Abstract
Alphaviruses are mosquito-transmitted RNA viruses that cause important diseases in both humans and livestock. Sindbis virus (SIN), the type species of the alphavirus genus, carries a 11.7-kb positive-sense RNA genome which is capped at its 5′ end and polyadenylated at its 3′ end. The 3′ nontranslated region (3′NTR) of the SIN genome carries many AU-rich motifs, including a 19-nucleotide (nt) conserved element (3′CSE) and a poly(A) tail. This 3′CSE and the adjoining poly(A) tail are believed to regulate the synthesis of negative-sense RNA and genome replication in vivo. We have recently demonstrated that the SIN genome lacking the poly(A) tail was infectious and that de novo polyadenylation could occur in vivo (K. R. Hill, M. Hajjou, J. Hu, and R. Raju, J. Virol. 71:2693–2704, 1997). Here, we demonstrate that the 3′-terminal 29-nt region of the SIN genome carries a signal for possible cytoplasmic polyadenylation. To further investigate the polyadenylation signals within the 3′NTR, we generated a battery of mutant genomes with mutations in the 3′NTR and tested their ability to generate infectious virus and undergo 3′ polyadenylation in vivo. Engineered SIN genomes with terminal deletions within the 19-nt 3′CSE were infectious and regained their poly(A) tail. Also, a SIN genome carrying the poly(A) tail but lacking a part or the entire 19-nt 3′CSE was also infectious. Sequence analysis of viruses generated from these engineered SIN genomes demonstrated the addition of a variety of AU-rich sequence motifs just adjacent to the poly(A) tail. The addition of AU-rich motifs to the mutant SIN genomes appears to require the presence of a significant portion of the 3′NTR. These results indicate the ability of alphavirus RNAs to undergo 3′ repair and the existence of a pathway for the addition of AU-rich sequences and a poly(A) tail to their 3′ end in the infected host cell. Most importantly, these results indicate the ability of alphavirus replication machinery to use a multitude of AU-rich RNA sequences abutted by a poly(A) motif as promoters for negative-sense RNA synthesis and genome replication in vivo. The possible roles of cytoplasmic polyadenylation machinery, terminal transferase-like enzymes, and the viral polymerase in the terminal repair processes are discussed.
Viruses belonging to the family Togaviridae carry positive-sense RNA genomes which are capped at their 5′ ends and polyadenylated at their 3′ ends (25, 41). Alphavirus and Rubivirus represent the two genuses grouped under the Togaviridae family (14, 41). The alphavirus genus includes some 26 different members that are transmitted primarily by mosquitoes to animals and humans (18, 25, 42). Rubella virus is the only member of the rubivirus genus and appears to infect only humans. The pathways of RNA synthesis in alphaviruses and rubiviruses are similar (14, 41). Upon entry into host cells, the incoming positive-sense genome is translated to produce viral RNA-dependent RNA polymerase (RdRp). The viral RdRp then copies the genomic RNA to produce a negative-sense intermediate. This negative-sense RNA intermediate serves as a template to produce a 49S genomic RNA and a 26S subgenomic RNA. Recognition of specific RNA motifs of viral positive-sense and negative-sense genomes by viral RdRp and host factors is thought to mediate virus-specific RNA synthesis. The availability of cDNA clones of alphaviruses and rubiviruses has facilitated the analysis of RNA motifs and proteins that regulate RNA synthesis and genome replication (6, 9, 14, 16, 22, 27, 29, 38, 42). In brief, cDNA clones of these viruses were altered to introduce specific nucleotide changes and transcribed in vitro to produce mutationally altered viral RNAs. These engineered viral RNAs were then introduced into cells in culture, and the biology of these mutations were studied (5, 42, 48).
Sequence analysis of several alphaviruses indicated the presence of a 19-nucleotide (nt) conserved element (3′CSE) adjacent to the poly(A) tail (33, 42). This 3′CSE was thought to serve as promoter for negative-sense RNA synthesis. As suggested for other eucaryotic mRNAs (17), the poly(A) tail of the alphavirus genome could contribute to genome stability and enhanced translation. Using Sindbis virus (SIN) defective interfering RNAs as templates for RNA synthesis, Levis et al. identified the importance of a 19-nt conserved element in genome replication in BHK cells (29). Using engineered SIN genomic RNA, Kuhn et al. indicated that three nucleotides located at the 5′ end of the 19-nt motif was not needed for replication (27). Kuhn et al. also reported that an insertion of 7 nt downstream of the 19-nt motif but preceding the poly(A) tail was tolerated by the polymerase (27). Recently, we reported that SIN RNA lacking the poly(A) tail (TT21s/Xho) was infectious and that the poly(A) tail was regained in vivo precisely downstream of the 3′CSE (22). We also reported that an in vitro-synthesized SIN RNA lacking the poly(A) tail (TT21s/Sst), but carrying a 1.8-kb nonviral sequence just downstream of the 3′CSE, was also infectious (22). Analysis of the virus recovered from TT21s/Sst RNA revealed the removal of the entire nonviral sequence and the addition of a poly(A) tail 3 nt downstream of the 3′CSE. These results indicated the existence of a cytoplasmic RNA processing and polyadenylation pathway in SIN-infected cells. Although the 3′ promoter elements of rubella virus have little or no homology to the alphavirus 3′ nontranslated region (3′NTR), it has recently been reported that transfection of the rubella virus genome lacking the poly(A)tail into mammalian cells resulted in the production of a poly(A)-carrying rubella virus genome (9).
To further understand the RNA repair pathway in SIN-infected cells, we developed a battery of SIN genome mutants with alterations within the 3′NTR and tested the sequence requirements for 3′-end repair. Here we report the existence of a novel 3′-end repair pathway involving the addition of AU-rich sequences to the damaged SIN 3′ terminus. The implications of these findings for RdRp-promoter interaction are discussed.
MATERIALS AND METHODS
Oligonucleotides.
A list of all oligonucleotides used in this study is given in Table 1.
TABLE 1.
List of oligonucleotides used in this study
| Name | Sequence | Locationa | Polarity |
|---|---|---|---|
| T11750 | AAAGGGAATTCCTCGAGGGGA | 3′Vec | + |
| JC 3259 | AATCAGCAGGGTCATCGC | 3′Vec | − |
| JC1000-1H | CTGCAGAAGCTTGCTGACTAGCACACGAAG | 3′S | + |
| T11350 | TAGTCAGCATCATGCTGC | 3′S | + |
| T11150 | GCGAACTTTATCGTA | 3′S | + |
| T11600B | GCAGCGTCTGCATAACTT | 3′NTR | + |
| T11200H | CTGCAGAAGCTTATGTAAACCACCAGCTGA | 3′S | − |
| 18TSac | CCTAGAGCTCAGTTTTTTTTTTTTTTTTTT | Poly(A) | − |
| 3′18Xho | CTAGCACTCGAGCAAATGTTAAAAACAAAAT | 3′CSE | − |
| 3′15Xho | CTAGCACTCGAGTGTTAAAAACAAAATTTT | 3′CSE | − |
| 3′18PA | TTTTTTTTTTAAATGTTAAAAAC | 3′CSE | − |
| 3′17PA | TTTTTTTTTTAATGTTAAAAAC | 3′CSE | − |
| 3′15PA | TTTTTTTTTTGTTAAAAACAAAAT | 3′CSE | − |
| 3′6PA | TTTTTTTTTTCAAAATTTTGTTG | 3′CSE | − |
| 3′0PA | TTTTTTTTTTTTGTTGATTAATAAA | 3′CSE | − |
| 3′PAA | CTAGCACTCGAGTTTTTTTTTTTTTTT | Poly(A) | − |
The region of the SIN genome to which the oligonucleotide belongs: 3′S, 3′ region of the S-coding region; poly(A), 3′ poly(A) tail; 3′CSE, 19-nt 3′CSE; 3′Vec, 3′ vector sequences downstream of the SIN genome in Tapa.
Plasmids.
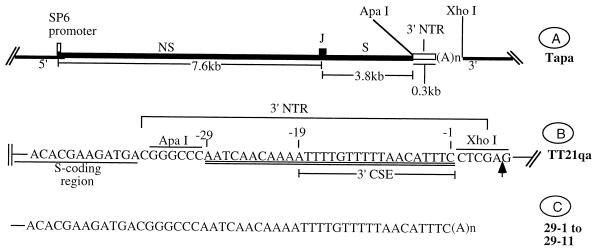
The plasmid Tapa (21–23) carries the complete protein coding region of SIN nonstructural (NS) and structural (S) genes positioned downstream of an SP6 promoter. The ApaI site located at the 5′ boundary of the 3′NTR sequence was used to introduce the desired alterations into the 3′NTR (Fig. 1A). The XhoI site located at the end of the poly(A) tail was generally used for linearizing the plasmid for in vitro synthesis of RNA transcripts. S3P (22) contains the complete 3′NTR of SIN, including the poly(A) tail flanked by ApaI and XhoI sites for easy manipulation. S3Ps is similar to S3P except that the poly(A) tail has been removed.
FIG. 1.
(A) Gene organization of Tapa, a SIN cDNA. (B) 3′ sequence of the TT21qa; NS, nonstructural genes; S, structural genes; (A)n, poly(A) tail. It is important to note that linearization of TT21qa with XhoI cleaves the DNA between A and G. Therefore, the RNA transcribed from the TT21qa/Xho construct may contain 1 to 5 bases of the XhoI motif. (C) 3′ sequence of the SIN genome recovered from BHK cells transfected with TT21qa/Xho RNA. The 11 PCR products corresponding to the viral RNA (Fig. 3A) were sequenced with JC1000-1H. The size of the poly(A) tail ranged from 17 to 39 nt. All 11 virus isolates were identical with respect to the 3′ terminal 73 nt.
(i) TT21qa.
Plasmid S3Ps was amplified with the primers JC3259 and 3′29 Apa, digested with SstI and ApaI, and ligated to the large ApaI and SstI fragment of the Tapa plasmid (22).
(ii) T3′18.
Plasmid S3Ps was amplified by PCR with primers T11750 and 3′18Xho. The resulting DNA was digested with XhoI and ligated to obtain S3P3′18. S3P3′18 was further digested with SstI and ApaI and ligated to the large SstI and ApaI fragment of Tapa to obtain T3′18.
(iii) T3′15.
Primers T11750 and 3′15Xho were used to amplify S3Ps by PCR, and the resulting DNA was digested with XhoI and ligated to obtain S3P3′15. The large SstI-ApaI fragment obtained from S3P3′15 was ligated to the corresponding large fragment of Tapa to obtain TT3′15.
(iv) S3P and T3′ derivatives carrying 3′-terminal deletions but carrying intact poly(A).
To delete bases from the 3′CSE, one of the negative-sense primers indicated below and T11750 were used to amplify the S3P plasmid. One percent of the final products was further amplified with primers T11750 and 3′PAA. These secondary PCR products were digested with XhoI and ligated. The plasmids derived from each of these ligations were sequenced to map the 3′ terminus of the 3′NTR. The ApaI-SstI fragment carrying each of the deletion mutations was ligated to the large ApaI-SstI fragment of Tapa to obtain full-length SIN cDNAs (T3′ clones) carrying 3′-terminal deletions but with intact poly(A). The deletion mutants of S3P and the negative-sense primers used in their construction were as follows: mutant S3P3′18(A)n, 3′18PA; S3P3′17(A)n, 3′17PA; S3P3′15PA, 3′15PA; S3P3′6(A)n, 3′6PA; and S3P3′0(A)n, 3′0PA.
In vitro synthesis of RNA transcripts.
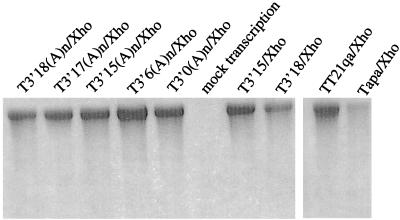
Approximately 5 μg of each plasmid was digested with an appropriate restriction enzyme and precipitated with ethanol. In vitro transcription of template DNA was carried out with 2 to 3 μg of DNA in a total volume of 30 μl as described previously (35–38). All the transcription reaction mixtures contained 1.5 mM cap analog and 1 μCi of [3H]UTP as a radioactive tracer. After 1 h of incubation at 39°C, the template DNA was digested with 1 μg of RNase-free DNase I (Life Technologies) and the RNA was purified by phenol-chloroform extraction and ethanol precipitation. The amount of RNA made was quantitated by trichloroacetic acid precipitation. Two percent of the RNA samples were denatured by glyoxal (35–38) and analyzed on a 1.25% agarose gel (Fig. 2).
FIG. 2.
Electrophoretic analysis of the in vitro-synthesized RNA transcripts. Two percent of the total RNA synthesized in a 30-μl reaction mixture was denatured with glyoxal and dimethyl sulfoxide and separated on a 1.25% gel. The gel was soaked in methanol–2,5-diphenyloxazole, dried, and fluorographed. The mock transcription reaction mixture contained no DNA template.
Cells, viruses, and infection.
BHK-21 cells and Vero cells were maintained in minimal essential medium (MEM) containing 10% fetal bovine serum. BHK cells were used for RNA transfection studies, preparation of virus stocks, and the analysis of gene expression from viruses. Since Vero cells form confluent cultures quickly and consistently, they were used for titration of virus stocks and to isolate individual virus plaques for further studies. Well-separated plaques were recovered and directly suspended in 600 μl of MEM. For virus infections, BHK cells grown in 35-mm-diameter petri plates were infected with 200 μl of the virus suspension at a multiplicity of infection (MOI) of 0.1 to 2 and incubated at 37°C for the desired times. Serial dilutions of virus stock were used for determining the virus titer on Vero cells.
Transfection of BHK cells with RNA.
As described previously (21, 22), semiconfluent BHK cells were washed twice with phosphate-buffered saline, layered with 0.2 ml of phosphate-buffered saline containing 2 to 800 ng of each of the in vitro-transcribed template RNAs and 25 μg of Lipofectin (Life Technologies) or Transfectace, and continually rocked for 30 min. At the end of transfection, the transfection mixture was removed and the cells were replenished with 2 ml of MEM containing 10% fetal bovine serum and incubated at 37°C. Cells were monitored for cytopathic effect (CPE), and culture supernatants were recovered after 2 to 3 days, clarified by centrifugation, and stored frozen. To determine specific infectivity of in vitro-synthesized RNAs, different amounts of RNA in the range of 20 ng to 2 μg were transfected into triplicate BHK cultures and allowed to incubate at 37°C for 4 h. The media were then removed and layered with 1% agarose, incubated at 37°C, and monitored for plaques over a period of 3 days. Transfection experiments which resulted in no virus production were repeated three times, and the culture supernatant derived from these transfections was passaged twice to confirm the absence of virus production.
In vivo labeling of viral RNAs and isolation and analysis of cytoplasmic RNA.
BHK cells were infected with plaque-purified viruses as described previously (22). At 2 to 3 h postinfection, 0.6 ml of MEM containing 5 μg of dactinomycin per ml was added to the plates. Twenty minutes later, 50 μCi of [3H]uridine (NEN) was added to each plate and the infection was continued at 37°C for 6 to 12 h. At the end of infection, cells were harvested and cytoplasmic RNA was isolated by disruption of cells with nonionic detergents and phenol-chloroform extraction of cytoplasmic extracts (22, 36). Approximately 4 to 6 μg of the isolated RNA was denatured with glyoxal, analyzed on a 1.25% agarose gel, and later fluorographed as previously described (21, 22).
Reverse transcription of cytoplasmic RNA, PCR amplification, and sequencing.
Cytoplasmic RNA isolated from BHK cells was reverse transcribed and amplified by PCR as previously described (21, 22). In brief, 5 to 8 μg of cytoplasmic RNA was annealed with 2 to 6 pmol of appropriate negative-sense primer in buffer containing 0.3 M NaCl and extended with murine leukemia virus reverse transcriptase. In addition to RNA and primer, the reaction mixture consisted of 50 mM Tris-HCl (pH 8.3), 70 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 0.35 mM deoxynucleotide triphosphates, and 400 U of reverse transcriptase in a total volume of 30 μl. The reaction mixture was incubated for 1 h at 37°C and subsequently for 20 min at 42°C. At the end of the incubation period, a 2-μl aliquot of the 10-fold-diluted first-strand reaction mixture was amplified by PCR. The PCR mixture consisted of 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 2 mM dithiothreitol, 100 μg of gelatin per ml, 2 to 5 pmol each of primers, 5 U of Taq polymerase, and 350 μM deoxynucleotide triphosphates in a volume of 50 μl. After 20 cycles of amplification, the reaction mixture was removed and 5% of the PCR products were analyzed on a gel. As previously described (21, 22), several control PCRs and nested PCRs were routinely used to ascertain the authenticity of the PCR products. Ten percent of the PCR products were isolated from a low-melting-point agarose gel and purified by phenol-chloroform extraction and precipitation with ethanol. The isolated DNA fragment was subjected to cycle sequencing by use of a Perkin-Elmer sequencing kit. The PCR products were also digested with suitable restriction enzymes encoded within the primers and cloned in plasmids Gem 3 or Gem 4 (Promega). DNA isolated from 1.5-ml cultures was subjected to cycle sequencing to confirm the sequences.
RESULTS
The 3′ 29 nt of the SIN genome contains signals for polyadenylation.
Previously, we showed that TT21qa/Xho RNA, which was composed of the entire protein coding sequence of the SIN genome and the terminal 29 nt of the 3′NTR, was infectious (22). The virus generated from TT21qa/Xho was plaque purified, and six representative plaques from two different virus stocks were used to infect BHK cells. [3H]uridine-labelled viral RNA was isolated from infected cells and analyzed on a denaturing agarose gel (Fig. 3A). Both genomic and subgenomic RNAs were demonstrable for each representative of the plaque-purified virus. The 12th plaque suspension did not produce any CPE on BHK cells, probably due to defective plaque recovery. Since all of the viable SIN isolates tested in our laboratory contained a poly(A) tail, we assumed that all these revertant viruses carried a poly(A) tail. Therefore, the cytoplasmic RNA was reverse transcribed with a poly(T) primer (18TSac) and amplified by PCR using primer 18TSac and primer T11150, which annealed to the 3′ region of the S coding region. The 0.25-kb amplification product which corresponds to the 3′ region of the SIN genome was purified to remove the primers and sequenced by using T11350. As shown in Fig. 1C, all the PCR products contained the 3′29-nt region and a poly(A) tail. None of the 11 individual virus isolates tested carried any other modifications within their 3′ terminus. RNA derived from uninfected cultures or the use of in vitro-made TT21qa/Xho lacking the poly(A) tail failed to produce any products upon RT-PCR (data not shown). These results indicated that the TT21qa/Xho RNA upon transfection into BHK cells underwent polyadenylation in vivo and released infectious virus. These results also indicated that the terminal 29-nt region of the SIN genome carried the minimal RNA signals required for polyadenylation, negative-sense RNA synthesis, and genome replication. Previously, we showed that a SIN genome carrying 24 nt of the 3′ terminus (TT21k/Xho) but lacking a poly(A) tail was noninfectious (22). Since a SIN genome carrying a 14-nt 3′CSE and a poly(A) tail was infectious (22), the lack of polyadenylation could have been the primary reason for the loss of infectivity of TT2k/Xho (22). These results also demonstrated that the de novo polyadenylation required specific 3′ motifs of the SIN genome.
FIG. 3.
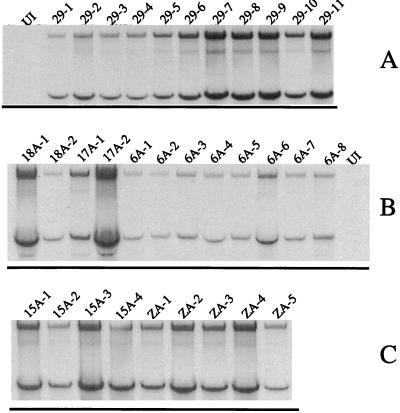
Expression of genomic and subgenomic SIN RNAs from representative virus isolates. Cultures of BHK cells were infected (MOI of 0.1 to 2.4) with the indicated virus isolates and labelled with [3H]uridine in the presence of dactinomycin. Approximately 4 to 6 μg of [3H]uridine-labelled cytoplasmic RNA was denatured with glyoxal, separated on a 1.25% agarose gel, and fluorographed. The upper band in each lane corresponds to the genomic RNA (49S), and the lower band corresponds to the subgenomic RNA (26S). The identity of the virus isolate is indicated at the top of each lane. UI, uninfected cell RNA. (A) Isolates 29-1 to 29-11, viral plaques derived from TT21qa/Xho; (B) plaques derived from T3′18(A)n, T3′17(A)n, and T3′6(A)n; (C) plaques derived from T3′15(A)n and T3′0(A)n. It is important to note that the amount of RNA made by each isolate is due mostly to the MOI, although base changes in recovered viruses could also be partly responsible.
Polyadenylation and repair of a SIN mutant carrying a G instead of C at the −1 position.
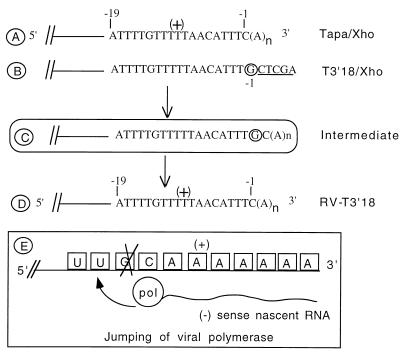
The 3′ end of the 3′CSE of most alphaviruses carries a C residue (27, 33, 34, 42). This nucleotide is numbered −1 in the 19-nt motif (Fig. 1A) (27). In addition, the 3′ conserved motif of all alphaviruses is composed of strictly an AU-rich region (27, 42). These observations suggest that the polyadenylation signal may involve the terminal C residue and probably other residues in the vicinity. To test if this C residue at the −1 position is critical for polymerase recognition and polyadenylation, the construct T3′18 was made. As shown in Fig. 4B, the linearized plasmid T3′18/Xho carries a G residue at the −1 position and a C residue at the +1 position. The +2 to +6 positions carried nonviral sequences instead of a poly(A) tail (Fig. 4). The resulting plasmid T3′18 was linearized with XhoI, transcribed in vitro (Fig. 2), and tested for its infectivity in BHK cells. Although the specific infectivity of this T3′18/Xho RNA was 127-fold lower than that of Tapa/Xho (wild-type RNA) (Table 2, experiment 16), infectious virus was consistently produced in all three transfections tested. BHK cells were infected with two virus isolates from each transfection experiment, and the expression of both genomic and subgenomic RNA was confirmed (Fig. 3B, lanes 1 and 2). The 3′ terminus of the genomic RNA of all six viral isolates derived from T3′18/Xho (RV-T3′18) indicated the reintroduction of a C residue at the −1 position and the elimination of a G residue at the −2 position (Fig. 4D). As described in Fig. 4E, the straightforward explanation of this observation would be polyadenylation of RNA just downstream of the C residue and removal of the G residue by polymerase jumping events during RNA replication steps. It should be noted that the TT21s/Xho RNA which carried a poly(A)-lacking SIN RNA with an intact 3′CSE and 1 to 5 nonviral bases (as in T3′18/Xho) downstream of the 3′CSE also gave rise to a SIN genome with wild-type 3′CSE and a poly(A) tail (22). These results indicated that polymerase recognition of the 3′ terminus of the SIN genome can occur even when the terminal C residue is displaced from its original context of UUUC to UUUGC followed by 1 to 5 nonviral bases at the extreme 3′ end.
FIG. 4.
Polyadenylation of the SIN genome carrying a G residue at the −1 position of the 3′CSE. (A) Sequence of the 3′CSE in Tapa. (B) Sequence of the 3′ terminus in T3′18/Xho. Insertion of a G residue in the −1 position of the 3′CSE and of a XhoI site at the 3′ terminus results in the introduction of a C residue in the +1 position. (C) Proposed intermediate during the formation of RV-T3′18 type revertant viruses. (D) Sequence of the 3′ terminus of all six viral isolates indicating the loss of the G residue and polyadenylation at the C residue. (E) Proposed polymerase jumping event during negative-strand synthesis and the loss of the G residue.
TABLE 2.
Structure and properties of SIN mutantsa
| Expt No. | Construct | 3′NTR | Poly(A) tail | Relative infectivityb | In vivo 3′ modificationsc | Reference |
|---|---|---|---|---|---|---|
| 1 | Tapa/Xho | −1 to −310 | Yes | 6,000 | None | 22 |
| 2 | TT21g/Xhod | −290 to −310 | No | 0 | None | 22 |
| 3 | TT21h/Xho | −290 to −310 | Yes | 0 | None | 22 |
| 4 | TT21i/Xho | −1 to −5 | Yes | 0 | None | 22 |
| 5 | TT21j/Xho | −1 to −10 | Yes | 0 | None | 22 |
| 6 | TT211/Xho | −1 to −14 | Yes | 79 | None | 22 |
| 7 | TT21m/Xho | −1 to −16 | Yes | 74 | None | 22 |
| 8 | TT21n/Xho | −1 to −20 | Yes | 328 | None | 22 |
| 9 | TT21q/Xho | −1 to −28 | Yes | 1,252 | None | 22 |
| 10 | TT21r/Xho | −1 to −63 | Yes | 3,671 | None | 22 |
| 11 | TTk/Xho | −1 to −24 | No | 0 | None | 22 |
| 12 | TT21s/Xho | −1 to −310 | No | 36 | PA | 22 |
| 13 | TT21ra/Xho | −1 to −63 | No | 41 | PA | 22 |
| 14 | TT21qa/Xho | −1 to −29 | No | 20 | PA | 22 |
| 15 | TT21Bgl-2 | −1 to −310 | Yes | 0 | None | 22 |
| 16 | T3′18/Xho | −2 to −310 | No | 47 | PA | Present work |
| 17 | T3′15/Xho | −5 to −310 | No | 24 | PA, R | Present work |
| 18 | T3′18(A)n/Xho | −2 to −310 | Yes | 19 | PA | Present work |
| 19 | T3′17(A)n/Xho | −3 to −310 | Yes | 11 | PA, R | Present work |
| 20 | T3′15(A)n/Xho | −5 to −310 | Yes | 26 | PA, R | Present work |
| 21 | T3′6(A)n/Xho | −14 to −310 | Yes | 43 | PA, R | Present work |
| 22 | T3′0(A)n/Xho | −20 to −310 | Yes | 37 | PA, R | Present work |
| 23 | Tapa/Xho (DNA control)e | −1 to −130 | Yes | 0 | None | Present work |
Duplicate cultures of BHK cells were transfected with 50 ng, 250 ng, or 2 μg of in vitro-synthesized RNA, overlaid with agarose, incubated at 37°C, and monitored for plaque formation over a period of 4 days. The number of plaques per microgram of RNA varied up to twofold among dilutions, suggesting an alteration in transfection efficiencies between diluted and undiluted RNA samples. Irrespective of the transfection efficiencies of individual test RNAs, the representative plaque-purified viruses tested yielded high-titer virus stocks (107 to 109 PFU/ml).
The number of plaques per microgram of test RNA relative to the value for Tapa/Xho. The value for the Tapa/Xho was set at 6,000 to depict relative infectivities of test RNA in whole numbers. The value represents the average of two experiments.
PA, polyadenylation; R, 3′ repair by AU additions.
TT21g/Xho carries a poly(U) at its 3′ terminus.
Mock transcription reaction carried out in the absence of SP6 RNA polymerase.
Repair of a terminally truncated SIN genome.
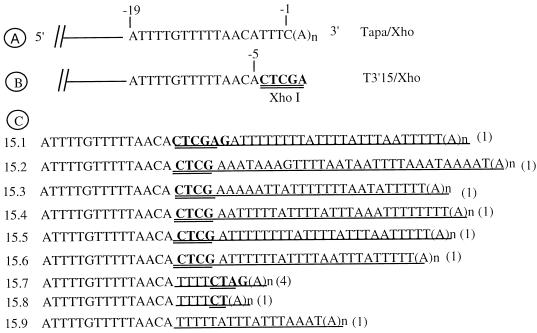
Studies reported in the previous section indicated that a SIN genome lacking a poly(A) tail and with a 3′-terminal C residue displacement was infectious. To further test the 3′ motifs necessary for polyadenylation, we designed a mutant (T3′15/Xho) in which the poly(A) tail and the terminal four nucleotides of the conserved 19-nt motif were deleted and fused to a XhoI sequence motif (Fig. 5B). The plasmid DNA was linearized with XhoI, and the corresponding RNA was synthesized in vitro. Triplicate cultures of BHK cells were transfected with 50 ng, 200 ng, and 1 μg of in vitro-synthesized RNA and monitored for CPE. Significant CPE was observed in all cultures by the third day of transfection. As expected, no virus was recovered from a control RNA lacking the entire 3′ end of the SIN genome (Table 2, experiment 2). Twelve virus suspensions corresponding to individual plaques were used to infect BHK cells, and the virus-specific RNA was isolated, amplified by reverse transcription (RT)-PCR using primers 18TSac and T11350, and sequenced.
FIG. 5.
Terminal repair of a 3′ truncated SIN genome. (A) Wild-type sequence of the 3′CSE in Tapa; (B) sequence of the 3′ terminus of T3′15/Xho indicating the deletion of the −1 to −4 positions and the insertion of a XhoI site at the terminus; (C) names and sequences of viral isolates generated from T3′15/Xho. The single underline indicates the AU-rich sequences and the poly(A) tail added at the 3′ terminus. The remnants of the XhoI motif are highlighted with a double underline. The number in parentheses at the end of each sequence corresponds to the number of viral isolates containing the same sequence. The sequences for isolates 15.1, 15.2, 15.3, and 15.7 (two separate isolates) were from experiment 1, those for isolates 15.4, 15.5, 15.7, 15.8, and 15.9 were from experiment 2, and those for isolates 15.6 and 15.7 were from experiment 3.
As shown in Fig. 5C, all of the viral isolates carried a poly(A) tail. A variety of AU-rich sequences were detected just upstream of the poly(A) tail. Different types of 3′ modifications were demonstrable from the sequence analysis. In the type I modification, as exemplified by isolate 15.1 (Fig. 5C), U-rich RNA motifs and a poly(A) tail were added without any loss of 3′-terminal sequences. In fact, the five bases corresponding to the XhoI motif at the 3′ end were retained (Fig. 5C, isolate 15.1). In the type II modification of the 3′ end, one or two terminal bases of the transfected RNA were lost with the concomitant addition of AU-rich motifs and a poly(A) tail. At least 5 of the 12 isolates (isolates 15.2 to 15.6) (Fig. 5C) belong to the type II modification. In both type I and type II modifications, the addition of bases occurred just downstream of the 3′ terminus of the parental RNA. Islands of oligo(A) and oligo(U) account for much of the nucleotide additions. In the type III modification, base deletion of the parental RNA was accompanied by the addition of AU-rich motifs internal to the 3′ terminus. For example, in isolate 15.7 (Fig. 5C), the internal bases CG of the terminal CTCGAG motif were lost, resulting in CTAG. As described previously (Fig. 4E), polymerase jumping events during viral RNA synthesis could have caused this loss. Interestingly, an oligo(U) motif was inserted 5′ to the CTAG motif, and a poly(A) tail was added downstream of the CTAG motif. Four separate isolates from three different transfections carried identical modifications of this kind. In the case of isolate 15.8, the deletion of the terminal CGAG motif was accompanied by the insertion of an oligo(U) motif 5′ to the CU terminus and the addition of a poly(A) tail downstream of the CU motif. In the type IV modification, as seen with isolate 15.9, the entire parental motif CTCGAG was lost and accompanied by the addition of AU-rich motifs and a poly(A) tail. These results indicate the existence of a pathway for polyadenylation and AU additions on terminally truncated SIN genomes in vivo.
Addition of AU-rich motifs to 3′-end mutants of polyadenylated SIN genomes.
From the studies reported in the previous section, it is clear that polyadenylation and addition of AU-rich motifs take place on SIN genomes lacking the 3′ poly(A) tail and a few of the adjoining conserved bases. Our previous results (22) (Table 2, experiment 12) showed that the SIN genome lacking only the poly(A) tail but carrying the entire wild-type 3′NTR (TT21s/Xho) was able to undergo polyadenylation but not any AU additions. Therefore, we hypothesized that the addition of AU-rich motifs occurs only when the 3′ 19-nt motif is significantly altered. But, it was not known whether AU additions could occur on 3′-end mutants of the SIN genome with an intact poly(A) tail. To test whether AU additions occur on polyadenylated 3′ SIN mutants, we developed a series of constructs carrying 1, 2, 4, 13, or 19 terminal base deletions of the 3′CSE but carrying an intact poly(A) tail (Fig. 6 and 7). BHK cells were transfected with different in vitro-synthesized RNAs and incubated for 3 days at 37°C. Mild to severe CPE was observed in all of the cultures. Control cultures transfected with TT21h/Xho (Table 2, experiment 3), a SIN genome lacking the 3′NTR but carrying a poly(A) motif, did not produce any CPE or virus. Similarly, cultures transfected with a mutant SIN RNA defective in polymerase expression also failed to produce any CPE or virus (Table 2, experiment 15). Plaque-purified virus isolates were used to infect fresh BHK cells, and viral RNA was labelled with [3H]uridine and analyzed (Fig. 3B and C). All virus isolates supported the synthesis of both 49S and 26S RNA. The 3′NTRs of these virus isolates were amplified by RT-PCR using primers 18TSac and T11350 and sequenced by using primer T11600B. As a control, all of the in vitro-synthesized template RNAs, pretreated with DNase, were subjected to RT-PCR and sequencing to rule out SP6 polymerase- or RT-PCR-mediated nucleotide additions or deletions.
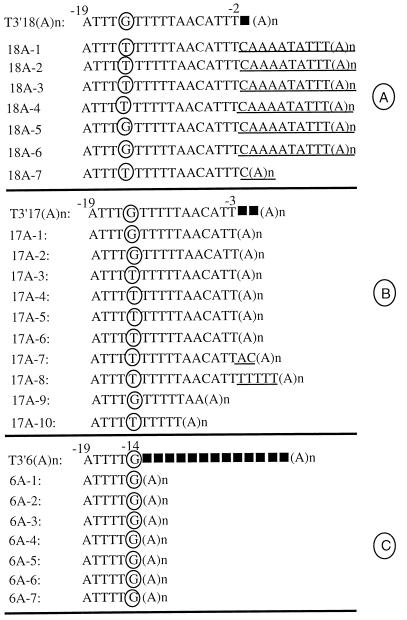
FIG. 6.
Addition of AU-rich motifs to 3′ mutants of polyadenylated SIN genome. The 3′ sequence of the template used for transfection of BHK cells and the 3′ sequence of the viral isolates recovered from the cells are given for each group. The positions of the base deletions introduced in the parental template are indicated as filled squares. The circled nucleotides highlight the base changes encountered in the revertant virus. The AU-rich sequences and poly(A) tail added to the 3′ end are marked with an underline. (A) T3′18(A)n and virus isolates derived from it. Isolates 18A-1 to 18A-3 correspond to experiment 1 and isolates 18A-4 to 18A-7 correspond to experiment 2. (B) T3′17(A)n and virus isolates derived from it. Isolates 17A-1 to 17A-4, 17A-7, and 17A-10 were derived from experiment 1. Isolates 17A-5, 17A-6, 17A-8, and 17A-9 were derived from transfection experiment 2. (C) T3′6(A)n and virus isolates derived it. The first three isolates were derived from experiment 1, and the rest were derived from experiment 2.
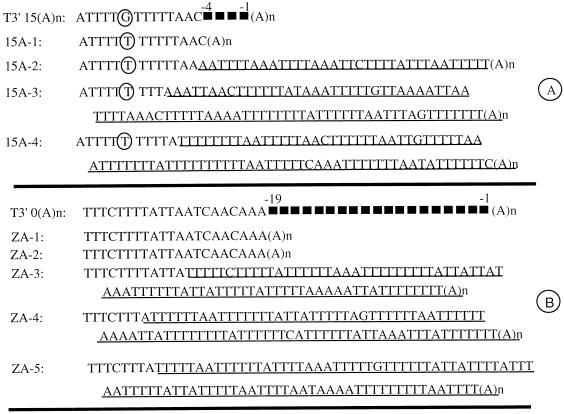
FIG. 7.
Addition of long AU-rich sequences of 3′ mutants of the SIN genome. (A) Sequences of T3′15(A)n and virus isolates generated from it. Isolates 15A-1, 15A-2, and 15A-3 were derived from experiment 1. Isolate 15A-4 was derived from experiment 2. (B) Sequences of T3′0(A)n and the virus isolates recovered from it. Isolates ZA-1, -2, and -3 were derived from experiment 1, and isolate ZA-4 was derived from experiment 2. The identification of residues and motifs is as described in the legend to Fig. 6.
The engineered T3′18(A)n/Xho RNA lacks the C residue from the −1 position of the 3′CSE (Fig. 6A). The 3′NTRs of seven virus isolates derived from two separate transfection experiments were sequenced. Six of the virus isolates (18A-1 to 18A-6) (Fig. 6A) carried a new motif, 5′-CAAAATATTT-3′, just 3′ of the poly(A) tail. Five of the isolates (18A-1 to 18A-4 and 18A-7) carried a U residue instead of a G residue at the −14 position. Since isolates 18A-5 and 18A-6 carried the wild-type G residue at the −14 position, the G-to-U change appears to be a compensatory change. Isolate 18A-7 carried a C insertion adjacent to the poly(A) tail in addition to carrying the G-to-U change at position −14, restoring the −1 position. Since isolate 18A-1, upon two passages in BHK cells, evolved into isolate 18A-7 (data not shown), it appears that polymerase jumping events have led to the formation of 18A-7-type genomes. These results indicate that the removal of the C residue at position −1 in a polyadenylated SIN genome results in in vivo repair by addition of a C residue or a short AU motif or both. The in vitro-synthesized T3′18(A)n template RNA carried the exact 3′ sequence found in the DNA template (Fig. 6A).
The T3′17(A)n/Xho RNA carries a 2-nt deletion adjacent to the poly(A) tail of the SIN genome (Fig. 6B). Viable viruses were recovered from BHK cells transfected with T3′17(A)n/Xho, and the 3′NTRs of 10 virus isolates were sequenced (Fig. 6B). Six of the isolates (17A-1 to 17A-6) retained the original 3′ mutations, indicating the viability of the mutant RNA. Since the infectivity of the T3′17(A)n RNA was severely affected (Table 2, experiment 19), compensatory mutations elsewhere in the genome must have occurred during the formation of viruses such as 17A-1 from the transfected T3′17(A)n RNA. Isolate 17A-7 (Fig. 6B) carried the dinucleotide AC as an insertion just 5′ to the poly(A) tail. Isolate 17A-8 carried an insertion of six U residues just upstream of the poly(A) tail. Isolates 17A-9 and 17A-10 carried 4- and 6-nt deletions in addition to the 2-nt deletion that was introduced in T3′17(A)n RNA. As found for the virus isolates recovered from T3′18(A)n/Xho, many of the isolates corresponding to T3′17(A)n also carried a G-to-U change at the −14 position. Thus, deletion of the dinucleotide UC from the −1 and −2 positions of the 3′NTR of the SIN genome results in the production of viable genomes carrying the original mutations or poly(U) additions. As suggested previously, polymerase jumping events may account for deletion events observed in isolates 17A-9 and 17A-10.
T3′6(A)n RNA carries a 13-nt deletion (−1 to −13) of the 19-nt 3′CSE located within the 3′NTR (Fig. 6C). Interestingly, BHK cells transfected with this RNA readily produced infectious virus, although the specific infectivity of this RNA was very low (Table 2, experiment 21). Sequence analysis of the 3′NTR of eight of the viruses isolates derived from T3′6(A)n demonstrated the preservation of the original 13-nt deletions and a poly(A) tail. No other modifications were found at the 3′ terminus. This observation indicated that not all 3′ deletions lead to terminal modifications. It is also possible that the 3′ sequence motif located adjacent to the poly(A) tail of T3′6(A)n is inhibitory for terminal AU additions. Since the parental RNA had low specific infectivity, the recovered viruses must carry some base changes within the genome which could facilitate efficient recognition of the mutant 3′ terminus.
The SIN mutant T3′15(A)n carried a 4-nt deletion (−1 to −4) adjacent to the poly(A) tail, thus making the C residue a terminal base abutting the poly(A) tail (Fig. 7A). Cytoplasmic RNA corresponding to nine different virus isolates obtained from two separate transfections were amplified by RT-PCR, and their 3′ regions were sequenced. The 3′ sequences of four representative isolates are given in Fig. 7A. Isolate 15A-1 displayed the original deletions of the T3′15(A)n sequence as well as the G-to-U change at the −14 position. The remaining five isolates had lost their terminal C residue (at the −6 position) and undergone extensive A (15 to 20%) and U (65 to 70%) additions. The C and G residues accounted for only 5 to 10% of the new bases. The size of the additions varied from 36 to 129 nt. These 3′ insertions consisted predominantly of oligo(U) motifs and a few oligo(A) motifs. Although the number of U residues in any given oligo(U) motif extended up to nine, only five A residues could be found maximally in any oligo(A) motif. As can be in seen in isolate 15A-4, a few residues 5′ to the C at the −6 position were deleted in three of the isolates sequenced (data not shown).
The SIN mutant devoid of the entire 19-nt 3′CSE, TT3′0(A)n/Xho, behaved much like to the 4-nt deletion mutant [T3′15(A)n]. Of the five virus isolates that were sequenced, two of them (ZA-1 and ZA-2) maintained the original deletions, indicating that the SIN genome lacking the entire 3′CSE is viable, and expressed both 26S and 49S RNA (Fig. 3C). The remaining three carried extensive AU additions. As described for T3′15(A)n/Xho, these additions involved predominantly oligo(U) motifs rather than oligo(A) motifs. These results indicate that cells transfected with a SIN genome lacking part or the entire 3′CSE are able to produce viable virus with or without AU additions within the 3′NTR.
DISCUSSION
The 3′ termini of all alphaviruses carry a poly(A) tail and a 19-nt conserved AU-rich RNA motif (3′CSE). The poly(A) tail and the 19-nt motif were thought to serve as viral promoters for negative-sense RNA synthesis and genome replication. Until recently, it was also believed that the poly(U) motif located at the 5′ end of a negative-sense copy of the alphavirus genome synthesized in vivo served as the template for the poly(A) tail (15, 27, 40, 41, 42). Kuhn et al. (27) reported that infectious virus could be generated from a SIN mutant whose poly(A) tail was displaced by 7 nt from the 19-nt motif. A SIN genome lacking a poly(A) tail but carrying one to five nonviral nucleotides at its 3′ end was shown to be infectious and polyadenylated in vivo (22). Although rubella virus is the only other known animal RNA virus which polyadenylates its tailless genomes, at least three plant RNA genomes were shown to undergo de novo polyadenylation in vivo (13, 20, 26). When the poly(A) tail of the SIN genome was replaced by a 1.8-kb-long nonviral sequence, infectious virus and the polyadenylated SIN genome could still be recovered from BHK cells (22). If the poly(A) tail is necessary for 3′-end recognition of the SIN genome, then polyadenylation of the transfected RNA should precede negative-strand synthesis. This would mean that long nonviral sequences located at the 3′ end would be removed by specific endonucleases, probably in the cytoplasm of host cells. Similar to the cleavage and polyadenylation of pre-mRNA transcripts in the nucleus (46), cleavage and polyadenylation of mRNA transcripts in the cytoplasm are also a well-studied process (references 45 and 47 and references therein). Although polyadenylation of procaryotic mRNAs is known, it does not appear to require specific RNA recognition motifs (39).
By oocyte injection of in vitro-synthesized capped mRNAs whose poly(A) tails were replaced with heterologous sequence, the cleavage and accurate cytoplasmic polyadenylation of these mRNAs were extensively studied (45, 47). The 3′NTRs of many eucaryotic mRNAs carry a number of AU-rich elements (8, 47). These 3′NTRs and probably the AU-rich elements in conjunction with several RNA-binding proteins play a role in mRNA stability, translation, localization, and polyadenylation (2, 3, 8, 10–12, 45). One of the several AU-rich elements and the canonical nuclear polyadenylation element AAUAA were required for cytoplasmic polyadenylation (45). The cytoplasmic polyadenylation elements located at the 3′UTR of mRNAs and the trans-acting protein factors involved in the recognition of these signals have been shown to be evolutionarily conserved from arthropods to vertebrates (45). Although the 3′NTRs of the SIN genome including the 19-nt conserved element carry several AU-rich elements, none of them are identical to these elements. Since polyadenylation can be effected by variant AU-rich motifs (45, 47), the precise nature of SIN RNA motifs that regulate cleavage and polyadenylation requires further investigation. Since the construct TT21qa/Xho, which carries the terminal 29 nt of the 3′NTR and the nonviral motif CUCGA at the 3′ end, was able to be polyadenylated at the precise location, the minimal signals for cleavage and polyadenylation must be located within this 29-nt region (Fig. 1).
The insertion of a G residue at the −1 position of 19-nt 3′CSE in T3′18/Xho moved the terminal C residue to the +1 position. In spite of these alterations, the T3′18/Xho RNA was polyadenylated at the C residue and the G residue was eliminated, probably by transcriptional skipping (Fig. 4C). No other modifications were observed at the 3′ terminus of the viral RNA corresponding to all eight virus plaques tested. On the contrary, deletion of the four terminal residues corresponding to −1 to −4 of the 3′ 19-nt motif and the poly(A) tail in the SIN genome (T3′15/Xho) (Fig. 5) gave rise to viral genomes with new AU-rich motifs and a poly(A) tail (Fig. 5C). Although the cytoplasmic polyadenylation machinery is known to add the poly(A) tail, it is not known if it plays a role in the addition of AU-rich sequences. As described for other viral systems (1, 19, 31), viral or cellular terminal transferases (adenylyl and uridylyl transferases) could have also added the AU-rich motifs. It is also puzzling to note that how the cytoplasmic polyadenylation system can add poly(A) tails to such a diverse group of AU-rich sequences. As described for the poliovirus system (1, 31), possible terminal uridylyl and adenylyl activity of the SIN RdRp should also be considered in these terminal repair processes. Although the poliovirus polymerase has been known to be associated with terminal adenylyl and possible uridylyl transferase activities (1, 31), addition of AU residues and a poly(A) tail at the 3′ terminus of the positive-sense genome has not been reported. Since cellular terminal transferases can add nucleotides to the 3′ end of RNAs without any template specificity, the 3′ terminus of templates such as TT21k/Xho (22) should have been extended and polyadenylated. Since TT21k/Xho, which carries a 24-nt AU-rich region (−1 to −24 of 3′NTR) as a 3′NTR, failed to undergo AU additions and polyadenylation (22) (Table 2, experiment 11), the role of template-independent terminal transferases in the addition of AU-rich motifs remains unclear. Although RNA-RNA recombination (4, 21, 22, 24, 28) and production of abortive transcripts by viral polymerase (7, 30) were implicated in the terminal repair of some RNA genomes, such events do not seem to explain polyadenylation and AU additions of the type reported here.
The sequences of isolates 15.7 and 15.8 (Fig. 5C) indicate further complexity of the 3′ repair process observed here. These two plaque-purified viruses retained their parental 3′ bases (the dinucleotide CT). The poly(A) tails were added just downstream of these parental terminal bases. Interestingly, a poly(U) motif was inserted just upstream of the CT residues. Since terminal transferases are not known to insert internal sequences, the viral polymerase must have introduced this sequence by some unknown mechanism. Curiously, a similar poly(U) motif and a poly(A) tail were reported to have been added during the terminal repair process of a beet yellow vein virus RNA (26). These results indicate the existence of a common pathway in both the plant and animal cells to repair AU-rich 3′ ends of alphaviral RNAs.
What are the sequence requirements for the addition of AU-rich motifs on SIN genome 3′ ends? The construct TT21s/Xho, which carries an intact 3′CSE and the entire 3′NTR, did not undergo AU additions but was accurately polyadenylated. Similarly, other SIN mutants lacking poly(A) which carry intact 3′CSE but with 3′NTR deletions of differing lengths (TT21ra and TT21qa) fail to undergo any AU additions although they undergo de novo polyadenylation (Table 2, experiments 13 and 14). Therefore, preservation of the 3′-terminal bases of the 3′CSE in the SIN genome seem to prevent AU additions. At least 14 bases from the 3′ terminus (−1 to −14 of the 3′CSE) of the SIN genome and a poly(A) tail were needed to generate infectious virus (22). Similarly, viruses carrying 16, 20, and 29 bases from the 3′ terminus in addition to the poly(A) tail were also infectious (22) (Table 2, experiments 7, 8, and 9, respectively). Sequence analysis of the viruses recovered from these experiments demonstrated the exact preservation of the parental sequence; no 3′ AU additions were observed in these virus isolates. A SIN mutant a carrying poly(A) tail but lacking one or more bases from the 3′ terminus of the 3′CSE (Fig. 5 to 7) undergoes addition of AU-rich motifs, indicating that AU additions are not inhibited by preexisting poly(A) tails on mutant SIN genomes.
On the basis of these findings, we propose a model in which polymerase recognition of the wild-type 3′CSE-poly(A) junction results in accurate copying of the positive-strand genome, whereas recognition of a mutant 3′CSE-poly(A) junction leads to polymerase stuttering on A- and U-rich residues and synthesis of AU-rich motifs which are added to the newly made negative-sense RNA. Viable genomes generated from this error-prone replicative process are amplified further. Recognition of a wild-type 3′ terminus even in the absence of a poly(A) tail (TT21s/Xho) leads to a poly(A) tail addition but little or no polymerase stuttering and, therefore, no AU additions. It is likely that the polyadenylated 3′ mutants (Fig. 6 and 7) are used directly as templates for negative-sense RNA synthesis and that the AU additions are introduced during new RNA synthesis. SIN genomes such as T3′15/Xho, which carries a mutant 3′CSE and lacks a poly(A) tail, could have been directly polyadenylated precisely at its 3′ end and polymerase stuttering events during negative- or positive-sense RNA synthesis might have incorporated the AU-rich elements. Evolution of these polyadenylated genomes might have led to the loss of some of the AU-rich sequences, resulting in the generation of more stable genomes. It is also possible that all SIN genomes lacking poly(A) which can undergo polyadenylation also undergo AU additions but that those genomes carrying the 19-nt 3′CSE (such as TT21s/Xho, TT21ra/Xho, or TT21qa/Xho) may effectively delete the extra AU motifs to quickly regain a wild-type 3′ terminus.
These studies also suggest that the addition of a 3′ poly(A) tail to a SIN genome lacking a poly(A) tail requires specific 3′NTR sequences, although the site of polyadenylation could vary. In the absence of RNA motifs that allow polyadenylation, SIN genomes fail to undergo AU additions. This would mean that poly(A) recognition by the viral RdRp may be important for AU additions. Since polymerase recognition of only mutant 3′CSE-poly(A) junction sites leads to AU additions, a loose binding of polymerase to the mutant 3′CSE motif appears to catalyze polymerization of A and U residues rather than C or G residues. However, it was curious that an extra G residue could be found in the isolates derived from T3′15/Xho (15.1 and 15.7) (Fig. 5C). The in vitro-synthesized T3′15/Xho RNA was expected to possess one of the following 3′ termini depending on how many of the bases from the XhoI motif were transcribed as RNA: C, CU, CUC, CUCG, or CUCGA. Once inside the cells, some of the terminal bases may be trimmed or new bases can be added, probably before polyadenylation. Isolate 15.1 carried the motif CTCGAG, even though the 3′ G could not have been copied from the DNA template during in vitro RNA synthesis because the template was digested with XhoI. It is unlikely that the 3′ G was derived by readthrough transcription of some undigested DNA template since T3′15 RNA carrying longer nonviral sequences at its 3′ end were noninfectious (unpublished data). Therefore, the terminal G within the CTCGAG motif must have been added during the repair process, although the bases A and U account for most of the additions downstream of this motif. The parental RNA template may serve as a template for the specific addition of G or C residues similar to the addition of A and U residues.
A significant outcome of this study is the generation of a battery of SIN genomes that lack the 3′CSE but which are nevertheless replication competent and stable for at least three passages in BHK cells. Although these SIN genomes lack the known 19-nt 3′CSE, all of them carry AU-rich motifs adjacent to the poly(A) tail. The finding that SIN genomes carrying one of the many 3′ AU-rich elements and a poly(A) tail can be effectively be recognized by viral RdRp is intriguing. Since many eucaryotic mRNAs carry AU-rich 3′ ends, it is hard to explain how the viral polymerase avoids copying cellular mRNAs. It is possible that secondary structures within the 3′NTR as well as sequences located elsewhere within the viral genome regulate initiation of negative-strand RNA synthesis. Rubella virus, another member of the togavirus family, also replicates well even in the presence of base deletions adjacent to the poly(A) tail (9). Poliovirus has also been shown to undergo replication in the absence of the entire 3′NTR (43). It was also reported that poliovirus polymerase can copy a variety of RNAs in a nonspecific manner, suggesting that initiation of RNA synthesis at least in vitro was less dependent on unique RNA promoter sequences (44). Coupling of genome translation and replication was in fact suggested to confer specificity during poliovirus RNA synthesis (32). But neither rubella virus nor poliovirus genomes seem to undergo any AU additions, although the SIN genome undergoes AU additions. These results have set a stage to reexamine our current understanding of 3′-end recognition in positive-stranded RNA viruses and particularly those genomes carrying a 3′ poly(A) tail.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM 57439 and by the RCMI program at Meharry Medical College.
We thank Teryl Frey and members of his laboratory for sharing some of their unpublished results on rubella virus.
REFERENCES
- 1.Andrews N C, Baltimore D. Purification of a terminal uridylyltransferase that acts as host factor in the in vitro poliovirus replicase reaction. Proc Natl Acad Sci USA. 1986;83:221–225. doi: 10.1073/pnas.83.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antic D, Keene J D. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 4.Biebricher C K, Luce R. In vitro recombination and terminal elongation of RNA by Q beta replicase. EMBO J. 1992;11:5129–5135. doi: 10.1002/j.1460-2075.1992.tb05620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer J C, Haenni A L. Infectious transcripts and cDNA clones of RNA viruses. Virology. 1994;198:415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- 6.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter C D, Simon A E. In vivo repair of 3′-end deletions in a TCV satellite RNA may involve two abortive synthesis and priming events. Virology. 1996;226:153–160. doi: 10.1006/viro.1996.0641. [DOI] [PubMed] [Google Scholar]
- 8.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 9.Chen M H, Frolov I, Frey T K. Fifth Southeastern Regional Virology Conference, Atlanta, Ga. 1998. Analysis of the 3′ cis-acting elements in the rubella virus genome; p. 7. [Google Scholar]
- 10.Decker C J, Parker R. Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr Opin Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 11.de Moor C H, Richter J D. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol Cell Biol. 1997;17:6419–6426. doi: 10.1128/mcb.17.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duggal R, Lahser F C, Hall T C. Cis-acting sequences in the replication of plant viruses with plus-sense RNA genomes. Annu Rev Phytopathol. 1994;32:287–309. [Google Scholar]
- 13.Eggen R, Verver J, Wellink J, De Jong A, Goldbach R, van Kammen A. Improvements of the infectivity of in vitro transcripts from cloned cowpea mosaic virus cDNA: impact of terminal nucleotide sequences. Virology. 1989;173:447–455. doi: 10.1016/0042-6822(89)90557-6. [DOI] [PubMed] [Google Scholar]
- 14.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey T K, Strauss J H. Replication of Sindbis virus. VI. Poly(A) and poly(U) in virus-specific RNA species. Virology. 1978;86:494–506. doi: 10.1016/0042-6822(78)90088-0. [DOI] [PubMed] [Google Scholar]
- 16.Frolov I, Hoffman T A, Pragai B M, Dryga S A, Huang H V, Schlesinger S, Rice C M. Alphavirus-based expression vectors: strategies and applications. Proc Natl Acad Sci USA. 1996;93:11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 18.Griffin D E. Molecular pathogenesis of Sindbis virus encephalitis in experimental animals. Adv Virus Res. 1989;36:255–271. doi: 10.1016/s0065-3527(08)60587-4. [DOI] [PubMed] [Google Scholar]
- 19.Grun J B, Brinton M A. Characterization of West Nile virus RNA-dependent RNA polymerase and cellular terminal adenylyl and uridylyl transferases in cell-free extracts. J Virol. 1986;60:1113–1124. doi: 10.1128/jvi.60.3.1113-1124.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guilford P J, Beck D L, Forster R L. Influence of the poly(A) tail and putative polyadenylation signal on the infectivity of white clover mosaic potexvirus. Virology. 1991;182:61–67. doi: 10.1016/0042-6822(91)90648-u. [DOI] [PubMed] [Google Scholar]
- 21.Hajjou M, Hill K R, Subramaniam S V, Hu J Y, Raju R. Nonhomologous RNA-RNA recombination events at the 3′ nontranslated region of the Sindbis virus genome: hot spots and utilization of nonviral sequences. J Virol. 1996;70:5153–5164. doi: 10.1128/jvi.70.8.5153-5164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill K R, Hajjou M, Hu J Y, Raju R. RNA-RNA recombination in Sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching. J Virol. 1997;71:2693–2704. doi: 10.1128/jvi.71.4.2693-2704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H V. Sindbis virus vectors for expression in animal cells. Curr Opin Biotechnol. 1996;7:531–535. doi: 10.1016/s0958-1669(96)80057-7. [DOI] [PubMed] [Google Scholar]
- 24.Jarvis T C, Kirkegaard K. The polymerase in its labyrinth: mechanisms and implications of RNA recombination. Trends Genet. 1991;7:186–191. doi: 10.1016/0168-9525(91)90434-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston R E, Peters C J. Alphaviruses. In: Fields B N, et al., editors. Field’s virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 843–898. [Google Scholar]
- 26.Jupin I, Bouzoubaa S, Richards K, Jonard G, Guilley H. Multiplication of beet necrotic yellow vein virus RNA 3 lacking a 3′ poly(A) tail is accompanied by reappearance of the poly(A) tail and a novel short U-rich tract preceding it. Virology. 1990;178:281–284. doi: 10.1016/0042-6822(90)90404-f. [DOI] [PubMed] [Google Scholar]
- 27.Kuhn R J, Hong Z, Strauss J H. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J Virol. 1990;64:1465–1476. doi: 10.1128/jvi.64.4.1465-1476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai M M. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levis R, Weiss B G, Tsiang M, Huang H, Schlesinger S. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell. 1986;44:137–145. doi: 10.1016/0092-8674(86)90492-7. [DOI] [PubMed] [Google Scholar]
- 30.Nagy P D, Carpenter C D, Simon A E. A novel 3′-end repair mechanism in an RNA virus. Proc Natl Acad Sci USA. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neufeld K L, Galarza J M, Richards O C, Summers D F, Ehrenfeld E. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 33.Ou J H, Strauss E G, Strauss J H. Comparative studies of the 3′-terminal sequences of several alpha virus RNAs. Virology. 1981;109:281–289. doi: 10.1016/0042-6822(81)90499-2. [DOI] [PubMed] [Google Scholar]
- 34.Pfeffer M, Kinney R M, Kaaden O R. The alphavirus 3′-nontranslated region: size heterogeneity and arrangement of repeated sequence elements. Virology. 1998;240:100–108. doi: 10.1006/viro.1997.8907. [DOI] [PubMed] [Google Scholar]
- 35.Raju R, Huang H V. Analysis of Sindbis virus promoter recognition in vivo, using novel vectors with two subgenomic mRNA promoters. J Virol. 1991;65:2501–2510. doi: 10.1128/jvi.65.5.2501-2510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raju R, Kolakofsky D. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J Virol. 1989;63:122–128. doi: 10.1128/jvi.63.1.122-128.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raju R, Subramaniam S V, Hajjou M. Genesis of Sindbis virus by in vivo recombination of nonreplicative RNA precursors. J Virol. 1995;69:7391–7401. doi: 10.1128/jvi.69.12.7391-7401.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice C M, Levis R, Strauss J H, Huang H V. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu Rev Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- 40.Sawicki D L, Gomatos P J. Replication of Semliki Forest virus: polyadenylate in plus-strand RNA and polyuridylate in minus-strand RNA. J Virol. 1976;20:446–464. doi: 10.1128/jvi.20.2.446-464.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlesinger S, Schlesinger M J. Togaviridae and their replication. In: Fields B N, et al., editors. Field’s virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 825–843. [Google Scholar]
- 42.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuschall D M, Hiebert E, Flanegan J B. Poliovirus RNA-dependent RNA polymerase synthesizes full-length copies of poliovirion RNA, cellular mRNA, and several plant virus RNAs in vitro. J Virol. 1982;44:209–216. doi: 10.1128/jvi.44.1.209-216.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verrotti A C, Thompson S R, Wreden C, Strickland S, Wickens M. Evolutionary conservation of sequence elements controlling cytoplasmic polyadenylation. Proc Natl Acad Sci USA. 1996;93:9027–9032. doi: 10.1073/pnas.93.17.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 47.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 48.Xiong C, Levis R, Shen P, Schlesinger S, Rice C M, Huang H V. Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science. 1989;243:1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]