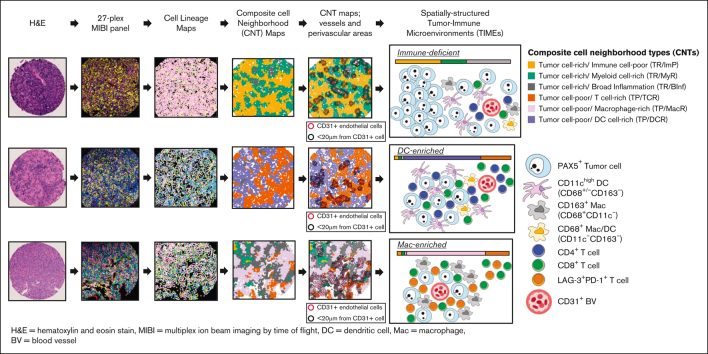
Key Points
-
•
DLBCLs are composed of local cell neighborhoods with distinct cellular, spatial, and functional features that define structured TIMEs.
-
•
DLBCL cell neighborhoods structurally define immune-deficient, DC-enriched, and macrophage-enriched immune microenvironments.
Visual Abstract
Abstract
Diffuse large B-cell lymphoma (DLBCL) not otherwise specified is the most common aggressive non-Hodgkin lymphoma and a biologically heterogeneous disease. Despite the development of effective immunotherapies, the organization of the DLBCL tumor-immune microenvironment (TIME) remains poorly understood.We interrogated the intact TIME of 51 de novo DLBCLs with triplicate sampling to characterize 337 995 tumor and immune cells using a 27-plex antibody panel that captured cell lineage, architectural, and functional markers. We spatially assigned individual cells, identified local cell neighborhoods, and established their topographical organization in situ. We found that the organization of local tumor and immune cells can be modeled by 6 composite cell neighborhood types (CNTs). Differential CNT representation divided cases into 3 aggregate TIME categories: immune-deficient, dendritic cell–enriched (DC-enriched), and macrophage-enriched (Mac-enriched). Cases with immune-deficient TIMEs have tumor cell–rich CNTs, in which the few infiltrating immune cells are enriched near CD31+ vessels, in keeping with limited immune activity. Cases with DC-enriched TIMEs selectively include tumor cell–poor/immune cell–rich CNTs with high numbers of CD11c+ DCs and antigen-experienced T cells also enriched near CD31+ vessels, in keeping with increased immune activity. Cases with Mac-enriched TIMEs selectively include tumor cell–poor/immune cell–rich CNTs with high numbers of CD163+ macrophages and CD8 T cells throughout the microenvironment, accompanied by increased IDO-1 and LAG-3 and decreased HLA-DR expression and genetic signatures in keeping with immune evasion. Our findings reveal that the heterogenous cellular components of DLBCL are not randomly distributed but organized into CNTs that define aggregate TIMEs with distinct cellular, spatial, and functional features.
Introduction
Diffuse large B-cell lymphoma (DLBCL) not otherwise specified (NOS) is an aggressive non-Hodgkin lymphoma with biological and clinical heterogeneity.1 Large-scale sequencing efforts have divided DLBCLs into genetic subgroups that provide insights into ontogeny and prognosis, and support the rational targeting of defined oncogenic signaling pathways.2, 3, 4, 5 Similarly, investigations into the cellular and molecular constituents of the DLBCL tumor-immune microenvironment (TIME) have revealed a diversity of infiltrating immune cells and putative targets for immune therapy.6,7 Nevertheless, little is known about the structured organization of the DLBCL TIME. In this study, we identify the diverse immune cell constituents of the DLBCL TIME. By modeling their local organization into composite cell neighborhood types (CNTs), we resolve the mutual exclusivity of dendritic cell–enriched and macrophage-enriched CNTs across cases and with respect to tumor vessels and functional biomarkers to define 3 aggregate TIME categories using multiplexed ion beam imaging-time of flight (MIBI-TOF) coupled with detailed, quantitative image analysis.
Methods
Tissue samples
Formalin-fixed, paraffin-embedded excisional biopsies from 51 de novo DLBCLs, diagnosed and classified based on the World Health Organization criteria as DLBCL NOS, and 8 tonsils or reactive lymph nodes were retrieved from the archives of Brigham and Women's Hospital (Boston, MA) with institutional review board approval (IRB# 2010P002736).1 All samples originated as excisional lymph node biopsies, taken for primary diagnosis before treatment and from an unselected patient population (aged 27-87 years) with nearly equal numbers of males and females (28 males and 23 females). Additional LBCL subtypes, including high-grade BCL, double-hit lymphoma, primary mediastinal (thymic) LBCL, primary DLBCL of the central nervous system, and others,1 were excluded from the series. Biopsies from patients with a history of lymphoma were excluded, as were core needle biopsies. All cases were genetically subtyped.2 The distribution of genetic cluster assignments was unselected and reflected the cases available for the series. Cases were arrayed as triplicate, 0.6 mm, randomly selected cores to capture the tumor heterogeneity in a tissue microarray (TMA) as described.8, 9, 10, 11 The cores were taken from areas with ample tumor, avoiding the edges of the tumor and areas adjacent to nontumor tissue. TMA was stained with a heavy metal isotope–labeled commercial 27-plex checkpoint panel with standard protocols (supplemental Table 1).12
Tissue imaging, image processing, and cell lineage assignments
TMA cores were imaged on a beta-iteration of the MIBIScope (IonPath, Palo Alto, CA) equipped with a research-grade oxygen duoplasmatron ion source and proprietary controller software at 1024 × 1024 pixels over a 500 μm × 500 μm field-of-view (FOV). The composition and organization of the TIMEs were determined from the acquired images using a stepwise analytical approach under expert pathologist review (K.T.W. and S.J.R.) (supplemental Figure 1; supplemental Methods).12 Final cell lineage assignments were remapped onto cell segmentation masks to generate cell lineage maps, which were compared with the original MIBI-TOF pseudoimages to confirm the accuracy of the assignments (by K.T.W. and S.J.R.) (supplemental Figure 1). The results from imaging individual FOVs (supplemental Figure 2) were combined for the final case-based data.
Composite CNT and aggregate TIME derivations
Using established methods, individual cell neighborhoods were defined by anchoring each cell in groups of 20 (self and 19 nearest neighbors).13,14 The collection of local cell neighborhoods was subjected to k-means clustering, as described (supplemental Figure 1).13 A supervised elbow plot suggested 6 composite CNTs as a reasonable, minimal number of clusters.
Samples were assigned to 1 of 3 aggregate TIME categories through a 1-dimensional k-means clustering based on the ratio of 1 + tumor cell–poor (TP) or macrophage-rich (MacR) to 1+ TP or dendritic cell–rich (DCR) cell neighborhood percent cellularity (supplemental Figure 3; supplemental Methods). In 50 of 51 cases, the k-means–based TIME categorization aligned with the presence of >3% of cells in either TP/MacR or TP/DCR neighborhoods, or ≤3% of cells in either of them (supplemental Figure 3). The 3% threshold definition was used to simplify the description of the 3 TIME categories. Additional details are provided in supplemental Materials.
CD31-proximity analyses
Twenty microns (the approximate diameter of 2 lymphocytes) was selected as a reasonable distance to survey for CNT enrichment proximal to vessels without losing the ability to detect steep changes in immune cell composition as the distance from vessels increased. The 20 μm distance also ensured that adequate numbers of cells remained outside the perivascular regions for meaningful comparisons within and across FOVs (further details are provided in supplemental Methods).
Statistical comparisons
Enrichment for defined cell populations within or outside the CD31-defined perivascular areas was tested using a 2-sided Wilcoxon sign-rank test with proximal and distant regions paired based on the sample (supplemental Methods). Statistics for relative biomarker expression and cell-type comparisons were derived from Mann-Whitney U tests.
Results
DLBCLs have heterogeneous immune cell infiltrates
To determine the orchestrated immune cell infiltration across biopsies from patients with DLBCL before treatment, we identified 51 diagnostic cases of de novo DLBCL NOS, and 8 tonsil or reactive lymph node samples that were found to contain high-quality tissue content upon expert histopathological review (Figure 1A). A TMA containing randomly selected triplicate cores from the biopsy samples was sectioned and stained with a panel of 27 metal isotope–labeled antibodies targeting cell lineage and functional and architectural biomarkers (supplemental Table 1; supplemental Figure 1A). MIBI-TOF mass spectrometry was used to acquire images with 1 FOV image per core. An expert review of individual biomarkers for each FOV revealed the expected patterns of biomarker expression, confirming the specificity of the multiplexed antibody panel (Figure 1A).
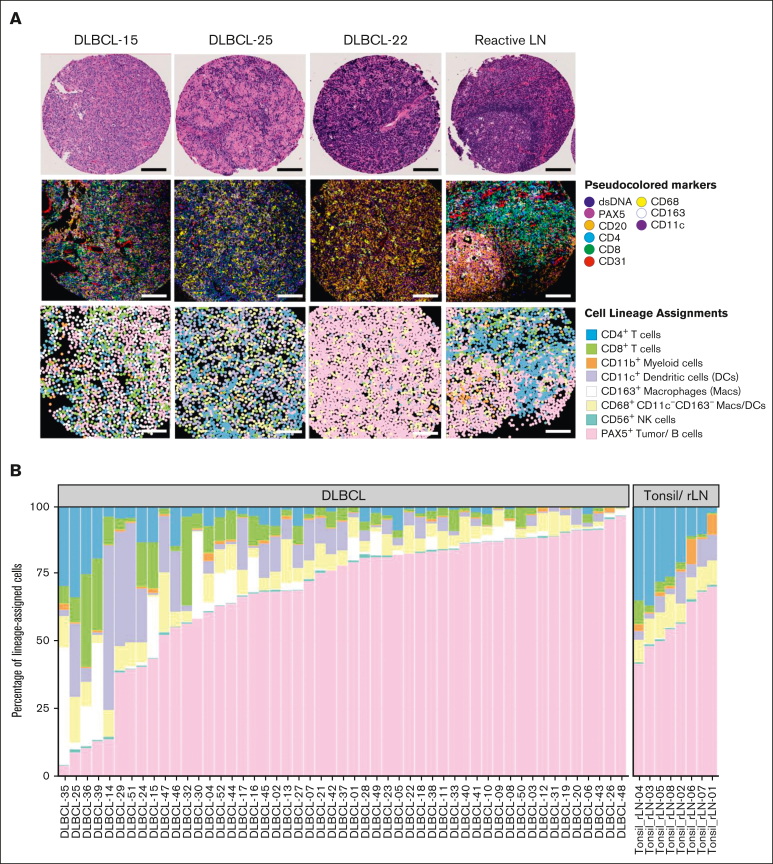
Figure 1.
The DLBCL TIME is heterogeneous.(A) Representative FOVs from 3 DLBCLs NOS and 1 reactive lymph node (rLN) demonstrating the heterogeneity of the TIME by hematoxylin and eosin staining (H&E) (top), MIBIscope pseudoimages colored per the indicated biomarkers (middle), and cell lineage maps colored per the indicated cell lineages (bottom). Bars represent ∼100 μm distance from 500 μm2 FOVs; digitized H&E visualized at an objective magnification ×4. (B) Percentage of lineage-assigned cells that are tumor cells or one of the indicated immune cell lineages from 51 DLBCLs and 8 rLN/tonsil controls colored as in panel A. Data are aggregated from 3 FOVs per case for most cases and a single FOV from controls. Cells without a defined cell lineage were classified as “other” and excluded. Data from individual FOVs are presented in supplemental Figure 2A.
We used computational clustering approaches (supplemental Figure 1A-F) to generate cell lineage maps based on the expression of 10 lineage-associated markers (CD3, CD4, CD8, CD68, CD163, CD11c, CD11b, CD56, CD20, and PAX5), which stratified individual cells into 8 defined cell lineages (CD3+CD4+ T cells, CD3+CD8+ T cells, CD11c+CD68+/−CD163– dendritic cells, CD163+CD68+/−CD11c– macrophages [with an M2-like phenotype],15 CD68+CD163–CD11c– macrophage/dendritic cell [DC] phagocytes [undifferentiated phenotype], CD11b+ myeloid cells, CD56+ natural killer cells, and CD20+PAX5+ tumor cells). Cells that did not express any of the 10 lineage-associated markers were excluded from the analysis. Expert pathologists’ review of the resulting cell lineage maps confirmed that they reflected the cell identities and their locations within tissues with high fidelity compared with the original MIBI-TOF pseudoimages (Figure 1A).
We then graphically represented each cell lineage as a proportion of the total lineage-assigned cells based on individual FOVs (given the multiple sampling for each case; supplemental Figure 2) and on a per-case basis (Figure 1B). Infiltrating immune cells, as a fraction of total lineage-assigned cells, varied little across FOVs within individual cases (mean standard deviation [SD] = 6%; supplemental Figure 2A) but varied considerably across total cases (SD = 24% [range, 4%-96%]. Similarly, the individual immune cell lineages, as a fraction of the total lineage-assigned cells, varied little across FOVs within individual cases (supplemental Figure 2A) but considerably across total cases (Figure 1B). In contrast, reactive lymph nodes and tonsils showed consistent cell lineage distributions across cases, as expected (Figure 1B). These findings indicate that the mixture of tumor and immune cells within diagnostic de novo DLBCL TIMEs is homogenous across randomly sampled areas within individual cases and highly heterogenous across total cases. The results also suggest a robust representation of the TIME for each case through the TMA approach using multiple, randomly selected tissue samplings.9, 10, 11
Tumor and immune cells are organized into 6 composite CNTs
To determine the local organization of tumor and immune cells in TIMEs, we first described an individual cell neighborhood (self and 19 nearest neighbors) for each lineage-assigned cell for a total of 337 995 individual, local cell neighborhoods across the 51 cases.13 We used k-means clustering and identified 6 composite CNTs into which the individual, local cell neighborhoods could be divided (Figure 2A).
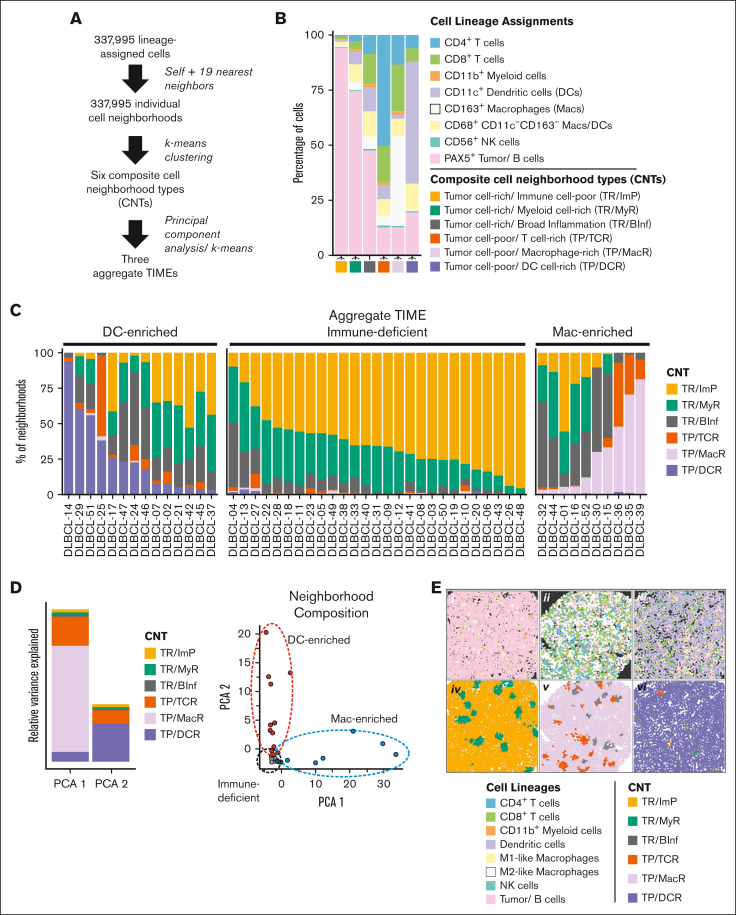
Figure 2.
Tumor and immune cells are organized into 6 composite CNTs that define 3 aggregate TIMEs. (A) Schematic flow of deriving 6 composite CNTs and their organization into 3 aggregate TIMEs. In brief, the lineage of each cell (self) and its 19 nearest neighbors was determined and defined as an individual cell neighborhood. The individual cell neighborhoods for all FOVs and cases (N = 337 995) underwent k-means clustering to arrive at 6 CNTs (supplemental Materials). Each cell was assigned to the best corresponding CNT based on the similarity of its cell neighborhood composition to each of the 6 CNTs. Cases, defined by their composite CNTs, were divided into 3 aggregate TIMEs distinguished by their unique CNT compositions, as determined by k-means clustering (supplemental Figure 3). (B) The mean compositions of each of the 6 indicated CNTs by cell lineages. (C) The percentage of total individual cell neighborhoods (1 neighborhood per cell) assigned to each of the 6 indicated CNTs by case. The data shown are summed from all FOVs. Data for individual FOVs are shown in supplemental Figure 2B. Cases were assigned to individual TIME categories according to the differential representation of cells in TP/DCR and TP/MacR neighborhoods after a k-means clustering analysis (supplemental Figure 3A). The k-means results were consistent with an assignment of cases with >3% of cells in TP/DCR neighborhoods as DC-enriched, cases with ≤3% of cells in both TP/DCR and TP/MacR neighborhoods as immune-deficient, and cases with >3% of cells in TP/MacR neighborhoods as Mac-enriched. In no case were >3% of cells in TP/DCR and TP/MacR neighborhoods (supplemental Figure 3B). (D) PCA of cases defined by aggregate CNT composition and showing that TP/MacR neighborhoods (in PCA 1) and TP/DCR neighborhoods (in PCA 2) are top contributors to the variance between cases and identify 3 aggregate TIMEs (red = DC-enriched, gray = immune-deficient, and blue = Mac-enriched; further details are presented in supplemental Methods). (E) Representative cell lineage maps (i, ii, iii) and their corresponding CNT maps (iv, v, vi, respectively) of select FOVs from cases in which most, but not all, cells reside in TR/ImP neighborhoods (DLBCL-22 with an immune-deficient TIME [i, iv]), TP/MacR neighborhoods (DLBCL-39 with a Mac-enriched TIME [ii, v]), and TP/DCR neighborhoods (DLBCL-14 with a DC-enriched TIME [iii, vi]). Note that CNT maps capture local cell heterogeneity within each TIME while maintaining spatial information. Additional comparisons are given in supplemental Figures 4-7.
Three CNTs were tumor cell–rich (TR) and composed primarily of tumor cells (Figure 2B). The cells in TR/Immune cell–poor (TR/ImP) neighborhoods (Figure 2B, yellow) were mostly tumor cells (mean = 94% of lineage-assigned cells), with rare intermixed immune cells (6%). Cells in the TR/ Myeloid cell–rich (TR/MyR) neighborhoods (Figure 2B, green) were also mostly tumor cells (mean = 75% of lineage-assigned cells), with more frequent intermixed immune cells (25%). Myeloid cells, including macrophages and DCs, predominated over T cells in this CNT (18% vs 7%). Cells in TR/Broad-inflammation (TR/BInf) neighborhoods (Figure 2B, gray) were a near-equal mixture of tumor cells and immune cells (mean = 48% vs 52% of lineage-assigned cells, respectively). Numbers of myeloid and T cells were comparable in this CNT (31% and 21% of lineage-assigned cells, respectively).
Three CNTs were tumor cell-poor (TP) and composed primarily of immune cells. The cells in TP/T-cell–rich (TCR) neighborhoods (Figure 2B, orange) were mostly of CD4+ and CD8+ T cells (mean = 63% of lineage-assigned cells), with few intermixed myeloid and tumor cells (20% and 14%, respectively). Cells in TP/Macrophage-rich (TP/MacR) neighborhoods (Figure 2B, lavender) were primarily CD163+ macrophages (CD163+CD68+/−CD11c–; mean = 32% of lineage-assigned cells), with fewer intermixed CD8+ T cells (24%), CD4+ T cells (13%), and tumor cells (14%). Cells in TP/Dendritic cell-rich (TP/DCR) neighborhoods (Figure 2B, purple) were primarily CD11c+ DCs (CD11c+CD68+/−CD163–; mean = 50% of lineage-assigned cells), with fewer intermixed tumor cells (19%) and CD8+ and CD4+ T cells (13%, combined). We additionally captured macrophage/DCs, consistent with a more undifferentiated phenotype (CD68+CD163–CD11c–) based on cell lineage assignment. However, these cells were observed in variable numbers across cell neighborhoods and did not define a specific CNT. Therefore, we found that cells residing in local cell neighborhoods can be modeled by 6 CNTs distinguished by (i) the relative proportion of tumor cells to nonmalignant immune cells when tumor cells predominate and (ii) the types of immune cells that predominate when tumor cells are a minority.
Differential representation of cells in TP/DCR and TP/MacR neighborhoods across cases define 3 aggregate TIMEs
Next, we examined the distribution of cells in their assigned CNTs across the cases. We found cells in the TR/ImP, TR/MyR, and/or TR/BInf neighborhoods in all cases (Figure 2C). We also found cells in TP/TCR neighborhoods in most cases (38 of 51 cases [75%]). Finally, we found more than minimal numbers of cells (>3%) in TP/DCR neighborhoods or TP/MacR neighborhoods in approximately half of the cases (25 of 51 cases [49%]). The differential representation of CNTs across cases did not appear to be a sampling effect because the relative proportions of cells assigned to each of the CNTs were remarkably consistent across randomly selected cores for each case (supplemental Figure 2B).
Intriguingly, we found that cases with cells in TP/DCR neighborhoods and cases with cells in TP/MacR neighborhoods were almost mutually exclusive (P <.001; Fisher exact test; Figure 2C). To further analyze the differential representation of CNTs across the case series, we performed a principal component analysis (PCA). Consistent with the observed mutual exclusivity, the first 2 principal components of the PCA were primarily driven by single microenvironmental features, with the TP/MacR neighborhood contributing the majority of variance to the first component and the TP/DCR neighborhood in the second component (Figure 2D). A PCA based on the differential representation of cell lineages across the case series did not identify individual features that were more discriminatory (supplemental Figure 3A).
We used k-means clustering to label cases based on the differential representation of cells in TP/MacR and TP/DCR neighborhoods. Using this approach, we divided cases into 3 TIME categories (supplemental Figure 3B; supplemental Methods). In 50 of 51 cases, the k-means results were consistent with the presence of >3% of cells in either TP/DCR or TP/MacR neighborhoods or were inconsistent in both (supplemental Figure 3C).
We labeled the 3 TIME categories by their primary distinguishing feature (Figure 2C). Cases with immune-deficient TIMEs (26 of 51 cases [51%]) generally had ≤3% cells in either TP/DCR or TP/MacR neighborhoods, cases with DC-enriched TIMEs (14 of 51 cases [27%]) had >3% of cells in TP/DCR neighborhoods, and cases with Mac-enriched TIMEs (10 of51 cases [20%]) had >3% of cells in TP/MacR neighborhoods (Figure 2C). As expected, cases with immune-deficient TIMEs had relatively high numbers of tumor cells and fewer immune cells (supplemental Figure 2). Cases with Mac-enriched TIMEs had relatively high numbers of immune cells, especially CD163+ macrophages. Cases with DC-enriched TIMEs also had relatively high numbers of immune cells, especially CD11c+ DCs (supplemental Figure 2).
CNT maps capture local cellular heterogeneity while maintaining spatial resolution
We next generated CNT maps to review CNT assignments for every cell, in every FOV, and for every case (Figure 2E; supplemental Figures 4-7). When we compared the CNT maps with their corresponding cell lineage maps from selected cases having immune-deficient, Mac-enriched, and DC-enriched TIMEs, we observed that the CNT maps efficiently captured overt and subtle differences in local cell populations in a spatially resolved manner (Figure 2E; supplemental Figure 4).
Cases with immune-deficient TIMEs are organized by cellular gradients anchored on CD31+ vessels
Next, we examined whether cells assigned to distinct CNTs were randomly distributed or had an orchestrated spatial organization. When we reviewed individual CNT maps for cases with immune-deficient TIMEs, we noticed that areas with cells in TR/ImP neighborhoods (yellow) bordered areas with cells in TR/MyR neighborhoods (green), which, in turn, bordered areas with cells in TR/BInf neighborhoods (gray) (Figure 3A, top left; supplemental Figure 5A). Areas with cells in TR/ImP neighborhoods (yellow) almost never bordered areas with cells in TR/BInf neighborhoods (gray). A chord diagram representing all contacts between cells in different CNTs for all cases with immune-deficient TIMEs confirmed these observations (Figure 3A, top right).
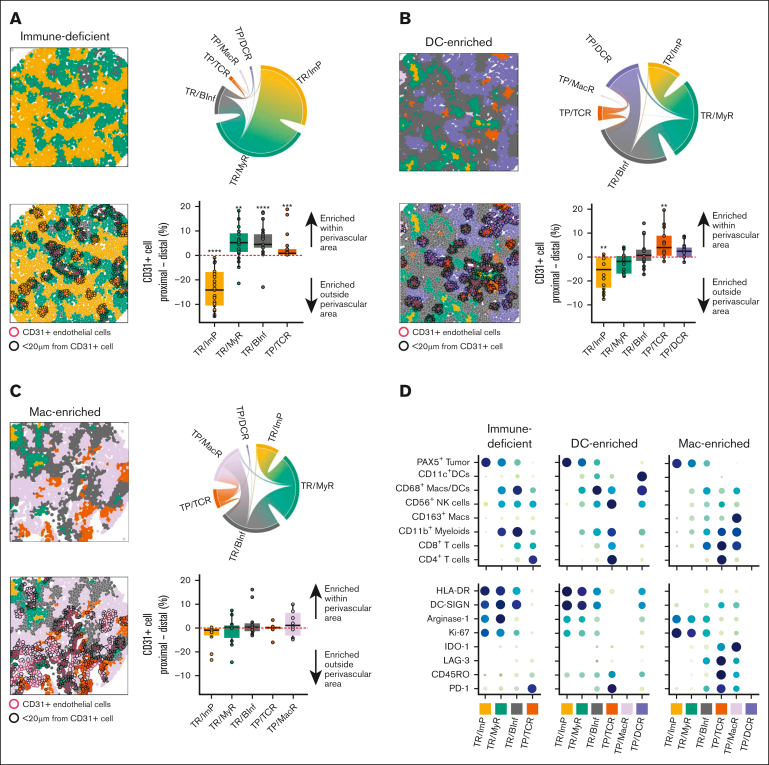
Figure 3.
Cases with immune-deficient, DC-enriched, and Mac-enriched aggregate TIMEs have distinct topographical structures and express distinct functional markers. (A) (Top left) Representative CNT map of a DLBCL with an immune-deficient TIME and colored per the CNT assignment of the cells as TR/ImP (yellow), TR/MyR (green), or TR/BInf (gray). (Top right) Chord diagram integrating all contacts between cells assigned to different CNTs for all cases with an immune-deficient TIME. Broader bands reflect greater number of contacts between cells. (Bottom left) Representative CNT map of a DLBCL with an immune-deficient TIME with an overlay of CD31+ endothelial cells (red outlines) and cells within a 20 μm radial distance from CD31+ cells (black outlines). (Bottom right) The differences in the percentages of cells assigned to the indicated CNT within (proximal) or outside (distal) a 20 μm radial distance from a CD31+ cell. Values for individual cases (dots), median values (horizontal line), and 25th and 75th centiles (boxes) are indicated. Positive values represent enrichment within the perivascular region, whereas negative values represent enrichment outside the perivascular area. Only significant P-values are annotated ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001 using the paired 2-sided Wilcoxon sign-rank test. (B) (Top left) Representative CNT map from a DLBCL with a DC-enriched TIME and colored per the CNT assignment of the cells as TP/DCR (blue), TP/TCR (orange), TR/BInf (gray), TR/MyR (green), or TR/ImP (yellow). (Top right) Chord diagram integrating all contacts among cells in neighborhoods assigned to different CNTs for all cases with a DC-enriched TIME. Broader bands reflect greater numbers of contacts between cells. (Bottom left) Representative CNT map from a DLBCL with a DC-enriched TIME with an overlay of CD31+ endothelial cells (red outlines) and cells within a 20 μm radial distance from CD31+ cells (black outlines). (Bottom right) The differences in the percentages of cells assigned to the indicated CNT within (proximal) or outside (distal) a 20 μm radial distance from a CD31+ cell. Values for individual cases (dots), median values (horizontal line), and 25th and 75th centiles (boxes) are indicated. Positive values represent enrichment within the perivascular region, while negative values represent enrichment outside the perivascular area. Only significant P-values are annotated. ∗∗P < .01, using the paired 2-sided Wilcoxon sign-rank test. (C) (Top right) Representative CNT map of a DLBCL with a Mac-enriched TIME that is color-coded per the CNT assignment of cells as TP/MacR (lavender), TP/TCR (orange), TR/ImP (yellow), TR/MyR (green), or TR/BInf (gray). (Top right) Chord diagram integrating all contacts between cells assigned to different CNTs for all cases with a Mac-enriched TIME. Broader bands reflect greater number of contacts between cells. (Bottom left) Representative CNT map of a DLBCL with a Mac-enriched TIME with an overlay of CD31+ endothelial cells (red outlines) and cells within a 20 μm radial distance from a CD31+ cell (black outlines). (Bottom left) The differences in the percentages of cells assigned to the indicated CNT within (proximal) or outside (distal) a 20 μm CD31+ cell. Values for individual cases (dots), median values (horizontal line), and 25th and 75th centiles (boxes) are indicated. Positive values represent enrichment within the perivascular region, while negative values represent enrichment outside the perivascular region. No P-values were significant using the paired 2-sided Wilcoxon sign-rank test. (D) The relative cell densities (in cells per mm2) of each indicated cell lineage (top) and the relative expression (in ion counts) of each indicated functional biomarker (bottom) for each indicated CNT (columns) and separated based on the aggregate TIME category (immune-deficient, DC-enriched, and Mac-enriched). Higher cell density (top) and ion counts (bottom) are indicated by larger circle size and darker color.
We then compared individual CNT maps with their corresponding MIBI-TOF pseudoimages. We noted that cells in TR/ImP neighborhoods (yellow) were frequently far from CD31+ vessels (Figure 3A red outlines, bottom left). In contrast, cells in TR/MyR (green) neighborhoods were often close to CD31+ vessels, and cells in TR/BInf (gray) neighborhoods were often immediately adjacent to CD31+ vessels (Figure 3A, bottom left).
To test whether cells in specific CNTs were enriched close to or far from vessels, we calculated the difference in the percentage of cells assigned to each CNT in areas ≥ 20 μm and < 20 μm from the nearest CD31+ cell across all cases with immune-deficient TIMEs (Figure 3A, bottom right). The 20 μm distance (diameters of ∼2 cells) ensured that most cells were counted outside the proximal region (supplemental Methods). We found that, on average, the percentage of cells in TR/ImP neighborhoods was significantly enriched in areas ≥ 20 μm from the nearest CD31+ cell (P < .0001). Conversely, we found that, on average, the percentage of cells in TR/BInf and TR/MyR neighborhoods were significantly enriched in areas <20 μm from the nearest CD31+ cell (P < .0001 and P < .001, respectively).
To further test this finding, we determined the relative percentage of cells in TR/ImP, TR/MyR, TR/BInf, and TP/TCR neighborhoods at defined distances from the nearest CD31+ cell for all cases with immune-deficient TIMEs. The percentage of cells in TR/ImP neighborhoods was lowest near CD31+ cells and increased with increasing distance from CD31+ cells (supplemental Figure 5B). In contrast, the percentage of cells in TR/MyR and TR/BInf neighborhoods were highest near CD31+ cells and decreased with increasing distance from CD31+ cells.
These data imply that the relative proportion of tumor cells to immune cells follows a gradient anchored on tumor vessels. Areas rich in tumor cells and deficient in immune cells are far from CD31+ vessels in TR/ImP neighborhoods (yellow). These areas border TR areas closer to CD31+ vessels with increased, but still minor numbers of myeloid cells in TR/MyR neighborhoods (green). Finally, these areas border TR areas very close or immediately adjacent to CD31+ vessels with increased, but still minor numbers of myeloid cells and T cells in TR/BInf neighborhoods (gray).
Cases with DC-enriched TIMEs are organized by cellular gradients distinct from cases with immune-deficient TIMEs
Next, we reviewed individual CNT maps for cases with DC-enriched TIMEs (Figure 3B, top left; supplemental Figure 6A). We noticed that areas with cells in TR/ImP neighborhoods (yellow) bordered areas with cells in TR/MyR neighborhoods (green), which, in turn, bordered areas with cells in TR/BInf neighborhoods (gray) in keeping with the stereotyped sequence found for cases with immune-deficient TIMEs. In addition, we found that areas having cells in TP/DCR (purple) and TP/TCR (orange) neighborhoods bordered one another and bordered areas with cells in TR/BInf (gray) neighborhoods. Areas with cells in TP/DCR (purple) and TP/TCR (orange) neighborhoods almost never bordered areas with cells in TR/MyR (green) or TR/ImP (yellow) neighborhoods (Figure 3B, top left; supplemental Figure 6A). A chord diagram representing all contacts between cells in different CNTs for all cases with DC-enriched TIMEs confirmed these observations (Figure 3B, top right).
We then calculated the difference in the percentage of cells assigned to each CNT in areas ≥20 μm and <20 μm from the nearest CD31+ cell across cases (Figure 3B, bottom right). We found that, on average, the percentage of cells in TR/ImP neighborhoods was significantly enriched in areas ≥20 μm from the nearest CD31+ cell (P < .001). Conversely, we found that, on average, the percentage of cells in TP/TCR neighborhoods was significantly enriched in areas <20 μm from the nearest CD31+ cell (P < .01). The percentage of cells in TP/DCR neighborhoods trended higher in areas <20 μm than in regions ≥20 μm from the nearest CD31+ cell also (P = .1; Figure 3B, bottom right).
To further test this observation, we determined the percentage of cells assigned to each CNT at defined distances from the nearest CD31+ cells for all cases with DC-enriched TIMEs. The percentage of cells in TR/ImP neighborhoods was lowest near CD31+ cells and increased with increasing distance from these cells (supplemental Figure 6B). In contrast, the percentage of cells in TR/BInf, TP/TCR, and TP/DCR neighborhoods was highest near CD31+ cells and decreased with increasing distance from these cells.
These data again imply that the relative proportion of tumor cells to immune cells follows a gradient anchored on tumor vessels. Areas rich in tumor cells and deficient in immune cells are far from CD31+ vessels in TR/ImP neighborhoods. These areas border TR areas with increased, but still minor numbers of myeloid cells and T cells in TR/MyR and TR/BInf neighborhoods. Unique to cases with DC-enriched TIMEs, areas with a minority of immune cells in TR/BInf neighborhoods border expanded areas that are preferentially located near CD31+ vessels and contain predominantly T cells in TP/TCR neighborhoods (orange) and DCs in TP/DCR neighborhoods (purple).
Cases with Mac-enriched TIMEs are organized by cellular gradients distinct from cases with immune-deficient TIMEs and DC-enriched TIMEs
Finally, we reviewed individual CNT maps for cases with Mac-enriched TIMEs (Figure 3C, top left; supplemental Figure 7A). We again observed that cells in TR/ImP (yellow) neighborhoods bordered cells in TR/MyR (green) neighborhoods, which, in turn, bordered cells in TR/BInf (gray) neighborhoods in keeping with the stereotyped border sequence noted previously. In addition, we observed that cells in TP/MacR (lavender) and TP/TCR (orange) neighborhoods bordered one another as well as cells in TR/BInf (gray) neighborhoods. Cells in TP/MacR (lavender) or in TP/TCR (orange) neighborhoods almost never bordered cells in TR/MyR (green) or TR/ImP (yellow) neighborhoods (Figure 3C, top left; supplemental Figure 7A). A chord diagram representing all contacts between cells in different CNTs for all cases with MacR-enriched TIMEs confirmed these observations (Figure 3C, top right).
We again calculated the difference in the percentage of cells assigned to each CNT in regions ≥20 μm and <20 μm from the nearest CD31+ cell across cases (Figure 3C, bottom right). We found no statistically significant enrichment for cells in specific CNTs in areas ≥20 μm or <20 μm from the nearest CD31+ cell (Figure 3C, bottom right). However, we found that the percentage of cells in TR/ImP neighborhoods was lowest close to CD31+ cells and highest far from CD31+ cells, and the percentages of cells in TR/BInf, TP/TCR, and TP/DCR neighborhoods were highest near CD31+ cells and lowest far from CD31+ cells (supplemental Figure 7B).
These data imply that the relative proportion of tumor cells to immune cells follows a gradient from areas with a high tumor cell to immune cell ratio and cells in TR/IP neighborhoods to areas with a low tumor cell to immune cell ratio and cells in TP/TCR (orange) and TP/MacR (lavender) neighborhoods. However, this gradient is more weakly associated with tumor vessels for cases with Mac-enriched TIMEs than for cases with immune-deficient and DC-enriched TIMEs. The distinct associations between CNTs and vessels were not attributable to significant differences in the density of CD31+ cells across TIME categories (supplemental Figure 8).
Cases with immune-deficient, DC-enriched, and Mac-enriched TIMEs differentially express functional markers of immune activity
Next, we wished to determine whether cases with distinct TIMEs showed differences in functional biomarker expression. Therefore, we examined the relative expression of 8 functional biomarkers included in the MIBI-TOF panel but not used to define cell lineages across cases (supplemental Table 1). These included biomarkers associated with immune stimulation (HLA-DR and DC-SIGN [CD209]), immune suppression (Arginase-1 [Arg-1], indoleamine 2,3-dioxygenase 1 [IDO-1]), T-cell activation (PD-1, in the absence of LAG-3),16 T-cell exhaustion (CD45RO, PD-1, and LAG-3),17 and cell proliferation (Ki-67) (Figure 3D).
Among cases with immune-deficient TIMEs, we found high HLA-DR, high DC-SIGN, moderate Arg-1, and low IDO-1 expression by cells in the TR neighborhoods (Figure 3D, left panel). We also found moderate-to-high PD-1, low CD45RO, and very low LAG-3 expression among the cells in the few TP/TCR neighborhoods found in cases with immune-deficient TIMEs. These data suggest a microenvironment in which tumor, myeloid, and T cells coordinately express at least a subset of functional biomarkers associated with immune activation.
Among cases with DC-enriched TIMEs, we found high HLA-DR, high DC-SIGN, low Arg-1, and very low IDO-1 expression by cells in TR neighborhoods (Figure 3D, middle). We also found moderate-to-high HLA-DR and high DC-SIGN expression by cells in TP/DCR neighborhoods. Finally, we found high PD-1, moderate CD45RO, and very low LAG-3 expression in cells in TP/TCR neighborhoods. These data suggest a microenvironment in which tumor, myeloid, and T cells coordinately express functional biomarkers associated with immune stimulation over immune suppression.
Among cases with Mac-enriched TIMEs, we found low HLA-DR, low DC-SIGN, and moderate-to-high Arg-1 expression by cells in TR neighborhoods (Figure 3D, right). We also observed low HLA-DR, low DC-SIGN, and high IDO-1 expression by cells in TP/MacR neighborhoods. Finally, we found high PD-1, high CD45-RO, and high LAG-3 expression by cells in TP/TCR neighborhoods. These data suggest a microenvironment in which tumor, myeloid, and T cells coordinately express functional biomarkers associated with immune suppression over immune stimulation. We also observed these differences in functional biomarker expression across TIME categories when we examined cases as a whole and without respect to individual CNTs (supplemental Figure 9).
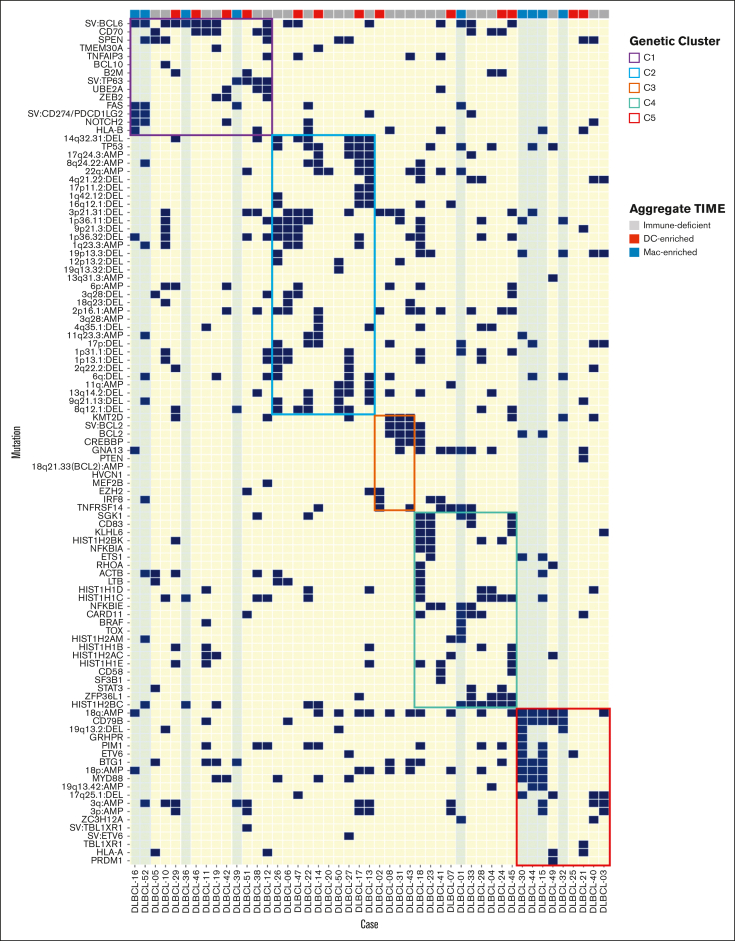
Cases with a Mac-enriched TIME have genetic signatures associated with immune evasion
The cellular constituents of TIMEs can be influenced by tumor genetics, although this has not been established at scale for BCLs.2,18 We, therefore, looked for potential associations among cases in each of the 3 TIME categories and the comprehensive genetic signatures of these tumors, as described by Chapuy et al (clusters C1-C5).2 We found that cases with immune-deficient or DC-enriched TIMEs were broadly represented in each of the cluster designations (Figure 4). In contrast, 9 of 10 cases with Mac-enriched TIMEs were assigned to C1 and C5, 2 genetic clusters associated with lymphomas with immune evasion phenotypes.2 Four of the 10 cases were assigned to C1, a cluster with mutation patterns found in transformed marginal zone lymphomas. Two of these 4 cases had CD274/PDCD1LG2 structural variants, and 3 of these 4 cases had FAS mutations (Figure 4). Four of the 10 cases were assigned to C5, a cluster characterized by mutation patterns found in immune-privileged lymphomas.2 One of the 10 cases was assigned to C4 and contained a FAS mutation (Figure 4). Therefore, we find that cases with Mac-enriched TIMEs often have genetic signatures associated with immune evasion, although this association did not reach statistical significance.
Figure 4.
DLBCLs with a Mac-enriched TIME have genetic signatures associated with immune evasion. Single nucleotide variants, structural variants, and copy number alterations in the indicated cases colored based on the aggregate TIME and arranged by genetic cluster designation as determined in the study by Chapuy et al.2 Note that cases with an immune-deficient or a DC-enriched TIME are seen across cluster designations, whereas cases with a Mac-enriched TIME have signatures found in tumors associated with immune evasion (ie, clusters C1 and C5 and cluster C4 associated with a FAS mutation).
Discussion
To better define the immune components of the DLBCL TIME and their cellular organization, we performed a quantitative spatial analysis of the DLBCL TIME using a 27-biomarker panel which we captured using MIBI-TOF and analyzed using a combination of commercial and novel image analysis software. Our accumulated findings indicate that the TIMEs of de novo, previously untreated DLBCLs are not devoid of immune infiltrates or topographically defined structures.1 Rather, we found that DLBCL tumor and immune cells are organized into distinct aggregate TIMEs that differ in their cellular constituents, local cellular organization, spatial structures, functional immune marker expression, and associations with defined genetic subtypes. Importantly, our results were highly concordant across multiple, randomly selected FOVs from each case (supplemental Figure 2A-B) and strongly suggest that we adequately sampled the tissues and that our results reflect intrinsic tumor properties.
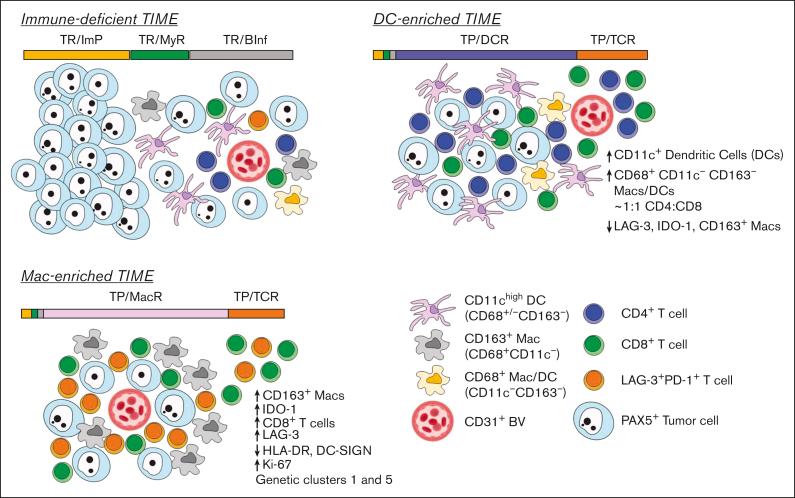
Taken together, our data suggest a working model of the 3 DLBCL microenvironments (Figure 5). DLBCLs with immune-deficient TIMEs are characterized by the predominance of tumor cells and a paucity of immune cells. Regions distant from blood vessels are especially devoid of immune cells, whereas regions approaching blood vessels have increasing, but still minimal, immune cell infiltrates. It stands to reason that the few immune cells that are present have exited from the adjacent vasculature and exhibit some immune activity. Cases with immune-deficient TIMEs are included in all genetic subtypes. DLBCLs with DC-enriched TIMEs are distinguished based on the regions in which immune cells predominate in DCR neighborhoods. Regions distant from blood vessels remained poor in the number of immune cells, whereas regions approaching blood vessels are increasingly immune cell rich and composed of T cells and DCs that express biomarkers supportive of immune activity. Cases with DC-enriched TIMEs are also included in all genetic subtypes. Finally, DLBCLs with Mac-enriched TIMEs are distinguished by regions in which immune cells predominate in MacR neighborhoods. Immune cell–rich neighborhoods are found both near and far from blood vessels, in keeping with a reduced gradient structure, and express biomarkers associated with immune suppression. Cases with Mac-enriched TIMEs are often found in genetic subgroups associated with immune evasion.
Figure 5.
Model showing the topographical organization of tumor and immune cells in DLBCLs with immune-deficient, DC-enriched, and Mac-enriched TIMEs. (Top left) DLBCLs with immune-deficient TIMEs are characterized by cells in tumor-rich (TR) CNTs. Individual cell neighborhoods furthest from blood vessels have very few immune cells compared with tumor cells and are organized as TR/ImP neighborhoods, consistent with immune exclusion. Individual cell neighborhoods closer to blood vessels are characterized by greater, but still a minor, number of immune cells compared to tumor cells and include greater numbers of myeloid and DCs than T cells organized as TR/MyR neighborhoods, suggestive of ongoing antigen sampling and limited T-cell stimulation. Regions nearest blood vessels show the highest numbers of immune cells relative to tumor cells. They include a variety of cell lineages organized as TR/BInf neighborhoods, consistent with extravasation of multiple immune cell types into the local microenvironment. (Top right) DLBCLs with DC-enriched TIMEs are characterized by cells organized as tumor-poor (TP) and TR CNTs. Uniquely to DLBCLs with DC-enriched TIMEs, regions nearest blood vessels show expanded numbers of T cells in TP/TCR neighborhoods and expanded numbers of DCs, CD4+ and CD8+ T cells in TP/DCR neighborhoods. Cells in TP/TCR and TP/DCR neighborhoods are enriched near blood vessels but also extend into the tumor parenchyma, suggestive of an active immune response. (Bottom left) DLBCLs with Mac-enriched TIMEs are characterized by cells organized as TP CNTs in addition to TR CNTs. Uniquely to DLBCLs with Mac-enriched TIMEs, regions close to and far from blood vessels show expanded numbers of T cells in TP/TCR neighborhoods and expanded numbers of CD163+ M2-like macrophages, IDO-1+ myeloid cells, CD8+ T cells, and PD-1+LAG-3+ exhausted T cells in the TP/MacR neighborhoods. In addition, there is lower HLA-DR expression in TP/ImP neighborhoods, suggestive of reduced immune stimulation. BV, blood vessel.
An important result of this study was our finding that cells in TP/DCR and TP/MacR neighborhoods were nearly mutually exclusive across cases. However, dividing cases based on this criterion was challenging because the percentages of cells in each TIME-defining CNT followed a continuum. We used a k-means clustering strategy to divide cases into the 3 TIME categories in an unbiased manner. The results were largely recapitulated using a threshold of >3% of cells in TP/DCR neighborhoods to identify cases with a DC-enriched TIME, >3% of cells in TP/MacR neighborhoods to identify cases with a Mac-enriched TIME, and ≤3% of cells in TP/DCR or TP/MacR neighborhoods to identify cases with an immune-deficient TIME. Although the 3% threshold appears low, it is important to recall that these are unique cell neighborhoods with a predominance of CD11c+ DCs and CD163+ macrophages, respectively, and few tumor cells. Moreover, most cases with DC-enriched or Mac-enriched TIMEs had cell percentages in these TIME-defining CNTs that far exceeded the threshold. Nevertheless, we acknowledge that the optimal threshold may change with the analysis of additional and larger datasets.
In addition, we have only captured a small subset of the cell lineage and functional biomarkers relevant to DLBCL. Therefore, it will be of great interest to expand the number of biomarkers analyzed in subsequent studies. Similarly, we have considered the spatial orientations of tumor and immune cells with respect to a single architectural feature. CD31 marks endothelial cells, high endothelial vessels, and lymphatics, so it will also be of interest to further phenotype and characterize the nature of the CD31+ cells and their relationships with the various immune cell populations.19 Finally, the regions of interest randomly selected for our TMA construction did not include tumor edges or extranodal tissue, which were likely to anchor additional spatial relationships. As such, the spatially defined model described here will undoubtedly evolve with further studies.
To date, most protein or immune cell–based biomarker studies of lymphoma TIMEs have focused on single biomarkers quantified independently and without regard to spatial geopositioning. Limitations with single-marker immunohistochemistry to evaluate immune cell infiltrates include an inability to identify critical immune cell subsets with distinct functional capabilities robustly. Nevertheless, such studies have repeatedly shown associations between single immune biomarker expression levels and clinical response to standard combination chemotherapy and novel immune therapies.18,20, 21, 22
Gene expression profiling and RNA sequencing of bulk or disaggregated single cells have the advantage of significantly increasing the number of biomarkers analyzed but at the expense of spatial and protein information and the types of tissues that can be analyzed.23, 24, 25 Nevertheless, these methods have revealed that DLBCL TIMEs are diverse in the degree of immune cell infiltration and the types of infiltrating cells.26,27
Multiplexed immunofluorescence has been used to bridge the gap between low-complexity protein analysis at single-cell resolution and RNA profiling and has been recently used to study BCL microenvironments and identify prognostic signatures and therapeutic targets.14,28,29 For example, a combination of multiplex assays derived from prognostically relevant gene expression signatures was used to show that high numbers of TIM-3+PD-L1+CD68+CD163– macrophages were associated with poor overall survival among patients with DLBCL enrolled in a clinical trial.28,29 We have used multiplex immunofluorescence (MIF) with spatially resolved image analysis to show that the TIME of TCR/histiocyte-rich large BCL (TCRLBCL) has an immunological and spatial signature closely resembling classic Hodgkin lymphoma, including a topographically defined immunosuppressive niche rich in PD-L1+ tumor-associated macrophages and PD-1+ T cells.14 In a small set of patients with relapsed/refractory TCRLBCL treated with pembrolizumab, objective clinical responses were observed. We also found that the TCRLBCL TIME objectively differs from the classic Hodgkin lymphoma (cHL) TIME by the differential representation of unique local cell neighborhoods associated with each tumor type.14 These observations raise the intriguing possibility that different lymphoma types may have distinct spatially defined TIME signatures, which may partly define the tumor.14
To date, to our knowledge, only 1 other highly multiplexed, spatially resolved protein-based analysis of the DLBCL microenvironment has been reported and for a limited number of cases.30 Our analyses showed both similarities and differences. Similar to the preceding study, we found that tumor and immune cells are organized into defined cellular groupings with geographically defined arrangements across tissues. Notable differences between our study and the prior include our use of a distinct antibody panel (11 of our 27 biomarkers [41%] are shared), a more extensive case series, aggregate TIME categories defined by the mutually exclusive CNT representation, and a spatial analysis that identified stereotyped geographic relationships with tumor vessels. It is important to note that CD11c, a biomarker critical for identifying DC-enriched TIME, was not included in the prior study panel. In the future, it will be essential to compare datasets through shared images and image analysis methods directly.
Sharing these complex data will also facilitate our understanding of how TIME categories associate with outcomes to standard and to novel therapies. In our series, all patients were treated with rixtuximab, cyclophosphamide, doxorubicin, vincristine, and prednisone. We did not find a statistically significant association between TIME category and progression-free survival, although the data set is small and was not powered to detect minor differences.
In conclusion, we find that DLBCL TIMEs are not random assortments of tumor and immune cells but are multicellular organizations with spatially defined structures that suggest distinct biologies. It is tempting to speculate that patients with DLBCLs and immune-deficient, DC-enriched, and Mac-enriched TIMEs may respond differently to various therapies. Indeed, it was recently reported that a subset of patients with DLBCLs and high T-cell content respond to PD-1 blockade, unlike the vast majority of patients with DLBCL,31 and that a subset of patients with DLBCLs and a transcriptionally-defined inflamed microenvironment respond poorly to chimeric antigen receptor T cells.32 It will be important to validate our observations in additional datasets and interesting to determine whether the stereotyped spatial organization of tumor and immune cells that we describe for de novo DLBCLs persists over time, with treatment, and at relapse with conventional and immunotherapies.
Conflict-of-interest disclosure: S.J.R. received research support from Affimed, Merck, and Bristol Myers Squibb (BMS) and is a member of the Scientific Advisory Board for Immunitas Therapeutics. K.T.W. presented a scientific seminar in collaboration with IonPath Inc (GenomeWeb Webinar series, 2020). M.A.S. has research funding from BMS, Bayer, AbbVie, and AstraZeneca. She is on advisory boards for AstraZeneca and BMS. The remaining authors declare no competing financial interests.
The current affiliation of Y.S. is Genentech, Inc., South San Francisco, CA.
Acknowledgments
K.T.W. was supported, in part, by the National Institutes of Health (NIH) under Ruth L. Kirschstein National Research Service Award T32HL007627 from the National Heart, Lung, and Blood Institute to the Department of Pathology, Brigham & Women's Hospital (BWH). S.J. is supported by NIH DP2AI171139, R01AI149672, a Gilead’s Research Scholars Program in Hematologic Malignancies, the Bill & Melinda Gates Foundation INV-002704, and previously, in part, by a Leukemia and Lymphoma Society of America Career Development Award. This work was supported, in part, by KITE pharmaceuticals (S.J.R.) and the Leukemia and Lymphoma Society of America (M.A.S.).
Authorship
Contribution: K.T.W. and S.J.R. contributed to the study design, data analysis, and manuscript preparation and review; J.L.W. and S.J. contributed to the data analysis and manuscript preparation and review; K.Z.K, A.G.-H., and Y.S. contributed to the data analysis and manuscript review; and M.A.S. contributed to the study design, data analysis, and manuscript review.
Footnotes
∗K.T.W, J.L.W., and S.J. contributed equally to this work.
Processed images for all markers and cell phenotype maps for all FOVs are available on request from the corresponding author, Scott J. Rodig (srodig@bwh.harvard.edu).
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171(2):481–494.e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson WH, Wright GW, Huang DW, et al. Effect of ibrutinib with R-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell. 2021;39(12):1643–1653.e3. doi: 10.1016/j.ccell.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Medeiros LJ, Young KH. Cancer immunotherapy in diffuse large B-cell lymphoma. Front Oncol. 2018;8(SEP):351. doi: 10.3389/fonc.2018.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. 2014;14(8):517–534. doi: 10.1038/nrc3774. [DOI] [PubMed] [Google Scholar]
- 8.Chen BJ, Chapuy B, Ouyang J, et al. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin Cancer Res. 2013;19(13):3462–3473. doi: 10.1158/1078-0432.CCR-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzankov A, Went P, Zimpfer A, Dirnhofer S. Tissue microarray technology: principles, pitfalls and perspectives - lessons learned from hematological malignancies. Exp Gerontol. 2005;40(8-9):737–744. doi: 10.1016/j.exger.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J Clin Oncol. 2008;26(34):5630–5637. doi: 10.1200/JCO.2008.17.3567. [DOI] [PubMed] [Google Scholar]
- 11.Giltnane JM, Rimm DL. Technology insight: identification of biomarkers with tissue microarray technology. Nat Clin Pract Oncol. 2004;1(2):104–111. doi: 10.1038/ncponc0046. [DOI] [PubMed] [Google Scholar]
- 12.Keren L, Bosse M, Marquez D, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174(6):1373–1387.e19. doi: 10.1016/j.cell.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schürch CM, Bhate SS, Barlow GL, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive. Front Cell. 2020;182(5):1341–1359.e19. doi: 10.1016/j.cell.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin GK, Weirather JL, Roemer MGM, et al. Spatial signatures identify immune escape via PD-1 as a defining feature of T-cell/histiocyte-rich large B-cell lymphoma. Blood. 2021;137(10):1353–1364. doi: 10.1182/blood.2020006464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch'ng ES. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front Oncol. 2019;9:1512. doi: 10.3389/fonc.2019.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang ZZ, Kim HJ, Villasboas JC, et al. Expression of LAG-3 defines exhaustion of intratumoral PD-1+ T cells and correlates with poor outcome in follicular lymphoma. Oncotarget. 2017;8(37):61425–61439. doi: 10.18632/oncotarget.18251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumeister SH, Freeman GJ, Dranoff G, Sharpe AH. Coinhibitory pathways in immunotherapy for cancer. Annu Rev Immunol. 2016;34:539–573. doi: 10.1146/annurev-immunol-032414-112049. [DOI] [PubMed] [Google Scholar]
- 18.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 19.Taskinen M, Jantunen E, Kosma VM, Bono P, Karjalainen-Lindsberg ML, Leppä S. Prognostic impact of CD31-positive microvessel density in follicular lymphoma patients treated with immunochemotherapy. Eur J Cancer. 2010;46(13):2506–2512. doi: 10.1016/j.ejca.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Xu-Monette ZY, Xiao M, Au Q, et al. Immune profiling and quantitative analysis decipher the clinical role of immune-checkpoint expression in the tumor immune microenvironment of DLBCL. Cancer Immunol Res. 2019;7(4):644–657. doi: 10.1158/2326-6066.CIR-18-0439. [DOI] [PubMed] [Google Scholar]
- 21.Coutinho R, Clear AJ, Mazzola E, et al. Revisiting the immune microenvironment of diffuse large B-cell lymphoma using a tissue microarray and immunohistochemistry: robust semi-automated analysis reveals CD3 and FoxP3 as potential predictors of response to R-CHOP. Haematologica. 2015;100(3):363–369. doi: 10.3324/haematol.2014.110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nam SJ, Go H, Paik JH, et al. An increase of M2 macrophages predicts poor prognosis in patients with diffuse large B-cell lymphoma treated with rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone. Leuk Lymphoma. 2014;55(11):2466–2476. doi: 10.3109/10428194.2013.879713. [DOI] [PubMed] [Google Scholar]
- 23.Tian L, Chen F, Macosko EZ. The expanding vistas of spatial transcriptomics. Nat Biotechnol. 2023;41(6):773–782. doi: 10.1038/s41587-022-01448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye X, Wang L, Nie M, et al. A single-cell atlas of diffuse large B cell lymphoma. Cell Rep. 2022;39(3):110713. doi: 10.1016/j.celrep.2022.110713. [DOI] [PubMed] [Google Scholar]
- 25.Staudt LM, Dave S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol. 2005;87:163–208. doi: 10.1016/S0065-2776(05)87005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monti S, Savage KJ, Kutok JL, et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105(5):1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 27.Kotlov N, Bagaev A, Revuelta M v, et al. Clinical and biological subtypes of B-cell lymphoma revealed by microenvironmental signatures. Cancer Discov. 2021;11(6):1468–1489. doi: 10.1158/2159-8290.CD-20-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Autio M, Leivonen SK, Brück O, Karjalainen-Lindsberg ML, Pellinen T, Leppä S. Clinical impact of immune cells and their spatial interactions in diffuse large B-cell lymphoma microenvironment. Clin Cancer Res. 2022;28(4):781–792. doi: 10.1158/1078-0432.CCR-21-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Autio M, Leivonen SK, Brück O, et al. Immune cell constitution in the tumor microenvironment predicts the outcome in diffuse large B-cell lymphoma. Haematologica. 2021;106(3):718–729. doi: 10.3324/haematol.2019.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colombo AR, Hav M, Singh M, et al. Single-cell spatial analysis of tumor immune architecture in diffuse large B-cell lymphoma. Blood Adv. 2022;6(16):4675–4690. doi: 10.1182/bloodadvances.2022007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kline J, Godfrey J, Ansell SM. The immune landscape and response to immune checkpoint blockade therapy in lymphoma. Blood. 2020;135(8):523–533. doi: 10.1182/blood.2019000847. [DOI] [PubMed] [Google Scholar]
- 32.Jain MD, Zhao H, Wang X, et al. Tumor interferon signaling and suppressive myeloid cells are associated with CAR T-cell failure in large B-cell lymphoma. Blood. 2021;137(19):2621–2633. doi: 10.1182/blood.2020007445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.