Abstract
The gut microbiota consistently shows strong correlations with lipid metabolism in humans and animals, and whether the gut microbiota contributes to muscle fatty acid (FA) deposition and meat traits in farm animals has not been fully resolved. In this study, we aimed to unveil the microbial mechanisms underlying muscle FA deposition in pigs. First, we systematically revealed the correlation between the gut microbiome and muscle FA levels in 43 obese Ningxiang pigs and 50 lean Duroc Landrace Yorkshire (DLY) pigs. Mutual fecal microbial transplantation showed that the obese Ningxiang pig-derived microbiota increased the muscle FA content and improved meat quality by reshaping the gut microbial composition in lean DLY pigs. Lactobacillus reuteri has been identified as a potential microbial biomarker in obese Ningxiang pig-derived microbiota-challenged DLY pigs. A gavage experiment using lean DLY pigs confirmed that L. reuteri XL0930 isolated from obese Ningxiang pigs was the causal species that increased the muscle FA content. Mechanistically, SLC22A5, a carnitine transporter, was downregulated in L. reuteri XL0930-fed DLY pigs, resulting in reduced muscle carnitine levels. Muscle and intestinal L-carnitine levels, which correlated with the muscle FA content, impeded fat synthesis and FA accumulation in in vitro and in vivo models. In conclusion, we uncovered an unexpected and important role of the obese Ningxiang pig-derived microbiota in regulating muscle FA metabolism via the SLC22A5-mediated carnitine system.
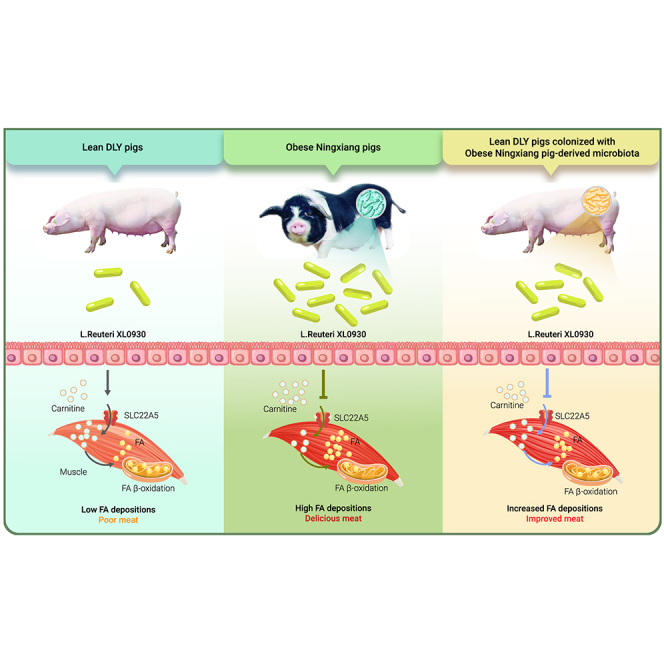
Graphical abstract
Public summary
-
•
The obese Ningxiang pig-derived microbiota increases the muscle FA content and improves meat quality in lean DLY pigs.
-
•
L. reuteri XL0930 isolated from obese Ningxiang pigs is the causal species that increases muscle FA levels.
-
•
The SLC22A5-mediated carnitine system contributes to the role of the obese Ningxiang pig-derived microbiota in muscle FA deposition.
Introduction
Meat quality, an economically important trait in the pig industry, is a determining factor in the overall acceptability of eating quality. Meat quality assessment includes the intramuscular fat content, marbling scores, moisture content, drip loss, color scores, and pH values, which are highly associated with the muscle fatty acid (FA) composition.1,2 The most notable correlation is observed in the positive relationship between the intramuscular fat content and saturated fatty acids (SFAs) in most pig breeds, while the intramuscular fat content, marbling score, and back fat thickness are negatively correlated with polyunsaturated fatty acids (PUFAs) due to their oxidizable nature.3 Thus, the FA composition determines meat quality and is closely related to the nutrition and eating quality of pork.4
Muscle FA composition is governed by heredity, age, diet, feeding environment, and the gut microbiota. In recent decades, the role of the gut microbiota has been widely confirmed in host metabolism, and germ-free and antibiotic-challenged animals are resistant to diet-induced lipid uptake and deposition.5,6,7 Previously, we also observed diverse gut microbial profiles between obese Shaziling pigs and lean Yorkshire pigs,8 which may be associated with metabolic phenotypes. Indeed, obese Jinhua pigs are characterized by a higher abundance of archaeal species related to more efficient fat deposition.9 However, it remains unclear whether and which gut microbiota species contribute to the gut microbiota-mediated muscle lipid accumulation and improvement in meat quality.
Here, we unveiled the relationship among the gut microbiota, muscle FA content, and meat quality by performing an association study between 43 obese Ningxiang pigs (a native Chinese breed) and 50 commercial lean Duroc Landrace Yorkshire (DLY) pigs. To further confirm whether the gut microbiome from Ningxiang pigs or DLY pigs governs swine fat metabolism, mutual fecal microbiota transplantation was conducted, and increased muscle FA contents and meat quality with shifts in the gut microbiome were observed in lean DLY pigs that received Ningxiang pig-derived microbiota. Lactobacillus reuteri has been identified as a potential microbial biomarker in obese Ningxiang pig-derived microbiota-challenged DLY pigs. Colonization of L. reuteri XL0930 isolated from obese Ningxiang pigs increased the muscle FA content in lean DLY pigs. Metabolomics indicated that L-carnitine and its derivatives were involved in muscle FA deposition and meat quality by targeting the SLC22A5 transporter.
Results
Muscle FA levels and the mucosal microbiota differed between obese Ningxiang pigs and lean DLY pigs
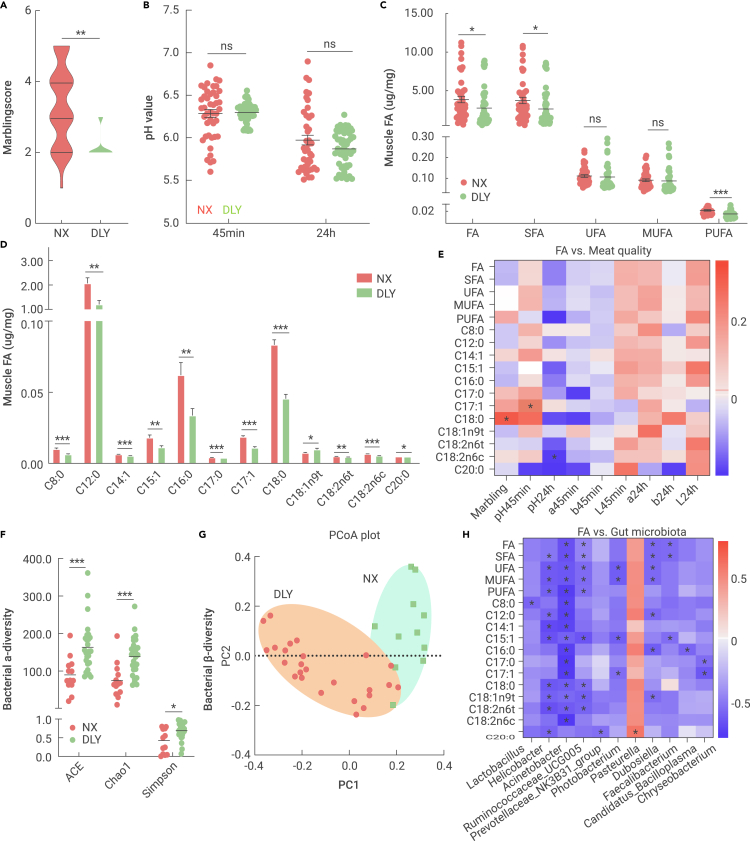
The marbling score (an index correlating intramuscular fat with meat quality) was first analyzed from 43 local fatty Ningxiang slaughtering pigs and 50 commercial lean DLY slaughtering pigs, and the Ningxiang pigs were predicted to have higher marbling scores (Figure 1A). The post-slaughter pH value of the muscle is a key index of meat quality reflecting oxidative rancidity, but no significant differences were noted for the pH values at 45 min and 24 h post slaughter between the two breeds (Figure 1C). Consistent with the marbling score, total FAs (TFAs), SFAs, and PUFAs were enriched in the longissimus muscle of Ningxiang pigs compared with the DLY subjects, but the unsaturated fatty acid (UFA) and monounsaturated fatty acid (MUFA) concentrations did not change (Figure 1C). Twelve FAs mainly contributed to the high TFA content in Ningxiang pigs, namely, C8:0, C12:0, C14:1, C15:1, C16:0, C17:0, C17:1, C18:0, C18:1n9t, C18:2n6t, C18:2n6c, and C20:0 (Figure 1D). We further performed Pearson correlation analyses between the differential FAs and meat quality in Ningxiang pigs and identified three associations: positive correlations for C17:1 (pH45min) and C18:0 (marbling score) and a negative correlation for C18:2n6c (pH24h) (Figure 1D). Together, obese Ningxiang pigs have higher muscle FA contents, which might partially explain the superior meat quality of Chinese indigenous pig breeds.
Figure 1.
Muscle FA and mucosal microbiota that were differentially expressed between obese Ningxiang pigs (NX) and lean DLY pigs
The longissimus muscle and ileal mucosa samples from 43 obese Ningxiang pigs and 50 commercial lean DLY pigs were collected for FA and gut microbiota analyses.
(A) Marbling score.
(B) pH value of muscle.
(C) Muscle FA concentrations.
(D) Differential FAs between Ningxiang pigs and DLY pigs.
(E) Pearson correlation analysis between FAs and meat quality.
(F) Bacterial α-diversity (ACE, Chao1, and Simpson indices).
(G) Bacterial β-diversity (PCoA plot).
(H) Marked associations between the top 100 genera and muscle FAs.
Our previous study indicated that the gut microbiota is involved in lipid metabolism in local fatty pigs.8 Thus, we further sequenced the mucosal microbiota in the ileum using full-length 16S rRNA between obese Ningxiang and lean DLY pigs. An average of 12,555 circular consensus sequence reads per sample were generated for species annotation and abundance analysis. Obese Ningxiang pigs exhibited a lowered α-diversity (Simpson, Chao1, and ACE indices) (Figure 1F), which was widely consistent in obese animal models. β-Diversity analysis revealed that the mucosal microbiota of the two pig breeds clustered separately along the principal coordinate axis (Figure 1G), indicating a diverse microbial community between the obese and lean breeds. Next, we analyzed the top 10 bacterial phyla and found that Firmicutes and Proteobacteria were the most predominant phyla in the two breeds (data not shown). The relative abundances of Firmicutes, Bacteroidetes, Tenericutes, Spirochaetes, Fusobacteria, and Kiritimatiellaeota were lower, whereas the abundance of Proteobacteria was markedly enriched in obese Ningxiang pigs (data not shown). We further used Pearson correlation analyses to identify the potential relationship between muscle FA levels and the gut microbiota at the genus level, and 11 out of the top 100 genera were markedly correlated with the muscle FA content (Figure 1H), including the differentially expressed Acinetobacter, Ruminococcaceae_UCG005, Prevotellaceae_NK3B31_group, Photobacterium, and Pasteurella (data not shown). Together, these results revealed a diverse gut bacterial profile between obese Ningxiang pigs and lean DLY pigs, which might be associated with the higher deposition of muscle FAs in Ningxiang pigs.
Obese Ningxiang pig-derived microbiota improved meat quality in lean DLY pigs
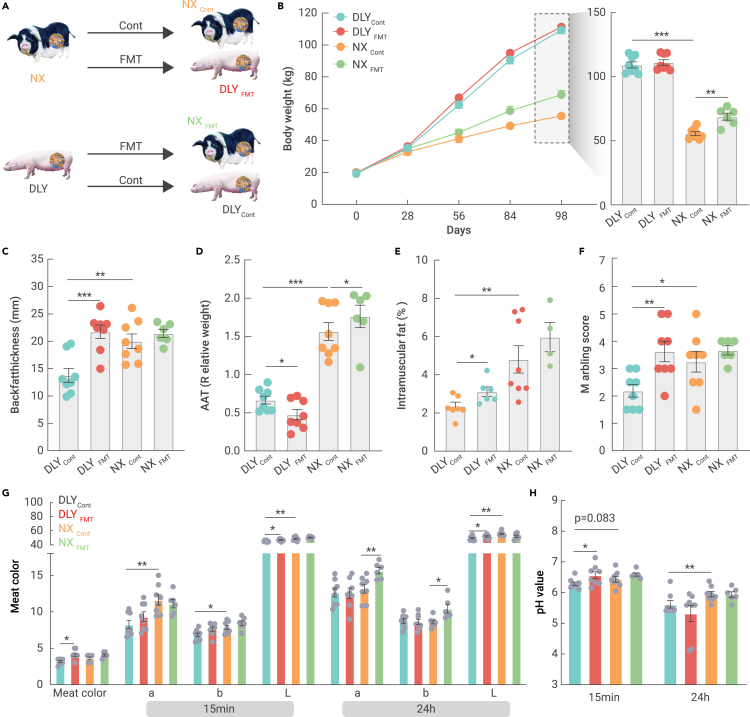
To validate the role of the gut microbiota in lipid deposition and meat quality, we conducted mutual fecal microbiota transplantation between obese and lean pigs for 16 weeks (Figure 2A). The body weight of Ningxiang pigs was lower than that of DLY pigs, while transplantation with DLY fecal microbiota markedly increased the body weight of Ningxiang pigs (Figure 2B). High lipid accumulation was observed in Ningxiang pigs, as evidenced by the increased backfat thickness (also named subcutaneous adipose tissue), abdominal adipose tissue (AAT) weight, and intramuscular fat compared to those of DLY pigs (Figures 2C–2E). We also noticed an enhancement of the backfat thickness and intramuscular fat content in fecal microbiota-transplanted DLY pigs (Figures 2C and 2E), while the relative weight of the AAT was reduced (Figure 2D).
Figure 2.
Obese Ningxiang pig-derived microbiota improved meat quality in lean DLY pigs
Sixteen male DLY and 14 male Ningxiang pigs underwent mutual fecal microbiota transplantation for 14 weeks.
(A) Experimental design for the mutual fecal microbiota transplantation.
(B) Body weight.
(C) Backfat thickness (mm).
(D) Relative weight of the AAT (%).
(E) Intramuscular fat (%).
(F) Marbling score.
(G) Meat color.
(H) pH values.
Next, we confirmed the better meat quality in Ningxiang pigs, as evidenced by the higher marbling score, meat color (a, b, and L), and pH (24 h) indices (Figures 2G and 2H). Interestingly, fecal microbiota transplantation from obese Ningxiang pigs markedly increased the marbling score, meat color, and pH (15 min) indices in the muscle of lean DLY pigs. We also noticed an improvement in the a24h and b24h indices (Figure 2G). Together, the current data revealed an effective means to improve meat quality, especially for commercial lean DLY pigs, via fecal microbiota transplantation.
Fecal microbiota transplantation shifted FA absorption and accumulation
FA accumulation in muscles is strongly associated with intramuscular fat and meat quality. Obese Ningxiang pigs had a higher FA content, including SFAs and USFAs (Figure S1A). Although meat quality was markedly improved after fecal microbiota transplantation, there was only a slight increase in the muscle FA content of fecal microbiota-transplanted DLY pigs. Next, we analyzed the FA types and compositions and found that seven SFAs (C6:0, C11:0, C13:0, C21:0, C22:0, C23:0, and C24:0) and five USFAs (C20:3n3, C20:5n3, C22:1n9, C22:6n3, and C24:1) (Figure S1A) were enhanced in response to fecal microbiota transplantation in DLY pigs. However, C21:0, C20:3n6, and C20:4n6 decreased in fecal microbiota-transplanted Ningxiang pigs. We further quantified lipid metabolism-related genes in the muscle but did not find any difference between the DLY and fecal microbiota-transplanted DLY pigs (Figure S1B).
The major source of body FA is dietary fat, which is digested and absorbed in the intestine. Thus, we analyzed intestinal genes related to FA uptake and found that CD36, an FA sensor and transporter,10 was expressed at a higher level in the jejunum and ileum of obese Ningxiang pigs (Figure S1C). Interestingly, fecal microbiota transplantation markedly upregulated DGAT1 in lean DLY pigs and inhibited jejunal DGAT1, MOGAT2, and ileal CD36 expression in the fecal microbiota-transplanted Ningxiang pigs. The unabsorbed FAs in the distal ileum were slightly reduced in the fecal microbiota of transplanted DLY pigs (Figure S1D), which might explain the higher FA content in the muscle. Indeed, receiving fecal microbiota from obese Ningxiang pigs tended to decrease unabsorbed FAs (such as C15:0) in lean DLY pigs, indicating an improvement in FA uptake. However, C22:6n3 and C24:1 were not detected in DLY pigs, while both were increased in microbiota-transplanted DLY pigs, and C22:6n3 was decreased in microbiota-transplanted Ningxiang pigs (data not shown). Further studies are needed to confirm the role of the gut microbiota in intestinal FA absorption in obese Ningxiang pigs, especially for C22:6n3 and C24:1.
Fecal microbiota transplantation shaped the gut microbiome in Ningxiang and DLY pigs
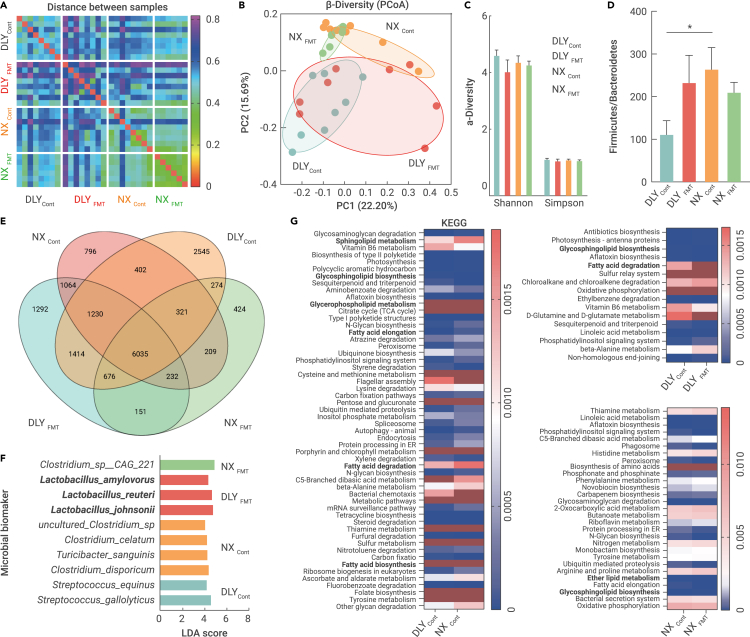
We further performed shotgun metagenomic sequencing to analyze the gut microbial profiles in the ileum after 16 weeks of fecal microbiota transplantation, and an average of 24,233,489 clean reads per sample were generated for splicing, assembly, and prediction of coding genes. We first performed a distance heatmap analysis using the binary Jaccard tool at the species level on the BMKCloud platform (www.biocloud.net), and distance differences were observed in the four datasets (Figure 3A). The β-diversity analysis also showed two different microbial clusters in the DLYcont and NXcont groups, while fecal microbiota transplantation tended to reprogram the microbial compositions according to the principal coordinate analysis (Figure 3B). However, we did not observe any differences in diversity (Shannon and Simpson indices) (Figure 3C). Here, the Firmicutes and Bacteroidetes abundances were not altered, but a higher ratio of Firmicutes to Bacteroidetes was observed in obese Ningxiang pigs (NXcont), and fecal microbiota transplantation tended to enhance this ratio in lean DLY pigs (DLYFMT) (Figure 3D). Venn analysis showed that obese Ningxiang pigs and lean DLY pigs co-colonized 7,988 species, whereas 2,301 species were present only in obese Ningxiang pigs. A total of 8,561 species were found in both Ningxiang pigs and fecal microbiota-transplanted DLY pigs; thus, the 573 species might be derived from Ningxiang pigs (Figure 3E). Next, linear discriminant analysis effect size (LEfSe) (LDA score >4) was further used to identify the microbial biomarkers and functional capacities of the gut microbiome with significantly different abundances between the four datasets, and Streptococcus gallolyticus, Streptococcus equinus (DLYcont), Clostridium disporicum, Turicibacter sanguinis, Clostridium celatum, uncultured Clostridium_sp (NXcont), Lactobacillus johnsonii, L. reuteri, Lactobacillus amylovorus (DLYFMT), and Clostridium_sp__CAG_221 (NXFMT) were enriched (Figure 3F). Microbial functional annotations identified by KEGG analysis revealed six lipid metabolic pathways, including sphingolipid metabolism, glycosphingolipid biosynthesis, glycerophospholipid metabolism, FA elongation, FA degradation, and FA biosynthesis (Figure 3G). Together, we proposed that obese Ningxiang pig-derived microbiota transplantation helped the conversion of the DLY gut microbiota to the microbial phenotype of Ningxiang pigs, which might further reprogram muscle FA deposition in DLY pigs.
Figure 3.
Fecal microbiota transplantation shaped the gut microbiome in Ningxiang and DLY pigs
Ileal chyme samples were collected from the fecal microbiota of obese Ningxiang and lean DLY pigs for metagenomic sequencing.
(A) Distance between samples.
(B) PCoA plot.
(C) α-Diversity (Shannon and Simpson indices).
(D) Microbial abundance.
(E) Venn analysis.
(F) Microbial biomarkers by LEfSe.
(G) Microbial functional annotations by KEGG analysis.
Obese Ningxiang pig-derived L. reuteri increased the muscle FA content in lean DLY pigs
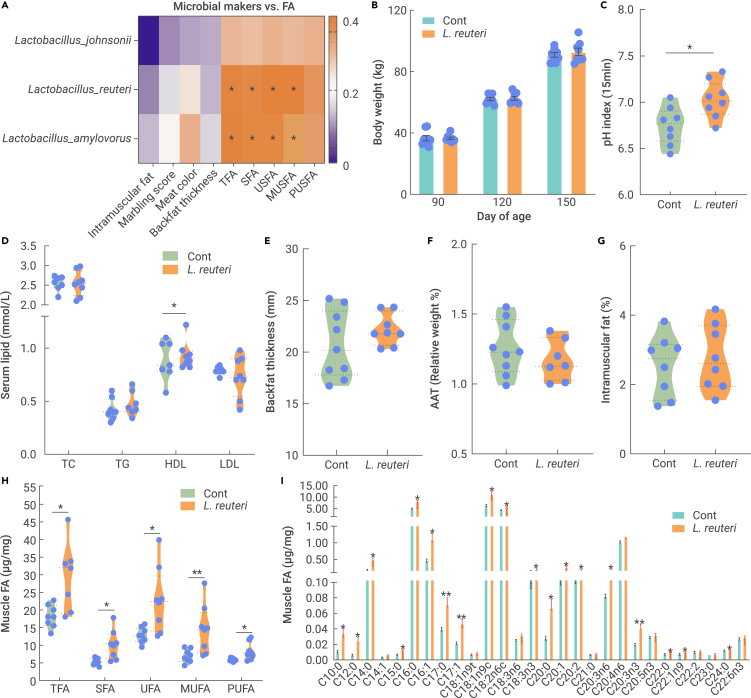
L. johnsonii, L. reuteri, and L. amylovorus were identified as microbial biomarkers for fecal microbiota-transplanted DLY pigs (DLYFMT) (Figure 3F), and Pearson correlation analysis (p value < 0.1) was performed to reveal the biomarker associations with meat quality and muscle FA levels. Eight associations were identified with positive correlations between L. reuteri and L. amylovorus and muscle FAs (TFAs, SFAs, USFAs, and MUSFAs) (Figure 4A). Since L. amylovorus failed to enhance muscle FA levels in a murine model (data not shown), the role of obese Ningxiang pig-derived L. reuteri was the main focus of this study.
Figure 4.
Obese Ningxiang pig-derived L. reuteri increased muscle fatty acids in lean DLY pigs
DLY pigs (90 days old) were orally administered 1 × 1011 CFU Ningxiang pig-derived L. reuteri XL0930 every day for 60 days (n = 8-9).
(A) Correlations between muscle FA levels and microbial biomarkers.
(B) Body weight of L. reuteri XL0930-fed pigs.
(C) pH value of muscle.
(D) Serum lipid levels.
(E) Backfat thickness (mm).
(F) Relative weight of the AAT (%).
(G) Intramuscular fat.
(H and I) Muscle FAs (μg/mg).
We colonized Ningxiang pig-derived L. reuteri XL0930 in DLY pigs (90 days of age) for 2 months, while body weight gain was not altered (Figure 4B). Interestingly, the pH value (15 min) and serum HDL level were markedly increased in L. reuteri XL0930-fed DLY pigs, indicating a role in the improvement in muscle oxidative rancidity and enhancement of lipid metabolism (Figures 4C and 4D). Backfat thickness, the relative weight of the AAT, and intramuscular fat were not enhanced (Figures 4E–4G), whereas L. reuteri XL0930 treatment significantly increased the muscle FA content (TFAs, SFAs, USFAs, MUFAs, and PUFAs) (Figure 4H), which was consistent with the correlations between L. reuteri and muscle FAs. Specifically, C10:0, C12:0, C14:0, C15:0, C16:0, C16:1, C17:0, C17:1, C18:1n9t, C18:2n6c, C18:3n3, C20:0, C20:1, C20:2, C20:3n6, C20:3n3, C22:0, C22:1n9, and C24:0 were increased in L. reuteri XL0930-fed DLY pigs (Figure 4I), indicating that obese Ningxiang pig-derived L. reuteri XL0930 promoted FA metabolism or absorption. Indeed, we found that supplementation with L. reuteri XL0930 downregulated AAC and LXRβ expression in the subcutaneous adipose tissue (SAT) but did not affect intestinal FA absorption-related genes (Figure S2).
Obese Ningxiang pig-derived microbiota reprogrammed L-carnitine metabolism in lean DLY pigs
Gut microbial metabolites and components act as signaling molecules to regulate the lipid-related endocrine system or act directly on metabolic organs.11 Thus, muscle metabolic profiles were further unveiled by LC-MS untargeted metabolomics (Shanghai Majorbio Bio-pharm Technology Co.). Partial least squares discriminant analysis (PLS-DA) was performed using the free online platform Majorbio Cloud Platform (www.majorbio.com), and the results revealed four clusters (Figure S3A). Seventy-six differentiated metabolites (p value < 0.05) were identified, with 46 enriched metabolites in obese Ningxiang pigs and 30 enriched metabolites in lean DLY pigs (Figure S3B). Considering that meat quality and the muscle FA content were notably improved in microbiota-transplanted DLY pigs, we mainly focused on the 37 differentiated metabolites between the DLYcont and DLYFMT datasets (Figure S3C). Interestingly, L-carnitine and its derivatives (i.e., 3-hydroxyhexadecanoyl carnitine, 3-hydroxyoctanoyl carnitine, (+/−)-myristoylcarnitine, (R)-3-hydroxybutyrylcarnitine, and DL-stearoylcarnitine) were enriched in obese Ningxiang pigs, while Ningxiang pig-derived fecal microbiota transplantation exhibited a markedly increased 3-hydroxyoctanoyl carnitine abundance in lean DLY pigs, which might be associated with lipid accumulation in microbiota-transplanted lean DLY pigs.
To evaluate whether muscle L-carnitine and its derivatives were derived from the gut microbiota, intestinal metabolic profiles were further studied (Figure S3D). The PLS-DA plot showed an obvious difference among the four clusters (Figure S3E), and 58 metabolites were differentially expressed between the Ningxiang and DLY pigs (Figure S3D). Consistently, L-carnitine and its derivatives were enriched in obese Ningxiang pigs (hydroxyisovaleroyl carnitine and acetylcarnitine) (Figure S3D) and microbiota-transplanted DLY pigs (3-hydroxydodecanoyl carnitine, DL-stearoylcarnitine, heptadecanoyl carnitine, 9,12-hexadecadienoylcarnitine, alpha-linolenyl carnitine, (+/−)-myristoylcarnitine, decanoyl-L-carnitine, and 3-hydroxyhexadecanoyl carnitine) (Figure S3F).
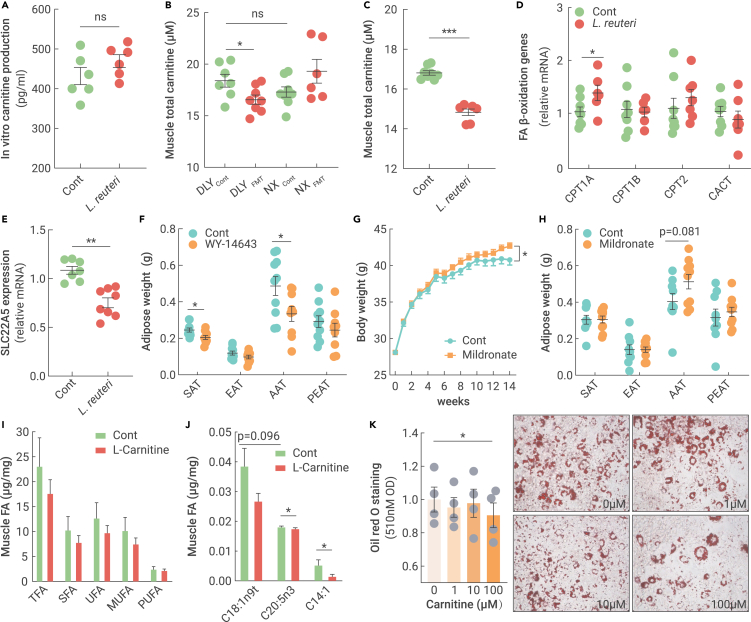
We next analyzed whether L-carnitine and its derivatives were from the gut microbiota, especially L. reuteri, and two positive associations between L. reuteri and intestinal L-carnitine derivatives (alpha-linolenyl carnitine and pentadecanoylcarnitine) were identified (Figure S4A). However, most muscle L-carnitine derivatives were negatively correlated with the abundance of L. reuteri, especially 2-methylbutyrylcarnitine and palmito-L-carnitine (Figure S4B). In vitro L-carnitine production from L. reuteri was further validated, and the L-carnitine content was not significantly increased in the culture medium (Figure 5A). Taken together, the results revealed that L. reuteri did not directly contribute to the changes in L-carnitine and its derivatives, and the alterations in muscle L-carnitine and its derivatives need to be further investigated.
Figure 5.
Fecal microbiota transplantation reprogrammed the muscle and intestinal metabolic profiles in Ningxiang and DLY pigs
(A) Carnitine production of L. reuteri XL0930 in vitro (n = 6).
(B) Muscle total carnitine content in fecal microbiota-transplanted obese Ningxiang pigs and lean DLY pigs.
(C) Muscle total carnitine content in L. reuteri XL0930-fed DLY pigs.
(D) FA β-oxidation-related genes in L. reuteri XL0930-fed DLY pigs.
(E) SLC22A5 expression in L. reuteri XL0930-fed DLY pigs.
(F) WY-14643 reduced fat deposition in mice (n = 10).
(G) Mildronate increased body weight in mice (n = 10).
(H) Mildronate tended to increase fat deposition in mice (n = 10).
(I and J) Muscle FA content in carnitine-fed DLY pigs (n = 8).
(K) Lipid accumulation in carnitine-incubated 3T3-L1 cells (n = 4).
L-carnitine and most derivatives, both in the muscle and intestine, showed a positive correlation with muscle FAs (Figure S4). While carnitine is generally assumed to be necessary for the transfer of long-chain FAs across the inner mitochondrial membrane for subsequent β-oxidation,12 the positive association cannot explain the enhanced muscle FA levels in L. reuteri-challenged DLY pigs. As metabolomics analysis determined only the free L-carnitine level but did not include the bound L-carnitine in organelles, such as mitochondria, we further used ELISA kits to measure the total L-carnitine concentration in the muscle. Compared with lean DLY pigs, the muscle total L-carnitine concentration was slightly lower in obese Ningxiang pigs, while Ningxiang pig-derived fecal microbiota transplantation markedly reduced the total muscle L-carnitine content in lean DLY pigs (Figure 5B). Similarly, oral L. reuteri treatment for two months also reduced the total muscle L-carnitine content (Figure 5C). In summary, total L-carnitine was reduced in response to the Ningxiang pig-derived microbiota.
Obese Ningxiang pig-derived microbiota targeted SLC22A5-mediated carnitine metabolism to govern muscle FA deposition
To identify the initiating events in muscle carnitine alteration, we examined the effects of obese Ningxiang pig-derived L. reuteri on carnitine-mediated FA β-oxidation. Carnitine palmitoyltransferases (CPTs) aid in the mitochondrial import of carnitine for mitochondrial FA β-oxidation, and CPT1 expression in the muscle is negatively correlated with FA deposition in pigs.13,14 However, CPT1 showed higher expression (Figure 5D), which was inconsistent with the increased FA level in L. reuteri-administered DLY pigs (Figure 4H). Next, we analyzed the expression of SLC22A5, a sodium-dependent carnitine transporter responsible for the cellular uptake of carnitine, and we found that oral L. reuteri significantly downregulated SLC22A5 (Figure 5E). Therefore, we proposed that the obese Ningxiang pig-derived microbiota blocked SLC22A5-mediated carnitine uptake, accompanied by a reduction in cellular carnitine.
To confirm that the obese Ningxiang pig-derived microbiota-mediated muscle FA deposition was controlled by SLC22A5 inhibition, we activated SLC22A5 in mice using a specific agonist (40 mg/kg WY-14643).15 Although WY-14643 treatment failed to affect body weight (data not shown), the SAT and AAT weights were markedly reduced, at least in part due to SLC22A5 activation and FA β-oxidation (Figure 5F). Mildronate, a competitive inhibitor of SLC22A5-mediated carnitine uptake,16 was further used to mimic the role of L. reuteri. Predictably, body weight and adipose weight tended to increase in response to the SLC22A5 inhibition-mediated carnitine reduction (Figures 5G and 5H).
Next, we tested the effects of dietary supplementation with 100 mg/kg L-carnitine on muscle FA metabolism in pigs. Dietary L-carnitine tended to reduce muscle FA (Figure 5I), especially UFAs (ie, C144:1, C18:1n9t, and C20:5n3) (Figure 5J). Indeed, 3T3-L1 cells incubated with 100 μM carnitine for 8 days showed markedly reduced lipid deposition without affecting cell viability (Figure 5K). However, we failed to observe a reduction in serum FA concentrations (Figure S5A), indicating that dietary L-carnitine only reduced muscle FA but did not affect gut FA absorption. In addition, the meat quality, pH value, and meat color were not changed (Figures S5B–S5D), and studies on high dosages of L-carnitine and its long-term effects are needed.
Discussion
Germ-free animals, antibiotic exposure, fecal microbiota transplantation, and probiotic treatments have widely confirmed the role of the gut microbiota in host lipid metabolism.17,18 We previously compared gut microbial development between obese Shaziling and lean Yorkshire pig breeds and observed a higher α-diversity and greater abundances of probiotics (i.e., L. johnsonii, L. amylovorus, and Clostridium butyricum) in fatty Shaziling pigs.8 Here, 43 obese Ningxiang pigs and 50 lean DLY pigs were screened, and 11 of the top 100 genera were identified to be correlated with muscle FAs. Fat deposition traits, intramuscular fat, and meat quality were the most relevant traits for FA composition in the genetic correlations of 2,448 pigs.3 These results indicated that the gut microbiota consistently showed strong correlations with muscle FA levels and meat traits in pigs.
Microbial differences have been confirmed between obese pigs and lean pigs, and whether the gut microbiota contributes to FA metabolism and meat traits is of great interest in animal science. Wu et al. transplanted the fecal microbiota of obese Jinhua pigs into antibiotic-treated mice, and the recipients exhibited a similar obese phenotype with elevated lipid and triglyceride levels.19 Here, we conducted mutual fecal microbiota transplantation between obese Ningxiang pigs and lean DLY pigs and found that backfat thickness and intramuscular fat were increased in fecal microbiota-transplanted DLY pigs with an improvement in meat quality. Zhang et al. reported positive correlations between the intramuscular fat content and the pH values of the longissimus muscle and SFAs.3 Consistently, we also observed a marked increase in seven SFAs (C6:0, C11:0, C13:0, C21:0, C22:0, C23:0, and C24:0) in obese Ningxiang pig-derived microbiota-challenged lean DLY pigs, which might explain the improvement in meat quality. The major source of body FA is dietary fat, which is digested and absorbed in the intestine. Obese Ningxiang pig-derived microbiota tended to enhance the intestinal transporting ability of FAs, with a reduction in the levels of unabsorbed FAs in the distal ileum. Germ-free mice are characterized by impaired fat digestion and absorption within the small intestine, while lipid transport is re-established after conventionalization with microbiota.20 Therefore, we proposed that by fecal microbiota transplantation, gut microbial cues in this region may have a great impact on the manipulation of lipid metabolism and meat quality in the pig industry.
Almost 1,000 species of gut microbiota inhabit the gastrointestinal tract. An unsolved question is which species contribute to obese Ningxiang pig-derived microbiota-mediated muscle lipid accumulation and meat quality improvement. Chen et al. systematically screened the correlation between the gut microbiome and lean meat percentage in 698 commercial Duroc pigs and identified that obese pigs had a significantly higher abundance of Prevotella copri, which was correlated with fat accumulation and the metabolites associated with obesity.21 In this study, we identified L. reuteri in obese Ningxiang pig-derived microbiota-challenged DLY pigs. Interestingly, a high relative abundance of L. reuteri was reported to be associated with a greater risk of obesity in several cross-sectional studies.22,23 Here, obese Ningxiang pig-derived L. reuteri XL0930 was first isolated, and its colonization was demonstrated to enhance the muscle FA content in lean DLY pigs. Based on these observations, we predicted that gut microbial manipulation, especially of L. reuteri, would be effective in improving meat quality. However, we failed to observe a marked improvement in the meat quality of L. reuteri-fed DLY pigs except for the pH index; thus, studies including high dosages and long-term effects are needed to validate the merit of L. reuteri in the pig industry. Several studies have also reported an anti-obesity effect of L. reuteri in diet-induced obesity via energy metabolic remodeling and gut microbial reshaping.24,25,26,27,28 The reason might be associated with strain specificity, and whole-genome sequencing of these strains is needed to unveil the inconsistent results and to guide clinical application.
The mechanism of the gut microbiota has been widely proposed to be the production of a myriad of metabolites.29 For example, lipid uptake and accumulation are enhanced in Desulfovibrio species cell-free supernatant-cultured intestinal epithelial cells, whose species have been demonstrated to favor intestinal FA absorption.30 Nevertheless, the specific metabolites that contribute to L. reuteri XL0930-mediated FA deposition are still elusive. Fortunately, free L-carnitine and its derivatives have been identified to correlate with muscle FAs, and lower contents of total L-carnitine have been confirmed in Ningxiang pig-derived microbiota or L. reuteri XL0930-challenged DLY pigs. Dietary L-carnitine tended to reduce muscle FAs in lean DLY pigs. Indeed, carnitine is generally assumed to be necessary for the transfer of long-chain FAs across the inner mitochondrial membrane for subsequent β-oxidation12; thus, the decreased level of L-carnitine seems to explain the higher FA accumulation due to the reduced FA β-oxidation.31 However, we did not observe a reduction in FA β-oxidation-related genes in L. reuteri-treated DLY pigs, indicating that there is no causality between the reduced muscle carnitine and FA β-oxidation in DLY pigs. Defects in the SLC22A5 carnitine transporter are characterized by decreased intracellular carnitine accumulation, increased loss of carnitine in the urine, and low serum carnitine levels.12,32 We also confirmed the role of SLC22A5 in obese Ningxiang pig-derived microbiota-mediated total carnitine reduction and FA accumulation in the muscle. Microbial-derived metabolites or miRNAs are widely reported to be the agents responsible for the interaction between the gut microbiota and host metabolism,29,33,34,35 and future studies are needed to identify the potential L. reuteri-derived molecules that regulate SLC22A5, which would provide new insights into the interaction among the obese Ningxiang pig-derived microbiota, SLC22A5-mediated carnitine uptake, and FA deposition in the muscle.
Conclusion
The discovery of a link between the gut microbiota, microbial metabolites, lipid deposition, and meat quality has broad implications for animal husbandry. Our study revealed a new potential correlation between the gut microbiota and muscle FA content (related to meat quality) in obese Ningxiang pigs and lean DLY pigs. Mutual fecal microbiota transplantation between obese Ningxiang pigs and lean DLY pigs suggests a new potential microbial strategy for muscle FA-based meat quality manipulation. Furthermore, our studies isolated obese Ningxiang pig-derived L. reuteri XL0930, which might serve as a novel dietary supplement and additive to improve meat quality in animal production. Our results also suggest that SLC22A5 downregulation resulted in lowered total muscle carnitine levels, which might be the key point between the gut microbiota and muscle FA levels.
Material and methods
See the supplemental information for details.
Data and code availability
The data supporting the conclusions of this article are available in the NCBI Sequence Read Archive (SRA) repositories: PRJNA855271 and PRJNA855286.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (U20A2055 and 32172761), the National Key Research and Development Program of China (2022YFD1301500), and the Hunan Youth Science and Technology Talent Project (2022RC1157).
Author contributions
Y.Y. and X.H. conceived the research idea. J.Y. designed the study and wrote the manuscript. Y.L., Y.T., F.Z., J.M., S.X., and T.Y. conducted the main analysis. L.M. and Q.Z. helped with the sample collection. G.L., J.Y., and Y.Y. reviewed and edited the manuscript.
Declaration of interests
The authors declare that they have no competing interests.
Published Online: July 26, 2023
Footnotes
It can be found online at https://doi.org/10.1016/j.xinn.2023.100486.
Contributor Information
Yulong Yin, Email: yinyulong@isa.ac.cn.
Xingguo Huang, Email: hxg68989@hunau.edu.cn.
Lead contact website
Xingguo Huang, https://ansci.hunau.edu.cn/xswyh/xxswyh/202008/t20200827_286577.html.
Supplemental information
References
- 1.Barbut S. Review: Automation and meat quality-global challenges. Meat Sci. 2014;96:335–345. doi: 10.1016/j.meatsci.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Wu Y., Zhao J., Xu C., et al. Progress towards pig nutrition in the last 27 years. J. Sci. Food Agric. 2020;100:5102–5110. doi: 10.1002/jsfa.9095. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y., Zhang J., Gong H., et al. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat Sci. 2019;150:47–55. doi: 10.1016/j.meatsci.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Wood J.D., Richardson R.I., Nute G.R., et al. Effects of fatty acids on meat quality: a review. Meat Sci. 2004;66:21–32. doi: 10.1016/S0309-1740(03)00022-6. [DOI] [PubMed] [Google Scholar]
- 5.Kimura I., Ozawa K., Inoue D., et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parséus A., Sommer N., Sommer F., et al. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017;66:429–437. doi: 10.1136/gutjnl-2015-310283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin J., Li Y., Han H., et al. Administration of exogenous melatonin improves the diurnal rhythms of the gut microbiota in mice fed a high-fat diet. mSystems. 2020;5:e00002–e00020. doi: 10.1128/mSystems.00002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J., Duan Y., Li R., et al. Gut microbial profiles and the role in lipid metabolism in Shaziling pigs. Anim. Nutr. 2022;9:345–356. doi: 10.1016/j.aninu.2021.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao G., Xiang Y., Wang X., et al. Exploring the possible link between the gut microbiome and fat deposition in pigs. Oxid. Med. Cell. Longev. 2022;2022 doi: 10.1155/2022/1098892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Huang X., Yang G., et al. CD36 favours fat sensing and transport to govern lipid metabolism. Prog. Lipid Res. 2022;88 doi: 10.1016/j.plipres.2022.101193. [DOI] [PubMed] [Google Scholar]
- 11.Han H., Yi B., Zhong R., et al. From gut microbiota to host appetite: gut microbiota-derived metabolites as key regulators. Microbiome. 2021;9:162. doi: 10.1186/s40168-021-01093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longo N., Frigeni M., Pasquali M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta. 2016;1863:2422–2435. doi: 10.1016/j.bbamcr.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao S.M., Ren L.J., Chen L., et al. Differential expression of lipid metabolism related genes in porcine muscle tissue leading to different intramuscular fat feposition. Lipids. 2009;44:1029–1037. doi: 10.1007/s11745-009-3356-9. [DOI] [PubMed] [Google Scholar]
- 14.Nong Q., Wang L., Zhou Y., et al. Low dietary n-6/n-3 PUFA ratio regulates meat quality, reduces triglyceride content, and improves fatty acid composition of meat in Heigai pigs. Animals. 2020;10:1543. doi: 10.3390/ani10091543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Vlies N., Ferdinandusse S., Turkenburg M., et al. PPAR alpha-activation results in enhanced carnitine biosynthesis and OCTN2-mediated hepatic carnitine accumulation. Biochim. Biophys. Acta. 2007;1767:1134–1142. doi: 10.1016/j.bbabio.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Grube M., Meyer zu Schwabedissen H.E.U., Präger D., et al. Uptake of cardiovascular drugs into the human heart: expression, regulation, and function of the carnitine transporter OCTN2 (SLC22A5) Circulation. 2006;113:1114–1122. doi: 10.1161/CIRCULATIONAHA.105.586107. [DOI] [PubMed] [Google Scholar]
- 17.Bäckhed F., Manchester J.K., Semenkovich C.F., et al. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin J., Li Y., Han H., et al. Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice. J. Pineal Res. 2018;65 doi: 10.1111/jpi.12524. [DOI] [PubMed] [Google Scholar]
- 19.Wu C., Lyu W., Hong Q., et al. Gut microbiota influence lipid metabolism of dkeletal muscle in pigs. Front. Nutr. 2021;8 doi: 10.3389/fnut.2021.675445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Guryn K., Hubert N., Frazier K., et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe. 2018;23:458–469.e5. doi: 10.1016/j.chom.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C., Fang S., Wei H., et al. Prevotella copri increases fat accumulation in pigs fed with formula diets. Microbiome. 2021;9:175. doi: 10.1186/s40168-021-01110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castañeda-Márquez A.C., Díaz-Benítez C.E., Bahena-Roman M., et al. Lactobacillus paracasei as a protective factor of obesity induced by an unhealthy diet in children. Obes. Res. Clin. Pract. 2020;14:271–278. doi: 10.1016/j.orcp.2020.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Million M., Maraninchi M., Henry M., et al. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. 2012;36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Chen L.H., Chen Y.H., Cheng K.C., et al. Antiobesity effect of Lactobacillus reuteri 263 associated with energy metabolism remodeling of white adipose tissue in high-energy-diet-fed rats. J. Nutr. Biochem. 2018;54:87–94. doi: 10.1016/j.jnutbio.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Choi S.I., You S., Kim S., et al. Weissella cibaria MG5285 and Lactobacillus reuteri MG5149 attenuated fat accumulation in adipose and hepatic steatosis in high-fat diet-induced C57BL/6J obese mice. Food Nutr. Res. 2021;65 doi: 10.29219/fnr.v65.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang B., Zheng F., Stanton C., et al. Lactobacillus reuteri FYNLJ109L1 attenuating metabolic syndrome in mice via gut microbiota modulation and slleviating inflammation. Foods. 2021;10:2081. doi: 10.3390/foods10092081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C., Fang R., Lu X., et al. Lactobacillus reuteri J1 prevents obesity by altering the gut microbiota and regulating bile acid metabolism in obese mice. Food Funct. 2022;13:6688–6701. doi: 10.1039/d1fo04387k. [DOI] [PubMed] [Google Scholar]
- 28.Zheng F., Wang Z., Stanton C., et al. Lactobacillus rhamnosus FJSYC4-1 and Lactobacillus reuteri FGSZY33L6 alleviate metabolic syndrome via gut microbiota regulation. Food Funct. 2021;12:3919–3930. doi: 10.1039/d0fo02879g. [DOI] [PubMed] [Google Scholar]
- 29.Krautkramer K.A., Fan J., Bäckhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 2021;19:77–94. doi: 10.1038/s41579-020-0438-4. [DOI] [PubMed] [Google Scholar]
- 30.Petersen C., Bell R., Klag K.A., et al. T cell-mediated regulation of the microbiota protects against obesity. Science. 2019;365:eaat9351. doi: 10.1126/science.aat9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeukendrup A.E., Randell R. Fat burners: nutrition supplements that increase fat metabolism. Obes. Rev. 2011;12:841–851. doi: 10.1111/j.1467-789X.2011.00908.x. [DOI] [PubMed] [Google Scholar]
- 32.Longo N., Amat di San Filippo C., Pasquali M. Disorders of carnitine transport and the carnitine cycle. Am. J. Med. Genet. C Semin. Med. Genet. 2006;142c:77–85. doi: 10.1002/ajmg.c.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agus A., Clément K., Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 2021;70:1174–1182. doi: 10.1136/gutjnl-2020-323071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu B.B., Gibson T.E., Yeliseyev V., et al. Dynamic Modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe. 2019;25:803–814.e5. doi: 10.1016/j.chom.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maruvada P., Leone V., Kaplan L.M., et al. The human microbiome and obesity: moving beyond sssociations. Cell Host Microbe. 2017;22:589–599. doi: 10.1016/j.chom.2017.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the conclusions of this article are available in the NCBI Sequence Read Archive (SRA) repositories: PRJNA855271 and PRJNA855286.