Abstract
Why the autosomal recombination rate differs between female and male meiosis in most species has been a genetic enigma since the early study of meiosis. Some hypotheses have been put forward to explain this widespread phenomenon and, up to now, only one fact has emerged clearly: In species in which meiosis is achiasmate in one sex, it is the heterogametic one. This pattern, known as the Haldane-Huxley rule, is thought to be a side effect, on autosomes, of the suppression of recombination between the sex chromosomes. However, this rule does not hold for heterochiasmate species (i.e., species in which recombination is present in both sexes but varies quantitatively between sexes) and does not apply to species lacking sex chromosomes, such as hermaphroditic plants. In this paper, we show that in plants, heterochiasmy is due to a male-female difference in gametic selection and is not influenced by the presence of heteromorphic sex chromosomes. This finding provides strong empirical support in favour of a population genetic explanation for the evolution of heterochiasmy and, more broadly, for the evolution of sex and recombination.
Measuring the opportunity for natural selection on gametes provides the first empirical evidence for a theory explaining why recombination at meiosis varies between males and females
Introduction
Sex differences in recombination were discovered in the first linkage studies on Drosophila [1,2] and Bombyx (Tanaka [1914] in [3]) almost one century ago. However, this observation remains today largely unexplained despite several attempts. Based on very limited observations (see Table 1), especially of Bombyx, in which the female is heterogametic, Haldane [3] suggested, as far as “these facts are anything more than a coincidence,” that the lower autosomal recombination rate in the heterogametic sex may reflect a pleiotropic consequence of selection against recombination between the sex chromosomes. Later, Huxley [4] showed that Gammarus males also recombined less than females. He gave the same evolutionary explanation, although he restricted it to cases of a marked sex difference.
Table 1. Data on Which the Haldane-Huxley Rule is Based.
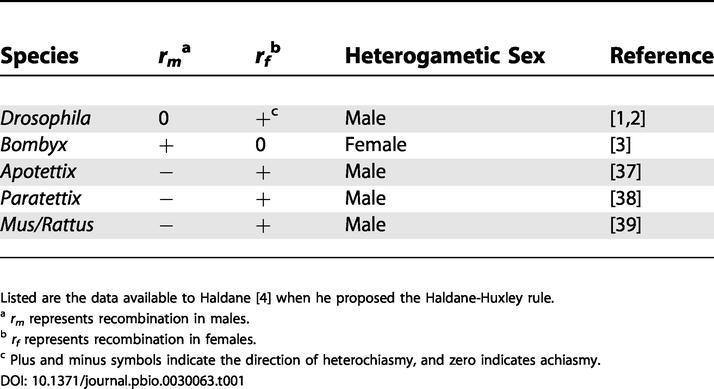
Listed are the data available to Haldane [4] when he proposed the Haldane-Huxley rule
a rm represents recombination in males
b rf represents recombination in females
c Plus and minus symbols indicate the direction of heterochiasmy, and zero indicates achiasmy
This conjecture has now been confirmed for achiasmate species (i.e., species in which only one sex recombines) and is referred to as Haldane-Huxley rule: Nei [5] showed theoretically that tight linkage should evolve on Y or W chromosomes, and Bell [6] compiled a large dataset showing that achiasmy evolved 29–34 times independently, each time with no recombination in the heterogametic sex.
However, for heterochiasmate species, three problems with the Haldane-Huxley pleiotropy explanation were discovered [7,8]. The first problem arose when substantial variation in male-female differences in recombination rate was found between pairs of autosomes within mice [8] and Tribolium [9,10], and between genotypes for the same pair of autosomes [11]. The second problem was the discovery that hermaphrodite species (the platyhelminth Dendrocoelum [12] and the plant Allium [13]) may present strong heterochiasmy between male and female meiosis despite having no sex chromosomes or even sex-determining loci. The third problem was the discovery of species in which the heterogametic sex recombines more than the homogametic one (e.g., in some Triturus species) [14]. Because of these contradictory observations, variation in heterochiasmy has remained difficult to explain because of the absence of an alternative theory as well as the lack of a clear pattern in the data.
In 1969, Nei [5] worked out the first “modifier” model to study the evolution of sex differences in recombination, and concluded for autosomes that “the evolutionary mechanism of these sex differences is not known at present.” Surveying an updated dataset, Bell [6] concluded that “female gametes experience more crossing over among hermaphroditic plants (and perhaps animals), but this is not invariably the case among gonochoric animals (…) certainly (this) has never received any explanation.” The idea that heterochiasmy may be explained by a sex rather than by a sex chromosome effect, which was ignored by Haldane because of Bombyx, was reconsidered. This led Trivers [15] to suggest that, because only males with very good gene combinations reproduce (relative to females, for whom reproduction success is often less variable), they should recombine less to keep intact these combinations. He accounted for exceptions by variation in the regime of sexual selection. The idea was criticized by Burt et al. [16], who also questioned the correlations—with an updated dataset—between heterochiasmy and either sex or heterogamety. These authors tried to correlate the level of heterochiasmy with the amount of “opportunity for sex-specific selection,” but failed to find an effect. They were tempted to advocate neutrality, but were puzzled by the positive correlation between male and female recombination rate and by evidence showing compensation (e.g., female mice tend to recombine more on the X, as if they were compensating for no recombination in males; similarly, no species is known with achiasmy in both sexes [16]). In 1994, Korol et al. [17] insisted on a possible role for gametic selection but did not give evidence in favour of this claim. Recently, Lenormand [18], using Nei's modifier approach, showed that it is very difficult to explain heterochiasmy by sex-specific diploid selection. Rather, a sex difference in selection during haploid phase, or a sex difference in diploid selection on imprinted genes, is a more likely explanation. He predicted that, as far as haploid selection is concerned, the sex experiencing the more intense haploid selection should recombine less. Indeed, when allelic effects interact to determine fitness (i.e., when there is “epistasis,” either negative or positive), recombining decreases mean fitness in the population of the next generation [19]. This effect occurs because recombination breaks up combinations of genes that have previously been built up by selection. For a given average recombination rate between sexes and for a given average epistasis between male and female haploids, it is always advantageous for the haploid population (male or female) with the greatest absolute value in epistasis to be produced with the lowest amount of recombination. In this way, the “recombination load” that the haploid population is exposed to is minimized.
In this paper, we would like to come up with a more quantitative evaluation of the possible role of haploid selection in shaping heterochiasmy. For that purpose, we first updated the dataset of Burt et al. [16] on heterochiasmy, focusing on genetic maps that have become available over the last 15 years. We then determined how fast heterochiasmy evolves, in order to measure the amount of phylogenetic inertia on this trait. Finally, we determined whether variables such as gender, heterogamety, or the opportunity for selection in the haploid phase, could explain variation in heterochiasmy. If there is selection with substantial epistasis on some genes during the haploid phase, we expect the sex with the greater opportunity for haploid selection to show less recombination. Alternatively, if selection during the haploid phase is weak or without substantial epistasis, we do not expect it to produce a directional bias in the amount of recombination displayed by either sex.
Results/Discussion
Sex Chromosomes
Heterochiasmy is a fast-evolving trait, and phylogenetic inertia does not satisfactorily explain its distribution. In contrast to achiasmy, we found that heterochiasmy is not influenced by the nature of the sex chromosomes. This is interesting, because it suggests that achiasmy and heterochiasmy are influenced by qualitatively different evolutionary forces, although they seem to differ only quantitatively. It would be useful to determine whether achiasmy evolved to reduce the average recombination rate or to change the relative amount of recombination between the sexes. The two situations may be discriminated by determining whether the homogametic sex in achiasmate species tends to recombine more than in closely related chiasmate species. Evidence for such compensation would indicate that achiasmy did not evolve to reduce the average recombination rate. In the absence of such compensation, however, achiasmy may simply reflect selection for tight linkage. In such a situation, we propose that Haldane-Huxley rule may be caused by the converse argument to the one previously considered: The presence of achiasmy only in the heterogametic sex may reflect selection to maintain nonzero recombination rate on X or Z chromosomes in the homogametic sex. In species in which the average autosomal recombination rate is selected against (i.e., towards a lower equilibrium value), loss-of-function (recombination) mutations with an effect restricted to one sex may spread only if they affect the heterogametic sex, because mutations suppressing recombination in the homogametic sex completely suppress recombination on the X or Z chromosome. The same argument applies to XO species and may explain why achiasmy is associated only with the heterogametic sex. In addition, this hypothesis does not require the existence of genes suppressing recombination between the sex chromosomes with autosomal pleiotropic effects. Under this hypothesis, there is no reason to find an effect of the presence of heteromorphic sex chromosome on the amount of heterochiasmy, as originally envisioned by Haldane and Huxley. Overall, this hypothesis would explain why heterochiasmy and achiasmy differ qualitatively and why we do not observe any effect of sex chromosomes on heterochiasmy.
Heterochiasmy in Animals
In animals, male-female dimorphism in haploid selection may also contribute to heterochiasmy. In general, there is no female haploid phase in animals, because meiosis is completed only at fertilisation. As far as at least some genes are expressed and under selection during the male haploid phase, this would tend to bias towards tighter linkage in males. Sets of genes responsible for male-specific meiotic drive systems would be good candidates and are often found in tight linkage. Measuring the opportunity for haploid selection in animals may be possible within some groups. Imprinting may, however, act as a confounding effect in many groups of animals while trying to measure the opportunity for “haploid” selection. Within-species comparisons of imprinted regions or of regions with sex-specific recombination using high-resolution maps [20] may be more fruitful to discriminate among potential causes of heterochiasmy in animals. In particular, there is evidence in humans that the reduction in crossing-over associated with imprinting is in the direction that theory predicts, even if this pattern is consistent with other explanations [21]. Finally, understanding exceptions within groups (e.g., male marsupials, contrarily to most mammals, recombine more than females of the species [22]) may also shed light on the different hypotheses.
Heterochiasmy in Plants
We found that plant heterochiasmy is correlated with the opportunity for male and female haploid selection. Female meiosis tends to exhibit lower recombination rates relative to male meiosis when selection is intense among female gametophytes (e.g., in Pinaceae) or mild among male gametophytes (e.g., in highly selfing species). This pattern is expected if heterochiasmy is determined by the relative magnitude of haploid selection in male and female individuals. Finding a pattern consistent with this general population genetic prediction is, of course, not firm evidence that male-female dimorphism in haploid selection is the evolutionary force generating heterochiasmy. Other correlates of selfing rates might have to be closely examined [23]. However, we consider this explanation the most parsimonious so far. Our finding provides, therefore, the first empirical evidence for a theory explaining male-female differences in the amount of recombination and contributes to our understanding of contradictory observations that have puzzled geneticists for almost a century. It also indicates that the amount of recombination may be shaped by indirect selection, and, therefore, corroborates theories based on selection and variation for the evolution of sexual reproduction.
Materials and Methods
An extended dataset
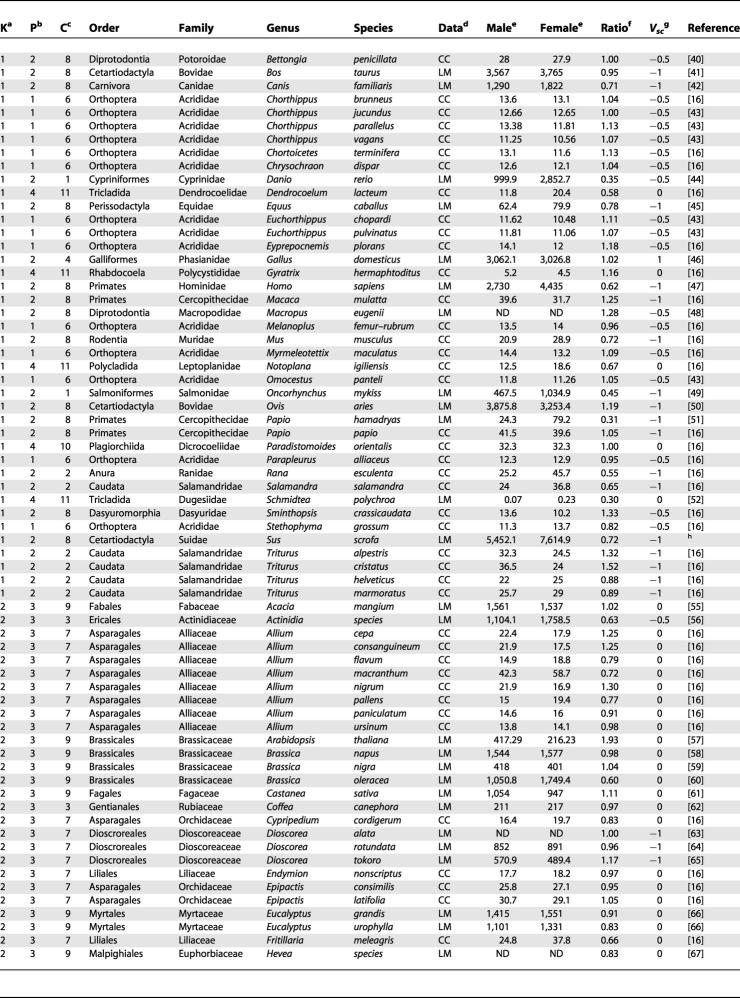
We measured heterochiasmy as the log of the male/-to-female ratio (ρ) of autosomal recombination rate measured either with chiasma number or map length. We log-transformed the ratio to avoid bias due to measurement error in the denominator. Chiasma-count data for different species were compiled by Burt et al. [16], and we used their dataset, adding a few recent studies. We compiled genetic map data and linkage studies in animals and plants for which both a male and a female map were available. Only homologous fragments (i.e., between shared markers) in male and female maps were considered (especially in low-resolution maps). Heterochiasmy data were available for 107 species, with 46 sets of data based on genomic maps (Table 2).
Table 2. Dataset Pooled by Species with Levels of Phylogenetic Grouping Used in the Analysis.
Note that references given in Burt et al. [17] were not repeated here
a K, kingdom. Numeric indicators in this column are: 1, Animalia; 2, Plantae
b P, phylum. Numeric indicators in this column are: 1, Arthropoda; 2, Chordata; 3, Embryophyta; 4, Platyhelminthes
c C, class. Numeric indicators in this column are: 1, Actinopterygii; 2, Amphibia; 3, Magnoliopsidae (subclass asterids); 4, Aves; 5, Coniferopsida; 6, Insecta; 7, Liliopsida; 8, Mammalia; 9, Magnoliopsidae (subclass rosids); 10, Trematoda; 11, Turbellaria
d Data refers to linkage map (LM) or chiasma count (CC)
e Male and female indicate the value for the chiasma count or map length for each sex
f Ratio refers to male/female recombination rate
g Vsc refers to the presence or absence of sex chromosome (see Materials and Methods, “Sex chromosome effect”)
h Data were obtained from maps DBNordic2 and NIAIJapan (http://www.genome.iastate.edu/pig.html) [54,55]
ND, no data
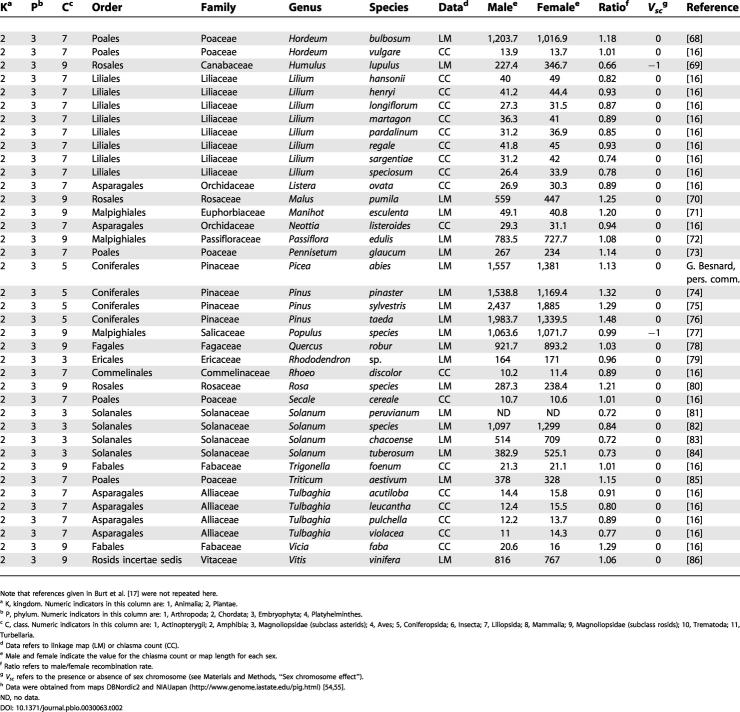
Table 2. Continued.
Phylogenetic inertia
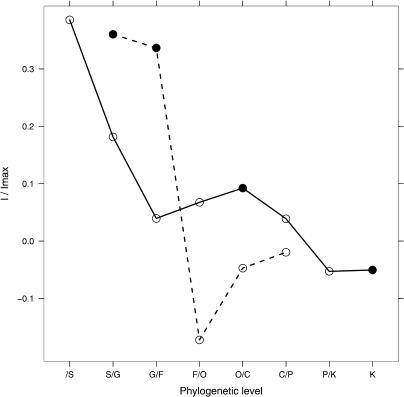
Heterochiasmy may evolve so slowly that there is important phylogenetic inertia. Alternatively, it may be so fast-evolving that the amount of heterochiasmy takes on nearly independent values among related species. In the same way, heterochiasmy may be so variable between genotypes within a species that it may be difficult to measure and irrelevant to analyse species specific effects. In order to get a picture of phylogenetic inertia on heterochiasmy, we estimated the phylogenetic autocorrelation of ρ using Moran's I spatial autocorrelation statistic [24]. When standardized, values of Moran's I vary from −1 to 1. Positive values indicate that heterochiasmy is more similar than random within a taxonomic level, whereas negative values indicate that it is more different. Because a few species had multiple estimates of heterochiasmy, we also estimated the within-species correlation. The resulting correlogram is shown in Figure 1. We found that heterochiasmy is a fast-evolving trait: Genotypes tend to be correlated within a species (I/Imax = 0.38, p = 7.9%), but this correlation is lower among species within genera (I/Imax = 0.18, P-value = 13%), and very low when comparing genera within families (I/Imax = 0.039, p = 63%). This pattern is very different from the one observed for highly autocorrelated traits using the same method (for instance, mammalian body size [25]). This analysis indicates that there is very little phylogenetic inertia overall on heterochiasmy, but that the species level is appropriate for our dataset. However, this low level of inertia may nevertheless inflate type-I error while testing the effect of independent variables on heterochiasmy. In order to avoid this problem, we tested the association between different variables and heterochiasmy using a generalized estimating equations linear model correcting for the full phylogeny (see below) [26].
Figure 1. Phylogenetic Correlogram of Heterochiasmy and Selfing Rate.
The y-axis represents Moran's I rescaled to enable comparisons between each taxonomic level for heterochiasmy (ρ, solid line) and selfing rate (Vm, dashed line). The x-axis represents the taxonomic level: /S is the correlation within species, S/G is the correlation of species within genera, etc. F, family; O, order; C, class; P, phylum; K, kingdom. Filled points indicate significance at p = 0.05.
Sex chromosome effect
For each species, we reported the presence of sex chromosomes. We defined the variable Vsc with the following values: −1 for XY/XX species, −1/2 for XO/XX or XY/XX without pseudoautosomal regions (marsupials), 0 for species without sex-chromosomes, and +1 for ZZ/ZW species. We distinguished the −1 and −1/2 cases to reflect the fact that, in the latter, recombination does not occur between sex chromosomes, so we expect a lower current selection pressure to suppress recombination. Under the Haldane-Huxley hypothesis, the presence of sex chromosomes is supposed to favour reduced recombination rate in the heterogametic sex. We therefore expect a positive effect of the variable Vsc on ρ. We did not find such an effect in animals or plants (the linear effect of Vsc on ρ is not significantly different from zero [p = 0.75 in animals and p = 0.52 in plants], assuming species were independent), and this result is unchanged if the −1 and −1/2 cases are not distinguished. Given this negative result, there was no need to do a phylogenetic correction.
Gametic selection
In animals from our dataset, there is no female haploid phase because the completion of meiosis occurs only at fertilisation (sperm triggers the end of meiosis). In male gametes, very few genes are expressed, and sperm phenotype is determined mostly either by the diploid genotype of the paternal tissue or by its mitochondrial genome. Imprinted genes, which can also affect the evolution of heterochiasmy [18,21], may be as numerous as haploid-expressed genes and act as a confounding factor while evaluating the “opportunity” for male or female gametic selection. As a consequence, we did not attempt to evaluate the opportunity for haploid selection in animals. Rather, we focused on plants, in which there is both a male (pollen) and female (ovule) haploid phase and during which many genes are expressed (e.g., as many as 60% of genes may be expressed in the male gametophyte [27,28]).
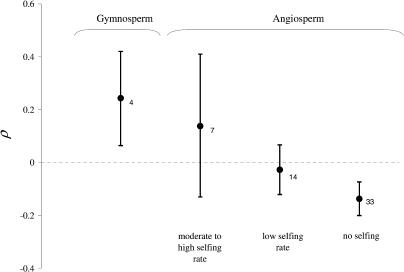
In order to evaluate the effect of the “opportunity for selection” for male haploid phase on ρ, we used selfing rate as an indirect variable estimating the degree of pollen competition. We assume that with high selfing rates, there is less genetic variation among competing pollen grains and, therefore, less scope for haploid selection. We defined Vm (the degree of male gamete competition in plants) using three values depending on the amount of selfing: 0 for dioecious, self-incompatible or largely outcrossing (less than 5% selfing reported) species; 1 for species exhibiting low selfing rates (less than 30% reported); and 2 for other species. We used these three broad categories to reflect the fact that selfing rate is often variable within species and that it is often measured indirectly and with low precision. We therefore expect a positive effect of the variable Vm on ρ if the opportunity for male gametic selection favours smaller ρ values, as predicted by the modifier model [18]. We tested this effect using the 57 species for which we were able to estimate Vm (Table 3). We used a linear model in R [29] assuming that all species are either independent or phylogenetically related. In the latter case, we used a generalized estimating equations linear model [26] with a plant phylogenetic tree to the family level using data from Davies et al. [30], and several calibration points, including the Picea/Pinus divergence approximately 140 million years ago [31], that are not included in the Davies et al. dataset. We found an effect in the right direction with or without correcting for the phylogeny (linear effect of ρ on Vm, p < 0.0002 in both cases, Figure 2). The fact that selfing plants exhibit higher recombination rates than their outcrossing relatives has been mentioned previously in the literature [32,33]. However, in most cases, recombination was measured only in male meiosis. It would be valuable to reexamine this trend in the light of our results that recombination in male meiosis is typically greater than in female meiosis among selfers.
Table 3. Plant Species Used to Test the Effect of Male and Female Opportunity for Selection.
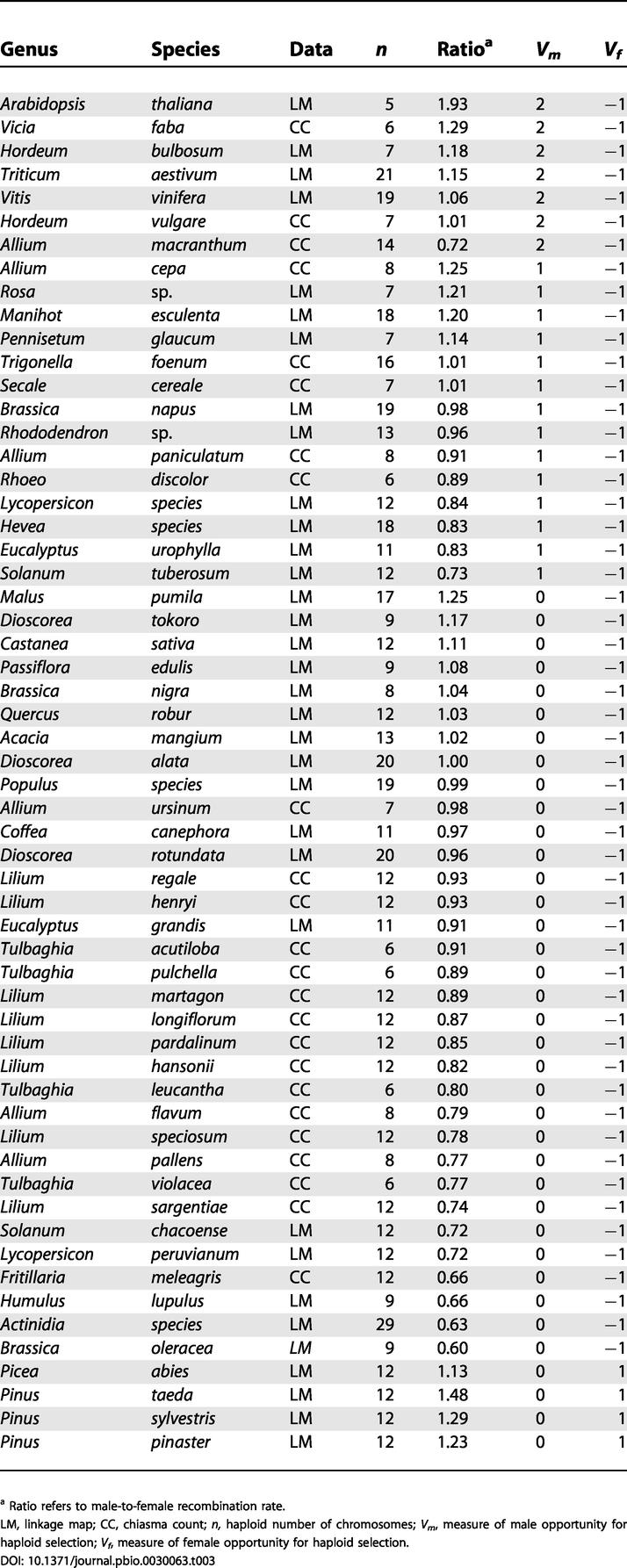
a Ratio refers to male-to-female recombination rate
LM, linkage map; CC, chiasma count; n, haploid number of chromosomes; Vm, measure of male opportunity for haploid selection; Vf, measure of female opportunity for haploid selection
Figure 2. Logarithm of Male-Female Ratio in Recombination Rate in Plants.
Mean and 95% confidence interval of ρ is shown for different groups of plants, assuming normality and independent data points The number of species in each group is indicated next to the mean.
In order to evaluate the effect of the “opportunity for selection” during the female haploid phase on ρ in plants, we contrasted angiosperms with gymnosperms. In angiosperms, ovules do not compete much with each other on a mother plant, because resource accumulation starts after fertilisation (i.e., during fruit development in the diploid phase). In Pinus (three species in our dataset; see Table 2), male meiosis, female meiosis, and pollination occur in the year prior to fertilisation, but the pollen tube stops growing until the next spring, while the female gametophytes continue to accumulate resources and compete with each other over the course of the year. The same situation occurs in Picea, although the period between female meiosis and fertilisation is only 2–3 mo [34]. Perhaps more importantly, the endosperm (which is the organ managing resources for the zygote) is haploid in Pinaceae, in contrast to the double fertilisation that occurs in angiosperms to produce at least a diploid (typically triploid) endosperm [35,36]. We therefore expect that ρ should be greater in Pinaceae, compared to angiosperms. We assigned Vf (the degree of female gamete competition in plants) the values 1 for gymnosperms and −1 for angiosperms. We expected a positive effect of the variable Vf on ρ according to the modifier model. An effect in the right direction was indeed detected (linear effect of Vf on ρ, p = 0.011 and p = 0.0001, with and without correcting for the phylogeny as above, respectively; see Figure 2).
Acknowledgments
We thank G. Besnard, J. Britton-Davidian, J.-B. Ferdy, S. Glémin, L. D. Hurst, P. Jarne, O. Judson, M. Kirkpatrick, S. P. Otto, J.-M. Prosperi, and C. Vosa for helpful comments, information, and stimulating discussions. This study was supported by the Centre National de la Recherche Scientifique and French Ministry of Research.
Competing interests. The authors have declared that no competing interests exist.
Author contributions. TL and JD conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, and wrote the paper.
Citation: Lenormand T, Dutheil J (2005) Recombination difference between sexes: A role for haploid selection. PLoS Biol 3(3): e63.
References
- Morgan T. Complete linkage in the second chromosome of the male of Drosophila . Science. 1912;36:719–720. [Google Scholar]
- Morgan T. No crossing over in the male of Drosophila of genes in the second and third pairs of chromosomes. Biol Bull Woods Hole. 1914;26:195–204. [Google Scholar]
- Haldane J. Sex ratio and unisexual sterility in hybrid animals. J Genet. 1922;12:101–109. [Google Scholar]
- Huxley J. Sexual difference of linkage in Gammarus chevreuxi . J Genet. 1928;20:145–156. [Google Scholar]
- Nei M. Linkage modification and sex difference in recombination. Genetics. 1969;63:681–699. doi: 10.1093/genetics/63.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. The masterpiece of nature: The evolution and genetics of sexuality. Berkeley: University of California Press; 1982. 685 pp. [Google Scholar]
- Callan HG, Perry PE. Recombination in male and female meiocytes contrasted. Philos Trans R Soc Lond B Biol Sci. 1977;277:227–233. doi: 10.1098/rstb.1977.0013. [DOI] [PubMed] [Google Scholar]
- Dunn L, Bennett D. Sex differences in recombination of linked genes in animals. Genet Res. 1967;9:211–220. doi: 10.1017/s0016672300010491. [DOI] [PubMed] [Google Scholar]
- Dawson P. Sex and crossing over in linkage group IV of Tribolium castaneum . Genetics. 1972;72:525–530. doi: 10.1093/genetics/72.3.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff A. Sex and crossing over in Tribolium castaneum . Genetics. 1964;50:491–496. doi: 10.1093/genetics/50.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DS. Different Frequency in the recovery of crossover products from male and female gametes of plants hypoploid for B-a translocations in maize. Genetics. 1984;107:117–130. doi: 10.1093/genetics/107.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor JB, Callan HG. Chiasma formation in spermatocytes and oocytes of the turbellarian. J Genet. 1952;50:449–454. [Google Scholar]
- Ved Brat S. Genetic systems in Allium. 2. Sex differences in meiosis. Chromosomes Today. 1966;1:31–40. [Google Scholar]
- Watson ID, Callan HG. Form of bivalent chromosomes in newt oocytes at first metaphase of meiosis. Q J Microsc Sci. 1963;104:281–295. [Google Scholar]
- Trivers R. Sex differences in rates of recombination and sexual selection. In: Michod RE, Levin BR, editors. The evolution of sex. Sunderland, Massachusetts: Sinauer Press; 1988. pp. 270–286. [Google Scholar]
- Burt A, Bell G, Harvey PH. Sex differences in recombination. J Evol Biol. 1991;4:259–277. [Google Scholar]
- Korol AB, Preygel IA, Preygel SI. Recombination variability and evolution. London: Chapman and Hall; 1994. 361 pp. [Google Scholar]
- Lenormand T. The evolution of sex dimorphism in recombination. Genetics. 2003;163:811–822. doi: 10.1093/genetics/163.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Barton NH. Recombination load associated with selection for increased recombination. Genet Res. 1996;67:27–41. doi: 10.1017/s0016672300033450. [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Hurst LD. Imprinted chromosomal regions of the human genome have unusually high recombination rates. Genetics. 2003;165:1629–1632. doi: 10.1093/genetics/165.3.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samollow PB, Kammerer CM, Mahaney SM, Schneider JL, Westenberger SJ, et al. First-generation linkage map of the gray, short-tailed opossum, Monodelphis domestica, reveals genome-wide reduction in female recombination rates. Genetics. 2004;166:307–329. doi: 10.1534/genetics.166.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koella J. Ecological correlates of chiasma frequency and recombination index of plants. Biol J Linn Soc Lond. 1993;48:227–238. [Google Scholar]
- Gittleman J, Kot M. Adaptation—Statistics and a null model for estimating phylogenetic effects. System Zool. 1990;39:227–241. [Google Scholar]
- Smith F. Similarity of mammalian body size across the taxonomic hierarchy and across space and time. Am Nat. 2004;163:672–691. doi: 10.1086/382898. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J. Analysis of comparative data using generalized estimating equations. J Theor Biol. 2002;218:175–185. doi: 10.1006/jtbi.2002.3066. [DOI] [PubMed] [Google Scholar]
- Mulcahy DL, SariGorla M, Mulcahy GB. Pollen selection—Past, present and future. Sex Plant Reprod. 1996;9:353–356. [Google Scholar]
- Bernasconi G, Ashman TL, Birkhead TR, Bishop JDD, Grossniklaus U, et al. Evolutionary ecology of the prezygotic stage. Science. 2004;303:971–975. doi: 10.1126/science.1092180. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, et al. Darwin's abominable mystery: Insights from a supertree of the angiosperms. Proc Natl Acad Sci U S A. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Tank DC, Sang T. Phylogeny and divergence times in Pinaceae: Evidence from three genomes. Mol Biol Evol. 2000;17:773–781. doi: 10.1093/oxfordjournals.molbev.a026356. [DOI] [PubMed] [Google Scholar]
- Stebbins G. Longevity, habitat, and release of genetic variability in the higher plants. Cold Spring Harb Symp Quant Biol. 1958;23:365–377. doi: 10.1101/sqb.1958.023.01.035. [DOI] [PubMed] [Google Scholar]
- Grant V. The regulation of recombination in plants. Cold Spring Harb Symp Quant Biol. 1958;23:337–363. doi: 10.1101/sqb.1958.023.01.034. [DOI] [PubMed] [Google Scholar]
- Owens JN, Johnsen O, Daehlen OG, Skroppa T. Potential effects of temperature on early reproductive development and progeny performance in Picea abies (L.) Karst. Scand J For Res. 2001;16:221–237. [Google Scholar]
- Raven P, Evert R, Eichhorn S. Biology of plants. New York: WH Freeman and Co; 1999. 875 pp. [Google Scholar]
- Baroux C, Spillane C, Grossniklaus U. Evolutionary origins of the endosperm in flowering plants. Genome Biol. 2002;3:reviews1026.1–reviews1026.5. doi: 10.1186/gb-2002-3-9-reviews1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabours R. Parthenogenesis and crossing-over in the grouse locust Apotettix . Am Nat. 1919;53:131–142. [Google Scholar]
- Haldane J. Note on a case of linkage in Paratettix . J Genet. 1920;10:47–51. [Google Scholar]
- Dunn L. Linkage in mice and rats. Genetics. 1920;5:325–343. doi: 10.1093/genetics/5.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman DL, Smith MJ, Rodger JC. A comparative study of chiasmata in male and female Bettongia penicillata (Marsupialia) Genetica. 1990;83:45–49. [Google Scholar]
- Barendse W, Vaiman D, Kemp SJ, Sugimoto Y, Armitage SM, et al. A medium-density genetic linkage map of the bovine genome. Mamm Genome. 1997;8:21–28. doi: 10.1007/s003359900340. [DOI] [PubMed] [Google Scholar]
- Neff MW, Broman KW, Mellersh CS, Ray K, Acland GM, et al. A second-generation genetic linkage map of the domestic dog, Canis familiaris . Genetics. 1999;151:803–820. doi: 10.1093/genetics/151.2.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano MI, Santos JL. Chiasma frequencies and distributions in gomphocerine grasshoppers—A comparative study between sexes. Heredity. 1990;64:17–23. [Google Scholar]
- Singer A, Perlman H, Yan Y, Walker C, Corley-Smith G, et al. Sex-specific recombination rates in zebrafish (Danio rerio) Genetics. 2002;160:649–657. doi: 10.1093/genetics/160.2.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson L, Sandberg K. Genetic linkage in the horse. II. Distribution of male recombination estimates and the influence of age, breed and sex on recombination frequency. Genetics. 1984;106:109–122. doi: 10.1093/genetics/106.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen MAM, Crooijmans R, Veenendaal A, Cheng HH, Siwek M, et al. A comprehensive microsatellite linkage map of the chicken genome. Genomics. 1998;49:265–274. doi: 10.1006/geno.1998.5225. [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: Individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenger KR, Mckenzie LM, Cooper DW. The first comprehensive genetic linkage map of a marsupial: The tammar wallaby (Macropus eugenii) Genetics. 2002;162:321–330. doi: 10.1093/genetics/162.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Danzmann RG, Gharbi K, Howard P, Ozaki A, et al. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics. 2000;155:1331–1345. doi: 10.1093/genetics/155.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox JF, Davies KP, Crawford AM, Hulme DJ, Vaiman D, et al. An enhanced linkage map of the sheep genome comprising more than 1,000 loci. Genome Res. 2001;11:1275–1289. doi: 10.1101/gr.135001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Witte SM, Kammerer CM, Hixson JE, Maccluer JW. Linkage mapping in Papio baboons: Conservation of a syntenic group of six markers on human chromosome 1. Genomics. 1995;28:251–254. doi: 10.1006/geno.1995.1138. [DOI] [PubMed] [Google Scholar]
- Pongratz N, Gerace L, Alganza AM, Beukeboom LW, Michiels NK. Microsatellite development and inheritance in the planarian flatworm Schmidtea polychroa . Belg J Zool. 2001;131:71–75. [Google Scholar]
- Archibald AL, Haley CS, Brown JF, Couperwhite S, Mcqueen HA, et al. The PiGMaP consortium linkage map of the pig (Sus scrofa) Mamm Genome. 1995;6:157–175. doi: 10.1007/BF00293008. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Chowdhary BP, Fredholm M, Hoyheim B, Johansson M, et al. A physically anchored linkage map of pig chromosome 1 uncovers sex- and position-specific recombination rates. Genomics. 1994;24:342–350. doi: 10.1006/geno.1994.1625. [DOI] [PubMed] [Google Scholar]
- Butcher PA, Moran GF. Genetic linkage mapping in Acacia mangium. 2. Development of an integrated map from two outbred pedigrees using RFLP and microsatellite loci. Theor Appl Genet. 2000;101:594–605. [Google Scholar]
- Testolin R, Huang WG, Lain O, Messina R, Vecchione A, et al. A kiwifruit (Actinidia spp.) linkage map based on microsatellites and integrated with AFLP markers. Theor Appl Genet. 2001;103:30–36. [Google Scholar]
- Vizir IY, Korol AB. Sex Difference in recombination frequency in Arabidopsis . Heredity. 1990;65:379–383. [Google Scholar]
- Kelly AL, Sharpe AG, Nixon JH, Evans EJ, Lydiate DJ. Indistinguishable patterns of recombination resulting from mate and female meioses in Brassica napus (oilseed rape) Genome. 1997;40:49–56. doi: 10.1139/g97-007. [DOI] [PubMed] [Google Scholar]
- Lagercrantz U, Lydiate DJ. Rflp mapping in Brassica nigra indicates differing recombination rates in male and female meioses. Genome. 1995;38:255–264. doi: 10.1139/g95-032. [DOI] [PubMed] [Google Scholar]
- Kearsey MJ, Ramsay LD, Jennings DE, Lydiate DJ, Bohuon EJR, et al. Higher recombination frequencies in female compared to male meisoses in Brassica oleracea . Theor Appl Genet. 1996;92:363–367. doi: 10.1007/BF00223680. [DOI] [PubMed] [Google Scholar]
- Casasoli M, Mattioni C, Cherubini M, Villani F. A genetic linkage map of European chestnut (Castanea sativa Mill.) based on RAPD, ISSR and isozyme markers. Theor Appl Genet. 2001;102:1190–1199. [Google Scholar]
- Lashermes P, Combes MC, Prakash NS, Trouslot P, Lorieux M, et al. Genetic linkage map of Coffea canephora Effect of segregation distortion and analysis of recombination rate in male and female meioses. Genome. 2001;44:589–596. [PubMed] [Google Scholar]
- Mignouna HD, Mank RA, Ellis N, Van Den Bosch N, Asiedu R, et al. A genetic linkage map of water yam (Dioscorea alata L.) based on AFLP markers and QTL analysis for anthracnose resistance. Theor Appl Genet. 2002;105:726–735. doi: 10.1007/s00122-002-0912-6. [DOI] [PubMed] [Google Scholar]
- Mignouna HD, Mank RA, Ellis N, Van Den Bosch N, Asiedu R, et al. A genetic linkage map of guinea yam (Dioscorea rotundata Poir.) based on AFLP markers. Theor Appl Genet. 2002;105:716–725. doi: 10.1007/s00122-002-0911-7. [DOI] [PubMed] [Google Scholar]
- Terauchi R, Kahl G. Mapping of the Dioscorea tokoro genome: AFLP markers linked to sex. Genome. 1999;42:752–762. [Google Scholar]
- Thamarus KA, Groom K, Murrell J, Byrne M, Moran GF. A genetic linkage map for Eucalyptus globulas with candidate loci for wood, fibre, and floral traits. Theor Appl Genet. 2002;104:379–387. doi: 10.1007/s001220100717. [DOI] [PubMed] [Google Scholar]
- Lespinasse D, Rodier-Goud M, Grivet L, Leconte A, Legnate H, et al. A saturated genetic linkage map of rubber tree (Hevea spp.) based on RFLP, AFLP, microsatellite, and isozyme markers. Theor Appl Genet. 2000;100:127–138. [Google Scholar]
- Devaux P, Kilian A, Kleinhofs A. Comparative mapping of the barley genome with male and female recombination-derived, doubled haploid populations. Mol Gen Genet. 1995;249:600–608. doi: 10.1007/BF00418029. [DOI] [PubMed] [Google Scholar]
- Seefelder S, Ehrmaier H, Schweizer G, Seigner E. Male and female genetic linkage map of hops, Humulus lupulus . Plant Breed. 2000;119:249–255. [Google Scholar]
- Maliepaard C, Alston FH, Van Arkel G, Brown LM, Chevreau E, et al. Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theor Appl Genet. 1998;97:60–73. [Google Scholar]
- Mba REC, Stephenson P, Edwards K, Melzer S, Nkumbira J, et al. Simple sequence repeat (SSR) markers survey of the cassava (Manihot esculenta Crantz) genome: Towards an SSR-based molecular genetic map of cassava. Theor Appl Genet. 2001;102:21–31. [Google Scholar]
- Carneiro MS, Camargo LEA, Coelho ASG, Vencovsky R, Leite RP, et al. RAPD-based genetic linkage maps of yellow passion fruit (Passiflora edulis Sims. f. flavicarpa Deg) Genome. 2002;45:670–678. doi: 10.1139/g02-035. [DOI] [PubMed] [Google Scholar]
- Busso CS, Liu CJ, Hash CT, Witcombe JR, Devos KM, et al. Analysis of recombination rate in female and male gametogenesis in pearl-millet (Pennisetum glaucum) using RFLP markers. Theor Appl Genet. 1995;90:242–246. doi: 10.1007/BF00222208. [DOI] [PubMed] [Google Scholar]
- Plomion C, Omalley DM. Recombination rate differences for pollen parents and seed parents in Pinus pinaster . Heredity. 1996;77:341–350. [Google Scholar]
- Lerceteau E, Plomion C, Andersson B. AFLP mapping and detection of quantitative trait loci (QTLs) for economically important traits in Pinus sylvestris A preliminary study. Mol Breed. 2000;6:451–458. [Google Scholar]
- Sewell MM, Sherman BK, Neale DB. A consensus map for loblolly pine (Pinus taeda L.). I. Construction and integration of individual linkage maps from two outbred three-generation pedigrees. Genetics. 1999;151:321–330. doi: 10.1093/genetics/151.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin TM, Zhang XY, Huang MR, Wang MX, Zhuge Q, et al. Molecular linkage maps of the Populus genome. Genome. 2002;45:541–555. doi: 10.1139/g02-013. [DOI] [PubMed] [Google Scholar]
- Barreneche T, Bodenes C, Lexer C, Trontin JF, Fluch S, et al. A genetic linkage map of Quercus robur L. (pedunculate oak) based on RAPD, SCAR, microsatellite, minisatellite, isozyme and 5S rDNA markers. Theor Appl Genet. 1998;97:1090–1103. [Google Scholar]
- Dunemann F, Kahnau R, Stange I. Analysis of complex leaf and flower characters in rhododendron using a molecular linkage map. Theor Appl Genet. 1999;98:1146–1155. [Google Scholar]
- Crespel L, Chirollet M, Durel CE, Zhang D, Meynet J, et al. Mapping of qualitative and quantitative phenotypic traits in Rosa using AFLP markers. Theor Appl Genet. 2002;105:1207–1214. doi: 10.1007/s00122-002-1102-2. [DOI] [PubMed] [Google Scholar]
- Vanooijen JW, Sandbrink JM, Vrielink M, Verkerk R, Zabel P, et al. An RFLP linkage map of Lycopersicon peruvianum . Theor Appl Genet. 1994;89:1007–1013. doi: 10.1007/BF00224531. [DOI] [PubMed] [Google Scholar]
- de Vicente MC, Tanksley SD. Genome-wide reduction in recombination of backcross progeny derived from male versus female gametes in an interspecific cross of tomato. Theor Appl Genet. 1991;83:173–178. doi: 10.1007/BF00226248. [DOI] [PubMed] [Google Scholar]
- Rivard SR, Cappadocia M, Landry BS. A comparison of RFLP maps based on anther culture derived, selfed, and hybrid progenies of Solanum chacoense . Genome. 1996;39:611–621. doi: 10.1139/g96-078. [DOI] [PubMed] [Google Scholar]
- Kreike CM, Stiekema WJ. Reduced recombination and distorted segregation in a Solanum tuberosum (2×) × S. spegazzinii (2×) hybrid. Genome. 1997;40:180–187. doi: 10.1139/g97-026. [DOI] [PubMed] [Google Scholar]
- Wang G, Hyne V, Chao S, Henry Y, Debuyser J, et al. A comparison of male and female recombination frequency in wheat using RFLP maps of homoeologous group-6 and group-7 chromosomes. Theor Appl Genet. 1995;91:744–746. doi: 10.1007/BF00220953. [DOI] [PubMed] [Google Scholar]
- Doligez A, Bouquet A, Danglot Y, Lahogue F, Riaz S, et al. Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor Appl Genet. 2002;105:780–795. doi: 10.1007/s00122-002-0951-z. [DOI] [PubMed] [Google Scholar]