders of this putative RNase H gene are not known, this domain was included in the PCR strategy (Fig. 1). We therefore analyzed the RNase H activity of the HERV-K GST-RT proteins of clones 7.1 and 10.1, which represent the short and long versions of the biologically active polymerase, respectively. This assay was performed with an internally labeled RNA transcript to which a complementary DNA oligonucleotide was annealed (schematic in Fig. 8). Treatment of this RNA-DNA duplex with the commercially available RNase H of E. coli yielded two RNA fragments of 139 and 176 nt (Fig. 8, lane 1). Such activity was not observed with the control GST sample (lanes 2 to 4), and efficient cleavage was obtained for the HIV-1 GST-RT protein (lanes 5 to 7). Most importantly, we measured low RNase H activity for the HERV-K RT enzyme of clone 10.1 (lanes 11 to 13). No RNase H activity was demonstrable for the RT enzyme of clone 7.1 (lanes 8 to 10), which may be related to the absence of C-terminal amino acids in this clone (Fig. 3). The RNase H activity measured for the HERV-K GST-RT enzyme of clone 10.1 is much lower than that of the HIV-1 GST-RT enzyme. These combined results indicate that this endogenous RT enzyme exhibits low polymerase and RNase H activities.
FIG. 8.
The HERV-K RT enzyme has RNase H activity. A schematic of the RNase H assay is indicated at the top of the figure. Cleavage of the internally labeled 344-nt RNA transcript at the site of the annealed DNA oligonucleotide will produce a 5′-terminal fragment of 176 nt and a 3′-terminal fragment of 139 nt. The positions of these signals are indicated in the lower panel, which shows an analysis of the RNA cleavage products on a denaturing sequencing gel. The mock-treated RNA is shown in lane 14. We tested increasing amounts (0.1, 0.2, and 1.0 μl) of the GST control (lanes 2 to 4), the HIV-1 GST-RT enzyme (lanes 5 to 7), and the HERV-K 7.1 and 10.1 enzymes (lanes 8 to 10 and 11 to 13, respectively). As a positive control, we used the E. coli RNase H enzyme (Boehringer; 1 μl of 1-U/μl stock) (lane 1).
The HERV-K RT gene has a mosaic genome structure due to frequent recombination.
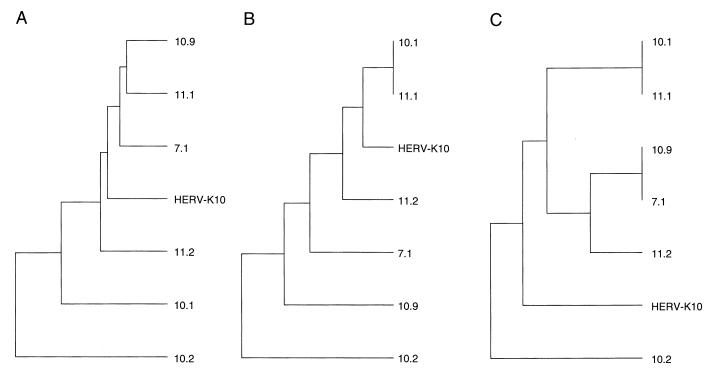
In the course of this study, we noticed that phylogenies for the HERV-K sequences differed significantly for the 5′ and 3′ parts of the RT gene (Fig. 9B and C, respectively; analysis of the complete RT gene is shown in Fig. 9A). The finding of discordant branching orders in the two topologies based on 5′ and 3′ sequences prompted us to inspect the actual gene sequences for signs of recombination. Indeed, the nucleotide sequence of the HERV-K genes suggested the presence of multiple crossovers. To analyze this genetic recombination pattern in a more systematic manner, we divided the 1.8-kb RT gene into 15 arbitrary segments around nucleotide positions that differed among the six new HERV-K isolates. Only the substitutions present in at least two isolates were included, thereby filtering out mutations that may have been introduced fortuitously during RT-PCR amplification. These informative nucleotide positions are shown at the top of Fig. 10 (e.g., position 4060, which is C in isolates 10.9, 10.2, and HERV-K10 but T in the other four isolates), and we subsequently marked the RT segments in an arbitrary manner (see the legend to Fig. 10). This analysis was performed for all 15 RT segments, and neighboring segments were marked so that genetic linkages were optimal. The pattern shown in Fig. 10 indicates a mosaic gene structure, suggesting that these sequences were the subject of multiple recombination events. Such mosaic genomes are likely to have been formed during reverse transcription, which is known to be a recombination-prone process (50).
FIG. 9.
The 5′ and 3′ domains of the HERV-K genes produce a different phylogeny. A phylogenetic analysis was performed by the Pileup program (GCG package). The distance along the vertical axis is proportional to the difference between sequences; the distance along the horizontal axis has no significance. We analyzed either the complete RT genes (positions 3929 to 5633) (A), the 5′ domains (positions 4134 to 4313) (B), or the 3′ domains (positions 5337 to 5633) (C). Several similarities are apparent; e.g., clone 10.2 is at the origin of all three trees, but some notable differences in the branching orders of the trees were observed.
FIG. 10.
Schematic of the mosaic structure of the HERV-K RT gene. The HERV-K RT genes were split in segments around the nucleotides that differ among the isolates. We excluded mutations that were present in a single isolate, thus filtering out potential sequence errors introduced in the RT-PCR amplification protocol. The informative nucleotide positions are indicated at the top. These segments were arbitrarily marked grey or white (e.g., segment 4060 C is white, segment 4060 T is grey). We optimized the genetic linkages, starting at the 3′ end, which may explain the absence of crossover sites in the extreme 3′ RT domain (positions 5353 to 5615).
DISCUSSION
We describe the identification of a functional RT enzyme encoded by endogenous retroviruses of the abundant HERV-K family that are integrated at multiple loci of the human genome. Such RT polymerase activity may have been instrumental in the evolution of the human genome, for instance in the formation of pseudogenes. Several candidate sources of RT activity in human cells have been reported previously. In addition to both exogenous and endogenous retroviral RTs, at least one “cellular” form of RT is the telomerase enzyme, an unusual DNA polymerase with an internal RNA template that encodes a repeat sequence, which is added to the 3′ end of chromosomes (4). However, telomerase is not active on exogenous templates and is therefore unlikely to have the properties required for pseudogene formation. There is convincing evidence that infection of cells with exogenous retroviruses can result in the formation of pseudogenes, although such cDNAs have unusual features compared with naturally occurring pseudogenes (10, 13, 23). Therefore, the focus has been primarily on endogenous sources of RT activity.
Our results indicate that several members of the extensive HERV-K endogenous retrovirus family may have provided this RT activity. The HERV-K virus family has been suggested previously to encode an active RT enzyme, because polymerase activity was measured in a variety of biological samples that contain HERV-K-like virion particles (6, 8, 40, 46, 47). Endogenous RT activity has been demonstrated experimentally in mammalian cells through de novo formation of pseudogene-like structures (27, 49). By using a newly developed in vivo assay, it was demonstrated recently that overexpression of the human endogenous LINE (L1) element yields RT activity that is able to generate reverse transcripts (11). It seems possible that both the HERV-K and LINE RT enzymes have played a role in shaping the human genome during evolution. An argument against the involvement of the HERV-K RT enzyme in pseudogene formation is that the RT enzymes of retroviruses prime reverse transcription in a highly specific manner, with regard to both the type of tRNA primer and the template RNA (9, 18, 22, 29, 38, 50). Although no details are currently available on the priming specificity of the HERV-K RT enzyme, it is unlikely that this endogenous retrovirus will be significantly different in this respect from the exogenous counterparts. For instance, the HERV-K RT enzyme is likely to use a specific tRNALys primer because of the presence of a fully complementary primer-binding site in the HERV-K genome (36). The activity of the LINE RT enzyme exhibits no template specificity. This seems the appropriate characteristic for an enzyme involved in the copying of random cellular transcripts (11), although there is also some evidence that particular cellular transcripts are more prone to pseudogene formation than others (41). Additional experimentation is required to establish the involvement of the HERV-K and/or LINE RT enzyme in pseudogene formation.
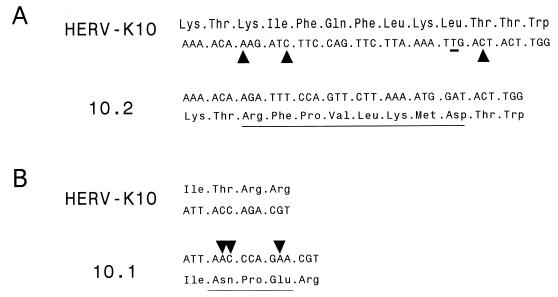
We and others previously found that purifying or negative selection seems to operate on the HERV-K genomes. At least in some of the subgroups, there is a remarkable conservation of the open reading frames (53, 55), and this result was confirmed in the present study for the HML-2 RT genes. Furthermore, of the mutations that are present, a strong prevalence of synonymous nucleotide substitutions was noted (54). The biological significance of this retrovirus family is substantiated further by the finding that multiple members are transcriptionally active. This also holds for the active RT species identified in this study, which were cloned by an RT-PCR strategy with cellular RNA as input. The cloning of several full-length HERV-K RT sequences allowed us to readdress some of the issues concerning the apparent conservation of these genes. Most strikingly, we noticed that some HERV-K elements have preserved their RT-encoding capacity despite the presence of deletions and/or substitutions that destroy the reading frame. Alignment of the RT sequence of clone 10.2 with that of HERV-K10 (Fig. 11A) indicates that three 1-nt deletions are present that are unique for this clone. Whereas any of the individual mutations would cause a frameshift during translation of the RT protein, the combination of all three mutations restores the reading frame and allows the expression of full-length RT with a mutant 8-amino-acid stretch (underlined in Fig. 11A). A somewhat similar situation is seen in clone 10.1, where a 2-nt insertion is combined with a 1-nt insertion to restore the RT reading frame, so that only 3 amino acids are read out of frame (Fig. 11B). In general, the identification of an enzymatically active HERV-K RT enzyme may help define new experiments to test the possible biological function of HERV-K elements in the host genomes and their contribution to disease induction. Perhaps most intriguing is the observation that HERV-K virus expression is induced in the pancreatic islets of diabetes type 1 patients (8), and it was suggested that the viral Env protein exhibits superantigen activity that may trigger this autoimmune disease. However, several recent reports put some of these results into question (21, 26, 35).
FIG. 11.
Multiple insertions and deletions are needed to restore the RT open reading frame. Part of the HERV-K10 nucleotide and RT amino acid sequences were compared with those of HERV-K isolate 10.2 (A) and clone 10.1 (B). (A) RT segment from positions 5186 to 5224. The three nucleotides deleted in clone 10.2 are marked by triangles in the HERV-K sequence. An additional T-to-A substitution is underlined. Compared with all other HERV-K RT genes, translation of clone 10.2 will shift transiently out of the regular reading frame, producing a unique 8-amino-acid stretch (underlined). (B) For clone 10.1, a shorter RT segment is shown (positions 5237 to 5249). The positions of a 2-nt insertion and a 1-nt insertion in the sequences of clone 10.1 are indicated. This results in a unique 3-amino-acid segment due to translation in a different reading frame. The complete amino acid sequences of clones 10.1 and 10.2 are shown in Fig. 3.
The polymerase and RNase H activities measured with the recombinant HERV-K GST-RT proteins were very low compared with those of the HIV-1 GST-RT protein. This may reflect the real biological activity of these endogenous RT enzymes. On the other hand, we analyzed only six RT enzymes of viruses that belong to the HML-2 subgroup, and it is possible that more active RT forms are encoded by other HERV-K elements, for instance in cell types other than the bone marrow cells used in this study. Furthermore, it cannot be excluded that this enzyme requires unique reaction conditions for optimal activity. The initial biochemical analyses suggest that the endogenous RT enzyme is not much different from that of exogenously replicating retroviruses like AMV or HIV-1. For instance, HERV-K enzyme has a marked preference for Mg2+ over Mn2+, which is not surprising because all retroviral RTs, excluding those from mammalian type C retroviruses, display such a preference. Nevertheless, this property should not be used to classify these viruses, because alteration of a single amino acid in the HIV-1 RT enzyme can result in a loss of the Mg2+ preference (42). It is possible that the GST-RT constructs are not optimally active because a suboptimal N- or C-terminus was chosen in our cloning strategy. To verify this, we constructed a nested set of N- and C-terminally truncated RT forms of clone 10.1. Although some increase in RT activity was measured for the 5′-shortened RT forms, no major increase in activity was obtained. We have also tested whether removal of the N-terminal GST domain did improve the polymerase properties, but we found no such effect. We did notice some spontaneous cleavage of the GST-RT proteins at the fusion site, suggesting that part of the RT protein may have been present in a GST-free form. It should also be mentioned that only a small inhibitory effect of the N-terminal GST extension was measured in the context of the HIV-1 RT enzyme (37) and the human T-cell leukemia virus type 1 RT enzyme (39).
Inspection of the HERV-K genomic sequences of different HERV-K family members revealed a high level of intergenic recombination. Initially, we noticed the lack of congruence in the topologies of phylogenetic trees constructed for different parts of the RT gene, which suggested the prevalence of recombination. This was verified by inspection of the nucleotide sequences. This result suggests that genetic recombination, a property of the RT enzyme that is observed regularly for contemporary viruses like HIV-1 (19, 43), is a characteristic of all retroviruses. Obviously, our results do not tell us when this recombination occurred. Recombination could have occurred many million years ago, during the exogenous life cycle of HERV-K precursor viruses. Alternatively, recombination may have occurred during the spread of endogenous HERV-K copies by intracellular retrotransposition. Further experimentation is required to provide more detailed information on the structure and function of the HERV-K RT enzyme. For instance, it will be of interest to test whether this “ancient” RT enzyme forms a dimeric complex, such as is seen for most contemporary RT enzymes (50).
ACKNOWLEDGMENTS
We thank Tonja van der Kuyl for critical reading of the manuscript and P. A. Voûte for support.
This research was sponsored in part by the ‘Stichting Kindergeneeskundig Kankeronderzoek’ (SKK).
REFERENCES
- 1.Andersson M-L, Medstrand P, Yin H, Blomberg J. Differential expression of human endogenous retroviral sequences similar to mouse mammary tumor virus in normal peripheral blood mononuclear cells. AIDS Res Hum Retroviruses. 1996;12:833–840. doi: 10.1089/aid.1996.12.833. [DOI] [PubMed] [Google Scholar]
- 2.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Oude Essink B B, van Kuilenburg A B P, Van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 3.Best S, Le Tissier P R, Stoye J P. Endogenous retroviruses and the evolution of resistance to retroviral infection. Trends Microbiol. 1997;5:313–318. doi: 10.1016/S0966-842X(97)01086-X. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn E H. Telomerases. Annu Rev Biochem. 1992;61:113–130. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 5.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–436. [PubMed] [Google Scholar]
- 6.Boller K, Konig H, Sauter M, Mueller-Lantzsch N, Lower R, Lower J, Kurth R. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196:349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 7.Boyd M T, Foley B, Brodsky I. Evidence for copurification of HERV-K-related transcripts and a reverse transcriptase activity in human platelets from patients with essential thrombocythemia. Blood. 1997;90:4022–4030. [PubMed] [Google Scholar]
- 8.Conrad B, Weissmahr R N, Boni J, Arcari R, Schupbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type 1 diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 9.Das A T, Klaver B, Berkhout B. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA(3Lys) J Virol. 1995;69:3090–3097. doi: 10.1128/jvi.69.5.3090-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derr K L, Strathern J N, Garfinkel D J. RNA-mediated recombination in S. cerevisiae. Cell. 1991;67:355–364. doi: 10.1016/0092-8674(91)90187-4. [DOI] [PubMed] [Google Scholar]
- 11.Dhellin O, Maestre J, Heidmann T. Functional differences between the human LINE retrotransposon and retroviral reverse transcriptases for in vivo mRNA reverse transcription. EMBO J. 1997;16:6590–6602. doi: 10.1093/emboj/16.21.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doolittle R F, Feng D-F, Johnson M S, McClure M A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 13.Dornburg R, Temin H M. cDNA genes formed after infection with retroviral vector particles lack the hallmarks of natural processed pseudogenes. Mol Cell Biol. 1990;10:68–74. doi: 10.1128/mcb.10.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin G C, Chretien S, Hanson I M, Rochefort H, May F E B, Westley B R. Expression of human sequences related to those of mouse mammary tumor virus. J Virol. 1988;62:1203–1210. doi: 10.1128/jvi.62.4.1203-1210.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris J M, Haynes R H, McIntosh E M. A consensus sequence for a functional human endogenous retrovirus K (HERV-K) dUTPase. Biochem Cell Biol. 1997;75:143–151. [PubMed] [Google Scholar]
- 16.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 17.Hottiger M, Gramatikoff K, Georgiev O, Chaponnier C, Schaffner W, Hubscher U. The large subunit of HIV-1 reverse transcriptase interacts with β-actin. Nucleic Acids Res. 1995;23:736–741. doi: 10.1093/nar/23.5.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isel C, Lanchy J-M, Le Grice S F J, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcriptase require the post-transcriptional modifications of primer tRNALys3. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 19.Jin M J, Hui H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura Y, Ayukawa T, Ishikawa T, Kanda T, Yoshiike K. Human endogenous retrovirus K10 encodes a functional integrase. J Virol. 1996;70:3302–3306. doi: 10.1128/jvi.70.5.3302-3306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan M S, Mason A, Coutant R, Chen Q-Y, Vargas A, Rao J, Gomez R, Chalew S, Garry R, Maclaren N K. HERV-K10s and immune-mediated (type 1) diabetes. Cell. 1998;95:14–16. doi: 10.1016/s0092-8674(00)81777-8. [DOI] [PubMed] [Google Scholar]
- 22.Lanchy J-M, Ehresmann C, Le Grice S F J, Ehresmann B, Marquet R. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J. 1997;15:7178–7187. [PMC free article] [PubMed] [Google Scholar]
- 23.Levine K L, Steiner B, Johnson K, Aronoff R, Quinton T J, Linial M L. Unusual features of integrated cDNAs generated by infection with genome-free retroviruses. Mol Carcinog. 1990;10:1891–1900. doi: 10.1128/mcb.10.5.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M D, Lemke T D, Bronson D L, Faras A J. Synthesis and analysis of a 640-bp pol region of novel human endogenous retroviral sequences and their evolutionary relationships. Virology. 1996;217:1–10. doi: 10.1006/viro.1996.0087. [DOI] [PubMed] [Google Scholar]
- 25.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lower R, Tonjes R R, Boller K, Denner J, Kaiser B, Phelps R C, Lower J, Kurth R, Badenhoop K, Donner H, Usadel K H, Miethke T, Lapatschek M, Wagner H. Development of insulin-dependent diabetes mellitus does not depend on specific expression of the human endogenous retrovirus HERV-K. Cell. 1998;95:11–14. doi: 10.1016/s0092-8674(00)81776-6. [DOI] [PubMed] [Google Scholar]
- 27.Maestre J, Tchenio T, Dhellin O, Heidmann T. mRNA retrotransposition in human cells: processed pseudogene formation. EMBO J. 1995;14:595–605. doi: 10.1002/j.1460-2075.1995.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mariani-Costantini R, Horn T, Callahan R. Ancestry of a human endogenous retrovirus family. J Virol. 1989;63:4982–4985. doi: 10.1128/jvi.63.11.4982-4985.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marquet R, Isel C, Ehresmann C, Ehresmann B. tRNAs as primer of reverse transcriptases. Biochimie. 1995;77:113–124. doi: 10.1016/0300-9084(96)88114-4. [DOI] [PubMed] [Google Scholar]
- 30.Mayer J, Meese E, Mueller-Lantzsch N. Multiple human endogenous retrovirus (HERV-K) loci with gag open reading frames in the human genome. Cytogenet Cell Genet. 1997;78:1–5. doi: 10.1159/000134614. [DOI] [PubMed] [Google Scholar]
- 31.Mayer J, Meese E, Mueller-Lantzsch N. Chromosomal assignment of human endogenous retrovirus K (HERV-K) env open reading frames. Cytogenet Cell Genet. 1997;79:157–161. doi: 10.1159/000134709. [DOI] [PubMed] [Google Scholar]
- 32.Mayer J, Meese E, Mueller-Lantzsch N. Human endogenous retrovirus K homologous sequences and their coding capacity in old world primates. J Virol. 1998;72:1870–1875. doi: 10.1128/jvi.72.3.1870-1875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medstrand P, Blomberg J. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol. 1993;67:6778–6787. doi: 10.1128/jvi.67.11.6778-6787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medstrand P, Linderskog M, Blomberg J. Expression of human endogenous retroviral sequences in peripheral blood mononuclear cells of healthy individuals. J Gen Virol. 1992;73:2463–2466. doi: 10.1099/0022-1317-73-9-2463. [DOI] [PubMed] [Google Scholar]
- 35.Murphy V J, Harrison L C, Rudert W A, Luppi P, Trucco M, Fierabracci A, Biro P A, Bottazzo G F. Retroviral superantigens and type 1 diabetes mellitus. Cell. 1998;95:9–11. doi: 10.1016/s0092-8674(00)81775-4. [DOI] [PubMed] [Google Scholar]
- 36.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oude Essink B B, Back N K T, Berkhout B. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 1997;25:3212–3217. doi: 10.1093/nar/25.16.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oude Essink B B, Das A T, Berkhout B. HIV-1 reverse transcriptase discriminates against non-self tRNA primers. J Mol Biol. 1996;264:243–254. doi: 10.1006/jmbi.1996.0638. [DOI] [PubMed] [Google Scholar]
- 39.Owen S M, Lal R B, Ikeda R A. Cloning and expression of a human T-lymphotropic virus type 1 protein with reverse transcriptase activity. J Virol. 1998;72:5279–5284. doi: 10.1128/jvi.72.6.5279-5284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patience C, Simpson G R, Colletta A A, Welch H M, Weiss R A, Boyd M T. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J Virol. 1996;70:2654–2657. doi: 10.1128/jvi.70.4.2654-2657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piechaczyk M, Blanchard J M, Sabouty S R E, Dani S, Marty L, Jeanteur P. Unusual abundance of vertebrate glyceraldehyde-3-phosphate dehydrogenase pseudogenes. Nature. 1984;312:469–470. doi: 10.1038/312469a0. [DOI] [PubMed] [Google Scholar]
- 42.Prasad V R, Lowy I, Santos de los T, Chiang L, Goff S P. Isolation and characterization of a dideoxyguanosine triphosphate-resistant mutant of human immunodeficiency virus reverse transcriptase. Proc Natl Acad Sci USA. 1991;88:11363–11367. doi: 10.1073/pnas.88.24.11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson D L, Hahn B H, Sharp P M. Recombination in AIDS viruses. J Mol Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- 44.Rosenberg N, Jolicoeur P. Retroviral pathogenesis. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 475–586. [PubMed] [Google Scholar]
- 45.Schommer J C, Sauter M, Krausslich H G, Best B, Mueller-Lantzsch N. Characterization of the human endogenous retrovirus K proteinase. J Gen Virol. 1996;77:375–379. doi: 10.1099/0022-1317-77-2-375. [DOI] [PubMed] [Google Scholar]
- 46.Seifarth W, Skladny H, Krieg-Schneider F, Reichert A, Hehlmann R, Leib-Mosch C. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J Virol. 1995;69:6408–6416. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simpson G R, Patience C, Lower R, Tonjes R R, Moore H D M, Weiss R A, Boyd M T. Endogenous D-type (HERV-K) related sequences are packaged into retroviral particles in the placenta and possess open reading frames for reverse transcriptase. Virology. 1996;222:451–456. doi: 10.1006/viro.1996.0443. [DOI] [PubMed] [Google Scholar]
- 48.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 49.Tchenio T, Segal-Bendirdjian E, Heidmann T. Generation of processed pseudogenes in murine cells. EMBO J. 1993;12:1487–1497. doi: 10.1002/j.1460-2075.1993.tb05792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telesnitsky A, Goff S P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 51.Tonjes R R, Boller K, Limbach C, Lugert R, Kurth R. Characterization of human endogenous retrovirus type K virus-like particles generated from recombinant baculovirus. Virology. 1997;233:280–291. doi: 10.1006/viro.1997.8614. [DOI] [PubMed] [Google Scholar]
- 52.Wilkinson D A, Mager D L, Leong J-A. Endogenous human retroviruses. In: Levy J A, editor. The Retroviridae. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]
- 53.Zsíros J, Jebbink M F, Lukashov V V, Voûte P A, Berkhout B. Evolutionary relationships within a subgroup of HERV-K-related human endogenous retroviruses. J Gen Virol. 1998;79:61–70. doi: 10.1099/0022-1317-79-1-61. [DOI] [PubMed] [Google Scholar]
- 54.Zsíros J, Jebbink M F, Lukashov V V, Voûte P A, Berkhout B. Biased nucleotide composition of the genome of HERV-K-related endogenous retroviruses and its evolutionary implications. J Mol Evol. 1999;48:102–111. doi: 10.1007/pl00006437. [DOI] [PubMed] [Google Scholar]
- 55.Zsíros J, Jebbink M F, Voûte P A, Berkhout B. Identification of novel human endogenous retroviral sequences belonging to the HERV-K family. AIDS Res Hum Retroviruses. 1998;14:1093–1098. doi: 10.1089/aid.1998.14.1093. [DOI] [PubMed] [Google Scholar]