Abstract
A short model genome RNA and also the genome RNA of influenza A virus bearing both 5′- and 3′-terminal common sequences activated the interferon-induced double-stranded-RNA-dependent protein kinase, PKR, by stimulating autophosphorylation in vitro. The activated PKR catalyzed phosphorylation of the alpha subunit of eucaryotic translation initiation factor 2 (eIF2α). The NS1 protein efficiently eliminated the PKR-activating activity of these RNAs by binding to them. Two mutant NS1 proteins, each harboring a single amino acid substitution at different regions, exhibited temperature sensitivity in their RNA binding activity in the mutant virus-infected cell lysates as well as when they were prepared as fusion proteins expressed in bacteria. The virus strains carrying these mutant NS1 proteins exhibited temperature sensitivity in virus protein synthesis at the translational level, as reported previously, and could not repress the autophosphorylation of PKR developing during the virus growth, which is normally suppressed by a viral function(s). As a result, the level of eIF2α phosphorylation was elevated 2.5- to 3-fold. The defect in virus protein synthesis was well correlated with the level of phosphorylation of PKR and eIF2α.
The genome of influenza A virus is composed of eight RNA segments with negative polarity. RNA segment 8 encodes two overlapping proteins, designated nonstructural protein 1 (NS1) and NS2, which are found abundantly in infected cells (reviewed in reference 20). The NS2 protein exists also in virions (32, 41), and the recent assignment of its function has led to a proposal of a new designation, NEP (26). So far there is no report indicating the presence of the NS1 protein in the virus particle. Various RNA binding activities of the NS1 protein have been reported and correlated to regulation operating at various posttranscriptional steps (8, 11, 12, 27, 28, 30, 31, 38), although no NS1 mutant viruses with the predicted phenotype have been reported. We previously demonstrated that the NS1 protein exhibits two modes of RNA binding activity (12, 13). One of them is a strong binding to double-stranded RNA (dsRNA) and a short model genome RNA (mini-vRNA) bearing both 5′- and 3′-terminal common sequences, which are expected to form a panhandle structure (5). However, the biological functions of these activities remain unknown.
The interferon-induced dsRNA-dependent protein kinase, PKR (for protein kinase, RNA activated) (also designated DAI, eIF2α kinase, or p68), is activated during virus growth, phosphorylates the alpha subunit of eucaryotic translation initiation factor 2 (eIF2α), and leads to inhibition of the translation initiation of the host cell as well as that of virus, thus contributing to the interferon-mediated antiviral response (reviewed in references 36 and 40). Many viruses, including influenza virus, have evolved various devices to overcome this host defense mechanism (reviewed in reference 16). Katze et al. (15) showed that PKR, which is first activated by some viral RNA products during influenza virus infection, is then suppressed, thus relieving the translational block exerted on the infected cells. As a suppressor of the PKR function, they purified a p58 cellular protein, which represses the phosphorylation of eIF2α by the already-activated PKR (21). Lu et al. (22) have reported that the NS1 protein can prevent the activation of PKR by binding to the dsRNA activator, thus suggesting the presence of another mechanism for the virus to escape the PKR translational block. On the other hand, the RNA species which activate PKR in influenza virus infection remain elusive, though they may be dsRNAs of virus origin.
In this study, we showed that the model mini-vRNA as well as the influenza virus genome RNA (vRNA) with their higher-order structure led to the activation of PKR in vitro, which was eliminated by preincubating them with the NS1 protein. We then analyzed two temperature-sensitive (ts) mutants affected in the NS1 gene to examine the possibility that the protein works as a suppressor of the activation of PKR in the virus-infected cells.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were grown in Eagle’s minimal essential medium (MEM) supplemented with 5% calf serum and were infected with viruses at 34 or 40°C as previously described (10). Wild-type influenza virus strain A/Udorn/72 (H3N2 subtype) and the ts mutants SPC45 and ICR1629 (37), which have a point mutation in the NS1 protein (10), were grown in the allantoic cavities of 11-day-old embryonated chicken eggs for 2 days at 34°C.
Construction of expression plasmids for GST-NS1 fusion proteins.
The unique BamHI site of the NS1 gene of strain PR8 was connected to the BamHI site of the pGEX-1 expression vector (Amersham Pharmacia Biotech), providing the glutathione S-transferase (GST)–NS1 fusion proteins. To introduce deletions into the NS1 gene, one of the HaeIII sites and the unique SspI site of the NS DNA were individually connected in frame to two tandem UAA codons of an oligonucleotide linker. The fragment carrying the linker-ligated HaeIII site encodes a truncated NS1 protein containing the N-terminal 82 amino acid residues (and two extra residues, E and F), providing NS82, while the fragment carrying the linker-ligated SspI site gives NS144, containing the N-terminal 144 residues. The plasmids for the NS1 mutant proteins with the inner residues deleted, NSΔ(34-39) and NSΔ(50-85) (the numbers in parentheses indicate the deleted amino acid residues), and those for the proteins having a point mutation corresponding to SPC45 or ICR1629 NS1s were constructed by in vitro mutagenesis (19) with the following oligonucleotide primers: 5′GATGCCCCATTCCTTCAGAATCCCTAAG3′ for NSΔ(34-39), 5′GGAAGGGGCAGCACTCCTGCGTCGCGTTACC3′ for NSΔ(50-85), 5′CCCATGTTGGAAATCAGATAGTAGAG3′ for SPC45, and 5′CATCATGTTGAAAACGAATTTCAGTG3′ for ICR1629.
Northwestern blotting.
By using the plasmids described above, the GST-NS1 fusion proteins were induced in Escherichia coli cells and purified as described previously (39). Influenza virus-infected HeLa cells were lysed with a lysis-loading buffer (0.125 M Tris-HCl [pH 6.8], 4.6% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 10% glycerol, 0.01% bromphenol blue). The purified GST fusion proteins (9.5 pmol each) or the virus-infected HeLa cell lysates were subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto nitrocellulose membranes (Bio-Rad) as described previously (17) with the following modifications. After blocking treatment with 5% bovine serum albumin and 0.1% Tween 20, the membrane was incubated in an RNA binding buffer (6.6 mM Tris-HCl [pH 7.6], 33 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 0.02% Tween 20) containing 32P-dsRNA or 32P-mini-vRNA probe (2 × 106 cpm/ml) (prepared as described previously [12]) and 10 μg tRNA per ml for 1 h at room temperature or 40°C. After being washed with the RNA binding buffer, the membrane was processed for autoradiography. The radioactivity of the binding complex was measured with a BAS 1000 image analyzer (Fuji film).
Preparation of PKR.
HeLa cells were grown in Eagle’s MEM supplemented with 5% calf serum and were treated with natural beta interferon (300 U/ml) (Torey) for 24 h at 37°C. From these cells, the PKR fraction was prepared and further purified by using a DEAE-cellulose column by a method described previously (35).
Preparation of eIF2.
eIF2 was prepared from rabbit reticulocytes as described previously (18) with some modifications.
In vitro phosphorylation assay.
Poly(A)-poly(U), poly(I)-poly(C), and tRNA were from Sigma, while the mini-vRNA (12) and the virion RNA (10) were prepared as described previously. Various RNAs were preincubated in the absence or in the presence of NS1 proteins (0.5 μg) for 30 min at 30°C in a standard reaction mixture containing 15 mM HEPES (pH 7.5), 63 mM KCl, 3 mM Mg acetate, 1 mM dithiothreitol, and 0.25% Nonidet P-40. ATP (to 10 mM) and [γ-32P]ATP (3 to 5 μCi) (Amersham Pharmacia Biotech) were then added to the mixture, followed by the PKR fraction (1 μg of protein), to carry out the PKR autophosphorylation reaction for 20 min at 30°C (final volume of 15 μl) essentially as described previously (34). To examine eIF2α phosphorylation by PKR (see Fig. 3), this PKR autophosphorylation reaction was carried out in the absence of labeled ATP. Thereafter, [γ-32P]ATP (3 to 5 μCi) and the eIF2 fraction (1 μg of protein) were added to the reaction mixtures, which were further incubated for 20 min at 30°C. The phosphorylated proteins were analyzed by SDS-PAGE followed by autoradiography.
FIG. 3.
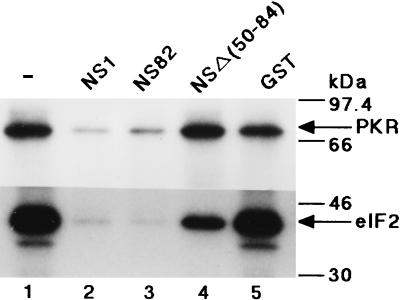
Effect of the GST-NS1 fusion proteins on PKR-catalyzed eIF2α phosphorylation. Mini-vRNA (1 μg/ml) was preincubated in the kinase reaction mixture for 30 min at 30°C without NS proteins (lane 1) or in the presence of GST–wild-type NS1 (lane 2), GST-NS82 (lane 3), GST-NSΔ(50-84) (lane 4), or the GST protein (lane 5). The PKR autophosphorylation reaction (upper panel) was carried out as described in the legend to Fig. 1A. For PKR-catalyzed eIF2α phosphorylation (lower panel), the PKR autophosphorylation reaction was performed as described above but without addition of the labeled ATP. Thereafter, [γ-32P]ATP and eIF2 were added to the reaction mixtures, which were further incubated for 20 min at 30°C. The phosphorylated proteins were subjected to SDS–15% PAGE as described in Materials and Methods. For protein markers on the right, see the legend to Fig. 1.
In vivo phosphorylation assay.
Confluent monolayers of HeLa cells in 35-mm-diameter dishes were infected with viruses at a multiplicity of infection (MOI) of 10 in Eagle’s MEM at 34 or 40°C, and after 1, 3, 4, and 5 h postinfection (p.i.), cells were washed with a phosphate-free buffer (118 mM NaCl, 4.75 mM KCl, 1.2 mM MgCl2, 0.26 mM CaCl2, 25 mM NaHCO3, 20 mM HEPES [pH 7.4]) and labeled with [32P] orthophosphate (500 μCi/ml) (American Radiolabeled Chemicals) in Dulbecco modified Eagle MEM lacking both phosphate and pyruvate. After being labeled for 2 h, the cells were washed with phosphate-buffered saline without Ca2+ and Mg2+ and disrupted by the procedure described previously by Katze et al. (15). The labeled cell lysates were reacted with a polyclonal antibody against human PKR (provided by H. Taira, Faculty of Agriculture, Iwate University) or with a polyclonal antibody against human eIF2α (C-20) (Santa Cruz Biotech) in the presence of protein G-Sepharose (Amersham Pharmacia Biotech) for 1 h at room temperature and then were processed as described previously (11). The immunoprecipitates were subjected to SDS-PAGE followed by autoradiography to examine PKR phosphorylation.
Western immunoblotting.
Total cell lysates were separated by SDS-PAGE. After electrotransfer from the gel to a nitrocellulose filter, the NS1 protein was reacted with a rabbit polyclonal antibody against NS1 (11) and detected by the enhanced chemiluminescence technique (ECL; Amersham Pharmacia Biotech).
RESULTS
NS1 eliminates the PKR activation induced by various RNAs, including the mini-vRNA.
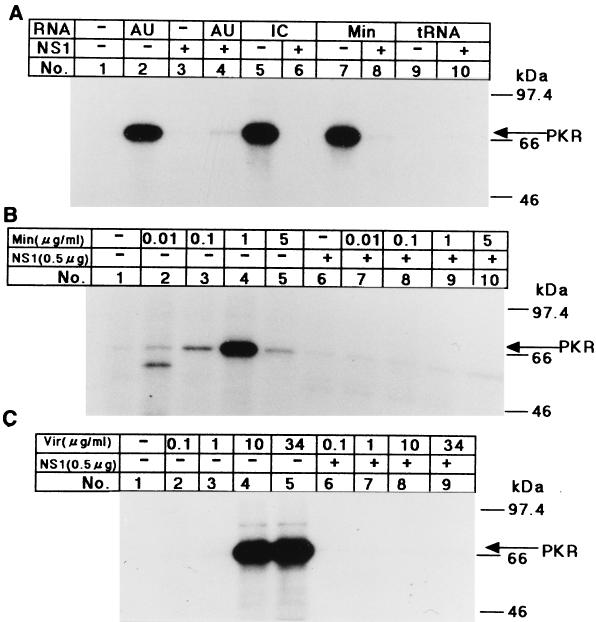
The stable panhandle structure of vRNA with a short stretch of double helix (1) was suspected to work as an activator of PKR. Therefore, we examined the effect of vRNA and a short (70-base) mini-vRNA (13) on the PKR autophosphorylation reaction in vitro. The PKR fraction was prepared from interferon-treated HeLa cells, and incubated with various RNAs. The mini-vRNA prepared in vitro activated the PKR autophosphorylation at a concentration of 1 μg/ml (4.3 × 10−8 M) but did not do so at 5 μg/ml (2.1 × 10−7 M) (Fig. 1B, lanes 4 and 5). Similarly, poly(I)-poly(C) and poly(A)-poly(U) dsRNAs, the commonly used activators of PKR, activated it at a concentration of 0.1 to 1 μg/ml (Fig. 1A, lanes 2 and 5) but did not do so at 10 μg/ml (data not shown). The vRNA isolated from virions activated PKR autophosphorylation at a concentration of 10 or 34 μg/ml (Fig. 1C, lanes 4 and 5), which was in the range of the activation concentration of the mini-vRNA on a molar basis (1.7 × 10−8 and 6.0 × 10−8 M, respectively). The mini-vRNA as well as poly(I)-poly(C) stimulated the phosphorylation of histones (data not shown) or eIF2α (see Fig. 3, lane 1), the substrates of PKR, which were added to the autophosphorylation reaction mixture. The mini-vRNA lost its PKR-activating ability when it was heat denatured or when either terminal common sequence was removed from it (data not shown).
FIG. 1.
Autophosphorylation of PKR stimulated by various RNAs and eliminated by preincubation with the NS1 protein. (A) Various RNAs were preincubated in the absence (−) or presence (+) of the GST-NS1 protein (0.5 μg) for 30 min at 30°C in the kinase reaction mixture as indicated above the lane number. ATP and [γ-32P]ATP were then added to the mixture, followed by the PKR fraction (1 μg of protein) for the PKR autophosphorylation reaction as described in Materials and Methods. The phosphorylated proteins were analyzed by SDS–10% PAGE. The tested RNAs (1 μg/ml each) were poly(A)-poly(U) (AU), poly(I)-poly(C) (IC), the mini-vRNA (Min), and tRNA. (B and C) Increasing amounts of the mini-vRNA (Min) (B) and virion RNA (Vir) (C) as indicated were preincubated in the absence (−) or presence (+) of 0.5 μg of GST-NS1 protein. PKR autophosphorylation was examined as described in for panel A. Migrations of the protein markers (Amersham Pharmacia Biotech) are indicated on the right of each panel.
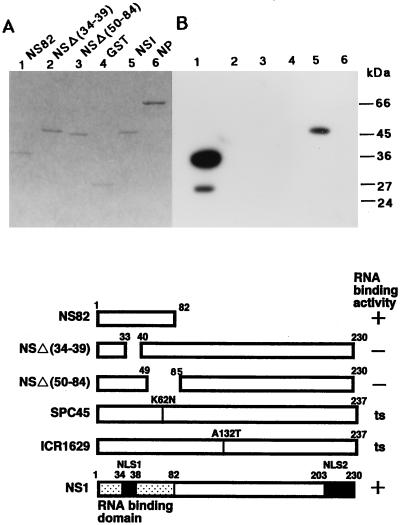
The NS1 protein extinguished the PKR autophosphorylation activated by vRNA and the mini-vRNA, as well as dsRNAs, when preincubated with them (Fig. 1B, lanes 7 to 10, and C, lanes 6 to 9, and data not shown). To investigate the relationship between the extinguishing activity and the RNA binding ability of NS1, we constructed four GST-NS1 fusion proteins carrying a deletion mutation in NS1: GST-NS144, GST-NS82, GST-NSΔ(34-39), and GST-NSΔ(50-84) (see Materials and Methods). The NS1 regions of these fusion proteins are shown in Fig. 2 (lower panel). Their RNA binding activities were examined by Northwestern blotting with 32P-labeled dsRNA as the probe (Fig. 2B). GST-NS144 (data not shown) and GST-NS82 retained the RNA binding activity, but GST-NSΔ(34-39) and GST-NSΔ(50-84) had lost it (Fig. 2B, lanes 1 to 3, and data not shown). The wild-type GST-NS1 and GST-NS82 inhibited PKR-catalyzed phosphorylation of eIF2α as well as the PKR autophosphorylation (Fig. 3, lanes 2 and 3), but the inhibition exerted by GST-NSΔ(50-84) was greatly diminished for both phosphorylations (Fig. 3, lane 4). These observations indicated that not only the nuclear localization signal (amino acid residues 34 to 39) but also residues 50 to 82 of NS1 were required for the RNA binding and inhibition of PKR activation by these RNAs, consistent with the results of Qian et al. (29).
FIG. 2.
Structures of NS1 deletion proteins and their RNA binding abilities. (Lower panel) Structures of NS1 deletion proteins NS82, NSΔ(34-39), and NSΔ(50-84). The amino acid substitutions in two ts mutant NS1s are also shown. (A and B) The GST-NS1 fusion proteins were induced in E. coli cells and purified by using a glutathione-agarose column. Equal molar quantities of the proteins (9.5 pmol) were subjected to SDS–12% PAGE, and the gel was stained with Coomassie brilliant blue (A). For Northwestern blotting (B), proteins in the gel were electroblotted onto a nitrocellulose membrane and subjected to the RNA binding reaction with a 32P-labeled dsRNA probe as described in Materials and Methods. Lanes 1, GST-NS82; lanes 2, GST-NSΔ(34-39); lanes 3, GST-NSΔ(50-84); lanes 4, GST protein; lanes 5, GST-NS1 (full length); lanes 6, GST-NP. Migrations of protein markers (Sigma) are indicated on the right of panel B.
RNA binding ability of ts mutant NS1 proteins.
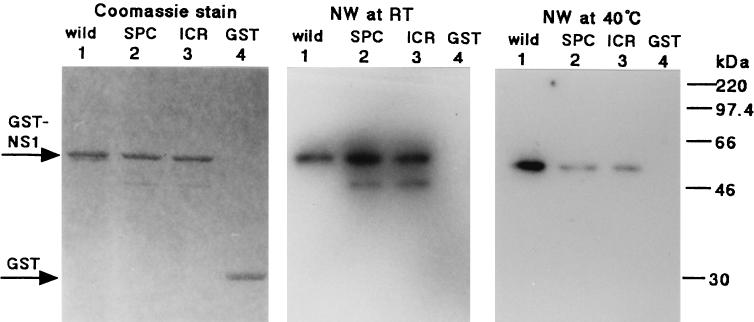
Previously we identified two ts mutants, SPC45 and ICR1629, which had single amino acid substitutions in the NS1 protein (10). We constructed GST-NS1 fusion proteins of each mutant NS1, and their RNA binding abilities were examined by Northwestern blotting as described above. Both proteins exhibited the same level of dsRNA binding activity as the wild-type NS1 fusion protein when the binding reaction was carried out at room temperature (Fig. 4, middle panel, lanes 1 to 3). In contrast, the binding activity greatly decreased for both mutant proteins at 40°C (Fig. 4, right panel, lanes 1 to 3).
FIG. 4.
RNA binding abilities of the NS1 ts mutant proteins. The GST-NS1 fusion proteins for the ts mutant NS1s (illustrated in Fig. 2) were expressed in E. coli and purified with a glutathione-agarose column. The temperature sensitivities of their RNA binding activities were examined by Northwestern blotting (NW) with 9.5 pmol of each protein as described in the legend to Fig. 2. The RNA binding reaction was performed with a 32P-labeled dsRNA probe at either room temperature (middle panel) or 40°C (right panel). The dye-staining pattern of these proteins is shown (left panel). GST–wild-type NS1, GST-SPC45 NS1, GST-ICR1629 NS1, and the GST protein were applied to lanes 1, 2, 3, and 4, respectively. For protein markers on the right, see the legend to Fig. 1.
Phosphorylation of PKR and eIF2α in cells infected with the NS1 ts mutant viruses.
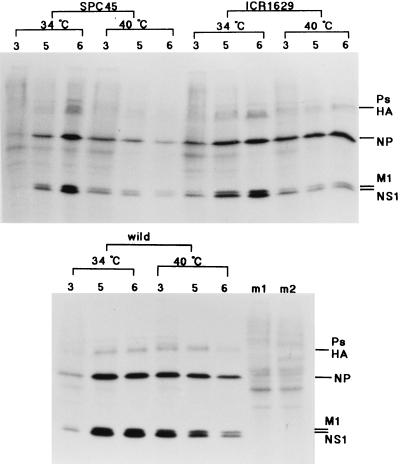
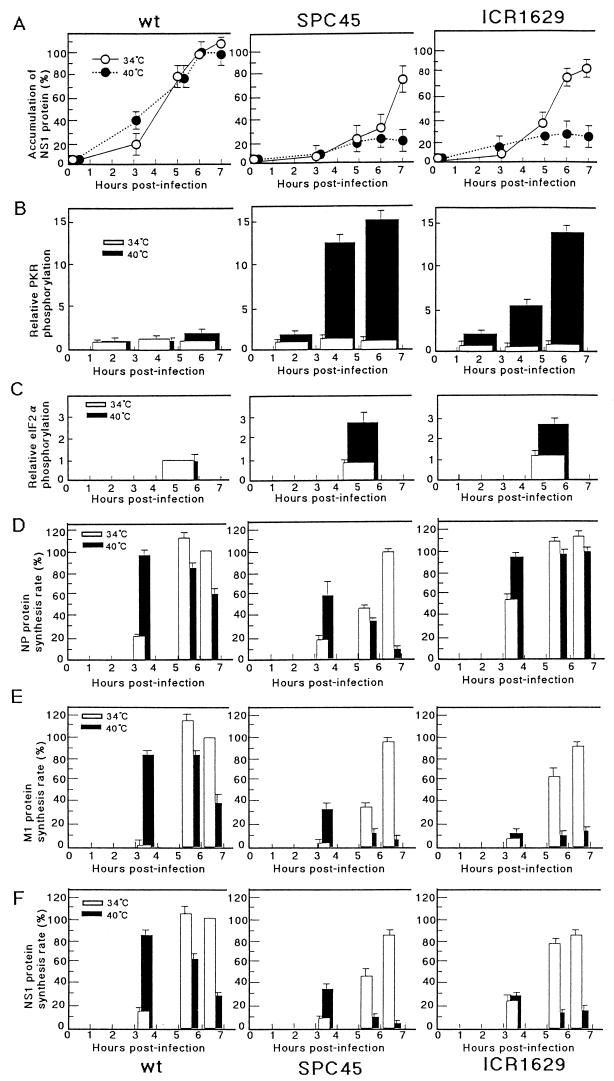
The above-described observation led us to examine the phosphorylation levels of PKR and eIF2α and their relation to the RNA binding activity of NS1 protein in the cells infected with these mutant viruses. For these experiments, we used HeLa cells as the virus host, because our PKR antibody could not cross-react with the canine PKR in MDCK cells, which were the host in the previous work on these ts mutant viruses. The virus production in HeLa cells at the nonpermissive temperature of 40°C was reduced to 50, <0.5, and 5% of that at 34°C for the wild-type, SPC45, and ICR1629 viruses, respectively. The syntheses of M1 and NS1 proteins at later times of growth for both mutants greatly decreased at 40°C, in agreement with the observations for the MDCK cells (10). In contrast, the synthesis of the nucleoprotein (NP), which was scarcely affected in MDCK cells for both mutants, was greatly depressed to 10% in the SPC45-infected HeLa cells but not in the ICR1629-infected cells (Fig. 5; see Fig. 8D to F).
FIG. 5.
Protein synthesis in virus-infected HeLa cells. Cells infected with the wild-type, SPC45, and ICR1629 viruses (MOI of 10) were grown at either 34 or 40°C and labeled with [35S]methionine (10 μCi/ml) for 30 min beginning from 3, 5, and 6 h p.i., as shown above the panels. For other experimental details, see reference 10. Lanes m1 and m2, cells mock infected and labeled during 6 to 6.5 h p.i. at 34 and 40°C, respectively. Ps, virus polymerase proteins. HA, hemagglutinin.
FIG. 8.
Quantitation of the results shown in Fig. 5, 6, and 7. All results shown are derived from at least three separate experiments and are the averages and standard deviations. (A) Amounts of NS1 accumulated in the infected cells grown at 34 or 40°C. For experimental details, see the legend to Fig. 7B. The intensities of the ECL bands were measured with a microdensitometer (dual-wavelength flying-spot scanner CS-9000; Shimadzu), and the relative values are shown, with NS1 in the wild-type lysates 6 h p.i. at 34°C set as 100%. (B to F) Radioactivities of gel bands were measured with a BAS1000 image analyzer. (B) Phosphorylation of PKR during virus growth. The radioactivity of the PKR band shown in Fig. 6A is represented as the relative value, with that of the wild-type virus-infected cells labeled from 5 to 7 h p.i. at 34°C set as 1.0. (C) Phosphorylation of eIF2α in virus-infected cells during 4 to 6 h p.i. (Fig. 6B). The relative values are represented, with the radioactivity obtained from the wild-type virus-infected cells at 34°C set as 1.0. (D to F) Rates of synthesis of NP (D), M1 (E), and NS1 (F) (30-min labeling) in the infected cells (Fig. 5), represented as the relative value, with that of the respective protein in the wild-type virus-infected cells labeled 6 h p.i. at 34°C set as 100%.
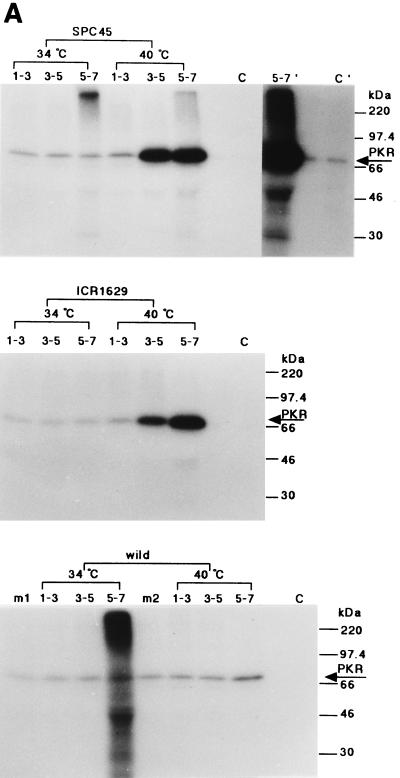
The virus-infected HeLa cells were incubated at 34 or 40°C and labeled metabolically with [32P]orthophosphate for 2 h starting from 1, 3, or 5 h p.i. The cell lysates were prepared and incubated with the anti-human PKR antibody, and the resulting immune complexes were subjected to SDS-PAGE and autoradiography as described in Materials and Methods. As shown in Fig. 6A (also see Fig. 8B), 32P incorporation into PKR in the cells infected with either of these mutant viruses at 34°C, as well as in the wild-type virus-infected cells at either 34 or 40°C, was at almost the same low level as in mock-infected cells during 1 to 7 h p.i. In contrast, 32P in PKR was greatly increased from 3 h p.i. in the cells infected with either of these mutant viruses at the nonpermissive temperature of 40°C, and more in the SPC45-infected cells than in the ICR1629-infected cells (11 and 8 times higher, respectively, during 3 to 5 h p.i.), consistent with the observation that the former virus protein synthesis was more severely depressed than the latter at 40°C.
FIG. 6.
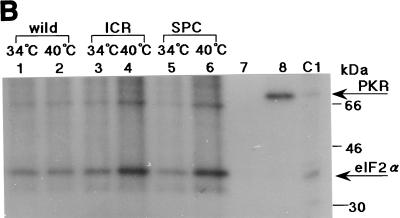
Phosphorylation of PKR and eIF2α in HeLa cells infected with the ts NS1 mutant viruses. Confluent monolayers of HeLa cells were infected with the wild-type or ts mutant viruses (MOI of 10) at either 34 or 40°C, labeled with [32P]orthophosphate (500 μCi/ml) (ARC) for 2 h, and harvested. Cell lysates were prepared and reacted with the polyclonal antibody against human PKR (A) or the polyclonal antibody against human eIF2α (C-20) (1 μg) (B) for 1 h at room temperature as described in Materials and Methods. The resulting immunoprecipitates were then analyzed by SDS-PAGE and autoradiography. (A) Phosphorylation of PKR. The labeling was from 1 to 3 h p.i. (lanes 1 to 3), from 3 to 5 h p.i. (lanes 3 to 5), and from 5 to 7 h p.i. (lanes 5 to 7), as shown above each panel. Lanes m1 and m2, cells mock infected and labeled during 5 to 7 h p.i. at 34 and 40°C, respectively. Lane C, 32P-PKR labeled in vitro. Longer exposures for lanes 5 to 7 and C are shown on the right of the upper panel (5-7′ and C′). For protein markers on the right, see the legend to Fig. 1. (B) Phosphorylation of eIF2α. The viruses were grown at 34 or 40°C as shown at the top, and the infected cells were labeled with 32P from 4 to 6 h p.i. The immunosupernatants of lanes 5 and 6 were treated with the anti-PKR antibody, and the immunoprecipitates (one-third of total) were analyzed in lanes 7 and 8, respectively. Lane C1, 32P-labeled purified PKR and rabbit eIF2α.
The level of 32P labeling in eIF2α was then examined. The infected cells were labeled for 2 h starting from 4 h p.i. (which gave the highest level of the eIF2α labeling). Cell lysates were prepared and treated with anti-human eIF2α antibody. The immunoprecipitates were analyzed as described above. As shown in Fig. 6B (also see Fig. 8C), 32P incorporation into eIF2α for these mutant viruses at 34°C was at the same level as that for the wild-type virus at 34 or 40°C. It was increased from 4 h p.i. in the cells infected with either of these mutant viruses at the nonpermissive temperature of 40°C (2.9 and 2.3 times more than that at 34°C for the SPC45 and ICR1629 viruses, respectively). It decreased in the mutant virus-infected cells labeled from 5 to 7 h p.i. at 40°C, probably because the subunit had already been phosphorylated at 5 h p.i. and the turnover of the incorporated phosphate was slow (data not shown).
dsRNA binding activity of NS1 in ts mutant-infected-cell lysates.
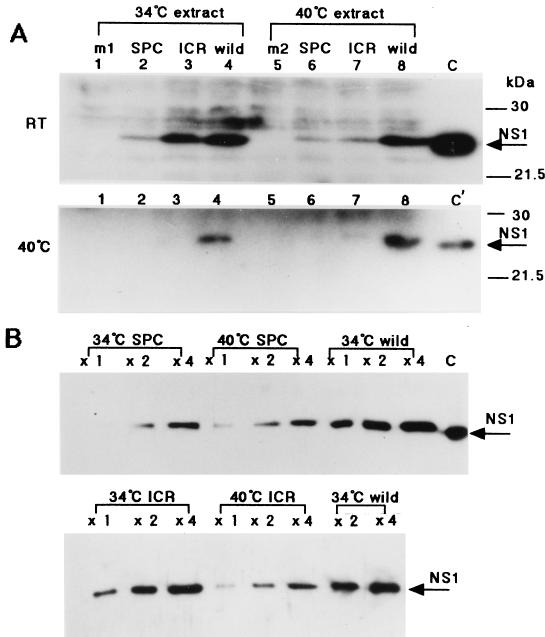
The level of dsRNA binding activity of NS1 protein in mutant virus-infected-cell lysates, which were prepared 6 h p.i. and separated by SDS-PAGE for Northwestern blotting as described above, were then examined (Fig. 7A). For estimation of the amount of NS1 protein in the same lysates, increasing amounts were subjected to SDS-PAGE for ECL immunodetection of NS1 protein with the anti-NS1 antibody. As shown in Fig. 7B, the amounts of NS1 used gave a linear ECL response. Within this linear range, the amount of NS1 in the infected-cell lysates was estimated for the mutant and the wild-type viruses grown at 34 or 40°C, as shown in Fig. 8A. It was greatly reduced for SPC45 even at 34°C and for both mutants at 40°C. At 6 h p.i., in the virus-infected-cell lysates grown at 34°C, the amounts of SPC45 and ICR1629 NS1 proteins were reduced to 30 and 80%, respectively, of the amount of wild-type NS1, and their RNA binding activities at room temperature were about 16 and 70%, respectively. When the infected cells were grown at 40°C, the amounts of both the SPC45 and ICR1629 NS1 proteins in the cell lysates were 25% of the amount of wild-type NS1, and their RNA binding activities at room temperature were about 15 and 23%, respectively. The RNA binding activities exhibited by these proteins at room temperature were roughly proportional to their amounts in the cell lysates whether grown at 34 or 40°C. In contrast, the binding ability of NS1 proteins in these mutant lysates was negligible when the binding reaction was performed at 40°C. It was thus shown that the RNA binding activities of these mutant NS1 proteins were also temperature sensitive in the virus-infected-cell lysates, in agreement with the observation made for the GST fusion proteins produced in E. coli (Fig. 4).
FIG. 7.
dsRNA binding activities of NS1 proteins in wild-type- or ts mutant-infected-cell lysates. The virus-infected cells (MOI of 10) were grown at either 34 or 40°C and lysed at 6 h p.i. as described in the legend to Fig. 6 and Materials and Methods. (A) dsRNA binding activities of NS1 proteins in cell lysates corresponding to 30 μl of the lysates (300× volume) (3 × 105 cells). Lysates from infected cells grown at either 34°C (lanes 1 to 4) or 40°C (lanes 5 to 8) were subjected to SDS-PAGE for Northwestern blotting with the 32P-dsRNA probe as described in the legend to Fig. 2. Lanes 2 and 6, SPC45-infected cells; lanes 3 and 7, ICR1629-infected cells; lanes 4 and 8, wild-type virus-infected cells, lanes 1 and 5, mock-infected cells (m1 and m2). The RNA binding reaction was performed at either room temperature (RT) or 40°C as indicated on the left. Lanes C and C′, 0.6 and 0.15 μg of purified NS1, respectively. (B) The amounts of NS1 in the cell lysates were compared. Increasing volumes (1× [0.1 μl of lysate; 103 cells], 2×, and 4×) of the lysates was subjected to SDS-PAGE for ECL immunodetection of NS1 protein with the anti-NS1 antibody. Lane C, 6 ng of purified NS1.
DISCUSSION
We have previously shown that the influenza virus NS1 protein exhibits stable binding to a model mini-vRNA, independently of temperature, which is very similar to its binding to dsRNA (12, 13). The binding depended on a higher-order structure of the mini-vRNA. Baudin et al. (1) have analyzed the secondary structure of a model vRNA, the panhandle domain of which is almost identical to ours. Based on their results, we assumed that our mini vRNA had the same panhandle structure. Hsu et al. (14) have shown that a panhandle structure is formed by the 5′- and 3′-terminal nucleotides of the virus ribonucleoprotein (RNP) in the infected cells as well as in the virions. The mini-vRNA and also vRNA stimulated the PKR activity at the same level and at the same concentration ranges as poly(I)-poly(C) or poly(A)-poly(U), the commonly used activators of PKR in vitro (9, 35) (Fig. 1). It was noteworthy that a dsRNA as short as 15 bp could effectively activate PKR, in light of the report showing that dsRNA shorter than 30 bp fails to bind stably to PKR and does not activate it (23). The bulges or some specific sequences in the panhandle structure might affect its PKR-stimulating activity.
The NS1 protein and its deletion proteins retaining the RNA binding ability effectively eliminated the PKR-activating activity of these panhandle RNAs as well as dsRNAs by interacting with them (Fig. 1 and 2). The RNA species activating PKR in influenza virus infection are thought to be highly structured viral RNAs or dsRNAs erroneously formed in the virus replication, with which the NS1 protein may interact and thereby eliminate their PKR-activating activity (reference 22 and this work). In addition, during the late phase of virus growth, the replicated vRNAs, with or without internal deletions, are transported through the nuclear pore to the cytoplasm in the form of an RNP complex (vRNP) associated with the NP protein and the viral RNA polymerases. NP could not eliminate the PKR-stimulating activity of mini-vRNA (our unpublished observations), in keeping with the observation that it cannot bind to the panhandle structure or to dsRNA (Fig. 2B) (reference 33 and our unpublished observations). Therefore, it may be possible that the panhandle structure of the vRNP complex is exposed in the case of a short supply of the polymerase proteins and tends to activate PKR in the cytoplasm especially in the later phase of the virus growth. The NS1 protein may interact with the panhandle structure and eliminate its PKR activation ability. Recently, Marion et al. (25) have reported evidence indicating the interaction of the NS1 protein with viral transcription-replication complexes in virus-infected cells. We also obtained similar results (our unpublished observations).
To determine the situation in the influenza virus-infected cells, we analyzed two ts mutants affected in the NS1 gene, SPC45 and ICR1629. With both NS1 mutants, the syntheses of two late proteins, the matrix protein (M1) and hemagglutinin, and also that of the NS1 protein were greatly reduced at the nonpermissive temperature of 40°C. In contrast, individual mRNAs, including those for the late proteins, remained almost at the same levels as those at 34°C, even when the syntheses of the late proteins and the NS1 protein were severely reduced (10). As one possible mechanism for these temperature-sensitive translational defects, we examined the phosphorylation level of PKR and its relation to the RNA binding abilities of NS1 proteins of these mutant viruses. At 40°C, the phosphorylation level of PKR was greatly increased from 3 h p.i. on the cells infected with these mutant viruses, and more so in SPC45-infected than in ICR1629-infected cells. The PKR thus activated led to an elevated phosphorylation level of eIF2α, the substrate of PKR, i.e., 2.5- to 3-fold the level with these mutant viruses at 34°C or with the wild-type virus at both temperatures, in agreement with their defect in virus protein synthesis (Fig. 8). The mutant NS1 proteins synthesized in the virus-infected cells, as well as those produced in E. coli cells as GST fusion proteins, exhibited temperature sensitivity in their dsRNA binding abilities (Fig. 4 and 7). The amount of SPC45 NS1 in the infected-cell lysates at 5 and 6 h p.i. was reduced to 25 to 30% of that of the NS1 protein in the wild-type virus-infected cells even when the cells were grown at 34°C, because its synthesis rate and stability were decreased (10). Yet, such a low level of NS1 could appreciably depress the activation of PKR. One of the reasons might be the lowered accumulation of vRNA (one of the candidates for PKR activators) in the mutant-infected cells (10) and also the underestimation of the mutant NS1 protein, which is degraded during the preparation of the cell lysates.
Recently, Egorov et al. (6) have succeeded in obtaining viruses with large deletions of the C-terminal part of the NS1 protein. One of these deletion mutants, containing the N-terminal 38 amino acids, exhibits the normal level of viral protein synthesis and grows to an high titer in Vero cells. In contrast, it cannot grow in MDCK cells, in which the level of its M1 protein is greatly reduced. As one mechanism of the differential virus growth, they suggested a deficiency in the expression of functional interferons in Vero cells, leading to lack of PKR induction. The virus with defective NS1 could grow in such cells.
As reported previously, Asn is substituted for Lys(62) in SPC45 NS1, while Thr is substituted for Ala(132) in ICR1629 NS1 (10). Both residues are conserved among influenza A viruses. According to the structure of the N-terminal RNA binding domain of the NS1 protein recently reported by Chien et al. (2), Lys(62) is located in helix 3, which is suggested to be one of the RNA binding domains. The procedure of Chou and Fasman (3) predicts that the substitution introduces a turn at the helix. This structural change may bring about unwinding of the helix, leading to instability of the whole NS1 molecule and temperature sensitivity of its RNA binding ability. As the N-terminal half of the NS1 protein exhibits the full range of activities hitherto reported for the full-length NS1 protein (24, 29), it was surprising that the substitution in ICR1629 affected the RNA binding ability of this protein. In this connection, it is remarkable that GST-NS82 exhibited a higher RNA binding activity than the full-length protein (Fig. 2). Somehow, the C-terminal half of the protein might interfere with the N-terminal RNA binding domain. In the mutant NS1 protein, the structural changes induced at high temperature might reinforce the interaction between the two domains, thus sterically inhibiting the RNA binding or the dimerization of NS1.
Several experiments in vivo have indicated that one functional role of the NS1 protein is the enhancement of viral mRNA translation, especially that of the late proteins (4, 7, 10, 27). Most of them have suggested that the NS1 protein enhances the recruitment of translation factors by interacting with cis-acting elements on the viral mRNA. Here, we have indicated another role of the NS1 protein in vivo, i.e., the inhibition of PKR activation by binding to virus RNAs, thus contributing to some part of the translational enhancement. Moreover, it was indicated that the translational block exerted by activated PKR in these NS1 mutant-infected cells could not be relieved by p58 alone, a cellular protein which represses the phosphorylation of eIF2α by the already-activated PKR (21).
ACKNOWLEDGMENTS
We thank K. Shimizu (Nihon University School of Medicine) for providing the virus strains, H. Taira for donation of anti-human PKR antibody, and Eiko Sakaguchi for technical assistance.
REFERENCES
- 1.Baudin F, Bach C, Cusack S, Ruigok R W H. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3159–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chien C, Tejero R, Huang Y, Zimmerman D E, Rios C B, Krug R M, Monteilon G T. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat Struct Biol. 1997;4:891–895. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- 3.Chou P Y, Fasman G D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 4.de la Luna S, Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNA. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desselberger U, Racaniello V R, Zazra J J, Palese P. The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 6.Egorov A, Brandt S, Sereinig S, Romanova J, Ferko B, Katinger D, Grassauer A, Alexandrova G, Katinger H, Muster T. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J Virol. 1998;72:6437–6441. doi: 10.1128/jvi.72.8.6437-6441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enami K, Sato T A, Nakada S, Enami M. Influenza virus NS1 protein stimulates translation of the M1 protein. J Virol. 1994;68:1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galabru J, Hovanessian A. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J Biol Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 10.Hatada E, Hasegawa M, Shimizu K, Hatanaka M, Fukuda R. Analysis of influenza A virus temperature-sensitive mutants with mutations in RNA segment 8. J Gen Virol. 1990;71:1283–1292. doi: 10.1099/0022-1317-71-6-1283. [DOI] [PubMed] [Google Scholar]
- 11.Hatada E, Takizawa T, Fukuda R. Specific binding of influenza A virus NS1 protein to the virus minus-sense RNA in vitro. J Gen Virol. 1992;73:17–25. doi: 10.1099/0022-1317-73-1-17. [DOI] [PubMed] [Google Scholar]
- 12.Hatada E, Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J Gen Virol. 1992;73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 13.Hatada E, Saito S, Okisio N, Fukuda R. Binding of the influenza virus NS1 protein to model genome RNAs. J Gen Virol. 1997;78:1059–1063. doi: 10.1099/0022-1317-78-5-1059. [DOI] [PubMed] [Google Scholar]
- 14.Hsu M-T, Parvin J D, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci USA. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katze M G, Tomita J, Black T, Krug R M, Safer B, Hovanessian A. Influenza virus regulates protein synthesis during infection by repressing autophosphorylation and activity of the cellular 68,000-Mr protein kinase. J Virol. 1988;62:3710–3717. doi: 10.1128/jvi.62.10.3710-3717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katze M G. The war against the interferon-induced dsRNA-activated protein kinase: can viruses win? J Interferon Res. 1992;12:241–248. doi: 10.1089/jir.1992.12.241. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi M, Toyoda T, Adyshev D M, Azuma Y, Ishihama A. Molecular dissection of influenza virus nucleoprotein: deletion mapping of the RNA binding domain. J Virol. 1994;68:8433–8436. doi: 10.1128/jvi.68.12.8433-8436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konieezny A, Safer B. Purification of the eukaryotic initiation factor 2-eukaryotic initiation factor 2B complex and characterization of its guanine nucleotide exchange activity during protein synthesis initiation. J Biol Chem. 1983;258:3402–3408. [PubMed] [Google Scholar]
- 19.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 20.Lamb R A. Genes and proteins of the influenza viruses. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 1–87. [Google Scholar]
- 21.Lee T G, Tomita J, Hovanessian A G, Katze M G. Characterization and regulation of the 58,000-dalton cellular inhibitor of the interferon-induced, dsRNA-activated protein kinase. J Biol Chem. 1992;267:14238–14243. [PubMed] [Google Scholar]
- 22.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 23.Manche L, Green S R, Schmedt C, Mathews M B. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marion R M, Aragon T, Beloso A, Nieto A, Ortin J. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 1997;25:4271–4277. doi: 10.1093/nar/25.21.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marion R M, Zurcher T, de la Luna S, Ortin J. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J Gen Virol. 1997;78:2447–2451. doi: 10.1099/0022-1317-78-10-2447. [DOI] [PubMed] [Google Scholar]
- 26.O’Neill R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park Y W, Katze M G. Translational control by influenza virus. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 28.Qian X-Y, Alonso-Capien F, Krug R M. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J Virol. 1994;68:2433–2441. doi: 10.1128/jvi.68.4.2433-2441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian X-Y, Chien C-Y, Lu Y, Montelione G T, Krug R M. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA. 1995;1:948–956. [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu Y, Krug R M. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu Y, Nemeroff M, Krug R M. The influenza virus NS1 protein binds to a specific region in human U6-U2 and U6-U4 sn RNA interactions during splicing. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 32.Richardson J C, Akkina R K. NS2 protein of influenza virus is found in purified virus and phosphorylated in infected cells. Arch Virol. 1991;116:69–80. doi: 10.1007/BF01319232. [DOI] [PubMed] [Google Scholar]
- 33.Ruigrok R W H, Baudin F. Structure of influenza virus ribonucleoprotein particles. II. Purified RNA-free influenza virus ribonucleoprotein forms structures that are indistinguishable from the intact influenza virus ribonucleoprotein particles. J Gen Virol. 1995;76:1009–1014. doi: 10.1099/0022-1317-76-4-1009. [DOI] [PubMed] [Google Scholar]
- 34.Saito S. Possible involvement of virus-induced protein kinase in the antiviral state induced with interferon-γ against Sindbis virus. J Interferon Res. 1989;9:23–34. doi: 10.1089/jir.1989.9.23. [DOI] [PubMed] [Google Scholar]
- 35.Samuel C E, Knutsonk G S, Berry M J, Atwater J A, Lasky S R. Purification of double-stranded RNA-dependent protein kinase from mouse fibroblasts. Methods Enzymol. 1986;119:499–517. doi: 10.1016/0076-6879(86)19070-7. [DOI] [PubMed] [Google Scholar]
- 36.Samuel C E. The eIF2 protein kinases, regulators of translation in eucaryotes from yeasts to humans. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 37.Shimizu K, Mullinix M G, Chanock R M, Murphy B R. Temperature-sensitive mutants of influenza A/Udorn/72 (H3N2) virus. II. Genetic analysis and demonstration of intrasegmental complementation. Virology. 1982;117:45–61. doi: 10.1016/0042-6822(82)90506-2. [DOI] [PubMed] [Google Scholar]
- 38.Skorko R, Summers D F, Galarza J M. Influenza virus in vitro transcription: roles of NS1 and NP proteins in regulating RNA synthesis. Virology. 1991;180:668–677. doi: 10.1016/0042-6822(91)90080-u. [DOI] [PubMed] [Google Scholar]
- 39.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 40.Staehli P. Interferon-induced proteins and the antiviral state. Adv Virus Res. 1990;33:147–200. doi: 10.1016/s0065-3527(08)60862-3. [DOI] [PubMed] [Google Scholar]
- 41.Yasuda J, Nakada S, Kato A, Toyoda T, Ishihama A. Molecular assembly of influenza virus: association of the NS2 protein with virion matrix. Virology. 1993;196:249–255. doi: 10.1006/viro.1993.1473. [DOI] [PubMed] [Google Scholar]