Abstract
Organs-on-a-chip, or OoCs, are microfluidic tissue culture devices with micro-scaled architectures that repeatedly achieve biomimicry of biological phenomena. They are well positioned to become the primary pre-clinical testing modality as they possess high translational value. Current methods of fabrication have facilitated the development of many custom OoCs that have generated promising results. However, the reliance on microfabrication and soft lithographic fabrication techniques has limited their prototyping turnover rate and scalability. Additive manufacturing, known commonly as 3D printing, shows promise to expedite this prototyping process, while also making fabrication easier and more reproducible. We briefly introduce common 3D printing modalities before identifying two sub-types of vat photopolymerization - stereolithography (SLA) and digital light processing (DLP) - as the most advantageous fabrication methods for the future of OoC development. We then outline the motivations for shifting to 3D printing, the requirements for 3D printed OoCs to be competitive with the current state of the art, and several considerations for achieving successful 3D printed OoC devices touching on design and fabrication techniques, including a survey of commercial and custom 3D printers and resins. In all, we aim to form a guide for the end-user to facilitate the in-house generation of 3D printed OoCs, along with the future translation of these important devices.
Graphical Abstract
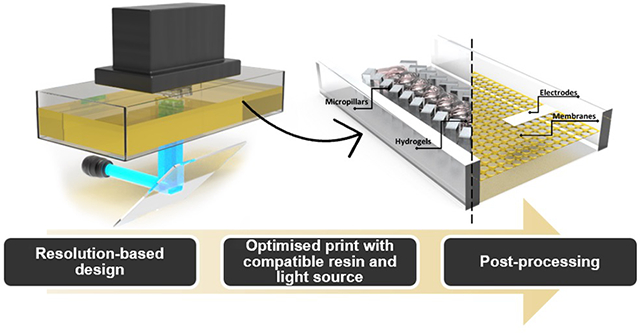
We provide a guide for 3D-printed microfluidic device fabrication in response to the recognition of 3D-printing as an accessible fabrication method.
1. Introduction
Organ-on-a-chip (OoC) platforms, typically comprising perfused cell cultures in micro-scale microfluidic devices, are quickly becoming the predominant pre-clinical in vitro testing method1,2. It is anticipated that as these devices continue to demonstrate translational outcomes, more researchers will develop and customize OoCs for specific applications within their laboratories. Polydimethylsiloxane (PDMS) replica molding of planar sheets with embedded microchannels is the current state of the art, and many researchers successfully utilize this material for custom OoCs. However, PDMS OoCs are limited by non-specific drug absorption issues, are difficult to democratize due to reliance on microfabrication infrastructure and are not compatible with processes for large-scale manufacturing3. To best democratize the in-house development of OoCs, additive manufacturing, better known as 3D printing, may be the fabrication method of the future, where researchers without access to cleanroom facilities may begin making their own OoCs. In the past five years, much work has been put into developing 3D printing techniques for microfluidic devices4,5 with a focus on increasing the resolution to achieve micro-scale features, developing unique, complex geometries, reducing fabrication times, and making prototype development accessible to more laboratories. Of the many forms of 3D printing available, material extrusion (fused filament fabrication (FFF)), material jetting (multi jet modeling (MJM)), and vat photopolymerization in the form of two-photon polymerization (TPP), stereolithography (SLA) and digital light processing (DLP) are the predominant modalities that have been explored for fabricating microfluidic devices.
Among the different 3D printing modalities that have been developed for microfluidic applications, vat photopolymerization methods, specifically SLA and DLP printing has made the most significant progress in recent years to achieve microfluidic voids with sufficiently small resolution (sub 100 μm) with good print fidelity. Moreover, significant efforts have been made to develop SLA/DLP printable photoresins that are biocompatible and transparent6–8.
Therefore, we expect the realization of 3D printed OoC devices will leverage recent advancements in vat-polymerization-based 3D printed microfluidic technology. However, achieving comparable functionalities to traditional PDMS-based OoCs through 3D printing is not a straightforward task. This challenge stems from the necessity of carefully selecting and aligning 3D printer configurations and operating parameters with the desired OoC functionalities. Consequently, this review aims to serve as a comprehensive guide for OoC developers interested in transitioning to 3D printing as a fabrication method, emphasizing crucial considerations for this endeavor.
We begin by providing an overview of vat polymerization-based 3D printing, exploring various common classes of 3D printing technologies. This understanding will enable us to grasp why SLA/DLP printing methods hold advantages for generating microfluidic devices. Subsequently, we delve into the motivations behind 3D printing OoCs, elucidate the shared features and functional requirements of such devices, address considerations specific to SLA/DLP 3D printing processes, and present an outlook on 3D printed OoC devices. Through this review, readers will gain the fundamental knowledge necessary to analyze their desired biological applications, prioritize resolution, biocompatibility, and other vital device functions, and make informed decisions when selecting an appropriate 3D printer and resin for generating and testing OoC devices.
2. Brief introduction to 3D printing modalities for microfluidic devices
This review focuses on SLA and DLP 3D printing as the primary 3D printing modality for future OoC generation. Though, in order to make this review accessible to researchers who are not familiar with 3D printing, this section provides a brief introduction to 3D printing modalities for microfluidic devices. As these modalities have been extensively reviewed and compared4,5,9,10, here we only provide a brief introduction to each, along with a summary of their respective advantages and limitations. This allows us to highlight SLA/DLP as the likely future of OoC 3D printing.
2.1. Fused deposition modeling
Fused Filament Fabrication (FFF), also known by the trademark Fused Deposition Modeling (FDM), is a material extrusion printing method and the most popular additive manufacturing technique across all 3D printing applications, owing to its cost-effectiveness and simplicity5. It involves the extrusion of a thermoplastic polymer through a heated nozzle onto a cooled plate5. While FDM printing has been successfully utilized to print OoCs, such as the polystyrene-based chips made by Mader et al.11, the complex, winding geometries achieved by Kotz et al.12, and the multi-material membrane-containing devices from Li et al.13, there remain significant limitations of this modality. Sun et al. demonstrated that printing 300 μm by 300 μm microchannels was possible14, though Mader et al.11 reported that they were unable to reproducibly print 200 μm by 200 μm channels due to sagging and clogging11. while this was an improvement from the minimum feature dimension of 321 μm shown by Macdonald et al.10, it remains nonetheless a barrier to creating accurate and effective microfluidic devices. Additionally, microfluidic devices fabricated from FDM printing often suffer from poor optical clarity with optical transmission of visible light ranging from 50–83% due to print surface roughness11,12,15. Moreover, the extruded polymer filaments often bond poorly to one another leaving voids in the prints that can limit the integrity of the microfluidic devices16. Consequently, other methods have been pursued with more success for 3D printing microfluidic devices.
2.2. Multi jet modelling
Material jetting, also known as multi-jet modeling and better known by the trade name PolyJet printing, involves the layer-by-layer deposition of photopolymer droplets that are immediately solidified with UV light. Interestingly, multiple different polymers can be used in one print, giving flexibility and adding functionality to the microfluidic devices. This method has been shown to effectively print a two-material device with transparent, hard plastic comprising most of the device, and a flexible rubber-like material being used for valving action areas17. Moreover, it has been effective in generating complex, multi-material vacuum pumps to mimic a miniaturized diaphragm18. Unfortunately, resolution remains an issue in PolyJet printing, with photopolymer flow before polymerization being attributed to most of this effect. For example, Ong et al. found that PolyJet printing was incapable of printing dimensions of 130 μm or 250 μm in the x-y dimension19, and the Breadmore group found the minimum printing resolution for PolyJet printers to be 205 μm10. Ultimately, it appears that sub-100 μm void features are difficult to achieve with this method4. Another key limitation of PolyJet printing is the clearance of internal voids. In order to make channels, a sacrificial matrix must be placed in the space where channels will be4. These sacrificial matrices can then be difficult to remove at later steps leading to poor print fidelity and poor reproducibility. As with FDM printing, transparency is a limitation due to print roughness. This forces researchers to consistently use post-processing work-arounds like the addition of nail polish17 or mineral oil20 to increase transparency.
2.3. Two-photon polymerization
Vat photopolymerization involves the curing, layer by layer, of a polymer base held in a vat, whereby the layered resin structure is generated on a build plate. The 3D printed device can be built from the bottom up using a ‘free-surface’ configuration, with the laser or bulb at the top of the printer, solidifying the first layer on a platform immersed in the vat, then moving the platform down and adding another layer on top. Alternatively, the laser or bulb can be configured at the bottom of the printer, such that the top layer of the device is printed first, and the subsequent layers are added below as the build platform is drawn out of the resin vat (Figure 1). One such method is two-photon polymerization (TPP).
Fig. 1.
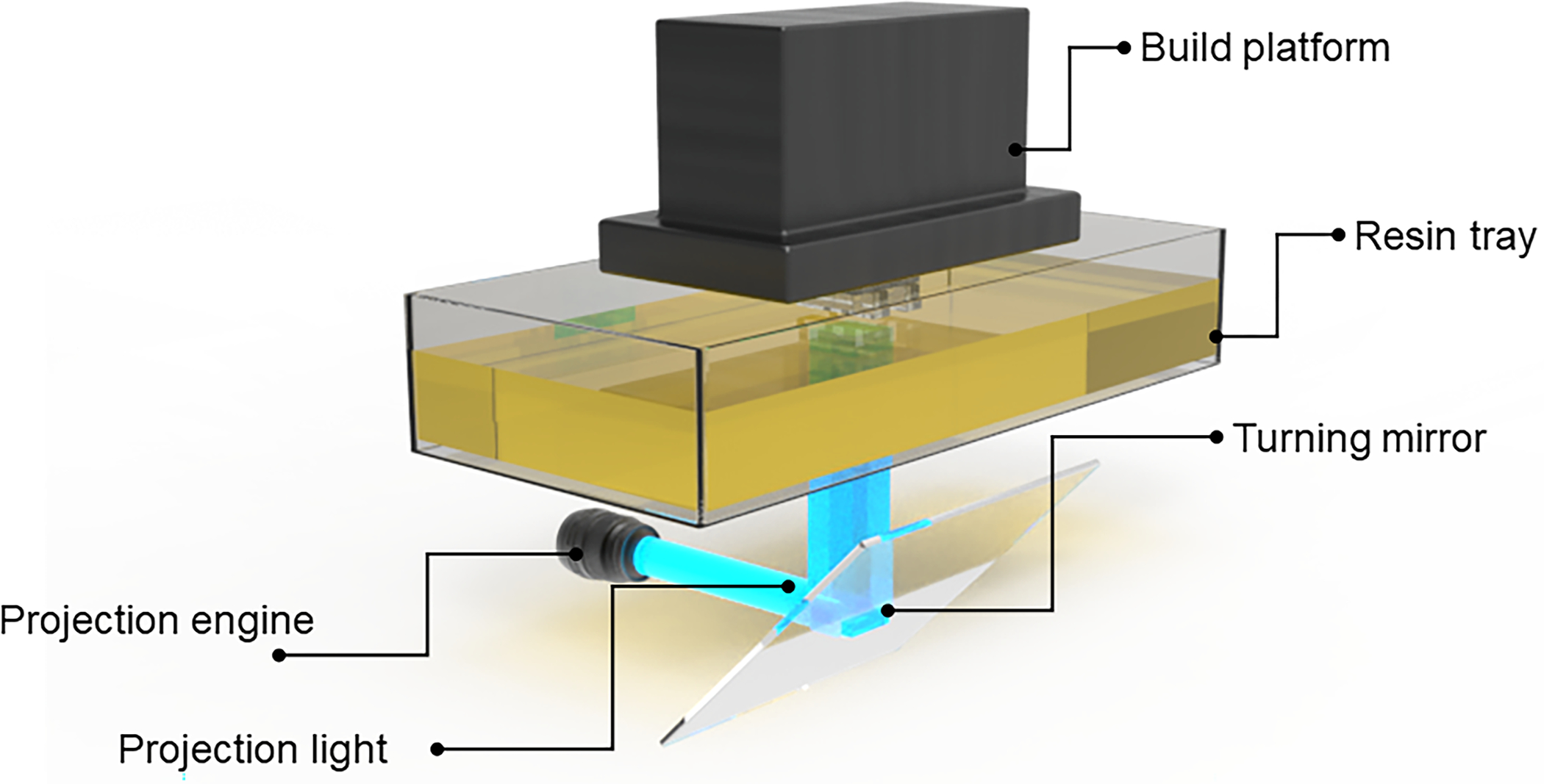
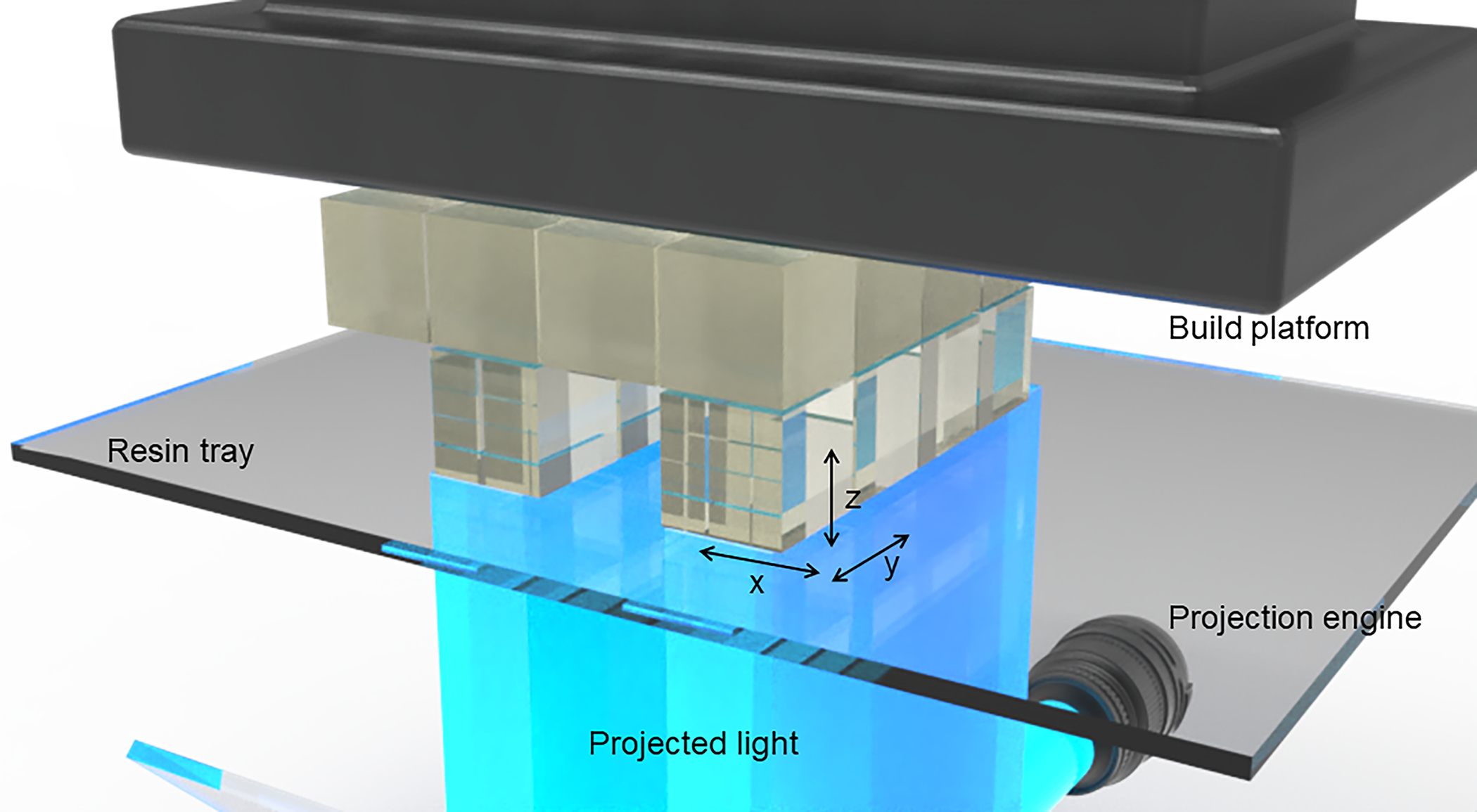
Anatomy of a DLP-SLA 3D printer: (A) Build platform for containing printed objects, a resin tray to house the printing resin, turning mirror to guide the light projection and optical engine as light source. (B) Close-up of an object being 3D printed through a bottom-up approach (‘bat’ configuration) by cured resin along a plane where the x-, y-resolution is determined by the pixel resolution of the light source and the z-resolution is determined by the optical penetration depth through the resin.
Two-photon polymerization (TPP) processes create solid structures by scanning a tightly focused laser throughout a 3D space containing a TPP resin and then removing the unpolymerized resin in a post processing step. Polymerization is only initiated within a confined focal volume called a voxel. The voxel is typically very small, ranging from 400–1000 nm in commercially available systems21 down to 100–300 nm in custom laboratory setups22. This allows for the fabrication of 3D printed parts with very intricate details. In fact, 3D printed voids with cross sections ranging from 40 μm × 100 μm down to 0.25 μm × 0.25 μm have been demonstrated23–29. There are a variety of TPP resins available, each with various strengths and limitations30. While TPP boasts extraordinary resolution, there are several major drawbacks for OoC applications. The first is long fabrication times and small print volumes due to the serial, scanned nature of the TPP process22,30. TPP is generally better suited for fabricating smaller, high resolution components that are integrated into larger devices31. Moreover, instrumentation for TPP printing is expensive owing largely to the use of a pulsed femtosecond laser and highly specialized optics. Commercial systems are available from NanoScribe for around 500,000 USD32, and from Microlight3D33 for around 200,000 USD, among others.
2.4. Stereolithography digital light processing
Both SLA and DLP can be referred to as ‘vat photopolymerization’ modalities, whereby a vat of photopolymer is cured, layer by layer, by UV light sourced from either a laser or LED for SLA and DLP, respectively. These methodologies have recently become the front-runners for the 3D printing of OoCs due to their ability to balance resolution, fabrication time, and cost-effectiveness. While they are distinct methods, their similarities in fabrication often lead them to be discussed as one – SLA/DLP. As such, this review will henceforth discuss SLA/DLP and mean it to be understood as different modalities with similar outputs. Much of the recent research into 3D printed microfluidic devices has focused on SLA/DLP printing20,34,35, owing primarily to advances in accuracy and resolution. In 2017, the Nordin group demonstrated effective SLA/DLP printing of 18 μm by 20 μm microfluidic channels using a custom 3D printer and custom resin8. In commercially available SLA/DLP printers, resolutions of 100 μm are readily accessible, but with optimization, resolutions down to as low as 27 μm35,36 have been reported. This allows the devices to reach ‘truly microfluidic’ levels and facilitates the inclusion of more complex geometries into micro-scale platforms. For example, Kuo et al. demonstrated the ability to SLA/DLP print a ‘3D router’ with stacked, overlapping microchannels, a passive chaotic micromixer and even a double-helix-like intertwining microchannel design35. Further, Gong et al. printed high density microfluidic devices including active components like pumps and valves with 20 μm SLA/DLP printed membranes37, and Sanchez Noriega et al. demonstrated the printing of very small fluidic components such as sub-20 μm valves used in a complex serial dilution assay38. Importantly, DLP printing in particular is high speed, allowing for rapid prototyping with print times ranging from minutes to a few hours depending on the number of layers. Each layer can be arbitrarily complex without increasing print time. Moreover, SLA/DLP printed devices can be highly transparent with appropriate printing set ups and post-processing techniques, making them more compatible with imaging-based measurements often used for microfluidic devices39–41. In all, these benefits position SLA/DLP as the most promising 3D printing modality to further develop microfluidic devices specifically for cell and tissue culture applications.
3. Motivations for using 3D printing to fabricate OoC devices
To appreciate the motivations behind 3D printing OoCs, one must understand the strengths and limitations of the current state of the art. The majority of researchers who generate lab-based OoCs utilize polydimethylsiloxane (PDMS) soft lithography. PDMS is a flexible and transparent elastomer, and soft lithography refers to the casting of a mix of PDMS base and crosslinker solution (often 10:1 base:crosslinker) into a microfabricated mold (most commonly fabricated on silicon) followed by PDMS crosslinking, whereupon the PDMS slab containing the fluidic network is demolded and plasma bonded to a glass slide or coverslip to create the microfluidic device. This fabrication process results in planar 2D microfluidic networks, although multiple PDMS layers can be aligned, stacked and bonded together to create 3D channel networks, albeit, this is a laborious process.
In many respects, PDMS microfluidic devices are well suited to support cell cultures for OoC development. PDMS is transparent, biocompatible, sterilizable, flexible, and gas permeable42. The many favorable characteristics of PDMS have allowed researchers to generate device architectures to grow cells in various tissue-mimetic configurations. For instance, Van der Meer et al.43 generated a PDMS device that successfully cultured an endothelial cell tube that aligned with the curvature of the designed channels, and were able to demonstrate the impact of cytokines on tube diameter and convolution. Moreover, Kim et al.44 utilized a PDMS membrane within a gut-on-a-chip device to demonstrate that the shear stress generated by flow is required for the polarization of gut epithelial cells. Many others have generated PDMS organ-on-a-chip devices like the lung-on-a-chip with stretchable alveoli45, the blood-brain-barrier device with integrated transendothelial resistance probe46 and the heart-on-a-chip for monitoring electrophysiology in acute hypoxia conditions47. While PDMS casting typically generates planar geometries, researchers have used stacked layers of PDMS to generate more complex geometries. For example, Kniazeva et al. fabricated a multilayer PDMS lung-on-a-chip with native-like gas permeance48. Companies like AIMBiotech49, Mimetas50 and CNBio51 have successfully translated PDMS-based devices into plastics for large-scale distribution, while TissUse52 and Emulate53 have effectively up-scaled the development of circulatory systems encompassing multiplePDMS devices. These devices continue to contribute to a wealth of knowledge in the OoC field. Importantly, the development and translation of PDMS-based OoC devices have made invaluable impacts on the scientific field and thus have been extensively reviewed in recent years1,54–57.
Despite the advantages and wide use of PDMS, there remain important limitations of its use, especially during the translational phase of the technology. These have been discussed at length3,42 and are a key barrier to the adoption of OoC technologies for routine clinical or biological in-vitro experimentation. The key barriers to translation are firstly, non-specific drug absorption limiting the utility of drug studies, secondly, difficulty democratizing the technology and finally, incompatibility with manufacturing processes. These, and some other minor limitations, will be discussed here.
A key application of OoC technologies is in-vitro drug testing, including toxicity screening58,59 and physiologically-based pharmacokinetic (PBPK) studies60–63. However, an often-cited disadvantage of PDMS OoC devices for drug testing applications is the non-specific absorption of small molecules. This is owing to PDMS porous and hydrophobic nature, and has precluded its use in many drug studies, and those looking to analyze secreted proteins or small molecules. In fact, Skardal et al.64 noted that PDMS could not be used in their multi-organ-chip for drug studies as the adsorption of the drugs would fundamentally confound their results. Media formulations can also be impacted by this non-specific absorption, with one study identifying the removal of estrogen, a small hydrophobic molecule, from cell culture media in a PDMS device65. While this effect can be accounted for using complex modeling of PDMS drug absorption66, this requires theoretical and experimental validation each time a new configuration, cell type or drug is introduced. It has also been noted that PDMS can leach unpolymerized polymers, damaging cell experiments. Regehr et al.65 identified uncured polymers in culture media and cells contained in PDMS microchannels, indicating that PDMS may alter media formulations and cell cultures. Uncrosslinked PDMS polymers on cell membranes may interfere with studies of cell membrane protein signaling, along with protein trafficking across a membrane3. However, no evidence has implicated PDMS in cytotoxicity, and researchers have consistently demonstrated the biocompatibility of PDMS42. In fact, researchers have reported long-term culture of typically sensitive cells such as neurons67 and primary cells68 in PDMS devices. Additionally, the hydrophobicity of PDMS does not lend itself to cell attachment or proliferation. As such, researchers must coat or treat PDMS to increase its hydrophilicity, adding yet another step to the fabrication process. Overall, the use of PDMS in cell culture can be impacted by absorption of small molecules, leaching, and its hydrophobic surface properties. These biological impacts can significantly interfere with in vitro drug-testing results and limit the scope of studies utilizing PDMS microfluidic devices.
PDMS OoC generation can be difficult to democratize because of the multi-step prototyping process and reliance on access to cleanrooms and skilled technicians to successfully and reproducibly fabricate devices. Prototyping starts with the generation of high resolution molds. This is typically achieved utilizing SU-8 soft lithography in a cleanroom. Access to cleanrooms and skilled researchers trained in optimizing photolithography processes (e.g. temperature ramping, exposure, baking and developing times of photoresist) can be a significant barrier for researchers outside the microfabrication field. A workaround is outsourcing although this can greatly increase prototyping time and cost. Once a mold is generated, PDMS replica molding poses as another time-consuming step. Firstly, PDMS must be mixed, degassed, poured into/onto a mold, prior to crosslinking. The PDMS prepolymer must then be crosslinked before the device is demolded carefully. Once demolded, the PDMS slab needs to be trimmed to the desired size and cleaned before being plasma bonded to a capping substrate, such as a glass slide. If the device design contains multiple microfluidic layers, the PDMS layers will need to be manually aligned and bonded together, which is operator-dependent and hence a source of inconsistency. This has motivated the development of techniques and tools to improve the efficiency of PDMS device fabrication. For instance, more efficient aligning methods have been developed, such as the stereomicroscope-based aligner created by He et al.69. Commercially available pick-and-place tools commonly utilized in microchip generation have potential to be adapted to the manufacturing of PDMS devices, though the uptake of such tools will rely on their adaptation to the flexible, elastic nature of PDMS. Several companies offer turn-key ‘soft-lithography workstations’ which is more affordable than setting up a conventional cleanroom. Nonetheless, it remains challenging to produce PDMS microfluidic devices at-scale in a highly reproducible manner, which may impact the quality of the OoCs and the biological data obtained from the model.
Since PDMS microfluidic devices are not compatible with large-scale manufacturing processes, owing to their manual fabrication and material properties being incompatible with scaling-up3, there is often a need to pivot to using manufacturing-compatible plastics like polystyrene (PS), polymethyl methacrylate (PMMA) and polycarbonate (PC) during the translational phase of OoC technologies. These thermoplastics have standardized and well-established manufacturing processes. Moreover, the well-established surface modification processes of tissue-culture plastic labwares can be adopted to have better control over protein and drug adsorption properties as compared to PDMS devices. However, plastic manufacturing techniques capable of producing high resolution features in microfluidic OoCs, such as injection molding, are only cost-effective at high production units (e.g. the Economic Batch Quantity (EBQ), a indicator for the number of production units to be cost-efficient for a particular manufacturing process, for injection molding is >10,00070). Therefore, it is cost-prohibitive to use these manufacturing-scale fabrication techniques during the product development phase or when smaller production units are desired, given that the demand for OoC devices is unlikely to reach the scale of a generic labware commodity. In addition, PDMS-based OoCs often contain mechanically active features, such as integrated Quake-style valves and pumps and thin flexible membranes, which are not always possible to fabricate with hard plastics. A redesign of an OoC device, initially conceptualized and developed based on PDMS, to match the requirements or constraints of manufacturing-compatible fabrication processes is costly and time-consuming, which greatly impedes their practical translation into the hands of end users. A way to facilitate and accelerate commercial translation of OoC technologies may be to conceptualize, design and develop OoC devices based on more manufacturable materials and processes. 3D printing provides an avenue to explore and test new design ideas to achieve similar functions to control the cell culture environment, such as cell compartmentalization and immobilization or hydrogel patterning, without reliance on micron-size structures and flexibility of the material.
Another impetus to use 3D printing to develop OoCs is the creation of new complex 3D geometries which can confer new functions to the device. 3D printing can generate geometries that are complex in the z-direction in a single print, greatly increasing the complexity of possible monolithic OoCs. This method of fabrication truly unlocks the use of the third dimension, allowing for structures that are much more complex and potentially more effective than ubiquitous 2D features. For example, Bertsch et al. 3D printed a micromixer with intersecting channels that ran in all three axes71, and Kuo et al. demonstrated the ability to print a 3D fluid router35. This is compared to PDMS microfluidics, which must be generated as 2D planar layers that are stacked and bonded together. Moreover, 3D printing reduces the turnaround time to take a device from ideation to a physical prototype ready for testing. This can be particularly valuable when researchers are looking to test a variety of different structures/topologies before determining a final design. As such, 3D printing has demonstrated potential to revolutionize the fast prototyping of OoC devices, and with more optimization from the OoC field, it is predicted to become the predominant method for generating lab-based in-vitro assays.
4. Common features and functional requirements for OoC applications
For 3D printed OoCs to be relevant and competitive with current fabrication techniques, they must fulfill functional properties achieved by the current gold-standard: PDMS microfluidic devices. These properties form a list of requirements and features important to 3D printing OoCs, which will be discussed in the context of 1) maintaining tissue culture within an OoC device; 2) stimulating cells in an OoC; and 3) measuring cell phenotype and functions in an OoC (Figure 2). Importantly, different features and requirements are of greater importance to different OoC-specific applications, and this is often defined by cell culture configuration and biological readouts required for specific organ systems, and the final application.
Fig. 2.
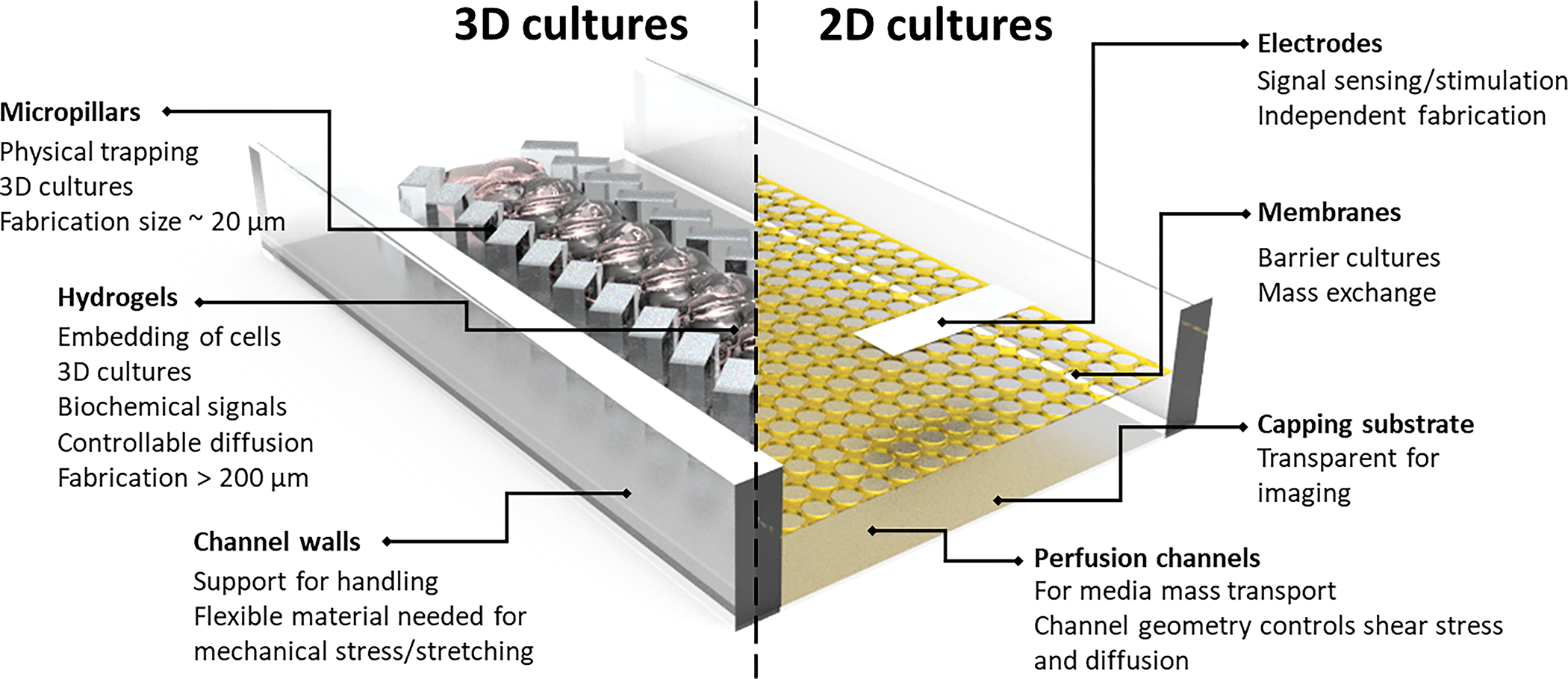
Key features and functional requirements of OoC microfluidic devices for 2D cultures relevant to barrier tissues e.g. epithelium and endothelium, and 3D cultures relevant to parenchyma and connective tissues e.g. liver, bone, heart. Connections between the fluidic networks to external pumps, valves and other devices are typically facilitated via commercial fitting (e.g. Luer fittings) at the cost of device footprint due to their larger liquid volume. Customized slip-fit connectors can be utilized to reduce device footprint.
4.1. Maintaining cells in OoCs
Cell culture configurations for OoC cultures are often distinct from other microfluidic applications like analytical chemistry and direct cell-based assays. Generally, they can be divided into 2 categories: 3D cell cultures and 2D monolayer or barrier tissue cultures57. 3D cell culture configurations using spheroids or cell-laden hydrogels are often employed to mimic parenchymal and connective tissues, such as liver, tumor, bone and cartilage. Spheroid and hydrogel cultures often rely on physical patterning using microstructures, such as micropillars72 or ‘phaseguides’73 to confine cells and hydrogels to specific locations within the microfluidic chamber. On the other hand, culture configurations to support barrier tissues, such as airway and intestinal epithelium tissues, involve an internal porous membrane facilitating a liquid-liquid or air-liquid interface. Devices designed for direct cell attachment to form 2D monolayer cultures may have different material requirements compared to those containing only non-adherent cells. Each of these factors must be considered during the initial conceptualization stage of the device.
The achievable resolution of the fabrication technique used to create an OoC device is a key consideration, and is clearly impacted by the desired culture configuration. For PDMS-based OoCs, this is generally not a limiting factor because soft lithography and PDMS replica molding can routinely generate features down to < 10 μm. However, since 3D printing cannot readily achieve such high resolution, it is important to understand the minimum feature sizes required to implement specific culture configurations. Table 1 summarizes common dimensions of different OoC features commonly found in existing OoC devices, which may help to inform corresponding designs in 3D printed counterparts.
Table 1.
Common structural features in OoC devices and their nominal dimensions
| Structure | Design | Dimensions | Fabrication Technique | Reference |
|---|---|---|---|---|
|
| ||||
| Device channels | Basic channel | 1 mm x 600 μm (width x height) | SLA/DLP | 74 |
| 2 mm x 300 μm (width x height) | Soft lithography | 75 | ||
| 300 μm x 1mm (width x height) | Soft lithography | 76 | ||
| 500 μmm x 75 μmm (width x height) | Soft lithography | 77 | ||
| 100 μm x 100 μm (width x height) | SLA/DLP | 78 | ||
| 18 μm x 20 μm (width x height) | SLA/DLP | 8 | ||
|
| ||||
| Microstructures for trapping cells/spheroids | Microwells | 400 μm x 600 μm (width x height) | Soft lithography | 79 |
| 1 mm (diameter) | Soft lithography | 80 | ||
|
| ||||
| Micropillars | 50 μm x 30 μm (width x length) | Soft lithography | 81 | |
| 40 μm (diameter) | Soft lithography | 82 | ||
| 14 μm (width) | SLA/DLP | 83 | ||
|
| ||||
| Structures for patterning gels | Micropillars | 100 μm (spacing) | Soft lithography | 84 |
| 250 μm x 250 μm (width x height) | Soft lithography | 85 | ||
| 250 μm x 250 μm (width x height) | Soft lithography | 86 | ||
|
| ||||
| Microchannels | 30 μm (width) | Soft lithography | 87 | |
| 27 μm (width) | SLA/DLP | 36 | ||
|
| ||||
| Barriers for cell migration, soluble factors or stimulation | Microchannels | 10 μm (width) | Soft lithography | 88 |
| 2 μm (width) | Soft lithography | 89 | ||
|
| ||||
| Porous membranes | 10 μm pores | Soft lithography | 90 | |
| 7 μm hexagonal pores | Soft lithography | 91 | ||
| 2.5–3.3 um pores | Soft lithography | 92 | ||
| 0.4 μm pores | Soft lithography with integrated PET or PTFE membrane | 93 | ||
| 0.4 μm pores | Soft lithography with integrated PET membrane | 94 | ||
For 3D spheroid-based cultures, micro-structures, such as micro-wells or micro-pillars, are used to physically immobilize cells that are introduced into the device. For this cell trapping mechanism, the microstructures and pitch (spacing between adjacent structures) sizes can vary from 10–30 μm81,95,96 to a few hundred μm97–99, depending on whether one wishes to immobilize single cells or pre-formed spheroids, respectively. Hence, there is more flexibility in the design of cell/spheroid trapping microstructures to meet the resolution limits of the fabrication technique. Similarly, there are multiple device architectures which allow for the incorporation of cell-laden hydrogels, and therefore allow for a higher degree of freedom in the device design to suit the achievable fabrication resolution. A common design is to use a micro-pillar array to pin a hydrogel track via surface tension as exemplified by OoC devices developed by Roger Kamm’s group99–101. The dimensions of such micropillars and inter-pillar gaps often need to be less than a few hundred microns to effectively “pin” the liquid hydrogel prepolymer99–101, and cannot be easily achieved by 3D printing. Alternatively, larger structural designs, such as “phase guides”73,102 or “rails”103,104 can be incorporated into larger channels. This change to channel geometry can create differential surface tensional forces on a liquid stream to direct its flow trajectory. Such structures may be more amenable to 3D printing.
PDMS-based barrier OoCs (e.g. lung, gut, blood brain barrier OoC) often incorporate prefabricated membranes, such as track-etched polycarbonate or polyester membranes used in TranswellTM inserts or pre-molded PDMS membranes. These membranes typically contain arrays of through-holes ranging from 0.4 μm (Transwell insert membrane) to 2.5–10 μm (PDMS membranes)90,92,105,106. The main function of the porous membrane is to provide a substrate on which epithelial or endothelial cells can attach and form a confluent layer while allowing diffusion of soluble molecules between the apical and basal compartments separated by the barrier tissue. Therefore, the size of single through-holes should ideally be smaller than a single cell (i.e. < 5 μm) while the cumulative characteristics of the through-hole array (i.e. size, pitch and density), which modulate the mass transport property of the membrane, should be specified by the intended biological application. Therefore, it is important to understand how critical the dimensions of the microchannel structures and/or barrier through-holes will contribute to the utility of the OoC of interest, so that the 3D printing hardware and operation process be optimized accordingly to recapitulate this resolution, or find sufficient work-arounds.
Since PDMS is extremely popular due to its biocompatible nature, 3D printed OoCs should be comparably biocompatible to be relevant. Biocompatibility not only refers to the ability of cell cultures to survive in shared media with the material, but also the ability of cells to attach appropriately to the surface. As such, in addition to not leaching toxic chemicals or polymers into solution, the material should also facilitate cell attachment, potentially with extracellular matrix coatings. There are many biocompatible 3D printable resins available, each with individual strengths and limitations. This is highly influenced by the composition, the photoinitiator, and the photo-absorber, which are discussed in section 5.2.
Due to the enclosed nature of microfluidic devices, equilibration between dissolved oxygen in the culture medium and atmospheric air would need to occur across the device. While PDMS is gas permeable, this process is not efficient enough once the device thickness exceeds 100 μm107. Hence, OoC devices rely on perfusion culture, where gas equilibration takes place outside in the medium reservoir, rather than static culture, to ensure optimal cell viability, thereby necessitating the need for world-to-chip connections. In 3D printed OoCs, this interface can control perfusion cultures, and is important for maintaining sterile culture conditions. PDMS-based OoCs interface with the world through tubing that can be connected to pumps or other flow controllers and sensors. As PDMS is self-sealing, the tubing can be directly inserted into the device without external manipulation. 3D printed resins, however, are rigid and do not self-seal, for the most part. While commercial OTS connectors like Luer fittings can assist in generating a uniform connecting interface, they also increase the footprint of the device thus limiting the OoC devices’ portability. One way to circumvent this issue is to integrate customized connections into the microfluidic OoC devices. The simplest approach is to directly fabricate Luer adaptors such as those fabricated by Microfluidic ChipShop108 or slip-fit connectors as part of the device. Many research groups have developed integrated, reversible interconnections not only to connect to tubing, but also to other microfluidic modules forming a modular microfluidic system. For example, Bhargava et al.109 demonstrated a 3D printed setup of microfluidic devices that allows for connection with other devices in x,y and z directions. Gong et al. developed 3D printed micro-gaskets that allow microfluidic chips to be sandwiched together110. Ong et al.111 and Yuen et al.112 have incorporated magnetic interconnects during the device fabrication process to allow for chip-to-chip as well as world-to-chip setups. Hence, the resin and resolution of the 3D printing process must be able to generate structures that can interact with OTS connectors or with existing fluidic equipment. This can also be important for the generation of Human-on-a-chip systems, where multiple OoC devices are required to interface to produce physiological systems for pharmacokinetic and pharmacodynamic studies.
4.2. Stimulating cells within OoCs
4.2.1. Biochemical stimulation.
OoCs often incorporate concentration gradient generators to control the spatio-temporal distribution of exogenous or endogenous cell signaling factors in the soluble microenvironment113. Passive gradient generators in the form of diffusion barriers are commonly employed to establish stable concentration gradients of secreted or applied soluble factors. These can be hydrogel barriers, whereby a track of hydrogel is patterned between as a “source” and “sink” compartments114–119. The porosity of the hydrogel will determine the diffusion characteristics of a drug or chemoattractant in the hydrogel, establishing a stable concentration gradient across the 2 compartments. The channel size for the hydrogel diffusive barriers is usually greater than 300 μm, which can be easily fabricated with current commercial 3D printers19. Another physical diffusive barrier takes the form of diffusion microchannel networks, which have been utilized in blood-brain barrier88,120,121 and liver-immune models122. These diffusive microchannels have dimensions around 2 to 10 μm in width so as to restrict cell migration while enabling diffusive transport of biomolecules across the barrier. With microchannel generation relying on sub-10 μm resolution, this feature remains a challenge for 3D printing fabrication. A gradient of soluble factors can also be applied by coupling the OoC to active gradient generators123, commonly referred to as “Christmas tree” gradient generators. This type of gradient generator consists of a series of 10–20 μm channels arranged in a hierarchical format to repeatedly split and combine laminar flow streams fed by 2 inlets containing the soluble factor and a diluent123–125. Considering that different types of gradient generators require the fabrication of different channel dimensions, researchers must take the printing resolution into account when adopting these functional designs.
4.2.2. Mechanical stimulation.
Tissue cultures involving hepatocytes81,111,126–129, endothelium130, adipocytes131, and chondrocytes132 have been shown to exhibit enhanced tissue-specific phenotype and functions when exposed to physiological fluid shear stress in microfluidic devices. Fluid shear stress in microfluidic OoCs is generally controlled by the perfusion flow rate and the microchannel’s dimensions, which typically range from 200 μm to 3 mm. The critical channel dimension to determine shear stress is the one along the axis of the flow velocity gradient acting on a cell culture (e.g. for a 2D cell culture, channel height will control shear stress magnitudes since velocity gradient is along y-axis). Most DLP 3D printers will have sufficient resolution to print channel geometries of 200 μm19 for shear stress applications. For OoCs where the cells are sensitive to shear stresses, such as hepatocytes and stem cells19,133, additional micro-structures or hydrogels may be needed to protect the cultured cells. The issues for fabricating these micro-structures are similar to those discussed for cell culture configurations previously.
In addition to shear stress, mechanical stresses, such as compression and stretching, are also important to maintain the physiological functions of intestinal epithelium, bone and cartilage134. To date, the generation of a mechanical stimuli in microfluidic tissue cultures has relied heavily on the elastic property of PDMS. For instance, compressive forces can be applied via pneumatic microvalve-like structures on underlying cells135. Alternatively, cyclic stretching can be applied on thin PDMS membranes on which intestinal or airway epithelial cells are grown on to mimic gut peristalsis or rhythmic breathing of the airway136. For 3D printed devices to generate compressive or stretching stimulation, the photoresin should be elastic, although this is not easily available with most commercial photo-resins. However, thin ( 50 μm thick) 3D printed poly(ethylene glycol) - diacrylate (PEGDA)37 can have a degree of flexibility, allowing researchers to generate pneumatically controlled Quake-style valves137,138. Pneumatic membrane valves 46 μm in diameter and “squeeze” valves measuring 15 × 15 μm have also been demonstrated38. Another approach is to incorporate plasticizers to create highly flexible 3D printed parts139. Note that plasticizers should be carefully selected as many plasticizers are not biocompatible140. 3D printing modalities other than SLA/DLP can provide even more flexible membranes141. Researchers must carefully consider how to best mechanically stimulate their cell cultures, given the rigidity and resolution of 3D printed devices.
4.2.3. Electrical stimulation.
The incorporation of electrodes into OoC devices allows for electrical stimulation to improve the electrophysiological functions of neural, skeletal muscle142 and cardiac tissues143. To date, the most commonly reported strategies involve patterning electrodes on a glass capping substrate, which is used to seal the microfluidic devices143,144. This process involves screen printing145 and/or vapor deposition146 of electrodes on the glass substrate. Since 3D printed resins cannot be readily bonded to the electrode-patterned glass substrates, researchers will require alternate methods of incorporating electrodes into 3D printed OoCs. One method is to design channels whereupon external electrodes can then be inserted into the microfluidic channels via access ports142,147. Alternatively, the access ports allow electrodes to be added in their liquid form and solidified prior to cell seeding148,149. It is important to note that SLA/DLP printing is inherently single-material. Utilizing other 3D printing modalities, however low in resolution, could facilitate more complex incorporations of electrodes and other stimulation materials.
4.3. Measuring cell phenotype and functions from OoCs
The integration of biological measurement apparatus is also critical to the utility of 3D printed OoCs. Many PDMS-based OoCs integrate microfabricated elements to measure electrical and mechanical signals generated by cells. These include electrodes to collect electrophysiological activities of neural or muscle tissues or measure the transepithelial electrical resistances (TEER) of barrier tissues, as well as mechanical cantilevers or micropillars to measure contractile forces generated by cardiac or muscle tissues57. The issues pertaining to the incorporation of recording electrodes in 3D printed OoC are similar to electrodes used for electrical stimulation discussed above. However, the placement of the electrodes are different depending on the desired measurement. For TEER, electrodes must be above and below the cell monolayer. This has been achieved in PDMS-based epithelial cell chips, where researchers utilized gold electrodes originating from each side of the device150, or with PDMS voids filled by platinum wires that were later sealed with UV-curable resin151. Microelectrode arrays (MEA) are often used to measure electrically active cells. These arrays can be integrated into PDMS devices in a similar manner to TEER electrodes, though they are usually patterned onto the capping glass substrate bonded to the microfluidic device.
Optical imaging (e.g. brightfield, epifluorescence, immunofluorescence) is an important modality to measure cell states in OoC devices. PDMS devices benefit from easy integration with such techniques, as they are often bonded to glass slides or coverslips. If imaging is a desired readout for a 3D printed OoC, several factors must be considered. An important criterion to obtain high quality images, which can be subsequently quantified by imaging processing, is the distance between the sample of interest (i.e. cell or tissues in the microfluidic device) and the objective lens. The objective lens of epifluorescence and confocal microscopes often have sub-millimeter working distances, which decrease with higher magnification objectives (e.g. the working distance of a 20x objective is typically 0.25–1 mm (maximum), while that of a 60x objective is around 0.1 mm to 0.3 mm). This issue can often be overlooked in PDMS OoC devices since the microfluidic channel can be bonded to a 170 μm thick glass coverslip, which has been optimized for high resolution bioimaging. 3D printed OoC devices must therefore be designed to either incorporate glass slides during the printing process or have viewing windows. The footprint of the microfluidic device should also fit into standard sample holders (i.e. typically designed to hold either glass slides, 35 mm petri-dishes or multi-well plates) of commercial microscopes so that they can be firmly secured during image acquisition. Optical properties, such as autofluorescence, of materials used to fabricate the OoC device must also be considered since they can potentially interfere with imaging of cellular structures or biomarkers that are labeled by fluorophores with overlapping emission spectrum. While PDMS does not autofluorescence, some commercial SLA/DLP resins are known to autofluoresce, especially when absorbing in the UV (100–400 nm) spectrum152.
5. Considerations for fabrication of 3D printed OoC devices
As discussed in the previous section, there is a set of requirements for 3D printed OoCs that will make them competitive with current PDMS-based OoCs. These surround the fabrication of structural elements for cell maintenance, stimulation and measurement at the desired resolution within a practical time period. In this section, we discuss how DLP/SLA 3D printing can achieve these functional features in an OoC device. DLP/SLA 3D printing can be characterized by three major steps: 1) device design, 2) the 3D printing process (fabrication), and 3) post processing. Each of these steps has significant implications for 3D printed OoC devices, and are summarized briefly in Figure 3.
Fig. 3.
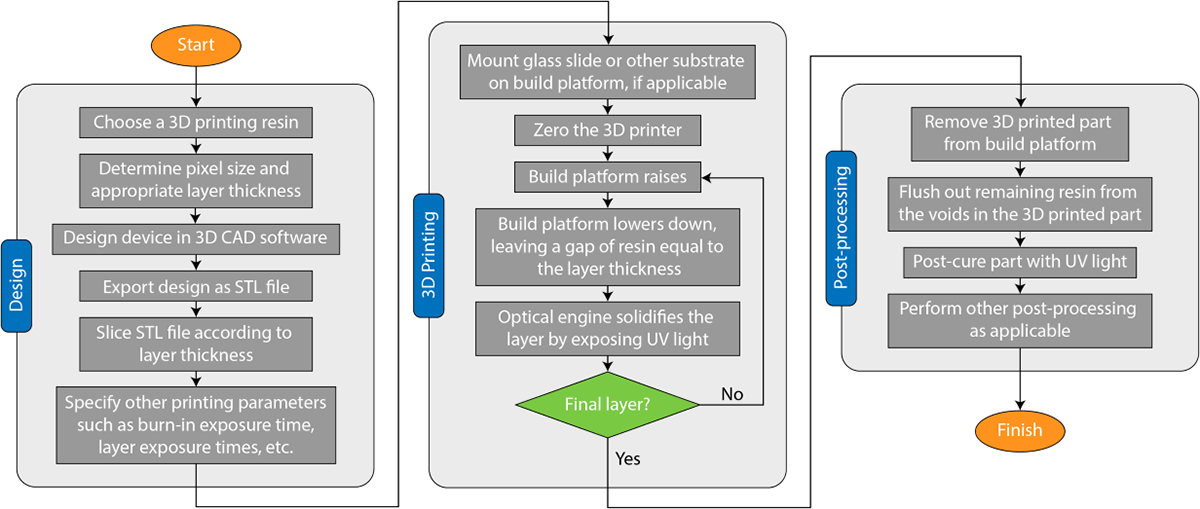
Process flow chart for SLA/DLP 3D printing for a microfluidic OoC device.
5.1. Device design
The general 3D printing flow is to generate a design in a 3D CAD software, export a Standard Tessellation Language (STL) file or other standard format, slice it, and then pass the slices to a 3D printer that handles fabrication. This creates a natural partition between the design and fabrication stages and these two stages are then consequently often viewed as wholly distinct. To truly unlock the full potential of DLP-SLA 3D printing, 3D printing process details should inform device design and resin selection, especially when the highest resolutions are needed as is common for OoC devices.
First, 3D printer pixel size should be considered during the design process. In a DLP/SLA 3D printer, pixel size (i.e. XY resolution, or image plane resolution) is determined by the size of the light beam reflected by micro-mirrors in the digital micromirror device (DMD)153 combined with the magnification/demagnification of the projection optics (Figure 1B). To achieve the best resolution in the XY plane, all features should be designed such that they fall on pixel boundaries. For example, if a 3D printer operates with a 10 μm pixel pitch and has 1:1 projection optics, features should be designed in increments of 10 μm. This is because the design will ultimately be converted to a stack of images before printing, which then will govern where UV light is exposed and where polymerization happens. When features do not align well with pixel boundaries, the slicing software that converts the 3D model into images will decide how to handle them, resulting in ambiguous rounding which can result in a loss of resolution. For example, in a 3D printing setup with 10 μm pixels, a 15 μm feature will either be rounded down to a single 10 μm pixel, or up to two 10 μm pixels, resulting in illumination across either 10 or 20 μm, usually without informing the user. Some small features close to the native resolution of the printer may be eliminated completely by this rounding process. Other features may turn out larger than expected. Such defects caused by rounding ambiguity have been observed by several groups35,154. Furthermore, the resolution of the sliced images should match the native resolution of the 3D printer DMD so each image pixel correlates to a single micromirror in the 3D printer. By designing in increments of pixel size and slicing to the proper resolution, rounding ambiguity is avoided and it is possible to fabricate features with smaller dimensions38.
Second, design in the Z dimension (see Figure 1B) should be guided by the target layer thickness in the 3D printed part. If a layer thickness of 10 μm is used in printing, then all features should be designed in 10 μm increments in the Z dimension, eliminating the ambiguity of how to handle designed features that do not align with physical layers. Additionally, contrary to the common practice of simply selecting a layer thickness on the 3D printer, an appropriate layer thickness should be determined by the optical properties of the 3D printing resin and illumination system, explained in greater detail in section 5.2.3.
Finally, researchers can consider how the true 3D nature of 3D printing can be leveraged to generate more complex devices in all three dimensions. Many existing microfluidic OoC designs are planar in nature due to the planar nature of conventional fabrication methods. With DLP-SLA 3D printing processes, devices can utilize hundreds of layers to accomplish more sophisticated fluidic routing, pack channels and other OoC structures into tighter volumes, and form complex 3D shapes and components that are not possible with traditional approaches. Gong et al. demonstrated a linear dilution mixer pump with several pneumatic valves, pumps, and mixing chambers packed into a 1.5 mm3 region, made possible by tightly packing the components and fluidic routing in 3D space155. Noriega et al. demonstrated a highly miniaturized pneumatic valve called a squeeze valve, where a pneumatic control channel wraps around both sides of a microfluidic channel, measuring 15 μm × 15 μm in the active area. These squeeze valves were then utilized to form a 10-stage two fold serial dilution module containing over 60 active pneumatic components with an area of just 2.2 mm × 1.1 mm38. These results demonstrate how better utilization of 3D space can enable devices that are impossible with traditional techniques.
5.2. Resin selection and biocompatibility
Resin selection is a critical step in designing and fabricating a successful OoC device. Most 3D printing resin formulations are somewhat similar, containing several major components: (1) A base monomer (and/or oligomers), (2) a photoinitiator, and often (3) photo absorber(s)34,156. The base monomers/oligomers provide the structural backbone of the formulation in liquid form via polymer chains that can be crosslinked to form a solid. The photoinitiator is responsible for converting energy from light into a reactive species, usually free radicals, which then interact with the monomers/oligomers to crosslink them, causing a phase change from liquid to solid157. The photo absorbers are responsible for limiting unwanted penetration of light deep into the resin which would negatively affect resolution (see section 5.2.3). Resins can typically be used across a variety of 3D printers, provided the optical sources of the printers are the same as the wavelengths the resin was designed for152,158.
An ideal resin should at least be 1) biocompatible, 2) high-resolution, 3) have good optical clarity with minimal autofluorescence, and 4) not be prohibitively viscous. Optimizing for one or more of these characteristics usually involves a tradeoff sacrificing the quality of the others. Here, we first briefly cover each of these four requirements and then discuss several classes of photo-resins that have been employed in SLA/DLP 3D printed microfluidics.
To begin, it is important to realize that the term “biocompatible” can have broad meaning in the context of 3D printing resins. For example, a commercial resin may be listed as biocompatible because it does not cause irritation to the skin with prolonged exposure, but may still be cytotoxic making it unsuitable for an OoC device. Additionally, many resins that advertise biocompatibility are only biocompatible under very specific conditions or with particular post-processing protocols6,156,159–166; thus, biocompatibility should be evaluated for each desired end use case. Biocompatibility also tends to decline the longer cells are in contact with the resin material156. Encouragingly, many commercial and custom resins can be rendered biocompatible with appropriate post processing techniques which will be discussed with each resin as applicable.
Second, “high-resolution” has a more nuanced meaning than is usually considered. Conventional 3D printing is primarily concerned with the fabrication of positive features. When 3D printing for microfluidics and OoC applications, the ability to produce small negative features (voids) is more important. Thus, high-resolution in this context means that a target resin can produce small enough voids when paired with the 3D printer of choice. In general, the creation of small voids is more difficult than the creation of small positive features due to over penetration of light. This is discussed in greater detail in section 5.2.3.
Third, the importance of optical clarity relates to the ability to image and quantify cells and tissues within the 3D printed device with conventional microscopy modalities (as previously discussed in section 4.3). There are two primary factors that influence optical clarity: surface roughness and the optical properties of a target resin. Surface roughness scatters light as it passes through the surface of a 3D printed device, blurring fine details and making observation more difficult. Post-processing techniques to improve optical clarity are discussed in section 5.4. The optical properties of a resin should also be well understood because even if a resin is visually clear, it is possible that certain wavelengths of light may be blocked or cause autofluorescence, thereby interfering with fluorescence imaging techniques.
Finally, the viscosity of a resin is important to consider because leftover unpolymerized resin must be cleared from the small voids that have been created during printing. The more viscous the resin, the harder it is to remove. Clearing strategies are discussed in section 5.4. Resin recoating during printing is also affected by resin viscosity158. If a resin is too viscous, it has a harder time flowing into the open space between the 3D printed part and the build tray in between layers, which can result in large defects.
These four primary resin properties can differ across the three common classes of resins that have been employed for 3D printing microfluidic devices, namely commercially available resins, biomaterial-based resins, and custom resins designed specifically for microfluidics. Here we highlight significant resins and their key characteristics for each of these resin classes.
5.2.1. Commercially available resins.
There are many commercially available 3D printing resins to choose from. Some have shown promise for 3D printed OoC devices. The two most widely reported properties of commercial resins in the literature are biocompatibility and resolution. A recent review by Guttridge et al. identified 130 commercially available photo-sensitive resins labeled as biocompatible167, however a number of studies have evaluated cell cytotoxicity and proliferation on some of these resins and shown varying degrees of cytotoxicity159,161,164–166. Cytotoxicity can often be reduced by appropriate post-processing steps which are also discussed here. These results and more are summarized for convenience in Table 2.
Table 2.
Commercially available DLP/SLA 3D printing resins that have been used for microfluidic or OoC applications. n.r. indicates that a particular specification was not mentioned in the corresponding references; likely indicates the specification was inferred from manufacturer specifications of the 3D printer used in the work. The notes next to each resin name indicate the suppliers. Similar resins are grouped together
| Resin name | Wavelength (nm) | Smallest features | Biocompatible? | Post processing and notes | Used in |
|---|---|---|---|---|---|
|
| |||||
| BioMed168 | 385 & 405 | 600 μm x 600 μm channels | Yes | UV post cure (60 min at 50°C), soak in PBS for 24 h at 50 °C. High autofluorescence in DAPI channel (~320–385, 445/50). Very stable at high temperatures (120 °C). | 152 |
| BioMed Amber V1168 | n.r., likely 405 | 500 μm channels | Yes | UV post cure (10–30 min). Channels printed orthogonal to the light source. | 165 |
|
| |||||
| 385 & 405 | 600 μm x 600 μm channels | Yes | UV post cure (20 min at 40°C), soak in PBS for 24 h at 50 °C. High autofluorescence in DAPI channel (~320–385, 445/50). Moderate autofluorescence in EGFP channel (470/40, 525/50). | 152 | |
| Clear 168 | 385 | Yes | UV post cure (6–30 min), baked, autoclaved. Minor visible damage from autoclaving | 169 | |
| n.r., likely 405 | 300 μm x 150 μm trenches | No | UV post cure (15 min), autoclaved. Did not survive autoclaving | 164 | |
| n.r. | No | 159 | |||
| n.r., likely 405 | 500 μm channels | No | UV post cure (10–30 min). Channels printed orthogonal to the light source. | 165 | |
|
| |||||
| MiiCraft Clear 170 | n.r. | 250 μm x 250 μm channels | n.r. | UV post cure (10 min) | 154 |
|
| |||||
| Dental LT Clear 168 | 385 | Yes | UV post cure (6 min), autoclaved | 169 | |
| Dental LT Clear 168 | n.r., likely 405 | 300 μm x 150 μm trenches | No | UV post cure (15 min), autoclaved. Did not survive autoclaving | 164 |
| Dental LT V1 168 | n.r., likely 405 | 500 μm channels | No | UV post cure (10–30 min). Channels printed orthogonal to the light source. | 165 |
| Dental SG 168 | n.r., likely 405 | 300 μm x 150 μm trenches | Yes (56 days) | UV post cure (15 min), autoclaved | 164 |
|
| |||||
| Elastic 50A V1168 | n.r., likely 405 | 500 μm channels | No | UV post cure (10–30 min). Channels printed orthogonal to the light source. | 165 |
|
| |||||
| Flexible 168 | 405 | Yes | UV post cure (6 min), autoclaved. Poor opacity. | 169 | |
| Flexible 80A V1168 | n.r., likely 405 | 500 μm channels | No | UV post cure (10–30 min). Channels printed orthogonal to the light source. | 165 |
|
| |||||
| Fototec 7150 Clear 171 | n.r., likely 354.7 | Yes | Extensive ethanol wash, air dry | 160 | |
| n.r., likely 354.7 | No | 159 | |||
|
| |||||
| FREEPRINT ortho 385172 | 385 | 1 mm x 600 μm channels | n.r. | UV post cure | 74 |
|
| |||||
| GR173 | n.r., likely 405 | 400 μm x 400 μm channels | n.r. | Sonication in ethanol (5 min), UV post cure (3 min) | 174 |
|
| |||||
| 385 | Yes | UV post cure (6 min), autoclaved. Cell viability was lower than expected | 169 | ||
| GR-10175 | 385 | 100 μm wide x 1.2 mm tall pillars | n.r. | UV post cure, baked. All channels were printed open-faced | 176 |
| n.r. | Yes | UV post cure (7 min) | 156 | ||
| 405 | 354 μm channels | Yes | UV post cure (10–30 min), surface polishing | 177 | |
|
| |||||
| 385 | Yes | UV post cure (36 min), autoclaved | 169 | ||
| High Temp168 | n.r., likely 405 | 300 μm x 150 μm trenches | Yes (56 days) | UV post cure (15 min), autoclaved. High autofluorescence in the UV range (ex/em 355/455 nm). | 164 |
|
| |||||
| KeyOrtho IBT178 | 385 | could not produce channels | n.r. | UV post cure. Not mechanically stable enough to produce channels | 74 |
|
| |||||
| 385 | 200 μm microneedles, no channels | Yes | UV bake and post cure (60 min) | 166 | |
| MiiCraft BV-007A170 | 385 | 200 μm x 200 μm channels | No | UV post cure (1 min), soak in PBS for 24 h at 37 °C. Negligible autofluorescence. High heat (>50 °C) delaminated the material over time. | 152 |
|
| |||||
| Model V2168 | n.r., likely 405 | 500 um channels | No | UV post cure (10–30 min). Channels printed orthogonal to the light source. | 165 |
|
| |||||
| PlasCLEAR179 | 385 | 1 mm x 600 μm channels | n.r. | IPA wash, UV post cure | 74 |
| 405 | No | 177 | |||
|
| |||||
| Tough 2000 V1168 | n.r., likely 405 | 500 μm channels | No | UV post cure (10–30 min). Channels printed orthogonal to the light source. | 165 |
|
| |||||
| VisiJet SL Clear 180 | n.r | No | 159 | ||
|
| |||||
| Watershed XC11122181 | n.r., likely 354.7 | No | 159,177 | ||
For example, Beckwith et al. used GR-10 resin (pro3dure medical GmbH, Germany) to fabricate a full microfluidic platform for perfusing and sustaining tumor fragments that included threaded connectors for world to chip connections, an in-line trap for removing air bubbles, and networks of channels as small as 354 μm177. The resin passed a 96-hour cytotoxicity test and only required the common post-processing techniques of flushing the microfluidic channels with IPA and a UV post cure. Bucciarelli et al. later used the same resin to fabricate pillars as small as 50 μm wide with aspect ratios up to 60176.
Piironen et al. evaluated four resins from FormLabs (Somerville, Massachusetts): Clear, High Temp, Dental SG, and Dental LT Clear. Dental SG and Dental LT Clear are both certified biocompatible according to the EN-ISO 10993–1:2009/AC:2010.24–27,28 standard. However, they found that none of the four resins could support cell culture as printed. Autoclaving the parts after printing improved biocompatibility, but only the Dental SG and High Temp resins could survive autoclaving without visible deformations. Once autoclaved, both the Dental SG and High Temp resins supported cell culture similar to plastic controls for at least 56 days, demonstrating a very high degree of biocompatibility164. Surface functionalization with Matrigel (Corning, New York, NY) was crucial for some of the cell types164,182. This study did not include any evaluation of the resolution limits of these resins.
Around the same time, Hart et al. also evaluated many of the same resins from Formlabs, as well as the previously reported GR-10 resin from Pro3dure176,177 and further investigated the effects of post processing on biocompatibility. Their biocompatibility tests were performed with HL-1 rat cardiomyocyte cells. They found that Clear Resin (FLGPCL04, Formlabs, Somerville, Massachusetts) had about 43% viability as printed, but almost 92% viability after sonication in 70 IPA. An added thermal bake showed similar viability. The addition of a six minute UV post cure after sonication in IPA increased viability to about 96%, autoclaving after sonication boosted viability to almost 99%, and the combination of sonication, UV post cure, and autoclaving showed over 99% viability. The high heat of autoclaving did however cause minor visible damage to the 3D printed parts made with the Clear resin169. They then evaluated High Temp resin (FLHTAM01, Formlabs, Somerville, Massachusetts) and found approximately 45% viability as printed and over 87% viability after sonication in IPA, UV post curing, and autoclaving. The High Temp resin withstood the autoclaving procedure much better showing no visible deformations. Extending the UV post cure from six minutes to 36 minutes further boosted cell viability to 92%169. Flexible Resin (FLFLGR02, Formlabs, Somerville, Massachusetts) showed 90% viability after sonication in IPA, UV post curing (six minutes) and autoclaving but had poor opacity. Dental LT Clear Resin (DLFLCL01, Formlabs, Somerville, Massachusetts) showed about 85% viability under the same post processing conditions. Finally, the GR-10 resin (Pro3dure, Iserlohn, Germany) showed about 83% viability under the same conditions. The authors mentioned they were still investigating why viability for the GR-10 resin was so low169. None of these devices evaluated resin resolution with respect to microfluidic features.
The following year, Carnero et al. evaluated seven resins from Formlabs: Clear V4, Dental LT V1, Tough 2000 V1, BioMed Amber V1, Flexible 80A V1, Elastic 50A V1, and Model V2 (Formlabs, Somerville, Massachusetts). They found that cells would adhere to the Dental and Clear resins, but culture did not progress past 24 hours. Encouragingly, the BioMed Amber V1 resin showed adequate biocompatibility in terms of cell adhesion and cell growth for human umbilical vein endothelial cells. None of the resins were able to produce channels with diameters smaller than 250 μm165.
Musgrove et al. evaluated the BioMed and Clear resins from Formlabs (Somerville, Massachusetts) and the MiiCraft BV-007A resin (CADworks3D, Canada). The BioMed resin devices were UV post cured for 60 minutes at 50°C and the Clear resin devices were post cured for 20 minutes at 45 °C. Both showed sufficient biocompatibility. The BV-007A based devices were post cured for one minute at room temperature and did not show biocompatibility. The BV-007A resin did however demonstrate superior resolution, allowing for the fabrication of channels as small as 200 μm × 200 μm, while the BioMed and Clear resins allowed for channels as small as 600 μm × 600μm. They also compared several post processing techniques and their effect on biocompatibility, finding that the most effective methods were soaking in sterile 1x phosphate-buffered saline without calcium and magnesium (PBS, Prod. No. 17–516 F, Lonza, USA) for 24 hours at 37 °C (for BV007a) or at 50 °C (for the BioMed and Clear resins), incubation at 37 °C for 24 hours, or a combination of the two (PBS soak at 37 °C for 24 hours). Autoclaving for 30 minutes at 120 °C using a gravity cycle was also found to be effective, but slightly less so than the previously mentioned methods152.
Tabriz et al. also used the MiiCraft BV-007A resin (Young Optics Inc., Hsinchu, Taiwan) to fabricate microneedle arrays and found 84% cell viability of human dermal fibroblasts after 2.5 h, suggesting good biocompatibility166.
There are a few high-level takeaways from these results. First, no commercial resins have been shown to be biocompatible as-printed. All require some level of post processing and several can be rendered biocompatible if post-processed correctly. The most common post processing techniques to improve biocompatibility include cleaning with isopropanol, UV post curing, autoclaving, baking, and soaking in solution to leach out cytotoxic components. For researchers concerned with biocompatibility, the GR-10 resin175 is a widely reported choice that has shown consistent biocompatibility. Second, the resolution limits of most commercial resins for negative features (i.e. voids) currently lie somewhere in the 200–800 μm range, with the best results being those of Musgrove et al. demonstrating flow channels as small as 200 × 200 μm in the MiiCraft BV-007A resin. This resin serves as a good starting point for researchers who are most concerned with resolution. These and the other results cited here show that commercially available 3D printing resins can serve as a viable starting point for 3D printed OoC devices. The best results however have been shown with custom formulated resins, discussed in section 5.2.3.
5.2.2. Biomaterial-based resins.
Next are the photo-crosslinkable extracellular matrix (ECM)-based biomaterials that have been developed for cell patterning or bioprinting applications, most notably gelatin methacrylate (GelMA) and polyethylene glycol diacrylate (PEGDA). These materials are typically highly biocompatible since they were formulated for tissue engineering applications where cells can be embedded and crosslinked with the resins183,184. Subsequent adaptations of these resins for SLA/DLP 3D printers have been used to create structurally complex microfluidic networks to mimic vasculatures and airway structure using both commercial185,186 and custom187 3D printers. Gonzalez et al. showed biocompatibility with directly adhered A549 cells in a device fabricated with a commercial 3D printer and a custom 3D printing resin consisting of a PEGDA (MW250 g/mol) monomer and BAPO photoinitiator163. Huh et al. investigated combinations of photoinitiators and UV absorbers in PEGDA and their effects on cell viability for DLP based bioprinting applications, and found that PEGDA with LAP photoinitiator and Maxgard® R1800 UV absorber had high initial cell viability for up to 14 days in culture, and could print complex cell-laden tissue constructs like a perfusable heart-shaped construct with open vesicles and atriums188. Ding et al. recently reviewed a wider array of light crosslinkable hydrogels that are compatible with DLP-SLA 3D printing systems189.
While most of these resins are highly biocompatible, their ability to make the small, enclosed microfluidic channels and thin membranes common in OoC applications is less well understood and has not been as widely reported. Some of the smallest such features using a biomaterial based resin are those of Grigoryan et al., which demonstrated 150 μm thick membrane-like fin elements used in their perfusable 3D static mixer and 200 μm channels as part of their distal lung subunit187. As such, this class of biomaterial based resin is best suited for device designs where cell biocompatibility is prioritized due to the use of sensitive cell types, or which do not have structures that require the highest resolution (< 100 μm). Also note that these resins tend to be less mechanically stable than other resin classes, introducing an additional tradeoff to consider.
5.2.3. Resins developed specifically for 3D printed microfluidics.
Finally, there are 3D printing resins which have been developed specifically for printing microfluidics. These resins are often formulated by optimizing and selecting specific combinations of base monomer, photoinitiator and photo absorber such that they are well aligned with the 3D printer’s optics to maximize printing resolution34,156. While common base monomers such as PEGDA may be used, extensive screening of both the photoinitiator and photo absorber must be performed to match the illumination spectrum of the 3D printer optical source. The photoinitiator should sufficiently overlap with the source spectrum to keep individual layer exposure times low, reducing print times190,191. The photo absorber must also overlap sufficiently with the optical source spectrum to enable high resolution in the Z direction if such resolution is required8,192. This is because as the 3D printing process progresses, voids filled with unpolymerized resin are created. Without an appropriate photo-absorber, the light from subsequent layers will penetrate into those previously printed layers and polymerize resin trapped inside of the voids, filling in previously fabricated negative features, effectively erasing them. Selectively filtering out certain wavelengths of light from the optical source can further enhance resolution by removing the higher wavelengths of light that are less readily absorbed by the photo absorber192. The chemistries and capabilities of several custom DLP/SLA 3D printing resins are summarized in Tables 3 and 4, respectively.
Table 3.
Custom DLP/SLA 3D printing resin chemistries. n.r. indicates that a particular specification was not mentioned in the corresponding references; likely indicates the specification was inferred from manufacturer specifications of the 3D printer used in the work
| Monomer(s) | Photoinitiator | Absorber(s) | Other ingredients | Wavelength (nm) | Used in |
|---|---|---|---|---|---|
| PEGDA (MW 250), HDDA (MW 226.3), BEDA (MW 512) | BAPO | 405 | 163 | ||
| RMS-083 (PDMS copolymer) | TPO-L (0.8% w/w) | Sudan I (0.2% w/w) | ITX (0.4% w/w, photosensitizer) | 385 | 194 |
| RMS-083 (PDMS copolymer) | TPO-L (0.8% w/w) | Sudan I (0.2% w/w) | ITX (0.4% w/w, photosensitizer), an unspecified diluent (80% w/w) | 385 | 195 |
| PEGDA (MW 700) | LAP | Quinoline yellow | 365 | 78 | |
| PEGDA, GelMA, Water | LAP | Tartrazine, curcumin, anthocyanin, gold nanoparticles (50 nm) | 405 | 187 | |
| GelMA suspended in PBS (20% w/v) | LAP (0.5% w/v) | Tartrazine (1.5 x 10−3 M) | n.r., likely 405 | 185,186 | |
| PEGDA (MW 250) | Irgacure 819 | 385 | 39 | ||
| PEGDA (MW 258) | Irgacure-819 (0.6% w/w) | Agarose (2% w/w), ITX (0.6% w/w, photosensitizer) | 385 | 35,36 | |
| PEGDA (MW 258) | Irgacure-819 (0.6% w/w) | ITX (0.6% w/w, photosensitizer) | 385 | 196 | |
| PEGDA (MW 258) | Irgacure 819 (1% w/w) | Sudan I (0.2% w/w) | 405 | 34,197 | |
| PEGDA (MW 258) | Irgacure 819 (1% w/w) | Sudan I (0.4% w/w) | AIBN (0.01% w/w, thermal initiator) | 405 | 37 |
| PEGDA (MW 258) | Irgacure 819 (1% w/w) | NPS (3% w/w) | 385 | 8 | |
| PEGDA (MW 258) | Irgacure 819 (1% w/w) | NPS (2% w/w) | 385 | 6,38,83,110,155 | |
| PEGDA (MW 258) | Irgacure 819 (1% w/w) | Avobenzone (0.38% w/w) | 385 | 6 |
Table 4.
Capabilities of custom DLP/SLA 3D printing resins as reported in literature. n.r. indicates that a particular specification was not mentioned in the corresponding references; likely indicates the specification was inferred from manufacturer specifications of the 3D printer used in the work
| Smallest Feaures | Biocompatible? | Post processing and notes | Used in |
|---|---|---|---|
| 27 μm x 1000 μm channels | Yes | Wash with water, soak in DI water (36 hr), UV post cure (12 hr). Channels use glass as bottom surface | 35,36 |
| ~800 μm channels | Yes | UV post cure and soak in water (12 h), oxygen plasma treatment. Coated with poly-D-lysine and Matrigel | 39 |
| 50 μm x ~250 μm channels (height x width) | n.r. | More gas permeable than PDMS | 195 |
| 60 μm tall x 540 μm wide channels 20 μm thick membranes | n.r. | IPA soak and flow-through. Transparent and gas permeable | 194 |
| 100 μm x 100 μm channels | Yes | Soak in DI water (overnight), UV post cure (22 min). Hydrogel | 78 |
| 150 μm thick mixer fins ~200 μm channels | Yes | Hydrogel | 187 |
| 5 mm channels | Yes | Wash in heated PBS. Cell-laden hydrogel | 185,186 |
| 750 μm diameter open faced wells | Moderate | Sonication in ethanol (5 min), UV post-cure (10 min), soak in ethanol (overnight), UV sterilization | 163 |
| 18 μm x 20 μm channels (height x width) 13 μm x 3 mm channels (width x height) | n.r. | UV post cure | 8 |
| 46 μm pneumatic valves (diameter) 46 μm x 50 μm channels 15 μm x 15 μm “squeeze” valves | n.r. | UV post cure | 38 |
| 300 μm x 50 μm pneumatic valves (diameter x height) 10 μm thick membranes 25 μm particle traps | n.r. | UV post cure | 83,110,155 |
| 1.08 mm x 60 μm pneumatic valves pnematic pumps, multiplexers | n.r. | Thermal cure at 80°C (30 min) | 37 |
| 108 μm x 60 μm channels 2 mm valves | n.r. | 34,197 | |
| 300 μm channels 500 μm valves | n.r. | 196 | |
| Spheriod culture plates | Yes (ISO 10993–5) | Plasma treatment to improve cell adherence | 6 |
| Spheriod culture plates | Yes (ISO 10993–5) | Soak in ethanol (12 hr), plasma treatment to improve cell adherence | 6 |
The Nordin group developed a custom DLP-SLA 3D printer which is paired with a custom resin formulation8,34,193 and demonstrated 3D printed microfluidic devices with pneumatic valves, pumps, and multiplexers34, 18 μm × 20 μm flow channels8, high density chip-to-chip interconnects to facilitate world-to-chip connections110, a complex mixer device155, pneumatic membrane valves 46 μm in diameter38, squeeze valves measuring 15 μm × 15 μm38, and a compact, highly integrated 2.2 mm × 1.1 mm 10-stage 2-fold serial dilution device38. The custom resin used consisted of poly(ethylene glycol) diacrylate (PEGDA, MW 258g/mol) as the monomer, phenylbis(2,4,6-trimethylbenzoyl)phos-phine oxide (Irgacure 819) as the photoinitiator, and 2-nitrophenyl phenyl sulfide (NPS) as the UV absorber8. Warr et al. demonstrated that the NPS based resin was not biocompatible as printed, but washing for 12 hours with 70% Ethanol rendered it cytocompatible6. Warr et al. also demonstrated that utilizing avobenzone as the UV absorber rather than NPS greatly improved the as-printed cell viability of the 3D printed microfluidic devices and then utilized it to generate spheroid cultures6.
Zhang et al. used custom 3D printer and resin consisting of PEGDA (MW 700), lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) photoinitiator, and quinoline yellow photo absorber to fabricate channels as small as 100 μm × 100 μm. They demonstrated biocompatibility by seeding primary human umbilical vein endothelial cells which reached near confluence over a 24 hour period78. Kuo et al. used a Pico 2 HD 3D printer (Asiga, Sydney, Australia) and a custom resin consisting of PEGDA (MW 258), Irgacure-819 photoinitiator and 2-isopropyl thioxanthone (ITX) photosensitizer to fabricate high aspect ratio microchannels measuring 27 μm wide by 1 mm tall35.
Several groups have demonstrated 3D printed microfluidic devices utilizing custom PDMS based resins. Femmer et al. formulated one of the first 3D printable PDMS based resins consisting of (methacryloxypropyl methylsiloxane)-dimethylsiloxane copolymer, ethyl(2,4,6-trimethylbenzoyl)phenyl phosphinate (TPO-L) as a photoinitiator and Orasol Orange as a photo-absorber198. Bhattacharjee et al. used a desktop DLP-SLA 3D printer to fabricate optically transparent submillimeter structures and microfluidic channels using a resin consisting of a blend of PDMS-methacrylate macromers with ethyl (2,4,6-trimethylbenzoyl) phenyl phosphinate (TPO-L) as the photoinitiator and isopropylthioxanthone (ITX) as the photo-absorber. The resulting structures had mechanical properties similar to thermally cured PDMS (Sylgard-184) and could be rendered suitable for mammalian cell culture with a solvent extraction post processing step162. More recently, Fleck et al. demonstrated channels 60 μm tall and membranes as thin as 20 μm using a resin consisting of (Methacryloxypropyl)methylsiloxane]-dimethylsiloxane copolymer (RMS-083) with 2, 4, 6-Trimethyl benzoyl diphenylphosphine oxide (TPO-L) as the photoinitiator, 2-Isopropylthioxanthone (ITX) as a photosensitizer, and Sudan I as a photo absorber194. Adding a diluent reduced the viscosity of this resin, enabling 50 μm channels and provided oxygen permeability better than PDMS (Sylgard-184)195.
Custom 3D printing resins clearly show the most promise for fabricating the highest quality OoC devices, in terms of both resolution and biocompatibility. Custom resins however are not as convenient and readily accessible as commercially available ones, so there is a tradeoff between ease of use and the best results. For groups well equipped to purchase, mix, and store resin components, or that require the highest resolution, the custom resins mentioned here could serve as a good starting point. For groups that are less concerned with resolution or do not want to mix resins on their own, one of the commercial resins noted above may prove convenient.
In addition to the three resin classes mentioned above, there are many resin materials used for OoC devices fabricated by traditional (non-3D printed) methods that may also have wider application in 3D printing. For researchers interested in investigating new resin chemistries, a recent review by Nahak et al. which includes a wide variety of OoC resins that could serve as a good starting point199.
5.3. 3D printer selection and process optimization
When selecting a 3D printer for an OoC application, there are several key specifications and features that should be considered including pixel size, layer thickness, optical illumination wavelength, range of exposure times, and build platform design, which are each discussed in detail below. We then discuss how to optimize 3D printing process parameters to achieve the best results.
5.3.1. Pixel size.
Pixel size serves as a rough measure of the minimum feature size a particular 3D printer can produce. When researching a 3D printer for a particular application, the advertised resolution may mean one of two things: 1) The size of a single micromirror on the internal Digital Micromirror Device (DMD) array or 2) the size of a single optical pixel in the projected image plane. DMD pixel dimensions are typically on the order of 5–25 μm, while the pixel size in the image plane can be significantly larger or smaller. While a 3D printer may advertise a particular resolution, this does not necessarily mean it is capable of producing features of that size, especially when negative features (voids) are desired; the actual manufacturable feature size is also dependent on resin properties, optical focus, image fidelity, and exposure time as discussed in section 5.2.38,188.
5.3.2. Layer thickness.
To select an appropriate layer thickness, the 3D printer optical source and the chemistry of the desired resin must be considered together. Layer thickness is primarily determined by the optical penetration depth of the selected resin at the wavelengths the 3D printer uses, and influenced to a lesser degree by the positional accuracy of the 3D printer’s vertical translation stage that moves the build platform up and down and the layer exposure time. As mentioned in greater detail in section 5.2.3, the 3D printer illumination spectrum should overlap sufficiently with both the resin photoinitiator and photo absorber spectra. Sufficient absorption by the photo absorber enables better resolution in the Z direction as it limits the penetration depth of the optical source to only the most recently fabricated layers8. To obtain minimal flow channel height for a given resin, the ratio of the build layer thickness to optical penetration depth should be in the range 0.3–1.034. The ability to control optical focus also plays an important role; the highest resolution can only be achieved when in good focus9.
5.3.3. Build platform.
It is also important to think about how your 3D printed devices will be attached to the build platform. Many DLP-SLA 3D printers operate by constructing the 3D printed part directly onto the surface of the build platform, which is typically rough to allow adhesion. This bond to the build platform plate is usually not very adhesive34, and can result in prints with poor optical clarity due to the rough surface. Better results have been shown by printing instead on bare or functionalized glass microscope slides which can then be attached to the build plate with two sided tape or some other adhesive, providing a stronger bond and a smooth surface for greater optical clarity34. More complex and capable build platforms are possible, such as those employed by Grigoryan et al. that include additional translational stages to enable multi-material bioprinting200. Ultimately, while the build stage can be altered to best suit the application, the method by which the print attaches to the platform should be considered when purchasing/utilizing a 3D printer. Also note that while most 3D printed parts use a consistent exposure time for most layers in a printing run, the first few layers often use a much higher exposure time called a burn-in exposure to ensure that the part is adhered well to the build platform or glass slides. These burn-in exposure times are typically anywhere from 5–20 times the normal layer exposure time.
5.3.4. Commercially available DLP-SLA 3D printers.
While commercially available DLP-SLA 3D printers currently cannot match the resolution of custom laboratory setups, there are several commercial options available that may prove adequate for certain applications. They are summarized in Table 5 for convenience.
Table 5.
Commercially available DLP/SLA 3D printers used in the literature. n.r. indicates that a particular specification was not mentioned in the corresponding references; likely indicates the specification as reported by the manufacturer when such a specification could be found
| Printer type | Supplier | Printer name | Wavelength (nm) | Pixel size (μm) | Image size (pixels) | Used in |
|---|---|---|---|---|---|---|
|
| ||||||
| DLP-SLA | Asiga, Sydney, Australia 179 | MAX X27 UV | 385 | 27 | 1920 x1080 | 166,169,176,194,5 |
| Pico 2 HD | 385 | 27 | n.r. | 35,36,6 | ||
| Pico Plus 27 | 405 | 27 | 1912 x1140 | 34,37,177 | ||
| Pico 2 DLP | 405 | 50 | n.r | 163 | ||
|
| ||||||
| B9 Creations, Rapid City, SD, USA201 | B9 Creator v1.1 | full color | 50 | 1024 x 768 | 197 | |
|
| ||||||
| Boston Micro Fabrication, Maynard, MA, USA173 | microArch™S140 | n.r., likely 405 | 10 | 1920 x1080 | 174 | |
|
| ||||||
| CELLINK, Gothenburg, Sweden202 | Lumen X | n. r., likely 405 | 50 | 1280 x1800 | 185,186 | |
|
| ||||||
| MiiCraft, CADworks3D, Canada170 | P110Y | 385 | 40 | n.r. | 152 | |
| M50–405 | 405 | 30 | n.r. | 152 | ||
| MiiCraft | n.r. | 56 | 854 x 480 | 154 | ||
|
| ||||||
| pro3dure medical, Iserlohn, Germany175 | FAB-12 | n.r. | n.r. | n.r. | 156 | |
|
| ||||||
| Laser SLA | 3D Systems, Valencia, CA, USA180 | ProJet 7000HD | n.r. | n.r. | N/A | 159 |
| Viper | n.r., likely 354.7 | n.r. | N/A | 177 | ||
| Viper Pro | n.r., likely 354.7 | n.r. | N/A | 159,160 | ||
|
| ||||||
| Formlabs, Somerville, MA, USA168 | Form 1 | n.r. | n.r. | N/A | 159 | |
| Form 2 | n.r. | n.r. | N/A | 164,169 | ||
| Form 3B | n.r., likely 405 | n.r. | N/A | 165 | ||
The most common suppliers reported are Asiga179, MiiCraft170, and formlabs168. Gonzalez et al. utilized an Asiga PICO 2 DLP-3D printer to fabricate their biocompatible 3D printed parts163. Fleck et al. used an Asiga MAX X27 UV printer to 3D print PDMS based microfluidic devices with 50 μm tall channels and 20 μm thick membranes194,195. Shan et al. used a Boston Microfabrication microArch™ S140 3D printer to fabricate a microfluidic device with channels as small as 400×400 μm that enabled high volumetric throughput nanoliposome preparation174. Shallan et al. achieved microfluidic channels measuring 250 μm × 250 μm with a MiiCraft 3D printer and a commercial resin154. Rogers et al. used a B9 Creator 3D printer v1.1 (B9 Creations201, Rapid City, SD) to fabricate microfluidic devices with 350 μm × 250 μm flow channels and 2 mm diameter pneumatic valves197. Tomov et al. and Cetnar et al. used the CELLINK Lumen X202 to fabricate cell-laden heart models185,186. The company Acrea 3D203 also supplies 3D printers with capabilities similar to some of the custom printers employed by the Nordin group8,34,37,38,83,110 with pixels as small as 7.6 μm and a variety of available optical wavelengths. While commercially available 3D printers currently don’t match the best results found in custom setups, they provide a lower barrier of entry and are capable of generating relevant microfluidic devices.
5.3.5. Process optimization.
Optimizing 3D printing parameters is critical to achieving the best results. The most common parameter that can be altered in a 3D printer is individual layer exposure time. A proper exposure time for a particular feature of interest is usually found by iteratively tuning it. Higher exposure times lead to more crosslinking in the resin, yielding to a stronger, stiffer material (as printed) but this can lead to overcurring that can erase small voids, especially in resins that do not employ a photo absorber34. Lower exposure times lead to a softer material during printing, which can cause the 3D printed part to break during printing152. For microfluidics, it is usually advantageous to use the lowest possible exposure time to minimize overcuring but one that is just high enough to maintain sufficient strength so the part will survive the mechanical stresses of the printing process.
Some 3D printers allow for control of other process variables such as the power level of the optical source and stage speeds, among others. When combined with other variables such as post cure time and cleaning methodologies, it can become difficult to identify which variables are most significant to success. In these cases, an appropriate Design of Experiments can help determine which variables are most significant. For example, Bucciarelli et al. created a statistical model to evaluate which 3D printing parameters had the largest effect on the formation of small, high-aspect ratio pillars. Interestingly, they found that both layer exposure time and post cleaning sonication power were significant176. Another effective approach is Sequential Process Optimization. This process involves iteratively tuning each step of the 3D printing process to achieve the best result, then moving to the next process parameter. Choi et al. have provided a guide that acts as a good starting point, addressing important issues such as deriving a working curve for a resin, determining exposure times, build plate adhesion and burn in exposure times, build plate leveling, resin recoating, and over and under-curing during printing158.
5.4. Post-processing
Post processing refers to all process steps performed after a 3D printed part is fabricated and removed from the 3D printer. For 3D printed microfluidic devices, the first post processing step is usually to clear unwanted unpolymerized resin from within the voids in the device. This is most commonly performed by pushing isopropanol through channels in the device or sonicating in isopropanol or another solvent. Another common post processing step is baking and/or UV curing to complete polymerization and improve overall material strength and biocompatibility. Other common methods to improve biocompatibility include baking, autoclaving, soaking in solution to leach out cytotoxic components, and coating with chemicals that promote cell adhesion such as fibronectin, collagen, and PDMS. Each of these methods tend to be unique to the resin formulations they apply to and are discussed in greater detail in line with each resin formulation in section 5.2 and in Tables 2 and 4.
As mentioned previously, surface roughness can be problematic when it comes time to image through a 3D printed device. Surface roughness can be improved by post-processing techniques such as surface polishing165,177, or coating with oil20,204, nail polish17,192 or resin192. Alternatively, excellent optical clarity can be achieved by printing on a glass substrate which provides a very smooth, transparent surface to image through37.
6. Outlook
Advances in SLA/DLP 3D printing have brought this technology to the forefront of microfluidic device fabrication. However, the task of optimizing the design, resin, 3D printer and post-processing technique for a specific OoC device is not trivial. Reviews like this act to assist researchers to best undertake SLA/DLP 3D printed OoC fabrication. As a final summary, we suggest that researchers pinpoint the important controllable parameters for their 3D printed OoCs, such that they can prioritize their choices and make changes to their plans accordingly. Figure 4 is designed to assist in this. The figure guides readers through the process of identifying their desired OoC characteristics in the first column, prioritizing key printing features in the second column, and adjusting the relevant controllable parameters in the third column accordingly. These controllable parameters align with Sections 5.1 (Design), 5.2 (Resin), 5.3 (3D Printing) and 5.4 (Post-processing), while the desired features are referenced and detailed in Section 4. It is our hope that reviews like this one are able to inform and assist researchers in fabricating their own 3D printed OoCs.
Fig. 4.
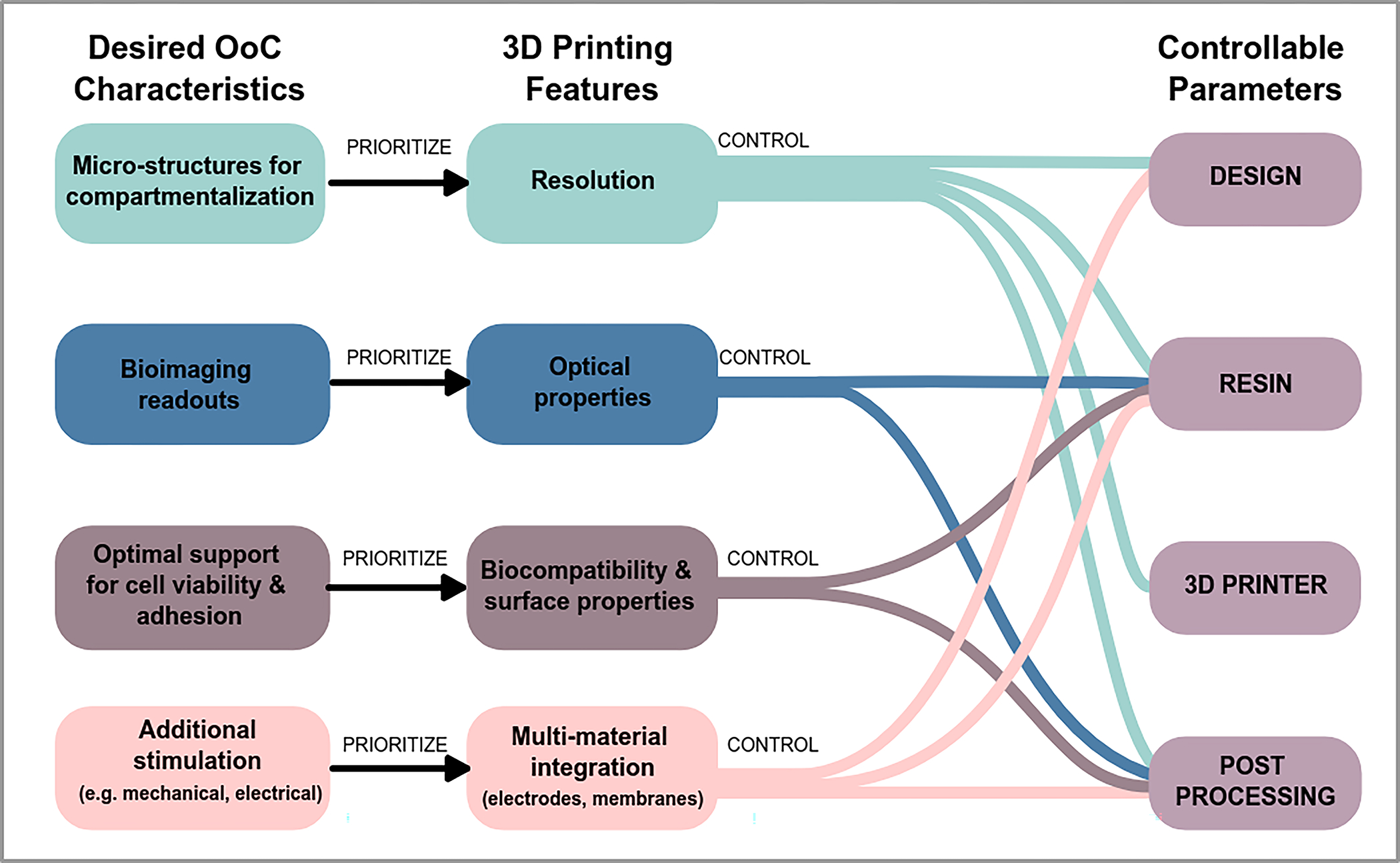
Identification of priority controllable parameters for SLA/DLP 3D printed OoC device design, fabrication and post-processing. To pinpoint the important controllable parameters for a SLA/DLP 3D printed OoC, researchers must identify their desired OoC characteristics in the first column, prioritize key printing features in the second column, and adjust the controllable parameters in the third column accordingly.
Importantly, there is still some work to be done before 3D printing can be widely adopted with ease. First, there are no commercial 3D printers that can match what is being accomplished in small research laboratories, making obtaining an appropriate 3D printer difficult. Researchers looking to generate useful, high-resolution, biocompatible OoC devices must either settle with the limitations of available commercial 3D printers (namely the lack of full control of the hardware, lower resolution), or undertake the sizable task of building a custom 3D printer. Once a 3D printer is acquired, there is much requisite process expertise that an end-user needs to obtain to use it to its full potential - though communications such as this review are aiding in sharing the required knowledge base. There is also a lack of a wide pool of spectrally matched, biocompatible, low viscosity resins with absorption profiles compatible with the highest resolution 3D printing. This remains one of the current high-interest areas of research.
Post processing methods also need standardization. Right now, most research groups use an iterative approach to identify methods that work for their particular applications, giving rise to many varied techniques whose underlying mechanisms aren’t always well understood. Standardizing post processing methods will make it easier to evaluate, compare, and use new resin chemistries consistently and will likely accelerate further development and wider adoption of 3D printing for OoC and other microfluidic applications.
Additionally, there is a lack of design tools and methodologies that integrate well with 3D printers. Current design strategies are disconnected from the fabrication process and are unnecessarily limiting. Almost all researchers take the route of using a CAD software that is ignorant of the 3D printing process to generate an STL file, then generate a stack of sliced images that are passed to a 3D printer along with a limited set of metadata for each layer like exposure time and layer thickness. As demonstrated by Noriega et al.38, more nuanced exposure and layer thickness strategies can produce better results even without improving the 3D printer. A way to design with 3D printing process details in mind and communicate more of this metadata to the 3D printer could shorten the design, fabricate, test cycle and enable new components and devices. More advanced design tools could also lower the barrier of entry to creating 3D printed microfluidics and OoC devices by incorporating the process details into the software, making it easier for a user to design and fabricate a device without having to be personally aware of all process details and interactions.
While each of these considerations provide a barrier to entry, commercially available 3D printers and resins can currently achieve meaningful results and many custom printers and resins have shown great promise. With the right 3D printer and resin combination, it is possible to achieve highly integrated devices, including built-in pneumatic valves, pumps, and membranes with internal dimensions comparable to those achievable with conventional PDMS devices. There is a small variety of resin materials to choose from, with more being developed every year, some of which have already shown to be compatible with cell culture or can be made biocompatible with post-processing processes. More and more research groups are investigating and improving 3D printing for microfluidic device fabrication, as represented by the number of publications over time (Figure 5).
Fig. 5.
Publications relating to 3D printed microfluidics over time. Search parameters on PubMed as follows, updated 04/14/23: “(microfluidic OR microfluidics) AND (“3D printing” OR “3D printed” OR “3D print” OR “additive manufacturing”), Last 10 years”.
Finally, 3D printing provides significant speed advantages for the design to test cycle. A device can be designed, fabricated, tested, and then redesigned, fabricated and tested again in quick succession. If a particular OoC device is small enough to fit several within one build area on a 3D printer, several could be printed at once, greatly speeding up fabrication and potentially offering a path towards high-throughput manufacturing. 3D printing also allows for an arbitrary level of complexity and integration that isn’t possible with traditional approaches - anything you can print and flush out can be integrated into a single device. Based on what has been shown in the literature and the clear promise 3D printing provides, we believe 3D printed microfluidics and OoC devices will continue to receive large research attention and that 3D printing could become the dominant method for generating such devices.
Acknowledgements
This work was supported by Australian Research Council grant numbers FT180100157 and DP200101658 awarded to Y.C.T and the National Institutes of Health grant number R15GM123405-02 awarded to G.P.N. L.M was supported by a QUT Postgraduate Research Award (QUTPRA).
Footnotes
Conflicts of interest
The authors declare no conflict of interest relating to this publication. One of the authors (G.P.N.) owns shares in Acrea 3D, a company commercializing microfluidic 3D printing.
Notes and references
- 1.Picollet-D’hahan N, Zuchowska A, Lemeunier I and Gac SL, Trends in Biotechnology, 2021, 39, 788–810. [DOI] [PubMed] [Google Scholar]
- 2.Strelez C, Jiang HY and Mumenthaler SM, Trends in Biotechnology, 2023, 41, 278–280. [DOI] [PubMed] [Google Scholar]
- 3.Berthier E, Young EWK and Beebe D, Lab on a Chip, 2012, 12, 1224. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen AV, Beauchamp MJ, Nordin GP and Woolley AT, Annual Review of Analytical Chemistry, 2020, 13, 45–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prabhakar P, Sen RK, Dwivedi N, Khan R, Solanki PR, Srivastava AK and Dhand C, Frontiers in Nanotechnology, 2021, 3, 1–16. [Google Scholar]
- 6.Warr C, Valdoz JC, Bickham BP, Knight CJ, Franks NA, Chartrand N, Ry PMV, Christensen KA, Nordin GP and Cook AD, ACS Applied Bio Materials, 2020, 3, 2239–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Männel M, Fischer C and Thiele J, Micromachines, 2020, 11, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong H, Bickham BP, Woolley AT and Nordin GP, Lab on a Chip, 2017, 17, 2899–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beauchamp MJ, Nordin GP and Woolley AT, Analytical and Bioanalytical Chemistry, 2017, 409, 4311–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macdonald NP, Cabot JM, Smejkal P, Guijt RM, Paull B and Breadmore MC, Analytical Chemistry, 2017, 89, 3858–3866. [DOI] [PubMed] [Google Scholar]
- 11.Mader M, Rein C, Konrat E, Meermeyer SL, Lee-Thedieck C, Kotz-Helmer F and Rapp BE, Micromachines, 2021, 12, 1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotz F, Mader M, Dellen N, Risch P, Kick A, Helmer D and Rapp B, Micromachines, 2020, 11, 873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, Smejkal P, Macdonald NP, Guijt RM and Breadmore MC, Analytical Chemistry, 2017, 89, 4701–4707. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z and Velasquez-Garcia LF, Journal of Microelectromechanical Systems, 2017, 26, 1356–1370. [Google Scholar]
- 15.Quero RF, da Silveira GD, da Silva JAF and de Jesus DP, Lab on a Chip, 2021, 21, 3715–3729. [DOI] [PubMed] [Google Scholar]
- 16.Rehmani MAA, Jaywant SA and Arif KM, Micromachines, 2020, 12, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castiaux AD, Selemani MA, Ward MA and Martin RS, Analytical Methods, 2021, 13, 5017–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor AP and Velasquez-Garcia LF, Journal of Microelectromechanical Systems, 2017, 26, 1316–1326. [Google Scholar]
- 19.Ong LJY, Islam A, DasGupta R, Iyer NG, Leo HL and Toh Y-C, Biofabrication, 2017, 9, 045005. [DOI] [PubMed] [Google Scholar]
- 20.Knowlton S, Yu CH, Ersoy F, Emadi S, Khademhosseini A and Tasoglu S, Biofabrication, 2016, 8, 025019. [DOI] [PubMed] [Google Scholar]
- 21.Anscombe N, Nature Photonics, 2010, 4, 22–23. [Google Scholar]
- 22.Geng Q, Wang D, Chen P and Chen S-C, Nature Communications, 2019, 10, 2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balakrishnan HK, Doeven EH, Merenda A, Dumée LF and Guijt RM, Analytica Chimica Acta, 2021, 1185, 338796. [DOI] [PubMed] [Google Scholar]
- 24.Wu D, Chen Q-D, Niu L-G, Wang J-N, Wang J, Wang R, Xia H and Sun H-B, Lab on a Chip, 2009, 9, 2391. [DOI] [PubMed] [Google Scholar]
- 25.Oellers M, Lucklum F and Vellekoop MJ, Microfluidics and Nanofluidics, 2019, 24, 4. [Google Scholar]
- 26.Giacomo RD, Krödel S, Maresca B, Benzoni P, Rusconi R, Stocker R and Daraio C, Scientific Reports, 2017, 7, 45897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Son AI, Opfermann JD, McCue C, Ziobro J, Abrahams JH, Jones K, Morton PD, Ishii S, Oluigbo C, Krieger A, Liu JS, Hashimoto-Torii K and Torii M, Scientific Reports, 2017, 7, 17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alsharhan AT, Acevedo R, Warren R and Sochol RD, Lab on a Chip, 2019, 19, 2799–2810. [DOI] [PubMed] [Google Scholar]
- 29.van der Velden G, Fan D and Staufer U, Micro and Nano Engineering, 2020, 7, 100054. [Google Scholar]
- 30.Liao C, Wuethrich A and Trau M, Applied Materials Today, 2020, 19, 100635. [Google Scholar]
- 31.Perrucci F, Bertana V, Marasso SL, Scordo G, Ferrero S, Pirri CF, Cocuzza M, El-Tamer A, Hinze U, Chichkov BN, Canavese G and Scaltrito L, Microelectronic Engineering, 2018, 195, 95–100. [Google Scholar]
- 32.Nanoscribe homepage, https://www.nanoscribe.com/en/.
- 33.Microlight3D homepage, https://www.microlight3d.com/.
- 34.Gong H, Beauchamp M, Perry S, Woolley AT and Nordin GP, RSC Advances, 2015, 5, 106621–106632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo AP, Bhattacharjee N, Lee Y-S, Castro K, Kim YT and Folch A, Advanced Materials Technologies, 2019, 4, 1800395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YT, Bohjanen S, Bhattacharjee N and Folch A, Lab on a Chip, 2019, 19, 3086–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong H, Woolley AT and Nordin GP, Lab on a Chip, 2016, 16, 2450–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noriega JLS, Chartrand NA, Valdoz JC, Cribbs CG, Jacobs DA, Poulson D, Viglione MS, Woolley AT, Ry PMV, Christensen KA and Nordin GP, Nature Communications, 2021, 12, 5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urrios A, Parra-Cabrera C, Bhattacharjee N, Gonzalez-Suarez AM, Rigat-Brugarolas LG, Nallapatti U, Samitier J, DeForest CA, Posas F, Garcia-Cordero JL and Folch A, Lab on a Chip, 2016, 16, 2287–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park HK, Shin M, Kim B, Park JW and Lee H, NPG Asia Materials, 2018, 10, 82–89. [Google Scholar]
- 41.Fritschen A, Bell AK, Königstein I, Stühn L, Stark RW and Blaeser A, Biomaterials Science, 2022, 10, 1981–1994. [DOI] [PubMed] [Google Scholar]
- 42.Tanyeri M and Tay S, in Viable cell culture in PDMS-based microfluidic devices, Elsevier, 2018, vol. 148, pp. 3–33. [DOI] [PubMed] [Google Scholar]
- 43.van der Meer AD, Orlova VV, ten Dijke P, van den Berg A and Mummery CL, Lab on a Chip, 2013, 13, 3562. [DOI] [PubMed] [Google Scholar]
- 44.Kim HJ, Huh D, Hamilton G and Ingber DE, Lab on a Chip, 2012, 12, 2165. [DOI] [PubMed] [Google Scholar]
- 45.Zamprogno P, Wüthrich S, Achenbach S, Thoma G, Stucki JD, Hobi N, Schneider-Daum N, Lehr C-M, Huwer H, Geiser T, Schmid RA and Guenat OT, Communications Biology, 2021, 4, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Booth R and Kim H, Lab on a Chip, 2012, 12, 1784. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Bolonduro OA, Hu N, Ju J, Rao AA, Duffy BM, Huang Z, Black LD and Timko BP, Nano Letters, 2020, 20, 2585–2593. [DOI] [PubMed] [Google Scholar]
- 48.Kniazeva T, Hsiao JC, Charest JL and Borenstein JT, Biomedical Microdevices, 2010, 13, 315–323. [DOI] [PubMed] [Google Scholar]
- 49.AIMBiotech homepage, https://aimbiotech.com/.
- 50.Mimetas homepage, https://www.mimetas.com/en/home/.
- 51.CNBio homepage, https://cn-bio.com/.
- 52.TissUse homepage, https://www.tissuse.com/en/.
- 53.Emulate homepage, https://emulatebio.com/.
- 54.Banik S, Uchil A, Kalsang T, Chakrabarty S, Ali MA, Srisungsitthisunti P, Mahato KK, Surdo S and Mazumder N, Critical Reviews in Biotechnology, 2022, 11, 1–19. [DOI] [PubMed] [Google Scholar]
- 55.Shakeri A, Khan S and Didar TF, Lab on a Chip, 2021, 21, 3053–3075. [DOI] [PubMed] [Google Scholar]
- 56.Morbioli GG, Speller NC and Stockton AM, Analytica Chimica Acta, 2020, 1135, 150–174. [DOI] [PubMed] [Google Scholar]
- 57.Leung CM, de Haan P, Ronaldson-Bouchard K, Kim G-A, Ko J, Rho HS, Chen Z, Habibovic P, Jeon NL, Takayama S, Shuler ML, Vunjak-Novakovic G, Frey O, Verpoorte E and Toh Y-C, Nature Reviews Methods Primers, 2022, 2, 33. [Google Scholar]
- 58.Gonçalves IM, Carvalho V, Rodrigues RO, Pinho D, Teixeira SFCF, Moita A, Hori T, Kaji H, Lima R and Minas G, Cancers, 2022, 14, 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu RXZ and Radisic M, Bioactive Materials, 2021, 6, 2801–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramadan Q, Fardous RS, Hazaymeh R, Alshmmari S and Zourob M, Advanced Biology, 2021, 5, 2100775. [DOI] [PubMed] [Google Scholar]
- 61.Sung JH, Expert Opinion on Drug Metabolism & Toxicology, 2021, 17, 969–986. [DOI] [PubMed] [Google Scholar]
- 62.Li Z, Hui J, Yang P and Mao H, Biosensors, 2022, 12, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee SH, Choi N and Sung JH, Expert Opinion on Drug Metabolism & Toxicology, 2019, 15, 1005–1019. [DOI] [PubMed] [Google Scholar]
- 64.Skardal A, Aleman J, Forsythe S, Rajan S, Murphy S, Devarasetty M, Zarandi NP, Nzou G, Wicks R, Sadri-Ardekani H, Bishop C, Soker S, Hall A, Shupe T and Atala A, Biofabrication, 2020, 12, 025017. [DOI] [PubMed] [Google Scholar]
- 65.Regehr KJ, Domenech M, Koepsel JT, Carver KC, Ellison-Zelski SJ, Murphy WL, Schuler LA, Alarid ET and Beebe DJ, Lab on a Chip, 2009, 9, 2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grant J, Özkan A, Oh C, Mahajan G, Prantil-Baun R and Ingber DE, Lab on a Chip, 2021, 21, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi M, Majumdar D, Gao Y, Brewer BM, Goodwin CR, McLean JA, Li D and Webb DJ, Lab on a Chip, 2013, 13, 3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Srigunapalan S, Eydelnant IA, Simmons CA and Wheeler AR, Lab on a Chip, 2012, 12, 369–375. [DOI] [PubMed] [Google Scholar]
- 69.He Y, Yu Y, Yang Y, Gu Y, Mao T, Shen Y, Liu Q, Liu R and Ding J, Bioactive Materials, 2022, 15, 288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ashby MF, Materials selection in mechanical design, Butterworth-Heinemann, 2011. [Google Scholar]
- 71.Bertsch A, Heimgartner S, Cousseau P and Renaud P, Lab on a Chip, 2001, 1, 56. [DOI] [PubMed] [Google Scholar]
- 72.Berthier J, Loe-Mie F, Tran V-M, Schoumacker S, Mittler F, Marchand G and Sarrut N, Journal of Colloid and Interface Science, 2009, 338, 296–303. [DOI] [PubMed] [Google Scholar]
- 73.Vulto P, Podszun S, Meyer P, Hermann C, Manz A and Urban GA, Lab on a Chip, 2011, 11, 1596. [DOI] [PubMed] [Google Scholar]
- 74.Subirada F, Paoli R, Sierra-Agudelo J, Lagunas A, Rodriguez-Trujillo R and Samitier J, Polymers, 2022, 14, 2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang W, Yan Y, Li CW, Xia HM, Chao SS, Wang DY and Wang ZP, Lab on a Chip, 2014, 14, 677–680. [DOI] [PubMed] [Google Scholar]
- 76.França CM, Tahayeri A, Rodrigues NS, Ferdosian S, Rontani RMP, Sereda G, Ferracane JL and Bertassoni LE, Lab on a Chip, 2020, 20, 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bruce A, Evans R, Mezan R, Shi L, Moses BS, Martin KH, Gibson LF and Yang Y, PLOS ONE, 2015, 10, e0140506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang R and Larsen NB, Lab on a Chip, 2017, 17, 4273–4282. [DOI] [PubMed] [Google Scholar]
- 79.Park J, Lee BK, Jeong GS, Hyun JK, Lee CJ and Lee S-H, Lab on a Chip, 2015, 15, 141–150. [DOI] [PubMed] [Google Scholar]
- 80.Kageyama T, Yoshimura C, Myasnikova D, Kataoka K, Nittami T, Maruo S and Fukuda J, Biomaterials, 2018, 154, 291–300. [DOI] [PubMed] [Google Scholar]
- 81.Ong LJY, Chong LH, Jin L, Singh PK, Lee PS, Yu H, Ananthanarayanan A, Leo HL and Toh Y-C, Biotechnology and Bioengineering, 2017, 114, 2360–2370. [DOI] [PubMed] [Google Scholar]
- 82.Xiao J, He W, Zhang Z, Zhang W, Cao Y, He R and Chen Y, RSC Advances, 2015, 5, 52161–52166. [Google Scholar]
- 83.Beauchamp M, Gong H, Woolley A and Nordin G, Micromachines, 2018, 9, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farahat WA, Wood LB, Zervantonakis IK, Schor A, Ong S, Neal D, Kamm RD and Asada HH, PLoS ONE, 2012, 7, e37333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chan JM, Zervantonakis IK, Rimchala T, Polacheck WJ, Whisler J and Kamm RD, PLoS ONE, 2012, 7, e50582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gregory CW, Sellgren KL, Gilchrist KH and Grego S, Biomicrofluidics, 2013, 7, 056503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mosadegh B, Huang C, Park JW, Shin HS, Chung BG, Hwang S-K, Lee K-H, Kim HJ, Brody J and Jeon NL, Langmuir, 2007, 23, 10910–10912. [DOI] [PubMed] [Google Scholar]
- 88.Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW and Jeon NL, Nature Methods, 2005, 2, 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP and Healy KE, Scientific Reports, 2015, 5, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mun KS, Arora K, Huang Y, Yang F, Yarlagadda S, Ramananda Y, El-Haija MA, Palermo JJ, Appakalai BN, Nathan JD and Naren AP, Nature Communications, 2019, 10, 3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Novak R, Didier M, Calamari E, Ng CF, Choe Y, Clauson SL, Nestor BA, Puerta J, Fleming R, Firoozinezhad SJ and Ingber DE, Journal of Visualized Experiments, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wei H, han Chueh B, Wu H, Hall EW, wing Li C, Schirhagl R, Lin J-M and Zare RN, Lab on a Chip, 2011, 11, 238–245. [DOI] [PubMed] [Google Scholar]
- 93.Sellgren KL, Butala EJ, Gilmour BP, Randell SH and Grego S, Lab on a Chip, 2014, 14, 3349–3358. [DOI] [PubMed] [Google Scholar]
- 94.Nalayanda DD, Puleo C, Fulton WB, Sharpe LM, Wang T-H and Abdullah F, Biomedical Microdevices, 2009, 11, 1081–1089. [DOI] [PubMed] [Google Scholar]
- 95.Sokolowska P, Zukowski K, Janikiewicz J, Jastrzebska E, Dobrzyn A and Brzozka Z, Biosensors and Bioelectronics, 2021, 183, 113215. [DOI] [PubMed] [Google Scholar]
- 96.Campisi M, Lim SH, Chiono V and Kamm RD, in 3D Self-Organized Human Blood–Brain Barrier in a Microfluidic Chip, ed. Ebrahimkhani MR and Hislop J, Springer; US, New York, NY, 2020, pp. 205–219. [DOI] [PubMed] [Google Scholar]
- 97.Mo SJ, Lee J-H, Kye HG, Lee JM, Kim E-J, Geum D, Sun W and Chung BG, The Analyst, 2020, 145, 3081–3089. [DOI] [PubMed] [Google Scholar]
- 98.Tu T-Y, Shen Y-P, Lim S-H and Wang Y-K, Frontiers in Bioengineering and Biotechnology, 2022, 10, 877480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jeon JS, Bersini S, Whisler JA, Chen MB, Dubini G, Charest JL, Moretti M and Kamm RD, Integr. Biol, 2014, 6, 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M and Kamm RD, Proceedings of the National Academy of Sciences, 2014, 112, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pavesi A, Tan AT, Koh S, Chia A, Colombo M, Antonecchia E, Miccolis C, Ceccarello E, Adriani G, Raimondi MT, Kamm RD and Bertoletti A, JCI Insight, 2017, 2, e89762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Trietsch SJ, Naumovska E, Kurek D, Setyawati MC, Vormann MK, Wilschut KJ, Lanz HL, Nicolas A, Ng CP, Joore J, Kustermann S, Roth A, Hankemeier T, Moisan A and Vulto P, Nature Communications, 2017, 8, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park D, Lee J, Lee Y, Son K, Choi JW, Jeang WJ, Choi H, Hwang Y, Kim H-Y and Jeon NL, Scientific Reports, 2021, 11, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Su C, Chuah YJ, Ong HB, Tay HM, Dalan R and Hou HW, Biosensors, 2021, 11, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Plebani R, Potla R, Soong M, Bai H, Izadifar Z, Jiang A, Travis RN, Belgur C, Dinis A, Cartwright MJ, Prantil-Baun R, Jolly P, Gilpin SE, Romano M and Ingber DE, Journal of Cystic Fibrosis, 2022, 21, 606–615. [DOI] [PubMed] [Google Scholar]
- 106.Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS and Ingber DE, Cell Reports, 2017, 21, 508–516. [DOI] [PubMed] [Google Scholar]
- 107.Kim M-C, Lam RHW, Thorsen T and Asada HH, Microfluidics and Nanofluidics, 2013, 15, 285–296. [Google Scholar]
- 108.Microfluidic ChipShop homepage, https://www.microfluidic-chipshop.com/.
- 109.Bhargava KC, Thompson B and Malmstadt N, Proceedings of the National Academy of Sciences, 2014, 111, 15013–15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gong H, Woolley AT and Nordin GP, Lab on a Chip, 2018, 18, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ong LJY, Ching T, Chong LH, Arora S, Li H, Hashimoto M, DasGupta R, Yuen PK and Toh Y-C, Lab on a Chip, 2019, 19, 2178–2191. [DOI] [PubMed] [Google Scholar]
- 112.Yuen PK, Lab on a Chip, 2016, 16, 3700–3707. [DOI] [PubMed] [Google Scholar]
- 113.Toh AGG, Wang ZP, Yang C and Nguyen N-T, Microfluidics and Nanofluidics, 2013, 16, 1–18. [Google Scholar]
- 114.Ayuso JM, Virumbrales-Munoz M, McMinn PH, Rehman S, Gomez I, Karim MR, Trusttchel R, Wisinski KB, Beebe DJ and Skala MC, Lab on a Chip, 2019, 19, 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rosser J, Bachmann B, Jordan C, Ribitsch I, Haltmayer E, Gueltekin S, Junttila S, Galik B, Gyenesei A, Haddadi B, Harasek M, Egerbacher M, Ertl P and Jenner F, Materials Today Bio, 2019, 4, 100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES and Jeon NL, Lab on a Chip, 2005, 5, 401. [DOI] [PubMed] [Google Scholar]
- 117.Ayuso JM, Virumbrales-Muñoz M, Lacueva A, Lanuza PM, Checa-Chavarria E, Botella P, Fernández E, Doblare M, Allison SJ, Phillips RM, Pardo J, Fernandez LJ and Ochoa I, Scientific Reports, 2016, 6, 36086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Frevert CW, Boggy G, Keenan TM and Folch A, Lab on a Chip, 2006, 6, 849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jeon NL, Baskaran H, Dertinger SKW, Whitesides GM, Water LVD and Toner M, Nature Biotechnology, 2002, 20, 826–830. [DOI] [PubMed] [Google Scholar]
- 120.Yang IH, Siddique R, Hosmane S, Thakor N and Höke A, Experimental Neurology, 2009, 218, 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peng B, Tong Z, Tong WY, Pasic PJ, Oddo A, Dai Y, Luo M, Frescene J, Welch NG, Easton CD, Thissen H and Voelcker NH, ACS Applied Materials & Interfaces, 2020, 12, 56753–56766. [DOI] [PubMed] [Google Scholar]
- 122.Chong LH, Li H, Wetzel I, Cho H and Toh Y-C, Lab on a Chip, 2018, 18, 3239–3250. [DOI] [PubMed] [Google Scholar]
- 123.Irimia D, Geba DA and Toner M, Analytical Chemistry, 2006, 78, 3472–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kwak TJ, Nam YG, Najera MA, Lee SW, Strickler JR and Chang W-J, PLOS ONE, 2016, 11, e0166068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ong LJY, Chia S, Wong SQR, Zhang X, Chua H, Loo JM, Chua WY, Chua C, Tan E, Hentze H, Tan IB, DasGupta R and Toh Y-C, Frontiers in Bioengineering and Biotechnology, 2022, 10, 952726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee PJ, Hung PJ and Lee LP, Biotechnology and Bioengineering, 2007, 97, 1340–1346. [DOI] [PubMed] [Google Scholar]
- 127.Inglebert M, Locatelli L, Tsvirkun D, Sinha P, Maier JA, Misbah C and Bureau L, Biomicrofluidics, 2020, 14, 024115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Choi J, Lee SY, Yoo Y-M and Kim CH, Cell Biochemistry and Biophysics, 2016, 75, 87–94. [DOI] [PubMed] [Google Scholar]
- 129.Smith RL, Donlon BS, Gupta MK, Mohtai M, Das P, Carter DR, Cooke J, Gibbons G, Hutchinson N and Schurman DJ, Journal of Orthopaedic Research, 1995, 13, 824–831. [DOI] [PubMed] [Google Scholar]
- 130.Booth R, Noh S and Kim H, Lab on a Chip, 2014, 14, 1880–1890. [DOI] [PubMed] [Google Scholar]
- 131.Yang F, Carmona A, Stojkova K, Huitron EIG, Goddi A, Bhushan A, Cohen RN and Brey EM, Lab on a Chip, 2021, 21, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yeatts AB and Fisher JP, Bone, 2011, 48, 171–181. [DOI] [PubMed] [Google Scholar]
- 133.Arora S, Srinivasan A, Leung CM and Toh Y-C, Current Stem Cell Research & Therapy, 2020, 15, 414–427. [DOI] [PubMed] [Google Scholar]
- 134.Lee D, Erickson A, Dudley AT and Ryu S, Experimental Mechanics, 2018, 59, 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee D, Erickson A, Dudley AT and Ryu S, Journal of Visualized Experiments, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huh D, Kim HJ, Fraser JP, Shea DE, Khan M, Bahinski A, Hamilton GA and Ingber DE, Nature Protocols, 2013, 8, 2135–2157. [DOI] [PubMed] [Google Scholar]
- 137.Weisgrab G, Ovsianikov A and Costa PF, Advanced Materials Technologies, 2019, 4, 1900275. [Google Scholar]
- 138.Unger MA, Chou H-P, Thorsen T, Scherer A and Quake SR, Science, 2000, 288, 113–116. [DOI] [PubMed] [Google Scholar]
- 139.Zhao C, Xia Z, Wang X, Nie J, Huang P and Zhao S, Materials & Design, 2020, 193, 108788. [Google Scholar]
- 140.Khizer Z, Akram MR, Sarfraz RM, Nirwan JS, Farhaj S, Yousaf M, Hussain T, Lou S, Timmins P, Conway BR and Ghori MU, Polymers, 2019, 11, 1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Taylor AP, Reyes JI and Velásquez-García LF, Journal of Physics D: Applied Physics, 2020, 53, 355002. [Google Scholar]
- 142.Shimizu K, Araki H, Sakata K, Tonomura W, Hashida M and Konishi S, Journal of Bioscience and Bioengineering, 2015, 119, 212–216. [DOI] [PubMed] [Google Scholar]
- 143.Yang IH, Gary D, Malone M, Dria S, Houdayer T, Belegu V, Mcdonald JW and Thakor N, NeuroMolecular Medicine, 2012, 14, 112–118. [DOI] [PubMed] [Google Scholar]
- 144.Taylor A and Jeon NL, Critical Reviews™ in Biomedical Engineering, 2011, 39, 185–200. [DOI] [PubMed] [Google Scholar]
- 145.Chmayssem A, Tanase CE, Verplanck N, Gougis M, Mourier V, Zebda A, Ghaemmaghami AM and Mailley P, Biosensors, 2022, 12, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Weltin A, Slotwinski K, Kieninger J, Moser I, Jobst G, Wego M, Ehret R and Urban GA, Lab on a Chip, 2014, 14, 138–146. [DOI] [PubMed] [Google Scholar]
- 147.Lee HU, Blasiak A, Agrawal DR, Loong DTB, Thakor NV, All AH, Ho JS and Yang IH, PLOS ONE, 2017, 12, e0179642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hallfors N, Khan A, Dickey MD and Taylor AM, Lab on a Chip, 2013, 13, 522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang Q, Gao M, Zhang L, Deng Z and Gui L, Sensors, 2019, 19, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Henry OYF, Villenave R, Cronce MJ, Leineweber WD, Benz MA and Ingber DE, Lab on a Chip, 2017, 17, 2264–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.van der Helm MW, Odijk M, Frimat J-P, van der Meer AD, Eijkel JCT, van den Berg A and Segerink LI, Journal of Visualized Experiments, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Musgrove HB, Catterton MA and Pompano RR, Analytica Chimica Acta, 2022, 1209, 339842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Monk DW and Gale RO, Microelectronic Engineering, 1995, 27, 489–493. [Google Scholar]
- 154.Shallan AI, Smejkal P, Corban M, Guijt RM and Breadmore MC, Analytical Chemistry, 2014, 86, 3124–3130. [DOI] [PubMed] [Google Scholar]
- 155.Gong H, Woolley AT and Nordin GP, Biomicrofluidics, 2019, 13, 014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Leonhardt S, Klare M, Scheer M, Fischer T, Cordes B and Eblenkamp M, Current Directions in Biomedical Engineering, 2016, 2, 113–116. [Google Scholar]
- 157.Bagheri A and Jin J, ACS Applied Polymer Materials, 2019, 1, 593–611. [Google Scholar]
- 158.Choi JW, Kim G-J, Hong S, An JH, Kim B-J and Ha CW, Scientific Reports, 2022, 12, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhu F, Friedrich T, Nugegoda D, Kaslin J and Wlodkowic D, Biomicrofluidics, 2015, 9, 061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Macdonald NP, Zhu F, Hall CJ, Reboud J, Crosier PS, Patton EE, Wlodkowic D and Cooper JM, Lab on a Chip, 2016, 16, 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schmelzer E, Over P, Gridelli B and Gerlach JC, Journal of Medical and Biological Engineering, 2016, 36, 153–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bhattacharjee N, Parra-Cabrera C, Kim YT, Kuo AP and Folch A, Advanced Materials, 2018, 30, 1800001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.González G, Baruffaldi D, Martinengo C, Angelini A, Chiappone A, Roppolo I, Pirri CF and Frascella F, Nanomaterials, 2020, 10, 1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Piironen K, Haapala M, Talman V, Järvinen P and Sikanen T, Lab on a Chip, 2020, 20, 2372–2382. [DOI] [PubMed] [Google Scholar]
- 165.Carnero B, Bao-Varela C, Gómez-Varela AI, Álvarez E and Flores-Arias MT, Materials Science and Engineering: C, 2021, 129, 112388. [DOI] [PubMed] [Google Scholar]
- 166.Tabriz AG, Viegas B, Okereke M, Uddin MJ, Lopez EA, Zand N, Ranatunga M, Getti G and Douroumis D, Micromachines, 2022, 13, 1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Guttridge C, Shannon A, O’Sullivan A, O’Sullivan KJ and O’Sullivan LW, Annals of 3D Printed Medicine, 2022, 5, 100044. [Google Scholar]
- 168.formlabs homepage, https://formlabs.com/. [Google Scholar]
- 169.Hart C, Didier CM, Sommerhage F and Rajaraman S, Biosensors, 2020, 10, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.MiiCraft homepage, https://miicraft.com/. [Google Scholar]
- 171.Dreve Otoplastik homepage, https://otoplastik.dreve.de/en/highlights/3d-material/.
- 172.Detax homepage, https://www.detax.de/en/content/3D-Produkte.php.
- 173.Boston Microfabrication homepage, https://bmf3d.com/.
- 174.Shan H, Lin Q, Wang D, Sun X, Quan B, Chen X and Chen Z, Frontiers in Bioengineering and Biotechnology, 2021, 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.pro3dure medical homepage, https://www.pro3dure.com/en/products/.
- 176.Bucciarelli A, Paolelli X, Vitis ED, Selicato N, Gervaso F, Gigli G, Moroni L and Polini A, Additive Manufacturing, 2022, 60, 103200. [Google Scholar]
- 177.Beckwith AL, Borenstein JT and Velasquez-Garcia LF, Journal of Microelectromechanical Systems, 2018, 27, 1009–1022. [Google Scholar]
- 178.keyprint homepage, https://keyprint.keystoneindustries.com/keyortho-ibt/.
- 179.Asiga homepage, https://www.asiga.com/.
- 180.3D Systems homepage, https://www.3dsystems.com/.
- 181.Somos WaterShed resin, https://www.stratasys.com/en/materials/materials-catalog/stereolithography-materials/somos-watershed-xc-11122/.
- 182.Lam MT and Longaker MT, Journal of Tissue Engineering and Regenerative Medicine, 2012, 6, s80–s86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tsang VL, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL and Bhatia SN, The FASEB Journal, 2006, 21, 790–801. [DOI] [PubMed] [Google Scholar]
- 184.Loessner D, Meinert C, Kaemmerer E, Martine LC, Yue K, Levett PA, Klein TJ, Melchels FPW, Khademhosseini A and Hutmacher DW, Nature Protocols, 2016, 11, 727–746. [DOI] [PubMed] [Google Scholar]
- 185.Tomov ML, Perez L, Ning L, Chen H, Jing B, Mingee A, Ibrahim S, Theus AS, Kabboul G, Do K, Bhamidipati SR, Fischbach J, McCoy K, Zambrano BA, Zhang J, Avazmohammadi R, Mantalaris A, Lindsey BD, Frakes D, Dasi LP, Serpooshan V and Bauser-Heaton H, Advanced Healthcare Materials, 2021, 10, 2100968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Cetnar AD, Tomov ML, Ning L, Jing B, Theus AS, Kumar A, Wijntjes AN, Bhamidipati SR, Do KP, Mantalaris A, Oshinski JN, Avazmohammadi R, Lindsey BD, Bauser-Heaton HD and Serpooshan V, Advanced Healthcare Materials, 2020, 10, 2001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, Johansson F, Randles A, Rosenkrantz JE, Louis-Rosenberg JD, Galie PA, Stevens KR and Miller JS, Science, 2019, 364, 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Huh J, Moon Y-W, Park J, Atala A, Yoo JJ and Lee SJ, Biofabrication, 2021, 13, 034103. [DOI] [PubMed] [Google Scholar]
- 189.Ding H, Dong M, Zheng Q and Wu ZL, Molecular Systems Design & Engineering, 2022, 7, 1017–1029. [Google Scholar]
- 190.Decker C, Macromolecular Rapid Communications, 2002, 23, 1067–1093. [Google Scholar]
- 191.Tasdelen MA, Lalevée J and Yagci Y, Polymer Chemistry, 2020, 11, 1111–1121. [Google Scholar]
- 192.van der Linden PJEM, Popov AM and Pontoni D, Lab on a Chip, 2020, 20, 4128–4140. [DOI] [PubMed] [Google Scholar]
- 193.Rogers CI, Pagaduan JV, Nordin GP and Woolley AT, Analytical Chemistry, 2011, 83, 6418–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fleck E, Sunshine A, DeNatale E, Keck C, McCann A and Potkay J, Micromachines, 2021, 12, 1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Fleck E, Keck C, Ryszka K, DeNatale E, Sunshine A and Potkay JA, ASAIO Journal, 2022, 68, 15–15. [Google Scholar]
- 196.Lee Y-S, Bhattacharjee N and Folch A, Lab on a Chip, 2018, 18, 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rogers CI, Qaderi K, Woolley AT and Nordin GP, Biomicrofluidics, 2015, 9, 016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Femmer T, Kuehne AJC and Wessling M, Lab on a Chip, 2014, 14, 2610. [DOI] [PubMed] [Google Scholar]
- 199.Nahak BK, Mishra A, Preetam S and Tiwari A, ACS Applied Bio Materials, 2022, 5, 3576–3607. [DOI] [PubMed] [Google Scholar]
- 200.Grigoryan B, Sazer DW, Avila A, Albritton JL, Padhye A, Ta AH, Greenfield PT, Gibbons DL and Miller JS, Scientific Reports, 2021, 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.B9 Creations homepage, https://www.b9c.com/.
- 202.CELLINK Lumen X, https://www.cellink.com/bioprinting/lumen-x/.
- 203.Acrea 3D homepage, https://acrea3d.com/. [Google Scholar]
- 204.Au AK, Lee W and Folch A, Lab on a Chip, 2014, 14, 1294–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]


