Abstract
Clonal tracking methods provide quantitative insights into the cellular output of genetically labelled progenitor cells across time and cellular compartments. In the context of gene and cell therapies, clonal tracking methods have enabled the tracking of progenitor cell output both in humans receiving therapies and in corresponding animal models, providing valuable insight into lineage reconstitution, clonal dynamics, and vector genotoxicity. However, the absence of a toolbox for analysis of clonal tracking data has precluded the development of standardized analytical frameworks within the field. Thus, we developed barcodetrackR, an R package and accompanying Shiny app containing diverse tools for the analysis and visualization of clonal tracking data. We demonstrate the utility of barcodetrackR in exploring longitudinal clonal patterns and lineage relationships in a number of clonal tracking studies of hematopoietic stem and progenitor cells (HSPCs) in humans receiving HSPC gene therapy and in animals receiving lentivirally transduced HSPC transplants or tumor cells.
INTRODUCTION
Genetic labelling permits quantitative tracking of clonal progeny via high-throughput sequencing (clonal tracking) and provides opportunities to interrogate clonal dynamics in a number of in vitro and in vivo contexts. The two most common clonal tracking approaches, cellular barcoding and viral integration site recovery, have been primarily leveraged to interrogate hematopoietic stem and progenitor cell (HSPC) or immune cell dynamics both in model animals1-6 and in humans7,8. In these methodologies, integrating retro- or lentiviruses are used to transduce individual HSPCs or other target populations such that individual cells each contain a unique, permanent genetic tag or integration site label that can be recovered from progeny cells via high throughput sequencing (Fig. 1, upper panel). These genetic tags are referred to as “barcodes” in the case of cellular barcoding. The genetically tagged cell and its progeny are referred to as a clone. Measurement of each label’s abundance in the pool of all recovered labels is directly associated with the abundance of that clone within the labelled population being assayed, for instance T cells, B cells or myeloid cells. These lineage abundance measurements can provide insights not only into the bias, stability, and ontogenetic relationships of HSPCs9, but also into the dynamics of clones within cell populations whose abundances are largely independent of HSPC behavior, such as certain T cell10 and natural killer (NK) cell populations11. Furthermore, such clonal tracking methods have also been leveraged to provide valuable insight into the clonal dynamics of cancer progression12, in vitro differentiation13, and CAR-T cells14. The biologic and clinical relevance of clonal tracking studies has recently been reviewed.15
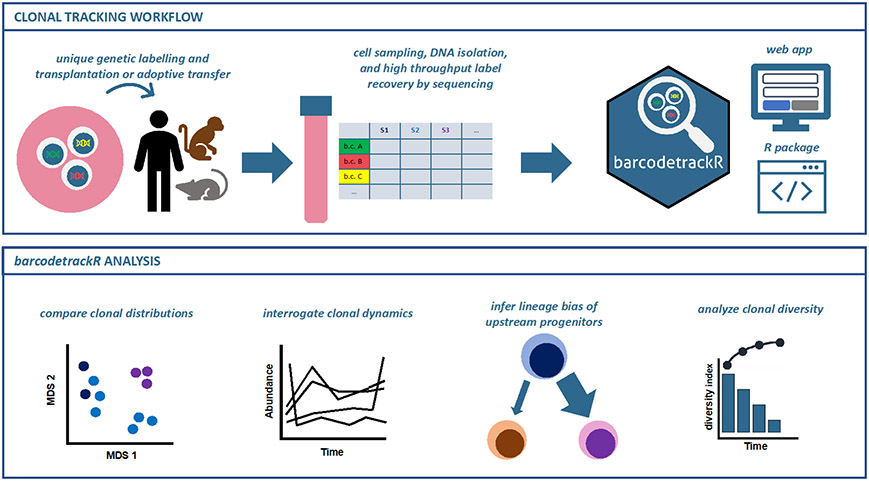
Figure 1: Clonal tracking experimental design and barcodetrackR analysis.
Clonal tracking experiments generally follow the depicted framework, which encompasses, in order, the genetic labelling of cells to create an integrated DNA tag, transplantation or adoptive transfer of these cells into a recipient, acquisition of cellular progeny from the recipient following transplantation or adoptive transfer, genetic tag retrieval from progeny cells followed by high-throughput sequencing, algorithmic quantification of detected individual unique tags, and finally, downstream analyses, where the barcodetrackR toolkit can be utilized (top panel). barcodetrackR contains functions for analyzing relationships between sample clonal distributions, interrogating clonal dynamics over time, inferring pairwise lineage bias of upstream progenitors, and analyzing longitudinal sample clonal diversity (bottom panel).
Given the diversity of labelling and recovery strategies, as well as underlying differences in vector constructs, a number of approaches for recovery of sequences from raw sequencing data and identification of “true” genetic tags as opposed to sequencing artifacts or other confounders have been developed and are largely approach-dependent16-26. However, tools with which to perform downstream analyses of the clonal abundances determined by these pipelines have not been published or made publicly available; as a result, flexible open-source tools, such as those that exist for single-cell RNA-sequencing27,28 have been sought after by those in the clonal tracking field in order to derive biological meaning in an accessible manner from these large datasets29. Such tools would also allow direct comparisons across datasets or meta-analyses.
Here, we present our open-source R package, barcodetrackR. barcodetrackR encompasses a variety of flexible tools that can provide insights into clonal dynamics and the relationships between cellular compartments starting with clonal abundance data (Fig. 1, lower panel). We illustrate the utility of barcodetrackR by analyzing publicly available clonal tracking datasets from studies in lentivirally transduced non-human primates9,11,30, immunodeficient mice transplanted with human cord blood cells31 or leukemic blast cells32, and lentiviral gene therapy patients7,33. More details on each dataset and access paths are summarized in Supplementary Table 1.
RESULTS
Infrastructure of the barcodetrackR package
barcodetrackR is an R package and an accompanying Shiny app for the analysis and visualization of clonal tracking data (Fig. 1). barcodetrackR depends on the Bioconductor SummarizedExperiment (SE) class34 for the organization of user-input clonal tracking data and accompanying metadata. By default, the SE object is instantiated with raw read counts, from which five assays are automatically calculated and stored in the object for downstream analysis and visualization: counts (the raw genetic tag counts), proportions (raw genetic tag counts divided by the total count for each sample), normalized (counts normalized to 1 * 106 or another specified scaling factor), logs (the log + 1 of the normalized assay), and ranks (the rank of each genetic tag per sample based on counts). A relative or absolute threshold can be specified to remove low abundance genetic tags which are more likely to arise from sequencing error or noise.2 We illustrate the distribution of genetic tag proportions and logs for three example datasets across various thresholding parameters in Supplementary Fig. 1 (see Methods for more detail on thresholding).
The barcodetrackR package contains 25 available functions, including functions for visualization of clonal tracking data, functions for conducting statistical tests on genetic tag abundances, and a function for estimating a minimum abundance threshold to determine reliable genetic tags. All visualization functions allow for the return of a plot object or a table of the plot data for storage and reproducibility. The barcodetrackR package also includes a graphical user interface or app (built using Shiny35) to allow researchers without programming experience to utilize these quantitative tools. In the following section, we highlight key functionalities of the barcodetrackR package by analyzing a set of publicly-available clonal tracking datasets (summarized in Supplementary Table 1).
Pairwise lineage relationships
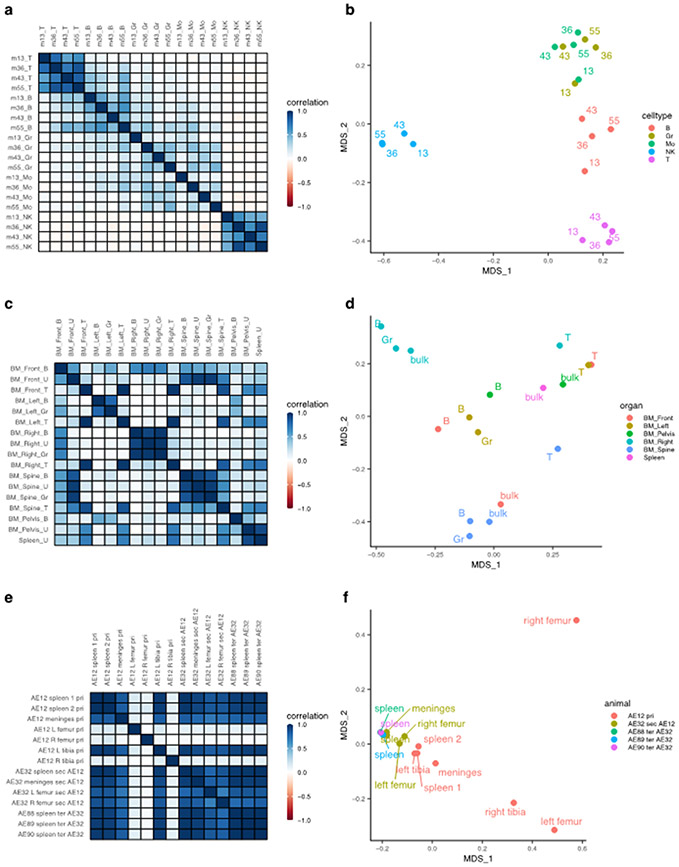
Pairwise comparisons of clonal abundance profiles in clonal tracking data provide insight into the relationships between upstream progenitor pools across cellular compartments. Here, we use barcodetrackR to determine and visualize the correlation values and dissimilarity indices (Fig. 2) between samples from three clonal tracking datasets (Six7 et al, Belderbos31 et al, Elder32 et al, Supplementary Table 1) as a means to interrogate the similarities of upstream progenitor pools contributing across cellular compartments. The Six dataset contains individual viral integration site read counts from longitudinally collected patient T cell, B cell, granulocyte (Gr), monocyte (Mo), and natural killer cell (NK) samples collected from patients following autologous lentiviral HSPC gene therapy. We find that the Gr and Mo samples share high correlation with one another, while the T cell, B cell, and NK samples show lower correlation with samples from other lineages, but high correlation between different timepoints within the same lineage (Fig. 2a). This trend is supported by hierarchically clustering the samples based on their correlation values (Supplementary Fig. 2a). A similar pattern is observed when plotting the Bray-Curtis dissimilarity indices between samples from the Six dataset, projected into two dimensions using principal coordinates analysis (PCoA), where the first axis of variation separates NK cells from other lineages based on their clonal abundances, and the second separates T cells, B cells, and myeloid (Gr and Mo) cells (Fig. 2b). These analyses suggest that the myeloid lineages are closely coupled and thus likely arise from shared pathways originating from the same HSPC pool, in comparison to disparate generation of mature T, B, and NK lineages.
Figure 2: Global clonal distributions.
(a) Pairwise Pearson correlation plots between longitudinal samples from the Six dataset. Row and column labels indicate months post-transplant (m) and cell type (T, T cell; B, B cell; Gr, Granulocyte; Mo, Monocyte; NK, Natural Killer cell). (b) Bray-Curtis dissimilarity indices between samples from the Six dataset, grouped by cell type and labeled based on months post-transplant. The x and y-axis represent the two main axes of variation after conducting principal coordinate analysis on the Bray-Curtis measures of dissimilarity (MDS). (c) Pairwise Pearson correlation plots between samples from different anatomical sites of a single transplanted mouse at euthanasia from the Belderbos dataset. Row and columns labels describe the anatomical site (BM, Bone Marrow) followed by the cell type (B, B cell; U, Unsorted samples; T, T cell; Gr, Granulocyte). (d) Bray-Curtis dissimilarity indices between samples from the Belderbos dataset grouped by the anatomical site and labeled by cell type. (e) Pairwise Pearson correlation values between samples of the same set of serial xenograft transplants from the Elder dataset. Row and column labels describe the animal code (e.g. AE12), followed by the anatomical site, then the serial transplant designation (pri, Primary; sec, Secondary, ter, Tertiary), followed by the donor animal code if it is a sec or ter sample. AE12 is the primary recipient of ALL blast cells, AE32 is the secondary recipient receiving cells from the primary animal AE12, and AE88, AE89, AE90 are tertiary recipients receiving cells from AE32. (f) Bray-Curtis dissimilarity indices between samples from the Elder dataset colored by the mouse of origin and labeled by the anatomical site.
These pairwise measures can be also used to compare clonal abundances across anatomical compartments. The Belderbos dataset contains clonal abundance information from a number of sorted and unsorted immune cell samples from bone marrow (BM) sites and the spleen of an immunodeficient mouse transplanted with lentivirally barcoded human cord blood CD34+ HSPCs (defined here, barcodes are high-diversity oligonucleotide sequences engineered into lentiviruses, typically flanked by known PCR primer sites to facilitate genetic tag abundance recovery). We observe high correlation between T cell samples across all anatomical sites, while B cell and Gr samples show high correlation to one another only within each anatomical site (Fig. 2c). This pattern is evident in the PCoA plot (Fig. 2d) and also supported by hierarchical clustering of the samples based on the similarity matrix of correlation values (Supplementary Fig. 2b). Unsorted cell samples from the spleen and pelvic BM vary in their clonal relationships to other samples, likely because of the underlying heterogeneity of the lineage composition of these bulk samples (Fig. 2c). These analyses support the notion that geographically isolated HSPC pools are responsible for the clonal composition of their respective geographic niches, and that the clonal composition of T cells across sites is largely independent from the output of these pools, supporting the canonical thymic-dependent developmental pathway for T cells followed by hematogenous dissemination.
Comparing clonal distribution across animals from serial transplant experiments can also provide insight into the self-renewal capacity of engrafted, clonally marked cells. The Elder dataset contains clonal abundance information from serial xenograft mouse transplants of lentiviral-transduced ALL blast cells. We observed high correlation of clonal abundances between samples collected from primary, secondary, and tertiary transplant recipient mice, excluding a few sites in the primary transplant (Fig. 2e). This aligns with the results presented by Elder32 et al noting equipotential functional capability of ALL cells with some variation between sites, based on random sampling of the population of engrafted ALL cells. Samples from the same “generation” of serial transplantation cluster together in principal coordinate space, supporting the notion presented in the study that ALL founder cells retain self-renewal capacity over several serial transplants (Fig. 2f). The distinct groupings of clones based on anatomic sampling site within the primary transplanted animals suggests that ALL clonal output also appears to be geographically compartmentalized, at least initially. This geographic compartmentalization may be reduced in secondary and tertiary recipients because the transplanted clonal pool is smaller and less diverse32, limiting the stochastic composition variation.
Altogether, these three examples illustrate the utility of barcodetrackR in probing global clonal relationships between samples collected from various lineages, locations or time points, providing valuable insights across a number of diverse biologic contexts.
Lineage clonality
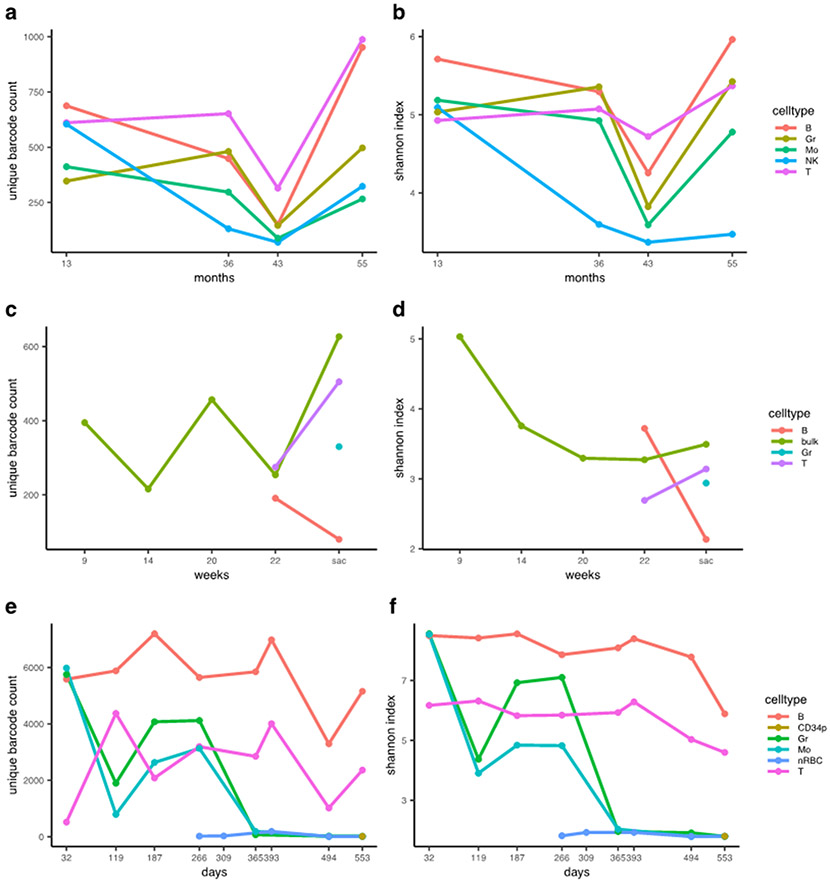
In clonal tracking studies, both clonal counts and diversity measures can provide insight into the clonality of progenitor cell pools. Here, we utilize barcodetrackR to assess clonality by visualizing the detected clone counts and Shannon diversities of samples from three datasets over time (Fig. 3). When quantifying clone numbers within the Six dataset, we show hundreds of unique integration sites retrieved across five purified peripheral blood lineage samples, with a larger number of clones detected in B cells and T cells as compared to Gr, Mo, and NK cells at most individual timepoints (Fig. 3a). Decreasing clonal diversity (Shannon index) in the NK cell lineage, as compared to other lineages, indicates that over time, a smaller number of clones account for a larger fraction of hematopoiesis in the NK cell compartment (Fig 3b). This implies a more oligoclonal population of contributing progenitors. The finding that the mature NK cell compartment is largely composed of a few high-contributing clones post-transplantation is in agreement with rhesus macaque autologous HSPC transplant studies11.
Figure 3: Measures of clonal diversity.
(a-b) The number of clones detected (a) and the Shannon diversity index (b) of each sample from the Six dataset, grouped by lineage. (c-d) The number of clones (c) and the Shannon diversity index (d) of each sample from the Belderbos dataset, grouped by lineage. (e-f) The number of clones detected (e) and the Shannon diversity index (f) of each sample from the Espinoza dataset, grouped by lineage. X-axes show months, weeks or days post-transplant with “sac” corresponding to the timepoint of euthanasia in the Belderbos dataset. The clone count reflects the number of unique clones detected in each sample, not the cumulative count at each timepoint. Shannon diversity is calculated on a per-sample basis based on the clonal population of each sample, not the cumulative number of clones. B, B cell; Gr, Granulocyte; Mo, Monocyte; NK, Natural Killer cell; T, T cell; bulk, unsorted population; CD34p, CD34 positive cell; nRBC, nucleated Red Blood Cell.
Next, we analyzed patterns in the number of detected clones and Shannon diversities over time in peripheral blood samples from a single mouse xenograft obtained from the Belderbos dataset. We show that more clones were detected from the bulk peripheral blood sample at sacrifice than at the first time point (green line, Fig. 3c). The Shannon diversity of bulk samples decreased after the 9-week time point before stabilizing (Fig. 3d), underscoring the notion that clone counts alone are not ideal measures of sample diversity. These findings suggest that within this xenograft transplantation model, the diversity of HSPC output becomes stable over time, in agreement with previous long term clonal tracking studies in macaque9 and human8.
Finally, we use barcodetrackR to quantify an extreme case of minimal clonal counts and Shannon diversities in the context of clonal hematopoiesis using the Espinoza30 dataset (Supplementary Table 1). In this study, multiple lentiviral insertions in a single HSPC eventually resulted in clonal erythroid and myeloid expansion and genotoxic abnormal dysplastic differentiation, while largely sparing the lymphoid lineages. In agreement with the findings of the study, we find that the longitudinal clone numbers contributing to the B and T cell lineages fluctuate, but that the Shannon diversity index of these lineages remains high, especially at early time points, indicating polyclonal contribution to the lymphoid lineages (Fig 3e-f). However, after day 266 post-transplant, we observe a massive drop-off in both the number of unique clones detected (Fig 3e) and the Shannon diversity (Fig 3f) within the myeloid lineages. This coincides chronologically with the development of dysplastic abnormal clonal hematopoiesis in the myeloid lineage.
While in the above examples we utilize unique detected clones at each time point, cumulative clone counts can also be calculated (Supplementary Fig. 3) and provide a complementary view of clone numbers over time. Altogether, these examples emphasize the utility of clonal counts and diversity measures in interrogating the clonal output of progenitor pools in a number of contexts.
Longitudinal clonal dynamics
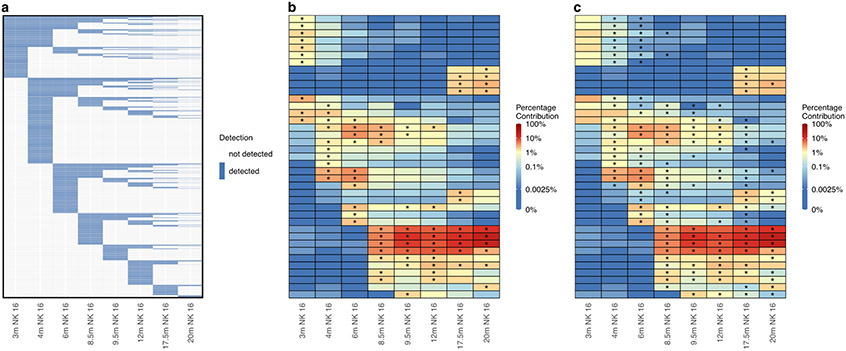
Longitudinal tracking of the abundance of individual clones can provide insight into clonal dynamics within lineages. We employed barcodetrackR to analyze longitudinal NK cell samples from an animal in the Wu11 et al study (Supplementary Table 1), in which NK cell clonal dynamics were interrogated over 3 years in rhesus macaques receiving lentivirally barcoded autologous HSPCs. The detected clones in the NK cell compartment remained largely independent from the HSPC pool responsible for the majority of non-NK hematopoiesis. We first visualize all individual NK cell clones from the Wu dataset in a binary heat map that depicts the presence or absence of all clones observed at 0.01% abundance or greater in at least one NK cell sample (Fig 4a). We find that new NK cell clones are detected at each time point, but that the number of newly detected clones decreases at later time points.
Figure 4: Longitudinal clonal patterns.
(a) Binary heat map showing the presence (blue) or absence (white) of 11,799 individual clones detected at a proportion of 0.01% or greater in at least one NK cell sample from the Wu dataset. Columns represent samples and rows represent individual clones ordered by their first time point of detection. (b) Heat map showing the log normalized counts of the top ten clones from each NK cell sample from the Wu dataset. Overlaid asterisks indicate which clone is one of the top ten contributing clones for each sample, and clones are ordered on the y-axis based on hierarchical clustering of their Euclidean distances between their log normalized counts across samples. (c) Heat map depicting the same log normalized count values as in (b) but with overlaid asterisks instead indicating which clones significantly changed in proportion from the previous sample based on a p-value of < 0.05 assessed by a chi-squared test of proportions with Bonferroni adjustment of p-values to account for multiple comparisons. m, months post-transplant; NK 16, CD3-CD14-CD20-CD56-CD16+ NK cells.
Next, we analyzed distinct clonal dynamics in individual NK cell clones using barcodetrackR to generate a heat map showing the abundance of the top ten NK cell clones from each sample over time (Fig. 4b). We visualize only the top clones in order to focus on the clones responsible for the majority of this cellular compartment’s clonal composition, with stars on the heatmap indicating the top ten contributing clones in each sample. This analysis reveals the waxing and waning patterns of high-abundance NK cell clones over time, which were further interrogated in the Wu study, and suggested underlying environmental stimuli such as a viral infection, further investigated in a subsequent study.36 Lastly, we utilized statistical testing within barcodetrackR to view changes in proportions of individual clonal contributions within the NK cell samples, marking clones with a star which had a statistically significant change (as assessed by a Chi-squared test with p-value adjustment for multiple comparisons) in abundance in the labelled sample in comparison to the previous sample (Fig. 4c). This type of visualization and analysis further highlights the highly dynamic clonal patterns within the NK cell compartment. Altogether, these results indicate that the longitudinal tracking of highly abundant clones within datasets can provide insight into clonal dynamics at a single-clone level. Such approaches would have numerous applications, such as assessing competitive clonal and subclonal dynamics within a tumor or premalignant state or response of specific immune cell clones to various stimuli.
Lineage bias
Clonal tracking studies measure HSPC clonal contributions to different mature blood cell lineages. Thus, the lineage bias of a single HSPC clone, such as one that skews towards a myeloid or lymphoid lineage, can be inferred from clonal tracking data by calculating a ratio of that clone’s abundances between two specific lineages. We define the log2 transformation of this ratio to be the log-bias for a particular clone. For example, a clone X with a normalized abundance of 100 in lineage A and normalized abundance of 50 in lineage B will have a log-bias of log2(100/50) = 1 towards lineage A (and conversely a log-bias of −1 towards lineage B), while a clone Y with a normalized abundance of 200 in lineage A and normalized abundance of 200 in lineage B will have a log-bias of log2(200/200) = 0 towards either lineage. Here, we use barcodetrackR to probe this concept of lineage bias in the Six clinical gene therapy trial dataset7 and the Koelle rhesus macaque dataset9(Supplementary Table 1), both based on lineage-purified samples following autologous transplantation with genetically tagged HSPCs.
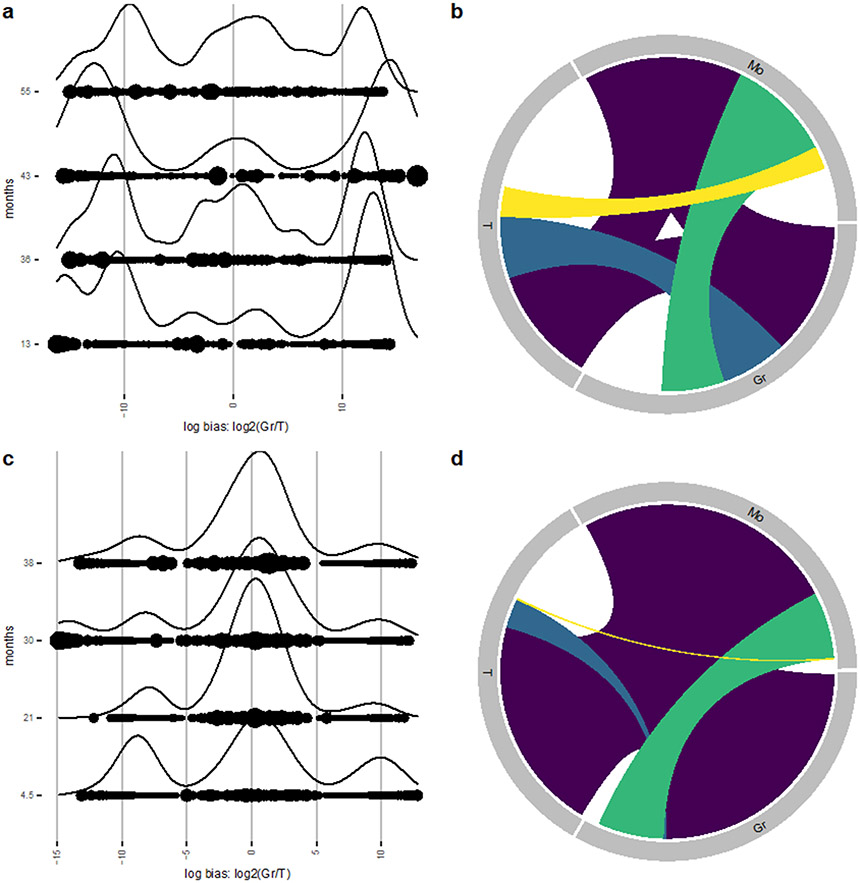
We first visualize the density of clones along the aforementioned log-bias axis for two chosen lineages over time (Gr and T) in the Six dataset (Fig. 5a). The ridge plot silhouettes depict kernel density estimators along this log-bias axis, weighted by the sum of their abundances in each lineage (to emphasize high-contributing clones), which enables us to find where high-contributing clones exist on the log-bias axis. We find the presence of three high-contributing sets of clones as determined by Gr/T lineage bias: Gr-biased (rightmost peak), balanced clones (middle peak), and T-biased (leftmost peak) (Fig. 5a). By systematically comparing each cell type in the dataset, we find that three sets of clones can be found when comparing Mo/T, Gr/B, or Mo/B lineages (Supplementary Fig. 4) further supporting differences in upstream progenitors accounting for myeloid versus lymphoid lineages. In contrast, when comparing Gr/Mo or T/B lineages, we find that a larger proportion of clones have balanced contribution to the two lineages, particularly at later time points (Supplementary Fig. 4). Interestingly, clones contributing to the NK cell samples are predominantly unilineage, sharing very little clonality with other lineages, including other lymphoid lineages such as T and B cells (Supplementary Fig. 4). This is in line with clonal tracking studies performed in a rhesus macaque animal model2,11. Conducting the same analysis on longitudinal samples from the Koelle dataset reveals the presence of Gr-biased, balanced, and T-biased clones at the 4.5-month timepoint (Fig. 5c) consistent with the Six dataset. However, there is an increase in abundance of balanced clones at later timepoints post-transplant, suggesting a shift away from hematopoiesis from downstream progenitor towards hematopoiesis from multipotent upstream progenitors capable of reconstituting both myeloid and lymphoid lineages. This is also the case when comparing the T cell lineage to the Mo lineage as the majority of clones contribute similar abundances to Gr and Mo lineages (Supplementary Fig. 5).
Figure 5: Lineage bias.
(a, c) Ridge plot shows clonal bias between Gr and T lineages at multiple timepoints of the Six dataset (a) and the Koelle dataset (c). Ridges indicate the abundance-weighted density at the value of log-bias on the x-axes, and dots indicate individual clones, sized by their overall abundance. Multiple ridge plots along the y-axes correspond to the time point of each sample in months post-transplant. (b, d) Chord diagram showing shared clonality between Gr, T, and Mo lineages from Six et al (b) and Koelle et al (d) datasets. Each uniquely colored chord represents a unique combination of lineages, and the width of each chord as it intersects with a lineage indicates the proportional contribution of that group of clones to that lineage. The space around the perimeter without a chord indicates the percentage contribution of unilineage clones. Gr, Granulocyte; T, T cell; Mo, Monocyte.
We next use barcodetrackR to construct an abundance-weighted chord diagram between three lineages in the Six and Koelle datasets. Chord diagrams show samples of interest as regions around a circle with links illustrating the shared clonality between each unique combination of samples. The thickness of the links illustrates the number of shared clones, or in the case of abundance-weighted chord diagrams (as shown in Fig. 5), the proportional contribution of that set of clones to each sample. For example, if there is a population of clones found in both samples A and B, with proportional contribution of 40% and 30% to the two samples respectively, this would be represented by a link between samples A and B which occupies 40% of sample A’s region of the perimeter and 30% of sample B’s region of the perimeter. We selected the Gr, T, and Mo lineages at the final 55-month time point of the Six dataset (Fig. 5b), finding that a large fraction of detected hematopoiesis at this time point is shared between all three lineages (purple). However, there also exist clones detected only in two lineages (yellow, blue, green), and biased clones only found in one lineage, indicated by the white space around the perimeter. Likewise, a similar pattern is observed in the Koelle dataset at the final 38-month time point (Fig. 5d) with a large fraction of detected hematopoiesis arising from clones detected in all three lineages (purple). The fraction of detected hematopoiesis arising from T-Gr or T-Mo restricted clones (blue, yellow respectively), however, is minimal compared to that arising from Gr-Mo restricted clones (green) in both datasets, suggesting that myeloid-biased upstream progenitors predominate within the total hematopoiesis of the Gr and Mo lineages at this time point. Thus, barcodetrackR provides a number of complementary functions useful for inferring the lineage biases of upstream progenitors from clonal tracking data.
Package versatility
barcodetrackR’s utility can extend to analyses of other data modalities and experimental settings outside of HSPC clonal tracking, provided that the input data is composed of genetic tag abundances that are likely to be shared to some degree across multiple samples of interest. One example of genetic tag abundance data arises from T-cell receptor (TCR) sequencing experiments, in which endogenous TCR sequences are sequenced from T cell samples and serve as genetic tags to mark individual T cell clones. Here, we highlight the versatility of our software package by extending our analyses to a TCR sequencing experiment of longitudinal samples from X-SCID patients treated with HSPC gene therapy33. barcodetrackR’s functions seamlessly extend to facilitate the analysis of T cell clone numbers and Shannon diversities across T cell samples in this experiment (Supplementary Fig. 6).
DISCUSSION
A recent gathering of over 30 researchers in the clonal tracking field (2018 StemCellMathLab workshop29) formalized a call for the development of open-source tools for the analysis of clonal tracking data in order to promote rigor and reproducibility within the field. Here, we provide and showcase our open-source R package and app barcodetrackR, which encompasses an extensive, flexible, and accessible set of tools in order to address these needs and serve as a critical foundation on which to build further analytical approaches in the clonal tracking field. While tools for the processing of the raw sequencing data from clonal tracking experiments have been previously developed18, barcodetrackR represents the first software package dedicated to interrogating the underlying biology represented by these clonal abundances. As shown, barcodetrackR is a multifaceted toolkit and has diverse applications, underscoring the utility of using complementary data analysis methods and visualizations to probe biological hypotheses. The development and implementation of a Shiny app further adds to the utility of the package by making it more accessible to the clonal tracking community, which continues to expand as sequencing costs decline and methodologies continue to improve.
Although we have designed barcodetrackR to accommodate any clonal tracking dataset with clonal abundance quantified per sample, different clonal tracking technologies have unique limitations and sources of uncertainty. For example, viral integration site analysis is subject to a low capture efficiency as compared to DNA barcode sequencing37, while increasing the number of amplification steps in recovery of DNA barcodes can increase the abundance of artifactual barcode sequences arising from PCR or sequencing error.38 Both technologies can be limited by under-sampling the clonal population of interest, which can be partially overcome by increasing sequencing depth or sampling more cells.37 To address potential pitfalls in analysis of clonal tracking data, we briefly recommend strategies for experimental design, genetic tag retrieval upstream of barcodetrackR, and data analysis within barcodetrackR in Supplementary Note 1, directing readers to more extensive referenced reviews when appropriate.
barcodetrackR to encompasses a large number of tools and methods; however, it is by no means an exhaustive toolbox, and we envision continuing to add to it in the future in order to address new biologic questions that arise. While the majority of prior clonal tracking experimental designs have precluded the acquisition of replicate samples and often encompassed small numbers of humans and/or animals, future studies will likely be able to acquire biological replicates in a number of different contexts to allow for more rigorous statistical testing of sample relationships and clonal dynamics. Clustering methods to identify populations of clones with similar properties have thus far been limited to hierarchical11 and k-means7 in the literature, but the growing development of clustering frameworks of single cells in the scRNA-seq field may provide a future basis by which to identify clusters of clones based on longitudinal behavior and distribution across compartments39. Similarly, the adoption of dimensionality reduction techniques such as t-SNE40 in the scRNA-seq field has already appeared to motivate novel clonal tracking visualizations41. And although barcodetrackR is designed for analysis of clonal abundance data from prospective lineage tracing techniques which involve introduction of an exogenous genetic tag, new technologies allow for retrospective lineage tracing based on somatic mutations. A number of software packages have been developed to infer clonal relationships from somatic mutations.42-46
As the clonal tracking field continues to grow and tools are further refined, we believe it is important as a field to continue to emphasize the aggregation and public availability of clonal tracking data. Ultimately, we believe barcodetrackR can be of high utility to the clonal tracking field and serve as an important step towards building a more robust and reproducible analytical framework in the field.
METHODS
Data collection and genetic tag retrieval
Multiple clonal tracking methodologies exist in the literature47, with the most recent methods relying on next-generation sequencing to retrieve lineage tracing elements. Several analysis pipelines exist for the retrieval and error-correction of lineage tracing elements from sequencing data16-19. The experimental techniques utilized, the number of cells sampled, the level of tagged cell within the population, the sequencing platform applied, and the computational method of genetic tag extraction affect the number and frequency of tags detected in a lineage tracing study.
The barcodetrackR package can operate on any dataset that contains rows as observations and columns as samples, regardless of which experimental method for genetic labelling and approaches for tag retrieval were used. In Supplementary Note 1, we highlight considerations for experimental design and genetic tag retrieval upstream of barcodetrackR and recommend strategies for reliable data analysis given some limitations and sources of uncertainty which are common across many clonal tracking technologies.
Instantiating an SE object
Instantiating an SE through the function create_SE requires two inputs: counts data with genetic tags as rows and samples as columns, and metadata providing information on each sample. For the Shiny app, count and metadata can be specified by uploading tabular data files. The following assays are created within the SE: counts: the raw values from the input dataframe, proportions: the per-column proportions of each entry in each column, ranks: the rank of each entry in each column, normalized: the normalized read values in counts-per million (CPM), and logs: the log of the normalized values. The default normalization is counts per million, and log-normalized values are calculated by taking the log of plus-one normalized data so that zeros are retained. Users can supply a custom scale factor and/or log base. The use of an SE object permits the addition of custom assays to the object to facilitate flexibility (e.g. custom normalization strategies). Additionally, users have the option of passing a relative or absolute minimum abundance threshold, described in the following section.
Applying a minimum abundance threshold
When creating an SE object, users have the option of including a relative or absolute threshold to exclude low-abundance occurrences that are more likely to come from noise or sequencing error. Using a relative threshold of 0.005, for example, retains genetic tags which are present at an abundance of 0.5% or greater in at least one sample. Likewise, an absolute threshold of 100 would only retain genetic tags which are present at a raw read count of 100 or greater in at least one sample. Supplementary Fig. 1 shows the effect of varying the threshold parameter on the number of unique clones detected and the distribution of genetic tag proportions and logs. In general, increasing the genetic tag abundance threshold decreases the prevalence of zero-count or low-abundance genetic tags in any given sample without significantly affecting the distribution of non-zero abundance genetic tags. However, increasing the threshold too much will result in lower number of detected clones and exclusion of real, confidently assigned genetic tags.
We include the function estimate_barcode_threshold to help users estimate an appropriate minimum abundance threshold for their data. The function estimates a relative threshold R (a percentage) based on the following formula:
Where R is the relative minimum abundance threshold (expressed as a percentage), N is the total number of cells/genomes analyzed in the sample, F is the Frequency of genetically modified cells within the sample, and N represents the minimum size of clone in the pool of cells/genomes that would be expected to be detected. N is calculated from the following equation:
Where P is the desired confidence level (e.g. 0.95 for 95% confidence level) and C is the efficiency of the method to capture a given clone. The value of C depends on the clonal tracking technology used and can be estimated by performing simulations or replicate sampling. Adair37 et al performed simulations and found that C values of 0.05 and 0.4 matched experimental data for viral integration site analysis and DNA barcode sequencing respectively.
The estimated threshold can be supplied to the create_SE command to exclude genetic tags below the threshold upon creation of the SE object. Alternatively, it can be applied to an existing SE through the function threshold_SE. In addition to estimating an appropriate minimum abundance threshold, we recommend exploring the distribution of genetic tag counts across thresholding parameters (as we have done in Supplementary Fig. 1) and ensuring that claims made from analyzing genetic tag abundances are not sensitive to the choice of thresholding parameter. Within this paper, the Six, Elder, Espinoza, Wu, and Koelle datasets were used with a relative threshold of 0.0001. The Belderbos and Clarke datasets were used with no threshold.
Global clonal distributions
The barcodetrackR package contains multiple tools for analyzing global clonal distributions between samples on the basis of correlation, pairwise distance measures, or dissimilarity indices. Users can view correlations between samples on a grid using the cor_plot function, or for two samples using the scatter_plot function.
The dist_plot function calculates pairwise distances between each sample-sample pair calculated on the specified assay using any similarity of distance measure included in the proxy R package. If desired, the samples can be hierarchically clustered, and a clustering tree can be displayed alongside the grid of distance values.
The mds_plot function calculates dissimilarity indices between samples using any distance metric within the vegdist function from the R package vegan48. The distance methods produce a matrix composed of the distances between samples, given the composition of genetic tags in each sample. Principal coordinates analysis is performed on the distance matrix in order to display dissimilarity between samples in two dimensions.
Clonal diversity
Three measures of within-sample diversity can be calculated by the function clonal_diversity: shannon diversity (H’) , simpson diversity (λ), and inverse-simpson, which is calculated as 1/λ. Their equations are as follows:
Where R is the total number of species (in this case genetic tags), and pi is the proportion of each genetic tag in the sample. Users can also display the Shannon count calculated as:
Additionally, nominal or cumulative counts of each sample can be displayed using the clonal_count function. To accurately compare counts or diversity between samples, the same number of labeled cells should be used as starting material for quantification. This is especially important when assessing diversity based on the nominal count of genetic tags retrieved. Belderbos et al showed that the Shannon count is stable with respect to filtering thresholds and stays below the theoretical library size upon re-sampling31. Therefore, in some cases, it may be beneficial to use Shannon count rather than nominal genetic tag counts when comparing diversity between samples.
Clonal patterns
Heat maps created by the function barcode_ggheatmap display clonal abundances across samples by coloring cells based on the log-normalized abundance of each clone. By default, the top clones from each sample are marked by a star. Individual clones are hierarchically clustered along the y-axis based on their log-abundance (or other specified assay) across samples. The dendrogram to depict hierarchical clusters of clones is drawn using ggdendro49.
To track the emergence of clones over time, the function barcode_binary_heatmap will display a binary heatmaps indicating the presence or absence of clones in each sample. A threshold is provided to specify the limit of detection. Clones with percentage abundance below this threshold in a given sample are set to “absent” and clones which do not pass this threshold in any sample are removed. Providing a minimum-abundance threshold limits the role that sampling bias may play on the detection or lack of detection of genetic tags.
Assessing changes in clonal abundance
Users can apply hypothesis testing to clonal abundances through the function barcode_stat_test. This function requires the sample size of cells or genomes for each sample which cannot be calculated from the genetic tag count data itself. The sample size should be the total number of labeled cells before amplification, because this is the population of cells which the clonal tracking data represent. To compare genetic tag abundances between samples, users can choose from a “chi-squared” or “fisher” exact test. The tests operate on a contingency table for each genetic tag to determine whether the tag changed in proportion based on its observed abundance and sample size. By default, each sample is compared to the previous, but users can also specify to compare each sample to a single reference sample (such as the initial time point). The p-values produced by the test can be adjusted for multiple comparisons using any adjustment method in R stats, such as the Bonferroni adjustment or the Benjamin and Hochberg false discovery rate adjustment.
When conducting hypothesis testing on clonal abundance data, it is important to consider the possibility of under-sampling the population of interest. If the two samples being compared, for example, represent two longitudinal blood samples meant to approximate the entire hematopoietic compartment of an animal, one must consider that the differences in genetic tag abundance could be due to both biological changes and sampling bias from two separate blood draws. One can estimate the level of sampling bias in an experiment through replicate sampling or simulations, but the possibility of under-sampling is likely to affect many clonal tracking studies.37 For this reason, we recommend users to use caution when interpreting p-values from the hypothesis testing. The different p-values for different genetic tags produced by the test give indication of each tag’s relative change from one sample to another. It is correct, therefore, to say that if clone A has a lower p value than clone B, it is more likely than clone B to have changed in abundance between samples given the provided total sample size. However, it is not advised to apply a certain p-value threshold such as 0.05 and assume that all changes in genetic tag abundance below this threshold represent any kind of biological ground truth.
Clonal bias (log-bias)
The bias_ridge_plot function calculates the log-bias and uses it as a continuous variable in order to display the density of clones at each level of log-bias. In order to calculate log-bias between clones that are only present in one sample, log-bias is calculated as followed:
The normalized value is taken from the SE slot which is scaled to counts per million. To display the distribution of log-bias measures in a ridge plot, the density of clones at each value of log-bias can be estimated using a kernel density estimator. The density estimator can be weighted by the combined abundance of the clone between the two samples, calculated as:
Users of the package can also opt to only analyze clones present in both samples which will minimize the possible effects of sampling bias but may exclude bona-fide lineage-restricted clones.
Chord diagrams
The chord_diagram function utilizes the circlize package in R50 to display shared clones between samples. Samples are shown as regions around a circle with their shared clonality shown as links between regions. With each unique combination, a new link is created with a unique. In the non-weighted setting, the function operates on the counts assay. Therefore, the length of each region around the circle represents the number of clones detected in each sample, and the width of links between regions is proportional to the number of shared clones. In the weighted setting, the function operates on the proportions slot. Each region around the circle has the same length corresponding to 100%, and the links between regions correspond to the fractional abundance of the shared clones within each sample. Therefore, in the weighted setting, the same link can have a different width at each connection to a region.
Shiny app
The launchApp function launches a local Shiny app that provides a graphical user interface for a number of barcodetrackR’s functions. The app is targeted to users with limited coding experience. An identical version of the app is available online at https://dunbarlabnih.shinyapps.io/barcode_app/ . Once the app is launched, it prompts users for 2 files of raw text data to be uploaded: the genetic tag counts table and the metadata table. There also exists a small example dataset that can be automatically loaded in the app for exploration of its capabilities using the button “Load Sample Data” (see screenshots in Supplementary Fig. 7). An example barcode genetic tag count table and metadata table are linked to in the app, as well as a link to the description and origin of the example dataset. Users can download figures or analysis results in tabular format using the download buttons.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank David Allan of the NHLBI Intramural Research Program for his contributions to approaches for statistical testing of genetic tag abundances. The authors thank members of the Dunbar lab for helpful feedback in revision of this manuscript. We also acknowledge reviewer #1, who provided the rationale and algorithmic explanation that directly motivated the creation of the estimate_barcode_threshold function. DAE was supported by NIH Medical Scientist Training Program T32 GM07170 and T32 G000046. RDM, SJK, CW and CED were supported by the Division of Intramural Research at the National Heart, Lung and Blood Institute.
Footnotes
Competing interests
Not applicable.
DATA AVAILABILITY
All clonal tracking datasets analyzed in this study are publicly available with accession instructions outlined in Supplementary Table 1. The Six dataset7 was downloaded from https://github.com/BushmanLab/HSC_diversity/tree/master/data. The Belderbos dataset31 was downloaded from its manuscript’s supplementary material. The Elder dataset32 was downloaded from GEO accession GSE149170. The Espinoza dataset30 was downloaded from GEO accession GSE153130. The Wu dataset11 was downloaded from its manuscript’s supplementary material. The Koelle dataset9 was downloaded from https://github.com/dunbarlabNIH/R-code-and-tabular-data. The Clarke dataset33 was downloaded from https://doi.org/10.5281/zenodo.1256169. Data were pre-processed in R to create tabular data files amenable for use with barcodetrackR. Source Data for Figures 2-5 are available with this manuscript.
CODE AVAILABILITY
The barcodetrackR package is freely available from GitHub under a Creative Commons 0 license and can be accessed at https://github.com/dunbarlabNIH/barcodetrackR. Additionally, the package is available through the Bioconductor repository [citation forthcoming once the package is approved by Bioconductor]. A frozen version of the package at the time of publication is available on Zenodo in ref 51. A frozen and interactive version of the package at the time of publication is available on Code Ocean in ref 52, allowing readers to reproduce all figures and the full barcodetrackR vignette within a pre-specified computational environment.
REFERENCES
- 1.Lu R, Neff NF, Quake SR & Weissman IL Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol 29, 928–933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu C et al. Clonal Tracking of Rhesus Macaque Hematopoiesis Highlights a Distinct Lineage Origin for Natural Killer Cells. Cell Stem Cell 14, 486–499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radtke S et al. A distinct hematopoietic stem cell population for rapid multilineage engraftment in nonhuman primates. Sci. Transl. Med 9, eaan1145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S et al. Dynamics of HSPC Repopulation in Nonhuman Primates Revealed by a Decade-Long Clonal-Tracking Study. Cell Stem Cell 14, 473–485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerrits A et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood 115, 2610–2618 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Wu C et al. Geographic clonal tracking in macaques provides insights into HSPC migration and differentiation. Journal of Experimental Medicine 215, 217–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Six E et al. Clonal tracking in gene therapy patients reveals a diversity of human hematopoietic differentiation programs. Blood 135, 1219–1231 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biasco L et al. In Vivo Tracking of Human Hematopoiesis Reveals Patterns of Clonal Dynamics during Early and Steady-State Reconstitution Phases. Cell Stem Cell 19, 107–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koelle SJ et al. Quantitative stability of hematopoietic stem and progenitor cell clonal output in rhesus macaques receiving transplants. Blood 129, 1448–1457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brugman MH et al. Development of a diverse human T-cell repertoire despite stringent restriction of hematopoietic clonality in the thymus. Proc Natl Acad Sci USA 112, E6020–E6027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C et al. Clonal expansion and compartmentalized maintenance of rhesus macaque NK cell subsets. Sci. Immunol 3, eaat9781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merino D et al. Barcoding reveals complex clonal behavior in patient-derived xenografts of metastatic triple negative breast cancer. Nature Communications 10, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter SN, Baker LC, Mittelman D & Porteus MH Lentiviral and targeted cellular barcoding reveals ongoing clonal dynamics of cell lines in vitro and in vivo. Genome Biology 15, R75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheih A et al. Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nature Communications 11, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordes S, Wu C & Dunbar CE Clonal tracking of haematopoietic cells: insights and clinical implications. Br J Haematol bjh.17175 (2020) doi: 10.1111/bjh.17175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry CC et al. INSPIIRED: Quantification and Visualization Tools for Analyzing Integration Site Distributions. Molecular Therapy - Methods & Clinical Development 4, 17–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman E et al. INSPIIRED: A Pipeline for Quantitative Analysis of Sites of New DNA Integration in Cellular Genomes. Molecular Therapy - Methods & Clinical Development 4, 39–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thielecke L, Cornils K & Glauche I genBaRcode: a comprehensive R-package for genetic barcode analysis. Bioinformatics 36, 2189–2194 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Bramlett C et al. Clonal tracking using embedded viral barcoding and high-throughput sequencing. Nat Protoc 15, 1436–1458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berry CC, Ocwieja KE, Malani N & Bushman FD Comparing DNA integration site clusters with scan statistics. Bioinformatics 30, 1493–1500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afzal S, Fronza R & Schmidt M VSeq-Toolkit: Comprehensive Computational Analysis of Viral Vectors in Gene Therapy. Molecular Therapy - Methods & Clinical Development 17, 752–757 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hocum JD et al. VISA - Vector Integration Site Analysis server: a web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC Bioinformatics 16, 212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spinozzi G et al. VISPA2: a scalable pipeline for high-throughput identification and annotation of vector integration sites. BMC Bioinformatics 18, 520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins TB et al. Identifying viral integration sites using SeqMap 2.0. Bioinformatics 27, 720–722 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zorita E, Cuscó P & Filion GJ Starcode: sequence clustering based on all-pairs search. Bioinformatics 31, 1913–1919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao L, Liu Z, Levy SF & Wu S Bartender: a fast and accurate clustering algorithm to count barcode reads. Bioinformatics 34, 739–747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf FA, Angerer P & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyne A-M et al. A track of the clones: new developments in cellular barcoding. Experimental Hematology 68, 15–20 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Espinoza DA et al. Aberrant Clonal Hematopoiesis following Lentiviral Vector Transduction of HSPCs in a Rhesus Macaque. Molecular Therapy 27, 1074–1086 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belderbos ME et al. Donor-to-Donor Heterogeneity in the Clonal Dynamics of Transplanted Human Cord Blood Stem Cells in Murine Xenografts. Biology of Blood and Marrow Transplantation 26, 16–25 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Elder A et al. Abundant and equipotent founder cells establish and maintain acute lymphoblastic leukaemia. Leukemia 31, 2577–2586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke EL et al. T cell dynamics and response of the microbiota after gene therapy to treat X-linked severe combined immunodeficiency. Genome Med 10, 70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan M, Obenchain V, Hester J & Pagès H SummarizedExperiment: SummarizedExperiment container. (2020). [Google Scholar]
- 35.Chang W, Cheng J, Allaire JJ, Xie Y & McPherson J shiny: Web Application Framework for R. (2020). [Google Scholar]
- 36.Truitt LL et al. Impact of CMV Infection on Natural Killer Cell Clonal Repertoire in CMV-Naïve Rhesus Macaques. Front. Immunol 10, 2381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adair JE et al. DNA Barcoding in Nonhuman Primates Reveals Important Limitations in Retrovirus Integration Site Analysis. Molecular Therapy - Methods & Clinical Development 17, 796–809 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thielecke L et al. Limitations and challenges of genetic barcode quantification. Sci Rep 7, 43249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiselev VY, Andrews TS & Hemberg M Challenges in unsupervised clustering of single-cell RNA-seq data. Nat Rev Genet 20, 273–282 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Maaten L. van der & Hinton G Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605 (2008). [Google Scholar]
- 41.Lin DS et al. DiSNE Movie Visualization and Assessment of Clonal Kinetics Reveal Multiple Trajectories of Dendritic Cell Development. Cell Reports 22, 2557–2566 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Jahn K, Kuipers J & Beerenwinkel N Tree inference for single-cell data. Genome Biol 17, 86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross EM & Markowetz F OncoNEM: inferring tumor evolution from single-cell sequencing data. 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zafar H, Navin N, Chen K & Nakhleh L SiCloneFit: Bayesian inference of population structure, genotype, and phylogeny of tumor clones from single-cell genome sequencing data. Genome Res. 29, 1847–1859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadeqi Azer E et al. PhISCS-BnB: a fast branch and bound algorithm for the perfect tumor phylogeny reconstruction problem. Bioinformatics 36, i169–i176 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vavoulis DV, Cutts A, Taylor JC & Schuh A A statistical approach for tracking clonal dynamics in cancer using longitudinal next-generation sequencing data. Bioinformatics 8 (2020) doi: 10.1093/bioinformatics/btaa672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kebschull JM & Zador AM Cellular barcoding: lineage tracing, screening and beyond. Nat Methods 15, 871–879 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Oksanen J et al. vegan: Community Ecology Package. (2019). [Google Scholar]
- 49.de Vries A & Ripley BD ggdendro: Create Dendrograms and Tree Diagrams using ‘ggplot2’. (2016). [Google Scholar]
- 50.Gu Z, Gu L, Eils R, Schlesner M & Brors B circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Espinoza DA, Mortlock RD, Koelle SJ, Wu C & Dunbar CE barcodetrackR: an R package for the interrogation of clonal tracking data (Zenodo Freeze). (Zenodo, 2021). doi: 10.5281/zenodo.4609410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Espinoza DA, Mortlock RD, Koelle SJ, Wu C & Dunbar CE barcodetrackR: an R package for the interrogation of clonal tracking data. (Code Ocean, 2021). doi: 10.24433/CO.6231752.v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All clonal tracking datasets analyzed in this study are publicly available with accession instructions outlined in Supplementary Table 1. The Six dataset7 was downloaded from https://github.com/BushmanLab/HSC_diversity/tree/master/data. The Belderbos dataset31 was downloaded from its manuscript’s supplementary material. The Elder dataset32 was downloaded from GEO accession GSE149170. The Espinoza dataset30 was downloaded from GEO accession GSE153130. The Wu dataset11 was downloaded from its manuscript’s supplementary material. The Koelle dataset9 was downloaded from https://github.com/dunbarlabNIH/R-code-and-tabular-data. The Clarke dataset33 was downloaded from https://doi.org/10.5281/zenodo.1256169. Data were pre-processed in R to create tabular data files amenable for use with barcodetrackR. Source Data for Figures 2-5 are available with this manuscript.
The barcodetrackR package is freely available from GitHub under a Creative Commons 0 license and can be accessed at https://github.com/dunbarlabNIH/barcodetrackR. Additionally, the package is available through the Bioconductor repository [citation forthcoming once the package is approved by Bioconductor]. A frozen version of the package at the time of publication is available on Zenodo in ref 51. A frozen and interactive version of the package at the time of publication is available on Code Ocean in ref 52, allowing readers to reproduce all figures and the full barcodetrackR vignette within a pre-specified computational environment.