Abstract
Point-of-care tests (POCTs) are increasingly being used in field settings, particularly outdoors. The performance of current POCTs—most commonly the lateral flow immunoassay—can be adversely affected by ambient temperature and humidity. We developed a self-contained immunoassay platform—the D4 POCT—that can be conducted at the POC by integrating all reagents in a capillary-driven passive microfluidic cassette that minimizes user intervention. The assay can be imaged and analyzed on a portable fluorescence reader—the D4Scope—and provide quantitative outputs. Here, we systematically investigated the resilience of our D4 POCT to varied temperature and humidity and to physiologically diverse human whole blood samples that span a wide range of physiological hematocrit (30–65%). For all conditions, we showed that the platform maintained high sensitivity (0.05–0.41 ng/mL limits of detection). The platform also demonstrated good accuracy in reporting true analyte concentration across environmental extremes when compared to the manually operated format of the same test to detect a model analyte—ovalbumin. Additionally, we engineered an improved version of the microfluidic cassette that improved the ease-of-use of the device and shortened the time-to-result. We implemented this new cassette to create a rapid diagnostic test to detect talaromycosis infection in patients with advanced HIV disease at the POC, demonstrating comparable sensitivity and specificity to the laboratory test for the disease.
Keywords: microfluidics, biosensor, infectious disease, immunoassay, point-of-care test, diagnostics
Graphical Abstract
Talaromycosis—an invasive fungal infection caused by the dimorphic fungus Talaromyces marneffei endemic in Southeast Asia—primarily affects immunocompromised individuals, particularly those with advanced HIV disease, in whom talaromycosis accounts for 18% of hospital admissions and is a leading cause of AIDS death.1–5 Mortality in talaromycosis patients is as high as 50% despite antifungal therapy and is driven primarily by late diagnosis.3,6 An impediment to reducing mortality is the reliance on decades-old diagnostic methods. The current gold-standard, culture isolation of T. marneffei from patient specimens, can take up to 28 days and has suboptimal sensitivity (<75%).2,3,5 Although several real-time polymerase chain reaction (PCR) assays have been developed with improved sensitivities,7,8 they require sophisticated laboratory facilities and skilled labor and thus are impractical in low-resource and out of hospital settings. As patient outcomes are highly correlated with the speed of diagnosis, a critical need exists for non-culture-based tests that can be conducted at the point-of-care (POC) rapidly and ideally with accuracies superior to that of the current culture method.
Rapid POC tests (POCTs), notably lateral flow immunoassays (LFIAs) have become an increasingly valuable tool for managing emerging infectious diseases—most recently highlighted during the coronavirus disease 2019 (COVID-19) pandemic. Their ease-of-use has enabled a pivotal shift toward untrained end users operating and interpreting POCTs outside of the clinic. However, existing tests have shortcomings that need to be addressed with the next generation of rapid POCTs. Notably, the performance of most LFIAs is susceptible to subjectivity, which can affect sensitivity and specificity. For example, a cohort of self-trained users detected 21.3% fewer PCR-confirmed COVID-19 positive cases when compared to laboratory scientists using the same LFIA platform.9 One potential source of this discrepancy is the misclassification of “weak band” positives, where the test line is barely visible, which has been shown to drastically change test sensitivity and specificity depending on how they are classified.10 There is also evidence that environmental factors such as the temperature and humidity affect LFIA performance. One study observed that Abbott BinaxNOW LFIAs operated on cool days (below <59 °F) had a decrease in sensitivity from 93.7 to 66.7% and a decrease in specificity from 100 to 95.2% when compared to >59 °F.11 Choi et al. have shown that varied humidity can impact flow in the nitrocellulose membrane of an LFIA and impact sensitivity and performance.12 As we continue to transition to POCTs that must be resilient to variations in the ambient conditions, it is critical to understand how the performance of POCTs changes in different operational conditions.
Next-generation POCTs should not only be easy to use and sensitive but also objective and reliable even in varied environmental factors. To address this challenge, we expanded upon our D4 platform technology13 to develop a passively driven microfluidic diagnostic POCT that provides sensitive, quantitative, and objective result to the operator with a workflow akin to an LFIA (Figure 1A). In this study, we first characterized the performance of the D4 in diverse operational environments (temperature and humidity) and then challenged with undiluted human whole blood using a model ovalbumin (OVA) system. Next, we engineered a new version of the microfluidic platform—the “EZ Load” cassette—to improve the ease-of-use and time-to-result (Figure 1B). Finally, we demonstrated the utility of this new, improved POCT for diagnosis of talaromycosis in patients with advanced HIV disease in a retrospective clinical study. This is an ideal testbed for the validation of this assay due to the need to test a marginalized patient population in areas with limited health care resources and in an environment with a wide variation in the expected operational conditions of the assay.
Figure 1.
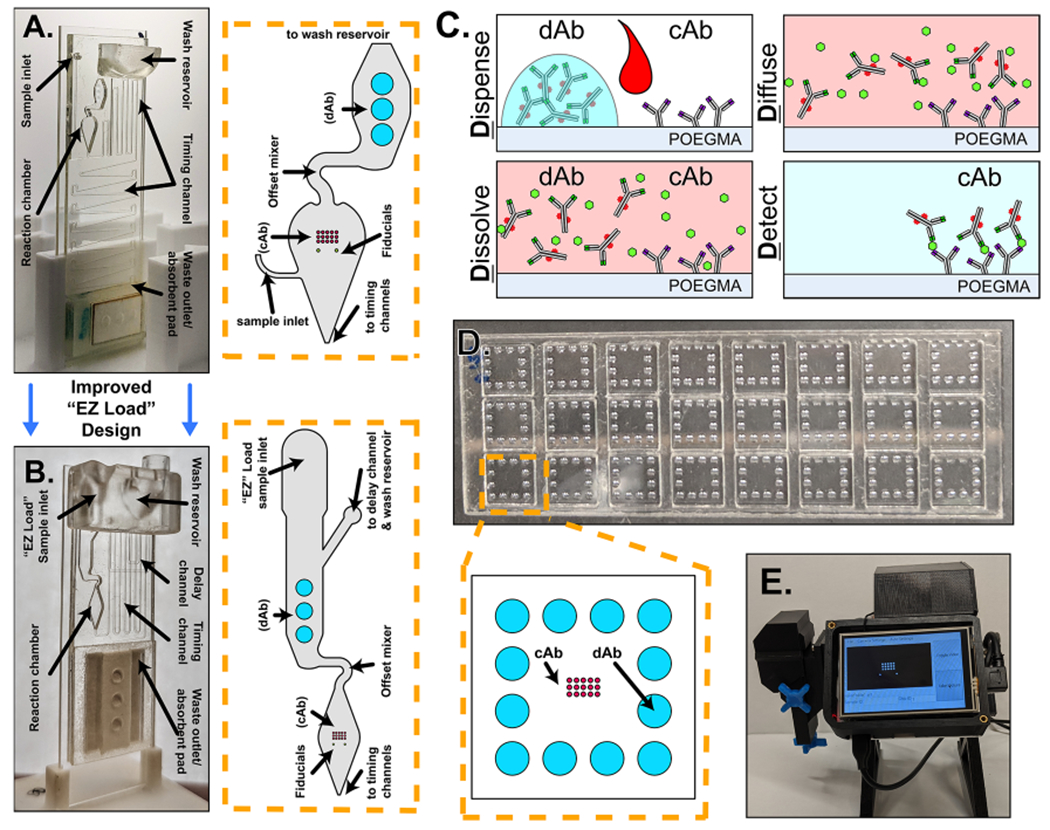
(A) Photograph of the first-generation microfluidic cassette with a zoomed in view of the reaction chamber to the right with annotations of notable features. (B) Photograph of the “EZ Load” microfluidic cassette with a zoomed in view of the reaction chamber to the right with annotations of notable features. (C) Schematic detailing the functional steps of the D4 assay. (D) Picture of an open format D4 slide showing 24 independent tests with a zoomed in view of the functional elements of a single test. (E) Photograph of the D4Scope used for fluorescence imaging of the cassettes.
RESULTS AND DISCUSSION
Development of the D4 Point-of-Care Test (POCT).
The D4 is a fluorescence-based POC immunoassay platform developed in our laboratory for diverse applications including breast cancer detection,14 Ebola virus diagnostics,15 and most recently for the serological profiling of SARS-CoV-213 and its variants.16 The D4 is a completely “self-contained” immunoassay built on a nonfouling poly(oligoethylene glycol methyl ether methacrylate) (POEGMA) brush grafted from silica glass, as described in more detail elsewhere.17,18 All antibodies are printed directly onto the POEGMA brush by inkjet printing. Capture antibodies (cAbs) are physically entangled without the need for covalent chemistry, while fluorescently labeled detection antibodies (dAbs) are inkjet-printed on an underlying dissolvable excipient pad. This allows a single drop of the sample to drive the sandwich fluorescent immunoassay to completion in a cascade of events that give the D4 its name (Figure 1C): a sample is Dispensed onto the chip, Dissolving the soluble dAb, which then Diffuse toward the immobilized cAb where a fluorescent sandwich is formed with the target analyte, which is Detected with a fluorescent reader by correlating the fluorescence intensity with known analyte concentration. The D4 was originally implemented in an “open format” (OF) configuration that enabled the parallelization of 24 tests on a single glass slide but required manual intervention in the form of timing, washing, and drying of the chip (Figure 1D). We subsequently developed a portable fluorescence reader—the D4Scope (D4S)—that eliminates the need for any centralized equipment for analysis of the D4 output, and importantly, it directly quantifies the fluorescence output of the antibody microarray (Figure 1E).13,15 The D4S replaces the bulky and expensive Genepix microarray scanner (G) that was previously used for imaging and analysis. Recently, we debuted an automated version of the D4 assay—the microfluidic D4 cassette (μF)—first used for quantitative serological profiling of SARS-CoV-2 antibodies from undiluted human serum and whole blood.13 In this work, the cassette has been adapted for the detection of antigens. The vertically oriented gravity- and capillary-driven microfluidic D4 cassette (Figure 1A) automates all user intervention steps of the D4 POCT—from the loading of the sample and running buffer at the start of testing to the readout of the test—while also preventing settling and binding of cellular and protein debris that can otherwise increase background fluorescence, allowing for operation in complex samples such as undiluted human whole blood without any preprocessing steps.
Resilience to Temperature and Humidity.
To characterize the impact of environmental extremes on the D4 POCT, we chose OVA as the model analyte and tested two different configurations of the D4 POCT: (1) the microfluidic D4 cassette imaged with the D4Scope (μF/D4S) and (2) the “open format” D4 imaged on a Genepix microarray scanner (OF/G). The μF/D4S configuration is our preferred POC configuration of the D4, while the manually operated OF/G is our more established configuration and will serve as a benchmark for performance.15,18,19 Assays were carried out at the following simulated environments: 5, 22, and 40 °C at 50% relative humidity (RH) (Figure 2A,D) and 10, 50%, and 80% RH at 22 °C (Figure 2B,E). Cassettes and slides were stabilized at each condition for 15–20 min prior to testing. The resulting data were fit to a five-parameter logistic regression for the analysis and calculation of the limit of detection (LOD) of the assay. The OF/G configuration achieved LODs between 0.04 and 0.33 ng/mL. The LOD of the μF/D4S configuration was between 0.10 and 0.41 ng/mL. This demonstrates that both configurations remain highly sensitive and functional under varied environmental conditions. Two likely sources for the minor difference in LOD are (1) the presence of light scattering acrylic on the periphery of the cAb spots when imaging on the D4S because of its larger field of view and area of illumination relative to the Genepix and (2) the inherent differences in the optical sensitivity of the two detectors. This is supported by the fact that only a ~5% increase in LOD was observed when the microfluidic cassettes were imaged on the D4Scope rather than the Genepix scanner (Table S1 and Figure S1). However, the D4Scope provides far greater ease-of-use, portability, low cost, and speed that justify its use.
Figure 2.
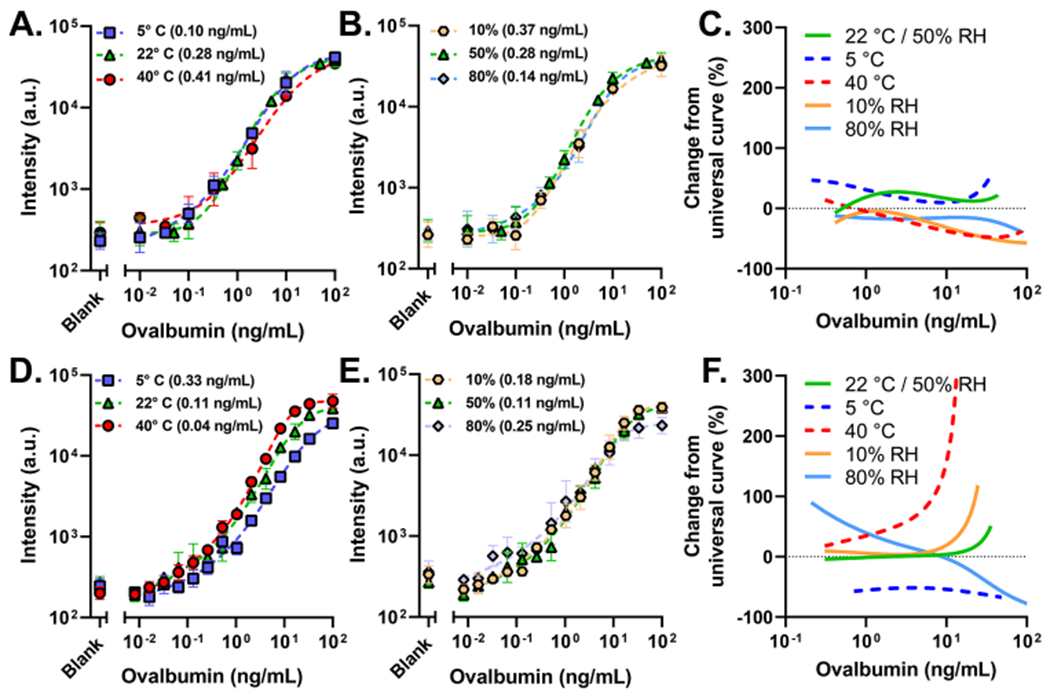
OVA dose–response curves run at 5, 22, and 40 °C/50% RH (A) and at 22 °C/10, 50, and 80% RH (B) spiked in FBS on μF/D4S. (C) Estimated percent change from the actual OVA concentration that μF/D4S is expected to report across concentrations. The same conditions for temperature (D) and humidity (E) run on OF/G. (F) Estimated percent change from the actual OVA concentration that the OF/G is expected to report across concentrations. OF/G format assays were run as a 15-point dose–response curve from 0.008 to 100 ng/mL and a blank with a 30 min incubation (n = 3, 4/225 points removed as outliers, and error bars are the standard deviation). μF/D4S configuration at the 22 °C/50% RH condition was run as a 10-point dose curve from 0.01 to 100 ng/mL and a blank with a ~23 min incubation (n = 4, no outlier removal, and error bars are the standard deviation). μF/G and μF/D formats at varied temperatures and humidity were run as an 8-point dose–response curve from 0.01 to 100 ng/mL and a blank with a 23 +/− 3 minute incubation (0, 0.1, 0.33, 2 ng/mL [n = 3], 0.01, 0.03, 10, 100 ng/mL [n = 2], 2/120 cassettes removed due to failure, and error bars are the standard deviation). LODs are in parentheses in the legends.
An extra sum-of-squares F test with a threshold P value of 0.05 was used to establish the relative sameness of the five tested conditions. A P value >0.05 would indicate that the datasets for each tested condition are similar enough to be described simply by one calibration curve. For the μF/D4S configuration, the P value was 0.0853 [F(20,77) = 1.561] when all five conditions were evaluated, indicating a high degree of sameness and suggesting resilience during operation in varied environmental conditions. For the OF/G configuration, the P value was <0.0001 [F(20, 180) = 15.13], indicating little to no sameness and suggesting perturbation as a function of varied environmental conditions, primarily temperature. As temperature decreases, there is attenuation in the fluorescence intensity for the OF/G configuration. This is a well-characterized phenomenon in ELISA and other ELISA-like assay formats,20–22 where higher temperatures result in faster antibody binding kinetics, and the assay thus reaches the steady state faster. However, this was not observed in the μF/D4S configuration. The microfluidic cassette is primarily driven by capillary pressure, which is modulated by temperature through the temperature dependence of viscosity.23 Therefore, as temperature increases, the incubation time decreases, which counterbalances the faster antibody kinetics at higher temperatures, and results in a more consistent performance, as indicated by the F test. The P value rises from 0.0001 to 0.8446 for the OF/G configuration when humidity is isolated, and the 80% RH condition is excluded. A possible explanation is that the highly hydrophilic nature of the POEGMA interface absorbed enough water during the 15–20 min stabilization time to adversely affect the performance of the assay.24,25 However, the curves remain concordant in the dynamic range above the LOD. This would also explain why the same behavior was not observed for the microfluidic cassette, which is sealed, preventing infiltration of high-humidity air into the reaction chamber during the stabilization period.
A more practical metric of resilience to environmental conditions is the likelihood of each configuration of the D4 to under- or over-report OVA concentrations at each condition. To test this, a universal five-parameter logistic regression was fit to the aggregate data of all five environmental conditions for each configuration of the D4 platform. Notably, the use of the universal fit had minimal impact on the calculated LOD of the assays (Table S1). Next, fluorescence intensities from the individual fits for each environmental condition—with associated known OVA values—were used to interpolate the concentration from the universal fits. The output concentration simulates a “measured” OVA value. The percent change in the two OVA concentrations can be used to calculate theoretical under- and over-reporting across the tested concentration range (Figure 2C,F). These results clearly demonstrate that the μF/D4S configuration is more resilient to environmental factors compared to the OF/G configuration, as evident by the tighter tolerance in over- and under-reporting for the former. Taken together, these data indicate that the μF/D4S assay is resilient to fluctuations in ambient temperature and humidity, which should enable successful deployment in diverse climatic settings, including outdoor testing or other locations that lack adequate climate control.
Performance in Undiluted Human Whole Blood.
Next, we investigated the performance when challenged with a more complex input sample—human whole blood. Blood is the ideal sample type to detect circulating biomarkers at the POC because it eliminates the need for sample preprocessing. We have demonstrated that the microfluidic cassette can be modified to operate with undiluted whole blood samples if they are collected in an anticoagulant tube (Figure 3A).13 Three changes were made to accomplish whole blood testing. (1) Due to the highly viscous nature of whole blood compared to serum, significantly shorter timing channels that lacked tall vertical loops were implemented in the microfluidic cassette. (2) A 30° slope was introduced to the offset in the reaction chamber of the cassette to prevent red blood cell (RBC) aggregation. (3) A 30° slope was also introduced at the junction between the channel outlet and the wicking pad to ensure consistent activation of wicking to initiate the washing and drying process. Figure 3A–C visualizes these changes from the original “serum” cassette to the one designed for whole blood.
Figure 3.
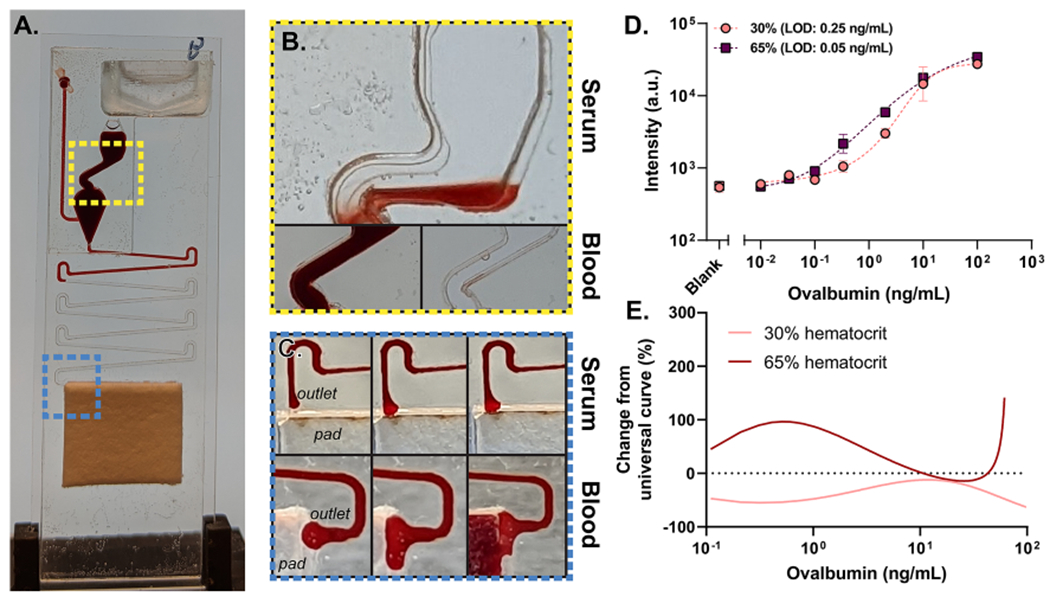
(A) Photograph of the modified blood cassette. (B) Photographs showing the modified 30° offset in the reaction chamber. (C) (Top) photograph of blood operating in the original “serum” cassette showing poor wicking of sample. (Bottom) Photograph of modified outlet with the improved wicking ability. (D) OVA dose–response curves in 30 and 65% hematocrit samples prepared from whole blood on the μF/D4S configuration of the D4; 8-point dose–response curves were generated from 0.01 to 100 ng/mL and a blank, with an incubation time of 23 min +/− 5 min. n = 3 for the 0, 0.01, 0.03, 0.1, 0.33, and, 2 ng/mL samples, whereas n = 2 for the 10 and 100 ng/mL samples. The error bars are the standard deviation. (E) Estimated percent change from the actual OVA concentration that μF/D4S is expected to report across concentrations.
In this study, we sought to understand the limitations of the cassette for quantitative diagnostic biosensing that can require near femtomolar sensitivities. In addition to the complexity of blood itself, variations in human hematocrit can play a large role in how a capillary- and gravity-driven system such as the microfluidic D4 assay performs in a clinical setting. Single-donor human whole blood was adjusted to hematocrits of 30 or 65% that span the physiologically expected range for humans across age, gender, and disease state and was tested on the optimized blood cassette.26,27 Figure 3D shows the resulting dose–response curves. The LODs for both conditions are comparable to or better than what was observed for OVA-spiked fetal bovine serum (FBS). The dose–response at 30% hematocrit is attenuated when compared to 65%, resulting in fits that are statistically distinct [F(5,28) = 8.669, P = <0.0001]. Despite the differences in the curves, analytical sensitivity remains comparable across all conditions tested. The differences, however, could result in modest over- and under-reporting of OVA concentrations within the dynamic range that would need to be considered in some applications that require precise quantitative capabilities (Figure 3E). One explanation for this shift may be the increased viscosity of blood at higher hematocrits which will increase incubation time. However, these results would represent the extremes of this behavior. Another explanation may be the presence of anti-OVA antibodies which are present in ~90% of humans28 and would be more abundant at lower hematocrits in our study because of how the hematocrit levels were adjusted. This effect would be OVA-specific and would not translate to other analytes.
Improved “EZ Load” Microfluidic Cassette.
Next, we sought to further improve the usability of the microfluidic cassette. This was first accomplished by identifying timing inefficiencies in “original” cassette’s design. The timing channel has two discrete sections. There are vertical loops that run parallel to the reaction chamber and downstream loops that snake below the reaction chamber toward the absorbent pad (Figure S2A). We theorized that the time-to-result could be vastly improved by programming all of the incubation times into the vertical loops, as they demonstrate increased incubation time as a function of channel length. To accomplish this, we redesigned the vertical loops of the cassette to be of identical height and then adjusted them from 0 to 20 mm (Figure 4A). The downstream loops were left in place to ensure that we can accurately identify where the efficiency gains came from. Using FBS, we tracked the time the sample spent in the vertical loops, downstream loops, and washing/drying (Figure 4B). We compared the results to the “original” unchanged cassette (left-most side of Figure 4B). Time spent in both vertical and downstream loops is considered the incubation time, while the time spent washing/drying represents the difference between incubation time and time-to-result. The ratio of the incubation time to time-to-result is the efficiency (listed above the bars in Figure 4B). Cassettes with downstream loops spent a consistent amount of time in the downstream loops (12.9 +/− 1.1 min) and washing and drying (15.8 +/− 2.2 min) while steadily increasing the time spent in vertical loops as the loop height increased, as expected. These results established that the incubation time could be programmed into the vertical loops. Next, two loop heights were selected to create cassettes without downstream channels. These designs achieved a superior efficiency and time-to-result while maintaining a stable incubation time (20.7 +/− 1.2 min for 17.6/no DS and 13.3 +/− 0.6 min for 13.2/no DS). More importantly, this more efficient cassette design achieves the same sensitivity and dynamic range as the “original” cassette while reducing the time-to-result by ~16 min (Figure 4C).
Figure 4.
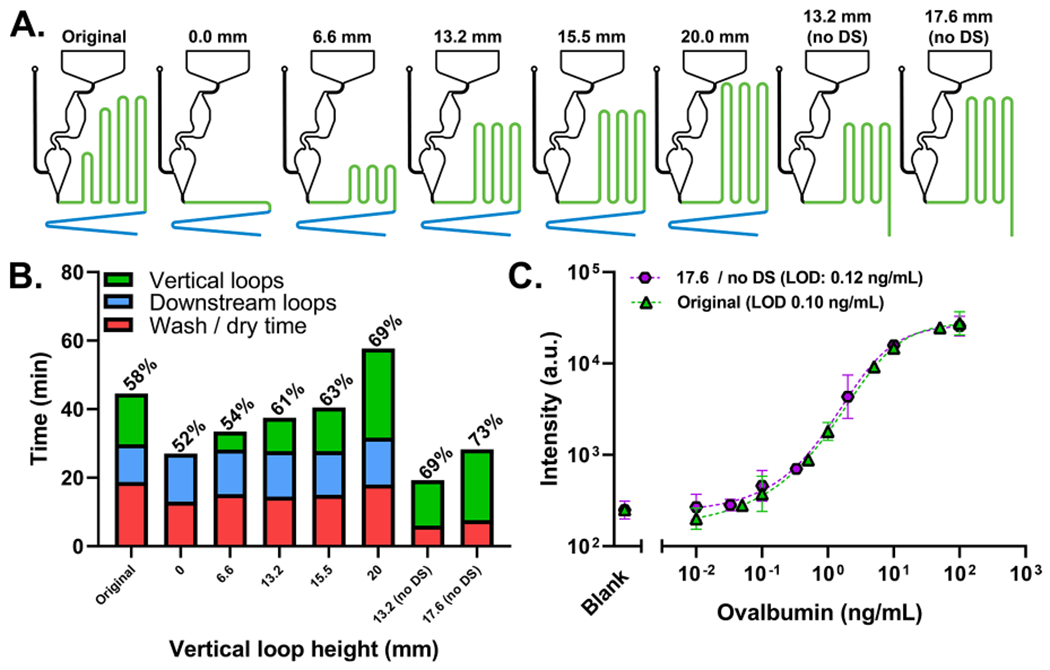
(A) Drawings showing modified cassette configurations. (B) Timing profile of the “original” version of the cassette (left), cassettes with modified vertical loops (middle six), and cassettes without vertical loops (right two). The timing efficiency is labeled above each bar. (C) OVA dose–response curves run in FBS using the “original” and “17.6/no DS” cassettes. The “original” was run as a 10-point dose curve from 0.01 to 100 ng/mL and a blank and has an incubation time of 23 ± 3 min (n = 4, error bars are the standard deviation). The “17.6 / no DS” cassette was run as an 8-point dose curve from 0.01 to 100 ng/mL and a blank and has an incubation time of 20.7 ± 1.2 min (n = 3, error bars are the standard deviation).
Next, we aimed to improve the ease of loading samples into the cassette. In the “original” cassette, a pipette is inserted perpendicular to the cassette, which requires an air-tight seal with the sample inlet to inject the sample into the reaction chamber (Figure S3A top). This requires a level of skill that can limit the scope of potential end users and increase the likelihood of user error. To improve the design, we widened the sample inlet and introduced the sample into the reaction chamber from above. This allows a simple dropper to be used to load the cassette (Figure 3SA bottom). This new inlet was used in conjunction with the more efficient timing channels to create the “EZ Load” microfluidic cassette (Figure 1B). To ensure the proper functionality of the cassette, two additional modifications were required. First, a delay channel was added to the wash buffer inlet to ensure proper sample loading prior to wash buffer release. Second, the reaction chamber shape was modified into a diamond to prevent air bubble formation when the sample settles into the reaction chamber. An exploded view of the cassette is shown in Figure S3B.
Diagnosis of Talaromycosis in Patients with Advanced HIV Disease.
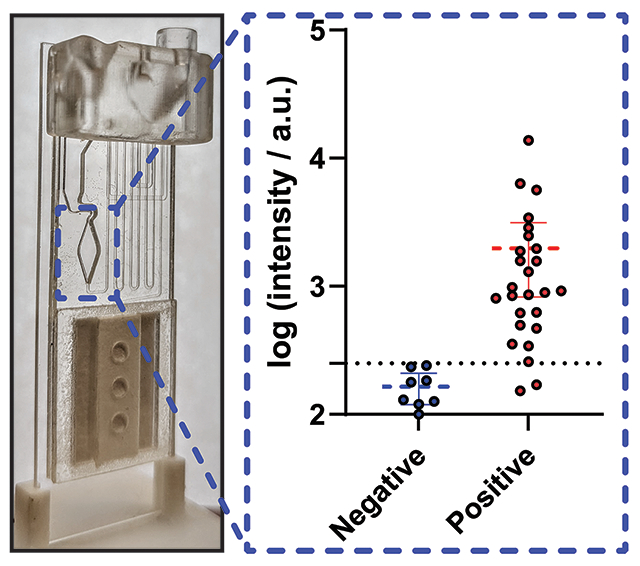
Finally, to demonstrate the clinical performance of the optimized μF D4 assay, we developed a POCT for talaromycosis diagnosis. We integrated a pair of previously discovered monoclonal and polyclonal antibodies into the D4 platform to detect Mp1p—a T. marneffei-specific cell wall mannoprotein that is abundantly secreted during talaromycosis infection.29 This pair demonstrated high analytical sensitivity in the μF/D4S configuration of the test (0.2 ng/mL) when spiked in pooled human serum (Figure S4A). Importantly, the cassettes remain functional and accurate when stored for 10 days at room temperature demonstrating the stability of the printed reagents (Figure S4B). The antibodies against Mp1p have been deployed in a laboratory-based enzyme immunoassay (EIA) format and have demonstrated a higher sensitivity than standard blood culture (86.3 vs 72.3%) and a high specificity (98.1%) in diagnosing talaromycosis in a large clinical study.30 Using the newly developed “EZ Load” microfluidic cassette, we carried out a case–control study using banked human plasma samples from a prospective talaromycosis screening cohort in Vietnam. Patients were adults hospitalized with advanced HIV disease as defined by a CD4 count <100 cells/μL who were determined to have culture-confirmed talaromycosis (26 cases) and no talaromycosis (8 controls). The OF/G and the μF/D4S configurations achieved a sensitivity of 92.3% (CI: 75.9%–98.6%) and a specificity of 100% (CI: 67.6–100.0%) (Figure 5). This is in comparison to the EIA that achieved a sensitivity of 96.2% (CI: 81.1–99.8%) and a specificity of 100% (CI: 70.1–100.0%) for the same samples (Figure S5), showing only a modest drop in sensitivity while gaining many benefits including testing at the POC without the need to carry out the test in a centralized facility, ease-of-use, fast time-to-result (30–45 min), and lower cost compared to ELISA and PCR. Importantly, this is a substantial improvement over the gold-standard blood culture method that can take a month to deliver a result with less sensitivity and is not a POCT. While a lateral flow POCT has been described for talaromycosis diagnosis by Pruksaphon et al.,31 our Mp1p test offers multiple advantages over an LFIA platform. Mp1p levels are detectable in both the blood and urine of infected patients1,30 and can be detected up to 16 weeks before patients develop symptoms. The described immunochromatographic test fails to function in serum and has an LOD of 0.6 μg/mL compared to our 0.2 ng/mL albeit for different target antigens. Additionally, the D4 assay has the ability to quantify and track antigen levels longitudinally in blood which for Mp1p would enable assessment of disease burden and severity and allow for monitoring the effectiveness of antifungal therapy and prognostication, which we have demonstrated can be done in other applications.13 The multiplexing capability of the D4 can enable additional tests to be integrated with the Mp1p assay to test for other AIDS-associated opportunistic infections.30,32 This can become invaluable, as the compromised immune system of those afflicted with talaromycosis means that there are often coinfections (e.g., other invasive mycoses, tuberculosis bacteria such as salmonellosis and other mycobacteria). These features of the D4 assay, as well as its evaluation in a larger clinical cohort, will be investigated in future studies, and we believe that they further differentiate our technology from existing tests.
Figure 5.
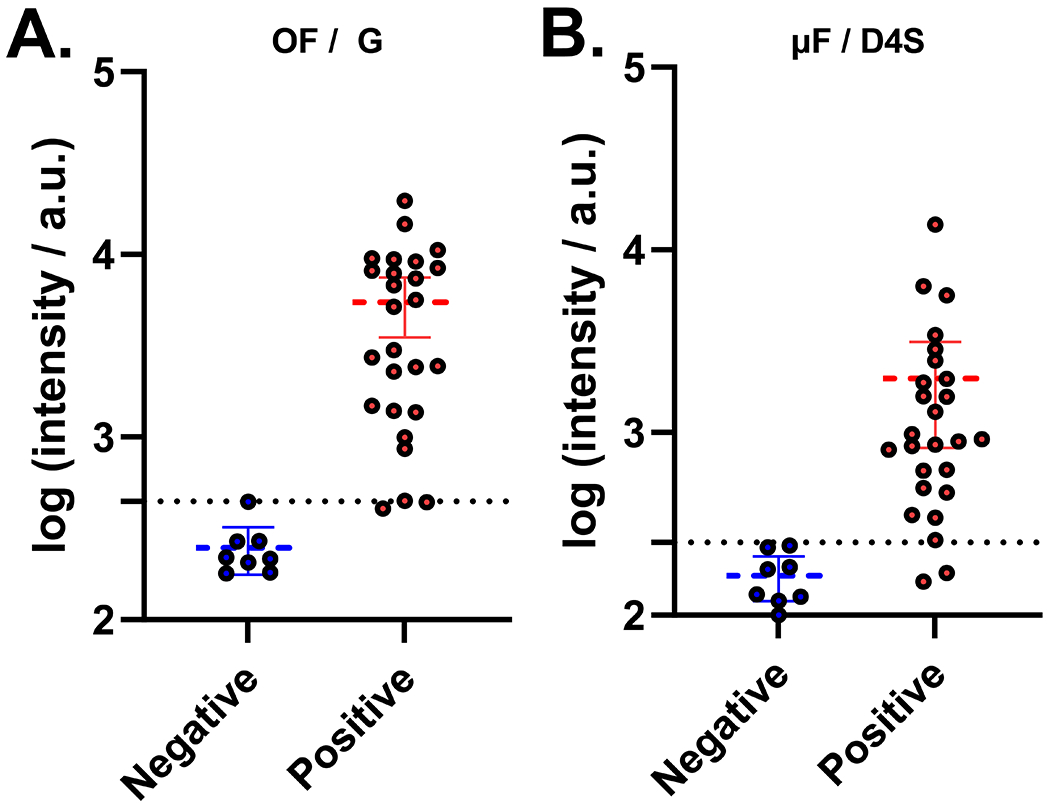
Data from 26 positive samples and 8 negative controls collected on the μF/D4S (A) and OF/G (B) configurations of the D4 assay. The dotted line represents the cutoff for the platform. 5/39 “EZ Load” cassettes removed due to failure from assembly error.
CONCLUSIONS
We demonstrated the resilience of a highly sensitive and passively automated microfluidic POCT to a wide range of environmental and physiological operational conditions. We showed that our new cassette architecture not only improves the portability of the platform but that it also shields the assay from environmental factors. Furthermore, we showed that the platform is easily amenable to functional upgrades that enhance the ease-of-use and reduce time-to-result without negatively affecting the performance of the device. As POCTs become more ubiquitous and move further away from the clinic, this level of systematic characterization, engineering safeguards, and user consideration is critical to improve the confidence in reported results of new POCTs and motivated in large part—this study, as we move forward to clinically translate this POCT. We also demonstrated the rapid adaptability of the platform by integrating the reagents to diagnose an emerging endemic fungal disease—talaromycosis—that has the potential to substantially improve prognosis of patients by enabling improved access to testing and more rapid intervention with antifungal therapies for symptomatic individuals. The talaromycosis D4 POCT has the potential to identify and treat patients before disease progression by providing a screening tool for at-risk individuals with advanced HIV disease where they live and work to detect the infection during the asymptomatic phase allowing for pre-emptive use of antifungal therapy. In conclusion, we believe that the D4 POCT platform is uniquely positioned to combine the ease-of-use of conventional LFIA tests with figures of merit typically associated with laboratory-grade tests, even in highly variable environmental test conditions.
MATERIALS AND METHODS
Operation of the Open Format D4 POCT.
Printed slides were loaded into microarray slide holders (Arrayit Co.). 60 μL of OVA (Sigma-Aldrich—A5503) in FBS (Sigma-Aldrich—F4135) was added into each well. Aluminum cover tape was applied to seal the arrays. The holders were then placed on an open-air rocking shaker for the desired incubation time. After incubation, the sample was removed, and each well was washed twice with 200 μL of wash buffer (0.1% v/v Tween-20 in 1× PBS). The slides were removed from the slide holder and completely submerged in a container of fresh wash buffer, before being moved to a container of deionized (DI) water. The slides were then promptly dried by centrifugation in a tabletop centrifuge at 2000g RCF for 10 s before imaging on an Axon Genepix 4400 tabletop scanner (Molecular Devices LLC).
Operation of the Microfluidic Cassettes.
The user places the microfluidic cassette vertically into the 3D-printed holder with the sample inlet oriented at the top and facing the user. Up to 22 additional cassettes are loaded onto the holder in the same way. For the “original” cassette, the user inserts the pipette firmly into the microfluidic cassette with the tip perpendicular to the cassette and pipets 60 μL of sample. Next, the user adds 135 μL of wash buffer to the wash reservoir. The user no longer needs to interact with the cassette until analysis on the D4Scope or Genepix. For the “EZ Load” microfluidic cassette, the sample volume and wash buffer volume are increased to 70 μL and 200 μL, respectively.
Variable Environmental Conditions.
For open format experiments, slides were affixed into microarray slide holders and stored in a plastic bag. For microfluidic experiments, cassettes were racked into a vertical slide holder and stored in a plastic bag. The temperature and humidity levels were measured using a Fisherbrand Traceable alarm relative humidity (RH)/temperature monitor by placing and sealing the monitor probe in the plastic bag. To simulate 10% RH at 22 °C, 80% RH at 22 °C, 50% RH at 4 °C, and 50% RH at 40 °C, an oven and refrigerator were used to modulate the temperature up and down, respectively, while the ambient room temperature was used to achieve 22 °C. To decrease the humidity to a desired level, 0.5 g packets of silica desiccant were added to the plastic bag incrementally until the desired humidity level remained stable. To increase humidity, small pieces of water-soaked paper towels were added to the plastic bag incrementally until the desired humidity level remained stable. Once the desired environmental condition was achieved and remained stable for >5 min, sample was added to the open format or microfluidic assays. For the open format assay, once the incubation time was complete, the slides were removed and processed, as discussed previously. The microfluidic cassettes were left at the specified environmental condition until the assay was completed. They were then removed and analyzed on the D4Scope and Genepix, as described previously.
Testing in Human Whole Blood.
Single-donor human whole blood treated with K2EDTA anticoagulant was purchased commercially (Innovative Research). A swinging bucket centrifuge was used to separate 50 mL aliquots of the blood into layers of plasma, buffy coat, and red blood cells (RBCs). The initial hematocrit percentage of the single donor was calculated as the percent ratio of RBCs to the total volume of the aliquot. Plasma was either removed or added from separate aliquots to achieve the desired percent hematocrit for testing.
Clinical Testing for Rapid Diagnosis of Talaromycosis.
Deidentified plasma samples were accessed from a talaromycosis screening cohort in Vietnam (Pro00101915, PI Thuy Le) approved by the Duke Health Institutional Review Board (IRB) via an exempted protocol approved by the Duke Health IRB (Pro00111992, PI Ashutosh Chilkoti). To facilitate operation in a biosafety cabinet, open format D4 assays were run using disposable adhesive gaskets. 1 mm CLAREX acrylic (Astra Products) backed with double-sided 9474LE adhesive (3M) was laser cut to form 24 separate wells matching the pattern of the open format D4 (Figure 1D). After incubation for 30 min, slides were submerged in wash buffer. Slides were washed an additional two times in fresh wash buffer and once in DI water. Tweezers were used to remove the gasket. The slide was washed one more time in DI water before being promptly dried by centrifugation in a tabletop centrifuge at 2000g RCF for 10 s before imaging on the Genepix scanner.
Data Analysis, Post-Processing, and Statistical Methods.
Genepix Pro analysis software, Microsoft Excel, and GraphPad were used for all data analysis and post-processing. More information regarding data processing, the objective outlier removal algorithm, and extra sum-of-squares F test can be found in the Supporting Information.
Supplementary Material
ACKNOWLEDGMENTS
A.C. acknowledges the support of the Defence Academy of the United Kingdom (Grant No. ACC6010469) and the NIH through Grants UH3CA211232, R01AI150888, and R01AI159992. D.S.K. acknowledges NIH Grant T32GM008555. T.L. acknowledges the following NIH grants: R01AI143409, U01AI169358, and 5P30AI064518.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssensors.3c00209.
Additional experimental methods, detailed table of LODs, dose–response curves for μF/G configuration, details on “EZ Load” cassette development, and data on the Mp1p implementation of the D4 (PDF)
Details of the manufacturing and assembly process of the microfluidic D4 POCT and demonstration of device operation and imaging on the D4Scope (MOV)
Source data (XLSX)
Complete contact information is available at: https://pubs.acs.org/10.1021/acssensors.3c00209
The authors declare the following competing financial interest(s): A.C., D.S.K., C.M.F., A.M.H., J.L., and J.T.H. are inventors on the patent filed by Duke University [PCT/US2021/046833, filed (20 August 2021) entitled Microfluidic assay device and describes the innovations used for the D4 microfluidic cassette in this work. A.C. and J.L. are inventors on a patent related to this work filed by Duke University [no. WO/2020/223713, filed (2 May 2020), published (5 May 2020)]. The patent is entitled Devices and methods for imaging microarray chips and describes innovations used for the D4Scope described in this work. Immucor Inc. has acquired the rights to the D4 assay on POEGMA brushes for in vitro diagnostics from Sentilus Inc. (cofounded by A.C. and A.M.H.). All other authors declare that they have no competing interests.
Contributor Information
David S. Kinnamon, Department of Biomedical Engineering, Pratt School of Engineering, Duke University, Durham, North Carolina 27708, United States
Jacob T. Heggestad, Department of Biomedical Engineering, Pratt School of Engineering, Duke University, Durham, North Carolina 27708, United States
Jason Liu, Department of Biomedical Engineering, Pratt School of Engineering, Duke University, Durham, North Carolina 27708, United States.
Thu Nguyen, Division of Infectious Diseases and International Health, Duke University School of Medicine, Durham, North Carolina 27708, United States.
Vo Ly, Hospital for Tropical Disease, Ho Chi Minh City 73009, Vietnam; University of Medicine and Pharmacy at Ho Chi Minh City, Ho Chi Minh City 72714, Vietnam.
Angus M. Hucknall, Department of Biomedical Engineering, Pratt School of Engineering, Duke University, Durham, North Carolina 27708, United States
Cassio M. Fontes, Department of Biomedical Engineering, Pratt School of Engineering, Duke University, Durham, North Carolina 27708, United States
Rhett J. Britton, Department of Biomedical Engineering, Pratt School of Engineering, Duke University, Durham, North Carolina 27708, United States
Jian-Piao Cai, State Key Laboratory of Emerging Infectious Diseases, Carol Yu Centre for Infection, Department of Microbiology, School of Clinical Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Pokfulam 000000, Hong Kong.
Jasper Fuk-Woo Chan, State Key Laboratory of Emerging Infectious Diseases, Carol Yu Centre for Infection, Department of Microbiology, School of Clinical Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Pokfulam 000000, Hong Kong; Hainan Medical University—The University of Hong Kong Joint Laboratory of Tropical Infectious Diseases, Hainan Medical University, Haikou 571101 Hainan, China.
Kwok-Yung Yuen, State Key Laboratory of Emerging Infectious Diseases, Carol Yu Centre for Infection, Department of Microbiology, School of Clinical Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Pokfulam 000000, Hong Kong; Hainan Medical University—The University of Hong Kong Joint Laboratory of Tropical Infectious Diseases, Hainan Medical University, Haikou 571101 Hainan, China.
Thuy Le, Division of Infectious Diseases and International Health, Duke University School of Medicine, Durham, North Carolina 27708, United States.
Ashutosh Chilkoti, Department of Biomedical Engineering, Pratt School of Engineering, Duke University, Durham, North Carolina 27708, United States.
REFERENCES
- (1).Jiang J; Meng S; Huang S; Ruan Y; Lu X; Li JZ; Wu N; Huang J; Xie Z; Liang B; et al. Effects of Talaromyces marneffei infection on mortality of HIV/AIDS patients in southern China: a retrospective cohort study. Clin. Microbiol. Infect 2019, 25, 233–241. [DOI] [PubMed] [Google Scholar]
- (2).Vanittanakom N; Cooper CR Jr; Fisher MC; Sirisanthana T Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin. Microbiol. Rev 2006, 19, 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Limper AH; Adenis A; Le T; Harrison TS Fungal infections in HIV/AIDS. Lancet Infect. Dis 2017, 17, e334–e343. [DOI] [PubMed] [Google Scholar]
- (4).Le T; Kinh NV; Cuc NTK; Tung NLN; Lam NT; Thuy PTT; Cuong DD; Phuc PTH; Vinh VH; Hanh DTH; et al. A Trial of Itraconazole or Amphotericin B for HIV-Associated Talaromycosis. N. Engl. J. Med 2017, 376, 2329–2340. [DOI] [PubMed] [Google Scholar]
- (5).Le T; Wolbers M; Chi NH; Quang VM; Chinh NT; Lan NPH; Lan NP; Lam PS; Kozal MJ; Shikuma CM; Day JN Epidemiology, seasonality, and predictors of outcome of AIDS-associated Penicillium marneffei infection in Ho Chi Minh City, Viet Nam. Clin. Infect. Dis 2011, 52, 945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kawila R; Chaiwarith R; Supparatpinyo K Clinical and laboratory characteristics of Penicilliosis marneffei among patients with and without HIV infection in Northern Thailand: a retrospective study. BMC Infect. Dis 2013, 13, No. 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Pornprasert S; Praparattanapan J; Khamwan C; Pawichai S; Pimsarn P; Samleerat T; Leechanachai P; Supparatpinyo K Development of TaqMan real-time polymerase chain reaction for the detection and identification of Penicillium marneffei. Mycoses 2009, 52, 487–492. [DOI] [PubMed] [Google Scholar]
- (8).Hien HTA; Thanh TT; Thu NTM; Nguyen A; Thanh NT; Lan NPH; Simmons C; Shikuma C; Chau NVV; Thwaites G; Le T Development and evaluation of a real-time polymerase chain reaction assay for the rapid detection of Talaromyces marneffei MP1 gene in human plasma. Mycoses 2016, 59, 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).UK COVID-19 Lateral Flow Oversight Team; Peto T; Affron D; Afrough B; Agasu A; Ainsworth M; Allanson A; Allen K; Allen C; Archer L; Ashbridge N; et al. COVID-19: Rapid Antigen detection for SARS-CoV-2 by lateral flow assay: a national systematic evaluation for mass-testing. eClinicalMedicine 2021, 36, 100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Whitman JD; Hiatt J; Mowery CT; Shy BR; Yu R; Yamamoto TN; Rathore U; Goldgof GM; Whitty C; Woo JM; et al. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat. Biotechnol. 2020, 38, 1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Pollock NR; Jacobs JR; Tran K; Cranston AE; Smith S; O’Kane CY; Roady TJ; Moran A; Scarry A; Carroll M; et al. Performance and Implementation Evaluation of the Abbott BinaxNOW Rapid Antigen Test in a High-Throughput Drive-Through Community Testing Site in Massachusetts. J. Clin. Microbiol 2021, 59, No. e00083–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Choi JR; Hu J; Feng S; Abas WABW; Pingguan-Murphy B; Xu F Sensitive biomolecule detection in lateral flow assay with a portable temperature–humidity control device. Biosens. Bioelectron 2016, 79, 98–107. [DOI] [PubMed] [Google Scholar]
- (13).Heggestad JT; Kinnamon DS; Olson LB; Liu J; Kelly G; Wall SA; Oshabaheebwa S; Quinn Z; Fontes CM; Joh DY; et al. Multiplexed, quantitative serological profiling of COVID-19 from blood by a point-of-care test. Sci. Adv 2021, 7, No. eabg4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Joh DY; Heggestad JT; Zhang S; Anderson GR; Bhattacharyya J; Wardell SE; Wall SA; Cheng AB; Albarghouthi F; Liu J; et al. Cellphone enabled point-of-care assessment of breast tumor cytology and molecular HER2 expression from fine-needle aspirates. npj Breast Cancer 2021, 7, No. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fontes CM; Lipes BD; Liu J; Agans KN; Yan A; Shi P; Cruz DF; Kelly G; Luginbuhl KM; Joh DY; et al. Ultrasensitive point-of-care immunoassay for secreted glycoprotein detects Ebola infection earlier than PCR. Sci. Transl. Med 2021, 13, No. eabd9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Heggestad JT; Britton RJ; Kinnamon DS; Wall SA; Joh DY; Hucknall AM; Olson LB; Anderson JG; Mazur A; Wolfe CR; et al. Rapid test to assess the escape of SARS-CoV-2 variants of concern. Sci. Adva 2021, 7, No. eabl7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hucknall A; Kim D-H; Rangarajan S; Hill RT; Reichert WM; Chilkoti A Simple Fabrication of Antibody Microarrays on Nonfouling Polymer Brushes with Femtomolar Sensitivity for Protein Analytes in Serum and Blood. Adv. Mater 2009, 21, 1968–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Joh DY; Hucknall AM; Wei Q; Mason KA; Lund ML; Fontes CM; Hill RT; Blair R; Zimmers Z; Achar RK; et al. Inkjet-printed point-of-care immunoassay on a nanoscale polymer brush enables subpicomolar detection of analytes in blood. Proc. Natl. Acad. Sci. U.S.A 2017, 114, E7054–E7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Fontes CM; Achar RK; Joh DY; Ozer I; Bhattacharjee S; Hucknall A; Chilkoti A. Engineering the Surface Properties of a Zwitterionic Polymer Brush to Enable the Simple Fabrication of Inkjet-Printed Point-of-Care Immunoassays. Langmuir 2019, 35, 1379–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wang GR; Yang JY; Lin TL; Chen HY; Horng CB Temperature effect on the sensitivity of ELISA, PA and WB to detect anti-HIV-1 antibody and infectivity of HIV-1. Zhonghua Yi Xue Za Zhi 1997, 59, 325–333. [PubMed] [Google Scholar]
- (21).McLaughlin MR; Barnett OW; Burrows PM; Baum RH Improved ELISA conditions for detection of plant viruses. J. Virol. Methods 1981, 3, 13–25. [DOI] [PubMed] [Google Scholar]
- (22).Sigma Millipore. ELISA Procedures. https://www.sigmaaldrich.com/US/en/technical-documents/protocol/protein-biology/elisa/elisa-procedures (accessed 2022.
- (23).Olanrewaju A; Beaugrand M; Yafia M; Juncker D. Capillary microfluidics in microchannels: from microfluidic networks to capillaric circuits. Lab Chip 2018, 18, 2323–2347. [DOI] [PubMed] [Google Scholar]
- (24).Chen S; Li L; Zhao C; Zheng J Surface hydration: Principles and applications toward low-fouling/nonfouling biomaterials. Polymer 2010, 51, 5283–5293. [Google Scholar]
- (25).Joh DY; McGuire F; Abedini-Nassab R; Andrews JB; Achar RK; Zimmers Z; Mozhdehi D; Blair R; Albarghouthi F; Oles W; et al. Poly(oligo(ethylene glycol) methyl ether methacrylate) Brushes on High-κ Metal Oxide Dielectric Surfaces for Bioelectrical Environments. ACS Appl. Mater. Interfaces 2017, 9, 5522–5529. [DOI] [PubMed] [Google Scholar]
- (26).HH B Clinical Methods: The History, Physical, and Laboratory Examinations; Butterworths, 1990. [PubMed] [Google Scholar]
- (27).Pagana K; Pagana T; Pagana T. Mosby’s Diagnostic and Laboratory Test Reference, 2020. [Google Scholar]
- (28).Kilshaw PJ; McEwan FJ; Baker KC; Cant AJ Studies on the specificity of antibodies to ovalbumin in normal human serum: technical considerations in the use of ELISA methods. Clin. Exp. Immunol 1986, 66, 481–489. [PMC free article] [PubMed] [Google Scholar]
- (29).Cao L; Chan CM; Lee C; Wong SS; Yuen KY MP1 encodes an abundant and highly antigenic cell wall mannoprotein in the pathogenic fungus Penicillium marneffei. Infect. Immun 1998, 66, 966–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Thu NTM; Chan JFW; Ly VT; Ngo HT; Hien HTA; Lan NPH; Chau NVV; Cai J-P; Woo PCY; Day JN; et al. Superiority of a Novel Mp1p Antigen Detection Enzyme Immunoassay Compared to Standard BACTEC Blood Culture in the Diagnosis of Talaromycosis. Clin. Infect. Dis 2021, 73, e330–e336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Pruksaphon K; Intaramat A; Simsiriwong P; Mongkolsuk S; Ratanabanangkoon K; Nosanchuk JD; Kaltsas A; Youngchim S An inexpensive point-of-care immunochromatographic test for Talaromyces marneffei infection based on the yeast phase specific monoclonal antibody 4D1 and Galanthus nivalis agglutinin. PLoS Neglected Trop. Dis 2021, 15, No. e0009058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ly VT; Thanh NT; Thu NTM; Chan J; Day JN; Perfect J; Nga CN; Chau NVV; Le T Occult Talaromyces marneffei Infection Unveiled by the Novel Mp1p Antigen Detection Assay. Open Forum Infect. Dis 2020, 7, No. ofaa502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


