Summary
The bacterial strain JCVI-syn3.0 stands as the first example of a living organism with a minimized synthetic genome, derived from the Mycoplasma mycoides genome and chemically synthesized in vitro. Here, we report the experimental evolution of a syn3.0- derived strain. Ten independent replicates were evolved for several hundred generations, leading to growth rate improvements of > 15%. Endpoint strains possessed an average of 8 mutations composed of indels and SNPs, with a pronounced C/G- > A/T transversion bias. Multiple genes were repeated mutational targets across the independent lineages, including phase variable lipoprotein activation, 5 distinct; nonsynonymous substitutions in the same membrane transporter protein, and inactivation of an uncharacterized gene. Transcriptomic analysis revealed an overall tradeoff reflected in upregulated ribosomal proteins and downregulated DNA and RNA related proteins during adaptation. This work establishes the suitability of synthetic, minimal strains for laboratory evolution, providing a means to optimize strain growth characteristics and elucidate gene functionality.
Subject areas: Evolutionary biology, Bioengineering, Synthetic biology
Graphical abstract
Highlights
-
•
The evolution of a minimal synthetic organism leads to growth improvements
-
•
Growth improvements are likely caused by targeted mutations to a transporter
-
•
Transcriptomic analysis revealed an upregulation of ribosomal proteins
-
•
This work establishes suitability of minimal strains for laboratory evolution
Evolutionary biology; Bioengineering; Synthetic biology
Introduction
The first successful attempt at creating (semi-) synthetic life was achieved with JCVI syn1.0, composed of a 1,079 kbp Mycoplasma mycoides-based artificial chromosome inserted into a Mycoplasma capricolum cell.1 The prefix “semi” applies due to basepair-for-basepair copying of the DNA sequence of an already extant organism. Subsequent significant efforts in genome minimization culminated in the JCVI-syn3.0 strain,2 the first synthetic life with a chromosome not existing elsewhere in nature and the smallest of any autonomously replicating cell. The remaining 531 kbp seemed essential for axenic life in rich medium, yet 79 of the 473 genes could not be assigned to any functional category. Additionally, genome reduction from JCVI-syn1.0 to JCVI-syn3.0 significantly reduced growth rate and resulted in pleomorphic cellular forms, making laboratory investigation difficult. A subsequent variant of JCVI-syn3.0, JCVI-syn3A, containing an extra 12 kbps encoding 16 protein coding genes, resulted in a growth rate increased by 73%3 and a nearly normal morphology.4 Much of this work helped reveal the function of the various genes added to and removed from these genomes.
All life, whether natural or synthetic, is subject to evolution due to unavoidable errors in DNA replication and intra-population competition among the ancestral and mutant strains. Adaptive laboratory evolution (ALE) thus stands as a powerful approach for both studying evolutionary processes in the lab and optimizing strain performance.5 Hence, we used ALE on JCVI-syn3A to investigate how a minimal, synthetic cell adaptively evolves and what biological insights can be extracted from the resultant data.
Results
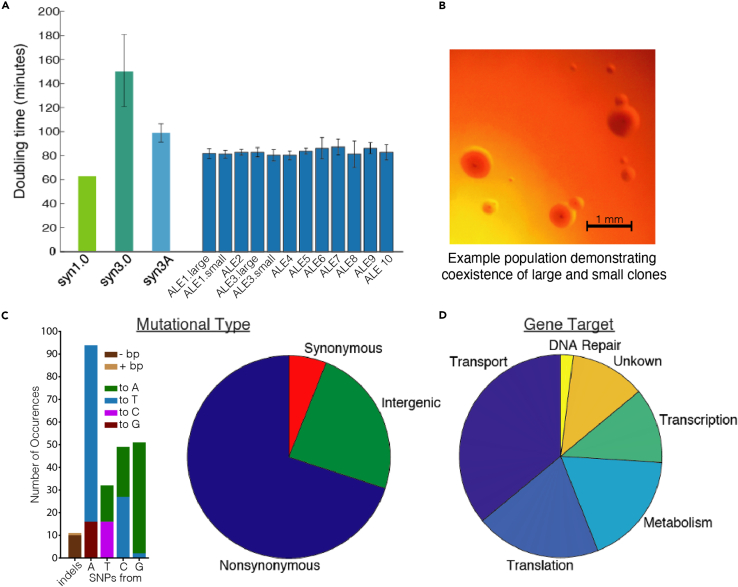
We evolved ten parallel lineages founded from JCVI-syn3A for ∼400 generations via serial propagation of batch cultures. All evolved strains grew at a rate ∼15% faster than the ancestral starting strain (Figure 1A), causing the growth rate to further approach that of strain JCVI-syn1.0, which has a 99% larger genome but grows only 38% faster. Mixed populations in two endpoint cultures were manifest as large and small colonies, something previously observed in adaptive evolution experiments6 (Figure 1B). Such large vs. small colony isolates nonetheless showed similar increase in growth rate when measured independently, despite possessing distinct mutations.
Figure 1.
Fitness improvements and mutational trends
(A) Growth rate improvements following evolution.
(B) Colony size variations observed in some evolved populations, a ∼500% increase in diameter between the smallest and largest. Culture of ALE 1 population diluted 10−6 and plated on SP4-KO agar medium plates, growth at 37°C for six days.
(C) Left; observed SNP bias for A/T incorporation and indel bias for genome reduction. Right; proportion of mutations changing protein amino acid sequences or falling between genes.
(D) Proportion of total observed mutations targeting genes falling into six broad functional categories.
We subjected evolved endpoint clones from each lineage, as well as midpoint clones for some populations, to whole genome sequencing. Phenotypic improvement after hundreds of generations of serial passage is unsurprising, but the multiple genetic alterations underlying such change are almost impossible to predict for such a unique organism lacking existing experimental evolution data. Laboratory workhorses such as E. coli are suitable for evolutionary interrogation due to the small number of genetic changes accrued even after hundreds of generations—with few mutations, causality can be easily established.7 This is not the case in hypermutable strains, where the hundreds of mutations they contain makes causal establishment significantly more difficult.8
We found that JCVI-syn3A has a distinct mutational spectrum but is not hypermutable, despite multiple DNA mismatch repair genes being removed from M. mycoides to form the JCVI-syn3A genome. Evolved strains had between 2 and 10 mutations, with an average of 8, composed of indels or SNPs of various types (Figure 1C). Certain mutations, such as rpoC were found across nearly all strains, which are likely the result of culturing of syn3A before the independent lineages and is a common phenomenon in ALE.9 Strains exhibited a pronounced mutational bias for A/T incorporation, with 36 of 38 SNPs causing a nucleotide change of X- > A/T; unsurprising given the Mycoplasma genus’ propensity for low GC content10 and the fact that JCVI-syn3A already has 76% A/T. Genome rearrangements were not observed and indels typically reduced genome size, ranging from between 1 and 175 bp in magnitude, while only a single +1 bp addition was observed. Mutations predominantly altered protein amino acid sequences while only two synonymous alterations were observed, in addition to several intergenic changes likely causing regulatory shifts for the downstream gene (Table S1). Such a distribution is characteristic of an organism undergoing selection and adaptively evolving rather than neutrally drifting.11 We also discovered several mutations found across all sequenced strains, likely reflective of mutations that were missed in the original GenBank reference genome sequence or that arose during culturing of syn3A before it was used to found the independent lineages, as is regularly seen in ALE experiments.9
The genetic targets of mutations fell into a range of functional categories (Figure 1D). Transporter genes were the most frequently mutated, likely influencing osmoregulation or to improve efficiency of nutrient uptake/inhibitory byproduct secretion, given that these would be particularly rate-limiting under selection for growth rate in an ALE environment so distinct from M. mycoides’ wild type growth in ruminants. Genes involved in translation, metabolism, and transcription were also frequent targets, something regularly observed when organisms are forced to optimize the functioning of their cellular subsystems for higher growth rate.12 Interestingly, multiple genes with completely unknown functions were clear targets for selection. These targets provide guidance on knock-in mutants to create for interrogation of the genes’ purpose, and further demonstrate the importance of these essential but uncharacterized proteins for proper cellular function. A single mutation in the DNA repair gene recO was also found, and though it was synonymous and thus potentially a neutral hitchhiker, it could also be adaptive, as has been observed in prior ALE experiments.9
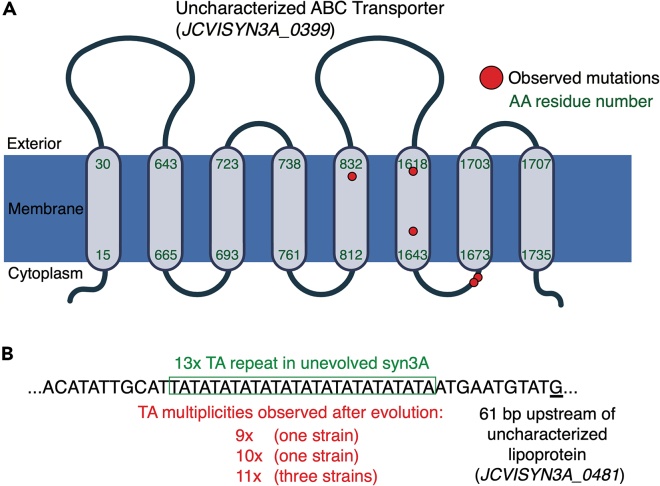
Evolved strains exhibited significant mutational overlap—eight genes acquired two or more mutations independently across all strains, a hallmark of regions most heavily under selection during ALE experiments.14 Noteworthy were uncharacterized ABC efflux transporter permeases, two of which mutated multiple times across the evolved strains. One of these acquired five separate amino acid changes and was found altered in eight of the ten evolved endpoints, with the targeted residues falling in both transmembrane and cytosolic regions (Figure 2A). Efflux transporters being under such heavy selection points to difficulties maintaining homeostasis at higher growth rates or the buildup of inhibitory molecules as a byproduct of intracellular metabolism. The need for altered efflux could potentially be elucidated with systems biology via metabolic modeling of the flux occurring within these minimal cells,3 paired with assays on the evolved strains.
Figure 2.
Regions under repeated selection
(A) An uncharacterized efflux ABC transporter permease (JCVISYN3A_0399) acquired multiple mutations independently across all but two of the ten evolved endpoints. Red dots signify altered residues. Transmembrane regions were estimated using TMHMM.13
(B) Shifts in upstream TA repeat multiplicity allowed for phase variable expression of an uncharacterized lipoprotein gene (JCVISYN3A_0481).
Genes involved in lipid metabolism and uptake were also under notable selection, including those encoding the catalytic subunit of fatty acid kinase FakA (four unique mutations) and an uncharacterized lipoprotein (three unique mutations). JCVI-syn3A relies on direct uptake to acquire several of its essential lipids and lipid metabolism is its third most energetically expensive metabolic process,3 thus it is unsurprising that we observe such optimizing mutations. Although the specific influence of FakA alterations is difficult to determine without experimental assays, the lipoprotein targeting mutations lend themselves to a simple explanation. All observed mutations were reductions in length of the simple sequence repeat (TA) 13 falling upstream of the lipoprotein gene (Figure 2B).
This variable repeat is located between the −10 and −35 region of the gene and undergoes high frequency expansion and contraction, linked to transcriptional phase variation in expression of the surface lipoprotein, depending on specific repeat length. The 13 repeats in the unevolved JCVI-syn3A correspond to an off expression state, whereas shortened lengths in the evolved populations reflect the on expression states.15,16
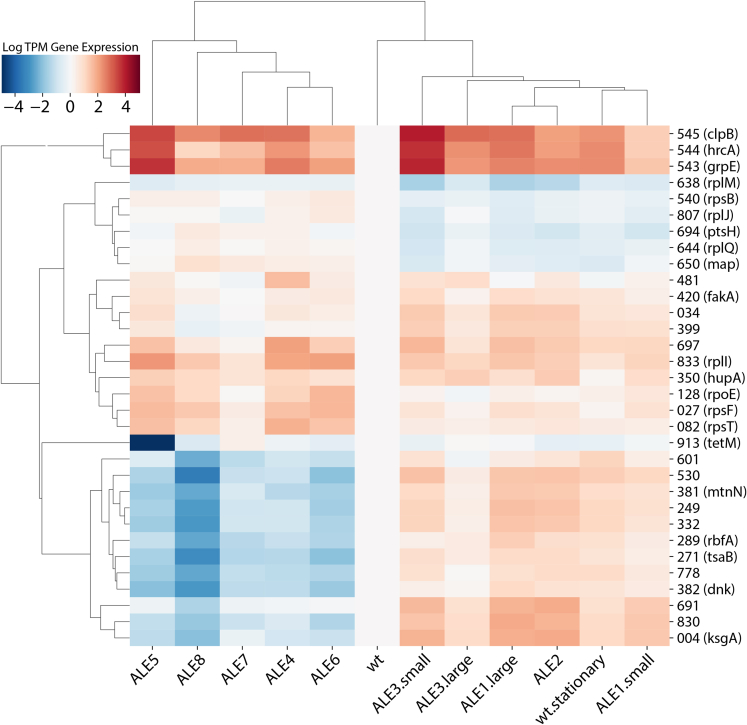
The tetracycline resistance protein TetM was another frequent target for selection, with six distinct mutations occurring across all strains. Several were upstream mutations that decreased expression (Figure 3), but deactivating mutations were also observed, such as an early frameshift and premature stop codon. Taken together with the fact that cells were not evolved in the presence of tetracycline and the strong nature of the TetM promoter, it appears TetM expression was energetically wasteful enough that it was repeatedly targeted for downregulation or deactivation, though there is the possibility that it serves some physiological function that becomes actively detrimental under these growth conditions. This demonstrates that cassettes used in strain construction can have a significant impact on ultimate strain phenotype, potentially warranting removal to optimize cellular fitness. Although it is hard to speculate on the reason for repeated mutations in genes with unknown function, one of the proteins (JCVISYN3A_0601) was clearly targeted for removal, acquiring both a change in start codon and a premature stop codon. This result indicates the potential for even further minimization of this already genomically compact organism.
Figure 3.
Differentially expressed genes
Expression values are in log tpm format centered on the JCVI-syn3A ancestor control (wt). Genes shown are those most differentially expressed compared to wt as well as those most upregulated/downregulated in ALE4-ALE8. The top and left dendrograms were produced using hierarchical clustering to group similar genes and samples together. RNAseq data were collected during exponential growth phase except for the wt.stationary sample, collected during stationary phase.
RNA-sequencing data were gathered for 12 isolates: the JCVI-syn3A ancestor in two different growth conditions, one small and large colony clone from the two populations exhibiting morphological heterogeneity, and the remaining six isolates from different endpoint strains. ALE5 has the majority of the unique upstream TetM mutations discussed previously and heavily downregulates expression of TetM, contrasting with other strains that acquired TetM frameshifts or stop codons that decreased protein rather than transcript expression. Strains fall into two main clusters of gene expression (ALE1-ALE3 vs. ALE4-ALE7) distinguished primarily by different regulatory patterns and overall higher expression for the ALE1-ALE3 group. It should be noted though that its similarity in expression to the stationary wild-type sample shows these TRN changes may be related to its growth phase rather than a possible evolutionary change. There are no mutations to transcription factors or other mutational differences that explain this clustering, but such an outcome is consistent with the stochastic nature of adaptive walks yielding different metabolic strategies in pursuit of the same fitness goal.12
When compared to the JCVI-syn3A control, all evolved strains tend to downregulate DNA replication/repair proteins and upregulate ribosomal proteins (Figure 3). This trend of upregulating ribosomal proteins (“greed”) at the cost of repair pathways (“fear”) is similar to the greed vs. fear tradeoff seen in many other ALEs and well analyzed via iModulons.17 The RNA polymerase mutations are the likely cause of this effect, as they are found in all but one of the evolved strains and are well known to favor growth-related functions in E. coli.17 Comparing the clusters, ALE4-ALE7 upregulate ribosomal proteins more and downregulate many uncharacterized proteins, DNA replication/repair proteins, and some rRNA/tRNA modifying proteins. ALE1-ALE3 not only upregulates some ribosomal proteins, but also downregulates others. The cause for this is unclear but these strains’ TRN’s do not appear to be as focused toward faster growth as ALE4-ALE7.
The top component in principle component analysis of the RNA-seq data explains 47% of the variance and is largely composed of ALE4-ALE7’s upregulated/downregulated genes. As data continue to accrue on the JCVI-syn3A and its derivatives4 it is likely that transcriptional and iModulon analysis will gain more explanatory power, particularly in combination with the mutational profiling herein.
Discussion
As synthetic life and designer organisms become more widespread and easier to produce18 it will become important to understand how these atypical species change over time. This work preliminarily establishes the suitability of laboratory evolution for addressing such questions. ALE provides a means not only to optimize strain phenotypes for laboratory culturing, but also to better understand the cellular processes at work. This includes exposing mutational trends, pointing to the rate-limiting systems most strongly under selection, and revealing specific genetic and transcriptomic alterations that can be studied in greater detail to elucidate gene functionality, either through knock-in mutant creation or assays on evolved strains. Moreover, minimal organisms such as JCVI-syn3A provide a fascinating insight into the smallest set of essential components necessary to qualify as life.19 With demonstrated suitability for evolutionary interrogation in the lab, we can proceed to better understand these systems and the evolutionary history of life on Earth.
Limitations of the study
No well-defined minimal medium is available for syn3A growth; the rich, not fully defined growth medium used limits the ability to interpret phenotypic outcomes with rigorous metabolic modeling.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| JCVI-syn3A | JCVI | NCBI accession no. CP016816.2 |
| Chemicals, peptides, and recombinant proteins | ||
| KnockOut™ Serum Replacement [GIBCO] | Thermo Fisher | |
| PicoGreen fluorescence | Thermo Fisher | |
| Critical commercial assays | ||
| Kapa HyperPlus Library Prep Ki | Kapa Biosystems | |
| Illumina HiSeq 4000 | Illumina | |
| Deposited data | ||
| Sequencing Data | this study | GSE205017 |
| RNA-seq Expression Data | this study | GSE205017 |
| Software and algorithms | ||
| Bowtie | https://bowtie-bio.sourceforge.net/index.shtml | Langmead and Salzberg24 |
| HTSeq | https://htseq.readthedocs.io/en/master/ | Anders et al.25 |
| pymodulon | https://pymodulon.readthedocs.io/en/latest/ | Sastry et al.17 |
| Sklearn | https://scikit-learn.org/stable/ | Pedregosa et al.26 |
| Pandas | https://pandas.pydata.org/ | McKinney27 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Bernhard Palsson (palsson@ucsd.edu)
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
JCVI-syn3A (NCBI accession no. CP016816.2) strains were used for all of this study.
Method details
ALE and growth characterization
ALE experiments were conducted with automated devices9 that serially propagated batch cultures of JCVI-syn3A between growth tubes with 15 mL of SP4-KO medium (containing 17% v/v KnockOut™ Serum Replacement [GIBCO] in lieu of fetal bovine serum in SP4), well aerated through mixing and kept at 37°C. Ten replicate lineages were started from the JCVI-syn3A ancestor and for each lineage 150 ul of culture was passed to a fresh tube once daily (∼6.6 generations/day), switching to 30 ul passage volume (9 generations/day) after 200 generations and continuing for a total of 400 generations.
Strains were isolated for characterization by plating of evolved populations and random selection of individual clones. Clones were inoculated into SP4-KO medium at 37°C and sample timepoints were taken regularly during the cultures’ period of exponential growth. Samples were centrifuged, cells were lysed, and DNA content was quantified via PicoGreen fluorescence (Thermo Fisher) as described previously.2 The slope of log(DNA fluorescence) vs. time was then calculated to determine culture growth rates; all growth curve data had a line of best fit with R2>0.99.
Whole genome sequencing
Evolved populations were streaked onto agar plates and single clones were selected for DNA sequencing. Genomic DNA was isolated using bead agitation in 96-well plates as outlined previously.20 Paired-end whole-genome DNA-seq libraries were generated with a Kapa HyperPlus Library Prep Kit (Kapa Biosystems) and run on an Illumina HiSeq 4000 platform with a HiSeq SBS Kit (150 base-pair (bp) reads). The generated DNA-seq FASTQ files were quality-controlled with AfterQC v.0.9.721 then processed with the breseq computational pipeline22 according to standard procedures (https://barricklab.org/twiki/pub/Lab/ToolsBacterialGenomeResequencing/documentation/) and aligned to the JCVI-syn3A genome (NCBI accession no. CP016816.2) to identify mutations. The lack of genome amplifications was determined with a custom script that identified discontinuities in read depth; all read depth coverage plots and marginal mutation calls were manually inspected. Genome sequence data supporting this study is publicly available at aledb.org (https://aledb.ucsd.edu/ale/project/52/) and in the NCBI SRA under GSE205017. All mutations found are included in Table S1.
Transcriptomics
RNAseq was performed using two biological replicates grown in log phase culture in the same environmental conditions as the ALE experiment. All cultures were immediately frozen at -80°C in growth medium, per standard protocols for mycoplasma handling. Log phase growth was assessed by taking DNA fluorescence measurements on the cultures and comparing results with those obtained for the growth rate characterization assay to check that they match log phase rather than stationary phase values. Total RNA isolation, rRNA removal, and sequencing library preparation was performed as previously described.23 Libraries were run on an Illumina NextSeq.
Quantification and statistical analysis
Raw transcriptomics sequencing reads were mapped using Bowtie,24 and reads were counted using HTSeq.25 Libraries were normalized using transcripts per million (TPM). Analysis of the RNA-sequencing data was done using pymodulon,17 sklearn,26 and pandas.27
Acknowledgments
Kim Wise and John Glass were funded by the following United States National Science Foundation grants: MCB 1840320, MCB 1818344, MCB 1840301 and MCB 2221237.
Author contributions
Conceptualization, J.I.G. and B.O.P.; Methodology, T.E.S. and K.S.W.; Formal Analysis, T.E.S., K.S.W., and C.D.; Investigation, T.E.S., K.S.W., and R.S.; Resources, A.M.F., J.I.G., and B.O.P.; Writing—Original Draft, T.E.S.; Writing—Review & Editing, T.E.S., K.S.W., C.D., J.I.G., and B.O.P.; Supervision, J.I.G. and B.O.P.
Declaration of interests
The authors declare no competing interests.
Published: July 28, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107500.
Supplemental information
Data and code availability
-
•
Genome sequence data supporting this study is publicly available at aledb.org (https://aledb.ucsd.edu/ale/project/52/) and in the NCBI SRA under GSE205017. The RNAseq data is submitted to NCBI as GSE205017 and is publicly available there.
-
•
Code used for this project is publicly available and cited in their relevant method sections. No additional custom code was created or used for this project.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact, Dr. Bernhard Palsson (palsson@ucsd.edu), upon request.
References
- 1.Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M., et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 2.Hutchison C.A., 3rd, Chuang R.Y., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L., et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 3.Breuer M., Earnest T.M., Merryman C., Wise K.S., Sun L., Lynott M.R., Hutchison C.A., Smith H.O., Lapek J.D., Gonzalez D.J., et al. Essential metabolism for a minimal cell. Elife. 2019;8 doi: 10.7554/eLife.36842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier J.F., Sun L., Wise K.S., Assad-Garcia N., Karas B.J., Deerinck T.J., Ellisman M.H., Mershin A., Gershenfeld N., Chuang R.Y., et al. Genetic requirements for cell division in a genomically minimal cell. Cell. 2021;184:2430–2440. doi: 10.1016/j.cell.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Sandberg T.E., Salazar M.J., Weng L.L., Palsson B.O., Feist A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019;56:1–16. doi: 10.1016/j.ymben.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozen D.E., Philippe N., Arjan de Visser J., Lenski R.E., Schneider D. Death and cannibalism in a seasonal environment facilitate bacterial coexistence. Ecol. Lett. 2009;12:34–44. doi: 10.1111/j.1461-0248.2008.01257.x. [DOI] [PubMed] [Google Scholar]
- 7.LaCroix R.A., Sandberg T.E., O’Brien E.J., Utrilla J., Ebrahim A., Guzman G.I., Szubin R., Palsson B.O., Feist A.M. Discovery of key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal media using adaptive laboratory evolution. Appl. Environ. Microbiol. 2014;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leon D., D’Alton S., Quandt E.M., Barrick J.E. Innovation in an E. coli evolution experiment is contingent on maintaining adaptive potential until competition subsides. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandberg T.E., Szubin R., Phaneuf P.V., Palsson B.O. Synthetic cross-phyla gene replacement and evolutionary assimilation of major enzymes. Nat Ecol Evol. 2020;4:1402–1409. doi: 10.1038/s41559-020-1271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson C.C., Vieira N.M., Vicente A.C.P., Thompson F.L. Towards a genome based taxonomy of Mycoplasmas. Infect. Genet. Evol. 2011;11:1798–1804. doi: 10.1016/j.meegid.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 11.Lang G.I., Desai M.M. The spectrum of adaptive mutations in experimental evolution. Genomics. 2014;104:412–416. doi: 10.1016/j.ygeno.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCloskey D., Xu S., Sandberg T.E., Brunk E., Hefner Y., Szubin R., Feist A.M., Palsson B.O. Evolution of gene knockout strains of E. coli reveal regulatory architectures governed by metabolism. Nat. Commun. 2018;9:3796. doi: 10.1038/s41467-018-06219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallgren J., Tsirigos K.D., Pedersen M.D., Armenteros J.J.A., Marcatili P., Nielsen H., Krogh A., Winther O. DeepTMHMM Predicts Alpha and Beta Transmembrane Proteins Using Deep. Neural Networks Preprint at. bioRxiv. 2022 doi: 10.1101/2022.04.08.487609. [DOI] [Google Scholar]
- 14.Bailey S.F., Rodrigue N., Kassen R. The effect of selection environment on the probability of parallel evolution. Mol. Biol. Evol. 2015;32:1436–1448. doi: 10.1093/molbev/msv033. [DOI] [PubMed] [Google Scholar]
- 15.Persson A., Jacobsson K., Frykberg L., Johansson K.-E., Poumarat F. Variable surface protein Vmm of Mycoplasma mycoides subsp. mycoides small colony type. J. Bacteriol. 2002;184:3712–3722. doi: 10.1128/JB.184.13.3712-3722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wise K.S., Foecking M.F., Röske K., Lee Y.J., Lee Y.M., Madan A., Calcutt M.J. Distinctive repertoire of contingency genes conferring mutation- based phase variation and combinatorial expression of surface lipoproteins in Mycoplasma capricolum subsp. capricolum of the Mycoplasma mycoides phylogenetic cluster. J. Bacteriol. 2006;188:4926–4941. doi: 10.1128/JB.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sastry A.V., Gao Y., Szubin R., Hefner Y., Xu S., Kim D., Choudhary K.S., Yang L., King Z.A., Palsson B.O. The Escherichia coli transcriptome mostly consists of independently regulated modules. Nat. Commun. 2019;10:1–14. doi: 10.1038/s41467-019-13483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredens J., Wang K., de la Torre D., Funke L.F.H., Robertson W.E., Christova Y., Chia T., Schmied W.H., Dunkelmann D.L., Beránek V., et al. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569:514–518. doi: 10.1038/s41586-019-1192-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lachance J.-C., Rodrigue S., Palsson B.O. Synthetic biology: minimal cells, maximal knowledge. Elife. 2019;8 doi: 10.7554/eLife.45379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marotz C., Amir A., Humphrey G., Gaffney J., Gogul G., Knight R. DNA extraction for streamlined metagenomics of diverse environmental samples. Biotechniques. 2017;62:290–293. doi: 10.2144/000114559. [DOI] [PubMed] [Google Scholar]
- 21.Chen S., Huang T., Zhou Y., Han Y., Xu M., Gu J. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinf. 2017;18(Suppl 3):80. doi: 10.1186/s12859-017-1469-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deatherage D.E., Barrick J.E. In: Engineering and Analyzing Multicellular Systems. Sun L., Shou W., editors. Vol. 1151. Humana Press; 2014. Identification of Mutations in Laboratory-Evolved Microbes from Next-Generation Sequencing Data Using breseq. (Methods in Molecular Biology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen K., Anand A., Olson C., Sandberg T.E., Gao Y., Mih N., Palsson B.O. Bacterial fitness landscapes stratify based on proteome allocation associated with discrete aero-types. PLoS Comput. Biol. 2021;17 doi: 10.1371/journal.pcbi.1008596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R. Dubourg V, et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12:2825—2830.
- 27.McKinney W. Data structures for statistical computing in python. Proceedings of the 9th Python in Science Conference. 2010;445:51–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Genome sequence data supporting this study is publicly available at aledb.org (https://aledb.ucsd.edu/ale/project/52/) and in the NCBI SRA under GSE205017. The RNAseq data is submitted to NCBI as GSE205017 and is publicly available there.
-
•
Code used for this project is publicly available and cited in their relevant method sections. No additional custom code was created or used for this project.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact, Dr. Bernhard Palsson (palsson@ucsd.edu), upon request.