Abstract
Background:
We aimed to evaluate and compare the performance of multiple myeloma (MM) selection algorithms for use in Veterans Affairs (VA) research.
Methods:
Using the VA Corporate Data Warehouse (CDW), the VA Cancer Registry (VACR), and VA pharmacy data, we randomly selected 500 patients from 01/01/1999 to 06/01/2021 who had (1) either one MM diagnostic code OR were listed in the VACR as having MM AND (2) at least one MM treatment code. A team reviewed oncology notes for each veteran to annotate details regarding MM diagnosis and initial treatment within VA. We evaluated inter-annotator agreement and compared the performance of four published algorithms (two developed and validated external to VA data and two used in VA data).
Results:
A total of 859 patients were reviewed to obtain 500 patients who were annotated as having MM and initiating MM treatment in VA. Agreement was high among annotators for all variables: MM diagnosis (98.3% agreement, Kappa = 0.93); initial treatment in VA (91.8% agreement; Kappa = 0.77); and initial treatment classification (87.6% agreement; Kappa = 0.86). VA Algorithms were more specific and had higher PPVs than non-VA algorithms for both MM diagnosis and initial treatment in VA. developed the “VA Recommended Algorithm,” which had the highest PPV among all algorithms in identifying patients diagnosed with MM (PPV = 0.98, 95% CI = 0.95–0.99) and in identifying patients who initiated their MM treatment in VA (PPV = 0.93, 95% CI = 0.90–0.96).
Conclusion:
Our VA Recommended Algorithm optimizes sensitivity and PPV for cohort selection and treatment classification.
Keywords: Veterans, Multiple Myeloma, Sensitivity, Algorithm, Cohort selection
INTRODUCTION:
Large research and administrative databases provide opportunities to study real-world treatment trends and outcomes in patients with multiple myeloma (MM).1he U.S. Veterans Affairs (VA) Healthcare System contains one of the largest, centralized MM databases available. Several studies have leveraged VA data to identify risk factors, develop prediction algorithms, and study novel treatments and interventions in a population that is under-represented in MM clinical trials.2,3,4 However, to date no algorithm to select newly-diagnosed and newly-treated patients with MM using VA’s database has been validated. Algorithms have been developed for case identification of patients with MM in other large databases such as Surveillance, Epidemiology, and End Results (SEER)-Medicare; Brandenburg and colleagues recently validated several of these algorithms using a local healthcare database and large pharmaceutical claims database.5 However, it is unknown whether these algorithms perform well in VA’s database,6 which differs from other databases in terms of the breadth and detail of data maintained.7,8
The objective of the current study was to evaluate and compare the performance of published MM selection algorithms used in VA databases and in external databases. Moreover, we aimed to validate not only these algorithms’ ability to identify cases of MM, but also to identify veterans with MM who initiated their treatment within the VA healthcare system. Finally, we aimed to validate the ability of VA’s pharmacy databases at classifying individual MM treatments and initial multi-drug treatment regimens. Our goal was to provide one or more high-performing selection algorithms for use in future studies of veterans with MM treated in VA, and to offer insights into the ability of algorithms for MM to generalize across datasets.
METHODS:
Data Sources and Population:
To identify veterans with potential MM, we used the VA Corporate Data Warehouse, which collects clinical, billing, and EHR information from Veterans treated in VA facilities throughout the United States.9 We also used the VA Cancer Registry (VACR), which collects information on cancer diagnosis and treatment compiled and submitted by local cancer registry staff at each of the 132 Veterans Affairs Medical Centers that diagnose and/or treat Veterans with cancer.10 We further used data from the Centers for Medicare and Medicaid Services (CMS).11From VA data, we randomly selected 500 patients from 01/01/1999 to 06/01/2021 who had (1) either one MM diagnostic code (International Classification of Diseases [ICD]-9: codes with prefix 203.0; ICD-10: codes with prefix C90.0) OR who were listed in the VACR as having MM, AND (2) at least one MM treatment code. MM treatment codes represented any class of medical therapy for MM, including proteasome inhibitors, immune-modifying drugs, and chemotherapy (see Supplemental Table 1). Treatment data were derived from the four separate sources in VA CDW: the Computerized Patient Record System (CPRS), which contains data on all orders for prescriptions written by physicians; RxOutpatFill, which contains data on all outpatient prescriptions filled by patients; Bar Code Medication Administration (BCMA), which contains data on all medications administered after scanning the patient’s identifying information; and Intravenous (IV), which contains data on all IV drugs administered.
Annotation of MM diagnosis, initial treatment in VA, and treatment classification:
Among the initial 500 patients selected above, we extracted all oncology, oncology pharmacy, and pathology notes 90 days prior to the Veteran’s date of their first MM treatment code and up to 90 days after. A team of annotators (C.D., H.H., M.A., K.V., G.F., and M.D.) reviewed all compiled notes to annotate the following for each Veteran. Annotators based their annotations on the documentation in the oncologists’ notes, which we considered “gold standard” given the expertise of the treating oncology team and their knowledge of each individual case:
Diagnosis of MM: “Yes”, “No” (monoclonal gammopathy of undetermined significance [MGUS], smoldering MM, amyloidosis, Waldenstrom macroglobulinemia were considered no), or “Information not available” (information in notes insufficient for annotation). Annotators also annotated the date of MM diagnosis.
Initiated MM treatment within VA: “Yes”, “No” (Veteran had previous or concurrent treatment for MM outside VA), or “Information not available.” Annotators also annotated the date of initial MM treatment.
Initial treatment regimen (if annotated as initiating treatment within VA): Individual MM treatments that made up initial treatment regimen (e.g., annotators documented lenalidomide, bortezomib, and dexamethasone if a Veteran was started on these three drugs as part of a triplet regimen).
We assigned 125 of the initial 500 patients to be reviewed by two independent annotators to assess for inter-annotator agreement. A third, separate adjudicator (C.E.) resolved any of the disagreements for the final dataset. After annotators completed their review of notes from the initial 500 patients, we randomly selected additional groups of patients using the same criteria above for further rounds of annotation until we reached 500 annotated cases of patients with MM who initiated treatment at VA. We included as controls the patients not marked as having a diagnosis of MM.
Comparison of Different Algorithms and Selection of Recommended Algorithm:
We performed a literature review to identify previously-developed selection algorithms to select patients newly diagnosed and treated for MM in large databases (Table 1). We chose to evaluate two non-VA algorithms—one for identifying cases with MM (henceforward referred to as “non-VA algorithm 1”) and one for identifying cases on treatment5 (“non-VA algorithm 2”). We also chose to evaluate two VA-specific algorithms—one using the cancer registry to identify patients with MM who initiated treatment in VA12(“VA algorithm 1”) and another using the entire VA CDW to identify patients with MM initiating treatment in VA2 (“VA algorithm 2”). Finally, we examined (1) different combinations of diagnostic and treatment codes and (2) the performance of algorithms restricted to the cancer registry versus algorithms selecting from the entire VA CDW. Based on the above comparisons, we modified one of the existing algorithms to optimize sensitivity and PPV for use in VA research.
Table 1:
Non-VA and VA-specific algorithms selected from literature to compare performance
| Algorithm | Notes |
|---|---|
|
NON-VA ALGORITHM 1 (FOR CASE IDENTIFICATION)5 Two-part algorithm. Both parts of the algorithm are required. Parts 1 and 2 have to be fulfilled separately and sequentially. Part 1: ≥2 ICD-9-CM codes for multiple myeloma (203.0x) AND ≥1 procedure code for bone marrow aspirate, biopsy, or interpretation OR ≥2 distinct procedure code for diagnostic tests (the tests must be different) Part 2: ≥2 ICD-9-CM codes for multiple myeloma (203.0x) 5 to 90 days after procedures identified in part 1 Index date: Date of diagnosis in Oncology EMR database without preceding treatment code. |
Original data sources: Henry Ford Health System, Optum Research Database Inclusion criteria: ≥18 years ≥2 ICD-9-CM(203.0) codes or ICD-10-CM (C90.0) for MM bone marrow aspirate, biopsy, or interpretation CPT 38220, 38221, 88305 HCPCS: G0364 diagnostic CPT: 82784, 83883, 86334, 84165, 84166, 77074 Chemotherapy: thalidomide, lenalidomide, bortezomib, cyclophosphamide, doxorubicin, melphalan, vincristine, carmustine, cisplatin, interferon, etoposide, thiotepa, and/or dexamethasone |
|
| |
|
NON-VA ALGORITHM 2 (FOR NEWLY DIAGNOSED AND INITATING TREATMENT)5 Two-part algorithm. Both parts of the algorithm are required. Parts 1 and 2 have to be fulfilled separately and sequentially. Part 1: ≥2 ICD-9-CM codes for multiple myeloma (203.0x) AND ≥1 procedure code for bone marrow aspirate, biopsy, or interpretation b OR ≥1 procedure code for two diagnostic tests c (the tests must be different) Part 2: ≥2 ICD-9-CM codes for multiple myeloma (203.0x) 5 to 90 days after procedures identified in part 1 AND ≥1 prescription claim for a multiple myeloma therapy Index date: Not explicitly discussed. Used date of first treatment for our study. |
Original data sources: Henry Ford Health System, Optum Research Database Inclusion criteria: ≥18 years ≥2 ICD-9-CM(203.0) or ICD-10-CM (C90.0) codes for MM Exclusion criteria: Patients without a documented height or weight bone marrow aspirate, biopsy, or interpretation CPT 38220, 38221, 88305 HCPCS: G0364 diagnostic CPT: 82784, 83883, 86334, 84165, 84166, 77074 Chemotherapy: thalidomide, lenalidomide, bortezomib, cyclophosphamide, doxorubicin, melphalan, vincristine, carmustine, cisplatin, interferon, etoposide, thiotepa, and/or dexamethasone |
|
| |
|
VA ALGORITHM 1 (FOR NEWLY DIAGNOSED AND INITIATING TREATMENT)12 (2) (ICD)-O3 codes 9732/3 within VACCR, excluded patients who didn’t receive chemotherapy within 6 months of diagnosis. Index date: Date of first treatment |
Original data sources: Cancer Registry Inclusion criteria: International Classification of Diseases (ICD)-O3 codes 9732/3 Exclusion criteria: Patients that didn’t receive chemotherapy within 6 months of MM diagnosis (We removed exclusion of patients who received transplant to make algorithm broader and more comparable to others in our study) |
|
| |
|
VA ALGORITHM 2 (FOR NEWLY DIAGNOSED AND INITIATING TREATMENT)2 (4) 3 ICD9–203.0 (or ICD10 C90) codes and at least 2 dates where MM treatment observed; 2nd treatment date must be within 6 months of 1st Index date: Date of first continuous treatment |
Data source: VA Corporate Data Warehouse Inclusion criteria: 3 dates for MM diagnosis, 2 dates on which MM treatment was observed. Veterans with at least 1 inpatient/outpatient visit each year within 3 years prior to start of MM treatment Exclusion criteria: Veterans with one or more MM treatment codes in the CMS data prior to or up to six months after the index date |
Analysis:
We performed descriptive statistics to summarize the characteristics of the study population. To assess for inter-annotator agreement among the annotation team, we performed Fleiss kappa analyses on the subset of patients reviewed by two independent annotators for (1) MM diagnosis, (2) initiation of treatment in VA, and (3) initial treatment regimens. We also evaluated the inter-annotator agreement for diagnosis date and initial treatment date by computing the median and interquartile ranges of the differences between dates annotated by each reviewer. To evaluate the performance of different selection algorithms in identifying (1) patients diagnosed with MM and (2) patients initiating their MM treatment within VA, we computed the sensitivity, specificity, and positive predictive value (PPV) for each algorithm in correctly classifying (1) and (2), using the annotated classifications as the true values.
Lastly, to evaluate the performance of VA pharmacy data in correctly classifying individual MM treatments, we computed the sensitivity, specificity, and PPV for three common MM treatments: bortezomib (V), lenalidomide (R), and dexamethasone (d). We compared performance across 7-, 14-, 30-, and 60-day treatment exposure windows after the initial diagnosis. Moreover, to evaluate the ability of VA pharmacy data to correctly classify initial multi-drug MM treatment regimens consisting of V, R, and d, we measured the presence of each treatment code across 7-, 14-, 30-, and 60-day treatment exposure windows from the first date of recorded V, R, or d treatment. Patients were classified as receiving Vd, Rd, or VRd based on the occurrence of these codes in a given window (e.g., patients with treatment codes for V, R, and d within 14 days of their first recorded treatment were classified as VRd) while excluding patients who received any other MM drugs within this time window (e.g., cyclophosphamide). We evaluated the agreement between the initial treatment classification based on VA pharmacy data and the initial treatment classification based on the annotations. All analyses were performed with R (version 4.0.3). This study was approved by the VA Boston Healthcare System Institutional Review Board/Office of Research and Development, protocol numbers 1582165–2 and 1578039–10.
RESULTS:
Annotations:
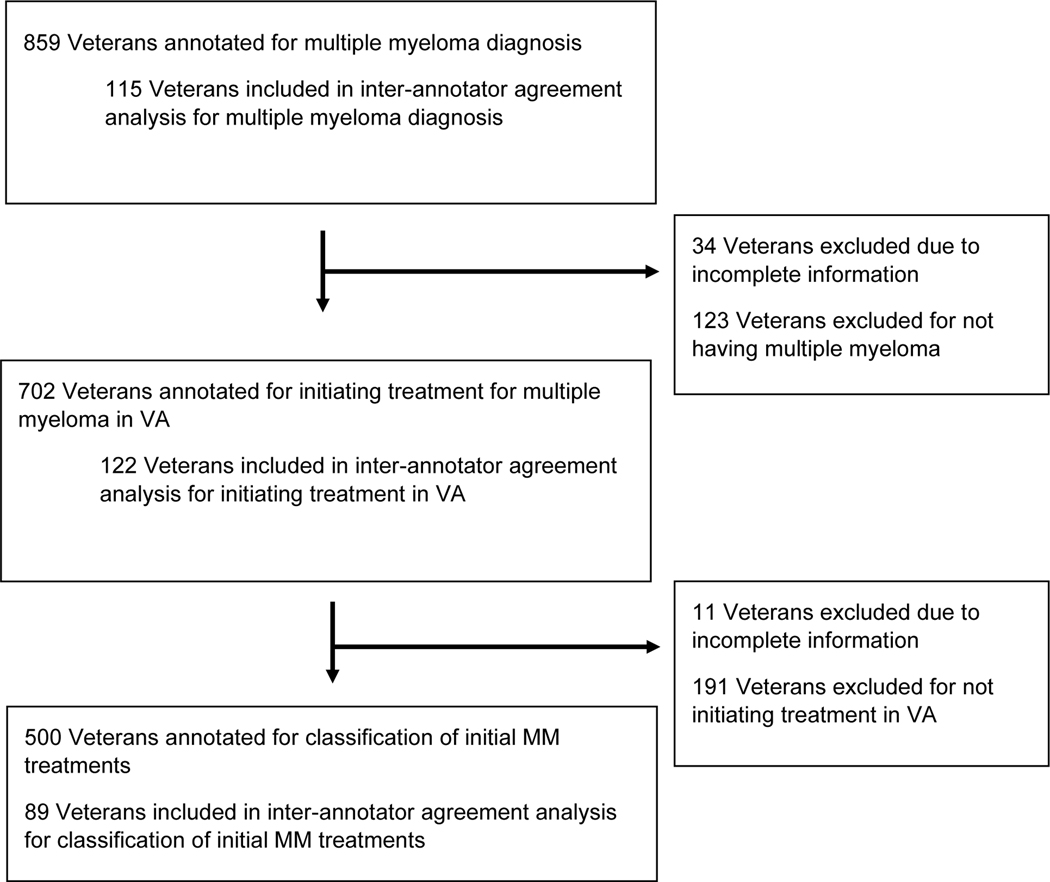
A total of 859 patients were reviewed to obtain 500 patients who were annotated as having MM and initiating MM treatment in VA (Figure 1). The median age of these 859 patients was 68.5 (Interquartile range [IQR] = 58.8, 74.7), 822 were male (95.7%), 495 (57.6%) were white and 239 (27.8%) were black (Table 2). Of these 859, 702 (81.7%) were annotated as having a diagnosis of MM. Five hundred of these 702 with MM were annotated as initiating their MM treatment within VA; 191 patients were excluded because they received part of or their entire initial treatment outside VA and 11 patients were marked as having incomplete information in notes.
Figure 1:
Flow diagram of selection of MM cases/controls
Table 2:
Baseline characteristics of population
| Characteristic | n=859 |
|---|---|
| Age (median [IQR]) | 68.53 [58.76, 74.68] |
|
| |
| Gender, M (%) | 822 (95.7) |
|
| |
| Race (%) | |
|
| |
| White | 495 (57.6) |
|
| |
| Black or African American | 239 (27.8) |
|
| |
| Native Hawaiian or other Pacific Islander | 5 ( 0.6) |
|
| |
| Asian | 4 ( 0.5) |
|
| |
| Unknown | 116 (13.5) |
|
| |
| Annotated as having diagnosis of multiple myeloma (%) | 702 (81.7) |
| Annotated as initiating treatment in VA (%) | |
|
| |
| Yes | 500 (58.2) |
|
| |
| No | 191 (22.2) |
|
| |
| Not multiple myeloma | 157 (18.3) |
|
| |
| Cannot tell | 7 ( 0.8) |
|
| |
| Info not explicit in notes | 4 ( 0.5) |
|
| |
| Year of initial treatment (%) | |
|
| |
| ≤ 2006 | 149 (17.3) |
|
| |
| 2007–2014 | 163 (19.0) |
| 2015–2021 | 188 (21.9) |
|
| |
| Not initially treated in VA, not true myeloma, or date not recorded | 359 (41.8) |
After the above exclusions and performing additional overlapping annotations to increase power, 115, 122, and 89 patients were included in the inter-annotator agreement analyses for MM diagnosis, initiating MM treatment in VA, and initial treatment classification, respectively. Agreement was high among annotators for all variables: MM diagnosis (98.3% agreement; Fleiss Kappa statistic = 0.93, 95% confidence interval [CI] = 0.84–1.00), initial treatment in VA (91.8% agreement; Fleiss Kappa statistic = 0.77, 95% CI = 0.63–0.91), and initial treatment classification (87.6% agreement; Fleiss Kappa statistic = 0.86, 95% CI = 0.78–0.94). The median difference between annotators’ annotated dates of MM diagnosis was 13 days (IQR = 0.5, 37.5 days), and the median difference between annotators’ annotated dates of initial MM treatment was 2 days (IQR = 0, 6.5 days).
Performance of Selection Algorithms:
In identifying (1) patients diagnosed with MM and (2) those who initiated their MM treatment in VA, the non-VA algorithms were more sensitive than the VA algorithms—MM diagnosis sensitivity (95% confidence Interval [CI]): Non-VA Algorithm 1 = 0.86 (0.83–0.88), Non-VA Algorithm 2 = 0.85 (0.82–0.88), VA Algorithm 1 = 0.28 (0.24–0.31), VA Algorithm 2 = 0.63 (0.60–0.67); initial MM treatment in VA sensitivity, (95% CI): Non-VA Algorithm 1 = 0.92 (0.89–0.94), Non-VA Algorithm 2 = 0.92 (0.89–0.94), VA Algorithm 1 = 0.35 (0.31–0.40), VA Algorithm 2 = 0.74 (0.70–0.78; Table 4). However, the VA Algorithms were more specific and had higher PPVs, especially for identifying patients initiating their MM treatment in VA—MM diagnosis PPV (95% CI): Non-VA Algorithm 1 = 0.92 (0.90–0.94), Non-VA Algorithm 2 = 0.92 (0.90–0.94), VA Algorithm 1 = 0.98 (0.94–0.99), VA Algorithm 2 = 0.93 (0.90–0.95); initial MM treatment in VA PPV (95% CI): Non-VA Algorithm 1 = 0.77 (0.73–0.80), Non-VA Algorithm 2 = 0.77 (0.74–0.81), VA Algorithm 1 = 0.93 (0.88–0.96), VA Algorithm 2 = 0.85 (0.81–0.88). Although both VA Algorithms had high PPVs, VA Algorithm 1 was more specific than VA Algorithm 2 in identifying patients diagnosed with MM—VA Algorithm 1 specificity (95% CI) = 0.96 (0.91–0.99), VA Algorithm 2 specificity (95% CI) = 0.71 (0.63–0.79)—and in identifying patients initiating their treatment in VA—VA Algorithm 1 specificity (95% CI) = 0.93 (0.88–0.96), VA Algorithm 2 specificity (95% CI) = 0.64 (0.57–0.71)—whereas VA Algorithm 2 was more sensitive (sensitivities provided above).
Table 4:
Sensitivity, specificity, and positive predictive value of the four main selection algorithms identified from the literature and our recommended algorithm for identifying Veterans diagnosed with multiple myeloma and for identifying Veterans with multiple myeloma who initiated treatment in VA
| Algorithm for multiple myeloma | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) |
|---|---|---|---|
| Non-VA Algorithm 1 | 0.86 (0.83–0.88) | 0.57 (0.48–0.66) | 0.92 (0.90–0.94) |
| Non-VA Algorithm 2 | 0.85 (0.82–0.88) | 0.57 (0.48–0.66) | 0.92 (0.90–0.94) |
| VA Algorithm 1 | 0.28 (0.24–0.31) | 0.96 (0.91–0.99) | 0.98 (0.94–0.99) |
| VA Algorithm 2 | 0.63 (0.60–0.67) | 0.71 (0.63–0.79) | 0.93 (0.90–0.95) |
| VA Recommended Algorithm | 0.45 (0.41–0.49) | 0.94 (0.88–0.97) | 0.98 (0.95–0.99) |
| Algorithm for initiation of treatment | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) |
| Non-VA Algorithm 1 | 0.92 (0.89–0.94) | 0.28 (0.22–0.35) | 0.77 (0.73–0.80) |
| Non-VA Algorithm 2 | 0.92 (0.89–0.94) | 0.29 (0.23–0.36) | 0.77 (0.74–0.81) |
| VA Algorithm 1 | 0.35 (0.31–0.40) | 0.93 (0.88–0.96) | 0.93 (0.88–0.96) |
| VA Algorithm 2 | 0.74 (0.70–0.78) | 0.64 (0.57–0.71) | 0.85 (0.81–0.88) |
| VA Recommended Algorithm | 0.58 (0.54–0.63) | 0.89 (0.84–0.93) | 0.93 (0.90–0.96) |
VA Recommended Algorithm includes selection criteria of VA Algorithm 1, but with the removal of height and weight restriction and the addition of requiring at least two billable visits in VA in the three years prior to initial treatment date (to define users of VA).
CI = Confidence Interval; PPV = Positive Predictive Value
When looking at the performance in identifying patients diagnosed with MM across different combinations of MM ICD and treatment codes, specificity was poor in algorithms containing only ICD codes but improved marginally with increasing number of ICD codes (Supplemental Table 2). Specificity improved further with the addition of MM treatments to algorithms and was highest when algorithms restricted selection to the VACR (versus being applied to entire VA CDW). These trends held true when looking at the performance of algorithms in identifying patients who initiated their MM treatment within VA (Supplemental Table 3).
Given the superior PPV in algorithms restricting to the VACR, we chose to modify VA Algorithm 1 to develop an algorithm with optimal sensitivity and PPV for use in VA research. To improve its sensitivity, we removed the requirement that patients needed to have values for height and/or weight prior to their diagnosis of MM. To identify patients who utilize the VA (and thus improve specificity), we required that patients had at least two separate years with at least one billable visit in the VA in the three years leading up to their diagnosis date. This “VA Recommended Algorithm” had the highest PPV among all algorithms in identifying patients diagnosed with MM (PPV = 0.98, 95% CI = 0.95–0.99) and in identifying patients who initiated their MM treatment in VA (PPV = 0.93, 95% CI = 0.90–0.96), while significantly increasing the sensitivity of VA Algorithm 1 for both purposes—sensitivity (95% CI) = 0.45 (0.41–0.49) vs. 0.28 (0.24–0.31) for identifying patients diagnosed with MM; 0.58 (0.54–0.63) vs. 0.35 (0.31–0.40) for identifying patients with MM who initiated their treatment in VA; Table 4). The median difference between initial MM diagnosis date indicated by the VACR among Veterans selected by the VA Recommended Algorithm and the diagnosis date annotated by reviewers was −5 days (IQR = −22 days, 6 days), and the median difference between the initial treatment date indicated by the VA Recommended Algorithm and the first treatment date annotated by reviewers was 0 days (IQR = 0 days, 3 days). We further analyzed algorithm performance by time period: for MM diagnosis in early years (1999–2009), sensitivity = 0.13, specificity = 0.99, and PPV = 0.99. In later years (2010–2021), sensitivity = 0.32, specificity = 0.94, and PPV = 0.97. For initiating treatment in early years (1999–2009), sensitivity = 0.15, specificity = 0.94, and PPV = 0.87. In later years (2010–2021), sensitivity = 0.43, specificity = 0.95, and PPV = 0.96.
Performance of VA Pharmacy Data in Initial Treatment Classification
Regarding classification of individual drugs, the PPV of VA pharmacy data was highest for V (PPV = 0.99) and R (PPV = 0.95) when using the 14-day exposure assessment window. The dexamethasone treatment codes were nonspecific (highest specificity = 0.62 using 14-day exposure assessment window); accordingly, we excluded dexamethasone from the codes used to classify initial multi-drug regimens. Regarding classification of initial multi-drug regimen, the exposure assessment window that provided the optimal balance between sensitivity and PPV for classifying initial MM treatment regimens was 14 days (Vd sensitivity = 0.90, PPV = 0.91; Rd sensitivity = 0.89, PPV = 0.89; VRd sensitivity = 0.83, PPV = 0.96; Table 5).
Table 5:
Sensitivity, specificity, positive predictive value of individual treatment codes and initial treatment regimens including lenalidomide and bortezomib measured within a 14-day exposure window starting on date of initial treatment from all five VA pharmacy databases and CPT/HCPCS procedure codes
| Classification | Sensitivity | Specificity | Positive Predictive Value |
|---|---|---|---|
| Lenalidomide, by itself or in a multi-drug regimen | 0.91 | 0.98 | 0.95 |
| Bortezomib, by itself or in a multi-drug regimen | 0.95 | 0.99 | 0.99 |
| Lenalidomide + Dexamethasone | 0.89 | 0.98 | 0.89 |
| Bortezomib + Dexamethasone | 0.90 | 0.97 | 0.91 |
| Bortezomib + Lenalidomide + Dexamethasone | 0.83 | 0.99 | 0.96 |
CPT = Current Procedural Terminology; HCPCS = Healthcare Common Procedure Coding System
DISCUSSION:
In our study comparing different selection algorithms identifying patients with MM in large databases, we found that algorithms developed specific for VA performed better than non-VA algorithms both in case identification and in identifying patients who initiated their MM treatment in VA. Algorithms that restricted to the VACR and those that incorporated MM treatment codes alongside MM ICD codes had the highest PPV, whereas algorithms that used the entire VA CDW had the highest sensitivity. We further found that VA pharmacy data performs well in classifying both individual treatments and initial treatment regimens. We modified an existing VA algorithm (VA Algorithm 1) to optimize sensitivity and PPV for cohort selection. When combined with VA pharmacy data, this “VA Recommended Algorithm” can accurately measure initial treatment date and MM drug regimens.
Our findings suggest a need for selection algorithms to be developed and validated in the databases in which they are intended to be used. We confirmed the conclusion of Brandenburg et al. that diagnostic (ICD) codes alone are insufficient to identify new cases of MM.5 This poor specificity is at least in part explained by the use of MM ICD codes for MM precursor conditions such as MGUS and smoldering MM. Currently, guidelines indicate that only active MM requires treatment,13,14 and therefore algorithms that incorporated MM treatment codes had much higher specificity.15 Moreover, the treatments commonly used for MM (e.g., lenalidomide and bortezomib) are themselves highly specific to MM and used only occasionally for other diseases.16.17 However, in contrast to Brandenburg et al.’s conclusion that an algorithm with certain core characteristics (like diagnostic procedures such as bone marrow biopsy) can perform well across datasets, we found that algorithms developed outside the VA (Non-VA Algorithms 1 and 2) performed worse than algorithms developed within VA (VA Algorithms 1 and 2) in both case identification and identification of patients who initiated their MM treatment within VA.
One of the unique strengths of VA data is the VACR, and to our knowledge we are the first to validate the VACR in correctly classifying cases of a particular cancer. Algorithms that included the VACR, including our VA Recommended Algorithm, were consistently more specific and had higher PPVs for identification of both MM cases and patients with MM initiating treatment in VA. Each VA medical facility is mandated to maintain a cancer registry with data on diagnosis and treatment of MM and other cancers in adherence with requirements set forth by several oversight bodies, including the North American Association of Central Cancer Registrations and the American Joint Commission on Cancer.18 However, we found that algorithms restricted to the VACR miss a proportion of patients (upwards of 30–40%) initiating treatment within VA, and future work should identify gaps in data collection and reporting to VACR to minimize disparities.19 More comprehensive capture of patients diagnosed with and initiating treatment within VA would not only improve the representativeness of health outcomes studies in veterans with MM but also improve the measurement of MM incidence. Moreover, performance of our VA Recommended Algorithm across treatment years was still excellent but varied slightly between early and later periods, and this should be taken into account depending on the era of focus for a given study.
Combined with our findings concerning the selection algorithms, we found that VA pharmacy data provide another unique strength that not only enhance performance of algorithms for MM case identification and treatment initiation in VA, but also provide accurate classification of initial MM treatments and accurate dates of first treatment. A large portion of Veterans with MM in our annotations were initially diagnosed and treated in external health systems—sometimes several years in advance of their first recorded MM treatment in VA data. Since previously treated patients have a substantially different prognosis than patients with newly diagnosed and newly treated MM, such misclassification could induce differential bias in studies that aim to measure survival and other outcomes in Veterans with MM.20 Moreover, many Veterans elect to receive primary management of their MM at external cancer centers but have VA pay for part of their treatment, e.g., R in someone on VRd.6 Since these Veterans will be recorded as only receiving R in VA data, selection algorithms may misclassify them as receiving Rd instead of VRd. This misclassification could induce another differential bias in studies that aim to compare the benefits and harms of different multi-drug treatment regimens such as VRd versus Rd.21 The need for algorithms and data sources that minimize immortal time bias and exposure misclassification is increasing given the plurality of three and now four-drug regimens in MM.
There are limitations to our study. First, both VA algorithms that we evaluated, as well as our Recommended VA Algorithm, require MM treatment codes for selection, precluding the possibility of studying patients diagnosed with MM but not undergoing treatment. However, this subgroup is likely small given that the treatment guidelines relevant to our study period all recommend immediate initiation of treatment upon diagnosis of active MM15; patients who do not initiate treatment are most likely too frail for treatment, decline treatment out of individual preference, or are entering hospice at end-of-life. Along with non-VA algorithm 1, other algorithms aimed at identifying MM cases independently of treatment have been developed and warrant further validation5,22 in VA and other databases. Moreover, we evaluated each algorithms’ ability to identify active, symptomatic MM, leaving a remaining need to develop and validate algorithms that identify smoldering MM and MGUS, as well as other conditions like Waldenstrom macroglobulinemia and amyloidosis.23 Finally, although we measured treatments in VA data using a current list of MM treatments, we evaluated VA pharmacy data in its ability to classify regimens containing V, R, and d. Future work should evaluate the ability of pharmacy data to evaluate other novel MM treatments, such as daratumumab.24
In conclusion, we found that VA-specific algorithms performed better than non-VA algorithms in identifying patients diagnosed with MM and patients initiating MM treatment with VA. Our Recommended VA Algorithm and findings will be of use to researchers that use VA and other large databases to study MM care and treatment outcomes. Moreover, we have shown the potential of integrating VACR, CDW, and pharmacy databases to create high-performing algorithms with which to phenotype patients with a particular disease. With further advances in tumor cytogenetics and biomarkers and greater synergy across structured and unstructured data,25 more sophisticated algorithms can be developed to advance precision medicine and redefine clinical phenotypes of patients with MM treated and followed in VA and other large databases. Future studies should also validate algorithms that measure important treatment outcomes—such as duration, discontinuation, and MM relapse—taking care to capture both internal information in VA data and external information from other data sources (e.g., CMS).
Supplementary Material
Table 3:
Results of inter-annotator agreement analyses
| Main variables | Total Annotated (n) | Percent Agreement | Fleiss Kappa Statistic (95% CI) |
|---|---|---|---|
| MM diagnosis (yes/no) | 115 | 98.3 | 0.93 (0.84–1.00) |
| Initial treatment in VA (yes/no) | 122 | 91.8 | 0.77 (0.63–0.91) |
| Initial treatment drug classification | 89 | 87.6 | 0.86 (0.78–0.94) |
| Dates of first diagnosis and treatment | Total Annotated (n) | Median, IQR | |
| Difference in days between annotators on date of MM diagnosis | 115 | 13 (0.5–37.5) | |
| Difference in days between annotators on date of initial MM treatment | 122 | 2 (0–6.5) |
CI = Confidence Interval; IQR = interquartile range
KEY POINTS:
The U.S. Veterans Affairs Healthcare System is one of the largest, centralized databases that provides opportunities to study real-world treatment trends and outcomes in patients with multiple myeloma, but no selection algorithm has been validated for selecting patients with newly diagnosed and newly treated myeloma
We validated different algorithms developed within and outside VA, and found that algorithms developed specific for VA performed better than non-VA algorithms both in case identification and in identifying patients who initiated their MM treatment in VA
We developed a “VA Recommended Algorithm” optimizes sensitivity and positive predictive value for cohort selection and treatment classification
PLAIN LANGUAGE SUMMARY:
Large research databases provide opportunities to study treatments and outcomes in patients with multiple myeloma (MM). The U.S. Veterans Affairs (VA) Healthcare System contains one of the largest, centralized MM databases available, but validated ways to identify newly diagnosed and treated patients are lacking. We brought together a team with clinical expertise to review oncology notes for each patient to annotate details regarding MM diagnosis and initial treatment within VA. We compared several methods of identifying patients with MM against these annotations in terms of the methods’ ability to identify patients who are diagnosed with MM, who began their treatment in VA, and what specific drugs with which the patients were treated. We found that methods originally developed using VA data outperformed methods originally developed in other databases, and we developed a new method which has superior accuracy among all methods compared. Our new method will be of use to researchers that use VA and other large databases to study MM care and treatment outcomes. Moreover, we have shown the potential of combining data from billing codes, pharmacy files, and other sources to accurately classify patients in large databases.
Funding:
This work was supported by the VA Office of Research and Development, Cooperative Studies Program (NRF, NVD, MTB); HCSRN-OAICs AGING Initiative Pilot (1R33AG057806) (CD, NRF), Harvard Translational Research in Aging Training Program (National Institute on Aging of the National Institutes of Health: T32AG023480) (CD); VA Career Development Award IK2CX002218 (CD); VA Merit Review Award 1I01BX001584 (NCM), and NIH grants P01-155258-07 and P50-100707 (NCM). Support for VA/CMS data is provided by the Department of Veterans Affairs, VA Health Services Research and Development Service, VA Information Resource Center (Project Numbers SDR 02-237 and 98-004)
Conflicts of Interest:
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. NCM is a consultant for Adaptive Biotechnology, Amgen,Bristol-Myers Squibb, Celgene, Glaxosmithkline, Janssen Biotech, Karyopharm Therapeutics, Legend, Pfizer Pharma GMBH, Novartis,Takeda Oncology and is a DSMB member at OncoPep, for work unrelated to the current study. NCM also has two grants(Description: NIH P01-155258 and NIH 5P50 CA100707) with the National Cancer Institute.
Footnotes
Disclaimer: The views expressed are those of the authors and do not represent the views of VA or the United States Government.
Institutional Review Board Statement: This study was approved by the VA Boston Healthcare System Institutional Review Board, numbers 1582165–2 and 1578039–10.
Informed Consent Statement: Patient consent was waived due no more than minimal risk to the privacy of individuals in this retrospective project.
Data Availability Statement:
The data underlying this article were accessed from the VA Corporate Data Warehouse. The derived data generated in this research may be shared on reasonable request to the corresponding author as permitted by VA policy.
References
- 1.Richardson PG, San Miguel JF, Moreau P, et al. Interpreting clinical trial data in multiple myeloma: translating findings to the real-world setting. Blood Cancer J. 2018;8(11):109. doi: 10.1038/s41408-018-0141-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DuMontier C, Fillmore NR, Yildirim C, et al. Contemporary Analysis of Electronic Frailty Measurement in Older Adults with Multiple Myeloma Treated in the National US Veterans Affairs Healthcare System. Cancers. 2021;13(12):3053. doi: 10.3390/cancers13123053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fillmore NR, DuMontier C, Yildirim C, et al. Defining Multimorbidity and Its Impact in Older United States Veterans Newly Treated for Multiple Myeloma. JNCI: Journal of the National Cancer Institute. 2021;113(8):1084–1093. doi: 10.1093/jnci/djab007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fillmore N, DuMontier C, Cheng D, et al. Multimorbidity patterns and their association with survival in a large national cohort of older veterans with multiple myeloma. JCO. 2019;37(15_suppl):8033–8033. doi: 10.1200/JCO.2019.37.15_suppl.8033 [DOI] [Google Scholar]
- 5.Brandenburg NA, Phillips S, Wells KE, et al. Validating an algorithm for multiple myeloma based on administrative data using a SEER tumor registry and medical record review. Pharmacoepidemiology and Drug Safety. 2019;28(2):256–263. doi: 10.1002/pds.4711 [DOI] [PubMed] [Google Scholar]
- 6.Aggarwal NK. Ramifications of the VA MISSION Act of 2018 on Mental Health: Potential Implementation Challenges and Solutions. JAMA Psychiatry. 2020;77(4):337–338. doi: 10.1001/jamapsychiatry.2019.3883 [DOI] [PubMed] [Google Scholar]
- 7.Deckro J, Phillips T, Davis A, et al. Big Data in the Veterans Health Administration: A Nursing Informatics Perspective. J Nurs Scholarsh. 2021;53(3):288–295. [DOI] [PubMed] [Google Scholar]
- 8.Singh JA, Holmgren AR, Noorbaloochi S. Accuracy of veterans administration databases for a diagnosis of rheumatoid arthritis. Arthritis Care Res. 2004;51(6):952–957. doi: 10.1002/art.20827 [DOI] [PubMed] [Google Scholar]
- 9.Price LE, Shea K, Gephart S. The Veterans Affairs’s Corporate Data Warehouse: Uses and Implications for Nursing Research and Practice. Nurs Adm Q. 2015;39(4):311–318. doi: 10.1097/NAQ.0000000000000118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein M, Scaria G, Ganti AK. Utilization of the Veterans Affairs Central Cancer Registry to evaluate lung cancer outcomes. Semin Oncol. 2019;46(4–5):321–326. doi: 10.1053/j.seminoncol.2019.10.001 [DOI] [PubMed] [Google Scholar]
- 11.Committee on Future Information Architectures Processes and Strategies for the Centers for Medicare and Medicaid Services; Shortliffe EH; Millet LI; National Research Council (U.S.); Division on Engineering and Physical Sciences; National Research Council (U.S.); Computer Science and Telecommunications Board; ebrary Inc. Strategies and Priorities for Information Technology at the Centers for Medicare and Medicaid Services; National Academies Press: Washington, DC, USA, 2012. Available online: https://yale.idm.oclc.org/login?URL=http://site.ebrary.com/lib/yale/Doc?id=10531101 [Google Scholar]
- 12.Sanfilippo KM, Luo S, Wang TF, et al. Predicting venous thromboembolism in multiple myeloma: development and validation of the IMPEDE VTE score. American Journal of Hematology. 2019;94(11):1176–1184. doi: 10.1002/ajh.25603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.KYLE RA, BUADI FI, RAJKUMAR SVE. Management of Monoclonal Gammopathy of Undetermined Significance (MGUS) and Smoldering Multiple Myeloma (SMM). Oncol Williston Park N. 2011;25(7):578–586. [PMC free article] [PubMed] [Google Scholar]
- 14.Landgren O, Kyle RA, Rajkumar SV. From Myeloma Precursor Disease to Multiple Myeloma: New Diagnostic Concepts and Opportunities for Early Intervention. Clin Cancer Res. 2011;17(6):1243–1252. doi: 10.1158/1078-0432.CCR-10-1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. American Journal of Hematology. 2020;95(5):548–567. doi: 10.1002/ajh.25791 [DOI] [PubMed] [Google Scholar]
- 16.Lenalidomide - NCI. Published October 5, 2006. Accessed May 31, 2022. https://www.cancer.gov/about-cancer/treatment/drugs/lenalidomide
- 17.Bortezomib - NCI. Published October 5, 2006. Accessed May 31, 2022. https://www.cancer.gov/about-cancer/treatment/drugs/bortezomib
- 18.Directive VHA 1412 Department of Veterans Affairs Cancer Registry System. Published online 2019:10. Available from: https://www.va.gov/vhapublications/ViewPublication.asp?pub_ID=8368
- 19.Yang DX, Khera R, Miccio JA, et al. Prevalence of Missing Data in the National Cancer Database and Association With Overall Survival. JAMA Netw Open. 2021;4(3):e211793. doi: 10.1001/jamanetworkopen.2021.1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernán MA, Sauer BC, Hernández-Díaz S, Platt R, Shrier I. Specifying a target trial prevents immortal time bias and other self-inflicted injuries in observational analyses. J Clin Epidemiol. 2016;79:70–75. doi: 10.1016/j.jclinepi.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prada-Ramallal G, Takkouche B, Figueiras A. Bias in pharmacoepidemiologic studies using secondary health care databases: a scoping review. BMC Med Res Methodol. 2019;19:53. doi: 10.1186/s12874-019-0695-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiala MA, Dukeman J, Tuchman SA, Keller M, Vij R, Wildes TM. Development of an Algorithm to Distinguish Smoldering Versus Symptomatic Multiple Myeloma in Claims-Based Data Sets. JCO Clin Cancer Inform. 2017;1. doi: 10.1200/CCI.17.00089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Development of an Algorithm to Distinguish Smoldering Versus Symptomatic Multiple Myeloma in Claims-Based Data Sets | JCO Clinical Cancer Informatics. https://ascopubs.org/doi/full/10.1200/CCI.17.00089 [DOI] [PMC free article] [PubMed]
- 24.Nooka AK, Kaufman JL, Hofmeister CC, et al. Daratumumab in multiple myeloma. Cancer. 2019;125(14):2364–2382. doi: 10.1002/cncr.32065 [DOI] [PubMed] [Google Scholar]
- 25.Inam S, Ross JA, Touzeau C, Moreau P, Kumar SK, Harrison SJ. Paving the way to precision medicine in multiple myeloma. Expert Rev Hematol. 2021;14(4):323–327. doi: 10.1080/17474086.2021.1905515 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article were accessed from the VA Corporate Data Warehouse. The derived data generated in this research may be shared on reasonable request to the corresponding author as permitted by VA policy.