Summary
Upon exposure to a bacterial pore-forming toxin, enterocytes rapidly purge their apical cytoplasm into the gut lumen, resulting in a thin intestinal epithelium. The enterocytes regain their original shape and thickness within 16 h after the ingestion of the bacteria. Here, we show that the regrowth of Drosophila enterocytes entails an inversion of metabolic fluxes from the organism back toward the intestine. We identify a proton-assisted transporter, Arcus, that is required for the reverse absorption of amino acids and the timely recovery of the intestinal epithelium. Arcus is required for a peak of amino acids appearing in the hemolymph shortly after infection. The regrowth of enterocytes involves the insulin signaling pathway and Myc. The purge decreases Myc mRNA levels, which subsequently remain at low levels in the arcus mutant. Interestingly, the action of arcus and Myc in the intestinal epithelium is not cell-autonomous, suggesting amino acid fluxes within the intestinal epithelium.
Subject areas: Natural sciences, Biological sciences, Zoology, Biochemistry, Parasitology
Graphical abstract
Highlights
-
•
Purged enterocyte regrowth relies on a reverse flux of amino acids from host to gut
-
•
A rapid upsurge in hemolymph amino acids (aa) requires the G/A/P Arcus transporter
-
•
Basally located Arcus may act in concert with MYC-mediated enterocyte regrowth
-
•
Clonal analysis of Arcus suggests a passage of aa between neighboring enterocytes
Natural sciences; Biological sciences; Zoology; Biochemistry; Parasitology
Introduction
The intestinal tract is one of the largest interfaces between the host body and the external environment, harboring a diverse microbial community that is mostly beneficial for the host.1 However, the proliferation of existing opportunistic pathogens as well as the exposure to food-borne pathogens can severely compromise host health.2 Resistance and resilience mechanisms in the host have been selected during evolution that ensure the homeostasis of the epithelium and limit damages inflicted by pathogens.3 Whereas resistance mechanisms usually reduce the pathogen burden, resilience/disease tolerance mechanisms allow the host to endure damages inflicted by pathogens or by its own immune response.4,5 The compensatory proliferation of intestinal stem cells (ISCs) is an example of a resilience response that maintains gut homeostasis by replacing damaged gut enterocytes.6
Amino acids are important in determining the outcome of an infection.7 Amino acid metabolism affects the host physiology and may serve as an energy source. In mammals, 59 amino acid transporters have been identified and categorized in 12 different solute carrier (SLC) families such as the proton-coupled amino acid transporters (SLC36 or PATs), the sodium-coupled neutral amino acid transporters (SLC38 or SNATs), the cationic (SLC7 or CATs) and the heterodimeric (SLC7/SCL3 or HATs) amino acid transporters.8,9 In Drosophila, 603 genes are predicted to encode putative transporter proteins, which correspond to 4% of the genome.10 Amino acid transporters may have redundant functions, as different transporters can carry the same amino acid. These characteristics allow adaptation to various environmental conditions.
Serratia marcescens (Sm) is a Gram-negative opportunistic human pathogen that secretes several virulence factors, such as proteases and pore-forming toxins. When injected directly in the hemolymph, Sm resists the systemic immune response essentially because of the O-antigen of its LPS and is highly virulent.11,12 In contrast, Sm is much less virulent in an intestinal infection model. Hemocyte-mediated phagocytosis in the body cavity is an effective response against the ingested Sm bacteria that manage to cross the intestinal barrier. Several genes required for susceptibility or for resistance against ingested Sm have been identified before in a genome-wide screen.13
An early epithelial defense against ingested Sm is the cytoplasmic purge of enterocytes triggered by hemolysin produced by SmDb11.14 Starting from 1 h after ingestion, the enterocytes of the midgut extrude a considerable portion of their cytoplasm through an aperture that is formed in the apical part of the cell. Damaged organelles such as mitochondria, and also likely toxins and invading bacteria, are actively extruded out of the exposed enterocytes. As a result of this purge, the epithelium becomes very thin and flat throughout the midgut. Here, we focus on enterocytes of the R2 region, which lose through the purge their dome-shaped apical domains typical of this portion of the midgut. The cytoplasmic purge limits damages to enterocytes as there is no increased enterocyte cell death nor compensatory stem cell proliferation during the early stages of the infection. Strikingly, the same cells are able to recover their normal shape and size within the first 20 h post infection.14 However, the processes required for the fast reestablishment of the epithelium remain unknown.
Here, we explore this question and show that a retrograde transport of metabolites occurs from the hemolymph to the enterocytes during the recovery of the epithelium. We found that the GLY, ALA, and PRO amino acid transporter CG1139 localizes basally to the labyrinth region of the enterocytes, a zone of intensive exchange between the enterocyte and the hemolymph, and is required for the retrograde transport of a MET analogue. A short-lived peak of amino acids in the hemolymph that occurs a few hours after challenge is dependent on this transporter. Moreover, CG1139 is required for the fast regrowth of the epithelium, but a recovery is still possible in its absence, albeit slower, suggesting a possible redundancy with other amino acid transporters shown to be potentially promoting also the regrowth of thinned enterocytes. Our data suggest that CG1139 effects may be mediated by MYC. Unexpectedly for a transmembrane protein, we found CG1139 to act non-cell autonomously and we suggest that amino acids likely circulate between wild-type and mutant enterocytes. We propose to name this transporter Arcus (ACS). Arcus is a female Roman deity homologous to the Greek Iris that personifies the rainbow and that replenishes rain clouds.
Results
A retrograde transport from the hemolymph to the gut takes place upon infection
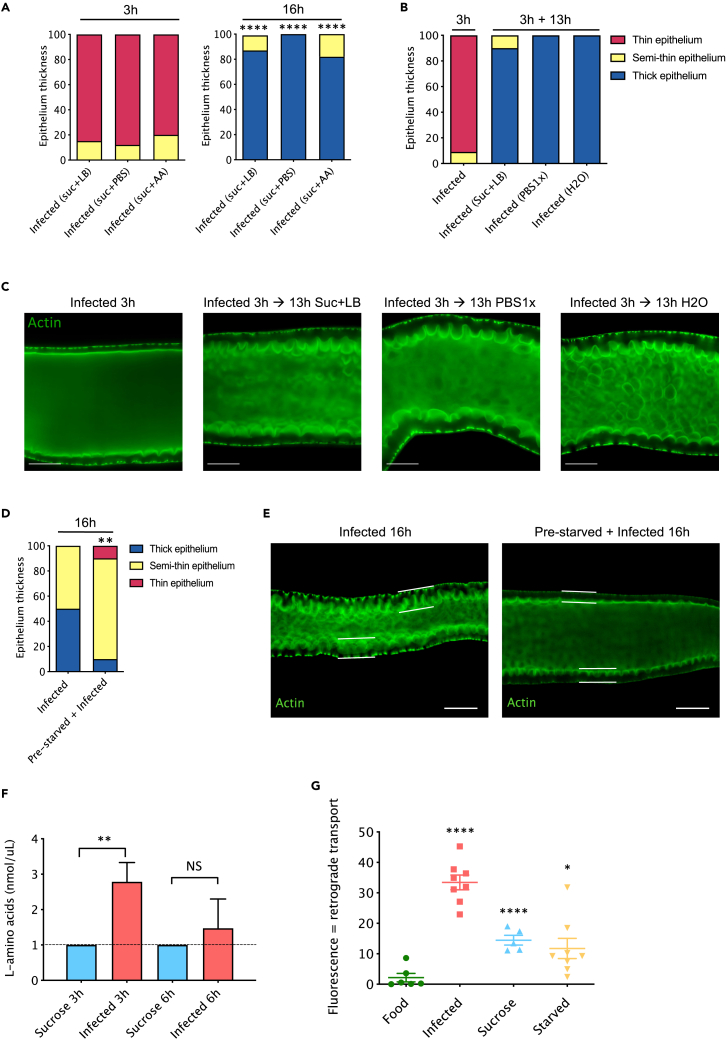
Upon SmDb11 ingestion, the gut epithelium becomes thin with a reduction of about two-fold in thickness.14 The elements required for the regrowth of enterocytes may either be derived from food or originate from the fly metabolic stores. In our usual infection solution, bacteria are mixed with 10% of lysogeny broth (LB) medium in 50mM sucrose. Removing LB from the solution did not prevent epithelial thinning at 3h nor recovery, which was also as efficient as a solution in which amino acids had been added instead of LB (Figure 1A). Next, we exposed the flies for 3 h to SmDb11 re-suspended in a PBS1x solution and then placed the flies for 13 h on sterile PBS1x or to H2O, or onto sucrose with LB as a control. Most flies were able to restore a thick epithelium in the absence of any external nutrients (Figures 1B, 1C, and S1A). These results suggest that upon infection, the fast recovery of the gut epithelial cells from the cytoplasmic purge is not dependent on newly acquired nutrients, but rather on metabolites stored in the insect body.
Figure 1.
Recovery of enterocytes after infection entails retrograde transport from body reserves
(A) % of epithelium thickness categories from flies infected with SmDb11 resuspended in sucrose solution with 10% LB, PBS1x or a mix of essential and non-essential amino acids (chi-square, ∗∗∗∗ = p < 0.0001, comparisons between 16h and 3h from corresponding samples). Dissected midguts were qualitatively assigned after microscopic examination to either of three categories: thin epithelium (red), semi-thin epithelium (yellow) or thick epithelium (blue) (n = 24–30 per experimental condition, pooled from two to three independent experiments).
(B and C) (B) % epithelium thickness categories from flies infected for 3h with SmDb11 in PBS1x solution, and after transfer to sucrose with 10% LB, PBS1x or H2O (3h + 13h). For each condition, representative pictures of gut enterocytes are shown in (C). The measured epithelial thicknesses of midguts issued from an independent experiment are presented in Figure S1A.
(D and E) (D) % epithelium thickness categories from flies pre-fed in standard food or pre-starved for 24h after SmDb11 infection. Representative pictures of gut enterocytes are shown in (E).
(F) Quantification of free amino acids in the hemolymph of flies 3h and 6h after sucrose or SmDb11 infection. Bars represent the mean and the error bars display Standard Deviation of the data (one-way ANOVA, ∗∗ = p < 0.001, n = 3 × 20 hemolymphs).
(G) Flies were injected with 50 μM AHA and exposed to regular food, SmDb11 (infected), sucrose or water (starved). Midguts were dissected 6h later and fluorescence from the incorporated amino acid was assessed by staining with an alkyne probe and measured using ImageJ (One-way ANOVA; ∗ = p < 0.05; ∗∗∗∗ = p < 0.0001, n = 5–8). Stainings are shown in Figure S2A. The median bars represent the mean while the error bars display Standard Deviation of the data. (C, E) Green = Actin. Scale bars: 50μm (C) and 100μm (E).
To explore this possibility, we pre-starved flies for 24 h in H2O to deplete their internal metabolic stores. Only 10% of the flies were able to recover to a normal epithelium, while the others 80% and 10% displayed respectively a semi-thin or thin epithelium (Figures 1D and 1E). These results show that the gut epithelium cannot recover efficiently 16h post-infection (p.i.) in pre-starved flies.
Next, we directly tested a possible retrograde transport of metabolites from the rest of the body to the gut epithelium. At 3h but not 6h p.i., we observed a transient increase of free amino acids in the hemolymph of flies (Figure 1F). We injected an L-azidohomoalanine (AHA) methionine analogue in the hemocoel of flies and exposed afterward the injected flies to either standard fly food, SmDb11, sucrose or to water (starved flies) for 6h. After reacting AHA with an alkyne fluorophore probe in a Click-iT reaction, the incorporated analog was detected by fluorescence in tissues in which translation is taking place.15 Hardly any fluorescence was measured in the gut of uninfected flies in contrast to infected ones, which displayed a large increase in midgut epithelium fluorescence 6 h p.i. Interestingly, flies that were placed on sucrose or starved on water also displayed an augmented midgut epithelium fluorescence (Figures 1G and S2A). These results show that the modified injected amino acid is being taken up from the hemolymph and incorporated into the enterocytes during the recovery phase of the cytoplasmic purge or upon amino acid starvation. Altogether, these experiments indicate the occurrence of a retrograde transport to the midgut epithelium in response to infection or starvation.
The amino acid transporter ACS/CG1139 is required for the fast recovery from the gut epithelium after the cytoplasmic purge
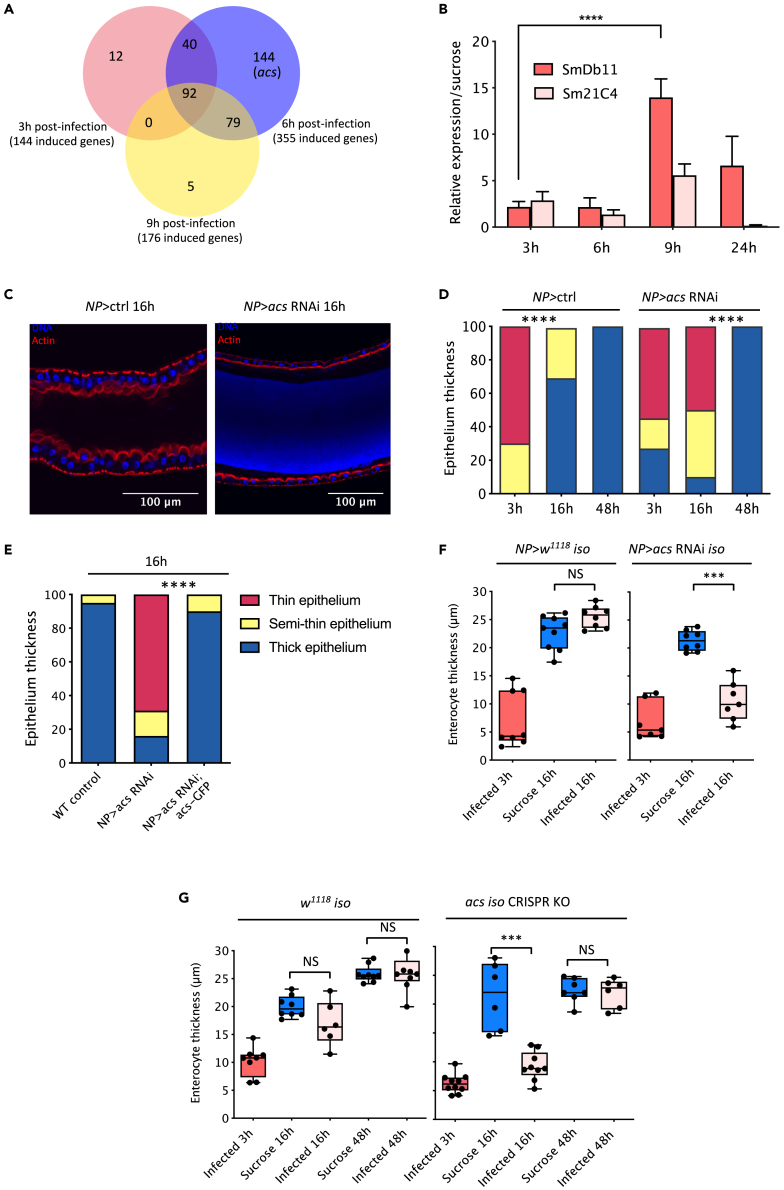
Given the results above, we next screened for altered recovery 28 putative amino acids transporter genes expressed in the Drosophila midgut by silencing their gene expression in enterocytes by RNA interference (RNAi) with a driver (NP1-Gal4, Gal80ts = NP) that is mostly expressed in enterocytes (Figure S1B). Among eight hits with a strongly impaired recovery phenotype at 16h p.i., the leading one is encoded by acs, which remarkably was the sole transporter gene found to be up-regulated 6h p.i. in an RNA-seq experiment that was previously performed in our laboratory (Figures 2A, 2C, and 2D). We validated the RNA-seq results by RT-qPCR and observed a more than 10-fold up-regulation of acs transcripts at 9 h p.i., which did not occur to the same extent upon a challenge with Sm21C4, an S. marcescens mutant for hemolysin that does not cause an accentuated cytoplasmic purge14 (Figure 2B).
Figure 2.
Fast recovery of gut epithelial thickness requires the ACS amino acid transporter
(A) Genes induced in the midgut 3h, 6h and 9h post SmDb11 infection identified by RNA-sequencing. acs was also induced at the 9-h time point but not significantly.
(B) Gene expression analyzed by RT-qPCR of acs after SmDb11 or Sm21C4 infection relative to sucrose. Bars represent the mean and the error bars display Standard Deviation of the data (one-way ANOVA, ∗∗∗∗ = p < 0,0001, n = 3 × 10 midguts).
(C) Midguts from control and NP>acs RNAi flies 16h post SmDb11 infection.
(D) % epithelium thickness categories after SmDb11 infection in control and NP>acs RNAi flies (chi-square, ∗∗∗∗ = p < 0.0001).
(E) % epithelium thickness categories 16h after SmDb11 infection in NP>ctrl, NP>acs RNAi and in the rescued line NP>acs-RNAi; UAS-acs-GFP (chi-square, ∗∗∗∗ = p < 0.0001).
(F) Control and NP > acs RNAi iso flies were exposed to sucrose or SmDb11 and enterocytes thickness was measured with ImageJ. Each dot represents the average of 10 enterocytes in one midgut (lmer, ∗∗∗ = p < 0.001, n = 7–9 midguts).
(G) w1118iso and a Knock-out CRISPR mutant for acs (acs iso CRISPR KO) were exposed to sucrose or SmDb11 and the thickness of the enterocytes was measured (lmer, ∗∗∗ = p < 0.001, n = 6–10 midguts). D-E: Qualitative quantification of intestines according to their epithelial thickness: Thin-red, Semithin-yellow, Thick-blue (n = 24–30). F-G: the middle bar of box plots represents the median and the upper and lower limits of the boxes indicate, respectively, the first and third quartile; the whiskers define the minima and maxima.
acs encodes an amino acid transporter similar to a mammalian PAT (Proton-assisted SLC36 Amino acid Transporter) (Figure S1C). It has previously been shown in D. melanogaster to promote cell growth and has been proposed to act as a transceptor involved in mTOR activation.16,17 The possibility that this transporter might regulate the recovery phase that follows the cytoplasmic purge prompted us to investigate further its role in enterocytes.
Having validated the efficiency of NP>acs RNAi (Figure S1D), we confirmed that acs function was indeed required in enterocytes by repeating the RNA silencing experiment with another enterocyte-specific driver, mex1-Gal4, and found also an impaired recovery in the acs-silenced midgut epithelia 16 h after Sm ingestion (Figure S1E). Next, we retested NP>acs RNAi flies 16 and 48 h p.i. Whereas the silenced flies displayed an impaired epithelial recovery as compared to control flies at 16h, they had fully recovered by 48h (Figure 2D). The 16h delayed recovery phenotype is indeed due to the silencing of the acs gene as it was observed with an independent RNAi line (Figure S1F) and was rescued by the ectopic expression of an acs-GFP transgene17 (Figure 2E).
To limit possible effects of the genetic background of acs RNAi, we isogenized this line into the w1118 iso background and measured more precisely the thickness of enterocytes. Overall, we obtained similar results to those obtained with the non-isogenized lines reported so far. In sucrose, both NP > iso and NP > acs RNAi iso harbored enterocytes of about 22μm thickness (the midgut epithelial thickness of wild-type flies feeding on food was roughly similar to that of flies feeding on sucrose solution when compared side-by-side). At 3 h p.i., when the cytoplasmic purge had occurred, the enterocytes had remarkably lost some 15μm thickness (median of 7μm) (Figure 2F). At 16 h p.i., while control flies had recovered their size (median of 25.5μm), enterocytes from NP > acs RNAi iso remained thin (median of 10.5μm), not different from SmDb11 at 3h (lm, p = 0.1023).
The silenced acs full recovery phenotype 48h post-infection may reflect a hypomorphic effect of RNAi. We therefore generated a null acs mutant by CRISPR-Cas9, which also displayed a delayed recovery phenotype (Figures 2G, S1G, and S1H), indicating that the absence of acs can be compensated, albeit slowly.
All together, these results show that acs encodes an amino acid transporter induced in the gut upon infection and that it is required in enterocytes for a fast recovery of the gut epithelium after the cytoplasmic purge.
A dynamic expression of ACS transcripts during infection
High-throughput expression data from FlyBase indicate that acs is expressed in the adult midgut, but it is also very highly expressed in the Malpighian tubules. In addition, data from Flygut-seq document an expression of acs in the gut visceral muscles.18 To visualize the tissues and the cell types in which acs is expressed, we used as a reporter a UAS-GFP transgene crossed to an acs CRISPR Knock-in mutant we have generated in which the entire CDS region of CG1139/acs has been deleted and replaced by Gal4 coding sequences (Figure S3A). Unexpectedly, although we observed the expression of acs in some enterocytes and mostly in the posterior midgut region, we also found it in cells with a shape characteristic of progenitor cells, in control or infection conditions (Figure S3B). Of note, the knock-in line may have deleted some regulatory elements present in the coding regions or introns and may therefore only partially reflect the endogenous expression of acs. Also, the transcript stability of Gal4 may differ from that of the endogenous acs gene.
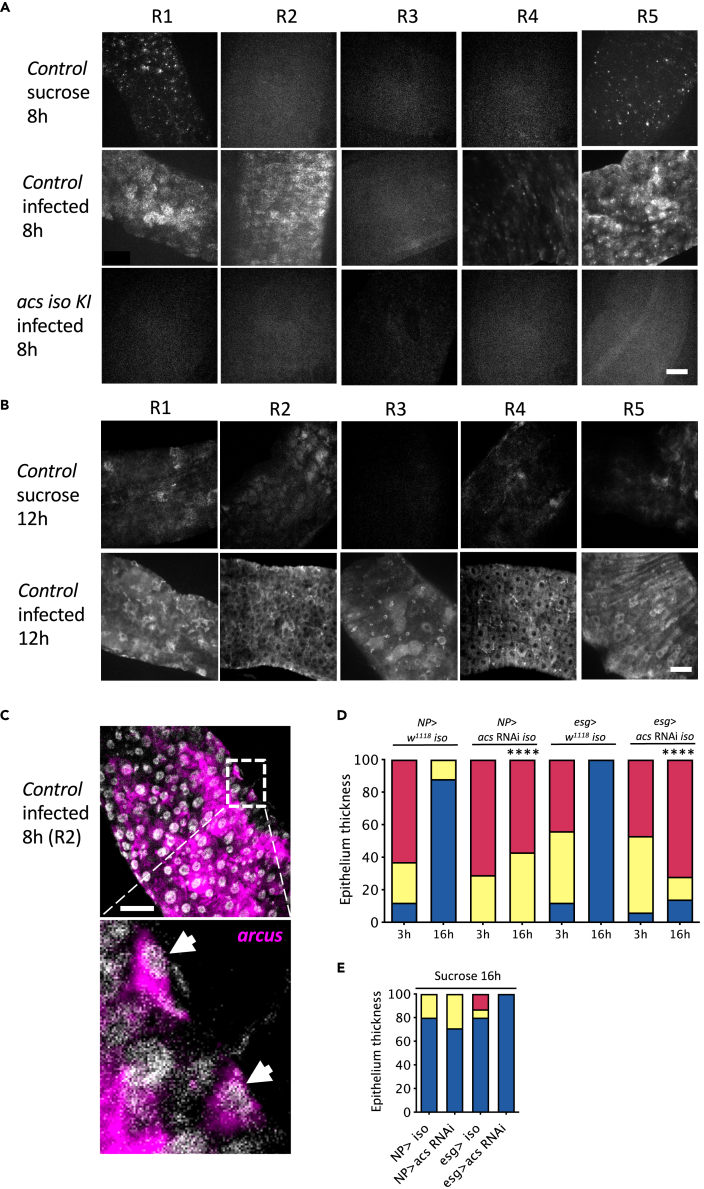
To verify the expression pattern of acs transcripts at different stages of the infection, we also performed in situ hybridization using the RNAscope technique. In control conditions (flies feeding on sucrose for 8 h), we observed some expression in scattered cells found mostly in the R1 and R5 regions, that is, the proximal and distal parts of the midgut. In contrast, the signal was stronger in these regions 8h p.i. and extended into the R2 region and to a lesser extent (weaker signal) in the R4 region. The hybridization was specific since no signal was detected in null acs KI mutants (Figure 3A). By 12h p.i., acs transcripts were observed throughout the midgut, including the R3 region (Figure 3B). Thus, the acs expression pattern is dynamic during infection and accounts for the increased steady-state expression measured initially by RT-qPCR (Figure 2B).
Figure 3.
acs expression is dynamic after the ingestion of SmDb11 bacteria
(A and B) acs dynamic expression was revealed by in situ hybridization using the RNAscope technique on time-matched uninfected and SmDb11 infected midguts at 8 and 12 h. The acs iso KI mutant is used as a negative control to probe the specificity of the signal. Scale bars 50μm.
(C) acs is expressed in progenitor cells that are identified through their basal position and small size of the nuclei (arrowheads). Scale bar 50μm.
(D and E) acs was knocked down in the enterocytes (NP > acs RNAi iso) or in the ISCs (esg> acs RNAi iso) and flies were infected with SmDb11 (D) or exposed to sucrose (E). % epithelium thickness categories were assessed at 3h and 16h (chi-square, ∗∗∗∗ = p < 0.0001, comparisons at 16h between knocked down flies and corresponding controls crossed with w1118iso). Qualitative quantification of intestines according to their epithelial thickness: Thin-red, Semithin-yellow, Thick-blue (n = 24–30).
To genetically assess the relevance of the acs expression pattern, we knocked down its expression by RNAi in a tissue-specific manner. Whereas silencing acs ubiquitously (ubi-Gal4-Gal80ts) led to an impaired recovery of the intestinal epithelium thickness, this phenotype was not observed upon silencing its expression in the fat body (yolk-Gal4), in the Malpighian tubules (uro-Gal4), in the visceral muscles (how-Gal4), and in the enteroendocrine cells (pros-Gal4) (Figures S3C–S3E). A strongly impaired recovery phenotype was also observed upon silencing acs in progenitor cells (esg-Gal4) in keeping with in situ hybridization data (Figures 3C–3E). These results suggest that acs is also required in the progenitor cells of the gut epithelium during recovery. Alternatively, enterocytes newly differentiated during the five to seven days RNAi induction period silence the induced expression of the transporter due to retention of the interfering acs dsRNAs in those cells. Future studies will determine whether this dynamic pattern of acs transcripts is mediated through transcriptional control or progressively enhanced transcript stabilization, a second possibility more in keeping with the unexpected data obtained with the knock-in reporter transgene.
ACS is required for increased fitness upon infection
We then assessed whether the impaired recovery phenotype might impact the fitness of the acs-silenced flies by monitoring them in several assays. Negative geotaxis assay did not reveal any difference between the silenced and control lines (Figure S4A). However, when chronically ingesting SmDb11, acs-silenced and mutant flies succumbed faster than control flies (Figures S4B and S4C). In addition, we found an increased bacterial burden in the crop and in the midgut of infected flies, even though the silenced flies did not ingest more food or defecate less after the infection (Figures S4D–S4G). Egg laying was also affected as the mutant flies did not present the increase in egg deposition observed in the control flies after infection (Figure S4H). Altogether, these results suggest that acs is required for increased fitness after infection, a likely indirect effect of its role in the timely recovery of the epithelium thickness.
ACS localizes to the basal part of the gut enterocytes and is required for the retrograde transport of amino acids
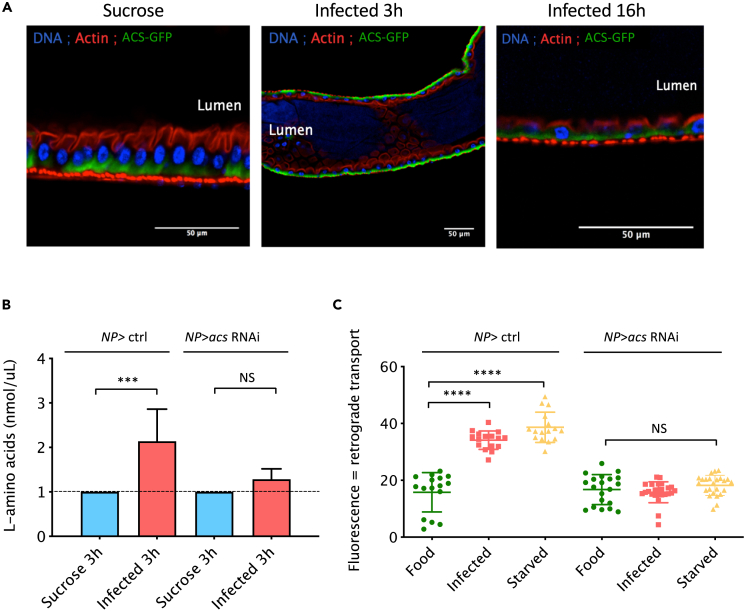
ACS has previously been shown to be localized at the cell surface and on the late endosomal and lysosomal surfaces in Drosophila S2 cells, as well as in the larval fat body.17 When its reporter transgene was expressed in enterocytes, the functionally active ACS-GFP fusion localized to the basal labyrinth region of the cell that allow intensive exchanges of enterocytes with the hemolymph19 in uninfected and infected flies (Figure 4A).
Figure 4.
ACS localizes to the basal side of the enterocytes in the midgut epithelium and is required for the retrograde transport
(A) Confocal pictures of dissected midguts from NP>UAS-acs-GFP flies exposed to sucrose or to SmDb11 for 3h and 6h. Blue:DNA; Red:actin; Green:ACS-GFP; n = 30.
(B) Quantification of free amino acids in the hemolymph of NP > ctrl and NP > acs RNAi 3h after sucrose exposure or SmDb11 infection. Bars represent the mean and the error bars display Standard Deviation of the data (one-way ANOVA, ∗∗∗ = p < 0.001, n = 3 × 20 hemolymphs).
(C) Flies were injected with 50 μM AHA and exposed to regular food, SmDb11 (infected) or water (starved). Midguts dissected 6h later and fluorescence from the incorporated amino acid was assessed by staining with an alkyne probe and measured using ImageJ. The median bars represent the mean while the error bars display Standard Deviation of the data(one-way ANOVA, ∗∗∗∗ = p < 0.0001, n = 16–24). Stainings shown in Figure S2B.
Unexpectedly, the free amino acid peak measured in the hemolymph of control flies 3 h post-infection was not detected in NP > acs RNAi flies (Figure 4B). Strikingly, the AHA methionine analogue was not taken up by the gut epithelium of these silenced flies after infection or starvation thus directly showing that ACS is required for the retrograde transport of amino acids into the intestinal epithelium (Figures 4C and S2B). These results suggest that ACS may act both directly and indirectly to support the retrograde transport to the gut epithelium.
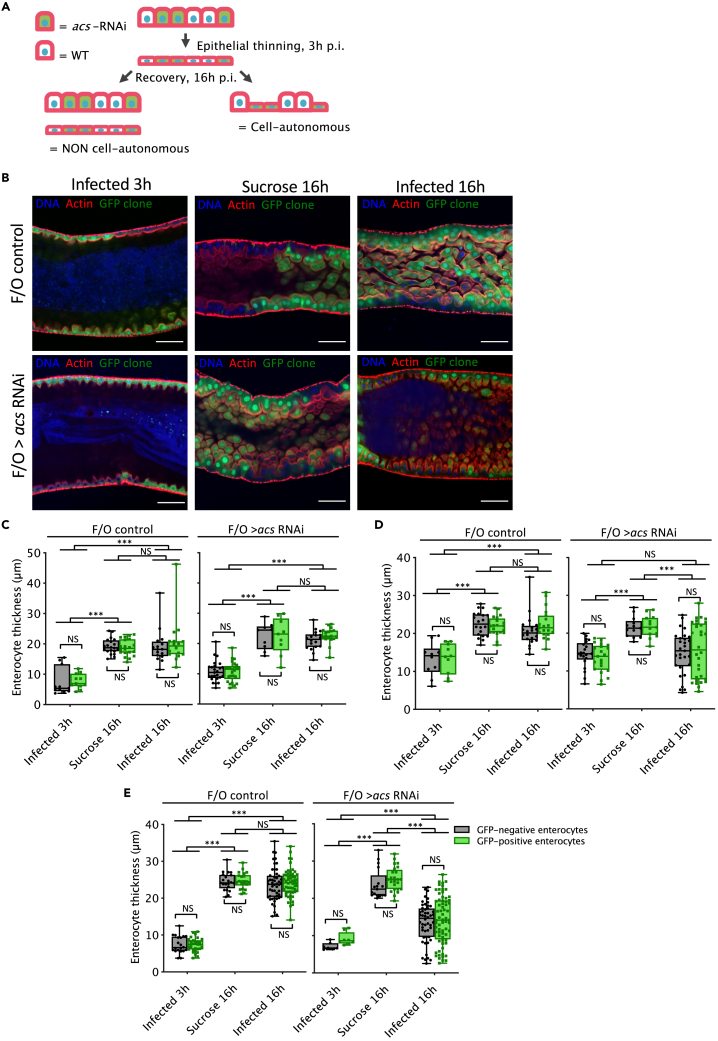
ACS is not required cell-autonomously for the recovery from the cytoplasmic purge
Whereas ACS localizes to the basal part of enterocytes in keeping with a direct function in taking up some amino acids from the hemolymph, the absence of an increased release of amino acids in the hemolymph was however perplexing as it suggested a signaling function for ACS. We therefore performed a clonal analysis in the gut epithelium using the escargot Flip Out technique (esgF/O) that marks with GFP silenced cells of a clone initially generated in progenitor cells20 (Figure 5A). We generated clones for 24, 48 or 72 h resulting in a variable proportion of silenced epithelial cells of the midgut, ranging from a limited number of clones at 24h to a much more extended proportion at 72h. Importantly, the extent of recovery between wild-type cells and acs-silenced (GFP-expressing) cells was similar in the three conditions, suggesting a non cell–autonomous function for acs (Figures 5B–5E). However, the midgut recovery was similar to that of uninfected controls when clones had been induced for 24 h (Figure 5C). In contrast, the recovery was impaired when the clones had been generated for 48 or 72 h (Figures 5D and 5E) but not as strongly as when acs silencing was driven in all enterocytes (compare Figure 2F to Figures 5D and 5E). Thus, the extent of recovery roughly correlates inversely to the proportion of acs-silenced epithelial cells.
Figure 5.
ACS is required non cell-autonomously for the recovery of the intestinal epithelium after SmDb11 infection
Clonal analyses performed using escargot Flp Out (esgF/O) system.
(A) w;esgGal4,tubGal80ts UAS-GFP;UAS-flp Act>CD2>Gal4 were crossed to UAS-acs-RNAi flies to generate clones of enterocytes expressing both GFP and RNAi against acs (F/O acs). A cell-autonomous mechanism implies that RNAi and wild type enterocytes recover at different rates, presenting a difference between their size 16h post-infection.
(B) Confocal pictures of dissected midguts from F/O control or acs F/O flies at 3h and 16h post-infection and at 16h post sucrose exposure. Clones had been generated for 72h. Blue:DNA; Red:Actin; Green:acs RNAi enterocytes GFP positive. Scale bar = 50μm. Quantification of GFP-positive and negative clones for each condition is represented in (E).
(C–E) Clones had been generated for 24h, 48h, and 72h, respectively; the middle bar of box plots represents the median and the upper and lower limits of the boxes indicate, respectively, the first and third quartile; the whiskers define the minima and maxima. (lmer, ∗∗∗ = p < 0.001, n = 4–7 midguts, merge of two independent replicates; NS: not significant).
The insulin signaling pathway and the transcription factor myc are required for the timely recovery of the gut enterocytes upon infection
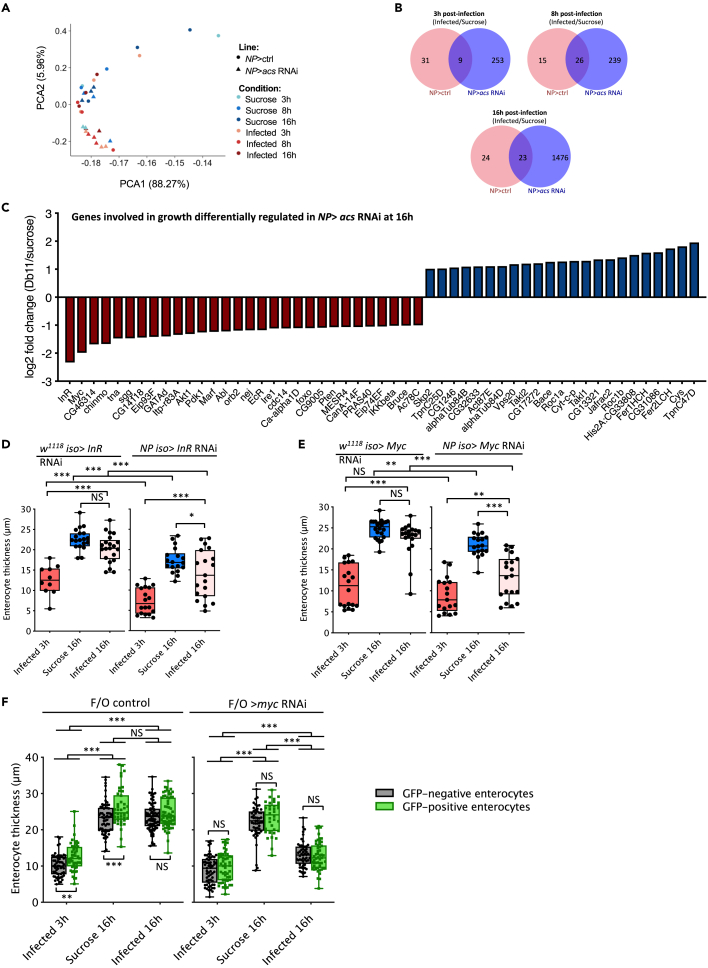
To understand how ACS functions, we performed an RNA-seq analysis in midguts of control flies or flies for which acs was silenced in the enterocytes at 3h, 8h, and 16h after SmDb11 infection. We performed a principal component analysis (PCA) and observed a clear-cut difference on the PCA2 axis between noninfected and infected NP > acs RNAi flies, especially at 8h and 16h p.i. This difference was less pronounced between uninfected and infected wild-type (Figure 6A). Accordingly, more genes were differentially expressed in NP > acs RNAi than in wild-type, when comparing the infected samples to the corresponding sucrose controls (Figure 6B). KEGG pathway enrichment analysis did not reveal a differential enrichment of a particular category between control and acs-silenced flies, not even at 16h after infection for which we found many categories being enriched in the NP > acs RNAi flies (Figure S5A). However, the hierarchical clustering revealed that NP > acs RNAi flies 16h p.i. had many genes down- and up-regulated and did not cluster with the other samples (Figure S5B). Thus, this analysis revealed that ACS has an important impact on the physiology of the midgut that is likely, however, to be indirect since the midgut epithelial cells of 16 h wild-type flies have recovered their initial morphology.
Figure 6.
InR and the transcription factor Myc is required for the recovery of the gut enterocytes after infection
NP > ctrl and NP > acs RNAi were exposed to sucrose or infected with SmDb11 for 3h, 8h and 16h. RNA-seq was performed on dissected midguts.
(A) Principal component analyses for gene expression of different samples.
(B) Venn diagram for each time-point comparing genes differentially expressed (fold-change≥2) in NP > ctrl and NP > acs RNAi after infection, relative to sucrose. The scattering of the NP > ctrl data points (A) may limit the power of the analysis and hence lowering the number of significantly differentially regulated genes displayed in the pink circles.
(C) Genes involved in growth that are up- or down-regulated in acs RNAi 16h after infection (relative to sucrose). All genes shown on the graph are differentially expressed with a log2-fold change≥2 and an adjusted p value≤0.05 (see also Table S6).
(D and E) (D) InR and (E) Myc were knocked down in the gut enterocytes (NPiso> InR RNAi and NPiso> Myc RNAi) and compared with the respective controls for the recovery of the gut enterocytes thickness 16h upon infection or exposure to sucrose.
(F) Midguts were dissected from F/O control or Myc F/O flies at 3h and 16h post-infection and at 16h post sucrose exposure. Thickness of GFP-positive and negative clones for each condition was quantified. Clones had been generated for 72h. D-F: the middle bar of box plots represents the median and the upper and lower limits of the boxes indicate, respectively, the first and third quartile; the whiskers define the minima and maxima (lmer, ∗∗∗ = p < 0.001, ∗∗ = p < 0.01, n = 10–22 midguts).
As the analyses above failed to provide us with significant insights, we decided to employ another method to help us identify gene categories enriched upon infection between the wild-type and NP > acs RNAi. Therefore, we performed a Gene Set Enrichment Analysis (GSEA) for all the time-points comparing control and mutant knock-down flies to each other.21 With respect to the gene sets that were significantly up-regulated in the wild-type compared to the acs KD flies, we focused on the 8 and 16h time-points, the period during which the intestinal epithelium recovers its initial shape and thickness. Strikingly, at 8h we found an enrichment for genes known to be involved in the positive regulation of organ growth, positive regulation of translation, and genes required for lamellipodia assembly (Figure S5C). Genes from the latter category were also up-regulated in the wild-type at 16h, like the related set of genes involved in actin nucleation (Figure S5D). Whether these two gene sets play a role in the formation of the basal labyrinth of enterocytes remains to be determined.
Next, we extracted the expression levels of growth-related genes from the RNAseq dataset and compared infected-acs RNAi flies versus their corresponding sucrose control at 16h when wild-type enterocytes have recovered and acs enterocytes remain thin (Figures 6C and S5E (3h and 8h p.i.)). Several genes that are down-regulated in the infected acs-silenced midguts relative to uninfected controls are related to the Insulin pathway (InR, Akt1, Pdk1, Dilp3, foxo, PTEN, PRAS40), cellular growth (Myc), and shaggy (sgg), a link between insulin signaling and MYC expression. Interestingly, silencing either the InR or Myc genes led to a significantly impaired recovery of enterocyte thickness (Figures 6D and 6E). Of note, both genes are also involved in regulating the basal size of enterocytes (compare sucrose 16h in control vs. silenced flies). The already thin InR-silenced enterocytes under basal conditions may account for the apparently higher partial recovery observed for this line as compared to Myc-silenced enterocytes. Consistent with an absolute requirement of insulin signaling for enterocyte growth, we failed to obtain InR null-mutant enterocytes in clonal analysis. Counterintuitively, Myc steady-state transcripts levels were found to be decreased upon infection at both 8 and 16h pi. Whereas this difference was significant for acs-silenced flies at both 8 and 16h pi., it was no longer significant at 16h pi. for the control flies, leading thereby to a relatively increased expression of Myc in wild-type vs. acs-silenced flies at 16h p.i. (Figure S5F). Thus, the levels of Myc may remain somewhat higher during recovery in wild-type as compared to the acs mutant enterocytes. In keeping with this finding, an esg-F/O clonal analysis with a Myc RNAi transgene revealed a non cell-autonomous phenotype similar to that of acs, which also led to an impaired recovery at 16h p.i. (Figure 6F).
The mTOR pathway is a major regulator of cell growth and ACS has been reported to interact with TOR in the fat body.17 Immunohistology stainings of midguts with antibodies against the phosphorylated form of 4EBP (P-4EBP), a direct TOR target, revealed an absence of signal in acs-silenced enterocytes 16h but not 3h pi. (Figure S6A). However, data obtained by directly silencing TOR, S6K or Tsc2 yielded ambiguous results with a mild impaired recovery phenotype observed both for TOR and its negative regulator TSC2 and a phenotype observed with silencing of S6K but not with a dominant negative allele (Figures S6B–S6E).
The EGFR pathway has been reported to promote enterocyte growth independently of Insulin and TOR signaling through the promotion of endoreplication in enteroblasts and newborn enterocytes after a Pseudomonas entomophila infection.22 Silencing EGFR expression in enterocytes did not affect the thickness of enterocytes under basal conditions but led to a mildly impaired recovery after exposure to SmDb11 (Figure S6F). In contrast, targeting the E2F transcription factors that regulate endocycling did not affect recovery (Figures S6G and S6H). Finally, Atg9 is also involved in regulating the thickness of enterocytes by negatively regulating the TOR pathway.23 As expected, the intestinal epithelium was thicker under basal conditions (sucrose) when Atg9 was silenced in enterocytes. The silenced enterocytes underwent a thinning of similar extent than wild-type controls and regained their original size 16h p.i. (Figure S6I).
In conclusion, the Insulin signaling pathway and Myc are involved in the regrowth of purged enterocytes and ACS appears to promote the recovery of the steady-state expression of Myc mRNA that is strongly reduced early on during the infection.
Discussion
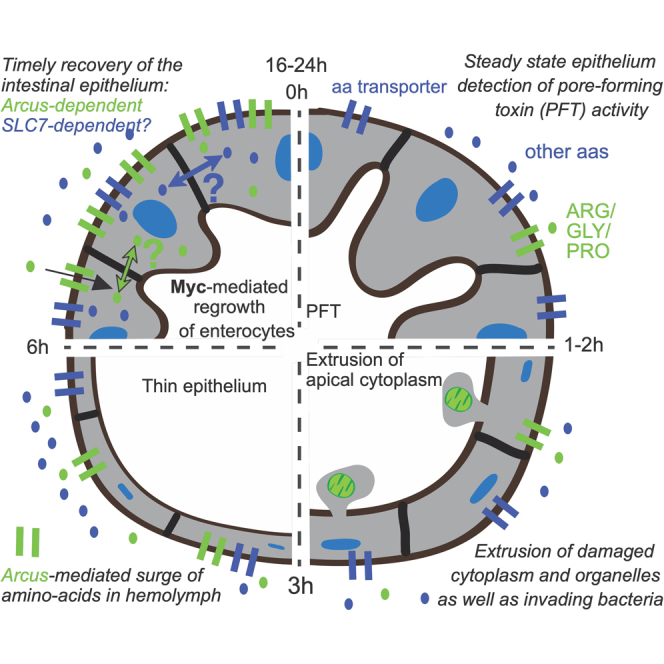
The links between nutrition and intestinal epithelium homeostasis have been documented in the Drosophila model in the past decade.22,23,24,25,26 Here, we demonstrate that the recovery of enterocyte shape and volume that follows their extensive thinning resulting from the purge of their apical cytoplasm upon exposure to SmDb11 pore-forming toxin does not involve the absorption of nutrients during or after the challenge. Rather, a reversion of metabolic fluxes from the organism to the intestinal epithelium is likely to occur, as documented by its uptake of an MET analogue injected in the hemocoel of the fly. We have identified the amino acid transporter Arcus/CG1139, known to transport ALA, PRO, and GLY, as being required for the retrograde transport of this MET analogue, and show that ACS is required for the timely regrowth of thinned enterocytes that may lose up to 90% of their volume. The study of ACS reveals that the intestinal epithelium is metabolically integrated, being highly sensitive to amino acid composition imbalance despite the likely exchange of amino acids between neighboring enterocytes. Besides its transporter function, ACS may play a role in interorgan communication by sharply increasing the levels of amino acids in the hemolymph 3 h p.i.
A retrograde transport of metabolites
Whereas one of the major gut physiological function is the absorption of nutrients for subsequent export to the rest of the organism, enterocytes may actually import amino acids baso-laterally in interdigestive periods and permanently as regards mammalian crypt enterocytes.27,28 The metabolic toll exerted by the enterocyte purge as well as that occurring upon starvation in enterocytes is compensated by the retrograde transport of amino acids from the rest of the organism, and likely of other metabolites, at least in the case of the purge since major organelles and cytoplasm are extruded in the gut lumen. Upon starvation, the gut epithelium does not become as thin as upon SmDb11 ingestion. Nevertheless, one may rationalize that the recovery of enterocyte functions is critical for survival so that the organism may quickly assimilate any food it might find while wandering. In the case of the purge, the simple delay of enterocyte recovery observed in acs mutants is likely related to the decreased fertility of females as well as the impaired survival upon infection, thereby testifying to the physiological relevance of fast recovery (Figures S4B, S4C, and S4H). As the purge corresponds to a defense reaction against ingested pathogens or xenobiotics, it is likely safer to reconstitute a normal epithelial barrier from internal stores rather than from potentially contaminated food.
ACS basal localization promotes the activity of this proton-assisted transporter: a low pH gradient from hemolymph to the basal side of the intestinal epithelium, peaking at 4.34 near the basal labyrinth, likely provides the proton-motive force for the import of amino acids.29 Thus, in contrast to other cell types, ACS may not necessarily function in the late endosomal/lysosomal compartment17 but rather at the membrane.
ACS mediates enterocyte regrowth through the insulin signaling pathway and Myc
The fast recovery of the intestinal epithelium after thinning can only be ascribed to regrowth since ISC compensatory proliferation is not involved.14 An alternative would be an increased ploidy of enterocytes to facilitate faster growth.22,25,26 Whereas we did find a mild requirement for EGFR function in the recovery phase, its role in growth may not be related to its role in the control of the ploidy of enterocytes since silencing the genes encoding the E2F transcription factors required for endocycling did not reveal any impaired recovery phenotype. Rather, it appears that the Insulin Signaling pathway and possibly the mTOR pathway mediate the regrowth of enterocytes through MYC, a major regulator of growth.30,31,32,33,34,35,36 Several studies have established a link between CG1139/ACS and mTOR activation,16,17,37,38 yet our genetic data do not allow us to draw an unambiguous conclusion on the existence of this link in recovering enterocytes.
Our results are consistent with a model in which MYC activity is decreased during the purge and subsequently progressively returns to original levels of expression in an acs-dependent manner. Indeed, we note that the expression of sgg is up-regulated at 3h (thin epithelium) in acs-silenced flies (Figure S5E). As sgg encodes GSK3β that negatively regulates MYC stability,39,40 we infer that MYC levels will take longer to be reinstated in acs mutant flies. Likewise, sestrin (sesn) encodes a negative feedback mTOR pathway inhibitor that works through an AMPK/Tsc2 axis.41 Its expression is also significantly upregulated at 3 and 8h but not 16h in acs mutants (Figure S5E) suggesting that it leads to a sustained repression of the mTOR pathway when the intestinal epithelium is thin. The delayed intestinal epithelium recovery phenotype in acs mutants may result from an impaired capacity to reactivate the Insulin signaling and possibly the mTOR pathways in a timely manner.
Our study highlights the importance of Myc in the recovery process given the similarity of its phenotype to that of acs. Its expression is regulated both at the transcriptional and post-transcriptional levels.42 We note that cMyc protein stability can be indirectly regulated by amino acids levels via system L transport of large, branched, aromatic or neutral amino acids in murine T lymphocytes and natural killer cells, specifically through the SLC7A5 transporter.40,43 Whereas the Drosophila SLC7A5 gene tested negative in our RNAi mini-screen, the SLC7A7 (minidiscs) and SLC7A9 (sobremesa) transporter genes yielded a phenotype nearly as pronounced as that of acs. Thus, an in-depth study of these two transporters is warranted and it will be especially interesting to determine whether their phenotypes are mediated by MYC.
ACS and homeostatic mechanism that counteract amino acid imbalance
ACS appears to function at several levels. Since it is required for the timely recovery of enterocyte shape and volume, it must be indirectly involved in the reverse metabolic imports not only of other amino acids but also of other basal metabolites to replenish the lost cytoplasm and organelles, a process that is likely coordinated. Indeed, ACS has been shown to transport ALA, GLY, and PRO. Thus, other amino acids, including methionine, are bound to be imported by other transporters. Candidates have been identified in our mini-screen (Figures S1B and S1C), with a prevalence of SLC7 family members transporting cations or belonging to the L-system as noted above. slif and mnd have been shown to be required for growth of the organism and imaginal discs respectively.44,45 MND also imports LEU into insulin-producing cells and regulates Dilp2 release and thus like SLIF plays a role in the systemic regulation of growth.46 It is also likely that other transporters fill in for ACS when mutated, albeit more slowly. Thus, when acs activity is missing, the likely decreased availability of ALA, GLY, and PRO likely limits the uptake of other metabolites in a regulatory process that remains to be delineated.
A second level of amino acid homeostasis throughout the midgut epithelium has been revealed by our clonal analysis in the midgut epithelium in which wild-type enterocytes fail to recover from the cytoplasmic purge to the same extent as acs enterocytes. Importantly, the level of recovery appeared to be approximately inversely proportional to the proportion of acs mutant cells in the epithelium. This correlation is unlikely to result from a persistence of the Arcus protein when clones are generated for a short time since the acs gene is initially expressed in only very few cells prior to infection. Whether clones have been generated for one or three days, the silencing of acs gene expression will occur upon the same time frame of the infection; only the number of epithelial cells in which acs will be silenced as its expression is induced will differ. This clonal analysis thus suggests that wild-type enterocytes are able to compensate the deficient uptake of ALA, GLY, or PRO in neighboring cells. The simplest hypothesis is to propose that amino acid can directly circulate between epithelial cells through gap junctions or alternatively by secretion and re-uptake by other amino acid transporters. A previous study in the posterior larval midgut did not reveal any electrical coupling between neighboring enterocytes, thus making unlikely the existence of gap junctions within this epithelium.47 However, the components of Drosophila gap junctions, the innexins, are expressed in the diverse types of the epithelial adult midgut48 and it will be interesting to determine whether the two major innexins expressed in enterocytes, Inx2 and Inx7, mediate the acs non cell-autonomous phenotype.
A function for ACS in interorgan communication?
acs function is required for the surge of amino acid concentration in the hemolymph shortly after exposure to SmDb11 (Figure 3C), before the strong induction of acs expression that occurs later on. The source of these amino acids has not been identified so far and hemolymphatic proteins are potential contributors that we failed to pinpoint reproducibly. As the increased concentration of amino acids in the hemolymph is temporally the first acs-dependent process we have identified, it may constitute the stimulus that coordinates the uptake of metabolites required for the regrowth of enterocytes.
Our transcriptomics analysis in wild-type guts did not allow us to identify obvious candidate signaling molecules. In acs mutants, hundreds of genes are differentially expressed upon SmDb11 ingestion, which suggests that the resilience response is perturbed and must be adapted to rise to the challenge. Given the biochemical nature of ACS, its potential signaling functions are likely mediated indirectly.
The finding of a role for acs in the generation of the increased amino acid concentration is perplexing as in unchallenged control midguts acs appears to be expressed in very few cells that might act as sentinels. Whether these cells are also required for the expanding expression of acs throughout the midgut epithelium remains to be determined. We have been unable to determine whether acs is required for its own increased expression since the frameshift KO mutant did not produce transcripts detectable by RT-qPCR or in situ hybridization, likely because of nonsense-mediated mRNA decay. How acs expression is regulated will be a major issue to address in future experiments. It will also be important to question the specificity of this ACS function, i.e., whether this phenotype is shared when mutating other transporters that appear to be also needed for recovery.
The size of a given cell type in an organ is usually rather uniform, implying there are inherent processes that regulate the control of final cell size that remain poorly understood at present.49 There are multiple mechanisms that contribute to the establishment of final cell size including nutrition, biogenesis, and most importantly cell division. Cytoplasmic extrusion as documented here provides a unique setting to study the regulation of cell size independently of most of the parameters cited above. Interestingly, we have recently established that the thickness of regrown enterocytes is slightly larger in enterocytes that have been exposed to SmDb11 hemolysin. This study establishes that cell size does not appear to be regulated intrinsically at the enterocyte level but may be a property set up at the levels of the organ or of the whole organism.
Limitations of the study
The present study has been performed under our conditions (food, exact temperatures, microbiota lacking Acetobacteraceae) and a limited number of genetic backgrounds. We have used our regular food for our experiments as a yeast-rich food we also use in our laboratory inhibits the apical cytoplasmic purge due to the presence of ethanol used to dissolve the methyl paraben preservative.50,51 The mini-genetic screen that identified Arcus relied on RNAi transgenic lines; except for acs, the other hits have not been validated using independent RNAi lines or mutants. The acs KI mutant deletes the whole locus, including the endogenous acs 3′ untranslated region, which may therefore affect the stability of the reporter transcripts as compared to the endogenous ones.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Fluorescein Isothiocyanate (FITC) | Sigma-Aldrich | #P5282 |
| Texas-Red labeled phalloidin | Invitrogen TM | #T7471 |
| Mouse anti-GFP antibody | Roche | #11814460001; RRID:AB_390913 |
| Goat anti-mouse FITC antibody | Abcam | #6785; RRID:AB_955241 |
| Rabbit anti-phospho-4EBP1 antibody | Cell Signaling | #2855; RRID:AB_560835 |
| Goat anti-rabbit FITC antibody | Abcam | #6717; RRID:AB_955238 |
| Vectashield | Vector Laboratories | H-1000 |
| Vectashield with DAPI | Vector Laboratories | H-1200 |
| Alexa Fluor 488 alkyne dye | Thermofisher | #A10267 |
| Bacterial and virus strains | ||
| Serratia marcescens Db11 (SmDb11) | Flyg et al.52 | N/A |
| Serratia marcescens 21C4 (Sm21C4) | Kurz et al.11 | N/A |
| Biological samples | ||
| Intestines | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Essential amino acids | Thermo Fischer Scientific | MEM Amino Acids Solution 50X #11130051 |
| Non-essential amino acids | Thermo Fischer Scientific | MEM Non-Essential Amino Acids Solution 100X, #11140076 |
| Chloramphenicol | Euromedex | 3886-B |
| Streptomycin | Sigma-Aldrich | S6501 |
| TRI Reagent RT | Molecular Research Center | TR118 |
| Bromoanisole | Molecular Research Center | BAN, 98%, |
| Critical commercial assays | ||
| iScript cDNA synthesis kit | Bio-Rad | #1708890 |
| iQ TM SYBR Green Supermix kit | Bio-Rad | #1525121 |
| L-amino acid quantitation kit | Sigma-Aldrich | #MAK002 |
| Click-it AHA (L-azidohomoalanine) kit | Thermofisher | #C10102 |
| Click-it Cell Reaction Buffer kit | Thermofisher | #C10269 |
| FLIC system (FLIC-MCU+DFM) | Sable Systems Europe | N/A |
| Deposited data | ||
| RNA sequencing WT 3h post-infection | Lee et al.14 | GEO Bioproject: GSE60504 |
| RNA sequencing WT 6h and 9h post-infection | This study | GEO Bioproject: PRJNA258425 |
| RNA sequencing WT vs. acs-RNAi at 3h, 8h and 16h post-infection | This study | GEO Bioproject: PRJNA821079 |
| Experimental models: Organisms/strains | ||
| Drosophila melanogaster wA5001 | Thibault et al.53 | N/A |
| D. melanogaster w1118iso | Laboratory of Luis Teixeira (Ryder et al.)54 | N/A |
| D. melanogaster NP-Gal4-tubGal80ts(also known as Myo31D) | Cronin et al., 2009; Nehme et al., 200712,13 | N/A |
| D. melanogaster NP-Gal4-tubGal80tsiso | This laboratory | N/A |
| D. melanogaster Ubi-Gal4tubGal80ts | This laboratory | N/A |
| D. melanogaster Yolk-Gal4 | This laboratory | N/A |
| D. melanogaster prospero-Gal4 | Bloomington stock center | #84276 |
| D. melanogaster esg-Gal4Gal80ts | Micchelli&Perrimon55 | N/A |
| D. melanogaster how>Gal4 | Bloomington stock center | #1767 |
| D. melanogaster mex1>Gal4 | Bloomington stock center | #91368 |
| D. melanogaster uro-Gal4 | Laboratory of Bruno Lemaitre (Li et al.)56 | N/A |
| D. melanogaster w; esgGal4tubGal80tsUAS-GFP; UAS-flp Act>CD2>Gal4 | Jiang et al.20 | N/A |
| D. melanogaster acs iso CRISPR KO | Sino-French Hoffmann Institute and this laboratory | N/A |
| D. melanogaster acs iso CRISPR KI | Well Genetics and this laboratory | N/A |
| D. melanogaster arcus-RNAi (CG1139 RNAi) | VDRC | #102363 |
| D. melanogaster arcus-RNAi iso | This laboratory | N/A |
| D. melanogaster UAS-CG1139-GFP | Gift from Prof. Deborah Goberdhan17 | N/A |
| D. melanogaster InR RNAi | Bloomington Drosophila Stock Center | #51518 |
| D. melanogaster Myc RNAi | Bloomington Drosophila Stock Center | #51454 |
| D. melanogaster S6K RNAi | Bloomington Drosophila Stock Center | #42572 |
| D. melanogaster S6K-KQ | Bloomington Drosophila Stock Center | #6911 |
| D. melanogaster TOR RNAi | Bloomington Drosophila Stock Center | #33627 |
| D. melanogaster Tsc2 RNAi | Bloomington Drosophila Stock Center | #31776 |
| D. melanogaster Atg9 RNAi | Bloomington Drosophila Stock Center | #28055 |
| D. melanogaster E2f1 RNAi | VDRC | #108837 |
| D. melanogaster E2f2 RNAi | VDRC | #100990 |
| D. melanogaster Egfr RNAi | VDRC | #107130 |
| Oligonucleotides | ||
| rp49 fw 5′GACGCTTCAAGGGACAGTATCTG-3′ | This laboratory | N/A |
| rp49 rv 5′- AAACGCGGTTCTGCATGAG3′ | This laboratory | N/A |
| acs fw 5′ACGTCAGCTTTTCGCAGGCCA-3′ | This publication | N/A |
| acs rv 5′-ACAACGCAGCCAGGGTGGAC-3′ | This publication | N/A |
| myc fw 5′ CAGTTCCAGTTCGCAGTCAA-3′ | This publication | N/A |
| myc rv 5′ AGATAAACGCTGCTGGAGGA-3′ | This publication | N/A |
| Software and algorithms | ||
| ImageJ/Fiji software | Fiji software | https://imagej.nih.gov/ij/ |
| GraphPad Prism v8.2.1 | GraphPad software | https://www.graphpad.com/ |
| CFX384 system | Bio-Rad | 1855484 |
| HISAT2 | Kim et al.57 | https://github.com/DaehwanKimLab/hisat2 |
| Bowtie2 | Langmead and Salzberg58 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| RSEM | Li et al.59 | https://github.com/deweylab/RSEM |
| DEseq2 algorithm | Love et al.60 | https://bioinformaticshome.com/tools/rna-seq/descriptions/DESeq2.html#gsc.tab=0 |
| GSEA v4.1.0 | Subramanian et al.21 | https://www.gsea-msigdb.org/gsea/index.jsp |
| Other | ||
| absorbent pads | Millipore | # AP1003700 |
| DNBseq platform | BGI | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dominique Ferrandon (D.Ferrandon@ibmc-cnrs.unistra.fr).
Materials availability
-
•
Drosophila melanogaster lines generated in this study are available upon request.
-
•
This study did not generate any unique new reagents.
Experimental model and study participant details
Drosophila melanogaster husbandry
Drosophila melanogaster flies were raised at 25°C and 60% humidity, 14h of daylight and fed a standard semi-solid cornmeal medium (6.4% (w/v) cornmeal (Moulin des Moines, France), 4.8% (w/v) granulated sugar (Erstein, France), 1.2% (w/v) yeast brewer’s dry powder (VWR, Belgium), 0.5% (w/v) agar (Sobigel, France), 0.004% (w/v) 4-hydroxybenzoate sodium salt (Merck, Germany)).
acs RNAi isso and NP iso were lines line isogenized to w1118 iso genetic background, as described in.61 The drivers were used either alone, crossed with w1118 iso as controls, or crossed with a RNAi or a reporter line. Crosses for Figures 5D, 5E, and S5B–S5I were performed by crossing the NP-Gal4-tubGal80ts driver isogenized to w1118 iso (referred as NP iso) or w1118 iso to the RNAi line of interest. Crosses were performed at 25°C except for Inr, Myc, Egfr, E2f1, E2f2 and Atg9, which were performed at 18°C. The F1 progeny of these crosses was then placed at 29°C with 70% humidity for 5 to 6 days in order to induce the expression of the Gal4tubGal80ts transgenes. For clonal analysis, the Flp-Out line w; esgGal4tubGal80ts UAS-GFP; UAS-flp Act>CD2>Gal4 from Jiang et al.20 was used. acs knock-out mutant was generated by the Sino-French Hoffmann Institute CRISPR-Cas9 platform and comprises a 5bp deletion in the sequence causing a presumably truncated non-functional protein (Figures S1E and S1F). acs knock-in mutant was generated by deleting and replacing the entire CDS region of CG1139/acs by cassette of Gal4 in Well Genetics (Figure S2A). Both mutants were isogenized to w1118 iso.
3-6 days old females were used for the experiments, unless otherwise specified in the figure legend.
Serratia marcescens
Serratia marcescens strain Db11 (SmDb11) was cultured on Luria-Bertani Broth (LB) agar plates with 100 μg/mL of streptomycin. The strain 21C4 Sm21C411 was cultured on LB-agar plates with 20 μg/mL chloramphenicol. The solid plates were placed overnight at 37°C to obtain colonies. For liquid cultures, one bacterial colony was taken off the solid plate and inoculated into 200 mL of liquid LB. These cultures were kept overnight at 37°C with agitation.
Method details
Infections
All infections were performed using a final bacterial OD 600 (Optical Density at 600 nm) of 10, except for the survival where OD = 1 was used. The OD was measured with a spectrophotometer and 50 mL of this solution was centrifuged at 4000 g/rcf for 10 minutes. The pellet was re-suspended in the appropriate volume of a 50mM sucrose solution containing 10% of LB, in order to reach a final OD = 10. 2 mL of this infection solution were added to two absorbent pads (Millipore AP1003700) that were placed at the bottom of medium-size vials (3.5 cm diameter). Twenty female flies of five to seven-day old were fed this infection solution, or sucrose 50mM as a control at 29°C.
For Figure 1A, essential amino acids (MEM Amino Acids Solution 50X, Thermo Fischer Scientific #11130051) were added to the infection solution to a final 1x concentration. The 1x solution is composed of: L-arginine hydrochloride (126.4 mg/L), L-cysteine (24 mg/L), L-histidine hydrochloride (42 mg/L), L-isoleucine (52.4 mg/L), L-leucine (52.4 mg/L), L-lysine hydrochloride (72.5 mg/L), L-methionine (15.1 mg/L), L-phenylalanine (33 mg/L), L-threonine (47.6 mg/L), L-tryptophan (10.2 mg/L), L-tyrosine (36 mg/L) and L-valine (46.8 mg/L). Non-essential amino acids (MEM Non-Essential Amino Acids Solution 100X, Thermo Fischer Scientific #11140076) were added to a final 1x concentration. The 1x solution is composed of: glycine (15 mg/L), L-alanine (17.8 mg/L), L-asparagine (26.4 mg/L), L-aspartic acid (26.6 mg/L), L-glutamic acid (29.4 mg/L), L-proline (23 mg/L) and L-serine (21 mg/L).
Fluorescent histochemical staining
Midguts were dissected in PBS and fixed for 30 minutes with 4% paraformaldehyde. Samples were washed three times with PBS-Triton X-100 0.1% (PBT 0.1%).
Actin staining
For actin staining midguts were incubated for 1h30 at room-temperature or overnight at 4°C in 10 μM Fluorescein Isothiocyanate (FITC) (Sigma-Aldrich #P5282) or Texas-Red labeled phalloidin (Invitrogen TM #T7471). Samples were then washed three times with PBT 0.1%.
GFP staining
To visualize the CG1139/ACS-GFP fusion protein, midguts were incubated with 1:500 anti-GFP (mouse) antibody (Roche #11814460001) for 3h. Samples were then washed three times with PBT 0.1% and incubated with a secondary goat anti-mouse FITC antibody (Abcam #6785) for 1h30. In the case of a co-staining with phalloidin and antibodies, the primary antibody was added alone for 3h, followed by 2h of incubation with a mix containing phalloidin and the secondary antibody. The visualization of acs KI> UAS-GFP was directly done without antibody staining.
Phospho 4EBP staining
Midguts were blocked 2h in PBT 0.1% with 2% bovine serum albumin (BSA). P-4EBP was detected with the anti-phospho-4EBP1 rabbit antibody from Cell Signaling (#2855). Midguts were incubated for 3h with 1:200 of the P-4EBP antibody, washed three times with PBT 0.1% and incubated with a secondary goat anti-rabbit FITC antibody (Abcam #6717).
All samples were mounted on diagnostic microscope slides (Thermo Fisher Scientific) with Vectashield plus DAPI (Vector Laboratories). Samples were observed using a LSM780 confocal microscope (Zeiss) or in Axioskop 2 microscope (Zeiss). All images were analyzed with the ImageJ/Fiji software.
RTqPCR
RNA was extracted from 5 or 10 midguts (without crop and Malpighian tubules) in triplicates. Midguts were crushed into 100 μL of TRI Reagent RT (Molecular Research Center) with 5% of bromoanisole (BAN, 98%, Molecular Research Center). Samples were vortexed, incubated 5 minutes at room temperature and centrifuged 10 minutes, 15 000rcf/g at 4°C. The upper phase of the samples was collected, mixed with 350 μL of isopropanol and vortexed. Tubes were centrifuged at 18 000rcf/g for 15 minutes at 4°C. The pellet was washed twice with 500μL of ethanol 70% and dried. RNAs were then re-suspended in 15μL of MilliQ water (Millipore). 1 μg of RNA was then used to generate cDNA by reverse transcription, using the iScript cDNA synthesis kit (Bio-Rad #1708890). The quantitative Polymerase Chain Reaction (qPCR) was performed with the Bio-Rad iQ TM SYBR Green Supermix kit and data were analyzed with the CFX384 system (Bio-Rad). mRNA quantitation was done by normalizing the amount of RNA detected for the gene of interest, with the control rp49 mRNA levels. The relative gene expression was calculated by normalizing the values obtained from sucrose fed flies (controls) versus challenged ones (infected with SmDb11). Primers used for rp49 were the forward 5′GACGCTTCAAGGGACAGTATCTG-3′ and the reverse 5′- AAACGCGGTTCTGCATGAG3’. Primers used for acs were the forward 5′ACGTCAGCTTTTCGCAGGCCA-3′ and the reverse 5′-ACAACGCAGCCAGGGTGGAC-3’. Primers used for myc were the forward 5′ CAGTTCCAGTTCGCAGTCAA-3′ and the reverse 5′ AGATAAACGCTGCTGGAGGA-3’.
Quantification of free amino acids
Free amino acids were measured using the L-amino acid quantitation kit from Sigma-Aldrich (#MAK002). Females flies were either starved on sterile water, kept on their normal food, fed sucrose 50mM or fed the usual infection solution for 3, 6 and 12h. The hemolymph from 20 flies were collected into 10μL of the kit buffer and processed according to the manufacturer’s instructions. Samples were run in biological duplicates.
Click-it assay
The Click-it reaction is a non-radioactive technique that allows the detection of nascent proteins. AHA corresponds to a modified amino acid mimicking methionine that is normally incorporated into proteins. The detection is based on the reaction between an azide group on the modified amino acid and the alkyne group of a fluorophore. The Click-it assay was performed with the Click-it AHA (L-azidohomoalanine) kit from Thermofisher (#C10102), the Alexa Fluor 488 alkyne dye (#A10267) and the Click-it Cell Reaction Buffer kit (#C10269). The reagents were prepared according to the manufacturer’s instructions. 50μM of AHA diluted in PBS was injected in the hemolymph of female flies, in order to observe we could later detect its presence in the midgut of flies. Flies were placed on different conditions: i) their normal food ii) sterile water iii) sucrose 50mM iv) SmDb11 diluted in sucrose 50mM. Midguts were dissected in PBS 6h post-treatment, fixed for 20 minutes in 4% paraformaldehyde and washed with PBT 0.1%. To detect the injected AHA, midguts were stained for 30 minutes (protected from light) with 0.5 mL of the following mix: 437.5μL of 1X Click-it Reaction Buffer, 10μL of CuSO 4, 50μL of Click-it Buffer Additive and 2.5μL of 1mM Alexa Fluor 488 alkyne. Midguts were washed three times with PBT 0.1% for 15 minutes and mounted on microscopy slides as previously described. Samples were immediately observed at the confocal microscope and the fluorescence intensity was measured with the ImageJ software.
Fitness parameters
The negative geotaxis assay was performed as described in Linderman et al.62: ten flies per vial were either infected with SmDb11 OD600 = 10 or fed in 50mM sucrose as a control. After 3h or 16h of feeding, they were transferred in the vials designed for the assay without anesthesia. A line was drawn at 8cm from the bottom of the tube. The number of flies crossing this line after 5 and 10 seconds was measured and plotted on a graph.
Survival assays were performed using 10 to 20 flies per vial in triplicates. The experiments were conducted with five to seven-day old adult females at 29°C with 70% humidity. The flies were fed either sucrose 50mM as a control or infected with an OD600 = 1 of SmDb11. Two absorbent pads were placed at the bottom of medium-size vials and 2mL of sucrose or bacterial solution were added to the filters. Each day, flies that were alive were counted and 200μL of 100mM sucrose was added to the vials.
Bacterial loads in the midguts or crop were accessed 16h post SmDb11 infection (OD600 = 10). Guts were dissected and pools of 3 midguts and crops were homogenized in 100μl of PBS1x and serial dilutions were performed to plate. Extracts were plated in LB agar containing 100μg/mL of streptomycin and incubated at 37°C overnight to count total number of colonies.
Food intake was accessed either by quantifying the level of ingested blue dye63 or using the FLIC system.64 To access the level of ingested blue dye, flies were either fed in 50mM sucrose or SmDb11, in a solution containing 5% of standard food blue dye. After 16h females were homogenized in 50μl of PBS1x and samples were centrifuged at maximum speed for 10min. Absorbance was accessed using the Varioscan and the wavelength was measured at 625nm. The absorbance for each sample was calculated to the relative controls, where flies were subjected to the same conditions but without blue dye. Each sample corresponds to a pool of 10 females. To access the food intake using the FLIC system we placed one fly per well feeding either in sucrose or SmDb11 solution (OD600 = 10). 12 females were accessed per condition in each experimental replicate. Number of events (groups of contiguous licks) was considered as a read-out for the food intake.
Defecation assay was adapted from a previously described assay,65 by feeding 10 females per vial for 16h either in 50mM sucrose or in SmDb11 (OD600 = 10) in a solution containing 5% of standard food blue dye. Fecal spots on the vial walls were counted.
Egg laying was measured using 1–2 days old w1118 iso and acs knock-out mutant females. Females and males were exposed for 16h to either 50mM sucrose or to SmDb11 (OD600 = 10), and after single females together with 2 males were transferred to vials with standard fly food for 24h, and flipped to new vials for another 24h. Number of eggs ovoposited was counted at 24h and at 48h after infection.
RNA sequencing
To analyze genes differentially regulated in NP > ctrl and NP > acs RNAi flies after infections, flies were infected with SmDb11 or exposed to sucrose and midguts were dissected 3h, 8h, or 12h post infection. Each sample comprised 10 midguts (without crop and Malpighian tubules) and three replicate samples were generated per condition.
Samples were sent to BGI and RNA was sequenced using the DNBseq platform. We used the Agilent 2100 Bio analyzer (Agilent RNA 6000 Nano Kit) to perform the total RNA sample quality control RNA concentration, 28S/18S, and the fragment length distribution. Firstly, we removed the reads mapped to rRNAs and obtained raw data; then, we filtered out the low quality reads (More than 20% of the bases qualities are lower than 10), reads with adaptors and reads with unknown bases (N bases more than 5%) to get the clean reads. We assembled those clean reads into Unigenes, followed with Unigene functional annotation and calculated the Unigene expression levels and SNPs of each sample. Finally, we identify DEGs (differential expressed genes) between samples and performed clustering analysis and functional annotations. After filtering the reads, clean reads were mapped to reference genome using HISAT257 and the average mapping ratio with reference genome was 94.08%. Clean reads were mapped to reference transcripts using Bowtie2,58 and gene expression level was calculated for each sample with RSEM.59 Based on the gene expression level, we used DEseq2 algorithms to identify the DEG (Differentially expression genes) between samples or groups. With DEGs we performed Gene Ontology (GO) classification, KEGG pathway classification and functional enrichment. We calculated false discovery rate (FDR) for each p value, in general, the terms which FDR not larger than 0.01 are defined as significant enriched. We extracted here genes involved in cell growth where we identified genes involved in insulin signaling and the transcription factor myc.
The average mapping ratio with reference genome was 94.08%, the average mapping ratio with gene was 80.06% and a total of 17,003 genes were detected.
Gene Set Enrichment Analysis (GSEA) was performed as described using GSEA v4.1.0.21 We used a gene set library from the FlyEnrichr that distributes the genes according to biological processes from their Gene Ontology, “GO_Biological_Process_AutoRIF_Predicted_zscore”.66,67
Quantification and statistical analysis
Classification of gut epithelium thickness
To determine the level of thinning and the recovery capacity, we used either qualitative or quantitative analysis. For qualitative analysis, midguts were classified into three different categories according to their epithelial thickness. Thick epithelium, which we represent in blue, corresponds to cells with the normal size, (around 20μm in length), with a clear dome shape characteristic of the intestine. Thin epithelium, which we represent in red, corresponds to cells that are very thin (around 5μm) and the normal dome shape of the cells is not observed. Semi-thin epithelium, which we represent in yellow, corresponds to cells with intermediate thickness (around 13μm), where the dome shape is still not completely defined. For quantitative analysis, pictures of midguts were acquired using Fluorescence Axioskope Zeiss and the length of gut enterocytes was measured using ImageJ/Fiji software. The measure for each midgut corresponds to the average of the measure between 10 enterocytes. Measures were taken every 5 to 10 enterocytes.
Statistical analyses
Statistical analyses were performed using GraphPad software Prism 6 and R. Statistical tests used are stated in the correspondent figure legends. For qualitative analysis of epithelium thickness, we used chi-square statistical tests. Survivals were analyzed using mixed effects cox models68; available at https://CRAN.R-project.org/package=coxme. Experiments including different factors were analyzed using Linear models (lm) or linear mixed-effect models (lmer, package lme4)69 in order to include the different factors of the experiment, such as the fly line or the treatment, and to include random factors such as the experimental replicates. Significance of interactions between factors was tested by comparing models fitting the data with and without the interactions, using analysis of variance (anova). Models were simplified when interactions were not significant. Pairwise comparisons of the estimates from fitted models were analyzed using lmerTest, lsmean, and multcomp packages. All experiments were performed at least three independent times and a representative experimental replicate was chosen for figure, except when indicated otherwise in the legend.
Acknowledgments
We thank Prof. Deborah Goberdhan for having kindly provided the UAS-CG1139-GFP line, the Vienna Drosophila Resource Center (VDRC, https://shop.vbc.ac.at/vdrc_store/) and Bloomington Drosophila Stock Center (NIH P40OD018537) for their resource. We are grateful to the SFHI CRISPR-Cas9 platform for generating the CG1139 KO strain. We thank Dr. Luis Teixeira for kindly providing the w1118 iso flies, as well as the isogenized balancers that allowed the isogenization of our lines. We thank Jenny Nguyen and Miriam Yamba for expert technical help in some experiments and to Dr. Roenick Olmo for the help with the GSEA analysis. We express our gratitude to Drs. Nicholas Sokol and Ishara Ariypala for expert advice on the use of the RNAscope technique in Drosophila. We are indebted to Dr. Diana Rodrigues for critical reading of the manuscript. This work has been funded by CNRS, University of Strasbourg, Agence Nationale de la Recherche (ANR) grant ANR-16-CE13-0011-01 (ENTEROCYTE_PURGE_RECOVERY), Fondation pour la Recherche Médicale (Equipe FRM DEQ20090515394 to DF) and fellowship FDT20170437224 to CS. Our work is also partially sponsored by Infinitus, Inc (China). The funding sources had no role in the design of the study nor in its execution, analyses, interpretation of the data or decision to publish the results.
Author contributions
Conceptualization: all authors except JL.
Investigation, validation: C.S., I.P., K.Z.L., S.L., M.L.
Formal analysis: I.P.
Methodology: C.S. and I.P.
Resources: J.L.
Visualization: C.S., I.P., and S.L.
Supervision: M.L. and D.F.
Writing-original draft-review and editing: I.P. and D.F. with inputs from other authors.
Project administration and Funding acquisition: D.F.
Declaration of interests
The authors declare no competing interest.
Published: July 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107490.
Supplemental information
Results from RNA-sequencing, with the log2FoldChange for all genes that are differently regulated after SmDb11 oral infection compared to respective sucrose controls for the correspondent time-points.
Data and code availability
-
•
RNA-sequencing data was deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Flint H.J., Scott K.P., Louis P., Duncan S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 2.Ryu J.H., Kim S.H., Lee H.Y., Bai J.Y., Nam Y.D., Bae J.W., Lee D.G., Shin S.C., Ha E.M., Lee W.J. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- 3.Ferrandon D. The complementary facets of epithelial host defenses in the genetic model organism Drosophila melanogaster: from resistance to resilience. Curr. Opin. Immunol. 2013;25:59–70. doi: 10.1016/j.coi.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R., Schneider D.S., Soares M.P. Disease tolerance as a defense strategy. Science. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soares M.P., Teixeira L., Moita L.F. Disease tolerance and immunity in host protection against infection. Nat. Rev. Immunol. 2017;17:83–96. doi: 10.1038/nri.2016.136. [DOI] [PubMed] [Google Scholar]
- 6.Bonfini A., Liu X., Buchon N. From pathogens to microbiota: How Drosophila intestinal stem cells react to gut microbes. Dev. Comp. Immunol. 2016;64:22–38. doi: 10.1016/j.dci.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Ren W., Rajendran R., Zhao Y., Tan B., Wu G., Bazer F.W., Zhu G., Peng Y., Huang X., Deng J., Yin Y. Amino Acids As Mediators of Metabolic Cross Talk between Host and Pathogen. Front. Immunol. 2018;9:319. doi: 10.3389/fimmu.2018.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thwaites D.T., Anderson C.M.H. The SLC36 family of proton-coupled amino acid transporters and their potential role in drug transport. Br. J. Pharmacol. 2011;164:1802–1816. doi: 10.1111/j.1476-5381.2011.01438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandasamy P., Gyimesi G., Kanai Y., Hediger M.A. Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 2018;43:752–789. doi: 10.1016/j.tibs.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Featherstone D.E. Glial solute carrier transporters in Drosophila and mice. Glia. 2011;59:1351–1363. doi: 10.1002/glia.21085. [DOI] [PubMed] [Google Scholar]
- 11.Kurz C.L., Chauvet S., Andrès E., Aurouze M., Vallet I., Michel G.P.F., Uh M., Celli J., Filloux A., De Bentzmann S., et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. Embo J. 2003;22:1451–1460. doi: 10.1093/emboj/cdg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nehme N.T., Liégeois S., Kele B., Giammarinaro P., Pradel E., Hoffmann J.A., Ewbank J.J., Ferrandon D. A Model of Bacterial Intestinal Infections in Drosophila melanogaster. PLoS Pathog. 2007;3:e173. doi: 10.1371/journal.ppat.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cronin S.J.F., Nehme N.T., Limmer S., Liegeois S., Pospisilik J.A., Schramek D., Leibbrandt A., Simoes R.d.M., Gruber S., Puc U., et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science. 2009;325:340–343. doi: 10.1126/science.1173164. 1173164 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K.Z., Lestradet M., Socha C., Schirmeier S., Schmitz A., Spenlé C., Lefebvre O., Keime C., Yamba W.M., Bou Aoun R., et al. Enterocyte Purge and Rapid Recovery Is a Resilience Reaction of the Gut Epithelium to Pore-Forming Toxin Attack. Cell Host Microbe. 2016;20:716–730. doi: 10.1016/j.chom.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Chakrabarti S., Liehl P., Buchon N., Lemaitre B. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe. 2012;12:60–70. doi: 10.1016/j.chom.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Goberdhan D.C.I., Meredith D., Boyd C.A.R., Wilson C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development. 2005;132:2365–2375. doi: 10.1242/dev.01821. [DOI] [PubMed] [Google Scholar]
- 17.Ögmundsdóttir M.H., Heublein S., Kazi S., Reynolds B., Visvalingam S.M., Shaw M.K., Goberdhan D.C.I. Proton-assisted amino acid transporter PAT1 complexes with Rag GTPases and activates TORC1 on late endosomal and lysosomal membranes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchon N., Osman D., David F.P.A., Fang H.Y., Boquete J.P., Deplancke B., Lemaitre B. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Shanbhag S., Tripathi S. Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J. Exp. Biol. 2009;212:1731–1744. doi: 10.1242/jeb.029306. 212/11/1731 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Jiang H., Patel P.H., Kohlmaier A., Grenley M.O., McEwen D.G., Edgar B.A. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. S0092-8674(09)00571-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang J., Bandura J., Zhang P., Jin Y., Reuter H., Edgar B.A. EGFR-dependent TOR-independent endocycles support Drosophila gut epithelial regeneration. Nat. Commun. 2017;8 doi: 10.1038/ncomms15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen J.K., Wang Y.T., Chan C.C., Hsieh C.W., Liao H.M., Hung C.C., Chen G.C. Atg9 antagonizes TOR signaling to regulate intestinal cell growth and epithelial homeostasis in Drosophila. Elife. 2017;6 doi: 10.7554/eLife.29338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Brien L.E., Soliman S.S., Li X., Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamamouna V., Panagi M., Theophanous A., Demosthenous M., Michail M., Papadopoulou M., Teloni S., Pitsouli C., Apidianakis Y. Evidence of Two Types of Balance between Stem Cell Mitosis and Enterocyte Nucleus Growth in the Drosophila Midgut. Development. 2020;147 doi: 10.1242/dev.189472. [DOI] [PubMed] [Google Scholar]
- 26.Bonfini A., Dobson A.J., Duneau D., Revah J., Liu X., Houtz P., Buchon N. Multiscale analysis reveals that diet-dependent midgut plasticity emerges from alterations in both stem cell niche coupling and enterocyte size. Elife. 2021;10 doi: 10.7554/eLife.64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boron W.F., Boulpaep E.L. Second edition Edition. Saunders; 2009. Medical Physiology. [Google Scholar]
- 28.Mastrototaro L., Sponder G., Saremi B., Aschenbach J.R. Gastrointestinal methionine shuttle: Priority handling of precious goods. IUBMB Life. 2016;68:924–934. doi: 10.1002/iub.1571. [DOI] [PubMed] [Google Scholar]
- 29.Shanbhag S., Tripathi S. Electrogenic H+ transport and pH gradients generated by a V-H+ -ATPase in the isolated perfused larval Drosophila midgut. J. Membr. Biol. 2005;206:61–72. doi: 10.1007/s00232-005-0774-1. [DOI] [PubMed] [Google Scholar]
- 30.Gallant P., Shiio Y., Cheng P.F., Parkhurst S.M., Eisenman R.N. Myc and Max homologs in Drosophila. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber-Agus N., Stein D., Chen K., Goltz J.S., Stevens L., DePinho R.A. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proc. Natl. Acad. Sci. USA. 1997;94:1235–1240. doi: 10.1073/pnas.94.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L., Edgar B.A., Grewal S.S. Nutritional control of gene expression in Drosophila larvae via TOR, Myc and a novel cis-regulatory element. BMC Cell Biol. 2010;11:7. doi: 10.1186/1471-2121-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teleman A.A., Hietakangas V., Sayadian A.C., Cohen S.M. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 2008;7:21–32. doi: 10.1016/j.cmet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Johnston L.A., Prober D.A., Edgar B.A., Eisenman R.N., Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierce S.B., Yost C., Britton J.S., Loo L.W.M., Flynn E.M., Edgar B.A., Eisenman R.N. dMyc is required for larval growth and endoreplication in Drosophila. Development. 2004;131:2317–2327. doi: 10.1242/dev.01108. [DOI] [PubMed] [Google Scholar]
- 36.Grewal S.S., Li L., Orian A., Eisenman R.N., Edgar B.A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 37.Goberdhan D.C.I., Wilson C., Harris A.L. Amino acid sensing by mTORC1: intracellular transporters mark the spot. Cell Metab. 2016;23:580–589. doi: 10.1016/j.cmet.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan S.J., Goberdhan D.C.I. PATs and SNATs: Amino Acid Sensors in Disguise. Front. Pharmacol. 2018;9:640. doi: 10.3389/fphar.2018.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parisi F., Riccardo S., Daniel M., Saqcena M., Kundu N., Pession A., Grifoni D., Stocker H., Tabak E., Bellosta P. Drosophila insulin and target of rapamycin (TOR) pathways regulate GSK3 beta activity to control Myc stability and determine Myc expression in vivo. BMC Biol. 2011;9:65. doi: 10.1186/1741-7007-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loftus R.M., Assmann N., Kedia-Mehta N., O'Brien K.L., Garcia A., Gillespie C., Hukelmann J.L., Oefner P.J., Lamond A.I., Gardiner C.M., et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat. Commun. 2018;9:2341. doi: 10.1038/s41467-018-04719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J.H., Budanov A.V., Park E.J., Birse R., Kim T.E., Perkins G.A., Ocorr K., Ellisman M.H., Bodmer R., Bier E., Karin M. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vervoorts J., Lüscher-Firzlaff J., Lüscher B. The ins and outs of MYC regulation by posttranslational mechanisms. J. Biol. Chem. 2006;281:34725–34729. doi: 10.1074/jbc.R600017200. [DOI] [PubMed] [Google Scholar]
- 43.Sinclair L.V., Rolf J., Emslie E., Shi Y.B., Taylor P.M., Cantrell D.A. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colombani J., Raisin S., Pantalacci S., Radimerski T., Montagne J., Léopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 45.Martin J.F., Hersperger E., Simcox A., Shearn A. minidiscs encodes a putative amino acid transporter subunit required non-autonomously for imaginal cell proliferation. Mech. Dev. 2000;92:155–167. doi: 10.1016/s0925-4773(99)00338-x. [DOI] [PubMed] [Google Scholar]
- 46.Manière G., Ziegler A.B., Geillon F., Featherstone D.E., Grosjean Y. Direct Sensing of Nutrients via a LAT1-like Transporter in Drosophila Insulin-Producing Cells. Cell Rep. 2016;17:137–148. doi: 10.1016/j.celrep.2016.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shanbhag S.R., Vazhappilly A.T., Sane A., D'Silva N.M., Tripathi S. Electrolyte transport pathways induced in the midgut epithelium of Drosophila melanogaster larvae by commensal gut microbiota and pathogens. J. Physiol. 2017;595:523–539. doi: 10.1113/JP272617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petsakou A., Perrimon N. Bioelectric regulation of intestinal stem cells. Trends Cell Biol. 2023;33:555–567. doi: 10.1016/j.tcb.2022.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ginzberg M.B., Kafri R., Kirschner M. Cell biology. On being the right (cell) size. Science. 2015;348 doi: 10.1126/science.1245075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terreri S., Sina-Rahme B., Pais I.S., Socha C., Lestradet M., Yamba W.M., Schirmeier S., Lee K.Z., Ferrandon D. A prior exposure to Serratia marcescens or xenobiotics primes Drosophila enterocytes against a recurring cytoplasmic purge. bioRxiv. 2021 doi: 10.1101/2021.10.31.466690. Preprint at. [DOI] [Google Scholar]
- 51.Franchet A., Niehus S., Caravello G., Ferrandon D. Phosphatidic acid as a limiting host metabolite for the proliferation of the microsporidium Tubulinosema ratisbonensis in Drosophila flies. Nat. Microbiol. 2019;4:645–655. doi: 10.1038/s41564-018-0344-y. [DOI] [PubMed] [Google Scholar]
- 52.Flyg C., Kenne K., Boman H.G. Insect pathogenic properties of Serratia marcescens: phage-resistant mutants with a decreased resistance to Cecropia immunity and a decreased virulence to Drosophila. J. Gen. Microbiol. 1980;120:173–181. doi: 10.1099/00221287-120-1-173. [DOI] [PubMed] [Google Scholar]
- 53.Thibault S.T., Singer M.A., Miyazaki W.Y., Milash B., Dompe N.A., Singh C.M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H.L., et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 54.Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., Drummond J., Webster J., Gubb D., Gunton N., Johnson G., et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Micchelli C.A., Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 56.Li X., Rommelaere S., Kondo S., Lemaitre B. Renal Purge of Hemolymphatic Lipids Prevents the Accumulation of ROS-Induced Inflammatory Oxidized Lipids and Protects Drosophila from Tissue Damage. Immunity. 2020;52:374–387.e6. doi: 10.1016/j.immuni.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 57.Kim D., Langmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome Bo. BMC Bioinf. 2011:12: 21–40. doi: 10.1201/b16589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferreira Á.G., Naylor H., Esteves S.S., Pais I.S., Martins N.E., Teixeira L. The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in Drosophila. PLoS Pathog. 2014;10:e1004507. doi: 10.1371/journal.ppat.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linderman J.A., Chambers M.C., Gupta A.S., Schneider D.S. Infection-Related Declines in Chill Coma Recovery and Negative Geotaxis in Drosophila melanogaster. PLoS One. 2012;7:e41907. doi: 10.1371/journal.pone.0041907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deshpande S.a., Carvalho G.B., Amador A., Phillips A.M., Hoxha S., Lizotte K.J., Ja W.W. Quantifying Drosophila food intake: comparative analysis of current methodology. Nat. Methods. 2014;11:535–540. doi: 10.1038/nmeth.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ro J., Harvanek Z.M., Pletcher S.D. FLIC: high-throughput, continuous analysis of feeding behaviors in Drosophila. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cognigni P., Bailey A.P., Miguel-Aliaga I. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 2011;13:92–104. doi: 10.1016/j.cmet.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128–619. doi: 10.1007/s00701-014-2321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmitt BMC Medical Research Methodology 2023: Therneau TM. coxme: Mixed Effects Cox Models. 2020. R package version 2.2-16. https://cran.r-project.org/web/packages/coxme/index.html.
- 69.Bates D.W., Mächler M., Bolker B., Walker S. Fitting Linear Mixed-Effects Models Using lme4. BMJ Qual. Saf. 2015;24:1–3. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results from RNA-sequencing, with the log2FoldChange for all genes that are differently regulated after SmDb11 oral infection compared to respective sucrose controls for the correspondent time-points.
Data Availability Statement
-
•
RNA-sequencing data was deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.