Abstract
The Steroidogenic acute regulatory protein (StAR)-related lipid transfer (START) domain represents an evolutionarily conserved superfamily of lipid transfer proteins widely distributed across the tree of life. Despite significant expansion in plants, knowledge about this domain remains inadequate in plants. In this work, we explore the role of cavity architectural modulations in START protein evolution and functional diversity. We use deep-learning approaches to generate plant START domain models, followed by surface accessibility studies and a comprehensive structural investigation of the rice START family. We validate 28 rice START domain models, delineate binding cavities, measure pocket volumes, and compare these with mammalian counterparts to understand evolution of binding preferences. Overall, plant START domains retain the ancestral α/β helix-grip signature, but we find subtle variation in cavity architectures, resulting in significantly smaller ligand-binding tunnels in the plant kingdom. We identify cavity lining residues (CLRs) responsible for reduction in ancestral tunnel space, and these appear to be class specific, and unique to plants, providing a mechanism for the observed shift in domain function. For instance, mammalian cavity lining residues A135, G181 and A192 have evolved to larger CLRs across the plant kingdom, contributing to smaller sizes, minimal STARTs being the largest, while members of type-IV HD-Zip family show almost complete obliteration of lipid binding cavities, consistent with their present-day DNA binding functions. In summary, this work quantifies plant START structural & functional divergence, bridging current knowledge gaps.
Keywords: START Domains, Oryza sativa, Lipid binding tunnels, Binding pockets, Fold prediction, Deep learning
Graphical Abstract
1. Introduction
The genomes of plants can endure small- and large-scale duplications far more successfully than any other kingdom and these duplication events are often combined with high rates of retention of extant pairs of duplicated genes, resulting in an abundance of duplicates, termed as ‘gene families’, with large, often hundreds of members. These gene families in turn, contribute to evolution of novel functions via sub- or neo-functionalization resulting in, for example, floral structure modifications, induction of disease resistance, and adaptation to stress [1], [2], [3]. Furthermore, whole-genome duplications as have been observed in several domesticated crop lineages (wheat, cotton and soybean), have contributed to important agronomic traits, such as grain quality, fruit shape, and flowering time. Because of the large number of such events in the plant kingdom, exploring the present-day diversity among gene family members, in terms of sequence, structure, and function has become a widely advancing field of investigation in plant biology.
Our lab has long focused on natural product biosynthesis and its regulation among plant genomes, successfully elucidating the evolution, diversification, and sub-functionalization of several gene families [4], [5], [6], [7]. One interesting discovery during these studies was the identification of a unique family of plant-amplified lipid transporters that may be involved in the crosstalk between two spatially separated natural product biosynthetic pathways [4]. This remarkable gene family encodes ‘START domains'; highly conserved proteins that have long been known as sterol transporters in mammals. However, there is limited information available on the START domains structure, binding cavity and its lining residues for ligand interaction in plants. This work attempts to investigate the START domains gene family with the aim to bridge current knowledge gaps, and to pave the way for future studies into whether and how these domains may serve as the hitherto unknown and unreported sensors or transporters of lipid/sterols in plants.
The term START is an acronym for ‘Steroidogenic acute regulatory protein (StAR) related lipid transfer’ domain, and it is an evolutionarily conserved domain of approximately 200–210 amino acids implicated in lipid/sterol binding and transport [8], [9]. The prototype START domain was first identified in a cholesterol-transporting mammalian StAR protein and later found to be significantly amplified across the plant kingdom [10], [11], [12]. In mammals, START domains perform lipid transport between intracellular compartments, lipid metabolism, lipid signalling modulation and many other physiological processes, and are reported to be involved in cancer, atherosclerosis, autoimmune diseases etc., thus forming potential targets for drug development [13], [14]. In contrast, functional investigations into plant STARTs have revealed vital roles for homeodomain associated START domains in developmental processes such as cell differentiation, organ polarity and shoot meristem embryonic patterning [15], [16], [17]. This feature of START domains associating with homeodomain (HD) transcription factors, appears to be evolutionarily distinct and unique to the plant kingdom, while other domains frequently found to be associated with START domains are bZIP, MEKHLA, PH and DUF1336 [12]. Very few studies are available for these non-HD classes of plant START domains, but some reports have implicated DUF1336-associated START proteins in pathogen resistance [18], [19]. Interestingly, plants have retained one class of START domains that have no other domain associated, and are therefore called ‘minimal START proteins’, and despite lacking any functional characterization, these domains show greatest homology to lipid transfer proteins of mammals, indicating a possible role in transfer of lipids in plants [20].
Currently, experimentally determined crystal structures are available only for mammalian (human and mouse) and invertebrate (silkworm) START domains, and these show a conserved structure with nine anti-parallel β-strands (ten in one case) and four α-helices arranged in a ‘helix grip’ manner, as depicted in Fig. 1 (panels A and B) [21], [22], [23]. The helices and sheets are all numbered from N- to C-termini, namely α1 to α4, and β1 to β9 (β10 in case of STARD4), respectively. Two α-helices (α1 and α4) are present at N and C terminal of proteins, respectively, while two short helices (α2 and α3) are present between the β3 and β4. In addition, two Ω loops have also been characterized; Ω1 between β5 and β6, and Ω2 between β7 and β8 [8], [24]. Two of the available human START domain tertiary structures were crystallized in ligand-bound forms, namely the Phosphatidylcholine transfer protein (STARD2 or PCTP; PDB id 1LN1) bound to a phosphatidylcholine analogue [25] and the START domain from the CERT (Ceramide transporter), bound to multiple synthetic ligands (PDB ID 2E3M, 2E3N, 2E3O, 2E3P, 2E3Q, 2E3R, 2E3S, 2Z9Y, and 2Z9Z) [26]. In both these structures, the α1 did not make any contact with ligand, rather, the binding of ligand required major conformational changes like unfolding or opening of C-terminal α helices (α4) and movements in the Ω1-loop. The central β strand and the α4 helix (acting as the lid) form a deep hydrophobic pocket, involved in ligand binding [25], [27]. Phosphatidylcholine, the ligand for PCTP, occupies approximately 723 Å3 within a solvent-accessible volume of 882 Å3 [25]. Ligand binding cavity volume of cholesterol-binding mammalian START domain STARD4 was found to be approximately 642 Å3, slightly larger than the volume of a cholesterol molecule (432 Å3) [24]. These two structures enabled the determination of a unique tunnel like cavity present deep within the helix-grip fold, as depicted in mesh representation in Figs. 1C and 1D.
Fig. 1.
The representative structures of human START domains (A) PCTP STARTD2 domain (PDB ID:1LN1) and (B) STARD4 (PDB ID: 6L1D) show the helix-grip fold. Ligand binding pockets are depicted as light blue mesh in (C) for PCTP (with ligand; red) and (D) blue mesh for STARD4.
There are reports suggesting multiple physiological functions of the plant START domain based on ligand preferences. For instance, START domains of Arabidopsis ATML and PDF2 proteins on interaction with sphingolipids and ceramides lead to positional signaling during epidermal differentiation [28], [29]. The START domain of the HD ZIP III protein binds with different phospholipids [30] which in turn increases transcriptional efficiency. The START domain of WHEAT KINASE START1 (WKS1) binds with phosphatidic acid (PA) and phosphatidyl inositol phosphates (PIPs) and provides resistance against Puccinia striiformis [31], [32]. All of this data suggests a functional shift in binding preferences that may have accompanied the expansion and huge abundance of this family in the plant kingdom. This functional divergence, combined with a lack of any structural data for plant STARTs has made it difficult to quantify or characterise these domains in plants.
The present work was undertaken to address the structural knowledge gap through deep learning, combined with a comprehensive analysis of ligand-binding tunnel architectures, to explore their role in predicting shape, size or chemical properties of cognate ligands, which may in turn assist in understanding the reported diversity in their function. The availability of several mammalian START structures and a large number of plants amplified START domain sequences offers a suitable case study for applying homology modelling and deep learning to this question and for correlating protein structure with function, as plant derived START domains are known to perform a variety of functions, while retaining a conserved structural core. Deep Learning not only identified tertiary structures, confirming the conserved structural fold of plant START domains, but also provided accurate data for identifying buried ligand binding cavities, which in turn allowed us to perform molecular surface based studies to understand the mechanistic differences between START sub-families and their correlation with functional evolution in this superfamily. Delineation of ancestral lipid binding tunnels within plant START domains and a detailed homology-based comparison of tunnel architectures in terms of pocket volumes, accessible surface area and shapes revealed that variability in ligand binding features may dictate the functional diversity of START domain-containing proteins. Our data reveals three distinct classes of pocket volumes in rice START domains, the largest being minimal STARTs, which appear to be the closest homologs of mammalian START domains, presenting a case for this class to be the hitherto unrecognized group of lipid transporters or sensors in plants.
2. Materials and methods
2.1. Data collection
A total of 28 START domains proteins were identified in cultivated rice variety (Oryza sativa var. japonica) from our earlier work on evolution of START domains across the rice pangenome [33]. These 28 START domains were classified into four major structural classes based on the arrangement of co-occurring domains (co-domains), namely (i) HD bZIP START MEKHLA; HZSM or Class III HD-Zip (ii) HD START; HS and HD bZIP START; HZS or Class IV HD-Zip (iii) PH START DUF1336; PSD and (iv) minimal START proteins; mS (lacking co-domains) [33]. The START domain regions from all proteins were extracted based on annotated border residues and were fed into the structure prediction pipeline as described below. Comparative residue-based analyses were performed using available crystal structures of mammalian START domains retrieved from the RCSB-PDB consortium (https://www.rcsb.org/) (using respective PDB ids of StAR (3P0L), PCTP (1LN1) and STARD4 (6L1D). Sequences were aligned using clustal omega at default parameters [34]. Structural element visualisation were done using ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) [35].
2.2. Tertiary structure prediction
The traditional methods of using threading [36] and position-specific iteration BLAST (psi-BLAST) against the protein databank (https://www.rcsb.org/) [37] to identify suitable structural templates for homology models do not work for START domains and that has been a major bottleneck in structural characterization of these proteins. Therefore, for this study, we used the most recent CASP (Critical Assessment of Structure Prediction) and CAMEO (Continuous Automated Model Evaluation) experiments to select three top scoring most widely used tools for structure prediction [38], [39]. These three tools, namely I-TASSER, C-I-TASSER and RoseTTAFold, were used for building tertiary structure models for plant START domains followed by validation as described in the next section. Five structural models were created for each of the 28 rice START domain sequences using these three methods.
I-TASSER uses profile-profile alignments (PPA) between the target and template to detect weak similarities using the multiple threading approach and full-length atomic model constructs by iterative template-based fragment assembly simulations [40], [41]. C-I-TASSER is an extended version of I-TASSER, that also adds deep-learning contact prediction to fragment assembly simulations [41], [42]. RoseTTAFold is based on a three-track neural network combining information across one-dimensional (sequence-level), two-dimensional (distance maps), and three-dimensional levels (3D atomic coordinates), is successively transformed and integrated [43]. This last method of three-track network-based structure predictions enables rapid solutions of structure modelling problems, providing insights into functions of proteins with currently unknown structures.
2.3. Model validation and quality assessment
Five structural models were generated for each of the 28 rice START domain sequences using the three methods described above. To identify which of the three prediction tools worked best, all 420 resulting START domain models were validated using VoroMQA [44] and Verify3D [45], [46] available through the SAVESv6.0: Structure Validation Server (https://saves.mbi.ucla.edu/). Verify3D fails models that do not fit minimal criteria, while VoroMQA cutoff score is 0.4 for accepting models. In addition, the overall quality of best models was evaluated using proSA [47] and ERRAT [48]. An ERRAT score of 50% or higher is considered to represent accurate high-quality models [48], [49], while ProSA maps results on available Z-score of all experimental structures (ranging from −10 to −3 for 200 amino acids). Finally, models passing criteria by both these methods were subjected to manual inspection via visualization in PyMOL to check for core structural integrity, where alpha helices and beta sheets were integrated and complete (The PyMOL Molecular Graphics System, Version 2.5.2 Schrödinger, LLC).
2.4. Cavity architecture studies
The selected set of START domain models was subjected to a search for potential ligand binding cavities or buried tunnels by using CASTp (Computed Atlas of Surface Topography of proteins) with a probe radius of 1.4 Angstroms [50]. CASTp identifies possible binding sites from 3D atomic coordinates of proteins and provides measurements of solvent-accessible surface (SA, Richards' surface) and molecular surface (MS, Connolly's surface) for each pocket and cavity. This tool uses an α-shape method developed in computational geometry to measure area and volume of each identified pocket and compute its imprint via pre-calculated grids of affinity potentials [51]. A number of surface concavities and/or buried pockets may be detected for a given structure and it is therefore important to manually inspect geometric and topological properties of all such cavities to identify the correct ones. For plant START domain models, the correct ligand binding tunnels were identified based on a combination of manual inspection and similarity to known cavities reported in the available crystal structures of mammalian START counterparts. This was followed by measurement of pocket volumes based on solvent-accessible surface (SA, Richards' surface) of the respective ligand-binding tunnels, and extraction of corresponding cavity lining residues (CLRs) using custom Python scripts. These CLRs were then classified using hydropathy and volume categories based on standardised IMGT (International ImMunoGeneTics information system) criteria [52]. The ‘hydropathy’ classes (hydrophobic, neutral and hydrophilic) were defined based on the amino acid hydropathy index [53], while the ‘volume’ classes were defined as very small, small, medium, large and very large based on the known residue volumes in angstrom units [54].
3. Results
3.1. Conserved core regions of plant and mammalian START domains
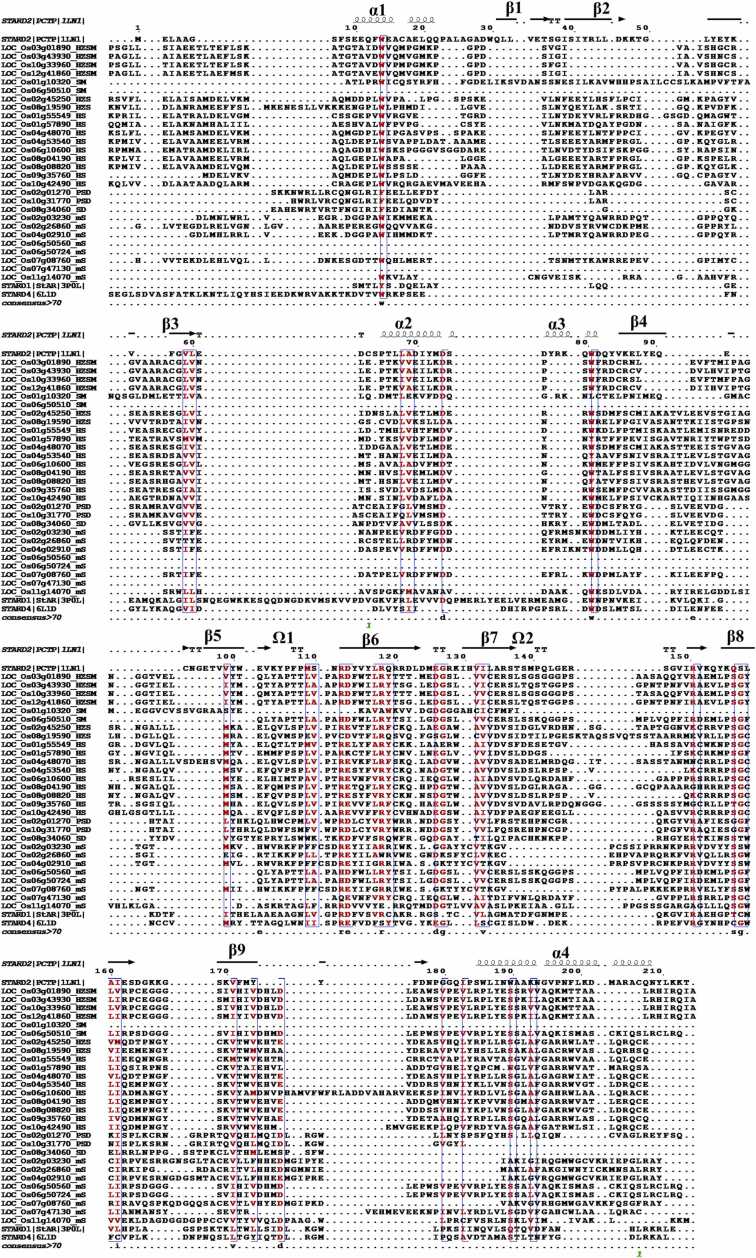
The extent of conservation between plant and animal START domain structural folds was assessed, first by sequence alignments, and then via deep learning, as described in the next section. Fig. 2 depicts all 28 rice START domains aligned with the three most representative mammalian structural counterparts, namely, StAR, PCTP and STARD4 representing three distinct subfamilies of human START domains [21]. In the absence of any reported tertiary structure for STARTs in the plant kingdom, a multiple sequence alignment enables an assessment of common features in sequence and/or structure, that may in turn reveal subtle or strong residue variations leading to changes in protein function through modulation of binding site architecture, ligand preference, ligand specificity and evolutionary conservation.
Fig. 2.
Alignment of 28 rice START domains with three human START domain sequences for which structure is known (first STAR2/PCTP (1LN1), and last two entries with StAR (3P0L) and STARD4 (6L1D), respectively.
Despite large gaps in the alignment between plant and human START domains, the key residues forming the helix-grip fold appear to be positionally conserved (Fig. 2). Moreover, the divergence is limited to N-terminal alpha-helix (α1) and initial β strands (β1 and β2), while conservation appears to be in the protein ‘core’ involved in cavity formation, namely the central β strands (β3 to β9) as well as the C-terminal alpha-helix (α4) (Fig. 2). Overall, while sequence similarity was observed in some regions of multiple sequence alignment, pair-wise similarity between the mammalian and plant START domains was nearly absent, making it difficult to identify suitable templates using traditional methods of homology modeling.
3.2. Deep learning approaches perform best for structure prediction
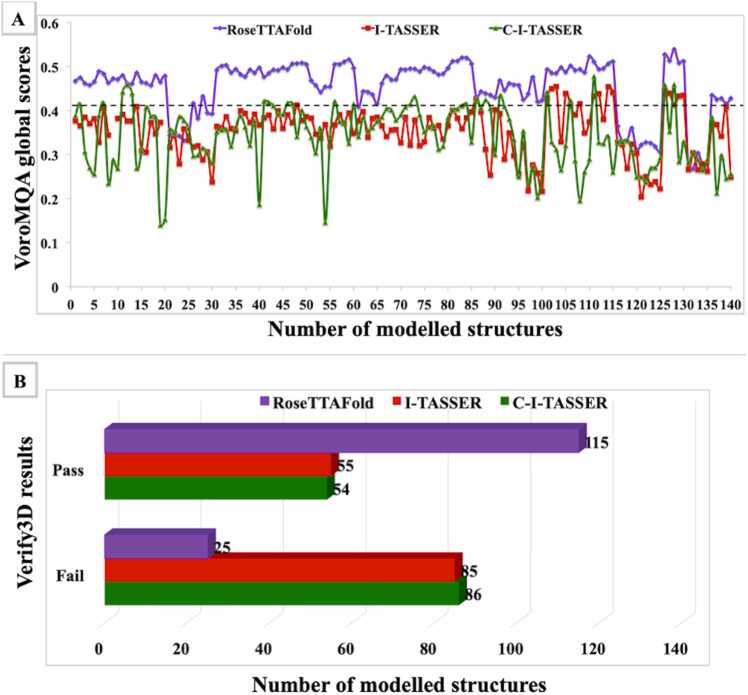
In the absence of suitable template structures for homology modeling, the tertiary structure prediction for rice START domains was undertaken using threading and deep learning approaches. Three modeling tools based on threading and deep learning algorithms were used for building the initial models, and for each tool, a total of five models were generated for each of the 28 rice START domains. In all, this resulted in the generation of 420 (i.e., 28 *3 *5) models that were further evaluated based on various parameters using Verify3D and VoroMQA as described in Materials and Methods. Fig. 3 represents the comparative VoroMQA global score, and Verify3D results for all 420 rice structural models.
Fig. 3.
Comparative Model validation for RoseTTAfold, I-TASSER and C-I-TASSER based rice START domain models. (A) VoroMQA global score (B) Verify3D scores. The X-axis represents the total number of initial models generated; Y-axis represents respective model quality and results.
VoroMQA (Voronoi tessellation-based Model Quality Assessment) is an all-atom knowledge-based protein structure validation/scoring method based on the statistics of inter-atomic contact areas instead of distances. It produces scores (ranging from 0 to 1) at atomic, residue and global levels, where scores greater than 0.4 indicate good models while lower scores indicate unreliable (0.3–0.4) models [44]. The comparative VoroMQA global scores for all 420 models reveal a clear preference for deep learning-based models (generated RoseTTAfold), as compared to models generated by the other two methods. Taken together, only 22 and 34 models generated by I-TASSER and C-I-TASSER, respectively showed acceptable scores > 0.4, but for each of these, the deep learning algorithm RoseTTAFold generated a higher scoring model. A total of 118 of the 140 models generated by RoseTTAFold showed scores better than 0.4, and these were selected for further analyses.
The superiority of deep learning-based models became clear not only from the VoroMQA scores, but also from Verify3D quality factors, as can be seen in Fig. 3 (Panel B). Verify3D helps in assessing structural models by calculating the compatibility of the constituent amino acids to the modelled protein. A 3D profile is built for each residue of the protein model, which characterizes the residue position in the model. Models that are passed by this tool possess at least 80% residue scores > = 0.2 in the 3D/1D profile [45], [46]. More than 80% (115 out of 140) plant START models generated by RoseTTAFold qualified Verify3D parameters, as compared to less than 40% (54–55 out of 140) models generated by threading methods I-TASSER and C-I-TASSER.
3.3. Validation and quality assessment of plant START models
Since deep learning models clearly outperformed threading-based models, these were selected for further evaluation. Out of the five models generated for each of the 28-rice START domains, the best model was selected based on the combined validation parameters described in the previous section, as well as additional scores measured by ERRAT and ProSA (Protein Structure Analysis) programs. ERRAT analyses the pattern of interactions to detect structural anomalies and can identify regions in structural models that may have been modelled incorrectly [48]. ProSA designates a specific Z-score to the input structure and depicts whether it is within the range of scores in the context of all known protein structures of similar size [47]. Table 1 depicts these scores, including the VoroMQA global scores, ERRAT quality factors, Verify3D results and ProSA scores of the 28 START domain models generated via deep learning. Of the 28 top scoring RosseTTAfold models, five failed the Verify3D parameters and had VoroMQA scores (< 0.4), leading to rejection of these models based on quality parameters. It has previously been emphasized how START domains display a characteristic conserved ‘helix-grip’ fold even in the absence of any significant sequence similarity. Presence of this fold feature was manually checked through rendition and visualization of each model in PyMOL. Table 1 also includes this filter and shows four models (marked by an asterisk) that lack intact alpha-beta helix grip fold configuration. Consequently, these nine models were rejected, and not included for further analyses.
Table 1.
Selection parameters for rice START domain models. The models that passed all validation parameters have a green ‘Yes’ in last column, while rejected models are denoted in red with reasons. *The α/β helix grip fold is not intact and therefore not included for further analysis.
The prefix “LOC_” in locus IDs was omitted for convenience.
Finally, a total of 19 rice START domain structural models qualified our strict validation parameters and these included four HZSM, eight HS, two HZS, single PSD and four minimal START domains. The ERRAT module of the SAVES v5.0 tool showed that the overall quality factor of these 19 RoseTTAFold models varied from 85 to 93 (In the range of 0–100), and VoroMQA scores of the START domain models were higher than 0.40. Further, the ProSA Z-scores were within the range of − 5.0 to − 7.0. Table 2 represents the Ramachandran statistics of these 19 selected START domain models and shows 84–93% of the residues in most favored regions with only 0–2.3% residues in the disallowed region. Taken together, from Table 1, Table 2, all validation parameters suggested high quality of the finally selected 19 rice START domain models. Since these 19 models include all major classes of START domain categories, these became the starting point for comprehensive structural evaluation of cavity architectures and for gaining insights into their tertiary folds, especially from the viewpoint of ligand binding.
Table 2.
The Ramachandran statistics and overall quality evaluation of 19 selected structural models.
| Ramachandran statistics |
||||||
|---|---|---|---|---|---|---|
| Locus ids | START classes | Common name | Total residues | Most Favoured (%) | Allowed regions (%) | Disallowed region (%) |
| Os03g01890 | HZSM | LF1 | 216 | 87.8 | 10.5 | 1.7 |
| Os03g43930 | HZSM | HB4 | 216 | 91.7 | 8.3 | 0 |
| Os10g33960 | HZSM | HOX9 | 216 | 88.9 | 10 | 1.1 |
| Os12g41860 | HZSM | HOX33 | 216 | 91.1 | 7.8 | 1.1 |
| Os02g45250 | HZS | ROC5 | 236 | 89.8 | 10.2 | 0 |
| Os08g19590 | HZS | TF1L | 250 | 88.9 | 10.6 | 0.4 |
| Os01g55549 | HS | ROC9 | 231 | 92.7 | 7.3 | 0 |
| Os01g57890 | HS | TF1 | 221 | 90.3 | 9.7 | 0 |
| Os04g48070 | HS | ROC4 | 245 | 83.6 | 15.4 | 0.9 |
| Os04g53540 | HS | ROC2 | 229 | 93.1 | 6.9 | 0 |
| Os08g04190 | HS | ROC7 | 230 | 93.3 | 6.7 | 0 |
| Os08g08820 | HS | ROC1 | 224 | 87.8 | 11.2 | 1 |
| Os09g35760 | HS | - | 224 | 91.2 | 8.3 | 0.5 |
| Os10g42490 | HS | ROC3 | 236 | 86.4 | 12.7 | 1 |
| Os10g31770 | PSD | - | 215 | 87.4 | 11.1 | 1.6 |
| Os02g03230 | mS | - | 206 | 87.4 | 11.5 | 1.1 |
| Os02g26860 | mS | - | 207 | 92.9 | 5.5 | 1.6 |
| Os04g02910 | mS | - | 207 | 88.6 | 9.1 | 2.3 |
| Os07g08760 | mS | - | 211 | 88.1 | 10.3 | 1.6 |
3.4. Ancestral Helix-grip Fold conserved among rice START domains
Fig. 4 depicts overall 3-D folds of representative models from each class of rice START domains while the individual structure of all 19 selected models is provided in Supplementary Figures 1A–S. Clearly, all models have a conserved α/β helix-grip fold typical of START domains, connected by short loops and turns. Despite overall core fold similarity, differences are apparent between structural classes, as can be seen in Fig. 4.
Fig. 4.
Top scoring deep-learning based models representing each structural class of rice START domains. (A&B) HZSM;HDbZIP START MEKHLA, (C) HZS;HD bZIP START, (D&E) HS;HD START, (F) PSD;PH START DUF and (G-I) mS;minimal START.
As can be seen in Fig. 4, the models enable color-based comparison of tertiary structures, and the similarity of the core regions is visibly clear. Apart from the conservation of the helix-grip fold, some common and unique structural features could be identified for each plant START structural class. For example, the two major α-helices at N and C terminals (α1 and α4) are arranged in similar configurations within each of the structural classes. Similarly, the β-strands that form the core of the helix-grip fold are also similar within a given class. These two features strongly impact the shape and architecture of the START internal tunnels/cavities, as has been discussed earlier.
Out of nine minimal START domains identified in rice, the structure of four minimal START domains has been modelled in this study (five were rejected, as shown in Table 1). Interestingly, the modelled structures of rice minimal START domains showed the highest similarity with mammalian START domains, with all nine-β strands and four α helices being fully conserved. The PH-associated plant START (PSD) models also showed the presence of nine β-strands and four α-helices, but the relative position of these secondary structural elements was unique to the PSD class, along with significant variations in loop regions (as shown in Fig. 4F). Among the other START domain class models, one consistent pattern is the slight distortion in the last alpha helix (α4), and the extent of this distortion appears to be unique to each class as, discussed below.
The four rice HZSM START models showed eight β- strands and four α-helices. The tertiary structure of these four START domains showed significant conservation, including a kink in the last alpha helix that connects via a long loop to the tunnel bordering the beta strand discernible in all models (as in the case of Fig. 4A and B). Although the central β-strands forming the core helix-grip fold are similar to mammalian START domains, variations can be observed in the N-terminal strands β1 and β2 (as per PCTP numbering). The former (β1 strand) was found to have a more extended loop region in all the HZSM START models whereas β2-strand was shorter in size within HZSM classes.
The sequences of START domains from the HS and HZS classes showed wide variability, but the deep learning-based structural models generated for these two sets of rice START domains appeared to be conserved within each class. Overall, the number of β strands varies from eight to nine in these classes, but the distortion in the terminal alpha helix (α4) is distinctive for each class as mentioned earlier. In case of HZS, this helix is highly extended with no kinks in secondary structural elements (Fig. 4C), whereas in case of HS models, this helix is much shorter and broken to produce a kink outward from the structure (Fig. 4 D and E), in contrast to the inwards-oriented kink observed in HZSM models. Further, the two β-strands (β3 and β9, as per PCTP numbering), which are part of the core helix-grip fold, were found to be comparatively longer in size, which may be due to sequence divergence.
In summary, tertiary structure analysis of rice START domains showed that, except for some insertion or deletions in loop regions, the core structures of all plant START domains retain the conserved β-strands and α-helices forming a helix-grip fold similar to mammalian counterparts. However, subtle but unique structural features were identified in each class of the rice START domain models and these features may have a bearing on the shape and architecture of the respective ligand-binding tunnels, which in turn may specify ligands or potential function of these START domains. These observations prompted us to analyse the cavity architecture in detail and investigate the cavity lining residues in each structural class, as described in the next section.
3.5. Cavity architecture and ligand binding tunnels of START domains
Topological and Geometric properties of protein structures, such as surface pockets, interior cavities, and cross channels, are critical for catalysis as well as ligand binding among proteins. Protein function is strongly dependent on molecular recognition, which is even more critical in case of ligand binding proteins, such as the START domains. The study of molecular surfaces of proteins can be helpful in the identification or prediction of potential binding sites. When the first X-ray crystal structure was determined for a (mammalian) START domain, it was postulated that shape of the START tunnel cavity played an important role in ligand specificity. As noted in the previous section, the overall structure of rice START domains remained conserved despite variations among different structural classes. Most importantly, α helices at both N and C termini are conserved, separated by eight to nine β sheets, and these form the typical helix-grip fold that surrounds the START domain cavity. The presence of this fold despite variations across START domain classes prompted us to explore conservation of key residues lining the binding site pockets, which may have a bearing on ligand binding and consequently the functions performed by START domains. Accordingly, ligand-binding tunnels were predicted for each of the 19 rice START domain models using a surface accessibility based method, followed by manual delineation of the cavity and pocket architecture analysis, as described in Materials and Methods. CASTp was selected as the method of choice as it performed well when tested against experimentally characterized and well known cavities of human START domain structures, namely PCTP (PDB:1LN1) and MLN64/STARD3. The START domain cavity for the PCTP with the bound ligand has been depicted in Fig. 1C as a reference for all plant domain assessments reported hereafter.
Proteins often have several internal pockets and tunnels of various sizes, emerging from the folds and spaces created during the folding process, thus, it is imperative to select the correct ligand binding cavity from amongst all predicted cavities. In order to find the true cavity for plant START domain models, and to avoid false predictions during cavity identification, a dual validation approach was adopted. Firstly, the true cavity should be present within the well conserved helix-grip fold, i.e., in close vicinity of the central beta-strands and two terminal helices (α1 and α4), as has been seen in known (mammalian) 3D structures. Secondly, there must be some overlap between CLRs (Cavity Lining Residues) of the true cavity, when superimposed with the CLRs reported for the mammalian START structures. The former clause was ensured manually for each cavity identified by means of visual rendering and inspection, while the latter was tested by in-house scripts to compare and match CLRs of each identified cavity with reported mammalian START domain CLRs from STARD4, as well as PCTP, that has been experimentally determined with bound ligand (PDB IDs 1LN1). Both experimentally determined START structures were therefore use to avoid bias in cavity selection. While additional experimental structures exist for mammalian START domains, it is important to note that many of these structures have been crystalized in apo-form without ligands, and therefore, information about experimental data such as cavity volume and CLR is not available for all available structure. Thus, for each rice START domain model, all identified cavities were subjected to (a) Extraction of cavity lining residues or CLRs (b) Testing of CLR coverage in comparison to mammalian CLRs and (c) Manual inspection of each selected cavity using PyMol. In summary, all rice START domain models were subjected to surface based cavity identification followed by analysis of their Cavity Lining Resides or CLRs.
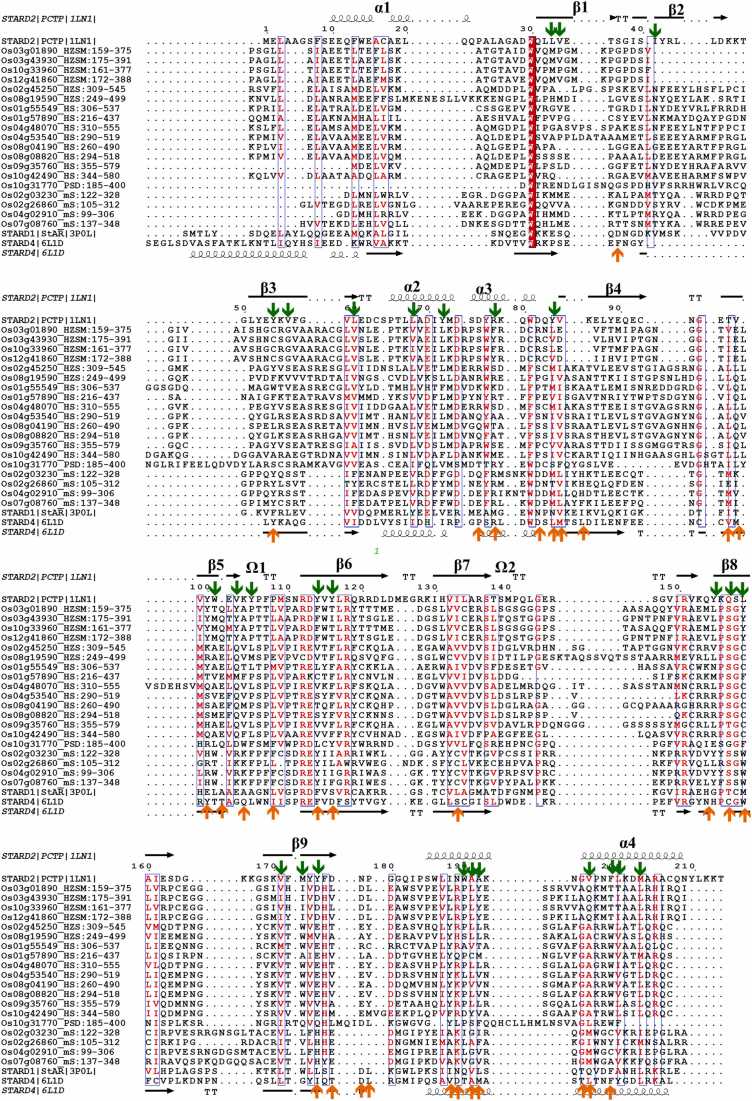
CASTp provides a number of potential or probable pockets for a given 3D structure along with a list of CLRs for each pocket and a detailed description of all atoms exposed to the cavity. Each cavity for each model was recorded, followed by validation of the cavities by means of testing coverage with known cavity lining residues of PCTP and STARD4. The co-crystal of PCTP/STARD2 bound with phosphatidylcholine identified 28 residues that were in contact with the ligand. Of these 28 CLRs, eighteen are hydrophobic, two hydrophilic and eight are neutral amino acids. Similarly, the hydrophobic tunnel of STARD4 is composed of eighteen hydrophobic residues, seven hydrophilic residues, and seven neutral residues. Fig. 5 shows the 19 rice START domains aligned with both PCTP and STARD4, and the reported CLRs for both PDB structures are highlighted on this Figure. Despite PCTP and STARD4 showing differences in the cavity volumes and binding to distinctive ligands, their CLRs have thirteen positional matches, as can be seen in Fig. 5, thus supporting our method for identifying the true cavities for plant-START domain models.
Fig. 5.
19 rice START domains aligned with human PCTP and STARD4 to map corresponding plant CLRs. All secondary structures for PCTP and STARD4 are highlighted at first and last positions, respectively. CLRs are marked as green (for PCTP) and orange (For STARD4) arrows. Note the 13 positional matches between PCTP and STARD4 cavities (Green and orange arrows on same column).
Plant START domain residues aligning with the CLRs of both known structures were extracted for each of the 19 rice START domains, retaining the original positional identity based on rice sequences. Supplementary Table 1 depicts the corresponding rice CLRs based on alignment with PCTP/STARD2 (PDB;1LN1), while supplementary Table 2 depicts the CLRs of the same 19 rice START domains with respect to STARD4 (PDB; 6L1D). For each rice START domain, the CLRs, extracted as described above, were individually matched with CLRs identified for each CASTp predicted pocket for that domain model. For example, if ten pockets were identified by CASTp for a given rice domain, then the CLRs corresponding to each of the ten potential cavities would be matched with the structure based CLRs for that domain listed in Supplementary Tables 1 and 2. Based on the number of matched residues, each predicted pocket would be ranked, leading to selection of the highest scoring pocket that showed the best positional conservation between the documented CLRs (based on PCTP and STARD4 structural alignment) and the CLRs surrounding the binding pockets detected by CASTp. If multiple pockets were identified with high scores, each of these would be retained and checked for veracity by manual visualisation. CASTp sometimes predicts multiple smaller cavities where a single large pocket should lie, and to overcome this, all the CASTp identified pockets were rendered and visualised individually in PyMOL to confirm if any sub-pocket or adjacent pockets were present in the known/expected cavity regions.
Cavities were successfully identified in all 19 rice START domain helix-grip regions, and Fig. 6 shows only the top ranked pockets in nine rice models corresponding to each of the five major structural classes, namely HZSM (panels A,B), HZS (panel C), HS (panel D-F), PSD (panel G) and minimal START (Panels H,I). The individual top ranked cavity of all 19 models is provided in Supplementary Figures 2 A–S. As can be seen from Fig. 6, the minimal START models appear to have single large and centrally located cavities that are closest in appearance to reported structures when compared with Fig. 1 (panel C and D). This observation supports patterns from earlier sections of this paper where minimal START proteins were found to have greater similarity to mammalian STARTs in terms of secondary structural features and sequence homology. All other rice START domain models appear to have diverged towards lower or smaller-sized cavities, as can be seen in Fig. 6. For example, predicted cavities in HZSM and PSD displayed a single sizeable pocket in the helix-grip fold regions (Fig. 6 A-B and G, respectively). The greatest divergence was observed in the case of the homeodomain-containing HS and HZS models. In one particular case (see the cavity in Fig. 6 Panel F), the entire HS domain cavity appears to have been obliterated, whereas in another case (panel 6E) the cavity remains very small despite being the combination of multiple adjacent cavities in the helix-grip region. A detailed cavity architecture analysis was conducted to quantify these patterns and this included pocket volume measurements & CLR comparison, as described in the next section.
Fig. 6.
Visual rendering (blue mesh) of putative ligand binding pockets identified in representative rice START domains.
3.6. Variability in binding pockets defined by cavity lining residues
Putative ligand binding cavities identified in each of the 19 rice START domains were structurally analysed in terms of pocket volume and shape, as described in Materials and Methods. The cavity volumes for each of the 19 rice START domains are depicted in Fig. 7, along with comparative cavity measurements for STARD4 and PCTP structures. The rice START cavities were classified into three categories based on their pocket volumes, namely (A) small (vol. <100 A3), (B) medium (vol. >100 to <400 Å3) or (C) large (vol. >400 Å3). As can be seen in Fig. 7, none of the rice domains have cavity volumes as large as the reported range of mammalian START structures. As expected, the largest rice cavities were observed for minimal START models with pocket volumes ranging from 430 to 550 Å3. In contrast, volumes of all other rice START models were much smaller in size, ranging from as low as 10 Å3 in case of the homeodomain models, to 220 Å3 for HZSM models. The pocket volume of MEKHLA associated START (HZSM) and PH START (PSD) fell under medium category. The pocket volumes of HZSM were less variable, all being close to 200 Å3, while the pocket volume of the standalone model of PH START domain was similar at 170 Å3.
Fig. 7.
Volumes of the rice START ligand-binding pockets.
As noted earlier in Fig. 6, the HD associated STARTs (HS START or HZS Class IV HD-Zip family) showed the most variation in pocket size. The two HZS class models showed contrasting pocket volumes, one (TF1L) being classified as medium with a volume of 136 Å3, while the other (ROC5) showed a very small pocket volume (25 Å3). Seven out of the eight modelled HS STARTs showed small pocket volumes (8–91 Å3), whereas only one (ROC9) showed medium volume of 148 Å3. Furthermore, the HD START models had fragmented pockets, i.e., most of these models showed two or more pockets in their expected helix-grip fold regions. Even after the multiple adjacent pockets were combined, these domains still showed very small cavity volumes. It may be noted here that all non-minimal plant STARTs are parts of multi-domain proteins and the drastically reduced cavities may reflect either a change in ligand preferences to smaller ligands, or a loss of binding function altogether. In any case, cavity volume measurements indicate evolutionary changes in binding abilities of the individual plant START structural classes and this needs to be investigated further in terms of residue level changes that may be responsible for these patterns. For example, the HS STARTs appear to have undergone near-complete obliteration of binding cavities despite retaining an overall conserved structure, and it would be interesting to explore how the residues in their core helix-grip fold region have changed to bring about such a drastic reduction in volume.
Similarly, the minimal STARTs are closest to mammalian structures in terms of having a single large central cavity, but with reduced volumes. These domains are similar in sequence to other multi-domain rice STARTs, and therefore, it would be interesting to explore whether and to what extent, subtle changes in individual CLRs may be responsible for the observed variations in cavity architecture. Minor amino acid substitutions often allow proteins to evolve over time, and modulate their binding abilities, resulting in changes in ligand preferences. The next section compares the CLRs individually across all rice START domains to identify class-specific residues that may influence putative pocket volumes across the various structural classes.
3.7. Physicochemical Variation in binding pockets of Plant STARTs
The cavity lining residues (CLRs) for all putative binding pockets were compared across the 19 rice STARTs and these are provided in Table 3, along with volumes for each model. A cursory glance at this table indicates that cavity volumes are roughly correlated with the number of residues lining the cavity, and the composition of CLRs is primarily hydrophobic. Both these features are expected, since larger cavities would be physically in contact with more residues and a tunnel placed within the hydrophobic core of a protein is expected to be composed of largely hydrophobic residues. Among the cavity regions, hydrophobic amino acids constitute almost half (50%) of total CLRs, and among these residues, aliphatic amino acids like Ala, Val, Leu and aromatic amino acids Trp and Phe are higher in number. Furthermore, the known role of START domains as lipid transporters would also require the ligand binding cavity to be mostly hydrophobic in nature, for example, the minimal START domains showed the highest number of CLR (52−58), followed by HZSM domain cavities with 33–41 CLRs, of which, almost half are hydrophobic, whereas the single PSD START showed 27 CLRs including 16 hydrophobic ones. However, the extent and impact of CLR hydrophobicity on pocket-binding patterns needs to be investigated individually, as this can be an important criterion in the kind of ligand that may bind to the rice domains.
Table 3.
Positional information of CLRs across different structural classes of START domains. The green letter colours indicate the positional match with respect to mammalian PCTP (PDB:1LN1), while the orange letter indicates the positional match with respect to STARD4 (PDB:6L1D). The residues which showed positional match for both (PCTP and STARD4) were indicated in bold red colour. The values in parentheses denote the number of total CLRs identified by CASTp and cavity volume in Å3. (*Two adjacent pockets detected in its known cavity regions **More than two adjacent/sub pockets were detected in its known cavity regions).
Another notable aspect of Table 3 is that majority of the HS-START domain cavities were identified as combined volumes of two or more very small CASTp identified cavities, and yet their joint volumes remain extremely small. For example, as discussed previously, most of the HS classes of STARTs showed multiple sub-pockets or/and adjacent pockets; the CLR of these pockets were combined. Among the HS-STARTs, the ROC1 showed the lowest CLRs (only 10 amino acids), ROC9 showed highest number of CLRs (38 amino acids) and CLRs among other HS STARTs varied from 23 to 29. Out of two HZS STARTs, the ROC5 has 21 CLRs, whereas TF1L showed 38 amino acids in CLRs. These contrasting CLR numbers combined with variance in cavity volumes measured for each of the 19 rice START domain models suggested a divergence of potential ligands across the five structural classes, as compared to the cholesterol/lipid-binding tunnels of mammalian START domains. All but the minimal rice START domains were found to have small and/or medium-sized cavities, and even the minimal STARTs with largest cavities were significantly smaller than their mammalian counterparts. The results for rice domains match similar findings earlier reported for Arabidopsis START domains [55] and taken together, these observations indicate that plant START domains may have evolved a novel class of previously unknown lipid binders/transporters with regulatory functions, mediated by the homeodomain (HD).
As noted in Fig. 1 panel C, the ligand binding cavities are often slightly larger than expected, suggesting the possibility and scope for binding additional types of ligands, but the consistent small sizes of all rice domains imply that putative ligands may be much smaller than expected/known for STARTs, and in the extreme scenario of the homeodomain (HD) associated START domains, the cavities may even have been obliterated completely during evolution. These observations require a careful and detailed examination of CLRs of the identified pockets. As can be seen from Table 3, there is variation in composition of individual pocket lining residues of different structural classes of START domains of rice. However, despite this variation, there were significant positional matches of CLRs when compared with the mammalian structures. Following the colour-coding pattern in Table 3, these positional matches among CLRs were marked in orange or green, depending on whether the match was found with STARD4 and PCTP START proteins, respectively. CLRs that matched with both mammalian structures are highlighted in dark red. It is clear from Table 3 that the maximum correspondence is between minimal START domains of rice and mammalian PCTP proteins, where 20–25 residues (out of 28 reported CLR in PCTP) were matched at their respective positions. Further, minimal STARTs also showed significant similarity with STARD4 domains, where almost two-third of the amino acids showed similar positional alignment in the pocket cavity.
In order to understand the variation in the cavity lining residues between START domains of humans and different structural classes of rice START domains, the residues forming the 19 tunnel pockets were compared with mammalian counterparts, as per the initial structure-based sequence alignment shown in Fig. 5. The most important residues among these CLRs are depicted in Fig. 8, where the first two rows represent CLRs in the mammalian structures, while all others rows depict the plant CLRs in corresponding positions. Further, these residues were compared in terms of their physicochemical properties using two characteristics, namely hydropathy and steric class, as described in Materials and Methods. Thus, Fig. 8 depicts the matching cavity lining residues and their physicochemical classification for the START domains of rice with respect to mammalian PCTP and STARD4 domains. The amino acids were classified into three classes based on hydropathy scales: hydrophobic, neutral, and hydrophilic. Similarly, the IMGT based steric class distributes the amino acids into five categories: very small, small, medium, large and very large. This Figure (Fig. 8) enabled a residue-based comparison of changes in hydropathy or steric nature of CLRs between mammalian and plant counterparts, which in turn, provided insights into subtle evolutionary changes in the cavity lining residues that may be responsible for the observed variation in the pocket size/volumes of mammalian START domains and rice START domains.
Fig. 8.
Comparison of important cavity lining residues (CLRs) of 19 START domains with respect to PCTP/STARD2 (first row) and STARD4 (second row). Panel (A) indicates the alignment and positional matches while (B) and (C) depict the changes in hydropathy and steric nature. Flat lines in B and C depict corresponding residues that are not involved in lining the respective domain cavity.
As expected, the CLRs of rice minimal START domains showed relatively few changes with respect to mammalian STARTs. Overall, hydrophobicity remained relatively similar with about 48% in mammalian structures and about 47% across minimal STARTs. Some of the notable residue changes in these STARTs include position V103 and A135 (STARD2 numbering), where a hydrophobic or neutral residue has mutated to a charged residue during evolution of plant domains as depicted in Fig. 9 (Panel A-D). In addition, it may be noted that very small residues at positions A135, G181 and A192 in the mammalian structures have mutated to larger CLRs in all plant domains, thereby contributing to smaller sizes of the plant cavities through obfuscation of tunnel space (Fig. 9, A and B). Class specific patterns were also identified in these CLRs, such as, for example, an alanine at position 191 in mammalian STARTs is substituted by significantly larger residues like Trp or Phe in most other plant cavities, but remains small in minimal STARTs. One on hand this patterns suggests a steric mechanism by which larger (mammalian) cavities got obliterated in most rice START domains, leading to smaller cavities, but it also points to minimal STARTs being the largest of all, and this closest in homology to ancestral counterparts that are able to bind to large lipids. The next section describes such class specific substation patterns in more detail.
Fig. 9.
Superimposition of cavities from a mammalian START (PCTP) (grey mesh) and representative START domain of rice (blue mesh) to illustrate the key CLR changes (side chains in red sticks) in pocket regions. The amino acid numbers shown as per the PCTP/STARD2 numbering. The panel A and B depicts the change in cavity volume for minimal START proteins (LOC_Os02g03230_mS). The panel C and D depicts the change in cavity volume for multi-domains START proteins, PH START DUF (LOC_Os10g31770_PSD).
The CLRs of HZSM class of START domains showed change in hydrophobic residues as compared to mammalian STARTs, with the amino acid substitutions at positions M73, A135, L159, A192 and F199 being most significant in terms of loss of hydrophobicity (STARD2 numbering). In terms of steric nature, positions A135, A191 and A192 are most critical in contributing to loss of space in the rice HZSM cavities by means of substitutions from very small amino acids (like Alanine), to larger CLRs like Aspartate, Leucine and Tyrosine.
As discussed previously, the CLRs of homeodomain HD containing START domains (HS and HZS) in rice showed comparatively lesser positional match with PCTP and STARD4 cavities, apart from pocket volume variations and presence of multiple adjacent sub-pockets. Overall hydrophobicity change compared to mammalian CLRs is not significant but there is a remarkable change in residues that are hydrophobic. For example, positions Y72, Y84, K104, Y114, Y116, Q157, and D177 have evolved from neutral/charged to hydrophobic, whereas positions V103 and A135 have become charged in regions where mammalian domains have hydrophobic residues. Similarly, the most drastic steric changes are also observed in this class of rice STARTs with CLRs at positions A135, M173, and N194 becoming much larger, thereby leaving much lesser space within the cavity of HS and HZS classes. Overall, these changes appear to be much more drastic in case of HS rather than HZS, thereby presumably obliterating the cavities of this domain during evolution. A similar case was observed and reported by our group earlier in Arabidopsis, where a single residue change was implicated in the shortened cavity size and volume of plant START domains [55]. The single PSD showed a change of CLR at V85, A135, L159 where smaller amino acids Alanine/Valine), were changed into very large CLR (Phenylalanine) as shown in Fig. 9 C and D.
In summary, comparative analysis of panels A, B and C in Fig. 8 illustrate the change in hydropathy or volume classes of START domains may affect ligand-binding via modulation of cavity tunnel architectures. Most importantly, we identify individual residues that have undergone drastic substitutions despite being in corresponding positions on the alignment, and contributing to overall fold conservation. Such a detailed residue level comparison is evidence for evolutionary change leading to functionally critical substitutions, thereby causing sub-functionalization to smaller ligands (as in case of minimal STARTs), or neo-functionalization in terms of loss of ligand binding ability altogether (as in case of HD containing STARTs). Supporting evidence for these changes has already been seen in our previous studies in terms of gene expression changes and GO enrichment [20], [33], [56]. The evolutionary change of HD START domains into new roles of transcriptional regulation has already been experimentally verified in case of Arabidopsis, and this provides support for the findings reported in case of rice domains [55]. Identification of exact ligands for plant START domains requires extensive molecular docking based on surface charge and shape complementarity for analysis of START ligand domains/protein-ligand complexes with the goal of explaining and ultimately predicting the stereo-specificity and ligand specificity. This work is currently underway in our laboratory along with several other leads for identification of potential ligands, including attempts to crystallise minimal STARTs with bound moieties.
4. Discussion and conclusion
Members of the steroidogenic acute regulatory (StAR)-related lipid transfer (START) domain family are known to function in binding and non-vesicular transport of lipids in mammals, although ligands have only been characterised for a few START domains (for e.g. cholesterol, 25-hydroxycholesterol, phosphatidylethanolamine, phosphatidylcholine, bile acids and ceramides). Detailed studies on these ligands give insights into the roles of these domains in various aspects of biology, including lipid biology, lipid metabolism, lipid trafficking and cell signalling, but from the plant perspective, this information is very limited, as no ligand bound structure has been identified for plants to date. The only report where potential ligands were tested for plants indirectly through metabolite–protein isolation protocol (affinity protein purification) followed by mass spectrometry [55] included potential plant lipids and other putative binders like Phosphatidylserine, Triacylglycerol, Sphingomyelin, Diacylglycerol, Phosphatidylcholine, Lysophosphatidylcholine, Protoporphyrin IX, Alpha-Linoleolcholine, and 9-Carboxy-Alpha-Tocotrienol [55]. Many plant START domains appear in multi-domain proteins, and they may serve as lipid sensors rather than transporters that signal biological responses. This assumption would also require the plant START domains to recognise and bind suitable ligand molecules. This work began with the premise that plant START domains do have internal cavities or hydrophobic tunnels that can bind ligands, whether for transport or to function as sensors.
The first part of this paper provides evidence for sufficient structural conservation between plant and mammalian STARTs to expect a binding tunnel within the helix-grip fold and the second part describes deep-learning and AI based generation of 19 reliable models for the 28 START domains of rice. The third part of this paper confirmed the presence of putative cavities in several of the 19 rice START domains, whose CLR patterns revealed the inherent structural flexibility and dynamic nature of plant START proteins that could explain the huge variation in binding pockets. Apart from the dynamic nature of binding sites, conformation of binding sites can often change upon ligand binding, and therefore, the size of the cavity cannot directly be related to the size of the ligands, as was noted in case of STARD2, where the structure was determined with bound ligand. However, taken together with a structure-based sequence alignment, the detailed cavity architecture analysis undertaken in this work led to the identification of key residues and structural determinants of ligand binding and selectivity in plant START domains.
Mammalian STARTs have been reported to bind and transport several lipid/sterol-based ligands such as cholesterol, PC (Phosphatidylcholine), oxy-cholesterol etc. The rice cavity architecture analysis in this work revealed that sterol-like ligands can be accommodated into only four of the 19 START domain models, and these are all minimal STARTs. Therefore, minimal STARTs appear to be the only class of present day plant STARTs domains that may have retained the capacity to bind and transport large lipid based molecules, similar to mammalian counterparts. This inference is also supported by transcriptomic and gene duplication studies conducted in our group, where we show that minimal STARTs are closest to mammalian homologs, and thus, have the highest potential for being lipid transporters [33], [55], [56]. As demonstrated in very recent reports [57], minimal START domains have played roles in transportation of C20 fatty acids from endoplasmic reticulum to chloroplast in Marchantia polymorpha. Deeper understanding of the structure of the START domain ligands and functions in plants could provide an entirely new avenue of research in plant specific cellular process. Detailed characterization and structural resolution of a ligand bound START domain in plants is currently underway in our laboratory.
Apart from minimal START domains, majority of the rice START domains were found to have extremely small tunnel regions. Interpretation of this pattern for multi-domain plant STARTs suggests that they may either be binding much smaller-sized, hitherto unidentified ligands, or they may have evolved into receptors for lipid-sensing rather than lipid-binding. This is further supported by available literature reports on signaling and transcriptional regulatory plant proteins containing START domains, that have already indicated that this domain may have a broader functional role than purely in lipid transport [18], [19], [58], [59], [60], [61]. In case of several HD containing START domains, our cavity architecture analysis shows a total obliteration of the tunnels, despite conservation of the helix-grip core structural fold, providing further evidence for evolution of new function/s. Furthermore, the paradigm that START domains only play a role in ligand binding and transportation of lipid sterols, does not stand in case of many rice START domains, in which, regardless of very small cavities detected, these were putatively expressed in different anatomical parts and developmental stages as shown in our previous studies [33]. Overall, our findings support earlier reports that majority of plant STARTs have evolved to perform other functions like regulation (through association with other domains like HD-ZIP etc), or at best these may serve as lipid sensors rather than transporters, that signal biological responses. For example, class III HD Zip protein has an additional domain at its C-terminal known as the MEKHLA domain. MEKHLA has been associated with regulating the response to light and redox activity. The evidence for the sensory module in class III HD-Zip protein in the form of MEKHLA domains supports the idea of the START protein-mediated signalling pathway in plants [58]. Similarly, earlier studies suggested that PH-START proteins (EDR2 protein) of Arabidopsis thaliana were found to express during early stages of seed germination as well as in different vegetative and floral tissues [18], [19]. These proteins cause the negative regulation of plant-type hypersensitivity, ethylene activated signalling pathways and leaf senescence.Association with HD has triggered directed evolution towards regulation in plant STARTs, but there is still atleast one class of plant minimal STARTs, that may have retained the role of lipid sensing and binding based on a large cavity identified via architectural variations (and lack of other domains). These inferences complement previous data from our group on the Arabidopsis START tunnels [55].
Taken together, our data reveals three distinct classes of pocket volumes in rice START domains, the largest being minimal STARTs, which appear to be the closest homologs of mammalian START domains, presenting a case for this class to be the hitherto unrecognized group of lipid transporters or sensors in plants. In contrast, the multi-domain plant START domains appear to have evolved regulatory functions, mediated by the homeodomain (HD), at the cost of ligand binding tunnel space. We hope that the current study quantifying the structural and functional divergence of START domains in plants will advance our knowledge of START domains and their roles in plant development and pave the way for further studies to resolve plant ligands and associated functional mechanisms.
Ethics approval and consent to participate
Not Applicable.
Funding
SKM received fellowship from the Department of Biotechnology (DBT) Government of India and National Institute of Plant Genome Research (NIPGR) during his Ph.D. KK received fellowship from the University Grant Commission (UGC), Government of India for her Ph.D. The publication charge of this article was covered from NIPGR Core Grant. These funding bodies do not have any role in design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
Authors acknowledge the support of National Institute of Plant Genome Research (NIPGR), New Delhi for infrastructure and DBT-eLibrary Consortium (DeLCON) for providing access to e-resources.
Consent for publication
Not Applicable.
Author contributions
GY and SKM conceived the idea and designed the work. SKM and KK performed the research work. GY, VG and SKM analysed the data and wrote the manuscript. All authors read the manuscript and approved it for final publication.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.07.039.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Data availability statement
All of the datasets supporting the results of this article are included within the article and its Supplementary Material.
References
- 1.Kejnovsky E., Leitch I.J., Leitch A.R. Contrasting evolutionary dynamics between angiosperm and mammalian genomes. Trends Ecol Evol. 2009;24:572–582. doi: 10.1016/J.TREE.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Panchy N., Lehti-Shiu M., Shiu S.H. Evolution of gene duplication in plants. Plant Physiol. 2016;171:2294–2316. doi: 10.1104/pp.16.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiao X., Li Q., Yin H., Qi K., Li L., Wang R., et al. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019;20:38. doi: 10.1186/s13059-019-1650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumari S., Priya P., Misra G., Yadav G. Structural and biochemical perspectives in plant isoprenoid biosynthesis. Phytochem Rev 2013 122. 2013;12:255–291. doi: 10.1007/S11101-013-9284-6. [DOI] [Google Scholar]
- 5.Yadav A., Thakur J.K., Yadav G. KIXBASE: a comprehensive web resource for identification and exploration of KIX domains. Sci Rep 2017 71. 2017;7:1–12. doi: 10.1038/s41598-017-14617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Priya P., Yadav A., Chand J., Yadav G. Terzyme: a tool for identification and analysis of the plant terpenome. Plant Methods. 2018;14:1–18. doi: 10.1186/S13007-017-0269-0/TABLES/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A., Yadav G. Shared ancestry of core-histone subunits and non-histone plant proteins containing the Histone Fold Motif (HFM). Https://DoiOrg/101142/S0219720021400011 2021;19. 10.1142/S0219720021400011. [DOI] [PubMed]
- 8.Tsujishita Y., Hurley J.H. Structure and lipid transport mechanism of a StAr-related domain. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 9.Iyer L.M., Koonin E.V., Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins Struct Funct Genet. 2001;43:134–144. doi: 10.1002/1097-0134(20010501)43:2<134::AID-PROT1025>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 10.Clark B.J., Stocco D.M. Expression of the steroidogenic acute regulatory (stAR) protein: a novel LH-induced mitochondrial protein required for the acute regulation of steroidogenesis in mouse leydig tumor cells. Endocr Res. 1995;21:243–257. doi: 10.3109/07435809509030440. [DOI] [PubMed] [Google Scholar]
- 11.Clark B.J., Wells J., King S.R., Stocco D.M. The purification, cloning, and expression of a novel luteinizing hormone- induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the Steroidogenic Acute Regulatory protein (StAR) J Biol Chem. 1994;269:28314–28322. doi: 10.1016/s0021-9258(18)46930-x. [DOI] [PubMed] [Google Scholar]
- 12.Schrick K., Nguyen D., Karlowski W.M., Mayer K.F.X. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004;5:R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soccio R.E., Breslow J.L. StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J Biol Chem. 2003;278:22183–22186. doi: 10.1074/jbc.R300003200. [DOI] [PubMed] [Google Scholar]
- 14.Clark B.J. The START-domain proteins in intracellular lipid transport and beyond. Mol Cell Endocrinol. 2020:504. doi: 10.1016/j.mce.2020.110704. [DOI] [PubMed] [Google Scholar]
- 15.Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Floyd S.K., Zalewski C.S., Bowman J.L. Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics. 2006;173:373–388. doi: 10.1534/genetics.105.054239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zalewski C.S., Floyd S.K., Furumizu C., Sakakibara K., Stevenson D.W., Bowman J.L. Evolution of the class IV HD-Zip gene family in streptophytes. Mol Biol Evol. 2013;30:2347–2365. doi: 10.1093/molbev/mst132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang D., Ade J., Frye C.A., Innes R.W. Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 2005;44:245–257. doi: 10.1111/j.1365-313X.2005.02523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkata B.P., Schirck K. START domains in lipid/sterol transfer and signaling in plants. 17th Inter Symp Plant Lipids. 2006:57–61. [Google Scholar]
- 20.Mahtha S.K. Computational analysis of the StAR-related lipid transfer (START) domains in plants [ Ph.D. thesis]. Jawaharlal Nehru Univ New Delhi 2022.
- 21.Alpy F., Tomasetto C. Give lipids a START: The StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 22.Thorsell A.G., Lee W.H., Persson C., Siponen M.I., Nilsson M., Busam R.D., et al. Comparative structural analysis of lipid binding START domains. PLoS One. 2011:6. doi: 10.1371/journal.pone.0019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sluchanko N.N., Slonimskiy Y.B., Egorkin N.A., Varfolomeeva L.A., Kleymenov S.Y., Minyaev M.E., et al. Structural basis for the carotenoid binding and transport function of a START domain. Structure. 2022:30. doi: 10.1016/J.STR.2022.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Tan L., Tong J., Chun C.J., Im Y.J. Structural analysis of human sterol transfer protein STARD4. Biochem Biophys Res Commun. 2019;520:466–472. doi: 10.1016/j.bbrc.2019.10.054. [DOI] [PubMed] [Google Scholar]
- 25.Roderick S.L., Chan W.W., Agate D.S., Olsen L.R., Vetting M.W., Rajashankar K.R., et al. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat Struct Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 26.Kudo N., Kumagai K., Tomishige N., Yamaji T., Wakatsuki S., Nishijima M., et al. Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc Natl Acad Sci USA. 2008;105:488–493. doi: 10.1073/pnas.0709191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanno K., Wu M.K., Scapa E.F., Roderick S.L., Cohen D.E. Structure and function of phosphatidylcholine transfer protein (PC-TP)/StarD2. Biochim Biophys Acta - Mol Cell Biol Lipids. 2007;1771:654–662. doi: 10.1016/j.bbalip.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagata K., Ishikawa T., Kawai-Yamada M., Takahashi T., Abe M. Ceramides mediate positional signals in Arabidopsis thaliana protoderm differentiation. Dev. 2021:148. doi: 10.1242/dev.194969. [DOI] [PubMed] [Google Scholar]
- 29.Wojciechowska I., Mukherjee T., Knox-Brown P., Hu X., Khosla A., Mathews G.L., et al. Arabidopsis PROTODERMAL FACTOR2 binds lysophosphatidylcholines and transcriptionally regulates phospholipid metabolism. BioRxiv. 2021 doi: 10.1101/2021.10.20.465175. 2021.10.20.465175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Husbands A.Y., Feller A., Aggarwal V., Dresden C.E., Holub A.S., Ha T., et al. The START domain potentiates HD-ZIPIII transcriptional activity. Plant Cell. 2023 doi: 10.1093/PLCELL/KOAD058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroda M. Phosphoinositides regulate chloroplast processes. Proc Natl Acad Sci USA. 2020;117:9154–9156. doi: 10.1073/PNAS.2004189117/ASSET/2EB682EE-B293-4E05-9E2F–38EB29A94921/ASSETS/GRAPHIC/PNAS.2004189117FIG01.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gou J.Y., Li K., Wu K., Wang X., Lin H., Cantu D., et al. Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell. 2015;27:1755–1770. doi: 10.1105/TPC.114.134296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahtha S.K., Purama R.K., Yadav G. StAR-related lipid transfer (START) domains across the rice pangenome reveal how ontogeny recapitulated selection pressures during rice domestication. Front Genet. 2021;12:1658. doi: 10.3389/fgene.2021.737194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larkin M.A., Blackshields G., Brown N.P., Chenna R., Mcgettigan P.A., McWilliam H., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server 2014;42:W320–4. 10.1093/NAR/GKU316. [DOI] [PMC free article] [PubMed]
- 36.Lobley A., Sadowski M.I., Jones D.T. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 2009;25:1761–1767. doi: 10.1093/BIOINFORMATICS/BTP302. [DOI] [PubMed] [Google Scholar]
- 37.Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/NAR/GKN201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kryshtafovych A., Schwede T., Topf M., Fidelis K., Moult J. Crit Assess Methods Protein Struct Predict (CASP)— XIV. 2021;89:1607–1617. doi: 10.1002/PROT.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robin X., Haas J., Gumienny R., Smolinski A., Tauriello G., Schwede T. Continuous automated model evaluatiOn (CAMEO)-perspectives on the future of fully automated evaluation of structure prediction methods. Proteins. 2021;89:1977–1986. doi: 10.1002/PROT.26213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy A., Kucukural A., Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J., Zhang Y. Protein structure and function prediction using I-TASSER. Curr Protoc Bioinforma. 2015;52 doi: 10.1002/0471250953.bi0508s52. 5.8.1-5.8.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng W., Zhang C., Li Y., Pearce R., Bell E.W., Zhang Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods. 2021;1 doi: 10.1016/J.CRMETH.2021.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., et al. Accurate prediction of protein structures and interactions using a three-track neural network. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olechnovič K., Venclovas Č. VoroMQA: Assessment of protein structure quality using interatomic contact areas. Proteins. 2017;85:1131–1145. doi: 10.1002/PROT.25278. [DOI] [PubMed] [Google Scholar]
- 45.Bowie J.U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional stucture. Science (80-) 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 46.Lüthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083A0. [DOI] [PubMed] [Google Scholar]
- 47.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35 doi: 10.1093/NAR/GKM290. W407–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/PRO.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Messaoudi A., Belguith H., Ben Hamida J. Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 β-lactamase. Theor Biol Med Model. 2013:10. doi: 10.1186/1742-4682-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky473. W363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Binkowski T.A., Naghibzadeh S., Liang J. CASTp: computed atlas of surface topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pommié C., Levadoux S., Sabatier R., Lefranc G., Lefranc M.P. IMGT standardized criteria for statistical analysis of immunoglobulin V-Region amino acid properties. J Mol Recognit. 2004;17:17–32. doi: 10.1002/jmr.647. [DOI] [PubMed] [Google Scholar]
- 53.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 54.Zamyatnin A.A. Protein volume in solution. Prog Biophys Mol Biol. 1972;24:107–123. doi: 10.1016/0079-6107(72)90005-3. [DOI] [PubMed] [Google Scholar]
- 55.Schrick K., Bruno M., Khosla A., Cox P.N., Marlatt S.A., Roque R.A., et al. Shared functions of plant and mammalian StAR-related lipid transfer (START) domains in modulating transcription factor activity. BMC Biol. 2014;12:70. doi: 10.1186/s12915-014-0070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahtha S.K., Citu, Prasad A., Yadav G. Springer; Cham: 2022. Complex Networks Reveal Biological Functions of START Domains in Rice: Insights from Computational Systems Biology. [DOI] [Google Scholar]
- 57.Hirashima T., Jimbo H., Kobayashi K., Wada H. A START domain-containing protein is involved in the incorporation of ER-derived fatty acids into chloroplast glycolipids in Marchantia polymorpha. Biochem Biophys Res Commun. 2021;534:436–441. doi: 10.1016/j.bbrc.2020.11.063. [DOI] [PubMed] [Google Scholar]
- 58.Mukherjee K., Bürglin T.R. MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 2006;140:1142–1150. doi: 10.1104/pp.105.073833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Umezawa T., Nakashima K., Miyakawa T., Kuromori T., Tanokura M., Shinozaki K., et al. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bürglin T.R., Affolter M. vol. 125. Springer; 2016. (Homeodomain Proteins: An Update). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukherjee K., Brocchieri L., Bürglin T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol. 2009;26:2775–2794. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Data Availability Statement
All of the datasets supporting the results of this article are included within the article and its Supplementary Material.