Abstract
Although the diversity, beauty, and intricacy of sexually selected courtship displays command the attention of evolutionists, the longevity of these traits in deep time is poorly understood. Population-based theory suggests sexual selection could either lower or raise extinction risk, resulting in high or low persistence of lineages with sexually selected traits. Furthermore, empirical studies that directly estimate the longevity of sexually selected traits are uncommon. Sexually selected signals—including bioluminescent courtship—originated multiple times during evolution, allowing the empirical study of their longevity after careful phylogenetic and divergence time analyses. Here, we estimate the first transcriptome-based molecular phylogeny and divergence times of Cypridinidae. We report extreme longevity of bioluminescent courtship, a trait important in mate choice and probably under sexual selection. Our relaxed-clock estimates of divergence times coupled with stochastic character mapping show luminous courtship evolved only once in Cypridinidae—in a Sub-Tribe, we name Luxorina—at least 151 millions of years ago from cypridinid ancestors that used bioluminescence only in antipredator displays, defining a Tribe we name Luminini. This time-calibrated molecular phylogeny of cypridinids will serve as a foundation for integrative and comparative studies on the biochemistry, molecular evolution, courtship, diversification, and ecology of cypridinid bioluminescence. The persistence of luminous courtship for hundreds of millions of years suggests that sexual selection did not cause a rapid loss of associated traits, and that rates of speciation within the group exceeded extinction risk, which may contribute to the persistence of a diverse clade of signaling species. [Ancestral state reconstruction; Biodiversity; co-option; divergence time estimates; macroevolution; Ostracoda; phylogenomics; sexual selection.]
By producing some of the most diverse, beautiful, and elaborate biological displays, the evolution of sexually selected traits is a topic of fundamental importance. While sexually selected traits in a handful of model clades (Prum 2017; Ryan et al. 2019) are the focus of much study, open questions remain, such as the longevity of such traits (Wiens and Tuschhoff 2020). Predicting longevity during macroevolution of sexually selected traits is difficult because population-based inferences that relate sexual selection to extinction are mixed (Kokko and Brooks 2003; Martínez-Ruiz and Knell 2017). On one hand, sexual selection could reduce mutation load, increase mean fitness, and decrease extinction (Tomkins et al. 2004; Long et al. 2012; Plesnar-Bielak et al. 2012; Lumley et al. 2015). On the other hand, costs imposed by sexual selection—through maladaptation of exaggerated traits and/or reduced effective population—could decrease population fitness and increase extinction (Promislow 1992; Tanaka 1996; Sorci et al. 1998; Doherty et al. 2003; Morrow and Fricke 2004). Together, these studies show many factors—especially population size, environmental variability, honesty of signal, and eco-evolutionary feedbacks—dictate relationships between sexual selection and extinction risk at population scales. If these factors vary across species during macroevolutionary radiations, the longevity of a sexually selected trait will not be predictable for individual populations or species. In addition, few empirical studies of trait duration are available, although examples of ancient signals, including those with modern sexual functions, date to at least 200 Ma. Examples include acoustic signals in tetrapods (Chen and Wiens 2020) and insects (Gu et al. 2012; Song et al. 2020), luminous courtship in terrestrial fireflies (Höhna et al. 2021; Powell et al. 2022), and the role of flowers in plant–pollinator interactions (Bao et al. 2019; van der Kooi and Ollerton 2020). Conversely, extinction rates increased in fossil ostracods that are sexually dimorphic in body size, where the dimorphism may be a strong proxy for postcopulatory sexual selection (Martins et al. 2018). Therefore, the longevity of sexually selected traits remains largely untested, despite implications for understanding the power of sexual selection to influence biodiversity on macroevolutionary timescales.
To study the origin and longevity of sexually selected traits in macroevolution, a valuable trait is bioluminescence, which evolved separately at least 94 times (Lau and Oakley 2021), providing replicative power for comparative analyses (Ellis and Oakley 2016). Bioluminescence has multiple organismal functions including defense and reproduction, which affect ecosystems (Morin 1983; Wilson and Hastings 1998; Haddock et al. 2010; Widder 2010). When used for courtship, bioluminescence is associated with more species compared to sister clades that lack luminous courtship, providing an example of a sexually selected trait that may increase rates of speciation (Ellis and Oakley 2016). Unfortunately, the specific number and timing of origins of bioluminescence remain uncertain for luminous groups and their relatives, despite some recent attention (see Ellis and Oakley 2016 for summary of ages). Here, we report new phylotranscriptomic analyses and divergence time estimates to understand the origins and longevity of bioluminescence and luminous courtship in cypridinid ostracods, a family of crustaceans that contains at least 250 species across 30 genera (Morin 2019), and which evolved bioluminescence separately from other animals.
Bioluminescent courtship in cypridinids is almost certainly influenced by sexual selection, as information rich, sexually dimorphic traits that allow for mate choice (Andersson 1994; Rivers and Morin 2013). While all known bioluminescent cypridinids can produce antipredatory displays by secreting defensive clouds of light in response to predation attempts (Morin and Cohen 2010; Rivers and Morin 2012; Morin 2019), only about 75 described and nominal species, restricted to the Caribbean, also produce bioluminescent courtship signals (Morin 1986, 2019; Morin and Cohen 2010; Reda et al. 2019). Beginning near twilight’s end (Gerrish et al. 2009), often in multispecies assemblages of 6–8 species that overlap in time and space (Gerrish and Morin 2016), male cypridinids produce patterns of ephemeral pulses of light, secreted while swimming in tight spirals (Rivers and Morin 2008). Like other radiations of sexually selected signals (Prum 2017), species-specific displays in signaling cypridinids vary considerably in many parameters (Morin 1986, 2019; Gerrish et al. 2009; Gerrish and Morin 2016; Hensley et al. 2019, 2021).
Previous phylogenetic analyses of morphology (Cohen and Morin 2003) and rDNA (Torres and Gonzalez 2007) both indicated a single origin of bioluminescence and suggested a single origin of luminous courtship in Cypridinidae (Morin 2019). However, beyond the topology itself, we have very little information about divergence times for these evolutionary transitions. One study of divergence times of Pancrustacea did include two luminous ostracod species but resulted in very large confidence intervals on the time of origin of their common ancestor, ranging from about 50 to 250 Ma (Oakley et al. 2013). Here, we present the first transcriptomic phylogenies and divergence time estimates for Cypridinidae, which indicate an ancient origin of bioluminescence and a long persistence of luminous courtship, a trait almost certainly under sexual selection (Morin and Cohen 2010). We conservatively estimate the origin of bioluminescent courtship in cypridinids to be at least 151 Ma. If researchers continue to identify the long persistence of traits under sexual selection, it would support these traits as often associated with increases in the rate of speciation relative to their effect on extinction.
Materials and Methods
Sampling Strategy and Collection
Based on previous morphological phylogenies and known distributions of species, we aimed
to balance cost-efficiency and diversity of sampling by choosing eight primary sampling
locations. We targeted species with luminous courtship mainly from five localities:
Jamaica, Honduras (Roatan), Belize, Panama, and Puerto Rico. We targeted species outside
the courtship signaling clade from three localities: Australia, Japan, and the United
States. We collected ostracods via net sampling during courtship displays, sediment
sampling, or carrion traps (see Cohen and Oakley
(2017) for sampling methods and Table S1 for taxa
sampled). We preserved samples in 95 ethanol for vouchers and
RNAlater for sequencing. To collect taxa with luminous courtship (where many species are
undescribed), we targeted unique courtship displays to isolate individual species because
ecologically overlapping courtship signals are distinct enough to distinguish species
(Gerrish and Morin 2016). We also measured the
length and height of carapaces to assign species to genera in the field, prior to
molecular analysis, and to affirm the single-species identity of pooled individuals
(Supplementary Table S2 available on Dryad at http://dx.doi.org/10.6076/dryad.D1PC7K). The ratio of carapace length to
height is usually diagnostic for genera of signaling cypridinids and distinguishes
sympatric species in combination with signals (Gerrish and
Morin 2016; Hensley et al. 2019; Reda et al. 2019).
ethanol for vouchers and
RNAlater for sequencing. To collect taxa with luminous courtship (where many species are
undescribed), we targeted unique courtship displays to isolate individual species because
ecologically overlapping courtship signals are distinct enough to distinguish species
(Gerrish and Morin 2016). We also measured the
length and height of carapaces to assign species to genera in the field, prior to
molecular analysis, and to affirm the single-species identity of pooled individuals
(Supplementary Table S2 available on Dryad at http://dx.doi.org/10.6076/dryad.D1PC7K). The ratio of carapace length to
height is usually diagnostic for genera of signaling cypridinids and distinguishes
sympatric species in combination with signals (Gerrish and
Morin 2016; Hensley et al. 2019; Reda et al. 2019).
Taxon Sampling and Sequencing
We generated new RNA-Seq data from 45 species of cypridinid ostracods and four species of
noncypridinid ostracods. We included outgroups from all four noncypridinid families of
Myodocopida, sequencing one species each of Rutidermatidae and Sarsiellidae, and two of
Philomedidae. We also used published sequence data from a cylindroleberid (SRX2085850)
(Schwentner et al. 2018) and predicted proteins
from the genome of Darwinula stevensoni (ENA PRJEB38362) (Schön et al. 2021). For transcriptome sequencing, we
combined RNA from whole bodies of up to 10 pooled individuals (collected by signal but
with identity corroborated by morphological features). Pooling is necessary due to the
small size of the animals ( 2 mm) and low RNA yield. For some
samples, we extracted RNA using Trizol, and for the rest, we used Qiagen RNEasy Kits. We
either used the Illumina or NEBNext Ultra RNA Library Prep Kits to prepare transcriptome
libraries (Supplementary Table S1
available on Dryad).
2 mm) and low RNA yield. For some
samples, we extracted RNA using Trizol, and for the rest, we used Qiagen RNEasy Kits. We
either used the Illumina or NEBNext Ultra RNA Library Prep Kits to prepare transcriptome
libraries (Supplementary Table S1
available on Dryad).
Quality Control and Data Set Assembly
We trimmed RNA-seq adapters and low-quality bases (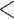 20) using
TrimGalore v0.4.1 (Krueger 2012), then assembled
trimmed reads using Trinity v2.2.0 (Grabherr et al.
2011). In cases of multiple transcriptomes from the same species, we combined
raw, trimmed reads to create single assemblies. We used CroCo v0.1 (Simion et al. 2018) to remove contigs in a given assembly both found
and expressed at a higher level in a transcriptome from another species (that was
sequenced on the same lane), which implies cross-contamination. We assessed completeness
using BUSCO v3.0.1 (Simão et al. 2015) using the
arthropod_odb9 single-copy ortholog database. We generally required transcriptomes to have
at least 100 complete BUSCO genes for further analyses. However, we made exceptions for
three species, (Photeros jamescasei, Jimmorinia gunnari, and
Melavargula japonica), to maintain these described and taxonomically
important species within the analyses. We produced protein predictions using TransDecoder
v3.0.1 (Haas et al. 2013), with default
parameters.
20) using
TrimGalore v0.4.1 (Krueger 2012), then assembled
trimmed reads using Trinity v2.2.0 (Grabherr et al.
2011). In cases of multiple transcriptomes from the same species, we combined
raw, trimmed reads to create single assemblies. We used CroCo v0.1 (Simion et al. 2018) to remove contigs in a given assembly both found
and expressed at a higher level in a transcriptome from another species (that was
sequenced on the same lane), which implies cross-contamination. We assessed completeness
using BUSCO v3.0.1 (Simão et al. 2015) using the
arthropod_odb9 single-copy ortholog database. We generally required transcriptomes to have
at least 100 complete BUSCO genes for further analyses. However, we made exceptions for
three species, (Photeros jamescasei, Jimmorinia gunnari, and
Melavargula japonica), to maintain these described and taxonomically
important species within the analyses. We produced protein predictions using TransDecoder
v3.0.1 (Haas et al. 2013), with default
parameters.
Orthology Determination
For analyses that relied on comparing orthologs (excluding paralogs), we used Orthofinder (Emms and Kelly 2019) plus phylopypruner (Thalén 2018). We grouped genes with Orthofinder 2.5.2 (Emms and Kelly 2019), using an MCL inflation parameter set to 2.1 and used Diamond as the similarity search program. This allowed us to cluster sequences based on similarity into “orthogroups” that contain a mix of orthologs and paralogs. Next, we created gene trees for the separate orthogroups by aligning each using the “auto” method of MAFFT v7.305 (Katoh and Standley 2013) and using maximum likelihood implemented in FastTree v2.1.9 (Price et al. 2010) for its speed, and we retained those results because they are congruent with concatenation methods using IQ-TREE2 (Minh et al. 2020). We used phylopypruner 1.2.3 (Thalén 2018) to remove paralogs, requiring a minimum of 20 species for each gene family, minimum support values of 0.7, and used the LS pruning method. We refer to this resulting set of orthologs as the “ppp orthologs.”
Previously Published Data Sets
We also added single-gene mitochondrial data (12S, 16S, CO1), summarized in Supplementary Table S5 available on Dryad and reported previously in conference proceedings (Torres and Gonzalez 2007) or publications (Ogoh and Ohmiya 2004; Wakayama and Abe 2006; Wang et al. 2019; Goodheart et al. 2020; Pham et al. 2020). We then identified these three mitochondrial genes from transcriptomes by finding the longest contigs with high similarity to published complete mitochondrial genomes of Vargula hilgendorfii and Vargula tsujii (Ogoh and Ohmiya 2004; Goodheart et al. 2020). We annotated mitochondrial data from transcriptomes using MITOS (Bernt et al. 2013), then aligned each mitochondrial gene using “auto” in MAFFT v7.305 (Katoh and Standley 2013).
Phylogenetic Reconstruction
We compared coalescence and concatenation-based analyses using maximum likelihood and Bayesian approaches. For coalescence-based approaches, we used ASTRAL-Pro (Zhang et al. 2020), which assumes the multispecies coalescent to estimate a species tree from multiple gene trees, including paralogs. We used the same gene trees as input for ASTRAL-Pro as phylopypruner (described above). Next, we determined the best-fit partitioning scheme for each concatenated data set using PartitionFinder v.2.1.1 (Lanfear et al. 2016) with RAxML (Stamatakis 2014) and rcluster (Lanfear et al. 2014). Using optimal partitioning schemes and models, we produced species trees in IQ-TREE v2.0.3, with additional options for -bnni and -alrt (set to 1000). We calculated gene Concordance Factors (gCF; Supplementary Table S4 available on Dryad) using IQ-TREE (Minh et al. 2020).
Divergence Time Estimation
We estimated divergence times of nodes in the cypridinid phylogeny using a Bayesian
relaxed molecular clock approach in MrBayes (Ronquist and
Huelsenbeck 2003), which offers a diversity of models, estimates tree topology,
and branch lengths in a Bayesian framework and has a scriptable command line interface,
leading to more repeatable science. Because Bayesian methods are computationally demanding
(Smith et al. 2018), we chose 15 orthologs to use
for divergence time estimation, as follows. We used sortadate (Smith et al. 2018) to rank all gene trees based on their consistency
with the species tree inferred from all genes in ASTRAL-Pro and based on the most
consistent root-to-tip branch lengths. To test the sensitivity of divergence time
estimates to the number of genes selected, we compared results from 15 gene trees to age
estimates from 20, 25, and 30 gene trees (Supplementary Fig. S7
available on Dryad). Although ostracods have a dense fossil record, most fossil ostracods
are podocopids and assigning fossils confidently to taxonomic groups is often challenging
(Tinn and Oakley 2008), restricting us to two
fossil constraints. The first was Myodocopida, for which we used an offset exponential
prior with a minimum age of 448.8 Ma and maximum of 509 Ma (Oakley et al. 2013; Wolfe et al.
2016). We also used a uniform prior between 443.8 and 509 Ma on the root of the
tree, which is the common ancestor of Ostracoda. Along with node constraints, we used the
Independent Gamma Rate (IGR) model (Lepage et al.
2007; Zhang 2016). We used a prior of
exp(10) for variation in the IGR model. For the prior on the overall rate of the molecular
clock (substitution rate/site/Myr), we assumed a lognormal distribution with a mean of
0.001 and standard deviation of 0.0007. For the model of amino acid substitution, we
assumed fixed-rate models and used the command aamodelpr mixed,
which explores multiple fixed-rate models in proportion to their posterior probabilities.
With these models and priors, we ran 500K steps of Markov Chain Monte Carlo (MCMC) in two
chains, using a starting phylogeny based on parsimony.
mixed,
which explores multiple fixed-rate models in proportion to their posterior probabilities.
With these models and priors, we ran 500K steps of Markov Chain Monte Carlo (MCMC) in two
chains, using a starting phylogeny based on parsimony.
Ancestral State Reconstructions
We reconstructed ancestral states for bioluminescence (presence/absence) and
bioluminescent courtship (presence/absence) using our tree generated with ASTRAL-Pro. In
order to create an ultrametric tree for ancestral state reconstruction analysis, we used
mean ages for nodes matched to those from our Bayesian relaxed-clock analysis. We set
matching branch lengths using the ph_bladj function in the R package phylocomr (Webb et al. 2008; Ooms
and Chamberlain 2019). We assessed fit for two models (for each character) using
corrected Akaike Information Criterion (AICc), where: (i) transition rates were equal (ER)
and (ii) forward and reverse transitions were different between states (all rates
different [ARD]). The ER model (AICc  24.02231) was slightly
better than the ARD model (AICc
24.02231) was slightly
better than the ARD model (AICc  25.72293) for
bioluminescence and for bioluminescent courtship (ER AICc
25.72293) for
bioluminescence and for bioluminescent courtship (ER AICc  23.799;
ARD AICc
23.799;
ARD AICc  24.70163), so we used ER for final
ancestral state reconstruction analyses in R using the rayDISC function in corHMM (Beaulieu et al. 2013). To test for significance of the
reconstruction at each node, we used proportional likelihood significance tests assuming a
log likelihood difference of two or greater represents a significant difference (Pagel 1999).
24.70163), so we used ER for final
ancestral state reconstruction analyses in R using the rayDISC function in corHMM (Beaulieu et al. 2013). To test for significance of the
reconstruction at each node, we used proportional likelihood significance tests assuming a
log likelihood difference of two or greater represents a significant difference (Pagel 1999).
To estimate timing of cypridinid bioluminescence and luminous courtship, we conducted time-tree stochastic character mapping (ttscm) (Alexandrou et al. 2013), which simulates character evolution with stochastic character mapping (Huelsenbeck et al. 2003) along time-calibrated phylogenies from the MCMC search to represent the posterior distribution of trees. While ancestral state reconstructions depict character state changes only at nodes, stochastic character maps allow state changes along branches. When branches are time-calibrated, this approach provides absolute estimates of the timing of character state changes. We used stochastic character mapping in phytools (Revell 2012) using the same character states and models as described for ancestral state reconstruction. We used 100 time-calibrated trees from the MCMC search to represent the posterior probability distribution of tree topologies and branch lengths, and summarized the timing character state changes in histograms.
Results
Assembly and Orthology Statistics
Our final matrices of transcriptomic data (Supplementary Table S3 available on Dryad) include 45 cypridinids from 15 of 25 genera, plus six outgroup taxa representing all four other myodocopid families and the previously published genome sequence of one podocopid (Schön et al. 2021). Some of these transcriptomes were used previously to identify luciferase genes (Hensley et al. 2021).
Evolution of Luminous Courtship and Cypridinid Bioluminescence
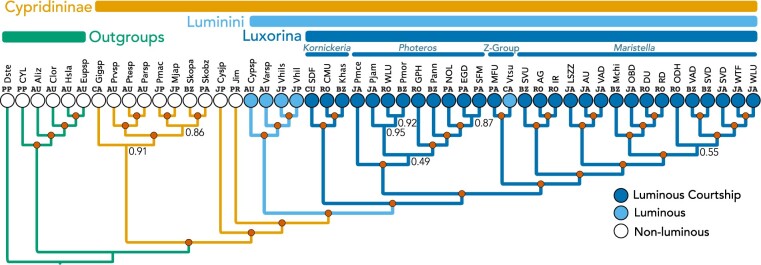
Our phylogenetic results support a single clade containing all Cypridinidae that use
bioluminescence for courtship signaling (Figs. 1 and
2, Supplementary Fig. S13
available on Dryad; Table 1). For this clade, we
propose the Sub-Tribe name Luxorina (lux light, uxoriae
light, uxoriae
 courtship). Although an important
consideration for defining Luxorina, our molecular data unfortunately does not contain the
monotypic genus Enewton (Cohen and Morin
2010). Despite returning to the same collection site in Jamaica as Cohen and Morin (2010), we could not find
Enewton harveyi (due to inclement weather). In previous parsimony
analyses of morphological traits (Cohen and Morin
2003, 2010), E. harveyi
is the sister of the rest of Luxorina. In the absence of molecular data,
Enewton as sister to the rest of Luxorina remains the best hypothesis
and we propose to include Enewton taxonomically within Luxorina.
courtship). Although an important
consideration for defining Luxorina, our molecular data unfortunately does not contain the
monotypic genus Enewton (Cohen and Morin
2010). Despite returning to the same collection site in Jamaica as Cohen and Morin (2010), we could not find
Enewton harveyi (due to inclement weather). In previous parsimony
analyses of morphological traits (Cohen and Morin
2003, 2010), E. harveyi
is the sister of the rest of Luxorina. In the absence of molecular data,
Enewton as sister to the rest of Luxorina remains the best hypothesis
and we propose to include Enewton taxonomically within Luxorina.
Figure 1.
Species phylogeny inferred using multispecies-coalescent and transcriptome data.
Circles at nodes indicate 1.0 posterior probability, and nodes with values less than
1.0 are indicated numerically. Circles at tips represent nonluminous (lightest),
luminous but noncourtship signaling (medium shade), and species with luminous
courtship (darkest shade). The three described genera of signaling species are labeled
at the top, along with “Z-group,” which is a clade of signaling species plus
Vargula tsujii. Vargula is polytypic (see also Cohen and Morin 2003; Morin 2019) and the type species is outside this group, so
formally, these should not be Vargula. Above the tree, we also
labeled noncypridinid Outgroups and Cypridinidinae. We herein suggest a name for the
bioluminescent Cypridinidae to be Tribe Luminini (for lumen  light and the typical Tribe suffix for zoology) and a name for the clade containing
courtship signaling cypridinids to be Sub-Tribe Luxorina (lux
light and the typical Tribe suffix for zoology) and a name for the clade containing
courtship signaling cypridinids to be Sub-Tribe Luxorina (lux
 light
light  uxoriae
uxoriae 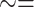 courtship
courtship
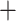 -ina
-ina  typical zoological subtribe suffix). Locality abbreviations for collecting sites of
species are above each tip as follows: AU
typical zoological subtribe suffix). Locality abbreviations for collecting sites of
species are above each tip as follows: AU  Australia, BZ
Australia, BZ
 Belize, CA
Belize, CA  California, USA, CU
California, USA, CU  Curacao, JA
Curacao, JA  Jamaica, JP
Jamaica, JP  Japan, PA
Japan, PA  Panama, PP
Panama, PP  Previously Published, PR
Previously Published, PR
 Puerto Rico, and RO
Puerto Rico, and RO
 Roatan, Honduras (see Supplementary Table S1 available on Dryad for specific localities).
Above locality names are abbreviations for species. Described species are abbreviated
as the first letter of genus and first three letters of species epithet; undescribed
species outside of Luxorina are labeled as the first three letters of genus followed
by sp. or two locality or collector letters when there are more than one undescribed
species from the genus; within Luxorina, we used 2–4 letter field codes for
undescribed species (see Supplementary
material available on Dryad for tree with full species names).
Roatan, Honduras (see Supplementary Table S1 available on Dryad for specific localities).
Above locality names are abbreviations for species. Described species are abbreviated
as the first letter of genus and first three letters of species epithet; undescribed
species outside of Luxorina are labeled as the first three letters of genus followed
by sp. or two locality or collector letters when there are more than one undescribed
species from the genus; within Luxorina, we used 2–4 letter field codes for
undescribed species (see Supplementary
material available on Dryad for tree with full species names).
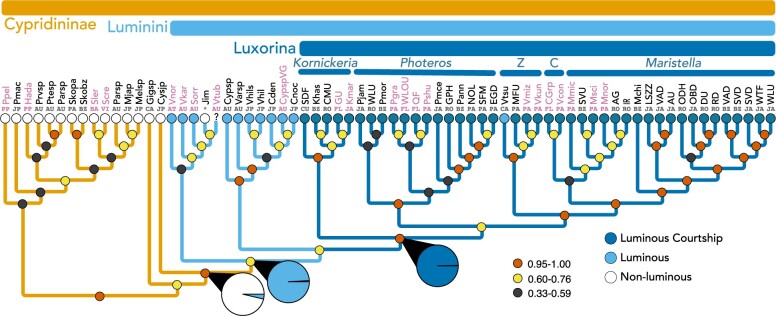
Figure 2.
Expanded taxon sampling by adding individual mitochondrial genes, mainly from Torres and Gonzalez (2007) to the transcriptome
data in Figure 1. We inferred this species
phylogeny using the multispecies coalescent. Taxa colored pink (light grey) are those
that contain only mitochondrial data. Small colored circles at nodes indicate
posterior probabilities low (0.33 0.59)
0.59)
 black, medium
(0.60
black, medium
(0.60 0.76)
0.76)  yellow (lightest shade), high (0.95
yellow (lightest shade), high (0.95 1.0)
1.0)
 red (medium shade). Locality
abbreviations are as in Figure 1 and also include
additional locations of VI
red (medium shade). Locality
abbreviations are as in Figure 1 and also include
additional locations of VI  US Virgin Islands
and FL
US Virgin Islands
and FL  Florida USA. For Jimmorinia (*), we
combined data from transcriptomes from a Jamaican species (J. cf
gunnari) with mitochondrial DNA from a specimen
(Jimmorinia sp. USVI) unidentified to species from VI. Pie charts
represent proportions of likelihood values for alternative ancestral states.
Bioluminescence was most likely absent (white) prior to Luminini, as supported by a
significantly high proportion of likelihood for the absence of bioluminescence
compared to the presence (medium shade). Significantly high likelihood proportions
first appear in Luminini for bioluminescence (medium shade) and Luxorina for
bioluminescent courtship signaling (darkest shade), compared to the absence of those
traits (black), which were analyzed separately as binary traits (see Supplementary material available on Dryad for full ancestral state
reconstruction results).
Florida USA. For Jimmorinia (*), we
combined data from transcriptomes from a Jamaican species (J. cf
gunnari) with mitochondrial DNA from a specimen
(Jimmorinia sp. USVI) unidentified to species from VI. Pie charts
represent proportions of likelihood values for alternative ancestral states.
Bioluminescence was most likely absent (white) prior to Luminini, as supported by a
significantly high proportion of likelihood for the absence of bioluminescence
compared to the presence (medium shade). Significantly high likelihood proportions
first appear in Luminini for bioluminescence (medium shade) and Luxorina for
bioluminescent courtship signaling (darkest shade), compared to the absence of those
traits (black), which were analyzed separately as binary traits (see Supplementary material available on Dryad for full ancestral state
reconstruction results).
Table 1.
Summary of support and age estimates for clades of interest from transcriptome data
| Clade | MSC-PP (Fig. 1 and Supplementary Figs. S1 and S2 available on
Dryad
|
ML-Boot  Supplementary Fig. S3 available on Dryad) Supplementary Fig. S3 available on Dryad) |
Gene concordance  Supplementary Fig. S4 available on Dryad Supplementary Fig. S4 available on Dryad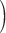
|
Bayes-PP  Supplementary Fig. S8 available on Dryad Supplementary Fig. S8 available on Dryad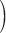
|
Age (Ma) 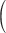 SupplementaryFig. S6 available
on Dryad SupplementaryFig. S6 available
on Dryad
|
|---|---|---|---|---|---|
| Luxorina | 1.0 | 100 |
49.8 (1111 of 2232) (1111 of 2232) |
1.0 | 181 (151–247) |
| Luminini | 1.0 | 100 |
45.3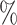 (77 of 170) (77 of 170) |
1.0 | 231 (197–293) |
| Kornickeria | 1.0 | 100 |
65.9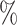 (1219 of 1850) (1219 of 1850) |
1.0 | 111 (47–193) |
| Maristella | 1.0 | 100 |
57.3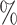 (1226 of 2138) (1226 of 2138) |
1.0 | 80 (49–107) |
| Z-Group | 1.0 | 100 |
63.4 (792 of 1249) (792 of 1249) |
1.0 | 66 (29–96) |
| Photeros | 1.0 | 100 |
83.3 (1730 of 2077) (1730 of 2077) |
1.0 | 53 (24–110) |
Our results also support a single clade containing all bioluminescent Cypridinidae
(Supplementary Fig. S12 available on Dryad; Table 1). For this clade of bioluminescent cypridindids, and noting this is a
separate origin of bioluminescence from halocyprid ostracods (Oakley 2005) that also involves a different chemistry (Campbell and Herring 1990), we propose the name Luminini
(lumen  light). While supported in each of our
analyses, the taxa available vary across data sets due to challenges obtaining
transcriptomic data from some species. In the transcriptome-based analyses, bioluminescent
cypridinids share a common ancestor in this clade with only luminous species. The data set
that combines mitochondrial and transcriptome data also supports Luminini (Fig. 2). However, the difference in taxon sampling
highlights an important result. Namely, four species (three luminous and one unknown) with
only mitochondrial data form a rather weakly supported clade (0.6 PP) with the
non-luminous Jimmorinia, which is the sister group of luminous taxa in
the transcriptome analyses. The three luminous species in this clade include
Vargula norvegica from the deep Atlantic Ocean, and Sheina
orri and Vargula karamu from Australia. Vargula
tubulata is also in this clade, but the status of bioluminescence in the
species is unknown. These species may form a clade within Luminini that also includes
non-luminous Jimmorinia, leading to an inference of loss of
bioluminescence in that genus (Fig. 2 and Supplementary
Fig. S10 available on Dryad).
light). While supported in each of our
analyses, the taxa available vary across data sets due to challenges obtaining
transcriptomic data from some species. In the transcriptome-based analyses, bioluminescent
cypridinids share a common ancestor in this clade with only luminous species. The data set
that combines mitochondrial and transcriptome data also supports Luminini (Fig. 2). However, the difference in taxon sampling
highlights an important result. Namely, four species (three luminous and one unknown) with
only mitochondrial data form a rather weakly supported clade (0.6 PP) with the
non-luminous Jimmorinia, which is the sister group of luminous taxa in
the transcriptome analyses. The three luminous species in this clade include
Vargula norvegica from the deep Atlantic Ocean, and Sheina
orri and Vargula karamu from Australia. Vargula
tubulata is also in this clade, but the status of bioluminescence in the
species is unknown. These species may form a clade within Luminini that also includes
non-luminous Jimmorinia, leading to an inference of loss of
bioluminescence in that genus (Fig. 2 and Supplementary
Fig. S10 available on Dryad).
Genus-Level Relationships within Cypridinidae
Our analysis included a number of undescribed species that we first assigned to genera using length to height ratio of the carapaces in the field (Supplementary Table S2 available on Dryad). Our phylogenetic reconstructions (Fig. 1) based on genetic data provide good support for these initial generic designations. Using these designations, both concatenated (Supplementary Fig. S3 available on Dryad) and coalescent approaches (Fig. 1) recovered four monophyletic genera (Kornickeria, Photeros, and Maristella and the not yet described “Z-Group”), all with strong support (Table 1). Additionally, Maristella (Reda et al. 2019), includes two distinct monophyletic clades, the small-bodied H-Group, which would remain in Maristella, and the larger-bodied U-Group (BZ-SVU, RO-AG, and RO-IR), which could be a new genus (Cohen and Morin 1989) (Fig. 1).
Furthermore, while strongly supported by transcriptome data, results of Maristella deserve more discussion because two other species (Vargula contragula and an undescribed species from Florida with only mitochondrial data) are closely related to Maristella, and may belong to yet another (undescribed) genus, provisionally called the C-group (Cohen and Morin 1986; Morin and Cohen 2017). Our results support the Maristella clade with high probability (1.0) to include both C-group and Maristella, but the position of the two C-group species compared to Maristella is unresolved (Fig. 2, Supplementary Fig. S10 available on Dryad ). Confidently resolving relationships of groups similar, but perhaps sister to Maristella will require more data.
Another group within Luxorina includes representatives of what is provisionally called “Z-group” (Cohen and Morin 1986; Morin and Cohen 1988, 2017), which will need formal description as a new genus. Our analyses that add taxa with only mitochondrial data include three Panamanian species in a clade with V. tsujii: undescribed MFU, “Vargula” kuna, and “Vargula” mizonoma. The latter two were previously called Z-group (Morin and Cohen 1988; Cohen and Morin 2003). Therefore, we report strong support for a clade that includes the Z-group and Vargula tsujii. Even though V. tsujii is a described species, the genus should not be named Vargula (Cohen and Morin 2003) because the type species is the distantly related V. norvegica (Fig. 2). These results reinforce the highly polyphyletic nature of Vargula (Cohen and Morin 2003) and uncover the closest relatives of the only Luxorina species known from the Pacific, V. tsujii.
We also find strong support (1.0) for the relationships among courtship signaling genera, with Kornickeria sister to a clade containing Photeros, Maristella, Z-Group, and C-group (Fig. 2). Next, Photeros is the sister clade (0.76) to Maristella, Z-Group, and C-group, with Z-group as the sister of Maristella plus C-group (1.0). The relationships among these genera in Luxorina varied among the previous studies cited above.
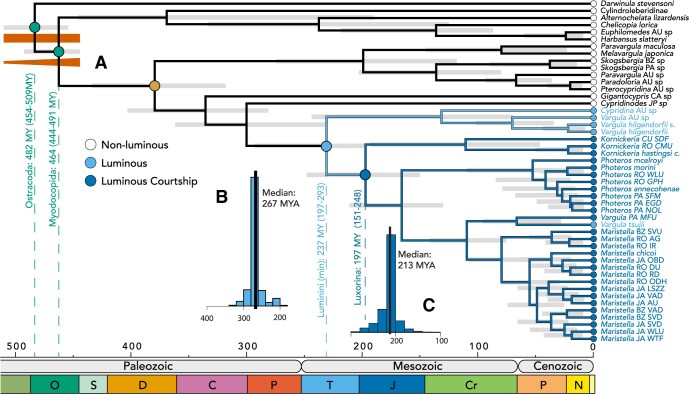
Divergence Time Estimates
Relaxed molecular clock analyses place the origin of the clade of species with
bioluminescent courtship between ( Luxorina) 151–248 Ma
(Bayesian Highest Posterior Density) with a median value of 197 Ma (Fig. 3a). Using time tree stochastic character mapping (ttscm) (Alexandrou et al. 2013), we estimate the character state
transition to luminous courtship occurred 213 Ma (Fig.
3c). The estimates for the origin of cypridinid bioluminescence
(
Luxorina) 151–248 Ma
(Bayesian Highest Posterior Density) with a median value of 197 Ma (Fig. 3a). Using time tree stochastic character mapping (ttscm) (Alexandrou et al. 2013), we estimate the character state
transition to luminous courtship occurred 213 Ma (Fig.
3c). The estimates for the origin of cypridinid bioluminescence
( Luminini) are less clear because of issues
with taxon sampling and the low quality of the transcriptome of
Jimmorinia. Namely, the data set used for divergence times does not
contain any species of the clade containing Jimmorinia, V.
norvegica, or the other luminous Australian species; these taxa form the sister
group to the rest of the luminous cypridinids (Fig.
1). Nevertheless, we can estimate the age of the common ancestor of all luminous
species with transcriptome data as a minimum estimate for the origin of cypridinid
bioluminescence to be 197–293 Ma with a median value of 237 Ma. Similarly, we can estimate
the age of the common ancestor of luminous cypridinids and their closest non-luminous
relative as a maximum age to be 233–361 Ma with a median value of 300 Ma (Fig. 3a). Using time tree stochastic character mapping
(ttscm) (Alexandrou et al. 2013), we estimate the
origin of bioluminescence occurred 267 Ma (Fig. 3b).
We also note our estimates of even the most closely related species are always more than 8
Ma (Supplementary Fig. S5 available on Dryad), even for two populations of
V. hilgendorfii. Although populations of V.
hilgendorfii are quite diverged (Ogoh and
Ohmiya 2005) our estimates seem very old at the tips of the tree, and we
speculate this could be an artifact of extrapolating young nodes from the deepest nodes in
the absence of other suitable fossils.
Luminini) are less clear because of issues
with taxon sampling and the low quality of the transcriptome of
Jimmorinia. Namely, the data set used for divergence times does not
contain any species of the clade containing Jimmorinia, V.
norvegica, or the other luminous Australian species; these taxa form the sister
group to the rest of the luminous cypridinids (Fig.
1). Nevertheless, we can estimate the age of the common ancestor of all luminous
species with transcriptome data as a minimum estimate for the origin of cypridinid
bioluminescence to be 197–293 Ma with a median value of 237 Ma. Similarly, we can estimate
the age of the common ancestor of luminous cypridinids and their closest non-luminous
relative as a maximum age to be 233–361 Ma with a median value of 300 Ma (Fig. 3a). Using time tree stochastic character mapping
(ttscm) (Alexandrou et al. 2013), we estimate the
origin of bioluminescence occurred 267 Ma (Fig. 3b).
We also note our estimates of even the most closely related species are always more than 8
Ma (Supplementary Fig. S5 available on Dryad), even for two populations of
V. hilgendorfii. Although populations of V.
hilgendorfii are quite diverged (Ogoh and
Ohmiya 2005) our estimates seem very old at the tips of the tree, and we
speculate this could be an artifact of extrapolating young nodes from the deepest nodes in
the absence of other suitable fossils.
Figure 3.
Time of taxon origins based on relaxed molecular clock analysis. a) We used two fossil constraints (green circles): Myodocopida, with offset exponential prior with a minimum of 448.8 Ma and maximum of 509 Ma and Ostracoda with a uniform prior between 443.8 and 509 Ma (Oakley et al. 2013; Wolfe et al. 2016) to estimate divergence times across our phylogeny; b) Results of time-tree stochastic character mapping (ttscm) estimates origin of bioluminescence to be 267 Ma because that is the median age of character state transitions to bioluminescence; c) Similarly, ttscm estimates the origin of luminous courtship to be 213 Ma.
Discussion
How sexual selection impacts biodiversity over long evolutionary timeframes is an open question. If sexual selection affects the relative rates of extinction and speciation, it would influence the longevity of traits and clades, ultimately impacting patterns of biodiversity. However, population-based results indicate sexual selection may either increase or decrease extinction risk (Kokko and Brooks 2003; Martínez-Ruiz and Knell 2017), leading to uncertainty about the long-term persistence of sexually selected traits. Furthermore, only if consistent and conserved during evolutionary radiations should microevolutionary processes fully dictate macroevolutionary patterns. Therefore, the long-term persistence of sexually selected traits on macroevolutionary timescales is an open empirical question (Wiens and Tuschhoff 2020). Our result in Luxorina ostracods of ancient co-option of bioluminescent antipredator signals and subsequent maintenance for courtship—lasting at least 151 Myr—provides a compelling example of a trait almost certainly under sexual selection with very long macroevolutionary persistence. The maintenance of bioluminescent courtship is counter to one line of population-level reasoning that the conspicuousness of many sexually selected traits should routinely lower fitness and increase rates of extinction and/or loss of traits under sexual selection (Promislow 1992; Tanaka 1996; Sorci et al. 1998; Doherty et al. 2003; Morrow and Fricke 2004). The sparse fossil record of this group makes detailed estimation of speciation and extinction rates very difficult, but future work could explore methods such as Bayesian model selection (Mitchell and Rabosky 2017) to investigate in more detail correlated increases in speciation or extinction with the onset of luminous courtship. Sexual selection can theoretically also reduce mutation load, increase mean fitness, and lead to lower extinction risks (Tomkins et al. 2004; Long et al. 2012; Plesnar-Bielak et al. 2012; Lumley et al. 2015).
Besides the effects of microevolutionary processes on extinction, other factors also
influence the longevity of sexually selected traits and the clades that contain them,
especially biogeography and life history. The biogeography of courtship signaling in
Luxorina ostracods appears restricted to the Caribbean. Therefore, Cohen and Morin (2003) hypothesized that Luxorina is a relatively recent
evolutionary radiation, yet it must be older than the formation of the Isthmus of Panama,
3.5–25 Ma (Coates et al. 1992; Farris et al. 2011; Bacon et al.
2015; O’Dea et al. 2016 and references
therein). Our molecular data push back the origin of Luxorina and their bioluminescent
courtship to about 197 (151–248) Ma (Fig. 3, Table 1), broadly consistent with the timing of initial
crust formation between the Americas 154-190 Ma (Baumgartner
et al. 2013). This suggests Luxorina originated and evolved on a time-scale very
similar to the Proto-Caribbean  Caribbean itself, while diversifying into
at least four major distinct clades (genera), each with an origin of tens of millions of
years ago (Table 1), with high species diversity, and
present-day distributions across most of the Caribbean (Cohen and Morin 1993, 2010; Reda et al. 2019).
Caribbean itself, while diversifying into
at least four major distinct clades (genera), each with an origin of tens of millions of
years ago (Table 1), with high species diversity, and
present-day distributions across most of the Caribbean (Cohen and Morin 1993, 2010; Reda et al. 2019).
The numerous islands of the Caribbean may have interacted with ecological and life history
characters in Luxorina to influence species diversity and therefore longevity of the entire
clade. Luxorina are shallow-water species with dispersal limited by internal fertilization,
brooding, and crawl-away instars (Cohen 1983; Wakayama 2007; Gerrish
and Morin 2008; Goodheart et al. 2020).
Dispersal is also limited in Luxorina by micro-habitat fidelity and depth specificity (Morin 2019). Dispersal limitation and habitat specificity
should increase endemic diversity in marine environments, especially in shallow waters
adjacent to islands (Pinheiro et al. 2017). As one
example, within the luxorine ostracod Photeros annecohenae, strong genetic
differentiation occurs in populations  100 km apart (Reda 2019). Consistent with studies in other systems like
rift lake cichlid fishes (Wagner et al. 2012), these
factors point to the strong possibility of ecological opportunity interacting with sexual
selection to diversify Luxorina and contribute to its longevity.
100 km apart (Reda 2019). Consistent with studies in other systems like
rift lake cichlid fishes (Wagner et al. 2012), these
factors point to the strong possibility of ecological opportunity interacting with sexual
selection to diversify Luxorina and contribute to its longevity.
While persistence of courtship signaling since its origin over 151 Ma is the rule across
dozens of Luxorina species, at least one exception does exist. Vargula
tsujii, with no known bioluminescent courtship (Cohen and Morin 2003; Goodheart et al.
2020), is a California species now separated by the Isthmus of Panama from its
closest relatives in the Caribbean, which all produce luminous courtship signals. While
Morin and others have searched for luminous ostracods along the North American Pacific
coast, Vargula tsujii remains the only known luxorine species outside the
Caribbean. Most known luxorine species ( 80
80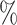 ) in the
Caribbean are collected only as adults using hand nets during brief periods of luminescent
courtship and are not found in baited traps or sand sweep collections. If Luxorina species
in the Pacific have lost their luminescent displays or have different seasonal, lunar or
diel timing for courtship behaviors, they could evade current sampling techniques.
Therefore, additional Luxorina species could exist along the Pacific coast of the Americas
such that extending sampling strategies could uncover additional Luxorina outside the
Caribbean. For the only known luxorine from outside the Caribbean, Vargula
tsujii, our transcriptome analyses infer a sister-group relationship to an
undescribed signaling species from Panama (MFU) we found in very shallow waters near
mangroves, within the provisional genus-level “Z-group.” Presumably, the V.
tsujii lineage was originally contiguously distributed with the rest of Luxorina,
until the closure of the Isthmus of Panama. Vargula tsujii may have lost
courtship signaling after this geological event. Why V. tsujii lost
courtship signaling is challenging to answer because it is a singular event.
) in the
Caribbean are collected only as adults using hand nets during brief periods of luminescent
courtship and are not found in baited traps or sand sweep collections. If Luxorina species
in the Pacific have lost their luminescent displays or have different seasonal, lunar or
diel timing for courtship behaviors, they could evade current sampling techniques.
Therefore, additional Luxorina species could exist along the Pacific coast of the Americas
such that extending sampling strategies could uncover additional Luxorina outside the
Caribbean. For the only known luxorine from outside the Caribbean, Vargula
tsujii, our transcriptome analyses infer a sister-group relationship to an
undescribed signaling species from Panama (MFU) we found in very shallow waters near
mangroves, within the provisional genus-level “Z-group.” Presumably, the V.
tsujii lineage was originally contiguously distributed with the rest of Luxorina,
until the closure of the Isthmus of Panama. Vargula tsujii may have lost
courtship signaling after this geological event. Why V. tsujii lost
courtship signaling is challenging to answer because it is a singular event.
Our inferences of very ancient transitions in bioluminescence traits, including long
persistence of courtship signaling, do depend on taxon sampling and could have been impacted
by a high percentage of missing data in our concatenated data set. However, given that our
coalescence and concatenated analyses resulted in extremely similar species trees, we expect
that more complete gene sampling should not affect our main conclusion. In fact, better
taxon sampling is likely to only make our dating estimates older; for example, if
Enewton is the sister group of all other Luxorina, it would push back the
origin of luminous courtship. Similarly, including species like V.
norvegica and S. orri in divergence estimates adds an older node
within Luminini to make the origin of bioluminescence even older. In addition, because the
ttscm technique does not estimate character state changes at nodes only, it may be less
affected by taxon sampling because it reconstructs distributions of ages. Therefore, the
most conservative interpretation of our results is to use the 95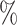 confidence intervals of critical nodes to make minimum estimates for character state
transitions. Assuming such minimum estimates, the origin of courtship signaling is at least
151 Ma and the origin of bioluminescence is at least 197 Ma in cypridinids. Even using these
very conservative minima, the origin of courtship signaling represents long persistence of a
sexually selected trait and Luminini as one of the oldest origins of bioluminescence
quantified thus far.
confidence intervals of critical nodes to make minimum estimates for character state
transitions. Assuming such minimum estimates, the origin of courtship signaling is at least
151 Ma and the origin of bioluminescence is at least 197 Ma in cypridinids. Even using these
very conservative minima, the origin of courtship signaling represents long persistence of a
sexually selected trait and Luminini as one of the oldest origins of bioluminescence
quantified thus far.
Summary
Bioluminescence evolved in the family Cypridinidae independently from other animals, and cypridinids use a substrate, cypridinid-luciferin (Morin 2011), that is endogenous and chemically different from that of other bioluminescent organisms (Hastings 1983; Thompson et al. 1989; Kato et al. 2004, 2007). Therefore, Luminini represent a critical lineage for understanding how bioluminescence evolved (Haddock et al. 2010; Morin 2019; Goodheart et al. 2020; Lau and Oakley 2021). A phylogenetic framework including divergence time estimates provides a foundation for important questions related to molecular evolution and features important for rapid diversification, including those related to behavior (Boake et al. 2002), sexual selection (Ritchie 2007), species diversification (Ellis and Oakley 2016), and the influence of the biochemical properties of bioluminescence on evolution (Hensley et al. 2019).
The macroevolutionary durations of traits under sexual selection are difficult to predict from population-level theory, and understudied empirically. We report the maintenance of bioluminescent courtship in Luxorina ostracods for at least 151 Ma, since it was originally co-opted from bioluminescence used in antipredator displays. Some other taxa also demonstrate long persistence of sexually selected traits (Gu et al. 2012; Bao et al. 2019; Chen and Wiens 2020; Song et al. 2020), but the macroevolutionary durations of such traits is understudied. As our understanding of the duration of sexually selected traits grows across systems, researchers will be able to address open questions about how sexual selection affects biodiversity, while disentangling other contributions such as ecological opportunity and life history characteristics.
Acknowledgments
We acknowledge funding support from the National Science Foundation to THO (DEB-1457754 and DEB-1146337), ET (DEB-1457462), GAG (DEB-1457439), JAG (BIO PRFB-1711201), and EAE (1515576 and 1702011). Permits from the Jamaican National Environment and Planning Agency (Permit Ref. no. 18/27), the Belize Fisheries Department (Permit no. 000003-16), the Honduran Department of Fish and Wildlife (Permit no. DE-MO-082-2016), the Puerto Rican Department of Natural and Environmental Resources (DRNA; Permit no. 2016-IC-113), and Panamanian Ministry of the Environment (MiAMBIENTE; Permit no. SE/A-33-17) were obtained for collections. We thank K. Osborn and J. Fergus for Gigantocypris. We thank Y. Mitani, Y. Ohmiya, and K. Ogoh for assistance collecting Japanese ostracods. Thanks to A. Parker for sending samples from Australia and A. Cohen for much wisdom shared through countless discussions with all of us and for confirming identification of some species.
Contributor Information
Emily A Ellis, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, Santa Barbara, CA 93106, USA.
Jessica A Goodheart, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, Santa Barbara, CA 93106, USA; Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California, San Diego, La Jolla, CA 92037, USA.
Nicholai M Hensley, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, Santa Barbara, CA 93106, USA; Department of Neurobiology and Behavior, Cornell University, Ithaca, NY 14850, USA.
Vanessa L González, Department of Invertebrate Zoology, Smithsonian Institution, National Museum of Natural History, 10th and Constitution NW, Washington, DC 20560-0105, USA.
Nicholas J Reda, Biology Department, University of Wisconsin–La Crosse, La Crosse, WI 54601, USA.
Trevor J Rivers, Department of Ecology and Evolutionary Biology, University of Kansas Lawrence, KS 66045, USA.
James G Morin, Department of Ecology and Evolutionary Biology, Cornell University, Ithaca, NY 14850, USA.
Elizabeth Torres, Department of Biological Sciences, California State University Los Angeles, Los Angeles, CA 90032, USA.
Gretchen A Gerrish, Biology Department, University of Wisconsin–La Crosse, La Crosse, WI 54601, USA; Trout Lake Station, Center for Limnology, University of Wisconsin – Madison, Boulder Junction, WI 54512, USA.
Todd H Oakley, Department of Ecology, Evolution, and Marine Biology, University of California, Santa Barbara, Santa Barbara, CA 93106, USA.
Supplementary Material
Data available from the Dryad Digital Repository: http://dx.doi.org/10.6076/dryad.D1PC7K.
Data Availability
Voucher samples can be accessed through the Australian Museum (AMS) (Promislow 1992; Tanaka 1996; Sorci et al. 1998; Doherty et al. 2003; Morrow and Fricke 2004), National Museum of Natural History (NMNH), Santa Barbara Museum of Natural History (SBMNH), and the Museum of Comparative Zoology (MCZ) (see Supplementary Table S1 available on Dryad for accession numbers). RNA-Seq sequence data can be accessed at the NCBI Sequence Read Archive (SRA) (see Supplementary Table S1 available on Dryad for accession numbers). Aligned data matrices and raw tree files can be accessed at the Dryad Digital Repository (DOI: 10.6076/D1PC7K).
References
- Alexandrou M.A., Swartz B.A., Matzke N.J., Oakley T.H.. 2013. Genome duplication and multiple evolutionary origins of complex migratory behavior in Salmonidae. Mol. Phylogenet. Evol. 69:514–523. [DOI] [PubMed] [Google Scholar]
- Andersson M.B. 1994. Sexual selection. Princeton (NJ): Princeton University Press. [Google Scholar]
- Bacon C.D., Silvestro D., Jaramillo C., Smith B.T., Chakrabarty P., Antonelli A.. 2015. Biological evidence supports an early and complex emergence of the Isthmus of Panama. Proc. Natl. Acad. Sci. USA 112:6110–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao T., Wang B., Li J., Dilcher D.. 2019. Pollination of Cretaceous flowers. Proc. Natl. Acad. Sci. USA 116:24707–24711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner P.O., Rojas-Agramonte Y., Sandoval-Gutierrez M., Urbani F., García-Delgado D., Garban G., Pérez Rodríguez M.. 2013. Late Jurassic breakup of the Proto-Caribbean and circum-global circulation across Pangea. EGU General Assembly Conference Abstracts. 13408. [Google Scholar]
- Beaulieu J.M., O’Meara B.C., Donoghue M.J.. 2013. Identifying hidden rate changes in the evolution of a binary morphological character: the evolution of plant habit in campanulid angiosperms. Syst. Biol. 62:725–737. [DOI] [PubMed] [Google Scholar]
- Bernt M., Donath A., Jühling F., Externbrink F., Florentz C., Fritzsch G., Pütz J., Middendorf M., Stadler P.F.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69:313–319. [DOI] [PubMed] [Google Scholar]
- Boake C.R.B., Arnold S.J., Breden F., Meffert L.M., Ritchie M.G., Taylor B.J., Wolf J.B., Moore A.J.. 2002. Genetic tools for studying adaptation and the evolution of behavior. Am. Nat. 160:S143. [DOI] [PubMed] [Google Scholar]
- Campbell A.K., Herring P.J.. 1990. Imidazolopyrazine bioluminescence in copepods and other marine organisms. Mar. Biol. 104:219–225. [Google Scholar]
- Chen Z., Wiens J.J.. 2020. The origins of acoustic communication in vertebrates. Nat. Commun. 11:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates A.G., Jackson J.B.C., Collins L.S., Cronin T.M., Dowsett H.J., Bybell L.M., Jung P., Obando J.A.. 1992. Closure of the Isthmus of Panama: the near-shore marine record of Costa Rica and western Panama. Geol. Soc. Am. Bull. 104:814–828. [Google Scholar]
- Cohen A.C. 1983. Rearing and postembryonic development of the myodocopid ostracode Skogsbergia lerneri from coral reefs of Belize and the Bahamas. J. Crustacean Biol. 3:235–256. [Google Scholar]
- Cohen A.C., Morin J.G.. 1986. Three new luminescent ostracodes of the genus Vargula (Myodocopida, Cypridinidae) from the San Blas region of Panama. Contrib. Sci. (Los Angel.). 373:1–23. [Google Scholar]
- Cohen A.C., Morin J.G.. 1989. Six new luminescent Ostracodes of the Genus Vargula (Myodocopida: Cypridinidae) from the San Blas Region of Panama. J. Crustacean Biol. 9:297–340. [Google Scholar]
- Cohen A.C., Morin J.G.. 1993. The cypridinid copulatory limb and a new genus Kornickeria (Ostracoda: Myodocopida) with four new species of bioluminescent ostracods from the Caribbean. Zool. J. Linn. Soc. 108:23–84. [Google Scholar]
- Cohen A.C., Morin J.G.. 2003. Sexual morphology, reproduction and the evolution of bioluminescence in Ostracoda. Paleontol. Soc. Pap. 9:37–70. [Google Scholar]
- Cohen A.C., Morin J.G.. 2010. Two new bioluminescent Ostracode genera, Enewton and Photeros (Myodocopida: Cypridinidae), with three new species from Jamaica. J. Crustacean Biol. 30:1–55. [Google Scholar]
- Cohen A.C., Oakley T.H.. 2017. Collecting and processing marine ostracods. J. Crustacean Biol. 37:347–352. [Google Scholar]
- Doherty P.F. Jr, Sorci G., Royle J.A., Hines J.E., Nichols J.D., Boulinier T.. 2003. Sexual selection affects local extinction and turnover in bird communities. Proc. Natl. Acad. Sci. USA 100:5858–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis E.A., Oakley T.H.. 2016. High rates of species accumulation in animals with bioluminescent courtship displays. Curr. Biol. 26:1916–1921. [DOI] [PubMed] [Google Scholar]
- Emms D.M., Kelly S.. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris D.W., Jaramillo C., Bayona G., Restrepo-Moreno S.A., Montes C., Cardona A., Mora A., Speakman R.J., Glascock M.D., Valencia V.. 2011. Fracturing of the Panamanian Isthmus during initial collision with South America. Geology 39:1007–1010. [Google Scholar]
- Gerrish G.A., Morin J.G.. 2008. Life cycle of a bioluminescent marine Ostracode, Vargula annecohenae (Myodocopida: Cypridinidae). J. Crustacean Biol. 28:669–674. [Google Scholar]
- Gerrish G.A., Morin J.G.. 2016. Living in sympatry via differentiation in time, space and display characters of courtship behaviors of bioluminescent marine ostracods. Mar. Biol. 163:190. [Google Scholar]
- Gerrish G.A., Morin J.G., Rivers T.J., Patrawala Z.. 2009. Darkness as an ecological resource: the role of light in partitioning the nocturnal niche. Oecologia 160:525–536. [DOI] [PubMed] [Google Scholar]
- Goodheart J.A., Minsky G., Brynjegard-Bialik M.N., Drummond M.S., Munoz J.D., Fallon T.R., Schultz D.T., Weng J.-K., Torres E., Oakley T.H.. 2020. Laboratory culture of the California Sea Firefly Vargula tsujii (Ostracoda: Cypridinidae): developing a model system for the evolution of marine bioluminescence. Sci. Rep. 10:10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A.. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J.-J., Montealegre-Z F., Robert D., Engel M.S., Qiao G.-X., Ren D.. 2012. Wing stridulation in a Jurassic katydid (Insecta, Orthoptera) produced low-pitched musical calls to attract females. Proc. Natl. Acad. Sci. USA 109:3868–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., MacManes M.D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C.N., Henschel R., LeDuc R.D., Friedman N., Regev A.. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddock S.H.D., Moline M.A., Case J.F.. 2010. Bioluminescence in the sea. Ann. Rev. Mar. Sci. 2:443–493. [DOI] [PubMed] [Google Scholar]
- Hastings J.W. 1983. Biological diversity, chemical mechanisms, and the evolutionary origins of bioluminescent systems. J. Mol. Evol. 19:309–321. [DOI] [PubMed] [Google Scholar]
- Hensley N.M., Ellis E.A., Gerrish G.A., Torres E., Frawley J.P., Oakley T.H., Rivers T.J.. 2019. Phenotypic evolution shaped by current enzyme function in the bioluminescent courtship signals of sea fireflies. Proc. Biol. Sci. 286:20182621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley N.M., Ellis E.A., Leung N.Y., Coupart J., Mikhailovsky A., Taketa D.A., Tessler M., Gruber D.F., De Tomaso A.W., Mitani Y., Rivers T.J., Gerrish G.A., Torres E., Oakley T.H.. 2021. Selection, drift, and constraint in cypridinid luciferases and the diversification of bioluminescent signals in sea fireflies. Mol. Ecol. 30:1864–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhna S., Lower S.E., Duchen P., Catalán A.. 2021. A time-calibrated firefly (Coleoptera: Lampyridae) phylogeny: using genomic data for divergence time estimation. bioRxiv:2021.11.19.469195. [Google Scholar]
- Huelsenbeck J.P., Nielsen R., Bollback J.P.. 2003. Stochastic mapping of morphological characters. Syst. Biol. 52:131–158. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S.-I., Oba Y., Ojika M.. 2007. Biosynthesis of cypridina luciferin in Cypridina noctiluca. Heterocycles 72:673–676. [Google Scholar]
- Kato S.-I., Oba Y., Ojika M., Inouye S.. 2004. Identification of the biosynthetic units of Cypridina luciferin in Cypridina (Vargula) hilgendorfii by LC/ESI-TOF-MS. Tetrahedron 60:11427–11434. [Google Scholar]
- Kokko H., Brooks R.. 2003. Sexy to die for? Sexual selection and the risk of extinction. Ann. Zool. Fennici. 40:207–219. [Google Scholar]
- van der Kooi C.J., Ollerton J.. 2020. The origins of flowering plants and pollinators. Science 368:1306–1308. [DOI] [PubMed] [Google Scholar]
- Krueger F. 2012. Trim Galore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) libraries. Available from: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Access date: April28, 2016. [Google Scholar]
- Lanfear R., Calcott B., Kainer D., Mayer C., Stamatakis A.. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 14:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R., Frandsen P.B., Wright A.M., Senfeld T., Calcott B.. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34:772–773. [DOI] [PubMed] [Google Scholar]
- Lau E.S., Oakley T.H.. 2021. Multi-level convergence of complex traits and the evolution of bioluminescence. Biol. Rev. 96:673–691. [DOI] [PubMed] [Google Scholar]
- Lepage T., Bryant D., Philippe H., Lartillot N.. 2007. A general comparison of relaxed molecular clock models. Mol. Biol. Evol. 24:2669–2680. [DOI] [PubMed] [Google Scholar]
- Long T.A.F., Agrawal A.F., Rowe L.. 2012. The effect of sexual selection on offspring fitness depends on the nature of genetic variation. Curr. Biol. 22:204–208. [DOI] [PubMed] [Google Scholar]
- Lumley A.J., Michalczyk Ł., Kitson J.J.N., Spurgin L.G., Morrison C.A., Godwin J.L., Dickinson M.E., Martin O.Y., Emerson B.C., Chapman T., Gage M.J.G.. 2015. Sexual selection protects against extinction. Nature 522:470–473. [DOI] [PubMed] [Google Scholar]
- Martínez-Ruiz C., Knell R.J.. 2017. Sexual selection can both increase and decrease extinction probability: reconciling demographic and evolutionary factors. J. Anim. Ecol. 86:117–127. [DOI] [PubMed] [Google Scholar]
- Martins M.J.F., Puckett T.M., Lockwood R., Swaddle J.P., Hunt G.. 2018. High male sexual investment as a driver of extinction in fossil ostracods. Nature 556:366–369. [DOI] [PubMed] [Google Scholar]
- Minh B.Q., Hahn M.W., Lanfear R.. 2020. New methods to calculate concordance factors for phylogenomic datasets. Mol. Biol. Evol. 37:2727–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.S., Rabosky D.L.. 2017. Bayesian model selection with BAMM: effects of the model prior on the inferred number of diversification shifts. Methods Ecol. Evol. 8:37–46. [Google Scholar]
- Morin J.G. 1983. Coastal bioluminescence: patterns and functions. Bull. Mar. Sci. 33:787–817. [Google Scholar]
- Morin J.G. 1986. Firefleas of the sea: luminescent signaling in marine Ostracode crustaceans. Florida Entomol. 69:105. [Google Scholar]
- Morin J.G. 2011. Based on a review of the data, use of the term “cypridinid” solves the Cypridina/Vargula dilemma for naming the constituents of the luminescent system of ostracods in the family Cypridinidae. Luminescence 26:1–4. [DOI] [PubMed] [Google Scholar]
- Morin J.G. 2019. Luminaries of the reef: the history of luminescent ostracods and their courtship displays in the Caribbean. J. Crustacean Biol. 39:227–243. [Google Scholar]
- Morin J.G., Cohen A.C.. 1988. Two new luminescent ostracodes of the genus Vargula (Myodocopida, Cypridinidae) from the San Blas region of Panama. J. Crustacean Biol. 8:620–638. [Google Scholar]
- Morin J.G., Cohen A.C.. 2010. It’s all about sex: bioluminescent courtship displays, morphological variation and sexual selection in two new genera of Caribbean Ostracodes. J. Crustacean Biol. 30:56–67. [Google Scholar]
- Morin J.G., Cohen A.C.. 2017. A guide to the morphology of bioluminescent signaling cypridinid ostracods from the Caribbean Sea, and a tabular key to the genera. Zootaxa 4303:301–349. [Google Scholar]
- Morrow E.H., Fricke C.. 2004. Sexual selection and the risk of extinction in mammals. Proc. Biol. Sci. 271:2395–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley T.H. 2005. Myodocopa (Crustacea: Ostracoda) as models for evolutionary studies of light and vision: multiple origins of bioluminescence and extreme sexual dimorphism. Hydrobiologia 538:179–192. [Google Scholar]
- Oakley T.H., Wolfe J.M., Lindgren A.R., Zaharoff A.K.. 2013. Phylotranscriptomics to bring the understudied into the fold: monophyletic ostracoda, fossil placement, and pancrustacean phylogeny. Mol. Biol. Evol. 30:215–233. [DOI] [PubMed] [Google Scholar]
- O’Dea A., Lessios H.A., Coates A.G., Eytan R.I., Restrepo-Moreno S.A., Cione A.L., Collins L.S., de Queiroz A., Farris D.W., Norris R.D., Stallard R.F., Woodburne M.O., Aguilera O., Aubry M.-P., Berggren W.A., Budd A.F., Cozzuol M.A., Coppard S.E., Duque-Caro H., Finnegan S., Gasparini G.M., Grossman E.L., Johnson K.G., Keigwin L.D., Knowlton N., Leigh E.G., Leonard-Pingel J.S., Marko P.B., Pyenson N.D., Rachello-Dolmen P.G., Soibelzon E., Soibelzon L., Todd J.A., Vermeij G.J., Jackson J.B.C.. 2016. Formation of the Isthmus of Panama. Sci. Adv. 2:e1600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogoh K., Ohmiya Y.. 2004. Complete mitochondrial DNA sequence of the sea-firefly, Vargula hilgendorfii (Crustacea, Ostracoda) with duplicate control regions. Gene 327:131–139. [DOI] [PubMed] [Google Scholar]
- Ogoh K., Ohmiya Y.. 2005. Biogeography of luminous marine ostracod driven irreversibly by the Japan current. Mol. Biol. Evol. 22:1543–1545. [DOI] [PubMed] [Google Scholar]
- Ooms J., Chamberlain S.. 2019. phylocomr: interface to “Phylocom.” Comprehensive R Archive Network (CRAN). [Google Scholar]
- Pagel M. 1999. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst. Biol. 48:612–622. [Google Scholar]
- Pham H.T.M., Tanaka H., Karanovic I.. 2020. Molecular and morphological diversity of Heterodesmus Brady and its phylogenetic position within Cypridinidae (Ostracoda). Zool. Sci. 37:240. [DOI] [PubMed] [Google Scholar]
- Pinheiro H.T., Bernardi G., Simon T., Joyeux J.-C., Macieira R.M., Gasparini J.L., Rocha C., Rocha L.A.. 2017. Island biogeography of marine organisms. Nature 549:82–85. [DOI] [PubMed] [Google Scholar]
- Plesnar-Bielak A., Skrzynecka A.M., Prokop Z.M., Radwan J.. 2012. Mating system affects population performance and extinction risk under environmental challenge. Proc. Biol. Sci. 279:4661–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell G.S., Saxton N.A., Pacheco Y.M., Stanger-Hall K.F., Martin G.J., Kusy D., da Silveira L.F.L., Bocak L., Branham M.A., Bybee S.M.. 2022. Beetle bioluminescence outshines aerial predators. Proc. R. Soc. B. 289:20220821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P.. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow D.E.L. 1992. Costs of sexual selection in natural populations of mammals. Proc. R. Soc. Lond. B 247:203–210. [Google Scholar]
- Prum R.O. 2017. The evolution of beauty: how Darwin’s forgotten theory of mate choice shapes the animal world - and us. New York: Knopf Doubleday Publishing Group. [Google Scholar]
- Reda N.J. 2019. Capturing speciation in action: rapid population divergence in the Caribbean bioluminescent ostracod Photeros annecohenae (Myodocopida: Cypridinidae). [Google Scholar]
- Reda N.J., Morin J.G., Torres E., Cohen A.C., Schawaroch V., Gerrish G.A.. 2019. Maristella, a new bioluminescent ostracod genus in the Myodocopida (Cypridinidae). Zool. J. Linn. Soc. [Google Scholar]
- Revell L.J. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3:217–223. [Google Scholar]
- Ritchie M.G. 2007. Sexual selection and speciation. Annu. Rev. Ecol. Evol. Syst. 38:79–102. [Google Scholar]
- Rivers T.J., Morin J.G.. 2008. Complex sexual courtship displays by luminescent male marine ostracods. J. Exp. Biol. 211: 2252–2262. [DOI] [PubMed] [Google Scholar]
- Rivers T.J., Morin J.G.. 2012. The relative cost of using luminescence for sex and defense: light budgets in cypridinid ostracods. J. Exp. Biol. 215:2860–2868. [DOI] [PubMed] [Google Scholar]
- Rivers T.J., Morin J.G.. 2013. Female ostracods respond to and intercept artificial conspecific male luminescent courtship displays. Behav. Ecol. 24:877–887. [Google Scholar]
- Ronquist F., Huelsenbeck J.P.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Ryan M.J., Akre K.L., Baugh A.T., Bernal X.E., Lea A.M., Leslie C., Still M.B., Wylie D.C., Rand A.S.. 2019. Nineteen years of consistently positive and strong female mate preferences despite individual variation. Am. Nat. 194:125–134. [DOI] [PubMed] [Google Scholar]
- Schön I., Rodriguez F., Dunn M., Martens K., Shribak M., Arkhipova I.R.. 2021. A survey of transposon landscapes in the putative ancient asexual Ostracod Darwinula stevensoni. Genes 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwentner M., Richter S., Rogers D.C., Giribet G.. 2018. Tetraconatan phylogeny with special focus on Malacostraca and Branchiopoda: highlighting the strength of taxon-specific matrices in phylogenomics. Proc. Biol. Sci. 285:20181524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- Simion P., Belkhir K., François C., Veyssier J., Rink J.C., Manuel M., Philippe H., Telford M.J.. 2018. A software tool “CroCo” detects pervasive cross-species contamination in next generation sequencing data. BMC Biol. 16:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.A., Brown J.W., Walker J.F.. 2018. So many genes, so little time: a practical approach to divergence-time estimation in the genomic era. PLoS One 13:e0197433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Béthoux O., Shin S., Donath A., Letsch H., Liu S., McKenna D.D., Meng G., Misof B., Podsiadlowski L., Zhou X., Wipfler B., Simon S.. 2020. Phylogenomic analysis sheds light on the evolutionary pathways towards acoustic communication in Orthoptera. Nat. Commun. 11:4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorci G., Møller A.P., Clobert J.. 1998. Plumage dichromatism of birds predicts introduction success in New Zealand. J. Anim. Ecol. 67:263–269. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y. 1996. Sexual selection enhances population extinction in a changing environment. J. Theor. Biol. 180:197–206. [DOI] [PubMed] [Google Scholar]
- Thalén F. 2018. PhyloPyPruner: tree-based orthology inference for phylogenomics with new methods for identifying and excluding contamination [Masters Degree]. Lund University. [Google Scholar]
- Thompson E.M., Nagata S., Tsuji F.I.. 1989. Cloning and expression of cDNA for the luciferase from the marine ostracod Vargula hilgendorfii. Proc. Natl. Acad. Sci. USA 86:6567–6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinn O., Oakley T.H.. 2008. Erratic rates of molecular evolution and incongruence of fossil and molecular divergence time estimates in Ostracoda (Crustacea). Mol. Phylogenet. Evol. 48:157–167. [DOI] [PubMed] [Google Scholar]
- Tomkins J.L., Radwan J., Kotiaho J.S., Tregenza T.. 2004. Genic capture and resolving the lek paradox. Trends Ecol. Evol. 19:323–328. [DOI] [PubMed] [Google Scholar]
- Torres E., Gonzalez V.L.. 2007. Molecular phylogeny of cypridinid ostracodes and the evolution of bioluminescence. Proceedings of the 14th International Symposium on Bioluminescence and Chemiluminescence: Chemistry, Biology, and Applications. p. 269–272. [Google Scholar]
- Wagner C.E., Harmon L.J., Seehausen O.. 2012. Ecological opportunity and sexual selection together predict adaptive radiation. Nature 487:366–369. [DOI] [PubMed] [Google Scholar]
- Wakayama N. 2007. Embryonic development clarifies polyphyly in ostracod crustaceans. J. Zool. 273:406–413. [Google Scholar]
- Wakayama N., Abe K.. 2006. The evolutionary pathway of light emission in myodocopid Ostracoda. Biol. J. Linn. Soc. Lond. 87:449–455. [Google Scholar]
- Wang X., Xu Q., Xiao J., Miao X., Liu P., Wang Z.. 2019. First record of the complete mitochondrial genome of Cypridina dentata (Myodocopida: Cypridinidae). Mitochondrial DNA B 4: 1607–1608. [Google Scholar]
- Webb C.O., Ackerly D.D., Kembel S.W.. 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24:2098–2100. [DOI] [PubMed] [Google Scholar]
- Widder E.A. 2010. Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science 328:704–708. [DOI] [PubMed] [Google Scholar]
- Wiens J.J., Tuschhoff E.. 2020. Songs versus colours versus horns: what explains the diversity of sexually selected traits? Biol. Rev. Camb. Philos. Soc. 95:847–864. [DOI] [PubMed] [Google Scholar]
- Wilson T., Hastings J.W.. 1998. Bioluminescence. Annu. Rev. Cell Dev. Biol. 14:197–230. [DOI] [PubMed] [Google Scholar]
- Wolfe J.M., Daley A.C., Legg D.A., Edgecombe G.D.. 2016. Fossil calibrations for the arthropod tree of life. Earth-Sci. Rev. 160:43–110. [Google Scholar]
- Zhang C. 2016. Molecular clock dating using MrBayes. arXiv.1603.05707v2. [Google Scholar]
- Zhang C., Scornavacca C., Molloy E.K., Mirarab S.. 2020. ASTRAL-Pro: quartet-based species-tree inference despite paralogy. Mol. Biol. Evol. 37:3292–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Voucher samples can be accessed through the Australian Museum (AMS) (Promislow 1992; Tanaka 1996; Sorci et al. 1998; Doherty et al. 2003; Morrow and Fricke 2004), National Museum of Natural History (NMNH), Santa Barbara Museum of Natural History (SBMNH), and the Museum of Comparative Zoology (MCZ) (see Supplementary Table S1 available on Dryad for accession numbers). RNA-Seq sequence data can be accessed at the NCBI Sequence Read Archive (SRA) (see Supplementary Table S1 available on Dryad for accession numbers). Aligned data matrices and raw tree files can be accessed at the Dryad Digital Repository (DOI: 10.6076/D1PC7K).