Abstract
The maturation of chemical synthesis during the 20th century has elevated the discipline from a largely empirical into a rational science. This ability to purposefully craft matter at the molecular level has put chemists in a privileged position to contribute to progress in neighboring natural sciences. Recently, we have witnessed another major advance in the field in which chemists use chemical and biological “synthetic” methods together to alter the structures and properties of biological macromolecules in ways heretofore unimagined. This interdisciplinary approach to synthesis has even allowed us to expand upon the defining characteristics of living organisms at the molecular level. In this perspective, we present a case study for the successful addition of new chemistries to the fundamental processes of the central dogma of molecular biology, exemplified by the expansion of the genetic code.
Graphical Abstract

INTRODUCTION
The central dogma of molecular biology defines the processes of DNA replication, transcription, and translation (Fig. 1).1 All genetic information is stored in a four-nucleotide genetic alphabet and is deciphered from sets of nucleotide triplets, or codons, according to the genetic code. Of the existing 64 codons (43), 61 code for one of the 20 canonical amino acids with the remaining three directing termination of translation. Both the genetic alphabet and code are understood to be universal to all life. The genetic information is first transcribed into messenger RNA (mRNA) and then translated into sequence-defined polypeptides (proteins) by the ribosome. Proteins constitute the primary macromolecular machinery of the cell. Their function is imparted by the molecular characteristics and tertiary arrangement of their polypeptide sequences and is thus intrinsically limited by the chemical space covered by the 20 proteinogenic amino acids whose side chains include carboxylic acids, amides, alcohols, amines, thiol and thiol ether, alkyl and aryl groups (and in a handful of archaea bacteria, selenocysteine and pyrrolysine amino acids)
Figure 1.
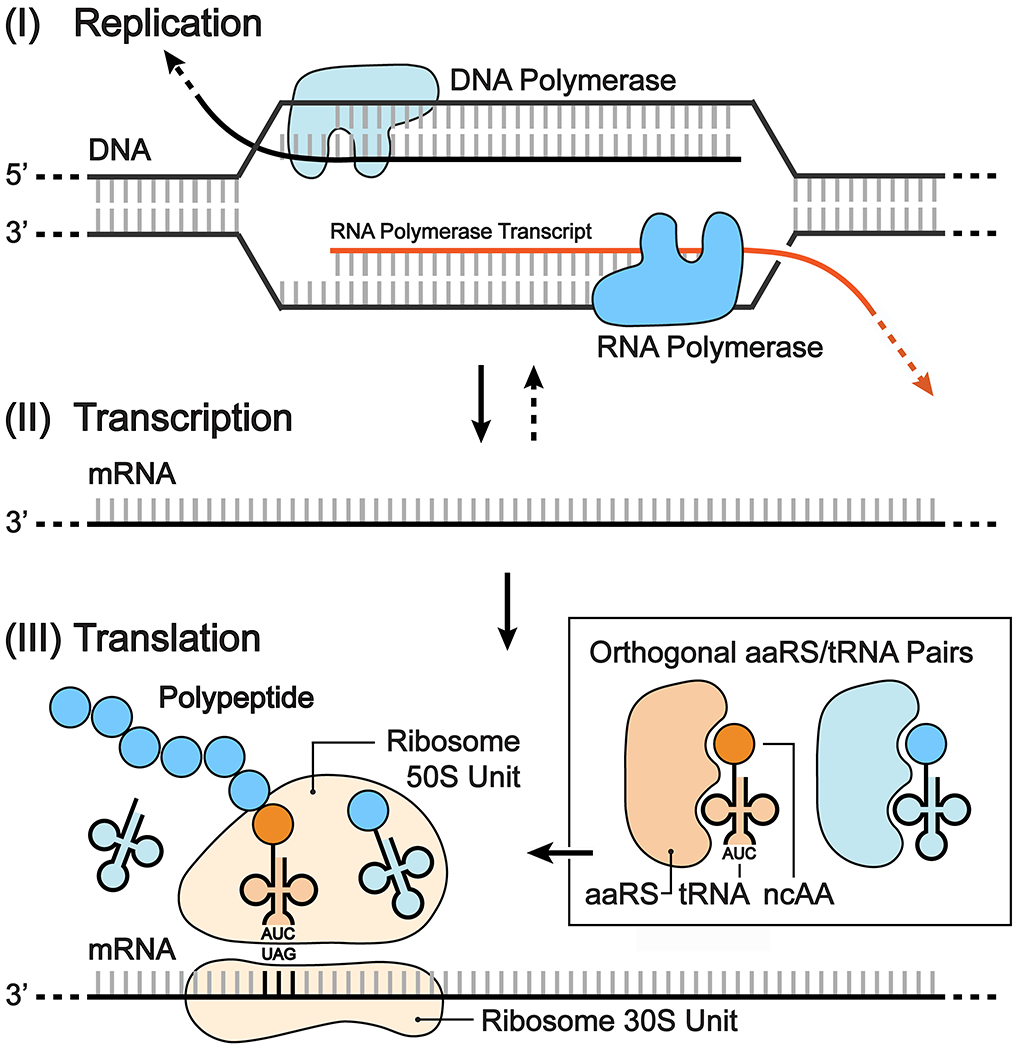
Expanding the chemical processes of the central dogma. The genetic code can be expanded to incorporate unnatural amino acids into proteins during translation using nonsense (illustrated in the schematic with the amber nonsense codon TAG), frameshift, repurposed synonymous or unnatural codons, and orthogonal aaRS/tRNA pairs.
The field of synthesis as exemplified by chemical synthesis and more recently the burgeoning field of synthetic biology, reflects our desire to control the three-dimensional structure of matter at the molecular level, be it small molecules or large complex molecules or systems of interacting molecules. The latter focus compelled chemists to ask whether one might be able to expand upon the chemistries contained within the central dogma itself to be able to create unnatural structures of unparalleled complexity that are generally inaccessible by conventional synthetic methodologies, be it chemical or by recombinant technologies. Herein, we outline how chemists have harnessed the processes of the central dogma of molecular biology toward this purpose.
EXPANSION OF THE GENETIC CODE
During translation, mRNA, the product of DNA transcription, serves as a template for the sequence-defined synthesis of polypeptides at the ribosome. The 61 sense codons are each assigned to one of the 20 canonical amino acids. This process is directed by a network of orthogonal sets of aminoacyl-transfer RNA synthetase (aaRS)/isoacceptor-transfer RNA (tRNA) pairs. Each amino acid is aminoacylated to its cognate tRNA by its requisite aaRS. Subsequently, the anticodon-containing charged tRNA is directed by the elongation factor (elongation factor thermo unstable, EF-Tu) to the ribosome, where it is paired with its corresponding mRNA codon. Successful anticodon-codon pairing directs peptidyl-transfer between the tRNA and the growing polypeptide chain until stop-codon-directed termination releases the protein (Fig. 1).
The addition of synthetic building blocks with new structures and chemistries to the genetic code of living cells requires reprogramming of the existing translation machinery to ensure orthogonality, that is the absence of crosstalk with the natural protein biosynthetic machinery. To this end, a number of prerequisites had to be considered: (i) cell-permeability or biosynthetic availability of metabolically stable non-canonical amino acids (ncAAs), (ii) compatibility of the ncAA with the elongation factor and the ribosome, (iii) the unique assignment of a codon with a corresponding tRNA and its cognate aaRS to a specific ncAA, and (iv) high selectivity for the allocated aaRS to its cognate tRNA and ncAA.2 Criteria (i) and (ii) are satisfied for many ncAAs. First, there is a vast chemical space of candidate ncAAs that are both chemically stable and cell permeable. Second, both the aminoacyl-binding site of EF-Tu, and the ribosome itself are amenable, without alteration, to a broad assortment of ncAAs. This was corroborated by the fact that modified amino acids had been incorporated into proteins using chemically aminoacylated tRNAs in vitro3,4,5
The major challenge to adding new amino acids to the code thus became the allocation of a unique codon with a cognate tRNA/aaRS pair and achieving its bioorthogonality to the host’s translation machinery. The first step toward this goal was the allocation of a specified codon. Here, the amber codon (TAG in DNA; UAG in mRNA) was chosen, as it is the least abundant (8%) stop codon in the genome of the model bacterium Escherichia coli genome, uncommon in essential genes, and because certain strains of E. coli naturally incorporate canonical amino acids at the amber codon (amber suppression).6 Mutating the anticodon of a ncAA-aminoacylated orthogonal tRNA to CUA, the amber anticodon, enables suppression of the amber codon. As a first step, a gene harboring an amber mutation at a specified locus was expressed in vitro using a cell-free transcription/translation system. The addition of amber suppressors (tRNACUA) that had been chemically aminoacylated with a variety of ncAAs enabled translation of the corresponding protein containing the respective ncAAs.7 This strategy could further be applied to in vivo applications by microinjection of such chemically aminoacylated tRNACUAs into the large Xenopus oocytes. While this allowed for the incorporation of ncAAs into proteins in oocytes, the use of chemically aminoacylated tRNAs for genetic code expansion was intrinsically limited by its stoichiometric nature and by the requirement for microinjection into cells.
To expand the capacity of the cell’s own translational machinery, an orthogonal translation machinery had to be introduced into cells that can autonomously aminoacylate ncAAs in a catalytic manner. Previous research had shown that the recognition of tRNAs by their cognate aaRSs can be specific to a domain of life or species.2 With this knowledge, the orthogonal MjtRNATyr/MjTyrRS pair from the archaea Methanococcus jannaschii was identified and introduced into E. coli. The MjTyrRS was an ideal first candidate because of the large number of interactions involved in Tyr side chain recognition by the RS (suggesting that the active site could be reconfigured to accommodate unnatural side chains), as well as a minimalist RS anticodon-loop-binding domain that was amenable to alteration of the tRNA anticodon to CUA. In addition, the MjTyrRS has no editing mechanism which could lead to deacylation of the ncAA, and it can be expressed in high levels in E. coli. To improve the orthogonality of the exogenous aaRS/tRNA pair, successive rounds of positive and negative selection were used to reduce crosstalk with the host endogenous tRNAs and aaRSs.8 Using this strategy several highly specific aaRS/tRNA pairs, many of which are mutually orthogonal, have been evolved to incorporate a plethora of ncAAs in bacteria, yeast, worms, mammalian cells, and in the hematopoietic system of mice.9,10,11,12,13,14 The fidelity rivals that of natural amino acids and yields up to 8 g/L in bacteria and up to 2 g/L in mammalian cells have been reached in large scale fermenters using the amber nonsense codon to specify the ncAA.15
EXPANDING THE CHEMICAL TOOLKIT OF NATURE
The above methodology has been used to introduce a wide array of ncAAs with diverse structures and properties into proteins in vivo, at current count over 200 unique ncAAs. These include metal-binding amino acids, fluorophores, IR probes, isotopically-labeled amino acids, redox-active amino acids, post-translationally modified amino acids and their stable analogues, photocaged amino acids, and protein-protein and protein-nucleic acid photocrosslinkers.2 One particularly useful application of an expanded genetic code is the use of ncAAs as bioorthogonal chemical handles for the highly selective modification of proteins with drugs, probes, nucleic acids and other agents. Living organisms post-translationally modify amino acids by acylation, glycosylation, methylation, phosphorylation, sulfation, etc. to extend the chemical repertoire of amino acids beyond the 20. ncAAs expand the scope of these modification reactions to the vast repertoire of organic methodologies and enable site-specific modification of individual proteins with exquisite chemical precision, as opposed to a residue-specific conjugation across the entire proteome that is observed with conventional protein labelling techniques (e.g. lysine-NHS ester or cysteine-maleimide conjugation).16 Site-specific protein conjugation has been used to attach a vast array of molecular and macromolecular (e.g., drugs, fluorophores, spin labels, polymers, polypeptides, nucleic acids) moieties to proteins containing ncAAs. This has enabled medicinal chemistry-like tuning of the physical and biological properties (biological activity, serum half-life, and stability/solubility) of protein conjugates and has become vital to the development of next-generation protein therapeutics.17
BIOORTHOGONAL PROTEIN MODIFICATION
Modification reactions for functionalization of ncAAs in vivo and in vitro include, but are not limited to Michael additions, cross couplings, metathesis reactions, 1,3-dipolar cycloadditions, isothiocyanate couplings, Diels-Alder reactions, and oxime formation. One of the first and foremost methods for bioorthogonal protein modification was based on the keto-functional group.18 Specifically, incorporation of p-acetylphenylalanine has enabled highly selective and efficient functionalization of proteins with alkoxy-amine derivatives yielding stable oxime bonds.19 This strategy benefits from facile synthetic access to derivatized (macro)molecules, excellent yields negligible perturbations in protein folding and function, and lack of immunogenicity caused by the ncAA. Oxime conjugation can be used for protein modification in vitro, but has only limited utility in vivo due to the lower pH requirements. Nonetheless this method has been applied to the development of six site-selective protein-drug or PEG conjugates in various stages of human clinical development and drug approval (ARX788, ARX517, BMS-986036, BMS-986259, CCW702, and SAR444245). Indeed, the high degree of selectivity possible with this method appears to afford improved efficacy and safety of antibody-drug conjugates (i.e., Herceptin-drug conjugates).
To enable bioorthogonal conjugation of proteins in vivo several criteria must be met: (i) the pre-and post-reacted reagents must be stable under physiological conditions, (ii) the reaction has to be high yielding with low reactant concentrations and display fast kinetics, and (iii) the labelled ncAA in both pre-and post-reacted form must not compromise protein function or solubility. Among the most common reactive ncAAs are propargyl and azide functionalized analogs.20,21,22,23 Both enable modification by Cu(I) catalyzed (CuAAC) or strain promoted (SPAAC) azide-alkyne cycloaddition.24,25 These reactions benefit from high selectivity and yield, benign reaction conditions, and chemical inertness of the 1,2,3-triazole linkage product in physiological conditions. However, conventional CuAAC is toxic to cells due to side reactions of Cu(I) with biomacromolecules. In contrast, SPAAC employs strained macrocyclic alkynes (i.e., cyclooctynes) to retain fast reaction kinetics in the absence of an exogenous catalyst. While SPAACs solve the in vivo cytotoxicity constraint, the potential impact of bulky cyclooctene constructs (e.g. cyclooctynyl lysine derivative ncAAs) on protein folding, solubility and immunogenicity, as well as the chemical stability of the azide species must be considered.26 Another common approach is conjugation by the inverse electron-demand Diels-Alder reactions of s-tetrazines (1,2,4,5-tetrazine) with strained alkenes or alkynes.27 This strategy is particularly adept for in vivo applications due to its fast reaction kinetics and physiological stability of both reactants and products. Because s-tetrazines can react with a variety of strained alkene and alkyne substrates (e.g. Nɛ-tert-butyloxycarbonyl-L-lysine) they provide opportunities to minimize the detrimental effects on protein folding or solubility (Fig. 2a and b).28 More recently, aryl fluorosulfate exchange has been developed for proximity-induced click chemistry in protein conjugation.29 Aryl fluorosulfates benefit from high chemical stability (negligible hydrolysis, thermolysis, and reduction), as well as compatibility with physiological conditions (relatively unreactive toward free amino acids). Fluorosulfate functionalized ncAAs (e.g. p-fluorosulfate-tyrosine) selectively form covalent bonds to proximal tyrosine, serine, and lysine residues of proteins through sulfur-fluoride exchange (SuFEx) which holds high promise for selective proximity-induced protein conjugation.29,30
Figure 2.
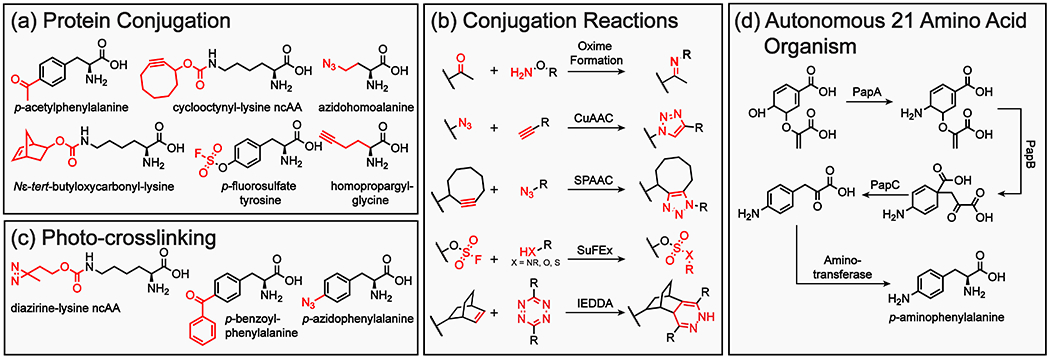
(a) Examples of ncAAs used for site-specific protein conjugation. (b) Chemical reactions used for site-specific protein conjugation. (c) Examples of ncAAs used for photo-crosslinking of proteins in vivo. (d) Heterologous pathway for the synthesis of the ncAA p-aminophenylalanine in E. coli.
Reactive ncAAs can also be used to investigate protein–protein and protein-nucleic interactions in their native environment. This is achieved through photo-crosslinking, which, in contrast to conventional approaches, is not limited to high affinity protein-protein interactions and introduces the ability to monitor events with spatiotemporal resolution. When developing candidate photocrosslinking ncAAs, the reactive functional groups need to fulfill a number of prerequisites: (i) the photocrosslinker should minimally perturb protein function, (ii) photoactivation can only minimally affect the cellular environment, (iii) in the inactive state (absence of irradiation) the photoactive group must be chemically inert, and (iv) in the activated state (upon UV radiation), the photo-crosslinker must form covalent bonds with a broad range of canonical amino acid side chains or with the peptide backbone (i.e. C–H, N–H and O–H bonds). Several photo-crosslinking ncAAs have been developed using azide (e.g. p-azidophenylalanine), diazirine (e,g, diazirine-lysine ncAA), and benzophenone (e.g. p-benzoylphenylalanine, pBzF) functional groups (Fig. 2c).22,31,32 Benzophenones are particularly useful due to their high chemical stability and inertness under ambient light. Photoexcitation of benzophenones (irradiation at 350-360 nm) does not compromise protein stability, and once activated, benzophenones preferentially react with unactivated C–H bonds, which are omnipresent on the exterior surface of proteins. The photoexcited triplet state of benzophenones readily relaxes in the absence of a reaction partner, which allows for repeated excitation and concomitant high crosslinking yields. This methodology has been used to probe a large number of protein interactions in the cell. We employed this technology to probe the interactome of short open reading frame (ORF)-encoded peptides (SEPs);33,34 other applications include understanding LPS (lipopolysaccharide) and phospholipid transport across bacterial membranes,35,36 the role of various ATPases in eukaryotic cellular processes,37,38 and how ubiquitylation of histones leads to chromatin decompaction,39 among others.
BIOLOGICAL CONSEQUENCES OF AN EXPANDED GENETIC CODE
In its current state, an organism with an expanded genetic code generally relies on exogenous feeding of the requisite ncAA. An alternative approach involves metabolically engineering organisms to produce a specific ncAA in vivo. We developed the first autonomous 21-amino acid E. coli variant by heterologous expression of genes from Streptomyces venezuelae to produce the bioorthogonal ncAA p-aminophenylalanine (pAF, Fig. 2d).40 By using an orthogonal aaRS/tRNA pair that adds pAF to the genetic code, we demonstrated that it could intracellularly be incorporated into proteins in bacteria. Similarly, a second autonomous organism has been generated in which 5-hydroxytryptophan is biosynthesized and added to the genetic code.41
An intriguing question is whether a 21-amino acid organism (like the above) has an organismal evolutionary advantage. We and others have begun to address this issue by placing bacteria with distinct 21 amino acid codes under selective pressure and exploring how they adapt.42 In one revealing experiment, E. coli encoding 21 amino acid (a different ncAA in each experiment) where grown at elevated temperatures-conditions where the survival of the bacteria depend on the thermal stability of the biosynthetic protein metA. Surviving clones replaced a canonical amino acid in metA with a genetically encoded keto-containing amino acid to crosslink the homodimeric protein and stabilize it by some 20 degrees.43 Other examples include increasing the fitness landscape of bacteriophage and the catalytic efficiency of enzymes for specific substrates.
Conversely, an organism’s dependence on ncAAs can be harnessed as a strategy for biological containment in applications such as live vaccines. Bacteria and viruses that are dependent on the availability of ncAAs have been generated by engineering genomically recoded organisms (GROs) harboring one or multiple amber codons in functionally conserved residues of the organism’s essential genes to make survival dependent on suppression by ncAAs.44,45 This has been applied to the conversion of influenza A viruses into virulent vaccines, a strategy that can in principle be extended to almost any virus.46 However, single point mutations enable high escape frequencies in GROs with only a single ncAA encoding amber codon, conversely multiple amber codon-ncAA substitutions can negatively affect growth rates. Alternatively, an essential gene of the organism can be evolved such that its function is strictly dependent upon a specified ncAA. This has been demonstrated for TEM-1 β-lactamase, which was evolved to be dependent on 3-iodo tyrosine or 3-nitro tyrosine.47 Another strategy to circumvent high escape frequencies is the incorporation of a ncAA on the exterior surface of an essential protein at a site at which it forms part of a protein–protein interface. Subsequent mutagenesis of the adjacent residues implicated in the recognition at the interface enables the formation of a strictly ncAA-dependent interaction. A ncAA-dependent protein–protein interface of the E. coli sliding clamp was engineered to be dependent on the presence of pBzF. This particular ncAA was chosen for its substantially larger side chain compared to canonical amino acids. Due to this size difference, multiple naturally-occurring mutations at the interface would be required to restore function when the pBzF residue of the new evolved protein interface is lost. This imparts a low escape frequency of 10−11, which holds high promise as a ncAA feeding based ‘kill switch’ for application in live bacterial vaccines.48
STRATEGIES FOR CODON REASSIGNMENT OR CREATION
A challenge in designating a codon for ncAA incorporation is the resulting impact that the suppression can have on the growth of the host organism. In an ideal ncAA incorporating biological system, orthogonal codons are assigned to the ncAA that do not interfere with the native biology of the host organism. E. coli encodes 2765 ochre (TAA, 64%), 1232 opal (TGA, 28%), and 321 amber (TAG, 8%) stop codons. Given the natural suppression of the amber codon in certain strains and its relatively low abundance, the amber codon is an obvious choice for non-orthogonal ncAA codon assignment (Fig. 3a). Amber codon suppression by ncAAs has been used to produce large quantities of proteins (yields of up to 8 g/L) in bacteria, and in mammalian cells (up to 2 g/L) where the TAG codon is more common.15 In an attempt to further improve yields in eukaryotic expression systems, engineered strains designed to circumvent nonsense mediated mRNA decay have been employed.49 A complementary strategy of encoding additional copies of the suppressor tRNA has proven helpful.50 Simultaneous suppression of the ochre codon and amber codon has been employed, to incorporate two distinct ncAAs into proteins.51,52 Furthermore, in E. coli, the tyrosine codon UAU can be repurposed as a dual-use codon to incorporate a third ncAA at the first position initiation codon, while not interfering with tyrosine decoding of UAU at later codons.53
Figure 3.
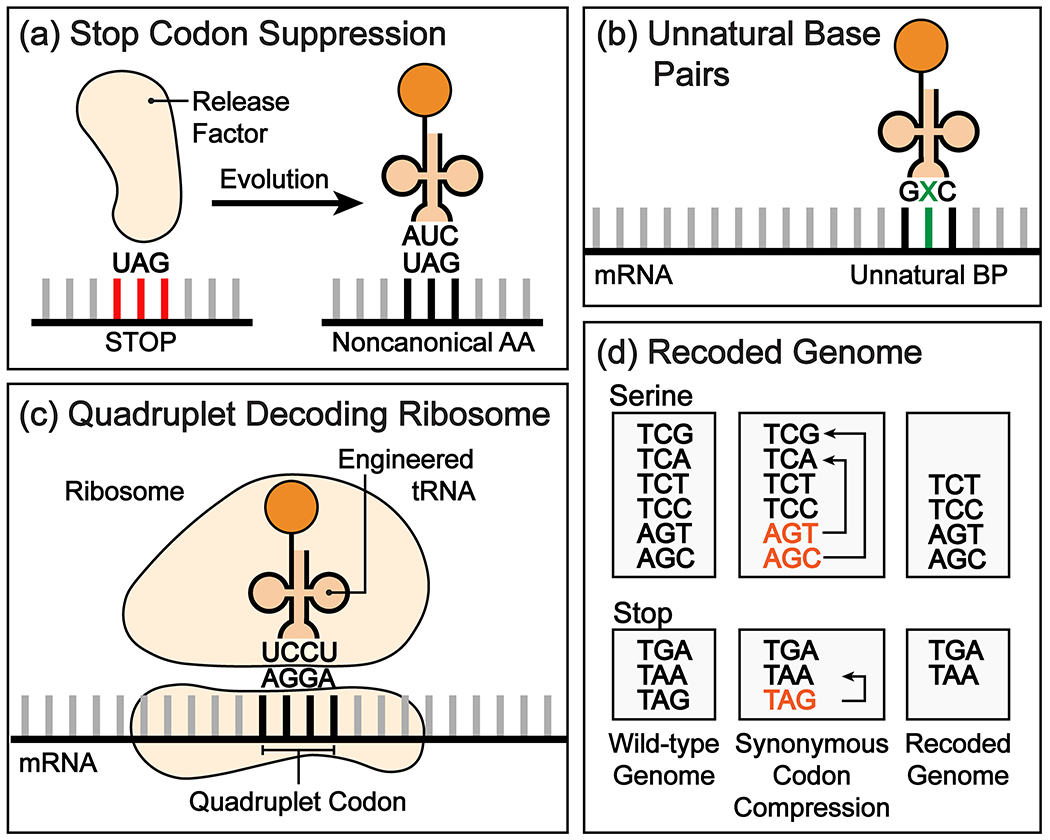
Strategies for codon reassignment and creation. ncAAs can be incorporated by (a) suppression of stop codons, by creation of additional codons using (b) an unnatural base pair or (c) quadruplet codons, or by (d) recoding of the entire genome of an organism in which synonymous codons are compressed.
However, to encode two or more ncAAs in an organism, one ideally wants unique codons that are orthogonal to termination factors and canonically assigned codons. One strategy that has been employed is the decoding of quadruplet base codons which can provide up to 256 additional codons (44).54 This approach relies on accommodation of extended anticodon tRNAs in the ribosome, which is well precedented with natural frameshift suppressors (Fig 3c). Indeed, engineered tRNAs with extended anticodon loops have been used to efficiently insert ncAAs into proteins and recently orthogonal ribosomes have been generated in an attempt to further improve the efficiency of four-base suppression.55 According to this strategy two (TAG and a single quadruplet codon, or two quadruplet codons) or three (TAG and two quadruplets codons) distinct ncAAs have been incorporated into proteins.55,56,57 These studies begin to raise the intriguing question whether one can entirely replace the three base genetic code with a four base code.
An alternative approach involves genome recombineering to create unique codons for ncAAs. In the first example all 321 amber codons in the genome of E. coli were mutated to the ochre codon to create an orthogonal amber codon for ncAA suppression.58 Termination factor RF1 terminates polypeptide synthesis at the amber codon. Deletion of the gene encoding RF1 enabled efficient four base suppression using codons of the form TAGN with no competition with termination factor recognition of the amber TAG stop codon.59 More recent efforts to reassign three codons in the genome of a single bacterium yielded a 59-codon organism, where both the amber codon and two serine codons (TCG and TCA) were successfully removed from the genome to allow for the incorporation of multiple distinct ncAAs (Fig 3d).60 Subsequent deletion of the corresponding tRNAs and RF1 rendered the strain completely resistant to viral infection and enabled the incorporation of three distinct ncAAs into a target protein in vivo.61 The recoded strain displayed an elongated phenotype and the growth rate of the organism decreased, but this direction holds promise.
The incorporation of multiple distinct ncAAs into a protein is contingent upon the development of mutually orthogonal aaRS/tRNA pairs. To date, a number of orthogonal pairs have been developed for the incorporation of ncAAs into proteins in bacteria. Of the developed aaRS/tRNA pairs several are mutually orthogonal and enable the incorporation of multiple UAAs, simultaneously. Mutually orthogonal pairs in bacteria include (TyrRS)/MjTyrtRNA with (PylRS)/MmPyltRNA and (EcTrpRS)/EcTrptRNA, (EcTyrRS)/Ec-TyrtRNA with (PylRS)/MmPyltRNA, as well as mutually orthogonal variants of (PylRS)/MmPyltRNAs with their own derivatives. In mammalian cells mutually orthogonal systems include mutually orthogonal PylRS/PyltRNA pairs, as well as combinations of PylRS/PyltRNA with (EcTyrRS)/Ec-TyrtRNA, (EcLeuRS)/Ec-LeutRNA, or (EcTrpRS)/EcTrptRNA. These pairs have been utilized to incorporate two or three ncAAs into proteins.52,62,63
ORTHOGONAL CODONS DERIVED FROM UNNATURAL BASE PAIRS
An elegant approach to generating orthogonal codons is the design and synthesis of orthogonal DNA base pairs that exist in the organism only at a specified site in a target plasmid (Fig 3b). Several requirements must be satisfied for implementation of an unnatural base pair into an in vivo system. Chemically and structurally, the UBP must (i) complement the geometry of the canonical base pairs to satisfy the minimal requirements to form a double helix, (ii) be chemically stable, (iii) have a melting temperature within range of those of the natural base pairs, (iv) have high pairing selectivity to ensure orthogonality with the canonical bases, (v) be efficiently and selectively replicated by DNA polymerases, and (vi) be transcribed to mRNA by RNA polymerases. Ideally, the cognate complementary base would be inserted into DNA with an error rate per base pair (fidelity) of at least 10−3 or 99.9% accuracy and must be transported into or biosynthesized in the cell.64
The first efforts toward expanding the genetic alphabet began in 1989 with constitutional isomers of C and G, disoC and disoG, which pair through complementary hydrogen-bonding interactions. In vitro studies with disoC and disoG confirmed their selective base-pairing interaction and demonstrated the base pairs’ use as a substrate for T7 RNA polymerase (Fig. 4). Subsequently, a chemically synthesized mRNA template comprising the unnatural ribonucleotide isoG–isoC pair enabled in vitro site-specific incorporation of a ncAA, 3-iodotyrosine, into a peptide.65 However, tautomerization of disoG and poor recognition of disoC by RNA polymerases limited the utility of this base pair, leading to decades of work on improved hydrogen-bonded UBPs. One example, the ds-dy UBP, was used to incorporate the ncAA 3-chlorotyrosine into proteins using a cell-free E. coli transcription and translation system, overcoming the challenge of the RNA polymerase recognition.66 Further synthetic improvements led to the UBP based on dZ and dP, which are more stable, do not epimerize, and are PCR amplified with a fidelity of 99.8%.67 The expansion of the genetic alphabet is not limited to one single UBP and recently the in vitro synthesis of DNA comprised of four orthogonal hydrogen-bonding base pairs was reported (Fig. 4).68
Figure 4.
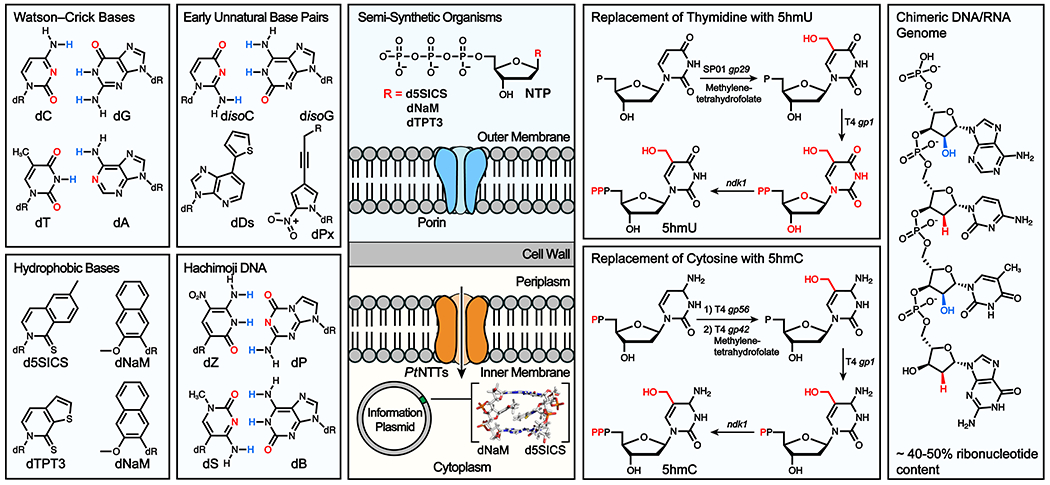
Chemical structure of exemplary hydrogen-bonded and hydrophobic unnatural base pairs (UBPs). Development of a semi-synthetic organism through UBP-import by heterologous nucleotide triphosphate transporters. Heterologous expression of SPO1 bacillus enzymes enables in vivo base modification of T to 5hmU and C to 5hmC in E. coli. Evolution of a strain of E. coli with a chimeric DNA/RNA genome.
Several alternatives to hydrogen-bonding base pairs have been developed including Cu(I) and Ag(I) metallo base pairs, and hydrophobic base pairs.64,69,70 The latter have the advantage of high orthogonality to the hydrogen bonded Watson-Crick base pairs, and high self-base pairing stability in water. The first hydrophobic isosteres of A and T, termed dQ and dF, were reported in 1998.71 Early UBPs were not broadly replicated with high fidelity by all DNA polymerases, thus motivating a proof-of-concept study to evolve the Stoffel fragment of Taq DNA Polymerase to replicate UBPs in vitro.72 Significant advances in hydrophobic base pair design led to the synthesis of dDs-dPa, which was the first hydrophobic base pair with PCR amplification fidelity >99%.73 Further development yielded dDs-dPx (Figure 4) with improved fidelity >99.9%.74 A substantial SAR of hydrophobic bases led to the discovery of lead candidates dNaM-d5SICS and dNaM-dTPT3, which were replicated with high fidelity (>99.9%) in any sequence context, were recognized by the A, B, and X families of DNA polymerases, and efficiently transcribed into RNA (Fig. 4).75
With the chemical and biological requirements for a UBP seemingly satisfied, the next step toward in vivo incorporation became nucleoside triphosphate (NTP) import. A variety of intracellular bacteria and algal plastids do not encode the enzymes necessary for NTP synthesis and rely on nucleoside triphosphate transporters (NTTs) to shuttle them across the cell membrane. A screen of NTTs from these organisms identified the Phaeodactylum tricornutum inner membrane protein NTT2 as an importer of both d5SICSTP and dNaMTP. The exogenous feeding of d5SICSTP and dNaMTP to E. coli coupled with plasmid-based overexpression of NTT2 resulted in the retrieval and insertion of UBPs into a secondary plasmid by host DNA polymerases in vivo.76 Subsequently, these UBPs were used to code for the incorporation of the ncAA N6-[(2-propynyloxy)carbonyl]-l-lysine (PrK) into GFP in live E. coli.77 To date, this represents the only example of a living organism encoding three orthogonal base pairs (Fig. 4).
The ability to incorporate unnatural base pairs raises the question of whether the E. coli genome is amenable to global replacement of native bases with modified variants. Previous efforts to modify genomic thymidines to chlorodeoxyuridines had yielded a genome with 90% of the intended conversion.78 Similarly, we engineered E. coli to replace 59% of genomic cytosines with 5hmC using the biosynthetic enzymes of bacteriophage T4.79 Efforts to mutagenize the E. coli genome, using methylnitronitrosoguanidine (NTG), to further increase the 5hmC content led to the serendipitous discovery of a strain with a chimeric DNA-RNA genome in which ~40-50% of the genome is comprised of ribonucleotides.80 Further we showed that the ribonucleotides are covalently linked to the deoxyribonucleotides in this strain, and there is a single replicating genome. This represents the first example of a modification to the DNA backbone in vivo, and the first example of an organism capable of replicating that has a high rNTP content in its genome. Surprisingly, a bottom-up approach to metabolically engineering an organism with a genome containing modified bases (to push evolution in a forward direction) may have revealed key aspects of natural evolution and the RNA world hypothesis (Fig. 4).81
OUTLOOK
At its core, the addition of new chemistries to the central dogma of molecular biology was enabled by the discovery and evolution of synthetic biological systems that are orthogonal to the endogenous macromolecular machinery of the cell. Looking forward, we believe that biorthogonal pathway engineering will remain at the center of future progress in synthetic biology. Thus far, a major focus of genetic code expansion has centered on appending new chemical functionality to the side chains of amino acids of proteins as probes of protein structure and functions and in the development of new therapeutics. Importantly, this advance is allowing engineering of therapeutic proteins with selectivity that rivals small molecule medicinal chemistry and promises to lead to a new generation of chemically modified protein with unprecedented molecular precision. The generation of multiple mutually orthogonal tRNA/aaRS pairs will further enable the incorporation of multiple modalities into proteins to form therapeutic proteins that are capable of activating or inhibiting multiple pathways to achieve a desired therapeutic effect. Multiple orthogonal pairs may also allow the creation of templated, ribosomally synthesized polymers composed entirely of unnatural building blocks. With respect to unnatural genetic materials, a plethora of work on the incorporation of unnatural nucleotides comprising unnatural bases, sugars, and backbone structures has been achieved in vitro. Moving these chemistries in to living organisms will require the elaboration of orthogonal DNA replication systems in both prokaryotes and eukaryotes, analogous to the use of orthogonal translation machinery for genetic code expansion.82 Furthermore, the development of an autonomous three base pair organism in which unnatural bases are derived biosynthetically will greatly enhance the utility of genetic alphabet expansion. In conclusion chemists have opened a new horizon in our ability to manipulate biological macromolecules and systems of molecules to better understand and ultimately reprogram the biology of the living cell to new ends. In the examples described herein, these advances have allowed us to remove in a very significant way a billion year constraint on the chemical nature of proteins imposed by the genetic code.83
ACKNOWLEDGMENTS
We acknowledge Kristen Williams for her assistance in manuscript preparation. This work is supported by NIH R01 GM062159
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interest.
REFERENCES
- 1.Crick F (1970). Central Dogma. Nature 227, 561–563. [DOI] [PubMed] [Google Scholar]
- 2.Liu CC, and Schultz PG (2010). Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem 79, 413–444. [DOI] [PubMed] [Google Scholar]
- 3.Krieg UC, Walter P, and Johnson AE (1986). Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl. Acad. Sci. U. S. A 83, 8604–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson AE, Woodward WR, Herbert E, and Menninger JR (1976). Nϵ-Acetyllysine Transfer Ribonucleic Acid: A Biologically Active Analogue of Aminoacyl Transfer Ribonucleic Acids. Biochemistry 15, 569–575. [DOI] [PubMed] [Google Scholar]
- 5.Heckler TG, Roesser JR, Xu C, Chang P in, and Hecht SM (1988). Ribosomal Binding and Dipeptide Formation by Misacylated tRNAphes. Biochemistry 27, 7254–7262. [DOI] [PubMed] [Google Scholar]
- 6.Cupples CG, and Miller JH (1988). Effects of amino acid substitutions at the active site in Escherichia coli β-galactosidase. Genetics 120, 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noren CJ, Anthony-Cahill SJ, Griffith MC, and Schultz PG (1989). A general method for site-specific incorporation of unnatural amino acids into proteins. Science (80-. ). 244, 182–188. [DOI] [PubMed] [Google Scholar]
- 8.Liu DR, and Schultz PG (1999). Progress toward the evolution of an organism with an expanded genetic code. Proc. Natl. Acad. Sci. U. S. A 96, 4780–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Brock A, Brad H, and Schultz PG (2001). Expanding the genetic code of Escherichia coli. Science (80-.). 292, 498–500. [DOI] [PubMed] [Google Scholar]
- 10.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, and Schultz PG (2003). An Expanded Eukaryotic Genetic Code. 301, 964–968. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto K (2002). Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 30, 4692–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao S, Koh M, and Schultz PG (2020). Expanding the genetic code of the human hematopoietic system. Proc. Natl. Acad. Sci. U. S. A 117, 8845–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianco A, Townsley FM, Greiss S, Lang K, and Chin JW (2012). Expanding the genetic code of Drosophila melanogaster. Nat. Chem. Biol. 8, 748–750. [DOI] [PubMed] [Google Scholar]
- 14.Parrish AR, She X, Xiang Z, Coin I, Shen Z, Briggs SP, Dillin A, and Wang L (2012). Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl-tRNA synthetase/tRNA pairs. ACS Chem. Biol 7, 1292–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang M, Lu Y, Chen S, and Tian F (2018). Harnessing the power of an expanded genetic code toward next-generation biopharmaceuticals. Curr. Opin. Chem. Biol 46, 123–129. [DOI] [PubMed] [Google Scholar]
- 16.Wong SS (1991). Chemistry of protein conjugation and cross-linking (CRC press; ). [Google Scholar]
- 17.Kim CH, Axup JY, and Schultz PG (2013). Protein conjugation with genetically encoded unnatural amino acids. Curr. Opin. Chem. Biol 17, 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dirksen A, and Dawson PE (2008). Rapid oxime and hydrazone ligations with aromatic aldehydes for biomolecular labeling. Bioconjug. Chem 19, 2543–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wangt L, Zhang Z, Brock A, and Schultz PG (2003). Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 100, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dieterich DC, Link AJ, Graumann J, Tirrell DA, and Schuman EM (2006). Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U. S. A 103, 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiick KL, Saxon E, Tirrell DA, and Bertozzi CR (2002). Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. U. S. A 99, 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin JW, Santoro SW, Martin AB, King DS, Wang L, and Schultz PG (2002). Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc 124, 9026–9027. [DOI] [PubMed] [Google Scholar]
- 23.Deiters A, and Schultz PG (2005). In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorganic Med. Chem. Lett 15, 1521–1524. [DOI] [PubMed] [Google Scholar]
- 24.Kolb HC, Finn MG, and Sharpless KB (2001). Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chemie - Int. Ed 40, 2004–2021. [DOI] [PubMed] [Google Scholar]
- 25.Jewett JC, and Bertozzi CR (2010). Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev 39, 1272–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Chen ZJ, Ren W, and Ai HW (2012). Reaction-based genetically encoded fluorescent hydrogen sulfide sensors. J. Am. Chem. Soc 134, 9589–9592. [DOI] [PubMed] [Google Scholar]
- 27.Devaraj NK, Weissleder R, and Hilderbrand SA (2008). Tetrazine-based cycloadditions: Application to pretargeted live cell imaging. Bioconjug. Chem 19, 2297–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, and Chin JW (2012). Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem 4, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong J, Sharpless KB, Kwisnek L, Oakdale JS, and Fokin VV (2014). SuFEx-based synthesis of polysulfates. Angew. Chemie - Int. Ed 53, 9466–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang N, Yang B, Fu C, Zhu H, Zheng F, Kobayashi T, Liu J, Li S, Ma C, Wang PG, et al. (2018). Genetically encoding fluorosulfate-l -tyrosine to react with lysine, histidine, and tyrosine via SuFEx in proteins in vivo. J. Am. Chem. Soc 140, 4995–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou C, Uprety R, Davis L, Chin JW, and Deiters A (2011). Genetically encoding an aliphatic diazirine for protein photocrosslinking. Chem. Sci 2, 480–483. [Google Scholar]
- 32.Chin JW, Martin AB, King DS, Wang L, and Schultz PG (2002). Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 99, 11020–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, and Saghatelian A (2013). Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol 9, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koh M, Ahmad I, Ko Y, Zhang Y, Martinez TF, Diedrich JK, Chu Q, Moresco JJ, Erb MA, Saghatelian A, et al. (2021). A short ORF-encoded transcriptional regulator. Proc. Natl. Acad. Sci. U. S. A 118, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherman DJ, Xie R, Taylor RJ, George AH, Okuda S, Foster PJ, Needleman DJ, and Kahne D (2018). Lipopolysaccharide is transported to the cell surface by a membrane-Tomembrane protein bridge. Science (80-.). 359, 798–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isom GL, Coudray N, MacRae MR, McManus CT, Ekiert DC, and Bhabha G (2020). LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope. Cell 181, 653–664.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodnar NO, and Rapoport TA (2017). Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell 169, 722–735.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna MJ, Sim SI, Ordureau A, Wei L, Wade Harper J, Shao S, and Park E (2020). The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science (80-.). 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debelouchina GT, Gerecht K, and Muir TW (2017). Ubiquitin utilizes an acidic surface patch to alter chromatin structure. Nat. Chem. Biol 13, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, and Schultz PG (2003). Generation of a bacterium with a 21 amino acid genetic code. J. Am. Chem. Soc 125, 935–939. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Tang J, Wang L, Tian Z, Cardenas A, Fang X, Chatterjee A, and Xiao H (2020). Creation of Bacterial Cells with 5-Hydroxytryptophan as a 21st Amino Acid Building Block. Chem 6, 2717–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu CC, Mack AV, Tsao ML, Mills JH, Hyun SL, Choe H, Farzan M, Schultz PG, and Smider VV (2008). Protein evolution with an expanded genetic code. Proc. Natl. Acad. Sci. U. S. A 105, 17688–17693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li JC, Liu T, Wang Y, Mehta AP, and Schultz PG (2018). Enhancing Protein Stability with Genetically Encoded Noncanonical Amino Acids. J. Am. Chem. Soc 140, 15997–16000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Amiram M, Patel JR, Gallagher RR, Rinehart J, et al. (2015). Recoded organisms engineered to depend on synthetic amino acids. Nature 518, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, Gregg CJ, Stoddard BL, and Church GM (2015). protein design. 518, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Si L, Xu H, Zhou X, Zhang Z, Tian Z, Wang Y, Wu Y, Zhang B, Niu Z, Zhang C, et al. (2016). Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science (80-.). 354, 1170–1173. [DOI] [PubMed] [Google Scholar]
- 47.Tack DS, Ellefson JW, Thyer R, Wang B, Gollihar J, Forster MT, and Ellington AD (2016). Addicting diverse bacteria to a noncanonical amino acid. Nat. Chem. Biol 12, 138–140. [DOI] [PubMed] [Google Scholar]
- 48.Koh M, Yao A, Gleason PR, Mills JH, and Schultz PG (2019). A General Strategy for Engineering Noncanonical Amino Acid Dependent Bacterial Growth. J. Am. Chem. Soc 141, 16213–16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q, and Wang L (2008). New methods enabling efficient incorporation of unnatural amino acids in yeast. J. Am. Chem. Soc 130, 6066–6067. [DOI] [PubMed] [Google Scholar]
- 50.Italia JS, Zheng Y, Kelemen RE, Erickson SB, Addy PS, and Chatterjee A (2017). Expanding the genetic code of mammalian cells. Biochem. Soc. Trans 45, 555–562. [DOI] [PubMed] [Google Scholar]
- 51.Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, and Liu WR (2010). A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew. Chemie - Int. Ed 49, 3211–3214. [DOI] [PubMed] [Google Scholar]
- 52.Xiao H, Chatterjee A, Choi SH, Bajjuri KM, Sinha SC, and Schultz PG (2013). Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew. Chemie - Int. Ed 52, 14080–14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tharp JM, Vargas-Rodriguez O, Schepartz A, and Söll D (2020). Genetic Encoding of Three Distinct Noncanonical Amino Acids Using Reprogrammed Initiator and Nonsense Codons. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, and Schultz PG (2004). An expanded genetic code in mammalian cells with a functional quadruplet codon. Proc. Natl. Acad. Sci. U. S. A 101, 7566–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neumann H, Wang K, Davis L, Garcia-Alai M, and Chin JW (2010). Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444. [DOI] [PubMed] [Google Scholar]
- 56.Dunkelmann DL, Willis JCW, Beattie AT, and Chin JW (2020). Engineered triply orthogonal pyrrolysyl–tRNA synthetase/tRNA pairs enable the genetic encoding of three distinct non-canonical amino acids. Nat. Chem 12, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang K, Sachdeva A, Cox DJ, Wilf NW, Lang K, Wallace S, Mehl RA, and Chin JW (2014). Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat. Chem 6, 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lajoie MJ, Rovner AJ, Goodman DB, Aerni H, Haimovich AD, Kuznetsov G, Mercer J. a, Wang HH, Carr P. a, Mosberg J. a, et al. (2013). Genomically Recoded Organisms Expand Biological Functions. Science (80-.). 342, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chatterjee A, Lajoie MJ, Xiao H, Church GM, and Schultz PG (2014). A bacterial strain with a unique quadruplet codon specifying non-native amino acids. ChemBioChem 15, 1782–1786. [DOI] [PubMed] [Google Scholar]
- 60.Fredens J, Wang K, de la Torre D, Funke LFH, Robertson WE, Christova Y, Chia T, Schmied WH, Dunkelmann DL, Beránek V, et al. (2019). Total synthesis of Escherichia coli with a recoded genome. Nature 569, 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robertson WE, Funke LFH, Torre D. De, Fredens J, Elliott TS, Spinck M, Christova Y, Cervettini D, Böge FL, Liu KC, et al. (2021). Sense codon reassignment enables viral resistance and Encoded Polymer Synthesis. Science (80-.). 3, 1057–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chatterjee A, Xiao H, and Schultz PG (2012). Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 109, 14841–14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Italia JS, Addy PS, Erickson SB, Peeler JC, Weerapana E, and Chatterjee A (2019). Mutually Orthogonal Nonsense-Suppression Systems and Conjugation Chemistries for Precise Protein Labeling at up to Three Distinct Sites. J. Am. Chem. Soc 141, 6204–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malyshev DA, and Romesberg FE (2015). The Expanded Genetic Alphabet. Angew. Chemie - Int. Ed 54, 11930–11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bain JD, Switzer C, Chamberlin AR, and Benner SA (1992). Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature 356, 537–539. [DOI] [PubMed] [Google Scholar]
- 66.Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, Nakayama H, Takio K, Yabuki T, Kigawa T, et al. (2002). An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol 20, 177–182. [DOI] [PubMed] [Google Scholar]
- 67.Yang Z, Chen F, Alvarado JB, and Benner SA (2011). Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J. Am. Chem. Soc 133, 15105–15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoshika S, Leal NA, Kim MJ, Kim MS, Karalkar NB, Kim HJ, Bates AM, Watkins NE, SantaLucia HA, Meyer AJ, et al. (2019). Hachimoji DNA and RNA: A genetic system with eight building blocks. Science (80-.). 363, 884–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Atwell S, Meggers E, Spraggon G, and Schultz PG (2001). Structure of a copper-mediated base pair in DNA. J. Am. Chem. Soc 123, 12364–12367. [DOI] [PubMed] [Google Scholar]
- 70.Zimmermann N, Meggers E, and Schultz PG (2002). A novel silver(I)-mediated DNA base pair. J. Am. Chem. Soc 124, 13684–13685. [DOI] [PubMed] [Google Scholar]
- 71.Matray TJ, and Kool ET (1998). Selective and stable DNA base pairing without hydrogen bonds. J. Am. Chem. Soc 120, 6191–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leconte AM, Chen L, and Romesberg FE (2005). Polymerase evolution: Efforts toward expansion of the genetic code. J. Am. Chem. Soc 127, 12470–12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, and Yokoyama S (2006). An unnatural hydrophobic base pair system: Site-specific incorporation of nucleotide analogs into DNA and RNA. Nat. Methods 3, 729–735. [DOI] [PubMed] [Google Scholar]
- 74.Kimoto M, Kawai R, Mitsui T, Yokoyama S, and Hirao I (2009). An unnatural base pair system for efficient PCR amplification and functionalization of DNA molecules. Nucleic Acids Res. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malyshev DA, Dhami K, Quach HT, Lavergne T, Ordoukhanian P, Torkamani A, and Romesberg FE (2012). Efficient and sequence-independent replication of DNA containing a third base pair establishes a functional six-letter genetic alphabet. Proc. Natl. Acad. Sci. U. S. A 109, 12005–12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, Corrêa IR, and Romesberg FE (2014). A semi-synthetic organism with an expanded genetic alphabet. Nature 509, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Ptacin JL, Fischer EC, Aerni HR, Caffaro CE, San Jose K, Feldman AW, Turner CR, and Romesberg FE (2017). A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551, 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marlière P, Patrouix J, Döring V, Herdewijn P, Tricot S, Cruveiller S, Bouzon M, and Mutzel R (2011). Chemical Evolution of a Bacterium’s Genome. Angew. Chemie - Int. Ed 8, 2011. [DOI] [PubMed] [Google Scholar]
- 79.Mehta AP, Li H, Reed SA, Supekova L, Javahishvili T, and Schultz PG (2016). Replacement of 2′-Deoxycytidine by 2′-Deoxycytidine Analogues in the E. coli Genome. J. Am. Chem. Soc 138, 14230–14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mehta AP, Wang Y, Reed SA, Supekova L, Javahishvili T, Chaput JC, and Schultz PG (2018). Bacterial Genome Containing Chimeric DNA-RNA Sequences. J. Am. Chem. Soc 140, 11464–11473. [DOI] [PubMed] [Google Scholar]
- 81.Neveu M, Kim HJ, and Benner SA (2013). The “strong” RNA world hypothesis: fifty years old. Astrobiology 13, 391–403. [DOI] [PubMed] [Google Scholar]
- 82.Ravikumar A, Arrieta A, and Liu CC (2014). An orthogonal DNA replication system in yeast. Nat. Chem. Biol 10, 175–177. [DOI] [PubMed] [Google Scholar]
- 83.Liu CC, Jewett MC, Chin JW, and Voigt CA (2018). Toward an orthogonal central dogma. Nat. Chem. Biol 14, 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]


