Abstract

Phytochemical study of the ethyl acetate root extract of Zygophyllum album has resulted in the isolation of a new saponin, Zygo-albuside D (1), along with two known compounds; (3-O-[β-D-quinovopyranosyl]-quinovic acid) (2), which is first reported in the root, and catechin (3), first reported in the genus. Their chemical structures were established by NMR and high-resolution mass spectrometry (HRMS). The new saponin (1) exhibited promising cytotoxicity with IC50 values of 3.5 and 5.52 μM on A549 and PC-3 cancer cell lines, respectively, compared to doxorubicin with IC50 values of 9.44 and 11.39 μM on A549 and PC-3 cancer cell lines, respectively. While it had an IC50 value of 46.8 μM against WISH cells. Investigating apoptosis-induction, compound 1 induced total apoptotic cell death in A549 lung cancer cells by 32-fold; 21.53% compared to 0.67% in the untreated control cells. Finally, it upregulated the pro-apoptotic genes and downregulated the antiapoptotic gene using gene expression levels. Compound 1 exhibited remarkable CDK-2 target inhibition by 96.2% with an IC50 value of 117.6 nM compared to Roscovitine. The molecular docking study further confirmed the binding affinity of compound 1 as CDK2 and Bcl2 inhibitors that led to apoptosis induction in A549 cancer cells. Hence, this study highlights the importance of compound 1 in the design of a new anticancer agent with specific mechanisms.
1. Introduction
Malignant neoplasms are among the major death causes worldwide which develop due to genome instability and mutations. Among the population, the most common types of malignancies are lung, colorectal, breast, and prostate cancer.1 Lung malignancies are the most prevalent cancer in men, as well as the fourth commonly diagnosed cancer among women.1,2 A variety of synthetic and semisynthetic anticancer medicines are accessible, but their therapeutic efficacy is limited by side effects and medication interactions. The majority of cancer chemotherapy medications are known to develop resistance and are limited by dose-limiting side effects. As a result, cancer treatment and drug discovery remain significant clinical problems.3,4
For thousands of years, many natural products, principally plants, have been used for the treatment of miscellaneous diseases.3 Natural compounds and their derivate analogues such as vinca alkaloids, paclitaxel, curcumin, and resveratrol have been recognized as successful alternatives for chemotherapeutic agents.5,6 In general, these phytochemicals act on multiple targets and can control various oncogenic transcription factors affecting tumor microenvironments that are commonly involved in cancer progression.4 The noticeable success achieved so far in utilizing natural compounds as anticancer agents has encouraged researchers to explore other secondary metabolites, such as saponins, for their antitumor activities.
Saponins have demonstrated outstanding anticancer potential both in vitro and in vivo.6−9 They exert their anticancer activities by different molecular mechanisms. Several saponins have been reported to induce cell cycle arrest in different cancer cells via modulation of cyclins and cyclin-dependent kinases (CDK) and check point proteins crucial for cell progression.6,8−11 Moreover, saponins can activate extrinsic and intrinsic pathways.6,8,9 The extrinsic pathway of apoptosis induced by saponins is mediated by the activation of Fas receptor which triggers the recruitment of pro-caspase 8 transformed into the active caspase 8. Consequently, activation of executioner caspase-3 occurred. Thus, poly-ADP-ribose polymerase was cleaved resulting in cellular components proteolysis.6 Meanwhile, saponins mediate the intrinsic route of apoptosis through the downregulation of Bcl-2 (an antiapoptotic protein) and upregulation of caspase-9 and caspase-3 (as pro-apoptotic proteins).6
Plants of genus Zygophyllum, among them Z. album L. which is an edible halophyte,12 have been praised by Egyptian traditional healers for their effectiveness for alleviating hypertension, diabetes, and rheumatism. Furthermore, evidence supporting the diverse pharmacological effects of Zygophyllum plants was acquired.13 For example, the antioxidant,14−17 anti-inflammatory,18 ganado-protective,19 and anticancer activities13,17,21 of Z. album aerial parts were proved. These reported medicinal properties of Z. album are attributed to its chemical constituents among which flavonoids,12,13,16,21−23 triterpenes,13 and saponins12,13,22,24−26 predominate.
This study investigates for the first time the root part of Z. album L. chemically and biologically, aiming to isolate natural chemical compounds with anticancer activity. The ethyl acetate fraction of the methanolic extract of Z. album roots was fractionated and chromatographed to yield three compounds. Two of them were saponins; a new one and a previously isolated one. While the third compound was catechin, which was reported for the first time in the genus Zygophyllum. The root extract of Z. album and the isolated compounds were tested for their anticancer activity.
2. Results and Discussion
2.1. Structure Elucidation of the Isolated Compounds
Structure elucidation of the isolated compounds 1, 2, and 3 was accomplished based on 1H NMR, 13C NMR, and HRMS spectral data besides their comparison with the literature. Figure 1 displays the chemical structures of isolated compounds 1–3.
Figure 1.

Chemical structures of isolated compounds.
Compound 1 was elucidated as a new compound, while compounds 2 and 3 were found to be known compounds; 3-O-[β -D-quinovopyranosyl]-quinovic acid26 and Catechin.27,28
Compound 1 (Figure 1) was isolated as a white powder. Deduced from its 1H and 13C NMR spectral data (Table 1), the molecular formula of compound 1 was C35H56O10S, which was further confirmed by 2D NMR spectroscopical analyses (Figure 2) and the HRMS which demonstrated a quasi-molecular ion [M-H]− at m/z 667.3473. The rhodizonate test gave yellow color as a positive result indicating the existence of sulfate group,20,29−32 which was further confirmed by The FTIR absorption bands in the ranges of 1330–1449 cm–1 and 1044–1086 cm–1 for S=O and C–O–S=O vibrations together with C–O–S stretch at 879 cm–1.31,33−36
Table 1. 13C NMR (125 MHz) and 1H (500 MHz) Measurements of New Compound 1 in MeOHa.
| No. | δC | δH (int., mult., JHz) | No. | δC | δH (int., mult., JHz) |
|---|---|---|---|---|---|
| 1 | 40.8 | 1.19 (1H, m) 1.76 (1H, m) | 19 | 38.5 | 2.11 (1H, dd, 15.0, 5.0) |
| 2 | 27.4 | 1.73 (1H, m) 1.08 (1H, m) | 20 | 142.6 | – |
| 3 | 78.4 | 3.15 (1H, dd, 15.0, 5.0) | 21 | 117.1 | 5.26 (1H, dd, 5.0, 10.0) |
| 4 | 41.5 | – | 22 | 37.0 | 1.82 (1H, m) 2.36 (1H, m) |
| 5 | 55.6 | 0.72 (1H, m) | 23 | 15.7 | 0.76 (3H, s) |
| 6 | 18.1 | 1.54 (1H, m) 1.36 (1H, m) | 24 | 29.3 | 0.98 (3H, s) |
| 7 | 33.8 | 1.36 (1H, m) 1.51 (1H, m) | 25 | 15.1 | 0.89 (3H, s) |
| 8 | 41.5 | – | 26 | 14.8 | 0.98 (3H, s) |
| 9 | 50.9 | 1.36 (1H, m) | 27 | 14.1 | 0.89 (3H, s) |
| 10 | 38.1 | – | 28 | 174.5 | – |
| 11 | 22.7 | 1.74 (1H, m) 1.82 (1H, m) | 29 | 21.5 | 1.08 (1H, d, 5.0) |
| 12 | 31.8 | 1.31 (1H, m) 1.22 (1H, m) | 30 | 20.8 | 1.62 (3H, s) |
| 13 | 40.8 | 2.36 (1H, m) | 1′ | 91.6 | 5.48 (1H, d, 5.2) |
| 14 | 41.5 | – | 2′ | 76.7 | 4.20 (1H, t, 5.2) |
| 15 | 27.3 | 1.22 (1H, m) 1.08 (1H, m) | 3′ | 75.8 | 3.75 (1H, dd, 8.5, 5.2) |
| 16 | 33.8 | 1.36 (1H, m) 2.32 (1H, m) | 4′ | 69.6 | 3.53 (1H, m) |
| 17 | 50.7 | – | 5′ | 60.7 | 3.72 (1H, m) 3.85 (1H, dd, 5, 10) |
| 18 | 49.2 | 1.22 (1H, m) |
Multiplicities were deduced from multiplicity-edited HSQC.
Figure 2.

Key COSY and HMBC correlations of compound 1.
Based on extensive inspection of 1D and 2D-NMR spectra, six tertiary methyl signals besides a secondary methyl one were observed at δH/C 0.76/15.7, 0.89/15.1, 0.89/14.1, 0.98/29.3, 0.98/14.8, 1.62/20.8 and 1.08 (d), J = 5.0/21.5 ascribed to H3-23/C-23, H3-25/C-25, H3-27/C-27, H3-24/C-24, H3-26/C-26, H3-30/C-30 and H3-29/C-29 respectively. Also, an oxygenated methine proton together with its corresponding carbon was observed at δH 3.15 (dd, J = 5.0, 10.0 Hz) and δC 78.4, respectively, which were attributed to H-3/C-3. An olefinic proton and its corresponding carbon were detected at δH = 5.26 (dd) and δC 117.1, respectively, which were attributed to H-21/C-21. Besides, the 13C NMR spectrum exhibited a quaternary sp2 carbon at δC 142.6 (C-20) and a carbonyl functionality at δC 174.5 (C-28). These resonances indicated an ursane aglycone with a double bond and an ester group at C-20 and C-28 respectively.37 The existence of one arabinose moiety was deduced from the 13C NMR spectral data which revealed the presence of the characteristic signal of an anomeric carbon at δC 91.6 along with four signals at δC 76.7, 75.8, 69.6 and 60.7.38 The anomeric carbon at δC 91.6 appeared shielded compared to that of l-arabinose (δC 100.7)37 due to the attachment of -SO3H functionality at C-2′. which shifts it upfield to 91.6.37,38 To ensure the identity of the sugar moiety, compound 1 was acid hydrolyzed.19,39 Then, the resulting sugar part of compound 1 was cochromatographed by PC with standard sugars. The glycone was assured to be arabinose (Rf = 0.18, 0.22, 0.29, and 0.52; eluent: BAW (n-butanol/AcOH/H2O; 4/1/5), BEW (n-butanol/EtOH/H2O; 4/1/2.2), BBPW (n-butanol/benzene/pyridine/H2O 5/1/3/3) and phenol satd. with H2O respectively).40 The α-configuration of l-arabinose was deduced from the 3JH-1′-H-2′ coupling constant (5.0 Hz) due to the diaxial interaction between the two protons in l-arabinose.41−45 Also, the FTIR spectrum further confirmed the α configuration of the arabinopyranose moiety. Since the characteristic absorption band of β-arabinopyranose in the region of 855–830 cm–1 was not observed.38
The 1H- and 13C NMR data of compound 1 were correlated with those for saponins previously reported in Zygophyllum(22,24,37,45) and confirmed by HMBC and COSY correlations (Figure 2). Accordingly, Zygo-albuside D (compound 1) was a new saponin that has not been previously isolated nor synthesized.
2.2. Biological Investigation
2.2.1. Cytotoxicity of Z. album Crude Extract and the Isolated Compounds
Samples of the crude extract of Z. album roots and pure compounds 1, 2, and 3 were tested by the MTT assay for their cytotoxicity on lung (A549) and prostate (PC-3) cancerous cells. Cytotoxicity results, as depicted in Table 2, for Z. album root extract exhibited moderate cytotoxicity on PC-3 (IC50 = 42.1 μg/mL) and A549 (IC50 = 36.4 μg/mL) cancer cells. Interestingly, the new compound (1) exhibited potent cytotoxicity with IC50 values of 3.5 and 5.52 μM, respectively, in comparison to doxorubicin as a standard (IC50 values of 9.44 and 6.19 μM, respectively). Additionally, compound (2) showed promising cytotoxicity, with IC50 values of 6.73 and 7.41 μM. On the other hand, compound (3) showed moderate cytotoxicity. Furthermore, Z. album crude extract and the isolated compounds 1–3 were not cytotoxic against normal (WISH) cells, with higher IC50 values than they were against cancer cells. Compound 1 had an IC50 value of 46.8 μM. These data demonstrated that the novel saponin, compound (1), was highly cytotoxic against A549 cells, justifying further investigation into its mechanism of action.
Table 2. Cytotoxic Activity of Crude Extract of Z. album and the Isolated Compounds against Lung, Prostate, and WISH Cell Lines Using MTT Assay.
| IC50 ± SDa |
|||
|---|---|---|---|
| Sample | Lung cancer (A549) | Prostate cancer (PC-3) | WISH (Normal cells) |
| Z. album crude extract | 36.4 ± 1.49b | 42.1 ± 2.14b | 86.9 ± 2.8a |
| Compound 1 | 3.5 ± 0.67e | 5.52 ± 1.04d | 46.8 ± 3.1b |
| Compound 2 | 6.73 ± 0.43d | 7.41 ± 0.64d | 42.6 ± 2.8c |
| Compound 3 | 66.22 ± 1.07a | 90.97 ± 1.47a | 37.8 ± 2.9d |
| Doxorubicin | 9.44 ± 0.64c | 11.39 ± 0.58c | 56.5 ± 2.7a |
Using GraphPad prism IC50 for the crude extract of Z. album are expressed as mean ± SD in terms of μg/mL. While IC50 for the isolated compounds was expressed as mean ± SD in terms of μM. The IC50 was recorded in triplicate for each sample. Means followed by different letters in the same column (vertically) are significantly different according to DMRTs at 0.05 level.
2.2.2. Investigation of the Compound 1 Apoptotic Effect on A549 Lung Cancer Cells
2.2.2.1. Annexin V/PI Staining Flow Cytometry
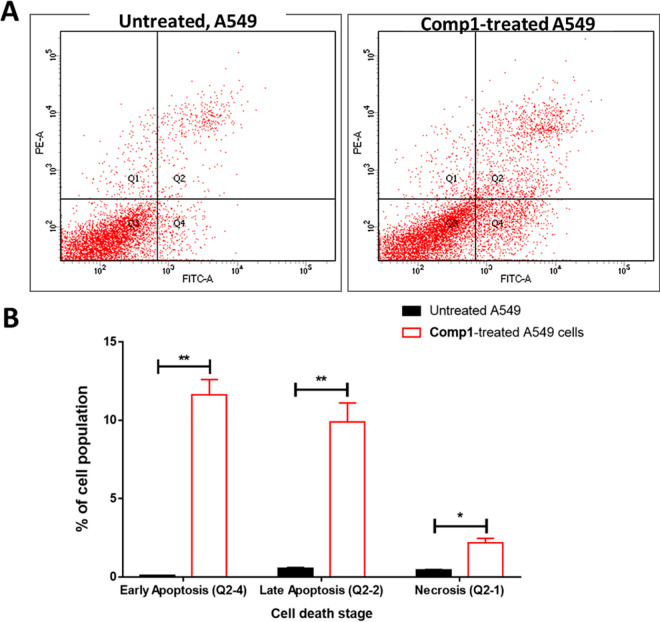
The mechanism of cytotoxicity of compound (1) against A549 cells was scrutinized by utilizing Annexin V/PI staining flowcytometry. As represented in Figure 3, compound (1) caused total apoptosis in A549 cancer cells by 21.53% compared to the untreated control cells (0.67%). These results demonstrated that treatment of A549 cells with compound (1) induced apoptotic cell death by a 32-fold change. Nonetheless, a 4.6-fold increase in necrotic cell death was observed. Our findings match earlier studies reporting the apoptotic activity of numerous naturally occurring triterpene saponins against cancer cells46−49
Figure 3.
Annexin V/PI labeling of untreated (control) and compound 1-treated A549 cells (IC50 = 2.34 μM, 48h) (A): Cytograms forapoptosis-necrosis assessment, “Q1: Necrosis, Q2: Late apoptosis, Q4: Early apoptosis’’. (B): Bar representation of apoptosis-necrosis assessment. Values are expressed as Mean ± SD of three independent trials, *(p ≤ 0.05) significant differences between treated and untreated cells employing unpaired t-test by GraphPad prism.
2.2.2.2. RT-PCR
To analyze the expression levels of apoptosis responsible genes (pro apoptotic and antiapoptotic) in the untreated and treated A549 cells, RT-PCR was employed to investigate the apoptotic effects of compound (1). As seen in Figure 4, compound 3-treatment upregulated the P53 gene by 9.28-fold; the Bax gene by 5.78-fold; and caspases 3, 8, and 9, by 6.42, 3.5, and 9.4-fold, respectively. Apoptosis-inducing activity in A549 cells after treatment was consistent with expected previous results, including a decrease in Bcl-2 gene expression by 0.42-fold.34,50,51
Figure 4.

Gene expression analysis of A549 cells treated with compound 1 (IC50 = 2.34 μM, 48h) and untreated cells. The house-keeping gene was β-actin. 2−ΔΔCT was employed to calculate the fold of change, where ΔΔCT is the difference between mean values of genes CT values in the treated and control groups. Values are expressed as the mean ± SD of three independent trials. Bars followed by different letters are significantly different according to DMRTs at the 0.05 level.
2.2.3. Investigation of Cyclin-Dependent Kinase-2 (CDK-2) Inhibition Activity of Compound 1
CDK-2 is another target for the induction of apoptosis in cancer cells by triterpene saponin. Numerous triterpene saponins inhibited and downregulated CDK-2 in various cancer cells including lung cancer cell lines.52,53 Consequently, compound 1 was evaluated for its CDK-2 inhibition activity.
As seen in Table 3, compound 1 exhibited remarkable CDK-2 target inhibition by 96.2% with an IC50 value of 117.6 nM compared to Roscovitine that caused 92.7% inhibition with an IC50 value of 140 nM. These results highlighted the CDK-2 inhibition of compound 1.
Table 3. IC50 of Compound 1 against CDK-2 Target Inhibition.
| Sample | Highest percentage of inhibition at [10 μM] | IC50 [nM] |
|---|---|---|
| Compound 1 | 96.2 ± 1.76 | 117.6a ± 1.97 |
| Roscovitine | 92.7 ± 1.45 | 140 ± 2.1 |
(p ≤ 0.05) statistically different using unpaired t test between two treated groups in GraphPad prism software. Values are expressed as mean ± SD of three independent trials.
2.3. Molecular Docking
The Bcl-2 family proteins, which include both anti- and pro-apoptotic proteins, are critical regulators of apoptosis, and a shift in the dynamic balance between these proteins can either block or promote cell death.54 B cell lymphoma 2 (Bcl-2) is a prominent apoptosis regulating protein from the Bcl-2 family. Acting as a negative regulator of apoptosis, Bcl-2 protein overexpression has been involved in tumor initiation and progression.54−56 Since it inhibits the hall marks of apoptosis including: blebbing of the plasma membrane, DNA cleavage, and nuclear condensation.54
On the other hand, CDK-2 (cyclin-dependent kinase-2) is a key protein in signaling pathways within the cell that regulate its proliferation and death. The phosphorylation of the FOXO1 protein is a key regulator of the cell cycle because it regulates the apoptotic response to DNA damage and is engaged in the transition between the G1 and G1-S phases.57 Apoptosis in cancer cells is induced by several chemotherapeutic agents through cell cycle arrest in G1 phase via CDK-2 downregulation.57,58
To put more emphasis on the apoptotic mechanism of compound (1) isolated from the Z. album root on A549 lung cancer cells, a computational molecular docking simulation was conducted to investigate the binding modes of the tested compound inside the Cyclin-dependent kinase (CDK2) and B-cell Lymphoma 2 (Bcl2). Docking results exhibited good binding affinities forming good interactive binding modes inside the protein active sites like their cocrystallized ligands. Roscovitine made arene-cation interaction with Lys 89 as the key amnio acid, while the cocrystallized ligand of Bcl-2 protein made H-bond with Arg 66 as the key amnio acid.
Interestingly, as displayed in Figure 5, compound (1) interacted with the CDK-2 protein (Binding energy = −16.48 kcal/mol), and it combined with Lys 89 via the formation of two H-bonds through the sulfate group as H-bond acceptors with distances of 2.8 and 1.3 Å, as well as it had one H-bond with Glu 8 through the hydroxyl group with distance of 1.29 Å as a H-bond donor. Moreover, it interacted with the Bcl-2 protein with two H-bonds with Arg 66 through the sulfate group as H-bond acceptors with distances of 2.48 and 2.1 Å, and the binding energy was −19.36 kcal/mol. So, molecular docking study explained the activity of compound (1) as dual inhibitors for CDK-2 and Bcl-2 proteins, and this led to apoptosis-induction in cancer cells as illustrated in anticancer activity.
Figure 5.
Ligand–receptor binding dispositions of docked compound 1 inside the CDK-2 and Bcl-2 proteins. A: Surface-view and B: Interactive view with the key amino acids Lys 89 (CDK-2) and Arg 66 (Bcl-2).
3. Materials and Methods
3.1. General Experimental Procedures
Using Bruker Avance DRX 500 MHz spectrometers (MA, USA), the 1D and 2D NMR spectral data were recorded. HRMS data were obtained utilizing a Thermo Scientific UPLC RS Ultimate 3000-Q Exactive (Thermo Fisher Scientific, Waltham, MA, USA) coupled with a Thermo Scientific Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer. Positive and negative detection was carried out independently. Solvents (analytical grade) were purchased from Fisher Scientific (Loughborough, UK). Normal phase silica gel (Fluka, St. Louis, Mo, USA, 230–400 mesh), Sephadex LH 20 (Sigma-Aldrich, St. Louis, Missouri, USA), Whatman cellulose chromatography papers, (Sigma-Aldrich, St. Louis, Missouri, USA) and TLC silica gel 60 F254 (0.2 mm, Merck, NY, USA) were employed for chromatographic investigations. The spots on the TLC were visualized first by a UV lamp at 255 and λ366 nm, and then sprayed with p-anisaldehyde/H2SO4. The paper chromatogram was sprayed with aniline phthalate reagent to visualize the sugar spots. Reference sugar standards (d, xylose (≥99%), d, ribose (≥98%), and l, arabinose (≥98%)) were obtained from Sigma-Aldrich (St. Louis, Missouri, USA).
3.2. Plant Material
During May 2019, Zygophyllum album roots were collected from Marsa Matrouh, located in the western Mediterranean coastal region of Egypt. Plant authentication was performed at Faculty of Science, Alexandria University. Under registration number of ZA-2019, a voucher specimen was placed in Pharmacognosy Department herbarium, Faculty of Pharmacy, Suez Canal University, Ismailia, Egypt.
3.3. Extraction and Isolation
About 900 g of the ground Z. album roots was extracted with methanol (3 × 2L) at 25 °C. The combined extracts were concentrated under reduced pressure to afford 30 g of brownish green crude extract.
For fractionation, about 25 g of the crude extract was dispersed in 1 L of distilled water (1L) and then extracted successively with n-hexane, CHCl3, EtOAc, and finally, n-BuOH (2L of each, three times). The fractions were concentrated under reduced pressure to obtain five fractions (ZAR-1 - ZAR-5) (5, 6, 10, and 15g respectively).
The ZAR-3 (100% EtOAc fraction, 5 g) was subjected to silica gel column chromatography initially eluted with 100% CHCl3 then gradients of CHCl3/MeOH were used until 50% MeOH in CHCl3, which yielded four subfractions (ZAR-3-a to ZAR-3-d). ZAR-3-a subfraction (1.5 g) was chromatographed on silica gel column applying gradient systems of 100% CHCl3 and MeOH until 50% MeOH in CHCl3, which yielded ZA-3-a-1 and ZA-3-a-2 subfractions. ZA-3-a-2 subfraction (260 mg) was subjected to column chromatography using silica gel as a stationary phase and gradients of CHCl3/MeOH as eluents until 100% MeOH. Then, final purification was achieved using a Sephadex LH-20 column and 50% MeOH in CHCl3 as an eluent to afford compound 1 as a colorless gum substance (60 mg) (Rf= 0.39; TLC eluent is 10% MeOH in CHCl3).
ZAR-3-b subtraction (1 g) was further fractionated applying silica gel column chromatography and CHCl3/MeOH gradients until CHCl3/ MeOH (1:1) to obtain three subfractions (ZAR-3-b-1 to ZAR-3-b-3). ZA-3-b-1 subfraction (240 mg) was rechromatographed on silica gel column and gradients of CHCl3/ MeOH initially with CHCl3/ MeOH (9:1) until 100% MeOH. For final purification, a Sephadex LH-20 column was used which was eluted by 50% MeOH in CHCl3 to give compound 2 as a colorless gum substance (25 mg) (Rf= 0.35; TLC eluent is 10% MeOH in CHCl3).
ZAR-3-c subfraction (1 g) was further fractionated by applying silica gel column chromatography and CHCl3/MeOH gradients, starting with 100% CHCl3 and ending with CHCl3/MeOH (1:1) to yield two subfractions (ZAR-3-c-1 and ZAR-3-c-2. The subfraction ZA-3-c-2 (280 mg) was further chromatographed using silica gel as a stationary phase and gradients of CHCl3/ MeOH as eluents using MeOH/CHCl3 (1:9) and culminating with 100% MeOH. A Sephadex LH-20 column eluted 50%MeOH in CHCl3 was used to achieve the final purification step, affording compound 3 as a white powder (11 m) (Rf= 0.30; TLC eluent is 10% MeOH in CHCl3).
3.4. Acid Hydrolysis of Compound 1
Compound 1 was acid hydrolyzed according to the procedure outlined in refs (19) and (39), and compound 1 (5 mg) was hydrolyzed at 95 °C using 2 N HCl (1 mL). After that, the reaction mixture was extracted with CHCl3 after dilution with H2O. Then, the aqueous phase left yielded l- arabinose, identified by comparison with authentic sugars using PC applying BAW (n-butanol/AcOH/H2O; 4/1/5), BEW (n-butanol/EtOH/H2O; 4/1/2.2), BBPW (n-butanol/benzene/pyridine/H2O 5/1/3/3) and phenol satd. with H2O respectively.40 Aniline phthalate reagent was employed for revealing the sugar spots.
3.5. Detection of the Sulfate Group
According to the method reported in refs (19 and 29), compound 1 (5 mg) was mixed with 2 N HCl (5 mL) then refluxed for 2 h followed by neutralization with dil. NaOH. After that, the reaction mixture was concentrated under reduced pressure. Then, the residue was analyzed by paper chromatography employing 90% MeOH as a developer. The chromatogram was then air-dried and treated with BaCl2 solution (100 mg dissolved in 50 mL of methanol (70%)). After drying, the chromatogram was finally sprayed with a methanolic solution of potassium rhodizoate (10 mg dissolved in 50 mL of methanol (50%)). The existence of a sulfate group was assured by the obtained yellow color.
3.6. Spectroscopic Data of the Isolated Compounds
3.6.1. Compound (1)
White powder; HRMS: m/z 667.3473 [M-H]−1H NMR (MeOH, 500 MHz) and 13C NMR (MeOH, 125 MHz) spectral data are presented in Table 1.
3.6.2. Compound (2) (3-O-[β-D-Quinovopyranosyl]-quinovic acid) [26]
White amorphous powder; 1H NMR (500 MHz, MeOD) δ 5.66 (m, 1H, H-12), 4.30 (d, J = 5.0 Hz, 1H, H-1′), 3.84 (m, 1H, 3′), 3.63 (m, 2H, 4′, 5′), 3.20 (m, 1H, 2′), 3.13 (dd, J = 5.0, 15.0 Hz, 1H), 2.98 (1H, m, H-16), 2.26 (1H, s, H-18), 2.25 (3H, m, H-9, H-15, H-16), 2.14 (1H, m, H-11), 2.08 (2H, m, H-2, H-15), 1.99 (1H, m, H-11), 1.88 (1H, m, H-2), 1.74 (1H, m, H-22), 1.71 (1H, m, H-7), 1.66 (2H, m, H-19, H-22), 1.68 (1H, m, H-7), 1.62 (1H, m, H-1), 1.48 (1H, m, H-6), 1.31 (1H, m, H-21), 1.28 (3H, s, H-23), 1.22 (1H, m, H-6), 1.19 (3H, s, H-25), 1.18 (3H, d, 5.0, H-29), 1.08 (1H, m, H-1), 0.94 (3H, s, H-24), 0.90 (1H, m, H-20), 0.85 (2H, s, H-5, H-26) and 0.77 (3H, d, 10.0, H-30).
13C NMR (125 MHz, MeOD): δC = 38.7 (C-1), 25.7 (C-2), 89.3 (C-3), 39.2 (C-4), 55.6 (C-5), 17.9 (C-6), 36.5 (C-7), 39.2 (C-8), 46.6 (C-9), 38.7 (C-10), 22.7 (C-11), 129.0 (C-12), 132.5 (C-13), 55.9 (C-14), 25.7 (C-15), 25.1 (C-16), 48.1 (C-17), 55.5 (C-18), 36.9 (C-19), 39.0 (C-20), 29.9 (C-21), 36.7 (C-22), 27.2 (C-23), 16.9 (C-24), 16.9 (C-25), 17.9 (C-26), 180.3 (C-27), 177.7 (C-28), 17.7 (C-29), 22.6 (C-30), 105.1 (C-1′), 74.5 (C-2′), 76.5 (C-3′), 75.6 (C-4′), 71.6 (C-5′), 17.8 (C-6′).
3.6.3. Compound (3) Catechin [27]
1H NMR (500 MHz, MeOD) δ 4.55 (1H, d, 6.5, H-2), 3.97 (1H, dd, 10.0, 4.5, H-3), 2.47 (1H, m, H-4a), 2.83 (1H, m, H-4b), 5.84 (1H, d, 1.5, H-6), 5.91 (1H, d, 2.0, H-8), 6.82 (1H, d, 1.5, H-2′), 6.69–6.76 (2H, m, H-5′, H-6′).
13C NMR (125 MHz, MeOD): δC = 82.9 (C-2), 68.8 (C-3), 28.6 (C-4), 157.3 (C-5), 96.2 (C-6), 157.8 (C-7), 95.5 (C-8), 156.9 (C-9), 100.8 (C-10), 132.2 (C-1′), 115.2 (C-2′), 146.3 (C-3′), 146.3 (C-4′), 116.1 (C-5′), 120.1 (C-6′).
3.7. Biological Investigation
3.7.1. MTT Assay for Evaluation of Cytotoxicity
Cell lines: A549, PC-3, and WISH were purchased from the Egyptian National Cancer Institute and grown on RPMI-1640/DMEM medium l-Glutamine (Lonza Verviers SPRL, Belgium, cat # 12-604F. Following standard tissue culture work, the cancer cells were cultured in a complete medium. Then, cells (5 × 104 per well) were seeded in triplicate in a 96-wel microplate followed by treatment with the test samples at concentrations of (0.1, 1, 10, and 100 μg/mL). The viability of the cells was determined on the second day using MTT solution (Promega, USA).59 For recording the absorbance, an ELISA microplate reader (BIO-RAD, model iMark, Japan) was utilized. GraphPad Prism 7 software was used to determine IC50 values in comparison to the control group viability, as was previously published in ref (60).
3.7.2. Investigation of Apoptosis
2.7.2.1. Annexin V/PI Staining and Cell Cycle Analysis
Treatment with compound 1 was continued for 48 h after A549 cells were incubated in 6-well culture plates (3–5 × 105 cells/well) in a humidified incubator the previous night. A suspension of the cells was prepared in 100 L of Annexin binding buffer solution 25 mM CaCl2, 1.4 M NaCl, and 0.1 M Hepes/NaOH, pH 7.4 after medium supernatants and cells were collected. Then the cell suspension was incubated with Annexin V-FITC solution (1:100) and propidium iodide (PI) at a concentration equals 10 μg/mL in the dark for 30 min. The Cytoflex FACS equipment was utilized to collect the labeled cells, and the cytExpert software was used to evaluate the results.60
3.7.2.2. Gene Expression Analysis (RT-PCR) for the Selected Genes
The expression levels of the proapoptotic genes: Caspases-3,8,9, P53 and Bax and the antiapoptotic gene: Bcl-2 was evaluated to scrutinize the apoptotic pathway in A549 cells induced by compound1; both forward and backward iterations of their sequences were depicted in Table 4. A549 cells were treated with compound 1 at its IC50 value and incubated for 48 h. A549 cells were then collected, total RNA was extracted using RNeasy Mini Kit (Qiagen, Hilden, Germany), and cDNA synthesis was performed with 500 ng of RNA using i-Script cDNA synthesis kit (BioRad, Hercules, USA) following manufacturer’s instructions. All reactions were performed for 35 cycles using the following temperature profiles 95 °C for 5 min (initial denaturation); 95 °C for 15 min (Denaturation), 55 °C for 30 min (Annealing), and 72 °C for 30 min (Extension). Findings were expressed as cycle thresholds (Ct) and Ct for estimating the gene abundance relative to β -actin (the housekeeping gene).61
Table 4. Sequences of Forward and Reverse Primers.
| Gene | Forward | Reverse | Accession number | Product size (bp) | Slope (PCR efficiency) |
|---|---|---|---|---|---|
| CASP3 | 5′- GGAAGCGAATCAATGGACTCTGG-3′ | 5′- GCATCGACATCTGTACCAGACC-3′ | NM_004346 | 304 | –3.54 (91.64%) |
| CASP8 | 5′- AGAAGAGGGTCATCCTGGGAGA-3′ | 5′- TCAGGACTTCCTTCAAGGCTGC-3′ | NM_001080125 | 263 | –3.34 (99.25%) |
| CASP9 | 5′- CGAACTAACAGGCAAGCAGC-3′ | 5′- ACCTCACCAAATCCTCCAGAAC-3′ | NR_102732 | 149 | –3.78 (83.89%) |
| TP53 | 5′- CCCCTCCTGGCCCCTGTCATCTTC-3′ | 5′- GCAGCGCCTCACAACCTCCGTCAT-3′ | NM_000546 | 1000 | –3.24 (103.54%) |
| BAX | 5′- TCAGGATGCGTCCACCAAGAAG-3′ | 5′- TGTGTCCACGGCGGCAATCATC-3′ | NM_004324 | 467 | –3.08 (111.19%) |
| BCL2 | 5′- CAAGGAGATGGAACCACTGGTG-3′ | 5′- CCGTATAGAGCTGTGAACTCCG-3′ | NM_004050 | 736 | –3.01 (114.89%) |
| BACT | 5′- CACCATTGGCAATGAGCGGTTC-3′ | 5′- AGGTCTTTGCGGATGTCCACGT-3′ | NM_001101 | 214 | –3.17 (106.76%) |
3.7.3. CDK-2 Inhibition Assay
Using a CDK2 Assay Kit (catalog no. 79599), the enzyme inhibition effect of compound 1 on CDK2 was estimated. The autophosphorylation inhibition percentage of the tested compound was measured for eight concentrations of compound 1, then the IC50 was calculated using the GraphPad prism7 software.62
3.8. Molecular Docking Study
AutoDock was used to dock the examined compounds (1–3) against the CDK-2 (PDB = 2a4l) and Bcl-2 (PDB = 4IEH) protein structures after standard procedures were followed.63 AutoDock Vina64 was employed to optimize protein and ligand structures and to favor them energetically. The binding activities were interpreted in terms of the binding energy together with ligand–receptor interactions. Then, Chimera software was utilized to visualize the binding modes.
3.9. Data Handling and Statistical Analyses
Data were handled and checked for normality using Shapiro-Wilk normality testing to check whether data were parametric or nonparametric. Data was presented as mean ± SD. Difference between untreated control and treated group was checked using an independent t test. Difference between treatments was performed using one- and two-way analysis of variance. Duncan’s Multiple Range test (DMRTs) were applied to further compare between groups at the 0.05 significance level. Data analysis was performed using IBM-SPSS version 29.0 for Mac OS.
4. Conclusion
In a pursuit of the isolation and identification of bioactive phytoconstituents, we report that the chemical investigation of Zygophyllum album roots resulted in the isolation of Zygo-albuside D (1); a new saponin together with compounds 2 and 3: 3-O-[β -D-quinovopyranosyl]-quinovic acid and catechin, which are known compounds, first reported in Z. album. The isolated compounds were screened for their cytotoxicity on PC-3 and A549 cancer cells. Among the tested compounds, compound (1) exhibited potent selective cytotoxicity with IC50 values of 3.5 and 5.52 μM on A549 and PC-3 cancer cell lines, while it had an IC50 value of 46.8 μM against WISH cells. Compound (1) exerted its cytotoxicity by triggering apoptosis. It increased the expression pro-apoptotic genes while decreasing the expression of the antiapoptotic ones. Furthermore, molecular docking study highlighted that compound (1) is an inhibitor of CDK2 and Bcl2. In brief, saponins could be regarded as gold mines of promising antineoplastic drugs; hence, compound 1 will be recommended to be further optimized and developed as target-oriented chemotherapeutic antilung cancer in future prospective.
Acknowledgments
This publication was supported by the Deanship of Scientific Research at Prince Sattam Bin Abdulaziz University, Al-Kharj, Saudi Arabia, as well as by Suez Canal University, Ismailia, Egypt.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c04314.
Figure S1: HRMS of Compound 1, Figure S2: FT-IR of Compound 1, Figure S3, 1H NMR spectrum of compound 1, Figure S4–S6: Partial expansion of 1H NMR spectrum of Compound 1, Figure S7: 13C NMR spectrum of Compound 1, Figures S8–S12: Partial expansion of 13C NMR spectrum of Compound 1, Figure S13: COSY spectrum of compound 1, Figure S14: Partial expansion of COSY spectrum of compound 1, Figure S15: HMBC spectrum of compound 1, Figures S16- S18: Partial expansion of HMBC spectrum of compound 1, Figure S19: HSQC spectrum of compound 1, Figures S19–S23: Expansion of HSQC of compound 1, Figure S24: HRMS of Compound 2, Figure S25, 1H NMR spectrum of compound 1, Figure S26–S28: Partial expansion of 1H NMR spectrum of Compound 2, Figure S29: 13C NMR spectrum of Compound 1, Figures S30–S33: Partial expansion of 13C NMR spectrum of Compound 2, S34–S36: COSY, HMBC and HSQC spectra of compound 2, Figure S37- MS of Compound 3, Figure S38 and S39: 1H NMR,13C NMR spectra of compound 3 (PDF)
Author Contributions
◆ (E.E.E., M.S.N.) Both authors share the first coauthorship.
The authors declare no competing financial interest.
Notes
The study protocol was approved by the ethical committee of the Faculty of Pharmacy at Suez Canal University (approval number: 202010PHDA1).
Supplementary Material
References
- Costea T.; Vlad O. C.; Miclea L. C.; Ganea C.; Szöllősi J.; Mocanu M. M. Alleviation of multidrug resistance by flavonoid and non-flavonoid compounds in breast, lung, colorectal and prostate cancer. Int. J. Mol. Sci. 2020, 21, 401. 10.3390/ijms21020401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi H.; Kimura R.; Matsunaga H.; Matsunaga T.; Yoshino Y.; Endo S.; Ikari A. Increase in Anticancer Drug-Induced Toxicity by Fisetin in Lung Adenocarcinoma A549 Spheroid Cells Mediated by the Reduction of Claudin-2 Expression. Int. J. Mol. Sci. 2022, 23, 7536. 10.3390/ijms23147536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S.; Dwivedi J.; Jain P. K.; Satpathy S.; Patra A. Medicinal plants for treatment of cancer: A brief review. Pharmacogn J. 2016, 8, 87. 10.5530/pj.2016.2.1. [DOI] [Google Scholar]
- Sethi G.; Shanmugam M. K.; Warrier S.; Merarchi M.; Arfuso F.; Kumar A. P.; Bishayee A. Pro-Apoptotic and Anti-Cancer Properties of Diosgenin: A Comprehensive and Critical Review. Nutrients 2018, 10, 645. 10.3390/nu10050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelhameed R. F.; Habib E. S.; Ibrahim A. K.; Yamada K.; Abdel-Kader M. S.; Ahmed S. A.; Ibrahim A. K.; Badr J. M.; Nafie M. S. Chemical constituent profiling of Phyllostachys heterocycla var. Pubescens with selective cytotoxic polar fraction through EGFR inhibition in HepG2 cells. Molecules 2021, 26, 940. 10.3390/molecules26040940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elekofehinti O. O.; Iwaloye O.; Olawale F.; Ariyo E. O. Saponins in Cancer Treatment: Current Progress and Future Prospects. Pathophysiology 2021, 28, 250. 10.3390/pathophysiology28020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.; Gao W.; Zhang Y.; Huang L.; Liu C. Chemical study and medical application of saponins as anti-cancer agents. Fitoterapia 2010, 81, 703. 10.1016/j.fitote.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Podolak I.; Grabowska K.; Sobolewska D.; Wróbel-Biedrawa D.; Makowska-Wąs J.; Galanty A. Saponins as cytotoxic agents: an update (2010–2021). Part II—Triterpene saponins. Phytochem Rev. 2022, 22, 113–167. 10.1007/s11101-022-09830-3. [DOI] [Google Scholar]
- Wang R. Current perspectives on naturally occurring saponins as anticancer agents. Arch Pharm. (Weinheim) 2022, 355, 2100469 10.1002/ardp.202100469. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Yang Y.; Lei L.; Tian M. Rhizoma paridis Saponins Induces Cell Cycle Arrest and Apoptosis in Non-Small Cell Lung Carcinoma A549 Cells. Med. Sci. Monit. 2015, 21, 2535. 10.12659/MSM.895084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T.; Hu G.; Wang A.; Sun X.; Yu X.; Jia J. Paris saponin VII induces cell cycle arrest and apoptosis by regulating Akt/MAPK pathway and inhibition of P-glycoprotein in K562/ADR cells. Phytother. Res. 2018, 32, 898. 10.1002/ptr.6029. [DOI] [PubMed] [Google Scholar]
- Ksouri W. M.; Medini F.; Mkadmini K.; Legault J.; Magné C.; Abdelly C.; Ksouri R. LC-ESI-TOF-MS identification of bioactive secondary metabolites involved in the antioxidant, anti-inflammatory and anticancer activities of the edible halophyte Zygophyllum album Desf. Food Chem. 2013, 139, 1073–1080. 10.1016/j.foodchem.2013.01.047. [DOI] [PubMed] [Google Scholar]
- Shawky E.; Gabr N.; El-gindi M.; Mekky R. A comprehensive review on genus Zygophyllum. J. Adv. Pharm. Res. 2019, 3, 1. 10.21608/aprh.2019.5699.1066. [DOI] [Google Scholar]
- Aboul-Enein A. M.; El-Ela F. A.; Shalaby E. A.; El-Shemy H. A. Traditional medicinal plants research in Egypt: Studies of antioxidant and anticancer activities. J. Med. Plant Res. 2012, 6, 689. 10.5897/JMPR11.968. [DOI] [Google Scholar]
- Kchaou M.; Ben-Salah H.; Mnafgui K.; Abdennabi R.; Gharsallah N.; Elfeki A.; Damak M.; Allouche N. Chemical composition and biological activities of Zygophyllum album (L.) essential oil from Tunisia. J. Agric Sci. Technol. 2016, 18, 1499. [Google Scholar]
- Bourgou S.; Megdiche W.; Ksouri R. The halophytic genus Zygophyllum and Nitraria from North Africa: A phytochemical and pharmacological overview. Med. Aromat Plants 2017, 3, 345. 10.1007/978-94-024-1120-1_13. [DOI] [Google Scholar]
- Ouffai K.; Azzi R.; Abboou F.; Lahfa F. B. Antihemolytic and antioxidant activities of aerial parts extracts of Zygophyllum album L. and Globularia alypum L. from Algeria. JNPRA 2022, 1, 41. 10.46325/jnpra.v1i03.27. [DOI] [Google Scholar]
- Feriani A.; Tir M.; Gomez-Caravaca A. M.; Contreras M. d. M.; Talhaoui N.; Taamalli A.; Segura-Carretero A.; Ghazouani L.; Mufti A.; Tlili N.; Allagui M. S.; et al. HPLC-DAD-ESI-QTOF- MS/MS profiling of Zygophyllum album roots extract and assessment of its cardioprotective effect against deltamethrin-induced myocardial injuries in rat, by suppression of oxidative stress-related inflammation and apoptosis via NF-κB signaling pathway. J. Ethnopharmacol 2020, 247, 112266. 10.1016/j.jep.2019.112266. [DOI] [PubMed] [Google Scholar]
- Abdelhameed R. F. A.; Fattah S. A.; Mehanna E. T.; Hal D. M.; Mosaad S. M.; Abdel-Kader M. S.; Ibrahim A. K.; Ahmed S. A.; Badr J. M.; Eltamany E. E. Zygo-Albuside A: New Saponin from Zygophyllum album L. with Significant Antioxidant, Anti-Inflammatory and Antiapoptotic Effects against Methotrexate-Induced Testicular Damage. Int. J. Mol. Sci. 2022, 23, 10799. 10.3390/ijms231810799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EL-attar M. M.; Abdel-tawab F. M.; Awad A.; Ahmad E. S.; Kamel H.; Hassan A. I. Anti-cancer Effects of Zygophyllum album and suaeda palaestina extracts on human liver cancer cell lines. Egypt J. Genet Cytol 2019, 48, 77–90. [Google Scholar]
- Mnafgui K.; Hamden K.; Ben Salah H.; Kchaou M.; Nasri M.; Slama S.; Derbali F.; Allouche N.; Elfeki A. Inhibitory Activities of Zygophyllum album: A Natural Weight-Lowering Plant on Key Enzymes in High-Fat Diet-Fed Rats. Evid.-Based Complement. Altern. Med. 2012, 2012, 620384. 10.1155/2012/620384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanean H.; El-Hamouly M.; El-Moghazy S.; Bishay D. 14-decarboxyquinovic and quinovic acid glycosides from Zygophyllum album. Phytochemistry 1993, 33, 667. 10.1016/0031-9422(93)85470-C. [DOI] [PubMed] [Google Scholar]
- Hussein S.; Marzouk M.; Ibrahim L.; Kawashty S.; Saleh N. Flavonoids of Zygophyllum album L. and Zygophyllum simplex L. (Zygophyllaceae). Biochem Syst. Ecol 2011, 39, 778. 10.1016/j.bse.2011.07.009. [DOI] [Google Scholar]
- Hassanean H. H.; Desoky E. K.; El-Hamouly M. M. A. Quinovic acid glycosides from Zygophyllum album. Phytochemistry 1993, 33, 663. 10.1016/0031-9422(93)85469-8. [DOI] [PubMed] [Google Scholar]
- Hasanean H. A.; El-Shanawany M. A.; Bishay D. W.; Franz G. Saponins from Zygophyllum album L. Bull. Pharm. Sci. Assiut 1989, 12, 117. 10.21608/bfsa.1989.71460. [DOI] [Google Scholar]
- Elgamal M. H. A.; Shaker K. H.; Pöllmann K.; Seifert K. Triterpenoid saponins from Zygophyllum species. Phytochemistry 1995, 40, 1233. 10.1016/0031-9422(95)00436-B. [DOI] [PubMed] [Google Scholar]
- Mrabti H. N.; Jaradat N.; Fichtali I.; Ouedrhiri W.; Jodeh S.; Ayesh S.; Cherrah Y.; Faouzi M. E. A. Separation, identification, and antidiabetic activity of catechin isolated from Arbutus unedo L. Plant roots. Plants 2018, 7, 31. 10.3390/plants7020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. J.; Liu Y. Q.; Li X. C.; Yang C. R. Chemical constituents of ecological tea from Yunnan. Acta Botanica Yunnanica 1995, 17, 204–208. [Google Scholar]
- Kitajima J.; Shindo M.; Tanaka Y. Two new triterpenoid sulfates from the leaves of Schefflera octophylla. Chem. Pharm. Bull. 1990, 38, 714. 10.1248/cpb.38.714. [DOI] [Google Scholar]
- Figueroa F. A.; Abdala-Díaz R. T.; Pérez C.; Casas-Arrojo V.; Nesic A.; Tapia C.; Durán C.; Valdes O.; Parra C.; Bravo-Arrepol G.; Soto L.; Becerra J.; Cabrera-Barjas G. Sulfated Polysaccharide Extracted from the Green Algae Codium bernabei: Physicochemical Characterization and Antioxidant, Anticoagulant and Antitumor Activity. Mar. Drugs 2022, 20, 458. 10.3390/md20070458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner Filho L. C.; Picão B. W.; Silva M. L. A.; Cunha W. R.; Pauletti P. M.; Dias G. M.; Copp B. R.; Bertanha C. S.; Januario A. H. Bioactive Aliphatic Sulfates from Marine Invertebrates. Mar Drugs 2019, 17, 527. 10.3390/md17090527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.-K.; Hsu C.-H.; Tsai M.-L.; Chen R.-H.; Drummen G. P. C. Inhibition of Oxidative Stress by Low-Molecular-Weight Polysaccharides with Various Functional Groups in Skin Fibroblasts. Int. J. Mol. Sci. 2013, 14, 19399. 10.3390/ijms141019399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou El Azm N.; Fleita D.; Rifaat D.; Mpingirika E. Z.; Amleh A.; El-Sayed M. M. H. Production of Bioactive Compounds from the Sulfated Polysaccharides Extracts of Ulva lactuca: Post-Extraction Enzymatic Hydrolysis Followed by Ion-Exchange Chromatographic Fractionation. Molecules 2019, 24, 2132. 10.3390/molecules24112132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyquist R. A.Interpreting infrared, Raman, and nuclear magnetic resonance spectra, Vol. 2; Academic Press: San Diego, CA, 2001; pp 85–117. 10.1016/B978-012523475-7/50185-6 [DOI] [Google Scholar]
- Ji A.; Yao Y.; Che O.; Wang B.; Sun L.; Li X.; Xu F. Isolation and characterization of sulfated polysaccharide from the Sargassum pallidum (Turn.) C. Ag. and its sedative/hypnotic activity. J. Med. Plants Res. 2011, 5, 5240–5246. [Google Scholar]
- Teodosio Melo K. R.; Gomes Camara R. B.; Queiroz M. F.; Jacome Vidal A. A.; Machado Lima C. R.; Melo-Silveira R. F.; Almeida-Lima J.; Oliveira Rocha H. A. Evaluation of Sulfated Polysaccharides from the Brown Seaweed Dictyopteris Justii as Antioxidant Agents and as Inhibitors of the Formation of Calcium Oxalate Crystals. Molecules 2013, 18, 14543. 10.3390/molecules181214543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smati D.; Mitaine-Offer A.-C.; Miyamoto T.; Hammiche V.; Lacaille-Dubois M.-A. Ursane-Type Triterpene Saponins from Zygophyllum geslini. Helv. Chim. Acta 2007, 90, 712–719. 10.1002/hlca.200790070. [DOI] [Google Scholar]
- Fernández P. V.; Quintana I.; Cerezo A. S.; Caramelo J. J.; Pol-Fachin L.; Verli H.; Estevez J. M.; Ciancia M. Anticoagulant activity of a unique sulfated pyranosic (1→ 3)-β-L-arabinan through direct interaction with thrombin. JBC 2013, 288, 223–233. 10.1074/jbc.M112.386441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. L.; Wu B.; Li H. R.; Li Y. Q.; Xu L. Z.; Yang S. L.; Kitanaka S. Triterpenoidal saponins from the barks of Zygophyllum fabago L. Chem. Pharm. Bull. 2008, 56, 858. 10.1248/cpb.56.858. [DOI] [PubMed] [Google Scholar]
- Harborne J. B.Sugars and their Derivatives. Phytochemical Methods, 2nd ed.; Chapman and Hall: London, 1984; pp 222–242. [Google Scholar]
- Silverstein R. M.; Webster F. X.; Kiemle D. J.. Spectrometric identification of organic compounds, 3rd ed.; John Wiley & Sons: New York, 1974. [Google Scholar]
- Zuo W.; Wang Q.; Li W.; Sha Y.; Li X.; Wang J. Structure elucidation and NMR assignments of an unusual triterpene saponin derivative from Ilex kudincha. Magn. Reson. Chem. 2012, 50, 325–8. 10.1002/mrc.2872. [DOI] [PubMed] [Google Scholar]
- Takeoka G. R.; Dao L. T.; Tamura H.; Harden L. A. Delphinidin 3-O-(2-O-beta-D-Glucopyranosyl-alpha-l-arabinopyranoside): a novel anthocyanin identified in Beluga black lentils. J. Agric. Food Chem. 2005, 53, 4932–4937. 10.1021/jf040493h. [DOI] [PubMed] [Google Scholar]
- Khalik S.M.A.; Miyase T.; El-Ashaal H. A.; Melek F.R. Triterpenoid saponins from Fagonia cretica. Phytochemistry 2000, 54, 853–859. 10.1016/S0031-9422(00)00168-0. [DOI] [PubMed] [Google Scholar]
- Tang L.; Jiang Y.; Chang H. T.; Zhao M. B.; Tu P. F.; Cui J. R.; Wang R. Q. Triterpene saponins from the leaves of Ilex kudingcha. J. Nat. Prod 2005, 68, 1169–1174. 10.1021/np050043z. [DOI] [PubMed] [Google Scholar]
- Zhong J.; Tan L.; Chen M.; He C. Pharmacological activities and molecular mechanisms of Pulsatilla saponins. Chinese Medicine 2022, 17, 59. 10.1186/s13020-022-00613-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthoni D. K.; Samarakoon S. R.; Piyathilaka P. C.; Rajagopalan U.; Tennekoon K. H.; Ediriweera M. K. Identification of 3-O-α-l-arabinosyl oleanolic acid, a triterpenoid saponin, as a new breast cancer stem cell growth inhibitor. Nat. Prod. Res. 2022, 36, 2923–2926. 10.1080/14786419.2021.1933971. [DOI] [PubMed] [Google Scholar]
- Simo L. M.; Messi L. M.; Mbing J. N.; Muller C. D.; Boyom F. F.; Begoudé A. B.; Pegnyemb D. E.; Haddad M.; Noté O. P. A New Triterpenoid Saponin from Albizia zygia Induced Apoptosis by Reduction of Mitochondrial Potential Status in Malignant Melanoma Cells. Planta Med. 2023, 89, 86–98. 10.1055/a-1806-2692. [DOI] [PubMed] [Google Scholar]
- Wang R. Current perspectives on naturally occurring saponins as anticancer agents. Arch. Pharm. (Weinheim, Ger.) 2022, 355, 2100469 10.1002/ardp.202100469. [DOI] [PubMed] [Google Scholar]
- Ellington A. A.; Berhow M.; Singletary K. W. Induction of macroautophagy in human colon cancer cells by soybean B-group triterpenoid saponins. Carcinogenesis. 2004, 26, 159–167. 10.1093/carcin/bgh297. [DOI] [PubMed] [Google Scholar]
- Li Q.; Li W.; Hui L. P.; Zhao C. Y.; He L.; Koike K. 13,28-Epoxy triterpenoid saponins from Ardisia japonica selectively inhibit proliferation of liver cancer cells without affecting normal liver cells. Bioorg. Med. Chem. Lett. 2012, 22, 6120–6125. 10.1016/j.bmcl.2012.08.027. [DOI] [PubMed] [Google Scholar]
- Leem D. G.; Shin J. S.; Kim K. T.; Choi S. Y.; Lee M. H.; Lee K. T. Dammarane-type triterpene ginsenoside-Rg18 inhibits human non-small cell lung cancer A549 cell proliferation via G1 phase arrest. Oncol. Lett. 2018, 15, 6043–6049. 10.3892/ol.2018.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.; Popovich D. G. Chemical and Biological Characterization of Oleanane Triterpenoids from Soy. Molecules 2009, 14, 2959. 10.3390/molecules14082959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ola M. S.; Nawaz M.; Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol. Cell. Biochem. 2011, 351, 41–58. 10.1007/s11010-010-0709-x. [DOI] [PubMed] [Google Scholar]
- Hanahan D.; Weinberg R. A. The hallmarks of cancer. Cell vol 2000, 100, 57–70. 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Vaux D. L.; Cory S.; Adams J. M. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 1988, 335, 440–442. 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- Vinod Prabhu V.; Elangovan P.; Niranjali Devaraj S.; Sakthivel K. M. Targeting apoptosis by 1,2-diazole through regulation of EGFR, Bcl-2 and CDK-2 mediated signaling pathway in human non-small cell lung carcinoma A549 cells. Gene 2018, 679, 352. 10.1016/j.gene.2018.09.014. [DOI] [PubMed] [Google Scholar]
- Li H.; Li J.; Su Y.; Fan Y.; Guo X.; Li L.; Su X.; Rong R.; Ying J.; Mo X.; Liu K.; Zhang Z.; Yang F.; Jiang G.; Wang J.; Zhang Y.; Ma D.; Tao Q.; Han W. A novel 3p22.3 gene CMTM7 represses oncogenic EGFR signaling and inhibits cancer cell growth. Oncogene 2014, 33, 3109. 10.1038/onc.2013.282. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol Methods 1983, 65, 55–63. 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Youssef E.; El-Moneim M. A.; Fathalla W.; Nafie M. S. Design, Synthesis and Antiproliferative Activity of New Amine, Amino Acid and Dipeptide-Coupled Benzamides as Potential Sigma-1 Receptor. J. IRAN CHEM SOC 2020, 17, 2515–2532. 10.1007/s13738-020-01947-6. [DOI] [Google Scholar]
- Livak K. J.; Schmittgen T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Nafie M. S.; Boraei A. T. A. Exploration of novel VEGFR2 tyrosine kinase inhibitors via design and synthesis of new alkylated indolyl-triazole Schiff bases for targeting breast cancer. Bioorg Chem. 2022, 122, 105708. 10.1016/j.bioorg.2022.105708. [DOI] [PubMed] [Google Scholar]
- Kishk S. M.; Eltamany E. E.; Nafie M. S.; Khinkar R. M.; Hareeri R. H.; Elhady S. S.; Yassen A. S. A. Design and Synthesis of Coumarin Derivatives as Cytotoxic Agents through PI3K/AKT Signaling Pathway Inhibition in HL60 and HepG2 Cancer Cells. Molecules 2022, 27, 6709. 10.3390/molecules27196709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.