Abstract
A 4.7-kb region of DNA sequence contained at the right end of the myxoma virus EcoRI-G2 fragment located 24 kb from the right end of the 163-kb genome has been determined. This region of the myxoma virus genome encodes homologs of the vaccinia virus genes A51R, A52R, A55R, A56R, and B1R; the myxoma virus gene equivalents have been given the prefix M. The MA55 gene encodes a protein belonging to the kelch family of actin-binding proteins, while the MA56 gene encodes a member of the immunoglobulin superfamily related to a variety of cellular receptors and adhesion molecules. A novel myxoma virus early gene, MST3N, is a member of the eukaryotic sialyltransferase gene family located between genes MA51 and MA52. Detergent lysates prepared from myxoma virus-infected cell cultures contained a virally encoded sialyltransferase activity that catalyzed the transfer of sialic acid (Sia) from CMP-Sia to an asialofetuin glycoprotein acceptor. Analysis of the in vitro-sialylated glycoprotein acceptor by digestion with N-glycosidase F and by lectin binding suggested that the MST3N gene encodes an enzyme with Galβ1,3(4)GlcNAc α2,3-sialyltransferase specificity for the N-linked oligosaccharide of glycoprotein. Lectin binding assays demonstrated that α2,3-sialyltransferase activity is expressed by several known leporipoxviruses that naturally infect Sylvilagus rabbits. The sialyltransferase is nonessential for myxoma virus replication in cell culture; however, disruption of the MST3N gene caused attenuation in vivo. The possible implications of the myxoma virus-expressed sialyltransferase in terms of the host’s defenses against infection are discussed.
The South American myxoma virus (MYXV) is the type virus of the genus Leporipoxvirus of the family Poxviridae (18). MYXV naturally infects the Brazilian tapeti (forest rabbit [Sylvilagus brasiliensis]), causing the development of a small localized tumor which can persist for several months. In contrast to the trivial symptoms in its natural host, infection of the European rabbit (Oryctolagus cuniculus) causes the often fatal disease myxomatosis. The Californian MYXV isolates which naturally infect the brush rabbit (Sylvilagus bachmani) also cause fulminant disease in the European rabbit (20). Other recognized members of the genus Leporipoxvirus include Shope fibroma virus (SFV; natural host, Eastern cottontail [Sylvilagus floridanus]), hare fibroma virus (natural host, European brown hare [Lepus europaeus]), and the squirrel fibroma virus (natural host, gray squirrel [Sciurus carolinensis]).
MYXV is known to encode a variety of cell-associated and secreted proteins which have been implicated in down-regulation of the host’s immune and inflammatory responses (37, 38) and inhibition of apoptosis of virus-infected cells (36). These virulence genes are encoded either within or adjacent to the 11.5-kb inverted terminal repeats present at either end of the 163.6-kb genome. As with other poxviruses, the central “conserved” region of the MYXV genome (∼140 kb) is presumed to encode genes primarily involved in nucleic acid synthesis and virion morphogenesis (25–27). Here we describe the identification of a new poxvirus gene encoded within the central region of the MYXV genome that belongs to the eukaryotic sialyltransferase family. The sialyltransferase family comprise more than 15 different membrane-bound glycosyltransferases of the trans-Golgi network (TGN) that catalyze the transfer of sialic acid (Sia) from CMP-Sia to the nonreducing terminal positions of N- and O-glycan of glycoproteins and oligosaccharide of glycolipids (49). Terminal Sia of glycoconjugates are known to play important biological roles in (i) maintenance of serum glycoproteins in circulation, (ii) receptor-ligand interactions between cells involved in immune and inflammation responses, (iii) enhanced metastatic capability of tumor cells, (iv) masking of receptors and antigens on tumor cells and microorganisms, and (v) viral attachment to target cells (for an extensive review of the biological roles of carbohydrates, see reference 60). Sialylation of virus- or host-encoded glycoproteins could thus have multiple influences on MYXV’s attempts to subvert the rabbit’s innate and acquired responses to infection.
MATERIALS AND METHODS
Cells and viruses.
SIRC (O. cuniculus cornea [ATCC CCL-60]), RK13 (O. cuniculus kidney [ATCC CCL-37]) and CV-1 (African green monkey [Ceropithecus aethiops] kidney [ATCC CCL-70]) were maintained in minimal essential medium (MEM) supplemented with 5% (vol/vol) fetal bovine serum. Poxviruses were grown on the cells described above in MEM supplemented with 0.5% (vol/vol) fetal bovine serum.
The following poxviruses were used in this study: (i) vaccinia virus (VACV) strain Western Reserve (WR [ATCC VR-1354]); (ii) Brazilian MYXV strains Lausanne (Lu [ATCC VR-115]), isolated in Campinas, Brazil, in 1949 (5), and Uriarra (Ur), isolated in Australian Capital Territory, Australia, in 1953 (41), which is a naturally attenuated derivative of the Moses strain (ATCC VR-116) isolated in São Paulo, Brazil, in 1909 (40); (iii) Californian MYXV strains MSD isolated in San Diego, Calif., in 1949 (21), and MSW, isolated near San Francisco, Calif., in 1950 (21); and (iv) Shope Fibroma virus (SFV) original A strain (ATCC VR-112), isolated in Princeton, N.J., in 1931 (53).
DNA sequencing and plasmid constructions.
In the MYXV Ur strain, a 7.3-kb EcoRI-G2 DNA fragment located approximately 131.9 to 139.2 kb from the left end of 163.6-kb viral genome (27) was ligated into the vector pGem7Zf(−) (Promega Corporation, Madison, Wis.). A 4.7-kb DNA sequence, including the 4.3-kb BamHI-N fragment plus sequences to the right end of the EcoRI-G2 fragment (GenBank accession no. U46577), was determined from both strands by using the ABI PRISM Dye Primer Cycle Sequencing Ready Reaction kit and an ABI DNA sequencer (Applied Biosystems, Perkin-Elmer, Foster City, Calif.).
The MST3N genes of the Brazilian MYXV strain Lu and Californian MYXV strain MSD were isolated by PCR using Pfu DNA polymerase (Stratagene, La Jolla, Calif.) and degenerate primers 5′-GGGATCCAT(A/T/C)TC(T/C)AG(A/G)CA(T/C)AA(A/G)CG (encoding an open reading frame [ORF] MA51-translated amino acid sequence, I-S-R-H-K-R, conserved between the Orthopoxvirus variola virus [VARV] and MYXV) and 5′-GGGATCCGC(A/G)CA(T/C)AA(A/T/C/G)CC(T/G/A)ATCAT (complementary sequence encoding an ORF A52-translated amino acid sequence, M-I-G-L-C-A, conserved between VARV, VACV, and MYXV) and ligated into the vector pCRscript (Stratagene). The DNA sequence of the entire 1.8-kb Lu PCR fragment (GenBank accession no. U46578) and the internal 1.2-kb EcoRV fragment containing the MSD-MST3N gene (GenBank accession no. AF030894) were determined.
The MYXV Ur SalI-R2 fragment, containing the majority of the MST3N gene, was ligated into the SalI site of pGem3Zf(+) (Promega Corporation) to generate pUrS-R2. The MYXV DNA fragment contained in pUrS-R2 was removed by digestion with EcoRI and HindIII and ligated between the respective sites of pGP7.5gpt (27), generating pUrST1. The MST3N gene sequences contained in pUrST1 were interrupted by the insertion of an XbaI-HindIII cassette containing a late promoter and the Escherichia coli lacZ gene isolated from pUrTK14L-lacZ (27), by blunt-end ligation into the unique BglII site located within the MST3N gene L-sialyl motif coding region, generating pUrST1-lacZ.
The VACV P11 late promoter was deleted from pUrTK11 (29) by digestion with XbaI and EcoRI, with the recessed 3′ ends filled in with Klenow fragment DNA polymerase and the blunt ends ligated to reconstitute an EcoRI site, generating the vector pUrTK11ΔP11. The MYXV Lu strain ClaI-EcoRV fragment containing the Lu MST3N gene together with its natural early promoter was isolated and ligated between the ClaI and HincII sites of pBluescript SK− (Stratagene), generating pBS-ST. The EcoRI-XhoI fragment containing the Lu MST3N gene and its promoter was excised from pBS-ST and ligated between the EcoRI and SalI sites of pUrTK11ΔP11, generating pUrTK11ΔP11/Lu-MST3N.
Recombinant myxoma viruses.
(i) Lu243Z (27) is a lethal grade I virus (>99% mortality, ≤13-day mean survival time [21]) which contains a synthetic late promoter and the E. coli lacZ gene inserted in the intergenic region between the MYXV β-subunit RNA polymerase (MA24) and the fusion protein (MA27) genes. (ii) MST3N gene knockout virus Lu(lacZ+/MST3N−) was constructed by the transient dominant selection procedure (19) using the plasmid pUrST1-lacZ and Lu-infected RK13 cells. This virus contains a synthetic late promoter and the E. coli lacZ gene interrupting the MST3N gene. (iii) Lu(lacZ+/Lu-MST3N+) was constructed by transfection of Lu(lacZ+/MST3N−)-infected RK13 cells with pUrTK11ΔP11/Lu-MST3N and selection for gpt gene expression by using mycophenolic acid (20 μg/ml), xanthine (250 μg/ml), hypoxanthine (15 μg/ml), aminopterin (2 μg/ml), and thymidine (10 μg/ml), which were added to the culture medium. Lu(lacZ+/Lu-MST3N+) contains an interrupted and inactive copy of the natural MST3N gene and a second intact copy inserted in the intergenic region between the thymidine kinase (tk, MJ2) and MJ2a genes (27). Expression of the MYXV-encoded α2,3-sialyltransferase by these recombinant viruses was determined with the lectin binding assay described below.
Viral mRNA preparation and Northern blot analysis.
RK13 cells were infected with MYXV at 10 PFU/cell and incubated in MEM for 16 h for the isolation of late viral mRNA or were incubated in MEM supplemented with 100 μg of cycloheximide per ml (Sigma Chemical Co., St. Louis, Mo.) for 10 h for early viral mRNA preparation. Poly(A)+ RNA was isolated from the infected cells by using the PolyATract System 1000 (Promega Corporation). A strand-specific 32P-labeled RNA probe complementary for the MST3N mRNA was prepared by using the Riboprobe Gemini II kit (Promega Corporation) and pUrS-R2. The Northern transfer, hybridization, and washing stringency procedures were performed as recommended by the manufacturer.
Cell lysate preparation.
Confluent monolayers of CV1 cells were infected with poxvirus at 1 PFU/cell in 80-cm2 culture flasks. Twenty-four hours postinfection, the cells were detached from the culture flasks with a cell scraper and washed three times by using phosphate-buffered saline (PBS) at 4°C. Cell lysates were prepared by suspension in 1 ml (per flask) of a mixture of 50 mM 2-[N-morpholino]-ethanesulfonic acid (MES)-OH (pH 6.1), 0.5% (vol/vol) Triton X-100, 100 mM NaCl, 1.5 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride, and 10 μg of aprotinin per ml and incubated at 4°C for 45 min. The lysate was clarified by centrifugation at 1,750 × g at 4°C for 15 min. Cleared lysates were stored at −70°C. Total protein concentrations were measured by using the bicinchoninic acid protein assay reagent (Pierce, Rockford, Ill.).
Sialyltransferase reactions.
Cell lysates, at concentrations of 30 μl (containing ∼50 μg of total protein), were mixed with 5 μl of asialofetuin {type I; Sigma Chemical Co. (10 mg/ml dissolved in MEST buffer, which was composed of 50 mM MES-OH [pH 6.1] and 0.5% [vol/vol] Triton X-100)} and 5 μl of CMP-[3H]Sia (DuPont NEN, Boston, Mass. [1 μCi/μl dissolved in MEST buffer]) and incubated for 90 min at 37°C. Reactions were terminated by heating at 90°C for 10 min. An assay containing a sample lysate prepared from VACV-infected cells was used as a negative control. A positive control consisted of the VACV-infected cell lysate including 5 mU of N-acetyllactosamine α2,6-sialyltransferase ST6Gal-I [rat liver; Boehringer-Mannheim GmbH, Mannheim, Germany]).
Peptide N-glycosidase F (PNGase F) digestion.
The total proteins from the sialyltransferase reactions were precipitated by overnight incubation with 8 volumes of acetone at −20°C and recovered by centrifugation for 30 min with a microcentrifuge. Protein pellets were dried and resuspended in 10 μl of H2O. The protein samples were denatured by the addition of 25 μl of 0.1 M β-mercaptoethanol–0.5% (wt/vol) sodium dodecyl sulfate (SDS) and heated at 100°C for 5 min. Samples were cooled to room temperature, and the total volume was adjusted to 40 μl with H2O. Denatured protein aliquots of 5 μl were mixed with 5 μl of 2× PNGase F buffer {40 mM sodium phosphate buffer [pH 7.2], 20 mM EDTA, 3% [wt/vol] 3-[(3-cholamidopropyl)-dimethyl-ammonio]-1-propanesulfonate CHAPS}. Reactions were initiated by the addition of 0.4 U of N-glycosidase F (Boehringer Mannheim) and incubated at 30°C for 16 h. The reactions were terminated by heating at 100°C for 5 min.
Fluorography.
Samples containing approximately 6 μg of the acceptor glycoprotein were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie blue to ensure equivalent loading of acceptor glycoprotein per lane. Fluorographic detection of the tritium-labeled sialoglycoproteins was achieved by impregnating the stained gels with Amplify (Amersham International Plc., Little Chalfont, United Kingdom), drying them at 80°C, and exposing them for 24 to 48 h to preflashed Hyperfilm-MP (Amersham International Plc.).
Determination of sialic acid linkage by lectin binding.
For sialylation of acceptor glycoprotein, 30 μl of Triton lysates prepared from poxvirus-infected cells (∼50 μg of total protein) was mixed with 5 μl of asialofetuin (10 mg/ml) and 10 μl of CMP-Sia (10 mM [CMP-NeuAc; Sigma Chemical Co.]) and incubated at 37°C for 3 h. Glycoprotein samples (∼12 μg of acceptor glycoprotein) were separated by SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride Hybond membrane (Amersham International Plc.). Determination of the bound Sia linkage to glycoprotein acceptor was accomplished with the digoxygenin (DIG) glycan differentiation kit (Boehringer-Mannheim), by using DIG-labeled lectins, Sambucus nigra agglutinin (SNA [for α2,6-linked Sia]), and Maackia amurensis agglutinin (MAA [for α2,3-linked Sia]) as recommended by the manufacturer.
Virulence assays.
Animal studies were conducted in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes. Outbred domestic rabbits raised in the CSIRO Wildlife and Ecology animal facility were housed in individual cages at a controlled ambient temperature of 22°C with a 12-h light cycle. Water and food were available at all times, and green feed was provided weekly. Virulence assays followed the format of Fenner and Marshall (21). For each virus [Lu243Z, Lu(lacZ+/MST3N−) or Lu(lacZ+/Lu-MST3N+)], six domestic male rabbits greater than 5 months old were inoculated by intradermal injection into the right flank with 100 PFU of virus in 100 μl of PBS. Animals were observed twice daily, and clinical signs were recorded. Times of death were determined to the nearest half day. Moribund animals were killed by an overdose of barbiturate intravenously, and the time of death increased by half a day. Once clinical signs of myxomatosis were present, all rabbits were injected twice daily subcutaneously with 0.75 to 0.9 mg of the analgesic buprenorphine as advised by the Institute Animal Ethics Committee. This has previously been demonstrated not to alter the survival time of rabbits infected with virulent myxoma virus. Results were determined as average survival times of rabbits infected with each virus, and statistical significance was examined by pairwise Student’s t tests.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this article have been deposited with the GenBank database and have been assigned accession no. U46577, U46578, and AF030894.
RESULTS
DNA sequence and protein similarities.
The deduced 4.7-kb DNA sequence (Fig. 1) contains six major ORFs. The MYXV ORFs have been termed MA51 (partial sequence), MA52, MA55, MA56, and MB1 (partial sequence), to correspond to the gene homologs encoded by the Copenhagen strain of VACV (23); and MST3N, designating the α2,3-sialyltransferase gene. Homologs of the VACV A53R (tumor necrosis factor receptor related), A54R, and A57R (guanylate kinase related) genes are not encoded in this region of the MYXV genome. The inferred amino acid sequences of the MYXV ORFs were compared to sequences in the nonredundant protein databases by using the gapped-BLASTp (V2.0.2) program (2).
FIG. 1.
Nucleotide sequence of the MYXV BamHI-N fragment plus additional sequences to the right end of the EcoRI-G2 fragment (GenBank accession no. U46577). The amino acid sequences encoded by ORFs MA51 (partial), MA52, MA55, MA56, and MB1 (partial) are indicated above, and that of MST3N is indicated below the sequence. Putative promoter sequences with similarity to the poxvirus early promoter motif (AAAAAATGAAAAAA[C/T]A) are indicated by solid arrows, and late promoters transcription initiation consensus sequences (TAAAT[G/A]) are underlined. Putative early transcription termination signals (TTTTTNT) are doubly underlined. Relevant restriction endonuclease sites used during the construction of plasmids described in this report are boxed: BamHI (GGATCC), BglII (AGATCT), ClaI (ATCGAT), EcoRI (GAATTC), EcoRV (GATATC), and SalI (GTCGAC).
ORFs MA51, MA52, and MB1.
The MA51- and MA52-encoded proteins only share significant similarity to the corresponding Orthopoxvirus homologs. The VACV A51R and A52R genes are nonessential for VACV replication (23), and in VARV, the A52 gene homolog is fragmented into multiple short ORFs (35). The MB1 gene encodes the MYXV homolog of the VACV serine/threonine protein kinase (23).
ORF MA55.
The MA55-encoded protein shares amino acid similarity with the actin-associated proteins kelch (66), MIPP (9), scruin (64, 65), and calicin (62); multiple poxvirus-encoded proteins, including VACV A55R, C2L, F3L (23), and MYXV T8, and T9 (57); and many Drosophila and mammalian zinc finger proteins. The MA55 protein contains an amino-terminal poxvirus zinc finger (POZ) domain, which is thought to be involved in the formation of hetero- and homoprotein dimers and which is usually found in the first 120 amino acids of actin-associated or zinc finger DNA binding proteins (1). The shared amino acid similarity between MA55 and the zinc finger proteins is restricted to the POZ domain. The MA55 protein does not contain consensus zinc finger motifs and is therefore unlikely to be directly involved in transcription regulation. The amino acid similarity between the MA55 protein and the actin-associated proteins extends beyond the POZ domain and includes six imperfect repeats (kelch repeats) of approximately 50 amino acids each (R1, amino acids 252 to 300; R2, 301 to 347; R3, 348 to 394; R4, 395–445; R5, 446–498; and R6, 499–545) found in the COOH-terminal region. Kelch repeats are predicted to form antiparallel β-strand “superbarrel” folds which have been implicated in actin binding (4, 52). This suggests that the MA55 early protein (see below) could be involved in altering the actin cytoskeleton during the early stages of the viral replication. Like the Drosophila kelch protein (47), the MA55 protein could bind actin through the kelch repeats and then cross-link the actin filaments via MA55 dimerization mediated through the POZ domain.
ORF MA56.
The results of a BLASTp similarity search indicated that the MA56 protein shares similarity to the Xenopus laevis neural cell adhesion molecule (N-CAM) isoforms (55) and the raccoonpox virus hemagglutinin (HA) (8); however, it is only distantly related to the VACV A56R HA protein. A similarity search conducted by using a BLITZ analysis identified significant amino acid similarity to additional cellular receptors and adhesion proteins, including yeast A-agglutinin attachment subunit (48), mouse type II interleukin 1 receptor (39), Orthopoxvirus HAs encoded by VACV (23) and VARV (35), and numerous other members of the immunoglobulin superfamily.
The MA56 protein contains an N-terminal type I signal sequence (amino acid residues 1 to 18), transmembrane domain (amino acids 197 to 213) predicted by using PSORT analysis (42), and an immunoglobulin domain (amino acid residues 33 to 105) predicted by using the Pfam-A HMM search (release 2.0) (54). Potential N glycosylation sites (NX[ST], residues 23, 32, 38, 56, 84, 92, 121, and 148) and phosphorylation sites (PKC [ST]X[RK]; residues 18, 51, 69, 117, 141, 147, 151; CK-2 [ST]-XX-[DE]; residues 26, 69, 74, and 94) are contained in the MA56 protein. The protein is rich in serine (18%) and threonine (14%) residues with multiple potential O glycosylation sites (Thr residues 110, 123, 124, 128, 129, 147, 151, and 189; Ser residues 113, 126, 150, 180, 181, 182, 185, 186, 187, 190, and 191), predicted by using NetOGlyc (V2.0) analysis (24). Like the Orthopoxvirus HA protein (46), the mature MA56 protein is likely to be expressed as a type I integral plasma membrane phosphoglycoprotein and is potentially a “viroreceptor,” with the oligosaccharide moiety involved in the binding to an uncharacterized ligand.
The MST3N gene.
The MST3N gene encodes a protein with significant amino acid similarity to a range of eukaryotic sialyltransferases with different enzymatic activity toward the N- and O-glycans of glycoproteins and oligosaccharide of glycolipids (49). The MST3N product contains a noncleavable signal-transmembrane sequence between amino acid residues 7 and 28. Like all known glycosyltansferases, the MST3N protein is predicted to be a type II integral membrane protein with the NH2-terminal region (residues 1 to 6) on the cytoplasmic side of the membrane with the catalytic domain of the protein in the TGN lumen. The MYXV MST3N protein lacks a large proteolytically sensitive stem region, which is commonly found between the type II signal and the catalytic domain of other sialyltransferases (49). Recognizable motifs in the MST3N protein include an L-sialyl motif (amino acids 83 to 127; C-I-V-V-G-N-S-Y-N-L-H-N-R-S-L-G-R-I-I-D-S-Y-N-V-V-F-R-L-N-D-A-P-V-R-A-F-E-R-D-V-G-T-K-T) involved in binding the CMP-Sia nucleotide donor (12) and an S-sialyl motif (amino acids 219 to 242; P-T-M-G-M-V-A-L-V-T-A-L-H-V-C-Q-G-V-T-I-T-G-F-G). The conserved cysteines of the two sialyl motifs are proposed to form a disulfide linkage that is required for correct folding and enzyme activity (13). The MST3N protein S-sialyl motif region is predicted to form an α-helical hydrophobic domain. The MYXV S-sialyl motif region is unlikely to be a second transmembrane domain, since the S-sialyl motif has been implicated in binding both donor and acceptor substrates (13) and would be unavailable if embedded in the TGN membrane. The MST3N protein contains several potential N-linked glycosylation sites (residues 34, 45, 95, 147, and 283); however, it is not predicted to contain O-linked oligosaccharide, as determined by using NetOGlyc analysis.
MYXV gene transcription.
All of the ORFs contained on the MYXV BamHI-N fragment (nucleotides 1 to 4278 [Fig. 1]) are likely to be transcribed early in infection. Upstream of the MST3N, MA52, MA55, and MA56 ORFs are found A-rich sequences that are similar to those of the poxvirus early promoter consensus motif [AAAAAATGAAAAAA(C/T)A] (14). The ORFs MA51 (partial sequence), MST3N, MA52, and MA55 do not contain early transcription termination motifs [TTTTTNT] (67); however, such motifs are found downstream of both the MA51 and MST3N ORFs, again suggesting these genes are transcribed early in infection. ORF MA56 has a TTTTTNT sequence located within the 3′-end region of the gene, suggesting the putative MA52, MA55, and MA56 early mRNAs have coterminal 3′ ends with transcriptional termination occurring downstream of the MA56 stop codon. The MA56 ORF does not contain an upstream late promoter transcription initiation motif [TAAAT(G/A)] (15); therefore, temporal expression of MA56 will differ from that of the early and late expressed VACV HA genes (6). Immediately upstream of the MB1 protein kinase gene is a late promoter initiation motif preceded by a potential early promoter, suggesting that the expression of MYXV protein kinase gene could be constitutive.
Direct evidence that the MST3N gene is transcribed during the early phase of infection was determined by Northern blot hybridization (Fig. 2). A strand-specific MST3N gene probe hybridized to a major early mRNA of approximately 1.3 kb, which is in agreement with the predicted positions of the proposed early promoter and transcription termination signal. The smaller early RNA species may have resulted from transcriptional interference from MA51 transcripts due to the artificial early RNA amplification in the presence of cycloheximide. Southern blot hybridization with the MST3N gene probe indicated there are no additional related sequences in the MYXV genome (data not shown).
FIG. 2.
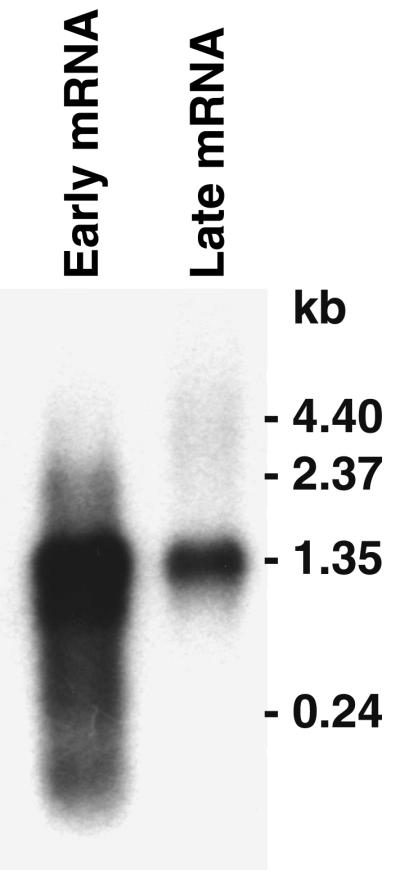
Northern blot [1 μg of poly(A)+ RNA per lane] of MYXV MST3N expression during the early and late transcription phases hybridized to an MST3N gene strand-specific RNA probe.
Hybridization of the MST3N gene probe to late mRNA gave a single transcript equivalent in size to the major early mRNA. The presence of a discrete species of MST3N mRNA in the late RNA preparation is unusual because normally a smear of hybridization to a higher-molecular-weight species is observed in Northern blots of poxvirus late RNA. It is unlikely that this mRNA is a late transcript, since there are no late promoter sequence motifs located immediately upstream of the gene. The most likely explanation is that the MST3N early mRNA either persists during the late phase of infection or results from reactivation of the early promoter, which has been observed with some Orthopoxvirus early genes (22). Alternatively, this early mRNA species may have resulted from asynchronous infection of the cell monolayer; however, this is unlikely, because the cells were infected at a multiplicity of 10 PFU/cell. The same early and late poly(A)+ RNA samples were probed previously (27) and displayed the typical pattern of hybridization to known early and late genes. The absence of a strong smear of hybridization to late mRNAs obtained with the MST3N gene-specific probe is probably due to the fact that poxvirus genes in this “conserved” region of the genome are generally oriented toward the right genome terminus (23) on the opposite strand from the MST3N gene. Conservation of this general gene organization in MYXV would result in the failure of the strand-specific MST3N probe hybridizing to late viral transcripts, since they would not be complementary sequences.
Myxoma virus-encoded sialyltransferase activity.
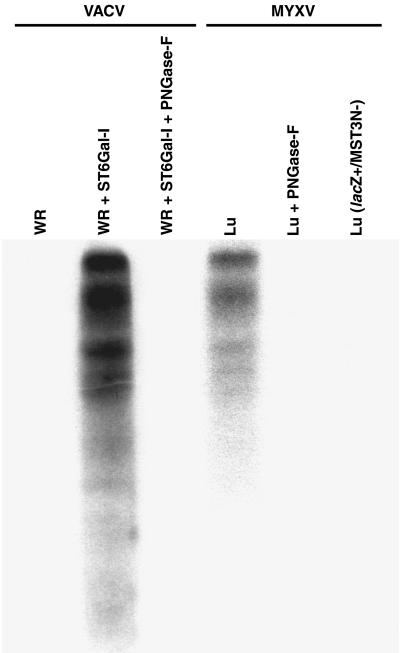
Triton X-100 extracts prepared from CV1 cells infected with either VACV, Lu, or Lu(lacZ+/MST3N−) were used for in vitro sialyltransferase reactions (Fig. 3). Lysates prepared from Lu-infected cells contained significant levels of sialyltransferase activity that transferred [3H]Sia to the asialofetuin acceptor, whereas, the VACV-infected cell lysates did not contain detectable levels of sialyltransferase activity. The addition of the N-glycan ST6Gal-I to the VACV-infected cell lysates indicated they contained no general inhibitors of sialyltransferase activity. Insertional inactivation of the MST3N gene in the recombinant virus Lu(lacZ+/MST3N−) abolished the observed sialyltransferase activity in MYXV-infected cells. This strongly suggests that the observed sialyltransferase activity was encoded by the MST3N gene and is unlikely to result from induction of a cellular activity resulting from MYXV infection.
FIG. 3.
Fluorograph of sialyltransferase reactions and PNGase F digestions using asialofetuin glycoprotein acceptor. Triton X-100 soluble lysates were prepared from tissue culture cells infected with VACV WR, MYXV Lu, or MYXV Lu(lacZ+/MST3N−) and reacted with CMP-[3H]Sia and asialofetuin, with the addition of rat liver sialyltransferase ST6Gal-I or N-glycosidase (PNGase F) as indicated.
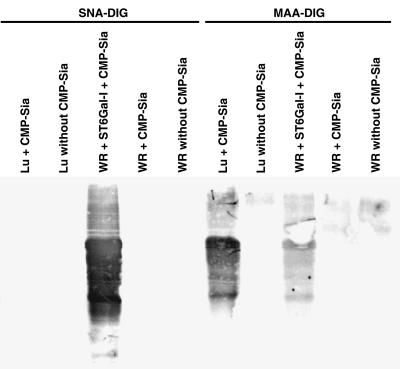
Removal of the [3H]Sia from the glycoprotein acceptor by digestion with PNGase F (Fig. 3) indicated that the transferred Sia was attached to the N-glycan, but not O-glycan, of the sialylated glycoprotein acceptor. The type of linkage of Sia to the acceptor resulting from the myxoma virus sialyltransferase was determined by lectin binding analysis (Fig. 4). The MAA lectin bound to the MYXV-sialylated acceptor, where SNA lectin failed to bind. In contrast, only the ST6Gal-I-sialylated glycoprotein bound SNA lectin. These results indicate that the MYXV-encoded sialyltransferase transfers Sia from CMP-Sia in an α2,3 linkage to N-glycan of the asialofetuin glycoprotein acceptor.
FIG. 4.
Lectin binding assay of asialofetuin glycoprotein acceptor sialylated by using virus-encoded sialyltransferase. Triton X-100 soluble lysates were prepared from tissue culture cells infected with VACV WR or MYXV Lu. Sialyltransferase reactions were performed with or without the addition of nucleotide-sugar donor CMP-Sia. The addition of rat liver sialyltransferase ST6Gal-I to the VACV samples was included as a positive control for α2,6-linked Sia. The blot was reacted with DIG-labeled lectin SNA or MAA followed by reaction with alkaline phosphatase-conjugated anti-DIG antibody and developed with X-phosphate–nitroblue tetrazolium.
α2,3-Sialyltransferase encoded by other leporipoxviruses.
RK13 cells were separately infected with different leporipoxviruses available in Australia; Brazilian MYXV Lu, two strains of Californian MYXV (MSD and MSW), and the SFV. Lectin MAA binding assays demonstrated that expression of α2,3-sialyltransferase activity is common to all of the leporipoxviruses tested (data not shown). Unfortunately viable samples of both hare and squirrel fibroma viruses were not available to determine if expression of α2,3-sialyltransferase is also a feature of leporipoxviruses which infect other lagomorphs and rodents. Isolation of the α2,3-sialyltransferase genes carried by these viruses was attempted by PCR. The Brazilian and Californian MYXV templates produced 1.8-kb PCR products of the expected size. The PCR products generated from the Lu and MSD virus templates were cloned, and the DNA sequences of the Lu-MST3N (GenBank accession no. U46578) and MSD-MST3N (GenBank accession no. AF030894) genes were determined.
Comparison of the 1,806-nucleotide Lu PCR product to the corresponding region of the Ur-derived DNA sequence identified only 4 nucleotide differences (Ur nucleotide positions 257, 538, 1140, and 1852 [Fig. 1]). This results in conservative amino acid substitutions in MA51 and MST3N, a semiconservative amino acid substitution in MA51, and a mismatched amino acid in the MA52 protein. The proofreading Pfu DNA polymerase used for the PCR has been reported to have a low rate of error per nucleotide (1.6 × 10−6) (34). It is possible that the minor nucleotide differences between the Lu and Ur sequences could be due to errors resulting from the PCR DNA amplification. The Lu and Moses strains of MYXV were both originally isolated approximately 40 years apart from the regions surrounding Rio de Janeiro and Sãn Paulo, Brazil. The Moses strain was propagated in laboratory rabbits for many years before being released in Australia in 1950 followed by isolation of the attenuated Ur strain in 1953. The DNA sequencing results indicate that the Lu and Ur strains of MYXV have highly conserved DNA sequences despite the years between the original isolation and the subsequent adaptation of the viruses to a new host in Australia. In contrast, with the Californian MSD strain, which evolved separately in its host, S. bachmani, the MSD-MST3N gene PCR product has only 84% nucleotide identity, 84% amino acid identity, and 92% similarity relative to the Ur-MST3N gene. A PCR product cold not be generated by using the SFV DNA template and degenerate primers described here. Southern blot hybridization of SFV DNA by using the Ur-MST3N gene-specific probe and low-stringency washing localized the SFV α2,3-sialyltransferase gene to the BamHI-F2 fragment (data not shown). This suggests that the SFV-ST3N gene is encoded in the equivalent location to MYXV, in the region between the genes encoding the SFV homologs of the A50R DNA ligase (45) and the B1R protein kinase (58).
Virulence assays.
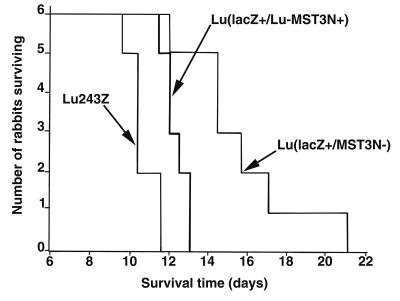
The results for rabbits infected with each virus are shown in Fig. 5 as stepwise survival graphs. Clinically, all three viruses induced classic myxomatosis in the inoculated rabbits. The clinical signs developed more rapidly in the Lu243Z control, which was clearly more virulent than the sialyltransferase knockout Lu(lacZ+/MST3N−) virus. Survival times were prolonged for rabbits infected with the sialyltransferase knockout, although all but one rabbit died. For humane reasons, this rabbit was euthanized 21 days postinfection. The sialyltransferase revertant virus Lu(lacZ+/Lu-MST3N+) was highly virulent and was difficult to distinguish from the Lu243Z virus clinically, although lesions at the primary inoculation site were recorded as black in the center a day later for the Lu(lacZ+/Lu-MST3N+) virus. In addition, deaths of the infected rabbits commenced later, suggesting the virus was slightly attenuated, possibly due to the extra genetic load resulting from the DNA insertions or additional uncharacterized mutations generated during cell culture passage.
FIG. 5.
MYXV virulence assays. Chart showing the survival times of laboratory rabbits infected with recombinant MYXV Lu243Z, Lu(lacZ+/MST3N−), and Lu(lacZ+/Lu−MST3N+).
Average survival times (mean ± standard deviation) of rabbits infected with the recombinant viruses were as follows: Lu243Z, 10.7 ± 0.7 days; Lu(lacZ+/MST3N−), 15.7 ± 3 days; and Lu(lacZ+/Lu-MST3N+), 12.3 ± 0.6 days. Based on the classification of Fenner and Marshall (21), viruses Lu243Z and Lu(lacZ+/Lu-MST3N+) are both of grade I virulence (>99% mortality, ≤13-days mean survival time). The virus Lu(lacZ+/MST3N−), while still virulent, would be classed as showing grade II virulence (95 to 99% mortality, 13- to 16-day mean survival time). The average survival time of each virus was significantly different from that of the others as follows: Lu243Z versus Lu(lacZ+/MST3N−), P < 0.001; Lu(lacZ+/Lu-MST3N+) versus Lu(lacZ+/MST3N−), P < 0.01; and Lu243Z versus Lu(lacZ+/Lu-MST3N+), P < 0.001. These analyses were performed by either including or excluding the survivor from the group of Lu(lacZ+/MST3N−)-challenged rabbits in the analysis; however, this did not affect the significance levels.
DISCUSSION
The identification and characterization of the Leporipoxvirus α2,3-sialyltransferase represent the first description of a natural virus-encoded glycosyltransferase involved in the biosynthesis of glycoproteins. The lack of strong amino acid similarity makes it difficult to assign the MYXV enzyme to any of the known α2,3-sialyltransferase classes ST3Gal-I to -IV (56), suggesting that the enzyme may have unique acceptor specificity. The closest similarities to the MYXV α2,3-sialyltransferase are the sialyltransferase classes ST3Gal-III (human [31], 36% identity and 54% similarity) and ST3Gal-IV (human [32], 43% identity and 60% similarity). Both ST3Gal-III and ST3Gal-IV enzymes utilize N-glycan terminated with Galβ1,3GlcNAc (type I; Lewisc [Lec]) and Galβ1,4GlcNAc (type II; N-acetyllactosamine [LacNAc]) disaccharides. However, ST3Gal-III enzymes preferentially utilize type I acceptors, while the ST3Gal-IV enzymes show the highest level of activity toward terminal type II disaccharides. Human ST3Gal-IV also utilizes Galβ1,3GalNAc (type III; Thomsen-Friedenreich [TF] disaccharide) acceptors of O-linked glycoprotein and glycolipids, whereas the ST3Gal-III enzymes show negligible activity toward these glycoconjugates (32). The asialofetuin glycoprotein contains three N-glycan chains terminated with type II disaccharide (43), which are acceptor substrates for the MYXV α2,3-sialyltransferase. Under the in vitro conditions described here, the MYXV α2,3-sialyltransferase does not contain appreciable substrate specificity toward the terminal O-linked type II or type III disaccharides of asialofetuin (17, 43). Therefore, the MST3N-encoded sialyltransferase is most similar in activity to the human ST3Gal-III, N-glycan Galβ1,3(4)GlcNAc α2,3-sialyltransferase (32).
Inactivation of the MST3N gene in the Lu grade I virulent virus resulted in generation of the mildly attenuated virus with grade II virulence, establishing that the sialyltransferase is not required for infection or induction of clinical myxomatosis in genetically susceptible laboratory rabbits. In the absence of sialyltransferase expression, disease symptoms are delayed, suggesting the MYXV α2,3-sialyltransferase may act synergistically with other virulence factors. The MYXV α2,3-sialyltransferase may be required when infecting certain cell types, for example, lymphocytes (44) complementing a cellular deficiency in ST3Gal activity (33), for correct sialylation of virus-expressed glycoproteins for optimal structure, stability, and biological function.
Terminal Sia of glycoconjugates are known to play important functions in cell-cell recognition (61). We speculate that MYXV-induced α2,3-sialylation of viral or host glycoproteins could also have some influence in regulating the host’s innate responses to virus infection. Many bacteria (e.g., Neisseria) and parasites (e.g., Trypanosoma) express sialyltransferase activities and salvage host Sia, attaching it to their own surface and inhibiting complement-mediated cell lysis and masking of antigenic sites (60). It has been proposed that macrophages contain a lectin-like activity that recognizes surface asialoglycoconjugates of apoptotic cells that are masked with Sia on normal cells (50). Increased expression of cell surface sialoglycoconjugates and the associated increase in negative charge have also been correlated with resistance to killing of target cells by activated NK cells (7). Enhanced expression of surface sialoglycoconjugates could mask MYXV-infected cells from components of the host’s innate responses, allowing increased viral replication and tissue dissemination prior to the development of a specific immune response.
Altered sialylation of glycoproteins expressed by MYXV-infected cells could also affect cell surface ligand-receptor interactions. The sialoadhesin family of I-type lectins are immunoglobulin superfamily proteins which act as Sia-dependent adhesion molecules. The sialoadhesin family includes sialoadhesin, expressed on macrophages in lymphoid tissues and at sites of inflammation, and CD33, expressed on myeloid cells and macrophages (10, 11). Both sialoadhesin and CD33 bind terminal trisaccharides Siaα2,3Galβ1,3GlcNAc and Siaα2,3Galβ1,4GlcNAc, which are the proposed products of the MYXV α2,3-sialyltransferase. In association with a GlcNAcα1,3/4-fucosyltransferase, the MYXV α2,3-sialyltransferase could also form the structures sialyl-Lewis a (sLea; Siaα2,3Galβ1,3[Fucα1,4]GlcNAc) and sialyl-Lewis x (sLex; Siaα2,3Galβ1,4[Fucα1,3]GlcNAc), which are counterreceptors for the C-type lectin family of Sia binding adhesion proteins, E-, P-, and L-selectins involved in regulation of cell trafficking in inflammation and immune responses (3, 61). The presence of α2,3-sialylated N-glycan on MYXV-expressed soluble glycoproteins could result in the binding and blockage of these Sia-dependent receptors, resulting in the inhibition of (i) inflammatory cell migration to the sites of virus replication, (ii) homing of lymphocytes to lymphoid tissues, and (iii) adhesion and stimulation of immune cells. Alternatively, expression of Siaα2,3-linked glycoproteins on viral particles or infected cells could mediate specific binding and infection of cells expressing Sia-dependent receptors assisting in the dissemination of the virus through the host’s tissues. Enhanced tumor cell surface sialylation of glycoconjugates has been correlated with increased metastatic capability mediated by Sia-dependent receptor binding (30).
In poxvirus-infected cells, the TGN membranes contribute to the outermost membrane of the secreted extracellular enveloped virus (EEV) (51). It has been demonstrated that treatment of tissue culture cells with sialidase enhanced purified VACV EEV binding and infectivity by 50%, whereas intracellular mature virus (IMV) binding was only marginally increased (59). It has also been shown that antiserum directed against the glycolipid asialo-GM1 inhibited VACV infection of mouse ovaries (28). Together these observations suggest that the VACV particles contain receptors for asialoglycoconjugates, possibly cellularly derived sialyltransferases that are usually resident in the TGN. Expression of the virus encoded α2,3-sialyltransferase on the outer membrane of the Leporipoxvirus EEV or on the surface of infected cells could result in enhanced binding to terminal type I or type II disaccharides of glycoproteins expressed on the surface of target cells.
The MYXV sialyltransferase is unlikely to be the only example of a virally encoded glycosyltransferase, because unusual virus-directed glycosylation has been observed with the Paramecium chlorella virus (63). Although a chlorella virus hyaluronan synthase gene has been characterized (16), genes encoding putative glycosyltransferases involved in the biosynthesis of the oligosaccharide of glycoproteins or glycolipids have not been identified. The list of known glycosyltransferase genes appearing in the databases is growing; however, there remain to be identified a large number of genes corresponding to the estimated >150 different glycosyltransferase activities. The complete nucleotide sequences of a number of viruses with large DNA genomes have been determined in recent years, and many ORFs have been identified which encode proteins of unknown function. Some of these uncharacterized viral proteins are potential type II integral membrane proteins characteristic of glycosyltransferases. With the future identification of new examples of glycosyltransferase genes, it is likely that some of these viral ORFs will also be shown to encode enzymes involved in the biosynthesis of oligosaccharide attached to glycoproteins or glycolipids.
ACKNOWLEDGMENTS
We thank Bob Seamark and Tony Robinson for critically reading the manuscript, Kelly White for technical assistance, and Robert Forrester for statistical advice.
This research was supported by the Australian Government’s Cooperative Research Centres Program.
REFERENCES
- 1.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevilacqua M P, Nelson R M. Selectins. J Clin Investig. 1993;91:379–387. doi: 10.1172/JCI116210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork P, Doolittle R F. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 5.Bouvier M. Quelques remarques sur la myxomatose. Epizootie de 1949 au Brésil (Région de Campinas, Est Sao-Paulo) Bull Off Int Epizoot. 1954;46:76–77. [Google Scholar]
- 6.Brown C K, Turner P C, Moyer R W. Molecular characterization of the vaccinia virus hemagglutinin gene. J Virol. 1991;65:3598–3606. doi: 10.1128/jvi.65.7.3598-3606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brutkiewicz R R, Welsh R M. Major histocompatibility complex class I antigens and the control of viral infections by natural killer cells. J Virol. 1995;69:3967–3971. doi: 10.1128/jvi.69.7.3967-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavallaro K F, Esposito J J. Sequences of the raccoon poxvirus hemagglutinin protein. Virology. 1992;190:434–439. doi: 10.1016/0042-6822(92)91229-n. [DOI] [PubMed] [Google Scholar]
- 9.Chang-Yeh A, Mold D E, Huang R C. Identification of a novel murine IAP-promoted placenta-expressed gene. Nucleic Acids Res. 1991;19:3667–3672. doi: 10.1093/nar/19.13.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crocker P R, Hartnell A, Munday J, Nath D. The potential role of sialoadhesin as a macrophage recognition molecule in heath and disease. Glycoconj J. 1997;14:601–609. doi: 10.1023/a:1018588526788. [DOI] [PubMed] [Google Scholar]
- 11.Crocker P R, Kelm S, Hartnell A, Freeman S, Nath D, Vinson M, Mucklow S. Sialoadhesion and related cellular recognition molecules of the immunoglobulin superfamily. Biochem Soc Trans. 1996;24:150–156. doi: 10.1042/bst0240150. [DOI] [PubMed] [Google Scholar]
- 12.Datta A K, Paulson J C. The sialyltransferase “sialylmotif” participates in binding the donor substrate CMP-NeuAc. J Biol Chem. 1995;270:1497–1500. doi: 10.1074/jbc.270.4.1497. [DOI] [PubMed] [Google Scholar]
- 13.Datta A K, Sinha A, Paulson J C. Mutation of the sialyltransferase S-sialylmotif alters the kinetics of the donor and acceptor substrates. J Biol Chem. 1998;273:9608–9614. doi: 10.1074/jbc.273.16.9608. [DOI] [PubMed] [Google Scholar]
- 14.Davison A J, Moss B. Structure of vaccinia virus early promoters. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 15.Davison A J, Moss B. Structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 16.DeAngelis P L, Jing W, Graves M V, Burbank D E, Van Etten J L. Hyaluronan synthase of chlorella virus PBCV-1. Science. 1997;278:1800–1803. doi: 10.1126/science.278.5344.1800. [DOI] [PubMed] [Google Scholar]
- 17.Edge A S, Spiro R G. Presence of an O-glycosidically linked hexasaccharide in fetuin. J Biol Chem. 1987;262:16135–16141. [PubMed] [Google Scholar]
- 18.Esposito J J, Baxby D, Black D N, Dales S, Darai G, Dumbell K R, Granados R R, Joklik W K, McFadden G, Moss B, Moyer R W, Pickup D J, Robinson A J, Tripathy D N. Poxviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: the classification and nomenclature of viruses. The Sixth Report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. pp. 79–91. [Google Scholar]
- 19.Falkner F G, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenner F. Myxoma virus. In: Osterhaus A D M E, editor. Virus infections of vertebrates. 5. Virus infections of rodents and lagomorphs. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 59–70. [Google Scholar]
- 21.Fenner F, Marshall I D. A comparison of the virulence for European rabbits (Oryctolagus cuniculus) of strains of myxoma virus recovered in the field in Australia, Europe and America. J Hyg. 1957;55:149–191. doi: 10.1017/s0022172400037098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcés J, Masternak K, Kunz B, Wittek R. Reactivation of transcription from a vaccinia virus early promoter late in infection. J Virol. 1993;67:5394–5401. doi: 10.1128/jvi.67.9.5394-5401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 24.Hansen J E, Lund O, Tolstrup N, Gooley A A, Williams K L, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 25.Jackson R J, Bults H G. The myxoma virus thymidine kinase gene: sequence and transcriptional mapping. J Gen Virol. 1992;73:323–328. doi: 10.1099/0022-1317-73-2-323. [DOI] [PubMed] [Google Scholar]
- 26.Jackson R J, Hall D F. The myxoma virus EcoRI-O fragment encodes the DNA binding core protein and the major envelope protein of extracellular poxvirus. Virus Genes. 1998;17:55–62. doi: 10.1023/a:1008005101787. [DOI] [PubMed] [Google Scholar]
- 27.Jackson R J, Hall D F, Kerr P J. Construction of recombinant myxoma viruses expressing foreign genes from different intergenic sites without associated attenuation. J Gen Virol. 1996;77:1569–1575. doi: 10.1099/0022-1317-77-7-1569. [DOI] [PubMed] [Google Scholar]
- 28.Karupiah G, Blanden R V. Anti-asialo-GM1 inhibits vaccinia virus infection of murine ovaries: asialo-GM1 as an additional virus receptor? Immunol Cell Biol. 1990;68:343–346. doi: 10.1038/icb.1990.46. [DOI] [PubMed] [Google Scholar]
- 29.Kerr P J, Jackson R J. Myxoma virus as a vaccine vector for rabbits: antibody levels to influenza virus haemagglutinin presented by a recombinant myxoma virus. Vaccine. 1995;13:1722–1726. doi: 10.1016/0264-410x(95)00113-f. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y J, Varki A. Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconj J. 1997;14:569–576. doi: 10.1023/a:1018580324971. [DOI] [PubMed] [Google Scholar]
- 31.Kitagawa H, Paulson J C. Cloning and expression of human Galβ1,3(4)GlcNAc alpha 2,3-sialyltransferase. Biochem Biophys Res Commun. 1993;194:375–382. doi: 10.1006/bbrc.1993.1830. [DOI] [PubMed] [Google Scholar]
- 32.Kitagawa H, Paulson J C. Cloning of a novel α2,3-sialyltransferase that sialylates glycoprotein and glycolipid carbohydrate groups. J Biol Chem. 1994;269:1394–1401. [PubMed] [Google Scholar]
- 33.Kitagawa H, Paulson J C. Differential expression of five sialyltransferase genes in human tissues. J Biol Chem. 1994;269:17872–17878. [PubMed] [Google Scholar]
- 34.Lundberg K S, Shoemaker D D, Adams M W, Short J M, Sorge J A, Mathur E J. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991;108:1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 35.Massung R F, Esposito J J, Liu L-I, Qi J, Utterback T R, Knight J C, Aubin L, Yuran T E, Parsons J M, Loparev V N, Selivanov N A, Cavallaro K F, Kerlavage A R, Mahy B W J, Venter J C. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature. 1993;366:748–751. doi: 10.1038/366748a0. [DOI] [PubMed] [Google Scholar]
- 36.McFadden G, Barry M. How poxviruses oppose apoptosis. Semin Virol. 1998;8:429–442. [Google Scholar]
- 37.McFadden G, Graham K. Modulation of cytokine networks by poxviruses: the myxoma virus model. Semin Virol. 1994;5:421–429. [Google Scholar]
- 38.McFadden G, Lalani A, Everett H, Nash P, Xu X. Virus-encoded receptors for cytokines and chemokines. Semin Cell Dev Biol. 1998;9:359–368. doi: 10.1006/scdb.1998.0245. [DOI] [PubMed] [Google Scholar]
- 39.McMahan C J, Slack J L, Mosley B, Cosman D, Lupton S D, Brunton L L, Grubin C E, Wignall J M, Jenkins N A, Brannan C I, Copeland N G, Huebner K, Croce C M, Cannizzarro L A, Benjamin D, Dower S K, Spriggs M K, Sims J E. A novel IL-1 receptor, cloned from B cells by mammalian expression, is expressed in many cell types. EMBO J. 1991;10:2821–2832. doi: 10.1002/j.1460-2075.1991.tb07831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moses A. O virus do mixoma dos coelhos. Mem Inst Oswaldo Cruz. 1911;3:46–53. [Google Scholar]
- 41.Mykytowycz R. An attenuated strain of the myxomatosis virus recovered from the field. Nature. 1953;172:448–449. doi: 10.1038/172448a0. [DOI] [PubMed] [Google Scholar]
- 42.Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsson B, Nordén N E, Svensson S. Structural studies on the carbohydrate portion of fetuin. J Biol Chem. 1979;254:4545–4553. [PubMed] [Google Scholar]
- 44.Opgenorth A, Graham K, Nation N, Strayer D, McFadden G. Deletion analysis of two tandemly arranged virulence genes in myxoma virus, M11L and myxoma growth factor. J Virol. 1992;66:4720–4731. doi: 10.1128/jvi.66.8.4720-4731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parks R J, Lichty B D, Karakis C, Evans D H. Characterization of the Shope fibroma virus DNA ligase gene. Virology. 1994;202:642–650. doi: 10.1006/viro.1994.1385. [DOI] [PubMed] [Google Scholar]
- 46.Payne L G. Characterization of vaccinia virus glycoprotein by monoclonal antibody precipitation. Virology. 1992;187:251–260. doi: 10.1016/0042-6822(92)90313-e. [DOI] [PubMed] [Google Scholar]
- 47.Robinson D N, Cooley L. Drosophila kelch is an oligomeric ring canal actin organizer. J Cell Biol. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy A, Lu C F, Marykwas D L, Lipke P N, Kurjan J. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol Cell Biol. 1991;11:4196–4206. doi: 10.1128/mcb.11.8.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasaki K. Molecular cloning and characterization of sialyltransferases. Trends Glycosci Glycotechnol. 1996;8:195–215. [Google Scholar]
- 50.Savill J, Fadok V, Henson P, Haslett C. Phagocyte recognition of cells undergoing apoptosis. Immunol Today. 1993;14:131–136. doi: 10.1016/0167-5699(93)90215-7. [DOI] [PubMed] [Google Scholar]
- 51.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmid M F, Agris J M, Jakana J, Matsudaira P, Chiu W. Three-dimensional structure of a single filament in the Limulus acrosomal bundle: scruin binds to homologous helix-loop-beta motifs in actin. J Cell Biol. 1994;124:341–350. doi: 10.1083/jcb.124.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shope R E. A filterable virus causing a tumor-like condition in rabbits and its relationship to virus myxomatosum. J Exp Med. 1932;56:803–822. doi: 10.1084/jem.56.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonnhammer E L, Eddy S R, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 55.Tonissen K F, Krieg P A. Two neural-cell adhesion molecule (NCAM)-encoding genes in Xenopus laevis are expressed during development and in adult tissues. Gene. 1993;127:243–247. doi: 10.1016/0378-1119(93)90727-k. [DOI] [PubMed] [Google Scholar]
- 56.Tsuji S, Datta A K, Paulson J C. Systematic nomenclature for sialyltransferases. Glycobiology. 1996;6:R5–R7. doi: 10.1093/glycob/6.7.647. [DOI] [PubMed] [Google Scholar]
- 57.Upton C, Macen J L, Wishart D S, McFadden G. Myxoma virus and malignant rabbit fibroma virus encode a serpin-like protein important for virus virulence. Virology. 1990;179:618–631. doi: 10.1016/0042-6822(90)90129-f. [DOI] [PubMed] [Google Scholar]
- 58.Upton C, Schiff L, Rice S A, Dowdeswell T, Yang X, McFadden G. A poxvirus protein with a RING finger motif binds zinc and localizes in virus factories. J Virol. 1994;68:4186–4195. doi: 10.1128/jvi.68.7.4186-4195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanderplasschen A, Smith G L. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J Virol. 1997;71:4032–4041. doi: 10.1128/jvi.71.5.4032-4041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varki A. Biological roles of oligosaccharides: all the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varki A. Sialic acids as ligands in recognition phenomena. FASEB J. 1997;11:248–255. doi: 10.1096/fasebj.11.4.9068613. [DOI] [PubMed] [Google Scholar]
- 62.von Bulow M, Heid H, Hess H, Franke W W. Molecular nature of calicin, a major basic protein of the mammalian sperm head cytoskeleton. Exp Cell Res. 1995;219:407–413. doi: 10.1006/excr.1995.1246. [DOI] [PubMed] [Google Scholar]
- 63.Wang I-N, Li Y, Que Q, Bhattacharya M, Lane L C, Chaney W G, Van Etten J L. Evidence for virus-encoded glycosylation specificity. Proc Natl Acad Sci USA. 1993;90:3840–3844. doi: 10.1073/pnas.90.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Way M, Sanders M, Chafel M, Tu Y H, Knight A, Matsudaira P. Beta-scruin, a homologue of the actin crosslinking protein scruin, is localized to the acrosomal vesicle of Limulus sperm. J Cell Sci. 1995;108:3155–3162. doi: 10.1242/jcs.108.10.3155. [DOI] [PubMed] [Google Scholar]
- 65.Way M, Sanders M, Garcia C, Sakai J, Matsudaira P. Sequence and domain organization of scruin, an actin-cross-linking protein in the acrosomal process of Limulus sperm. J Cell Biol. 1995;128:51–60. doi: 10.1083/jcb.128.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xue F, Cooley L. Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 67.Yuen L, Moss B. Oligonucleotide sequence signaling transcriptional termination of vaccinia virus early genes. Proc Natl Acad Sci USA. 1987;84:6417–6421. doi: 10.1073/pnas.84.18.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]