Abstract
A deep learning (DL) network for 2D-based breast mass segmentation in unenhanced dedicated breast CT images was developed and validated, and its robustness in radiomic feature stability and diagnostic performance compared to manual annotations of multiple radiologists was investigated. 93 mass-like lesions were extensively augmented and used to train the network (n=58 masses), which was then tested (n=35 masses) against manual ground truth of a qualified breast radiologist with experience in breast CT imaging using the Conformity coefficient (with a value equal to 1 indicating a perfect performance). Stability and diagnostic power of 672 radiomic descriptors were investigated between the computerized segmentation, and 4 radiologists’ annotations for the 35 test set cases. Feature stability and diagnostic performance in the discrimination between benign and malignant cases were quantified using intraclass correlation (ICC) and multivariate analysis of variance (MANOVA), performed for each segmentation case (4 radiologists and DL algorithm). DL-based segmentation resulted in a Conformity of 0.85±0.06 against the annotated ground truth. For the stability analysis, although modest agreement was found among the four annotations performed by radiologists (Conformity 0.78±0.03), over 90% of all radiomic features were found to be stable (ICC>0.75) across multiple segmentations. All MANOVA analyses were statistically significant (p≤0.05), with all dimensions equal to 1, and Wilks’ lambda ≤0.35. In conclusion, DL-based mass segmentation in dedicated breast CT images can achieve high segmentation performance, and demonstrated to provide stable radiomic descriptors with comparable discriminative power in the classification of benign and malignant tumors to expert radiologist annotation.
Keywords: Breast Cancer Diagnosis, Breast CT, Convolutional Neural Networks, Deep Learning, Radiomics
1. INTRODUCTION
With the advancements in medical image analysis, clinical images are now being considered not only as graphical representations intended for visual perception alone, but as mineable, multidimensional data [1]. Extracting relevant data from medical images is referred to as radiomics. For this purpose, images may be analyzed by high-throughput computing algorithms that extract several quantitative features, which can be used to develop mathematical models and classifiers for diagnostic decision support [1]. Automated medical image analysis has seen a rapid growth in the past few years, and is motivated by the fact that intrinsic characteristics contained in medical images can be quantified and subsequently related to specific physiological and pathological conditions [2].
The pipeline of quantitative radiomics involves several steps, including the identification of the region of interest in the image, the segmentation of the structure to be analyzed (which is, in most cases, performed manually by expert readers), and the extraction of quantitative features [2]. Once obtained, these features can be statistically analyzed, and used to develop classification models to predict the investigated diagnostic outcome.
One of the main areas where radiomics has been applied is breast cancer imaging, due to its high incidence rate [3]. Classification models based on quantitative descriptors have been proposed for digital mammography [4,5,6], digital breast tomosynthesis [7], breast ultrasound [8], and breast MRI [9,10], with the objective of assessing the risk of breast cancer development [6], differentiating benign versus malignant lesions [4,5,8,9], and predicting cancer recurrence and survival rates [10].
Among the most recently developed technologies for breast imaging, dedicated breast CT has been proposed to overcome the problem of tissue superposition in mammography. Breast CT, optimized for the contrast and spatial resolution requirements of breast cancer imaging, can provide real 3D images of the breast, allowing for a complete characterization of breast tissue and, especially, of lesions [11]. Without tissue superposition, tumor features such as shape, heterogeneity, and degree of infiltration might be obtained with higher accuracy compared to when using mammography, potentially leading to more predictive radiomic descriptors of malignancy and aggressiveness.
Since in morphological imaging (such as in unenhanced breast CT), malignant and benign tumors may appear differently in the image mainly according to shape, definition of boundaries, and heterogeneity in voxel intensity [12], radiomic biomarkers should investigate tumor shape, margin, and texture. In order to quantify these characteristics, numerous radiomic features should be calculated, leading to a huge amount of data extracted from each image. This poses major difficulties in the development of robust diagnostic models, especially when datasets are limited [1], as is the case with breast CT imaging, a modality still under research and not yet implemented in the daily clinical routine. Furthermore, many radiomic features are of considerable complexity, a fact which makes their computational cost high, especially in the case of 3D descriptors calculated in tomographic imaging techniques [13].
This can be partially solved by considering the tomographic image as a stack of 2D slices, and performing any radiomic analysis on a 2D basis. A 2D radiomics approach has been shown to provide, in some studies, similar performance compared to 3D radiomic analyses [13], with the additional advantage of a much simpler mathematical formulation of radiomic features (and corresponding lower computational cost). In parallel, a 2D approach allows for the development of more advanced and robust diagnostic classifiers through the augmentation of the dataset (for example, through the collection of multiple image slices, multiple image views, or affine transformations of each tumor image [14–15]). However, this approach makes the manual segmentation process of all regions of interest highly time consuming and, therefore, not sustainable in clinical practice, especially if tens (or hundreds) of 2D images need to be annotated from each case.
Therefore, automated tumor segmentation methods are needed, especially where the volume to be segmented is usually of considerable size and complexity in shape (e.g. in mass-like lesions). With the advancements in artificial intelligence, deep learning algorithms can be trained to perform the segmentation task in a supervised fashion, which have demonstrated to achieve high performance with low computational times, as reported in previously conducted studies on mass segmentation in digital mammography [16–18], breast ultrasound [19,20], and breast MRI [21].
For breast CT, to the best of our knowledge, only unsupervised segmentation methods have been proposed [22–23], which report an average DICE similarity performance of 0.8 [22], with some cases where the DICE drops to below 0.7 [23]. Therefore, the application of deep learning in breast CT images for lesion segmentation remains to be investigated.
Moreover, while the superior performance of deep learning over traditional segmentation methods has been repeatedly demonstrated, the viability of computerized segmentation as input for radiomic models has not be studied to a large extent. Some previous works evaluated the stability of radiomic features across different annotations for head and neck squamous cell carcinoma [24], pleural mesothelioma [24], lung [24–27] and liver [28, 29] cancer, but radiomic feature stability among radiologist annotations and deep learning-based segmentation in breast cancer imaging remains to be investigated in depth. In a single publication (to the best of our knowledge) on radiomics robustness in dynamic contrast-enhanced breast MRI [30], only radiomic-based classification performance was evaluated, without investigating the stability of the descriptors.
Therefore, in this work, we implemented a deep learning-based method for breast mass segmentation and classification in unenhanced dedicated breast CT images, and we validated it against a ground truth dataset in terms of segmentation performance, and against the annotation of multiple breast radiologists in terms of radiomic feature stability and diagnostic power in the classification of benign and malignant masses.
The proposed study therefore aims to investigate the validity of engineered solutions for breast mass segmentation (in the perspective of radiomic analyses) compared to human expert annotations, with future application for computer-aided diagnosis in dedicated breast CT imaging.
2. MATERIALS AND METHODS
2.A. Breast CT Images
The unenhanced dedicated breast CT images used in this study were prospectively collected as part of an ethics-board approved patient trial being performed at our institution, with all women providing written informed consent. Women 50 years of age or older with a suspicious finding detected at mammographic screening were eligible for this study.
Exclusion criteria were suspected or confirmed pregnancy, bilateral mastectomy, the presence of the suspicious lesion in the axillary tail, prior breast cancer or breast biopsy in the recalled breast in the last 12 months, presence of palpable lesions, breastfeeding, frailty or inability to cooperate.
For each patient, as part of the clinical routine, the presence of the lesion was assessed by the combined use of digital breast tomosynthesis and/or breast ultrasound, and all masses were identified and localized on the breast CT images by an experienced breast radiologist.
2.B. Breast CT Scan Protocol
Images were acquired by trained radiographers with a dedicated breast CT clinical system (Koning Corp., West Henrietta, NY) [31, 32]. The system has an x-ray tube with a tungsten target and aluminum filter, and the tube voltage was set to 49 kV for all acquisitions. The x-ray source has a half-cone beam geometry, and the resulting spectrum has a nominal focal spot of 0.3 mm and a half-value layer of 1.39 mm Al. The breast CT system has a source-to-imager distance of 92.3 cm, a source-to-isocenter distance of 65 cm, and is equipped with an energy-integrating detector (4030CB, Varian Medical Systems, Palo Alto, California, USA) with dimensions 397 mm × 298 mm (1024 × 768 elements) and nominal pixel size of 0.194 mm. Tomographic image reconstruction was performed through a filtered backprojection algorithm, with a reconstructed voxel size of 0.273 mm (isotropic).
A complete breast CT scan is achieved through the acquisition of 300 projections during a full revolution of the x-ray tube and detector around the patient breast, in a total time of 10 seconds. The x-ray tube operates in pulsed mode, with a constant 8 ms pulse; the tube current is automatically set for each patient breast by acquiring two scout images normal to each other (16 mA, 2 pulses of 8 ms each per projection). According to the signal level in the two scout images, the tube current is selected between 12 mA and 100 mA. The dose varied for each patient breast, with the average level (for a breast of mean composition and size) being 8.5 mGy [32].
2.C. Data Collection and Annotation
Within this study, 69 patient images containing a total of 93 mass-like lesions were collected, with patient age ranging between 50 and 86 years, (mean 61.1 years). The distribution of the image dataset within the four BI-RADS® breast density categories was 5.85% (n=4, BI-RADS® a), 42.0% (n=29, BI-RADS® b), 47.80% (n=33, BI-RADS® c), and 4.35% (n=3, BI-RADS® d).
The size of the lesions, given by the major diameter, ranged between 4.8 mm and 27.0 mm (mean 10.2 mm, median 8.5 mm). 59 masses were benign (49 cysts, 5 fibroadenoma, 3 lymph nodes, 1 hamartoma, 1 atypical papilloma), 25 biopsy-proven malignant (12 ductal carcinoma in situ (DCIS), 8 invasive ductal carcinoma (IDC), 5 combinations of tumor types), and for 9 cases the lesion type was not available. All cysts (n=49) were diagnosed through ultrasound examination, while all solid masses (n=35) were biopsyproven (through stereotactic or ultrasound-based biopsy).
All lesions were divided into training (n=50: 24 cysts, 2 fibroadenoma, 2 lymph nodes, 6 DCIS, 3 IDC, 2 combinations of tumor types, 1 atypical papilloma, 1 hamartoma, 9 with unknown lesion type), validation (n=8: 4 cyst, 1 fibroadenoma, 1 DCIS, 1 IDC, 1 combination of tumor types), and test sets (n=35: 21 cysts, 2 fibroadenoma, 1 lymph node, 5 DCIS, 4 IDC, 2 combinations of tumor types). For each lesion type, the number of cases to be assigned to each dataset was defined a priori (to allow for case stratification), and then the lesions were assigned to each dataset randomly.
2.D. Data Augmentation
Since this study aims to perform breast mass segmentation on a 2D basis for subsequent radiomic analyses, a single image patch was collected for each mass in the coronal plane intersecting the mass center. Each patch had fixed dimensions of 128 × 128 voxels, so as to fully encompass the largest mass in our dataset (27.0 mm, equivalent to 99 voxels).
Given the strong dependency of deep learning performance on dataset size, different augmentation strategies were performed for the 58 breast masses included in the training and validation sets , in order to maximize the deep learning model training effectiveness. In the first augmentation step, an additional 8 patches (still of 128 voxel side length) were collected from each training-validation mass, in addition to the coronal ones. Two of these were generated from the other planes perpendicular to the coronal view (sagittal and axial). The other six were extracted from the planes of symmetry that cut two opposite faces of an imaginary cube (circumscribing the mass) into its diagonals (that is, each plane contains two opposite edges of the cube, and four vertices). This process resulted in a first dataset of 522 training and validation patches (some of which are shown in Figure 1), and 35 test patches. All training, validation, and test patches were manually segmented under the supervision of a qualified breast radiologist with experience in breast CT imaging, providing a labelled dataset considered as the ground truth. Three other breast radiologists segmented the 35 patches of the test set, which were used to evaluate the radiomic feature stability across different annotations (as explained in Section 2.I).
Fig. 1.
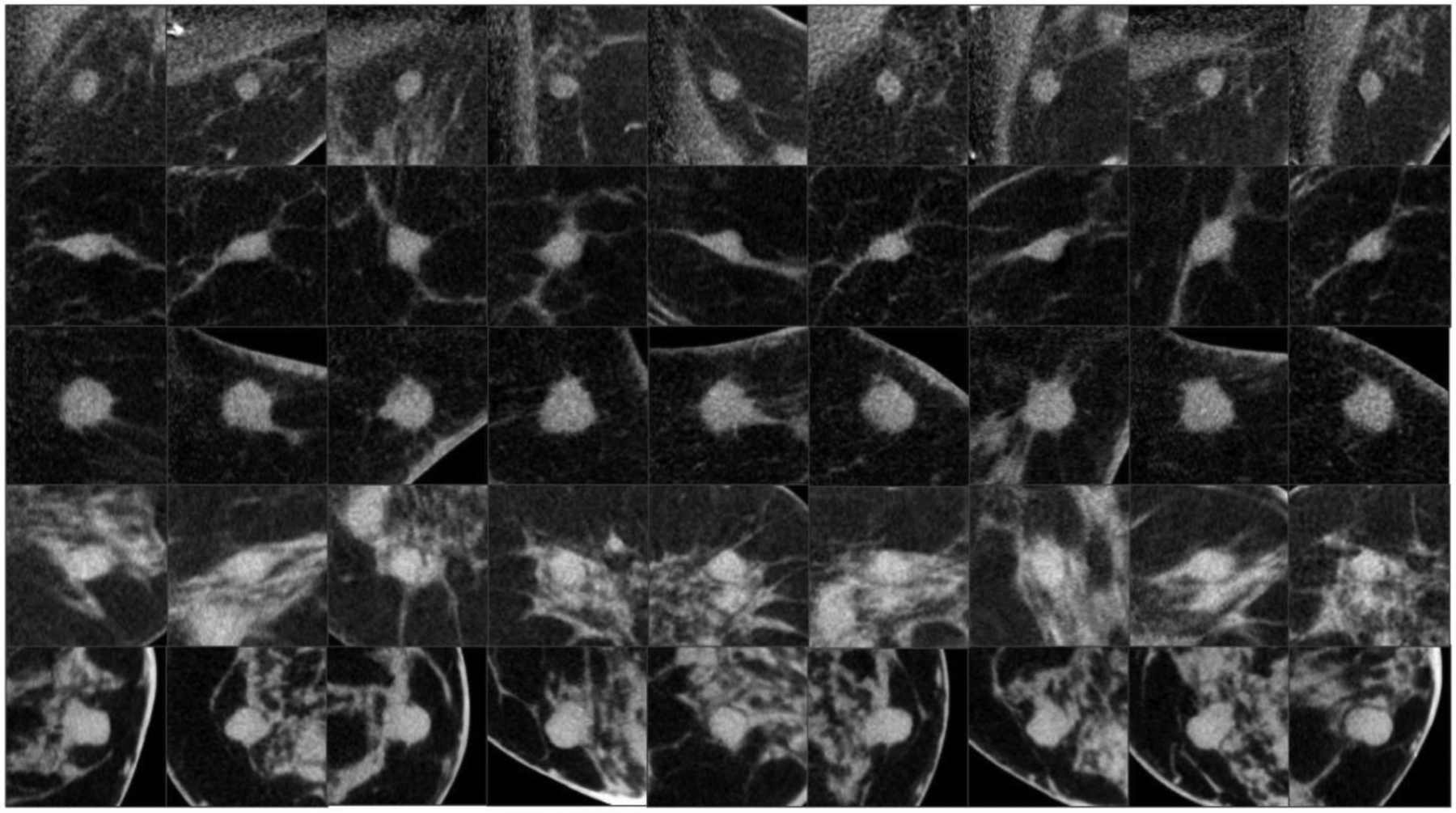
Examples of training image patches generated with the first augmentation strategy. Each row shows the same mass captured in a multi-view manner along 9 different planes. Three planes correspond to the coronal, sagittal and axial view, while the other six to the planes of symmetry that cut two opposite faces of an imaginary cube, circumscribing the mass, into its diagonals.
In the second augmentation strategy, traditional rotation (three rotations, with random angles ranging among 1°–20°, 10°–30°, and 20°–40°), mirroring (horizontal and vertical), and shearing (along the horizontal and vertical axis, with a shear ratio varying randomly between 1% and 20%) were performed. All these affine transformations were performed in a cumulative manner, resulting in a total of 324 patches for each mass.
Finally, a third augmentation strategy was performed using a Generative Adversarial Network (GAN). This deep learning model was used to synthesize additional training data through the generation of new, realistic images of breast masses. A GAN architecture is composed of two main blocks: a generator and a discriminator, which are trained to compete against each other. Given an input noisy vector, the former generates synthetic images which are fed to the discriminator. This latter is trained in parallel to recognize between real breast masses, and synthetic ones. During training, gradients from the discriminator decision are propagated to the generator (which never directly sees the real mass images), allowing to adjust the parameters of the model to generate, at each iteration, more realistic synthetic images.
The implemented GAN model [33] was trained to output synthetic image patches of breast masses, and respective annotations, and was used to generate 450 pairs of patches, which were then further augmented using the second augmentation strategy described above. As a result of this last augmentation method (summed with the two previously described), a total of 34,992 image patches (and respective annotations) were available for training and validation.
A scheme of the GAN model is shown in Figure 2, and details about architecture and training parameters are described in the following Subsection.
Fig. 2.
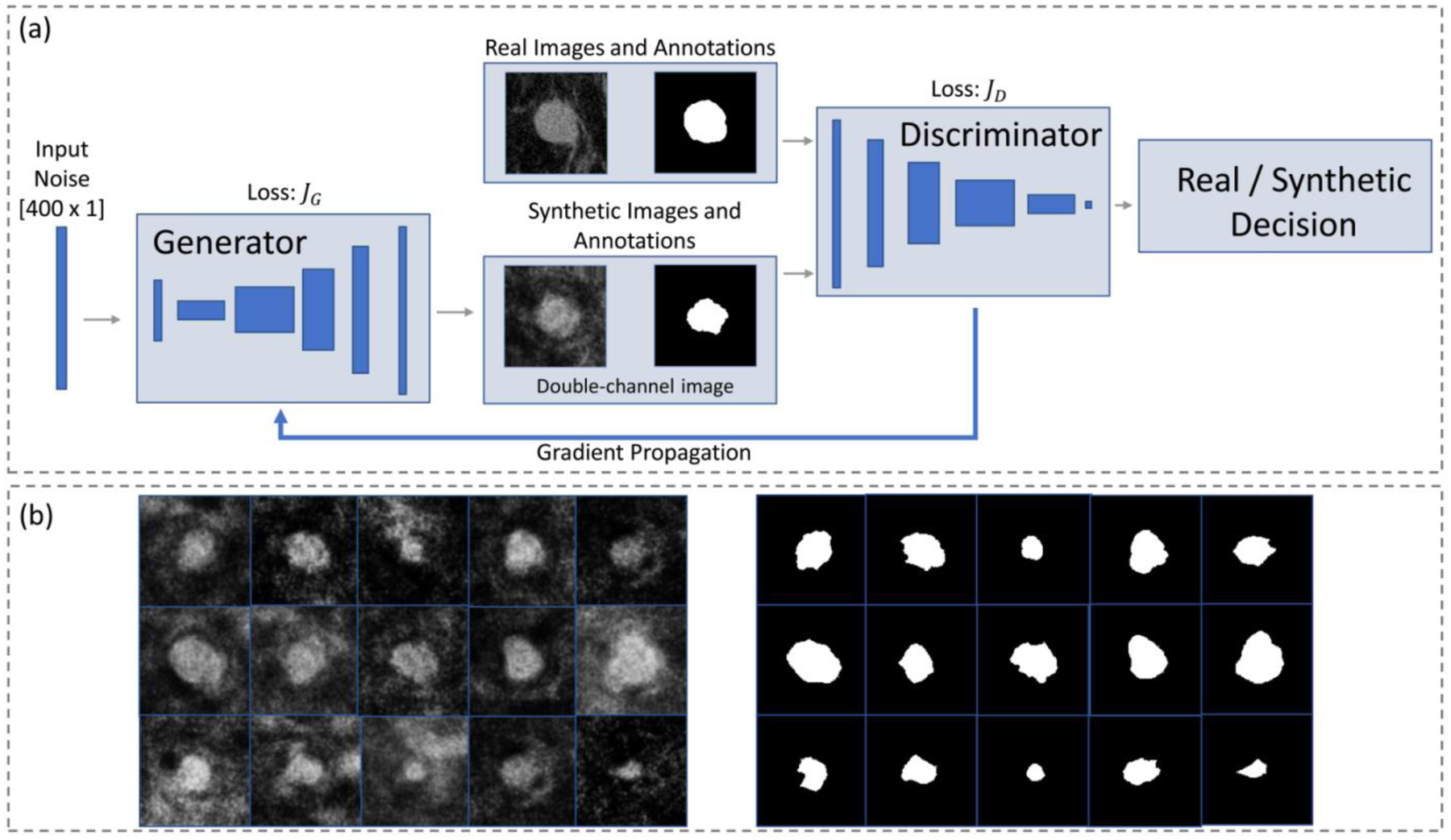
(a) Scheme of the implemented GAN [33] used as an augmentation strategy to generate synthetic images and respective annotations. (b) Some examples of the generated synthetic images, and respective annotations.
2.E. GAN for Data Augmentation: Architecture and Training
The implemented model [33] is a modification of the standard GAN [34], which forces the generator to create annotation masks in addition to synthetic images. The discriminator then judges the results from the generator on a pair basis, allowing the whole GAN to implicitly learn about the structure of both real mass images and ground truth labels. The model is based on the DCGAN [35] architecture, which uses a fully convolutional generator and discriminator without pooling layers. The generator takes a noisy vector x as input (dimensions kept fixed to 400 × 1, uniform noise ranging between −1 and 1), and outputs the synthetic images. The discriminator takes both and the real images and provides a binary output classifying each image as either real (y = 1) or fake (y = 0) using binary cross-entropy as the loss function (jD):
| (1) |
where m is the mini-batch size. The generator loss function (JG) is similar to equation (1), but it only evaluates the output from the discriminator (i.e. real training images are not directly fed to the generator):
| (2) |
By minimizing JD, the discriminator can recognize correctly between real and fake images, while minimizing JG allows the generator to create realistic synthetic images.
The size of the feature maps of the generator were [512; 256; 128; 128; 128], while the discriminator feature map dimensions were set to [128; 128; 256; 512; 512]. All weights were normally initialized, and batch normalization was implemented to reduce overfitting.
The GAN was trained using the Adam (adaptive moment estimation) optimization method [36], an algorithm that adapts the learning rate for each network weight by using first and second moments of the gradient, with an initial learning rate of 0.0001 and an exponential decay rate for the first and second moment estimates of β1= 0.5,β2 = 0.999, and a mini-batch size of 64 examples. The model was trained using the image patches from the training set (50 masses and respective annotations), after extracting 9 patches from each mass (450 patches in total) using the first data augmentation strategy (as explained above).
So as to process both the original and the annotated image, the DCGAN architecture was modified to include two input channels [33]. The first channel corresponds to the original image, while the second to the respective manually annotated mask. During training, the discriminator judges the quality of the image-annotation pairs, instead of evaluating only the original image. After training, the generator creates synthetic examples which are composed by a mass patch, and the associate segmentation mask; these pairs can then be used as additional examples for the training of supervised deep learning models aiming at automatic segmentation.
2.F. Breast Mass Segmentation through Deep Learning
For the segmentation of breast mass patches, a U-net architecture [37] composed of an encoder-decoder structure was implemented (Figure 3). This model performs pixel-wise mapping between the original image and the manually annotated mask, learning the segmentation task in a supervised fashion. The encoder reduces the input feature space dimensions through 3×3 convolutions and max pooling operations (kernel size 2×2, stride of 2), while the decoder recovers the information through 2×2 nearestneighbor up-sampling followed by two 3×3 convolutional kernels. All results of convolutional blocks from the encoding part are concatenated with each corresponding decoding step, allowing to preserve the high detail of the original input image. The final layer consists of a 1×1 convolution followed by a sigmoid activation function, which outputs the segmentation result in the form of a pixel-wise probability. The network was trained using mini-batches of 16 examples and the Adam optimization method. The initial learning rate was set to 0.001, and decayed exponentially every 10 epochs (over a maximum of 50 epochs). The energy function was computed by a pixel-wise softmax (equation 3) over the final feature map combined with the cross-entropy loss function (equation 4) [37]:
| (3) |
| (4) |
Fig. 3.
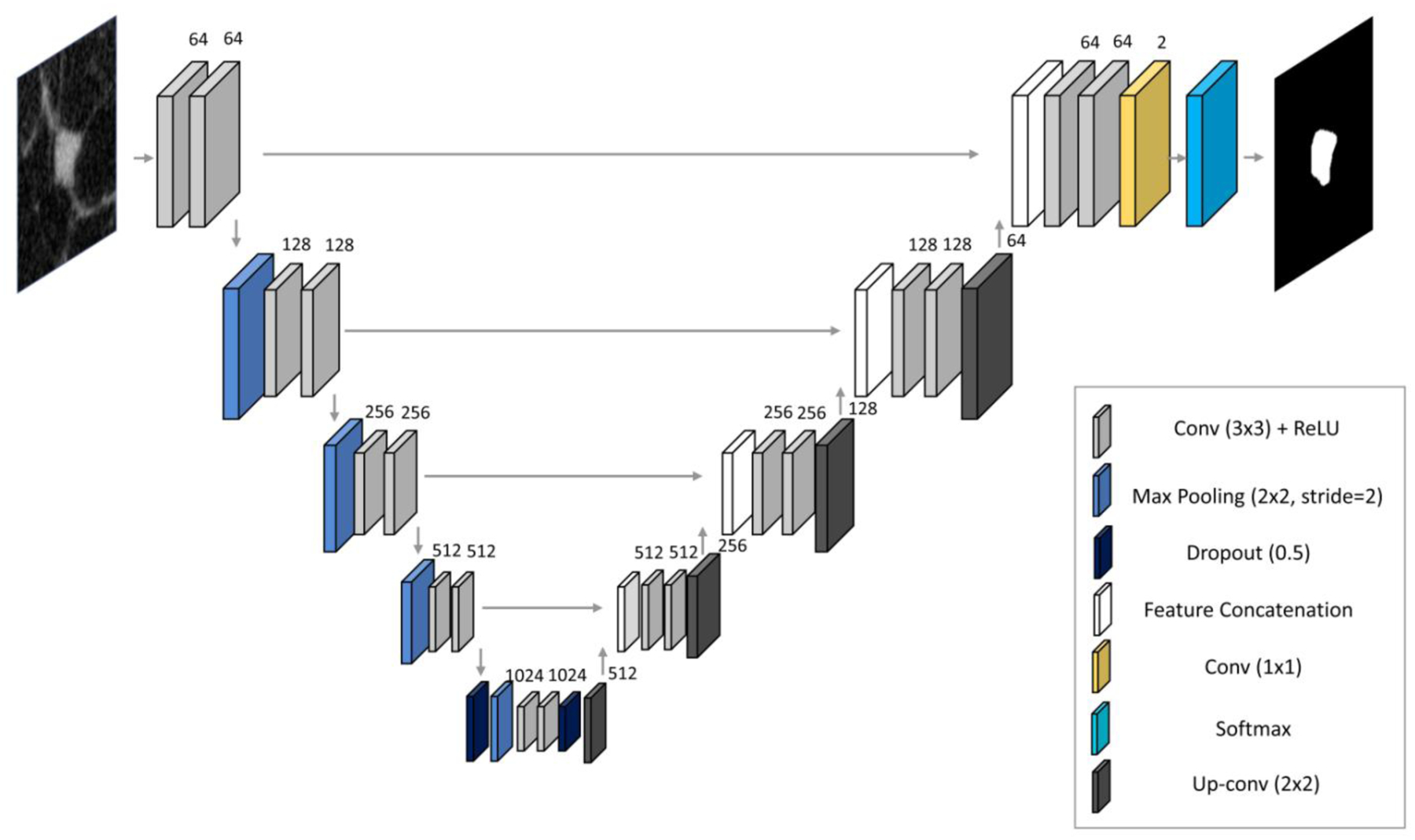
U-net architecture implemented for breast mass segmentation.
Equation (3), in which Xi represents the activation value for an input pixel i, and K the number of possible classes (two in case of binary segmentation), maps the non-normalized output of the network to a probability distribution over the predicted output class. Equation (4) penalizes wrong network predictions, by comparing the ground truth labels ti with the network predictions pi(x).
When implementing the network, the validation set was used both for hyperparameter tuning, and for overfitting prevention, by evaluating the network accuracy on the validation set, and stopping the training if the validation accuracy did not increase after 5 epochs. Accuracy was calculated as for binary classification, but was applied pixel-wise: the sigmoid activation function in the last layer outputs a pixel-wise probability map between 0 and 1, which is rounded and compared with the ground truth annotated images.
2.G. Radiomics Descriptors
A pipeline for the automatic extraction of radiomic features (327 texture-based, 18 shape- and contour-based) was implemented. Texture was quantified through different descriptors which can be divided into five major categories: histogram-based (first order moments of the image patch gray-level distribution) [38], Haralick (second order moments, which recognize frequency patterns of neighboring pixels) [39], run length (which capture the coarseness of texture over different linear orientations) [40], structural and pattern (which characterize tissue architectural complexity, possible directionality of structures, and local intensity variations) [41–45], and Gabor filters (which analyze the frequency content within the image in specific directions and in localized regions) [46]. Texture analysis was performed for each mass both inside the segmented boundary, and within an annular region whose centerline is given by the edge of the segmented area, and whose total thickness along the radial direction equals 10 voxels, to capture the texture of the mass margins.
Shape and contour analysis were performed through the extraction of 18 features calculated from each segmented mass. Features include regional descriptors based on geometrical characteristics [47], and more complex measurements based on Fourier descriptors applied to the centroid-distance function [48], to the shape contour [49], and moments of the mass boundaries [50].
A detailed mathematical formulation of all implemented radiomic features is reported in Tables 3 and 4 (Online Appendix).
2.H. Segmentation Performance
To evaluate the segmentation performance of the deep learning algorithm (A) using the manual annotations as ground truth (B), four similarity metrics were used:
- DICE similarity, defined as the intersection between the two samples A and B over their union, ranging between 0 (no overlap) and 1 (perfect overlap)
(5) - Sensitivity, which measures the proportion between positive voxels which are correctly segmented by the algorithm (TP) to the total number of ground truth positive voxels (PgroundTruth)
(6) - Precision, defined as the ratio between TP and all voxels which are segmented by the algorithm (Palgorithm)
(7) - Conformity, which considers the ratio between the total number of incorrectly classified voxels (Vmisclassified) and TP:
(8)
This latter metric varies within a much wider range compared to the other three, spanning between −∞ (no overlap between A and B), and 1 (perfect overlap). A value of zero indicates that the number of correctly segmented voxel equals the number of misclassified voxels.
Evaluation was performed against the ground truth annotation for four different models, which were trained using the different data augmentation strategies (as described in Section 2.D):
Using only the 9 multi-view patches collected from each training and validation mass (augmentation 1)
Using the 9 multi-view patches, and traditional affine transformations (augmentation 2)
Using the 9 multi-view patches, and synthetic images generated with the GAN (augmentation 3)
Using the 9 multi-view patches, traditional affine transformations, and synthetic images generated with the GAN (augmentation 4)
To evaluate the stability of radiomic features among the algorithm, the considered ground truth, and the three additional expert manual annotations, the model with the highest performance was selected.
2.I. Radiomic Feature Stability
The stability of radiomic features between the annotations performed by the four radiologists included in this study and the segmentation resulting from the deep learning algorithm was quantified using the intraclass correlation coefficient (ICC(3,1)), a statistical indicator that measures the consistency of feature descriptors [51]. ICC indicates the degree of variability of feature values that is due to a real difference among the cases, as opposed to disagreement between annotations. It varies between 0 and 1, with a value above 0.75 often considered as a good threshold to indicate that the descriptors are stable across different segmentations [52].
The analysis of feature stability for the DL segmentations was performed considering both one radiologist at a time (to evaluate the algorithm performance over different radiologists’ annotations), and all four radiologists together (to evaluate the actual stability of the descriptors accounting for all the annotations at a time). The process was repeated multiple times, each time eliminating features with high inter- correlation (with a correlation threshold varying from 1 to 0.7), and for multiple ICC threshold levels (from 0.9 to 0.7).
Finally, to investigate the differences in diagnostic performance in mass classification based on radiomic feature descriptors, multivariate analysis of variance (MANOVA) was performed for all five segmentation cases (four manual, one computerized). MANOVA was used to test the equality of the means of the two groups, i.e. benign vs malignant lesions. Therefore, radiomic features of the test set masses were tested against their pathology ground truth, which represents a nominal variable assuming only two values (0 and 1). To avoid multicollinearity, highly correlated variables were removed prior to the analysis (with a correlation threshold of 0.7) [53].
For each analysis performed, the MANOVA dimension, the Wilk’s Lambda, and the p-value were reported. The dimension of the MANOVA (d) was used to assess whether the two groups (benign, malignant) were separable in the MANOVA canonical hyperplane. In fact, d is an estimate of the dimension of the group means, and a value equal to 1 indicates that the means of the two groups can be considered as different (with a statistical significance given by the p-value). Finally, the Wilk’s Lambda expresses the ratio between the determinant of the variance within each of the group, and the sum between the determinants of the variance within and between each group [53]. All MANOVA analyses were performed using the Statistical Toolbox available in MATLAB (The MathWorks, Natick, MA, USA).
3. RESULTS
3.A. Segmentation Performance
The algorithm resulted in the best performance when trained with all augmented data (9 multi-view planes, affine transformations, and synthetic images), achieving an average DICE for the test set images of 0.93±0.03, a sensitivity of 0.92±0.03, a precision of 0.93±0.05, and a conformity of 0.85±0.06. Some example results are shown in Figure 4. Segmentation performance using only the 9 planes and affine transformations resulted in comparable performance, while the training using only the 9 planes resulted in significantly lower performance. Between affine transformations and synthetic images, the former provided a higher increase in segmentation performance (Table 1).
Fig. 4.
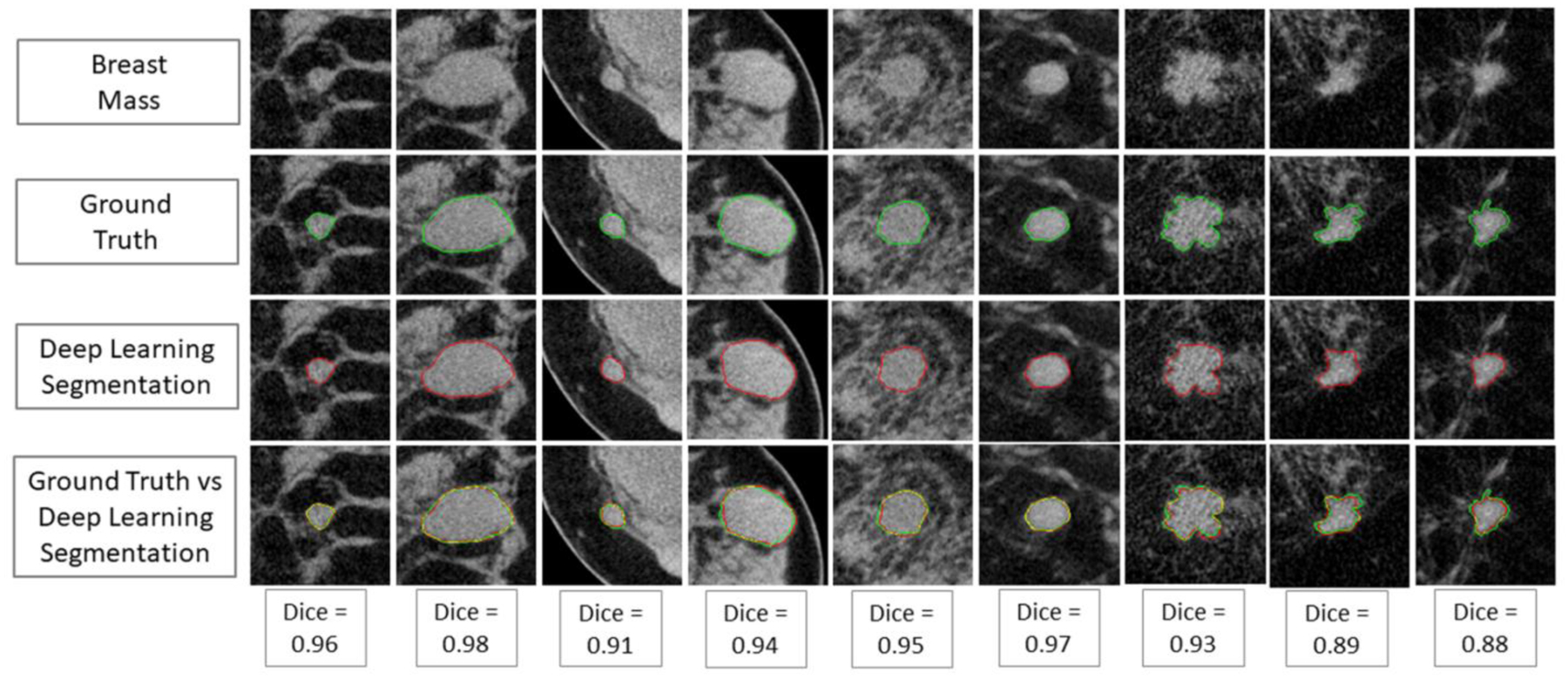
(First row) Examples of original test masses; (second row) ground truth (single manual) annotation; (third row) deep learning-based segmentation; (last row) graphical comparison between ground truth and automated segmentation.
Table 1.
Results of the segmentation performance metrics (mean, standard deviation) for the four different data augmentation strategies implemented.
| DICE | Sensitivity | Precision | Conformity | |
|---|---|---|---|---|
| Augmentation 1 | 0.70 (0.16) | 0.70 (0.19) | 0.72 (0.18) | 0.59 (0.18) |
| Augmentation 2 | 0.92 (0.03) | 0.92 (0.03) | 0.92 (0.03) | 0.83 (0.07) |
| Augmentation 3 | 0.87 (0.09) | 0.85 (0.13) | 0.91 (0.11) | 0.65 (0.37) |
| Augmentation 4 | 0.93 (0.03) | 0.92 (0.03) | 0.93 (0.05) | 0.85 (0.06) |
3.B. Radiomic Feature Stability
Overall, modest agreement was found among the four annotations performed by radiologists (Conformity: 0.78±0.03), and between all radiologists and the DL-based segmentation (Conformity: 0.78±0.04). A few examples of different segmentations are shown in Figure 5.
Fig. 5.
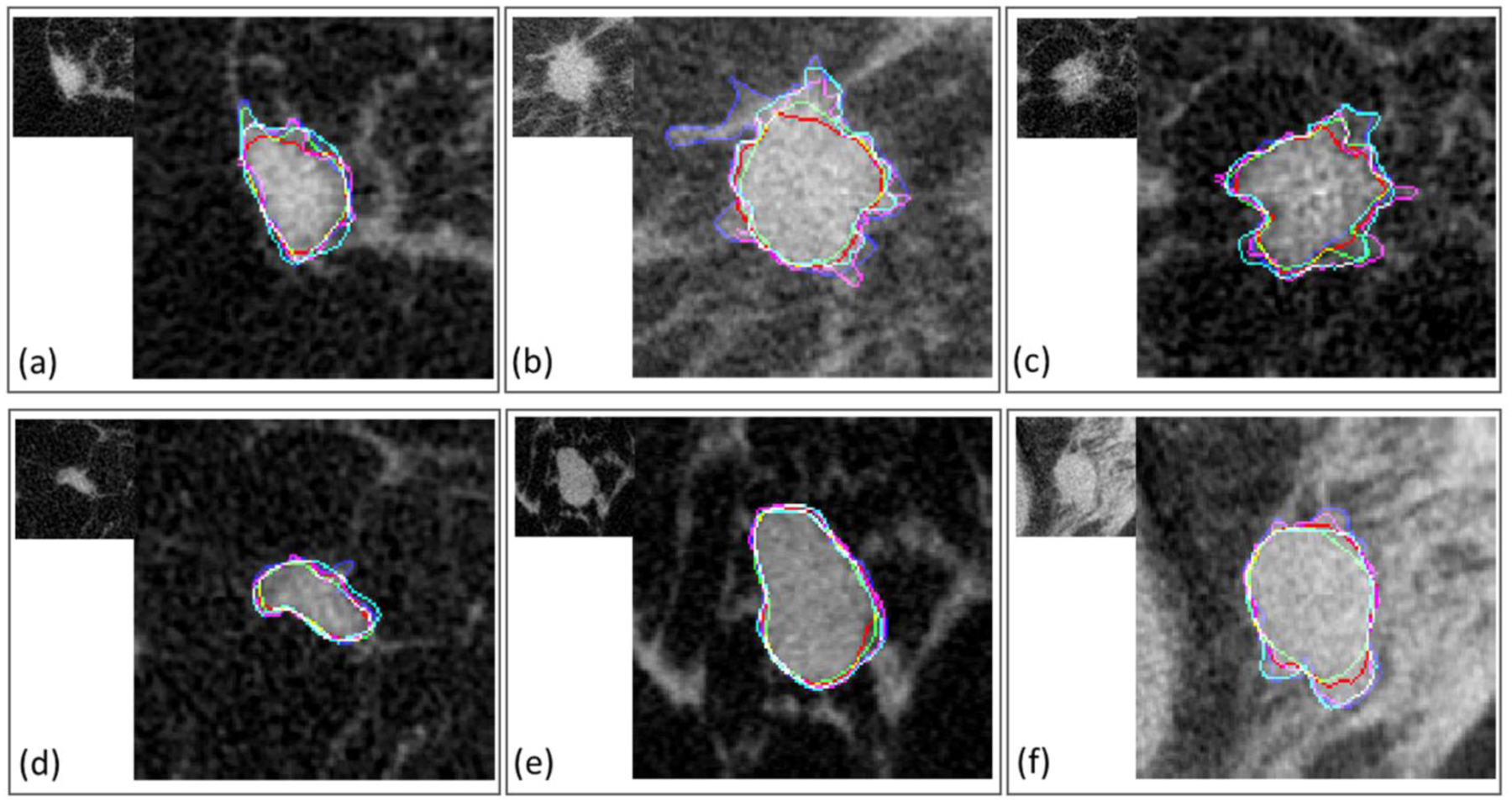
Examples of breast masses included in the test set, with different segmentations overlaid. (a-c) are malignant, (d-f) are benign cases.
Results from the stability analysis are shown in Figure 6 and 7. Overall, comparisons between each radiologist and the deep learning segmentation resulted in the majority of the radiomic features being stable. When all radiomic features were analyzed (i.e. highly correlated features not eliminated), at least the 90% of descriptors were stable (ICC>0.75) for all comparisons between each radiologist and the algorithm. When lowering the correlation threshold to eliminate highly correlated features, the percentage of stable features decreased, but the majority of features were still stable (ICC>0.75) for all comparisons (Figure 6, a–d).
Fig. 6.
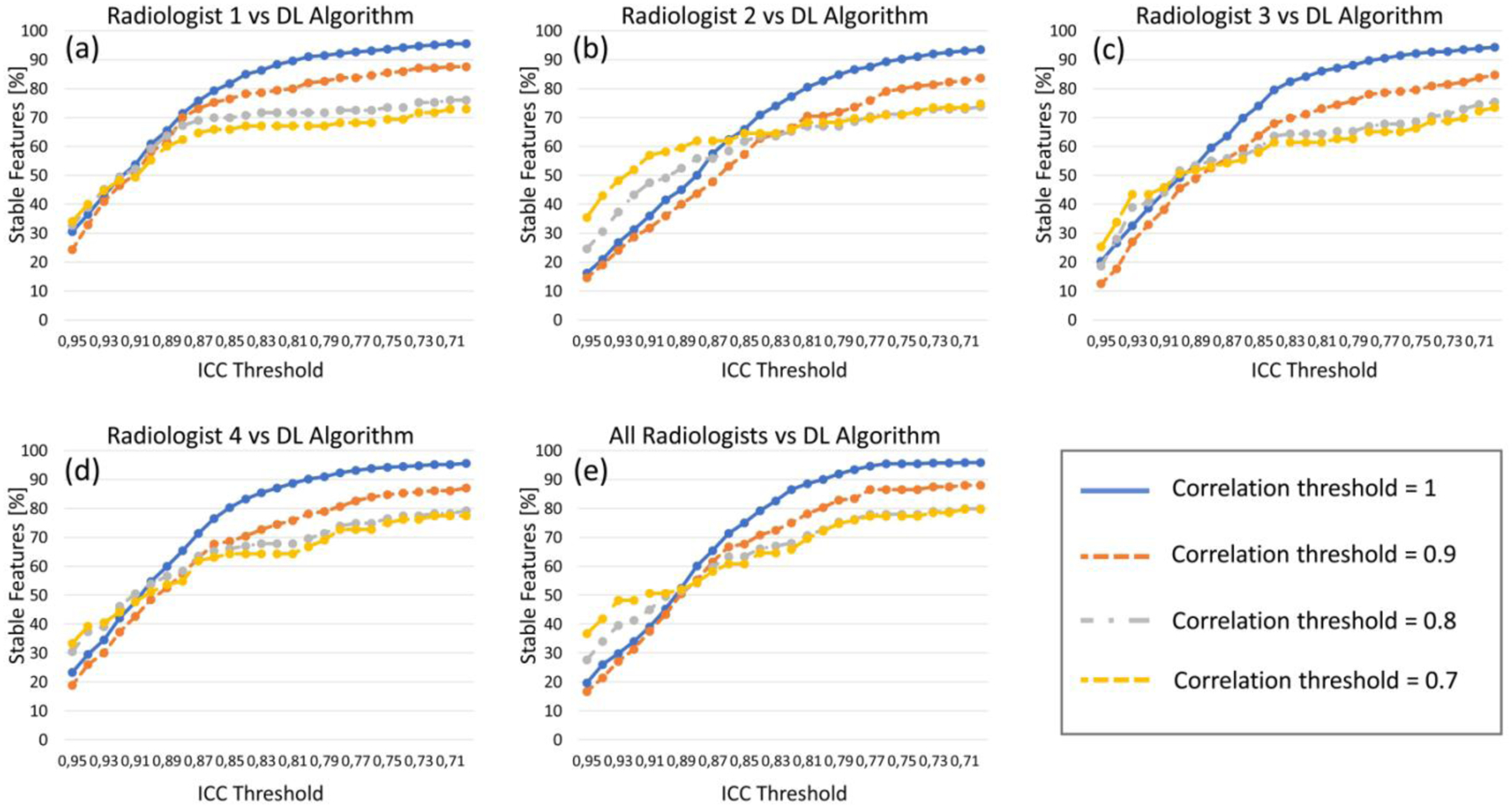
Results of radiomic feature stability analysis. Each graph shows the percentage of features (y axis) having different ICC values (x axis), after eliminating the highly correlated features for four thresholds of correlation (1, 0.9, 0.8, 0.7). (a)-(d) show the feature stability for the four radiologists’ annotations (each compared with the DL algorithm), while (e) shows the stability for the deep learning-based segmentation compared to all radiologists together.
Fig. 7.
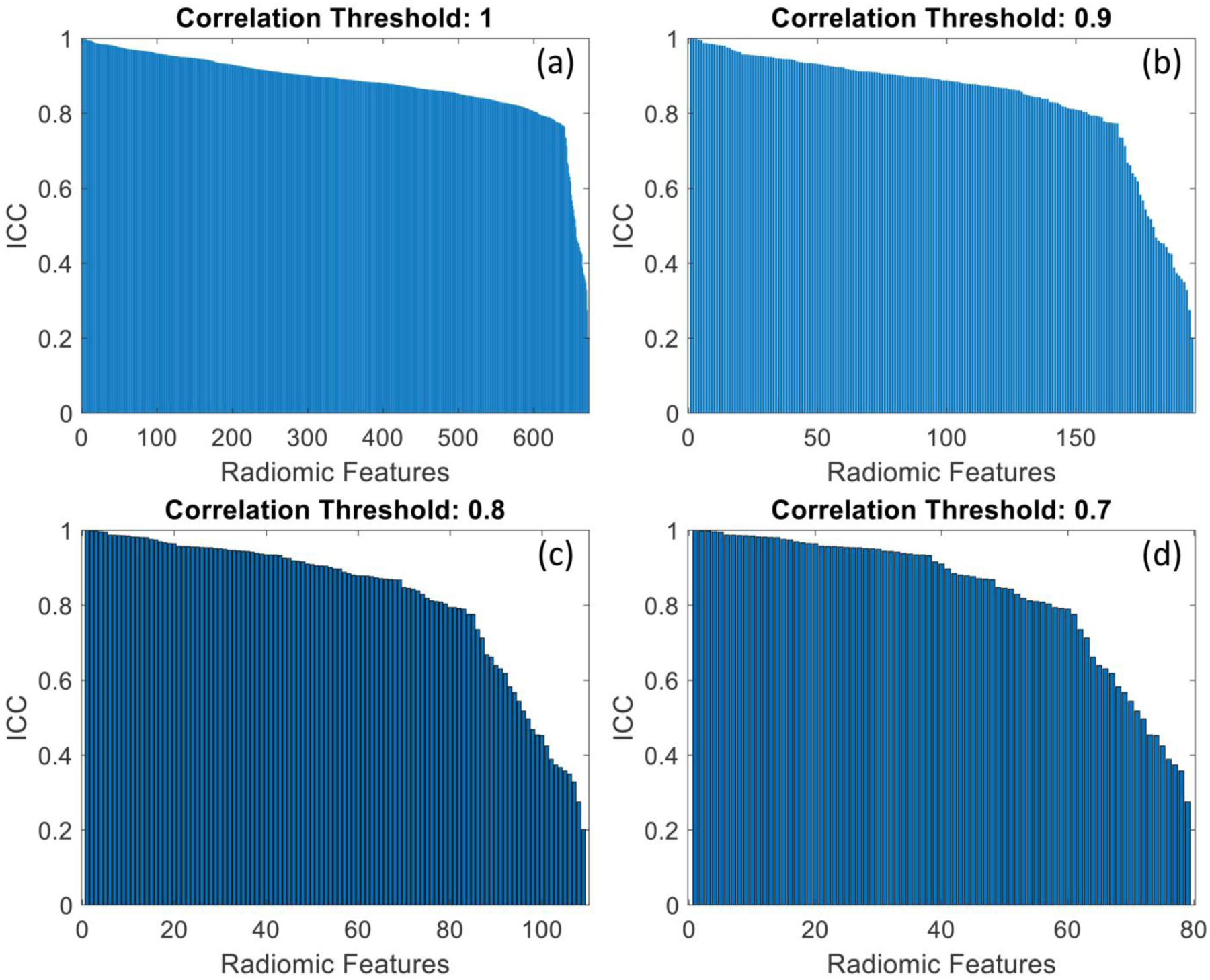
Graphs showing the ICC distribution for all radiomic features, when the annotation of all radiologists was simultaneously compared with the deep learning segmentation. Each plot shows the results of the stability analysis for different correlation thresholds, used to eliminate highly correlated features (a: 1; b: 0.9; c: 0.8; d: 0.7).
By comparing all five segmentations together, the percentage of stable features (ICC>0.75) was 95.4%, 86.5%, 78.0%, and 77.2%, after eliminating correlated features with a threshold of 1, 0.9, 0.8, and 0.7, respectively (Figure 6, e).
Overall, texture features extracted from the masses and from their margins were the most stable (322 on 327, and 310 on 327, respectively), while only half of the shape and contour features showed high stability (9 on 18).
MANOVA analyses resulted in similar discrimination between benign and malignant masses for all five segmentations (Figure 8). All three groups of radiomic descriptors (texture, margin, shape) were found to provide discriminant features. All analyses were statistically significant (P<0.05), with MANOVA dimension (d) of 1, and all Wilks lambda were below 0.35. Complete findings for all analyses are reported in Table 2.
Fig. 8.
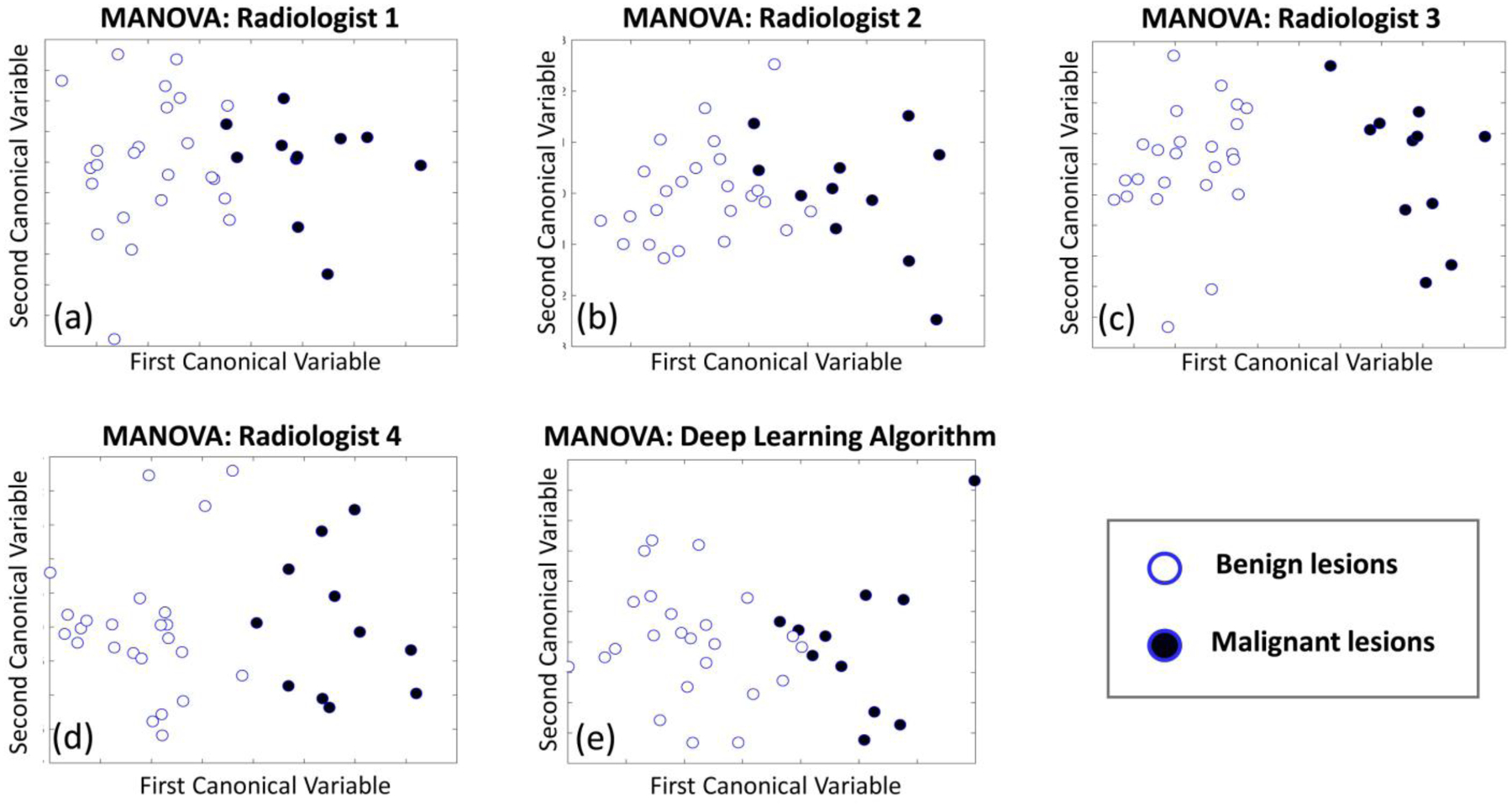
Graphs displaying the statistical analysis performed using MANOVA on the radiomic features extracted from the test set masses. Each plot shows the results of the analysis based on a different annotation (a-d), and on the deep learning segmentation (e).
Table 2.
Results of the MANOVA for the four radiologists and for the deep learning-based segmentation in the discrimination between benign and malignant masses based on radiomic features.
| p-value | d | Wilks Lambda | |
|---|---|---|---|
| Radiologist 1 | 0.009 | 1 | 0.321 |
| Radiologist 2 | 0.003 | 1 | 0.276 |
| Radiologist 3 | 0.001 | 1 | 0.129 |
| Radiologist 4 | 0.005 | 1 | 0.233 |
| DL algorithm | 0.015 | 1 | 0.342 |
A list of all features used, their ICC value, and the features that were selected after the correlation analysis (with a threshold of 0.9, 0.8, and 0.7) are reported in the Online Supplemental Material.
4. DISCUSSION
In this work, we developed a deep learning-based algorithm for 2D breast mass segmentation in unenhanced dedicated breast CT imaging, and we validated it in terms of segmentation performance and stability of radiomic feature descriptors across multiple expert manual annotations.
Although, in the past few years, radiomic approaches based on convolutional neural networks have been proposed to directly analyse the lesions without the need for mass contouring, segmentation remains an important and critical aspect in radiomics, as confirmed by the number of studies involving lesion segmentation and handcrafted radiomic features being still much increasing, due to the advantage of handcrafted features to capture different physiological phenomena without an excessive increase in feature space size [54].
The best segmentation performance was achieved using extensive data augmentation, which includes the use of synthetic images generated by a GAN. However, while traditional augmentation strategies (rotations, mirroring, and shearing) improved the segmentation results considerably compared to only using 9 views from each mass (DICE increased by over 30%), the additional inclusion of synthetic cases only increased the DICE results by an additional 1%. This highlights that traditional augmentation methods are still valuable, and that synthetic images generated through GANs have a lower impact in performance increase, and this could be due to the fact that synthetic images possess features which are not fully representative of real cases. However, when added to the 9 views only, synthetic images could increase the DICE by 20%, suggesting that, for small datasets, GANs could still be helpful in increasing segmentation performance. While previous studies on chest x-ray lung segmentation showed a negligible benefit in segmentation performance when adding synthetic images to original cases [33], our findings could be due to the increased difficulty of segmenting size- and shape-varying structures (e.g. breast masses) as opposed to organs. Given the higher difficulty in the segmentation task, in case of very limited datasets there seems to be a benefit when using synthetic images for training a supervised segmentation model, as also previously reported in [55]–[56]. However, our findings are related to the specific model implemented in this work [33], and might therefore be different when other architectures are used. Therefore, also accounting for the very low increase in segmentation performance by including the synthetically generated images, further statistics with additional mass cases are required in future for a more meaningful performance comparison assessment. Furthermore, due to the limited number of masses available for this study, the same dataset was used to train both the U-net and the GAN. This limitation might reduce the impact of synthetic images on the segmentation performance, which could increase if the GAN was trained on different training examples compared to the model used for segmentation. The limited size of the dataset used in this study could be addressed in future work by the acquisition and inclusion of additional patient images. This could allow for improvement of the realism of the synthetically generated images, and consequently the performance of the automatic segmentation. The availability of larger image datasets could also allow for implementation of a conditional GAN architecture, where the input is not given by a simple noise vector, but by mask priors. While this approach is generally harder to train due to the larger dimension of the input to the generator and to the pixel level constraints given by the input mask [57], it could help improve the quality of the generated images. Therefore, it could help produce more realistic synthetic examples to be used to ameliorate the performance of the subsequent segmentation model.
While the implemented GAN can generate an arbitrary number of images, we chose to generate 450 synthetic mass patches to match the number of original training examples deriving from the first augmentation strategy (9 view augmentation). This was done to evaluate the potential increase in segmentation performance when the network was trained with only the 9 mass views and the synthetic samples, with a number of synthetic cases equal to the number of real images. While a larger number of synthetic cases could be included in this step, we do not expect further significant improvements, as the GAN was trained with the same image patches used to train the U-Net. With a larger number of examples available to train the GAN, especially different examples from the ones used to train the U-Net, additional insights may be achieved, and, in this case, a larger number of generated synthetic images could provide further benefit in the segmentation performance.
Deep learning applied for breast mass segmentation in dedicated breast CT images achieved higher performance compared to traditional, unsupervised methods (DICE of 0.8) [22], [23], although former analysis were performed on different datasets, which may impact results. The algorithm also demonstrated better results compared to some deep learning-based segmentation algorithms applied to breast ultrasound (DICE of 0.82 [19], and DICE of 0.89 [20]), and similar results compared to digital mammography (DICE of 0.91 [16]), and DICE of 0.93 [17]). This could be due to the better contrast of dedicated breast CT (as opposed to ultrasound), and to the possibility to perform extensive data augmentation thanks to the images being in fully three dimensions, which allows for the increase in the U-net training set size to a large number of examples (resulting in data set sizes similar to those in mammography). However, despite the promising results, the experiment should be repeated when a larger number of patient cases is available, possibly with images acquired with different breast CT systems, and for different radiation dose levels, to evaluate the effect of different noise magnitudes and frequencies on the segmentation performance.
The majority of radiomic features were found to have good stability, and features from each group were selected in all MANOVA analyses, indicating that a strong radiomic signature is obtained by the combination of radiomic descriptors belonging to different categories (mass and margin texture, and shape). Considering the non-negligible variability observed in the annotations of the four radiologists (Conformity of 0.78), this finding demonstrates a considerable robustness in radiomic descriptors extracted from breast CT mass-like lesions, suggesting that reliable radiomic signatures could be obtained even with different segmentation results. Moreover, the percentage of stable descriptors remained high when highly correlated features were eliminated. This is in line with previous findings conducted on liver masses [29], and indicates that only a small subset of radiomic features could be used to draw diagnostic conclusions. This is a desirable outcome, since dealing with too many features (compared to the number of available cases) usually requires correction for multiple testing, and increases the risk of overfitting of any predictive model designed upon the feature values.
MANOVA was chosen as a statistical test to evaluate the discrimination between benign and malignant cases due to its appropriateness in handling multi-dimensional data simultaneously. In fact, as opposed to univariate analyses (e.g. ANOVA or t-test), single statistical indicators are provided without the need to correct for multiple comparisons [58]. However, little information is provided about the power of single descriptors. Therefore, in future and with larger datasets, further insights could be achieved by applying additional statistical tests, to better evaluate the discriminant power of each radiomic descriptor and provide a more reliable analysis from a diagnostic perspective.
MANOVA analyses resulted to be statistically significant, and all presented a dimensionality of 1, indicating that the differences observed in the two samples (benign and malignant masses) is not due to random chance. Furthermore, except for a single radiologist whose annotation led to a much improved separation of the two classes in the MANOVA hyperplane (Radiologist 3, Figure 8.c), all segmentations led to similar discriminations between benign and malignant cases.
These results confirmed not only the power of automatic segmentation for radiomic purposes, but also highlighted a significant difference between the two mass types. However, these findings should be confirmed with future studies, with an enlarged test set, additional experts’ annotations, and further statistics to better assess all comparisons among all segmentation results.
With a larger test set, future studies will also analyze each non-stable radiomic descriptor, to evaluate whether the low stability is due to high variability among segmentations, or low variability across the image cases. Although results from the MANOVA show a good discriminant power (and therefore suggest a variability between the two classes of interest, benign and malignant), some features may show low stability because our dataset is not representative enough. Therefore, the analysis should be repeated with a larger number of cases, to understand which features should be avoided prior to radiomic analysis. The main limitation of this study is the relatively limited dataset size, due to breast CT still being in the clinical research realm, and not yet implemented in daily clinical routine. With an increased number of test cases, especially in terms of different lesion types, further insights could be achieved, both in terms of segmentation performance, and on its effect on radiomic feature stability. Furthermore, the training set was annotated by a single radiologist, and this can potentially bias the segmentation performance towards this single expert. However, from a radiomics perspective, this does not seem to significantly affect the diagnostic power, while the segmentation performance could be further increased by using the entire dataset annotated by multiple readers.
In future work, with an expanded dataset, this study will be included in the development of an automated computer-aided diagnosis system for dedicated breast CT images, with the goal of predicting breast mass malignancy grade and, consequently, attempt to reduce the number of negative biopsies.
5. CONCLUSIONS
Deep learning-based 2D segmentation of breast masses in unenhanced dedicated breast CT images can achieve high performance against manually annotated ground truth. Furthermore, it demonstrated to provide stable radiomic feature descriptors, with a discriminative power in the classification of benign and malignant tumors comparable to expert manual annotation.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by grant R01CA181171 from the National Cancer Institute, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute, or the National Institutes of Health. The authors are grateful to Linda Appelman, Mechli Imhof-Tas, and Roel Mus for providing the additional manual annotations used in this study.
Footnotes
DISCLOSURE OF CONFLICTS OF INTEREST
The authors have no relevant conflicts of interest to disclose.
REFERENCES
- [1].Gillies RJ, Kinahan PE, and Hricak H, “Radiomics: Images Are More than Pictures, They Are Data.” Radiology, vol. 278, no. 2, pp. 563–77, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lambin P, Rios-Velazquez E, Leijenaar R, et al. , “Radiomics: extracting more information from medical images using advanced feature analysis.” Eur J Cancer, vol. 48, no. 4, pp. 441–6, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. , “Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012,” European Journal of Cancer, vol. 49, pp. 1374–1403, 2013. [DOI] [PubMed] [Google Scholar]
- [4].Drukker K, Giger ML, Joe BN, et al. , “Combined Benefit of Quantitative Three-Compartment Breast Image Analysis and Mammography Radiomics in the Classification of Breast Masses in a Clinical Data Set,” Radiology, vol. 290, no. 3, pp. 621–628, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li H, Mendel KR, Lan L, Sheth D, and Giger ML, “Digital Mammography in Breast Cancer: Additive Value of Radiomics of Breast Parenchyma,” Radiology, vol. 291, no. 1, pp. 15–20, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zheng Y, Keller BM, Ray S, et al. , “Parenchymal texture analysis in digital mammography: A fully automated pipeline for breast cancer risk assessment,” Med Phys, vol. 42, no. 7, pp. 4149–60, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tagliafico AS, Valdora F, Mariscotti G, et al. , “An exploratory radiomics analysis on digital breast tomosynthesis in women with mammographically negative dense breasts,” Breast, vol. 40, pp. 92–96, 2018. [DOI] [PubMed] [Google Scholar]
- [8].Lee SE, Han K, Kwak JY, Lee E, and Kim EK, “Radiomics of US texture features in differential diagnosis between triple-negative breast cancer and fibroadenoma,” Sci Rep, vol. 8, no. 1, pp. 13546, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, and Kuhl C, “Radiomic versus Convolutional Neural Networks Analysis for Classification of Contrast-enhancing Lesions at Multiparametric Breast MRI, “ Radiology, vol. 290, no. 2, pp. 290–297, 2019. [DOI] [PubMed] [Google Scholar]
- [10].Xiong Q, Zhou X, Liu Z, et al. , “Multiparametric MRI-based radiomics analysis for prediction of breast cancers insensitive to neoadjuvant chemotherapy,” Clin Transl Oncol. 2019. doi: 10.1007/s12094-019-02109-8. [DOI] [PubMed] [Google Scholar]
- [11].Lindfors KK, Boone JM, Nelson TR, Yang K, Kwan AL, and Miller DF, “Dedicated breast CT: initial clinical experience,” Radiology, vol. 246, no. 3, pp. 725–33, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Surendiran B, Ramanathan P, and Vadivel A, “Effect of BIRADS shape descriptors on breast cancer analysis,” Int. J. Medical Engineering and Informatics, vol. 7, no. 1, pp. 65–79, 2015. [Google Scholar]
- [13].Shen C, Liu Z, Guan M, et al. , “2D and 3D CT Radiomics Features Prognostic Performance Comparison in Non-Small Cell Lung Cancer,” Transl Oncol., vol. 10, no. 6, pp. 886–894, 2017. 31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Setio AA, Ciompi F, Litjens G, et al. , “Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks,” IEEE Trans Med Imaging, vol. 35, no. 5, pp. 1160–1169, 2016. [DOI] [PubMed] [Google Scholar]
- [15].Kumar D, Chung AG, Shaifee MJ, Khalvati F, Haider MA, and Wong A, “Discovery Radiomics for Pathologically-Proven Computed Tomography Lung Cancer Prediction,” Image Analysis and Recognition ICIAR; vol. 10317, pp. 54–62, 2017. [Google Scholar]
- [16].Zhu W, Xiang X, Tran TD, Hager GD, and Xie X, “Adversarial deep structured nets for mass segmentation from mammograms,” IEEE ISBI 2018; doi 10.1109/ISBI.2018.8363704. [DOI] [Google Scholar]
- [17].Al-Antari MA, Al-Masni MA, Choi MT, Han SM, T. and Kim S “A fully integrated computer-aided diagnosis system for digital X-ray mammograms via deep learning detection, segmentation, and classification,” Int J Med Inform., vol. 117, pp. 44–54, 2018. [DOI] [PubMed] [Google Scholar]
- [18].De Moor T, Rodriguez-Ruiz A, Mérida AG, Mann R, and Teuwen J, “Automated soft tissue lesion detection and segmentation in digital mammography using a u-net deep learning network,” Proc. of IWBI 2018; doi 10.1117/12.2318326. [DOI] [Google Scholar]
- [19].Kumar V, Webb JM, Gregory A, et al. , “Automated and real-time segmentation of suspicious breast masses using convolutional neural network,” PLoS One., vol. 16, no. 13(5):e0195816, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hu Y, Guo Y, Wang Y, et al. , “Automatic tumor segmentation in breast ultrasound images using a dilated fully convolutional network combined with an active contour model,” Med Phys., vol. 46, no. 1, pp. 215–228, 2019. [DOI] [PubMed] [Google Scholar]
- [21].Levman J, Warner E, Causer P, and Martel A, “Semi-automatic region-of-interest segmentation based computer-aided diagnosis of mass lesions from dynamic contrast-enhanced magnetic resonance imaging based breast cancer screening,” J Digit Imaging, vol. 27, no. 5, pp. 670–8, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kuo HC, Giger ML, Reiser I, et al. , “Level set segmentation of breast masses in contrast-enhanced dedicated breast CT and evaluation of stopping criteria,” J Digit Imaging, vol. 27, no. 2, pp. 237–47, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee J, Nishikawa RM, Reiser I, J. and Boone M. Boone, “Optimal reconstruction and quantitative image features for computer-aided diagnosis tools for breast CT,” Med Phys., vol. 44, no. 5, pp. 1846–1856, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pavic M, Bogowicz M, Würms X, et al. , “Influence of inter-observer delineation variability on radiomics stability in different tumor sites,” Acta Oncol., vol. 57, no. 8, pp. 1070–1074, 2018. [DOI] [PubMed] [Google Scholar]
- [25].Van Griethuysen JJM, Fedorov A, Parmar C, et al. , “Computational Radiomics System to Decode the Radiographic Phenotype,” Cancer Res., vol. 77, no. 21, pp. e104–e107, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Parmar C, Rios Velazquez E, Leijenaar R, et al. , “Robust Radiomics feature quantification using semiautomatic volumetric segmentation,” PLoS One, vol. 9, no. 7, pp. e102107, 2014. 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Leijenaar RT, Carvalho S, Velazquez ER, et al. , “Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability,” Acta Oncol. Vol. 52, no. 7, pp. 1391–7, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Echegaray S, Gevaert O, Shah R, et al. , “Core samples for radiomics features that are insensitive to tumor segmentation: method and pilot study using CT images of hepatocellular carcinoma,” J Med Imaging (Bellingham), vol. 2, no. 4, pp. 041011, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Moltz Jan Hendrik, “Stability of radiomic features of liver lesions from manual delineation in CT scans,” Proc. of SPIE Med Im., vol. 10950, no. 109501W-1, 2019. [Google Scholar]
- [30].Spuhler KD, Ding J, Liu C, et al. , “Task-based assessment of a convolutional neural network for segmenting breast lesions for radiomic analysis,” Magn Reson Med., vol. 82, no. 2, pp. 786–795, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Boone JM, Nelson TR, Lindfors KK, and Seibert JA, “Dedicated breast CT: radiation dose and image quality evaluation,” Radiology, vol. 221, no. 3, pp. 657–67, 2001. [DOI] [PubMed] [Google Scholar]
- [32].Sechopoulos I, Feng SS, and D’Orsi CJ, “Dosimetric characterization of a dedicated breast computed tomography clinical prototype,” Med Phys., vol. 37, no. 8, pp. 4110–20, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Neff T, Payer C, Stern D, and Urschler M, “Generative Adversarial Network based Synthesis for Supervised Medical Image Segmentation,” Proc. of the OAGM&ARW Joint Workshop, pp. 140–145, 2017. [Google Scholar]
- [34].Goodfellow IJ, Pouget-Abadiey J, Mirza M, et al. , “Generative Adversarial Nets,” NIPS 2014. [Google Scholar]
- [35].Radford A, Metz L, and Chintala S, “Unsupervised representation learning with deep convolutional generative adversarial networks,” arXiv preprint arXiv, 1511.06434, 2015. [Google Scholar]
- [36].Kingma DP, and Ba JL, “Adam: A method for stochastic optimization,” arXiv preprint arXiv, 1412.6980, 2014. [Google Scholar]
- [37].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” Proc. Med. Image Comput. Comput.-Assisted Intervention, pp. 234–241, 2015. [Google Scholar]
- [38].Caballo M, Teuwen J, Mann R, and Sechopoulos I, “Breast parenchyma analysis and classification for breast masses detection using texture feature descriptors and neural networks in dedicated breast CT images,” Proc. of SPIE Med Im., vol. 10950, no. 109500J, 2019. [Google Scholar]
- [39].Haralick RM, Shanmugam K, and Dinstein I, “Textural Features for Image Classification,” IEEE Transactions on Systems, Man, and Cybernetics, vol. SMC-3, no. 6, pp. 610–621, 1973. [Google Scholar]
- [40].Galloway MM, “Texture Analysis Using Gray Level Run Lengths,” Computer Graphics and Image Processing, vol. 4,pp. 172–179, 1975. [Google Scholar]
- [41].Ojala T, Pietikainen M, and Maenpaa T, “Multiresolution Gray-Scale and Rotation Invariant Texture Classification with Local Binary Patterns,” IEEE Trans. Pattern Analysis and Machine Intelligence, vol. 24, no. 7, pp. 971–987, 2002. 33 [Google Scholar]
- [42].Laws KI, “Rapid Texture Identification,” Proc. SPIE 0238, Image Processing for Missile Guidance 1980; doi: 10.1117/12.959169. [DOI] [Google Scholar]
- [43].Tencer L, Reznakova M, and Cheriet M, “A new framework for online sketch-based image retrieval in web environment,” Information science, signal processing and their applications (ISSPA), 11th international conference on. IEEE, Montreal, QC, pp.1430–1431, 2012. [Google Scholar]
- [44].Keller JM, Chen S, and Crownover RM, “Texture description and segmentation through fractal geometry,” Computer Vision, Graphics, and Image Processing, vol. 45, no. 2, pp. 150–166, 1989. [Google Scholar]
- [45].Iwahoria Y, Hattoria A, Adachia Y, Bhuyanb MK, Woodhamc RJ, and Kasugai K, “Automatic Detection of Polyp Using Hessian Filter and HOG Features,” Procedia Computer Science, vol. 60,pp. 730–739, 2015. [Google Scholar]
- [46].Haghighat M, Zonouz S, and Abdel-Mottaleb M, “CloudID: Trustworthy cloud-based and cross-enterprise biometric identification,” Expert Systems with Applications, vol. 42, no. 21, pp. 7905–7916, 2015. [Google Scholar]
- [47].Liu J, and Shi Y, “Image Feature Extraction Method Based on Shape Characteristics and Its Application in Medical Image Analysis,” Applied Informatics and Communication. ICAIC 2011. Communications in Computer and Information Science, vol. 224, pp. 172–178, 2011. [Google Scholar]
- [48].Yang M, Kpalma K, and Ronsin J, “A Survey of Shape Feature Extraction Techniques,” Yin PY (ed.) Pattern Recognition IN-TECH, pp. 43–90, 2008. [Google Scholar]
- [49].Shen L, Rangayyan RM, and Desautels JL, “Application of shape analysis to mammographic calcifications,” IEEE Trans Med Imaging, vol. 13, no. 2, pp. 263–74, 1994. [DOI] [PubMed] [Google Scholar]
- [50].Gupta L, and Srinath MD, “Contour sequence moments for the classification of closed planar shapes,” Pattern Recognition, vol. 20, no., 3, pp. 267:272, 1987. [Google Scholar]
- [51].Shrout PE, and Fleiss JL, “Intraclass correlations: uses in assessing rater reliability,” Psychol Bull., vol. 86, no. 2, pp. 420–8, 1979. [DOI] [PubMed] [Google Scholar]
- [52].Koo TK, and Li MY, “A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research,” J Chiropr Med., vol. 15, no. 2, pp. 155–163, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Weinfurt KP, “Multivariate analysis of variance,” In Grimm LG& Yarnold PR (Eds.), Reading and understanding multivariate statistics, pp. 245–276, Washington, DC: American Psychological Association, 1995 [Google Scholar]
- [54].Afshar P, et al. , “From Handcrafted to Deep-Learning-Based Cancer Radiomics: Challenges and opportunities,” IEEE Sign. Proc. Magazine, vol. 36, no. 4, pp 132–160, 2019 [Google Scholar]
- [55].Tang Y-B., et al. , “CT-Realistic Data Augmentation Using Generative Adversarial Network for Robust Lymph Node Segmentation,” Proc. of SPIE Med Im., vol. 10950, no. 109503V, 2019. 34 [Google Scholar]
- [56].Majurski M, et al. , “Cell Image Segmentation using Generative Adversarial Networks, Transfer Learning, and Augmentations,” Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops (2019) [Google Scholar]
- [57].Ren Y, et al. , “Mask Embedding for Realistic High-Resolution Medical Image Synthesis,” Shen D et al. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2019. MICCAI 2019. Lecture Notes in Computer Science, vol 11769, pp 422–430, Springer, Cham. [Google Scholar]
- [58].Warne RT, “Primer on Multivariate Analysis of Variance (MANOVA) for Behavioral Scientists,” Practical Assessment, Research & Evaluation, vol. 19, no. 17, 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


