Abstract
As the number of heart transplants performed annually in the United States and worldwide continues to increase, there has been little change in graft longevity and patient survival over the past two decades. The reference standard for diagnosing acute cellular and antibody mediated rejection includes histologic and immunofluorescence evaluation of endomyocardial biopsy (EMB) samples, despite its invasiveness and the high interrater variability for grading histologic rejection. Circulating biomarkers and molecular diagnostics have shown significant predictive value for rejection monitoring, and emerging data support their ability to diagnose other post-transplant complications. Genomic (cell-free DNA), transcriptomic (profiling of messenger RNA and microRNA), and proteomic (protein expression quantitation) methodologies to diagnose these post-transplant outcomes have been evaluated with varying levels of evidence. In parallel, growing knowledge about the genetically mediated immune response leading to rejection (immunogenetics), has enhanced our understanding of antibody-mediated rejection, associated graft dysfunction, and mortality. Antibodies to donor human leukocyte antigens (HLA), and the technology available to evaluate these antibodies continues to evolve. This review aims to provide an overview of biomarker and immunologic tests used to diagnose post-transplant complications. This includes a discussion of pediatric heart transplantation and the disparate rates of rejection and mortality experienced by Black transplant patients. This Frontiers review describes diagnostic modalities currently available and utilized after transplant and describes the landscape of future investigations needed to enhance patient outcomes after heart transplantation.
Keywords: heart transplantation, biomarkers, immunogenetics, cell-free DNA, microRNAs, HLA
Introduction
The number of heart transplantations (HTs) performed within the United States has nearly doubled in the past decade due to advances in donor utilization and the ongoing opioid epidemic, and contemporary data indicate that the number of transplants yearly exceeds 4,000 in the US and 7,000 worldwide. Despite the increased availability of this definitive therapy, clinical outcomes after HT have changed minimally in the past two decades, with a median post-transplant survival of 13 years1. Within the first year after HT, 13–24% of patients experience cardiac allograft rejection (CAR), and, as time from transplant increases, chronic rejection, cardiac allograft vasculopathy (CAV), and late graft failure become more prevalent1,2. The endomyocardial biopsy (EMB) was developed 50 years ago to diagnose CAR and remains the reference standard; however, it is limited by invasiveness, complication rate (approximately 4%), requirement of general anesthesia (in children), and variability in histopathologic interpretation which can lead to under- and over-treatment3–5.
In the past two decades there has been extensive investigation into molecular (-omic) diagnostics and other circulating biomarkers to facilitate the non-invasive diagnosis of CAR. Contemporary methodologies, including blood-based cell-free DNA (genomics) and gene expression profiling (GEP, transcriptomics) are now used clinically to reduce surveillance EMB frequency (Table 1, Figure 1)6–10. Research into the use of immunogenetics in HT has paralleled this growth in understanding of molecular biomarkers. Through more precise genotyping and enhanced detection of antibodies to human leukocyte antigens (HLAs), there has been heightened recognition of the consequences of pre-transplant sensitization that informs desensitization strategies, organ selection, and has all but eliminated hyperacute rejection at the time of transplant. After transplant, the contribution of donor-specific antibodies (DSAs) to antibody-mediated rejection (AMR), which often manifests with more significant graft dysfunction and has a predilection for recurrence, is widely recognized12–14. With the clinical adoption of these technologies, a better understanding of differences in recipient age and race/ethnicity as well as other factors affecting the immune response and risk of post-transplant adverse events is needed15–18. This state-of-the-art review aims to provide an overview of the current and emerging modalities utilized to manage HT patients.
Table 1.
Description of Molecular Diagnostics, Circulating Biomarkers, and Immunogenetic Modalities
| Technology | Description | Commercially Available Assay (Cost in USD*) |
|---|---|---|
| Donor-Derived Cell-free DNA (dd-cfDNA) | Circulating ~150 base pair nucleotide fragments measured as ratio of donor to recipient DNA or %dd-cfDNA | Allosure ($2,790) Prospera ($2,790) |
| Gene Expression Profiling (GEP) | 11-gene panel scored from 0 to 40, with 9 genes for normalization | AlloMap ($3,240) |
| Intragraft mRNA Transcripts | Gene expression profiling of endomyocardial biopsy tissue samples (includes nCounter research assay) | MMDx, Molecular Microscope ($3,159) |
| MicroRNA (miRs) | Circulating ~22 base pair non-coding RNA molecules | |
| Soluble Protein Biomarkers | Natriuretic peptides (BNP, NT-proBNP); markers of myocyte injury (troponin, high-sensitivity troponin); inflammatory markers (CRP, IL-6, others) | BNP and NT-proBNP ($39) cTnI, cTnT, hs-cTnI, hs-cTnT($12) CRP($13) IL-6 ($17) |
| Protein Expression Profiling | Multi-protein assay that can be evaluated for clinically relevant expression profiles (includes Olink® and Somascan® research assays) | |
| Exosomes | Extracellular vesicles that can be isolated and their encapsulated proteins and nucleic acids can be quantitated. | |
| Human Leukocyte Antigen (HLA) Antibodies | Pre- or post-transplant (donor-specific) antibodies to HLA antigens | HLA Typing ($780) HLA Antibody Screen ($64) Luminex HLA DSA Quantification ($680) |
| Non-HLA Antibodies | Angiotensin II type-I-receptor (AT1R), endothelin type A receptor, and major histocompatibility-complex class I-related chain A (MICA) | AT1R ($450) |
BNP, brain type natriuretic peptide; CRP, C-reactive protein, cTn, cardiac Troponin; PRA, panel reactive antibody; DSA, donor-specific antibody
cost data from 2023 Centers for Medicare & Medicaid Services Clinical Laboratory Fee Schedule11.
Figure 1. Non-Invasive Diagnostics and Respective Evidentiary Support to Identify Rejection Phenotypes.
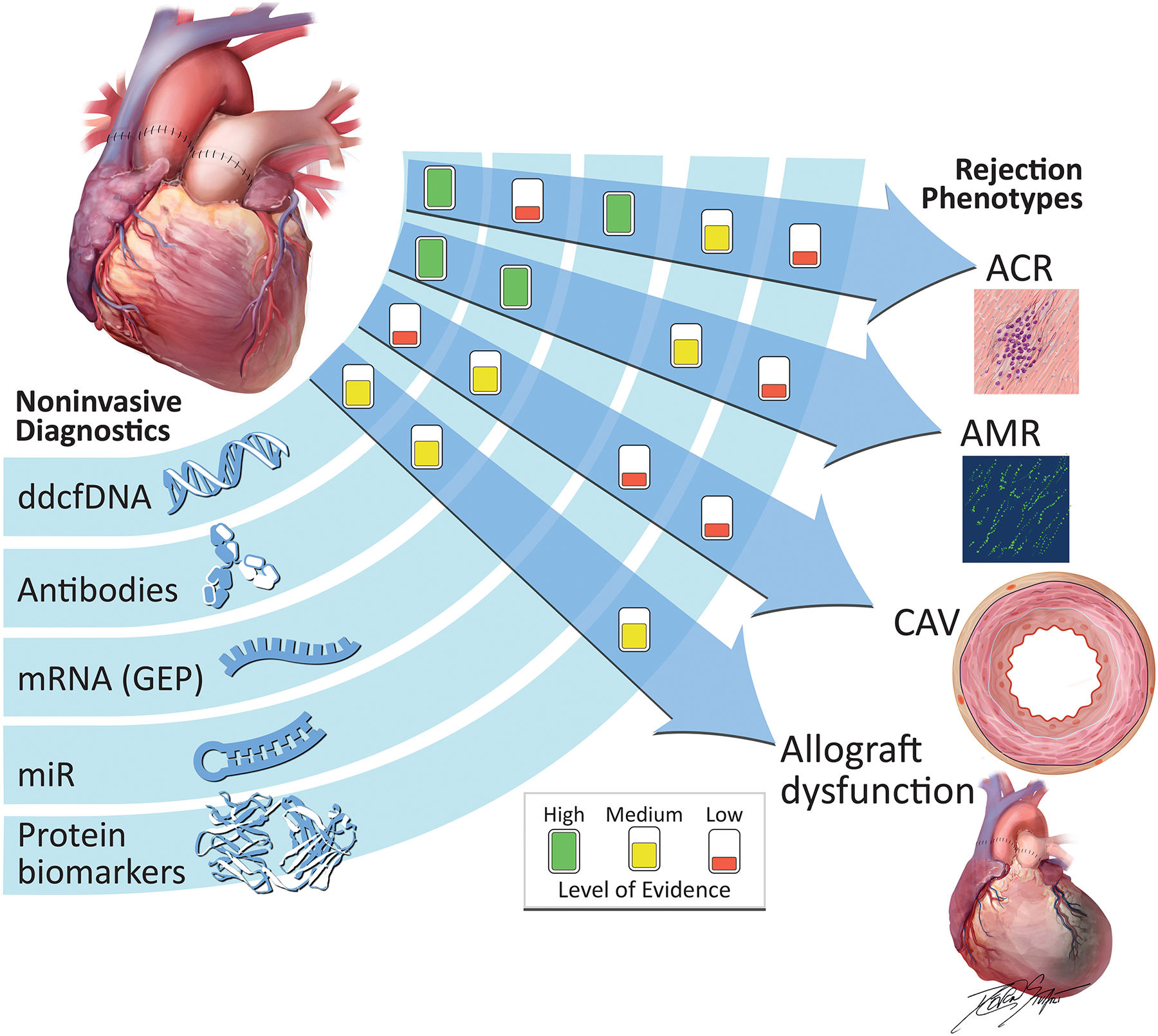
Differing levels of evidence exist for non-invasive diagnostic modalities to detect post-transplant rejection, per recommendations of a recent consensus guideline. Associations are denoted as clinical discovery, clinical validity and clinical utility. Only the association between GEP and ACR meets clinical utility criteria, given the IMAGE randomized, controlled trial. The GRAfT, multicenter prospective study resulted in clinical validity for dd-cfDNA’s ability to detect ACR and AMR. The presence of HLA donor-specific antibodies have been associated with AMR, CAV, and allograft dysfunction. Recent microRNA work has provided evidence of its role in diagnosis of ACR and AMR. Finally, proteomic evaluation has identified circulating biomarkers of allograft dysfunction and CAV.
ACR, acute cellular rejection; AMR, antibody-mediated rejection; CAV, coronary allograft vasculopathy; dd-cfDNA, donor-derived cell-free DNA; HLA, human leukocyte antigen; mRNA, messenger RNA; miR, microRNA.
Donor-Derived Cell-Free DNA
Cell-free DNA describes extracellular double-stranded DNA, 20–166 base pairs in length that reside in most bodily fluids and originate from nuclear, mitochondrial, or non-human (bacterial or viral) sources19. With cell death due to disease processes such as CAR, as well as with natural cell turnover (apoptosis), cell-free DNA is released into the bloodstream. The utilization of cell-free DNA in solid organ transplantation stems from knowledge gained in its prior applications, in prenatal diagnostics (fetal DNA found in maternal circulation) as well as cancer surveillance (tumor DNA found in bloodstream). The initial work demonstrating that cell-free DNA can be used for CAR evaluation was performed at Stanford University: donor and recipient pre-transplant DNA were evaluated with whole genome sequencing to identify ~1.5 million distinguishing single nucleotide polymorphisms (SNPs)20. However, only a subset of the donor-recipient SNPs remain informative in identifying the donor fraction. Informative SNPs represent those where the recipient is homozygous (2 alleles with the same nucleotide) and different from the donor, who is either heterozygous or homozygous for a different nucleotide; thus permitting differentiation of donor and recipient DNA. The ratio of donor-derived to recipient DNA allows for calculation of the percentage of donor-derived cell free DNA (dd-cfDNA). Given the small concentration of donor (as compared to recipient) cell-free DNA, dd-cfDNA assays require high sensitivity for the detection of donor vs. recipient cell-free DNA. A prospective study from Stanford of 21 pediatric and 44 adult recipients showed that a dd-cfDNA threshold of 0.25% was associated with the composite outcome of biopsy-proven acute cellular rejection (ACR ≥ Grade 2R) or AMR, with area under the curve (AUC) of 0.83 (Figure 2)20.
Figure 2. Featured Donor-Derived Cell-Free DNA Publications and their Contributions to the Evaluation of Cardiac Allograft Rejection.
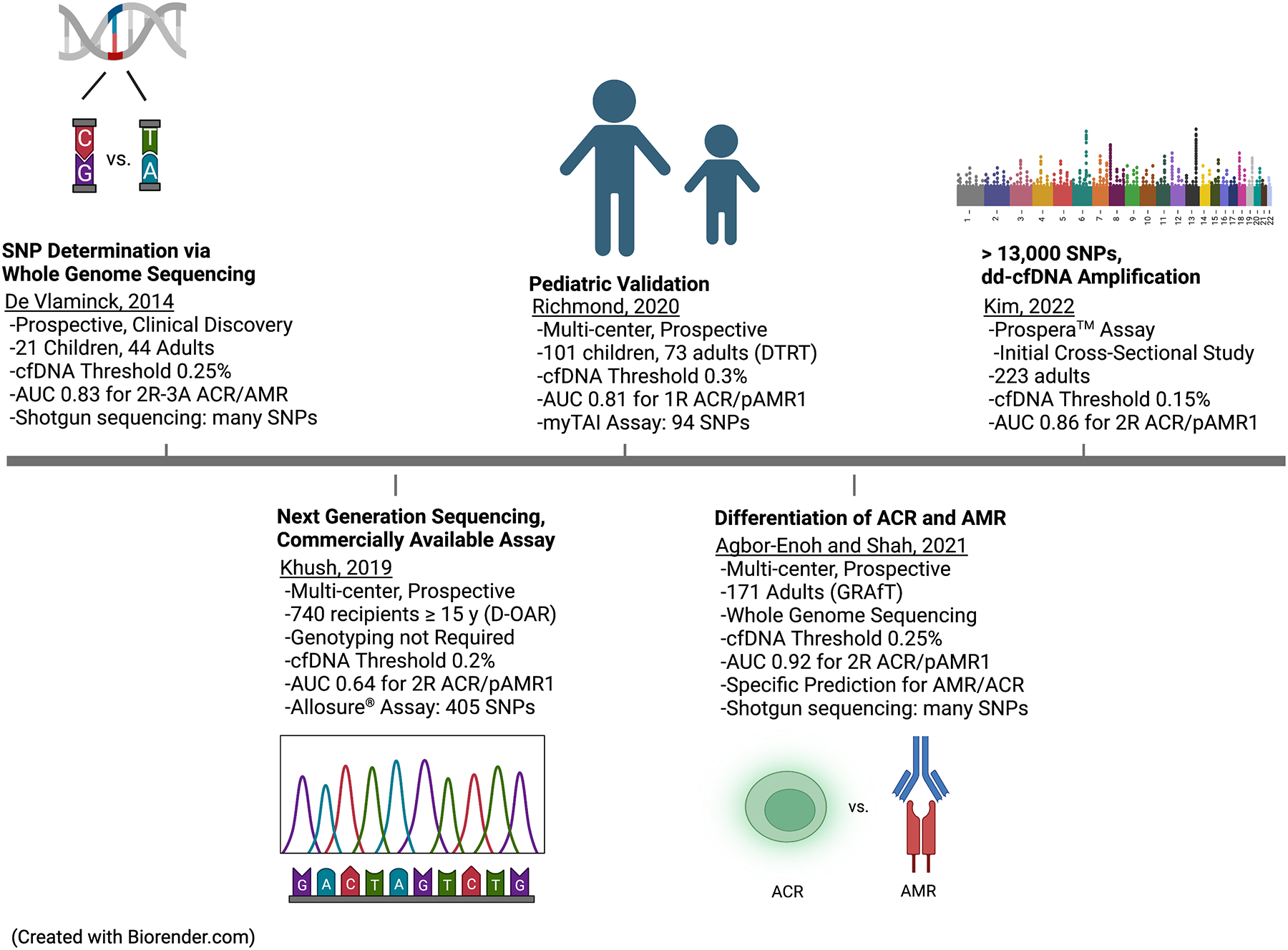
This represents the time course of major studies which contributed to our understanding of the potential clinical applications of dd-cfDNA in the diagnosis of cardiac allograft rejection. The contributions include the initial research assay or clinical discovery work performed at Stanford University in 2014. Clinical validation studies by the GRAfT consortium showed high diagnostic performance of dd-cfDNA when considering ACR and AMR separately. Expansion of research into a large cohort of pediatric patients by the DTRT group. And finally, use of commercial assays that do not require donor or recipient genotyping.
ACR, acute cellular rejection; AMR, antibody-mediated rejection; AUC, area under the curve; cfDNA, cell-free DNA; DTRT, DNA-Based Transplant Rejection
Test; GRAfT, Genomic Research Alliance for Transplantation; SNP, single nucleotide polymorphisms.
A contemporary publication from the Genomic Research Alliance for Transplantation (GRAfT), a multicenter, prospective, longitudinal cohort study, which required donor and recipient genotyping in 165 adults, demonstrated that a dd-cfDNA threshold of 0.25% was associated with a composite of ACR ≥ 2R or pathologic AMR (pAMR) ≥1, with AUC of 0.92, sensitivity of 81%, specificity of 85%, positive predictive value (PPV) of 19.6%, and negative predictive value (NPV) of 99.2%7. The GRAfT study had adequate power to evaluate dd-cfDNA diagnostic performance for both ACR and AMR, with individual AUCs of 0.89 and 0.95, respectively. Importantly, the cohort was diverse and included a higher proportion (44%) of Black patients than prior dd-cfDNA studies, as these patients have historically worse post-transplant outcomes. Given the interrater variability in EMB interpretation and the frequent clinical observation of allograft dysfunction by echocardiography with a negative EMB, the GRAfT investigators evaluated the ability of EMB to detect rises of dd-cfDNA (i.e., utilizing dd-cfDNA rather than EMB as the reference standard). The EMB had 19.6% sensitivity and 99.2% specificity in detecting elevated cell-free DNA. Of the 135 dd-cfDNA elevations that were missed by the EMB (i.e., a ‘false negative’ EMB), 21% had concurrent allograft dysfunction (decline in ejection fraction ≥ 5%) and 44% preceded a diagnosis of CAR (more frequently AMR than ACR). Clinical management of patients with a negative EMB and abnormal dd-cfDNA (or other molecular marker) is challenged by a lack of sufficient data to guide clinicians. The long-term trajectory of these patients is unclear, and the risks of over-immunosuppression need to be considered carefully.
To enable broad applicability and quick turnaround of dd-cfDNA results, approaches targeting prevalent SNPs in the general population have been developed and are now commercially available. These approaches eliminate the need for donor and recipient genotyping and utilize multiplex polymerase chain reaction (PCR) coupled with targeted next-generation sequencing of informative SNPs that permit dd-cfDNA quantification6,7,21. The most widely studied and clinically employed assay is Allosure® (CareDx, Inc., Brisbane, CA), which currently includes a panel of 405 SNPs and was clinically validated in the Donor-Derived Cell-Free DNA-Outcomes AlloMap Registry (D-OAR) study6. This observational, prospective study enrolled 740 HT recipients ≥ 15 years of age (at multiple time points post-transplant) from 26 centers, utilizing a dd-cfDNA threshold of 0.2% to predict a composite outcome of ACR ≥ 2R or pAMR ≥1, resulting in a sensitivity of 53.8%, specificity of 76.1%, PPV of 11.6%, NPV of 96.6%, and AUC of 0.64 (which is moderate and highlights the lack of precision of this modality as compared to its previous pre-clinical evaluations). Another commercial assay is Prospera™ (Natera, Inc., Austin, TX) which interrogates ~13,000 SNPs. In an initial two-center case-control evaluation of this assay, there was a sensitivity of 78.5, specificity of 76.9, PPV of 25.1%, NPV of 97.3%, and AUC Of 0.86 for the detection of ACR ≥ 2R or pAMR ≥ 122. Currently, dd-cfDNA tests are optimally used to avoid biopsies when there is low pre-test probability and clinical suspicion for ACR and AMR.
Gene Expression Profiling
Gene expression is an intriguing biomarker as it may not only permit surveillance and diagnosis of CAR but may provide insights into molecular mechanisms implicated in the alloimmune response. From peripheral blood mononuclear cells, the quantity of messenger RNA (mRNA) transcripts for genes involved in inflammation, tissue injury, or other known CAR processes can be reverse transcribed (RT) to DNA, then amplified via PCR, allowing quantification. The eight-center Cardiac Allograft Rejection Gene Expression Observational (CARGO) trial utilized RT-PCR methodology to quantify expression of candidate genes (Table 2)8. This study initially performed a microarray analysis of over 7,000 candidate genes, and, with training and validation testing, resulted in an 11-gene classifier test that became AlloMap® (CareDx, Inc., Brisbane, CA). The CARGO study included a validation cohort distinguishing ACR ≥ 3A among samples with a score >20 (maximum score = 40) as well as a prevalence analysis, including samples taken at a longer (≥ 1-year) post-transplant time and utilizing a higher (>30) rejection score, resulting in a PPV of 6.8% and NPV of >99% for this assay. This was followed by one of the only randomized, controlled clinical trials in CAR diagnosis: the Invasive Monitoring Attenuation through Gene Expression (IMAGE) study, which was designed to test the non-inferiority of surveillance with GEP as compared to EMB9. In this study, 602 low-risk HT patients (those with normal cardiac function and without history of AMR or CAV) who were ≥ 6 months post-transplant were randomized to a traditional EMB or GEP surveillance strategy. There were similar two-year event rates (hemodynamically significant ACR or AMR, graft dysfunction, or mortality) between EMB (15.3%) and GEP (14.5%) groups, suggesting that this non-invasive approach may be reasonable in low-risk patients >6 months post-transplant.
Table 2.
Blood-Based GEP of Messenger RNA and MicroRNA Transcripts
| Publication | Study Name | Patient Population & Study Design | Description | Outcome |
|---|---|---|---|---|
| Messenger RNA | ||||
| Deng 8 , 2006 | CARGO | 8 U.S. centers, 629 patients, Prospective cohort study | Derivation and validation of GEP | AUC of 0.72 to detect ACR ≥ 3A with GEP score >20, prevalence analysis resulting in 99% NPV using score >30 |
| Pham 9 , 2010 | IMAGE | 13 U.S. centers, 602 patients, Randomized clinical trial | Noninferiority analysis of EMB vs. GEP starting at 6-months | With GEP score <34 to rule out rejection, similar event rates in EMB and GEP groups |
| Moayedi 23 , 2019 | OAR | 35 U.S. centers, 1504 patients, Cross-sectional | Clinical validation with long-term follow-up of patients who underwent GEP surveillance | 2% prevalence of ACR both before and after six months post-transplant, 94% 5-year survival |
| MicroRNA | ||||
| Duong Van Huyen 24 , 2014 | 4 French centers, 113 patients, Case-control study | Pre-selected evaluation of 14 miR transcripts by RT-PCR, from both EMB and serum samples | Four miRs were differentially expressed in tissue and serum and validated in ACR and AMR as compared to controls | |
| Dewi 25 , 2017 | 6 Canadian centers, 63 patients, Case-control study | Pre-selected miRs evaluated by RT-PCR evaluation in serum | Two miRs were associated with ACR, after controlling for immunosuppressive drug levels, kidney function, and C-reactive protein | |
| Constanso-Conde 26 , 2020 | 1 Spanish center, 66 patients, Prospective Cohort study | Pre-selected evaluation of differential miR expression in ACR by RT-PCR | miR-181a-5p was differentially expressed in ACR ≥ 2R with rise and fall pattern associated with development/treatment of rejection | |
| Shah 27 , 2022 | GRAfT | 5 U.S. centers, 157 patients, Case-control study | NGS evaluating miR expression | Distinct miR scores between 0–100 were able to distinguish ACR and AMR. Developed and validated for ACR ≥ 2R and pAMR ≥ 1 |
AUC, area under the curve; ACR, acute cellular rejection; AMR, antibody-mediated rejection; EMB; endomyocardial biopsy; GEP, gene expression profiling; mRNA, messenger RNA; miR, microRNA; NGS, next generation sequencing; NPV, negative predictive value; RT-PCR, reverse transcriptase polymerase chain reaction;
Post-transplant surveillance strategies at many HT centers have incorporated dd-cfDNA and GEP with obviation of EMB in patients with normal molecular testing results. Important considerations regarding this strategy are the following: 1) GEP has only been validated for the detection of ACR, while dd-cfDNA has been validated in ACR and AMR6,7, 2) GEP scores vary with time post-transplant, rise as corticosteroids are weaned, and can only be implemented after 55 days post-transplant, while dd-cfDNA can be implemented as early as 14 days post-transplant, allowing for resolution of early graft injury associated with ischemia-reperfusion injury and surgery6–8,28, 3) the interpretability of GEP scores is unreliable in the setting of high-dose prednisone, cytomegalovirus infection, receipt of hematopoiesis stimulating agents, and blood transfusions29,30, and 4) the PPV for GEP is 7%, while for dd-cfDNA it is nearly 20%6–8. Importantly, systemic infection and inflammation can lead to cellular apoptosis and raise total cell-free DNA levels, thus reducing the donor fraction. Quantitative PCR is one method to more accurately calculate the absolute quantity of cell-free DNA to improve calculation of the donor fraction22,31. In clinical practice, GEP and dd-cfDNA scores are often used concordantly to surveil post-transplant status and decide on the utility of performing EMB. In a single-center analysis evaluating this strategy among 153 HT patients, 495 combined GEP/dd-cfDNA tests were performed, leading to cancellation of 84% of surveillance biopsies, as opposed to 71% if GEP alone was used32. Critically important is that in the subgroup of tests where the GEP was positive and dd-cfDNA was negative (23% of combined testing results), none of the patients went on to develop CAR. In a similar evaluation from another center, ~20% of combined testing results had a positive GEP and negative dd-cfDNA also with none of those patients having CAR33. These findings support the higher specificity of dd-cfDNA over GEP and question the clinical necessity of using both dd-cfDNA and GEP in contemporary practice.
MicroRNAs
Aside from protein-coding genes or mRNA transcripts, the genome encodes for microRNAs (miRs), which are small (~22 nucleotide) RNA sequences that regulate protein expression34. With the development of next generation sequencing, over 2,000 miRs have been described in humans and more than half of all protein-coding genes are known to be regulated by miRs34. MicroRNAs are intriguing biomarkers as they are often contained within vesicular bodies (such as exosomes) or bound to lipoproteins, which confers both in vivo and in vitro stability35. In CAR, Duong Van Huyen and colleagues evaluated the potential of 14 pre-selected miRs and annotated their expression by PCR in EMB and serum samples36. They were able to identify four miRs that were associated with ACR and AMR in both the tissue and blood with AUCs > 0.90, as well as specific expression patterns that differentiated ACR from AMR.
In a recent publication from the GRAfT investigators, next generation sequencing was utilized to identify differential expression of ACR- and AMR-associated miRs in 157 patients27. The GRAfT investigators developed a panel of ACR and AMR-specific miRs which was used to create a clinical rejection score for ACR and AMR. These scores were then internally and externally validated in distinct patient cohorts from GRAfT and Stanford University, leading to a GRAfT ACR AUC of 0.94 and AMR AUC of 0.82, and Stanford ACR AUC of 0.72. MicroRNA score elevations preceded clinical rejection and improved with therapy, suggesting that they could be used as a quantitative marker. The results of this study suggest the potential of miRs as a true ‘liquid biopsy’ with the ability to detect ACR and AMR; however further clinical validation studies are needed.
Intragraft Gene Expression Profiling
While molecular methodologies have allowed for the obviation of many surveillance biopsies, tissue diagnosis remains necessary when rejection is clinically suspected (i.e., for-cause biopsy). When EMB is obtained, GEP (mRNA evaluation) of tissue samples can enhance precision in identifying the mechanisms of allograft injury, rejection and graft dysfunction. The Molecular Microscope® Diagnostic system (MMDx, Transcriptome Sciences, Alberta, Canada) is a GEP analysis wherein RNA transcripts are extracted from EMB samples preserved in RNAlater® (Thermo Fisher Scientific, Waltham, MA) and hybridized to human genome microarrays. The initial work in this area was published by a multi-center group from Canada, Italy, and France, where 331 EMB samples were evaluated to define archetypes, or clusters of gene transcripts, based on unsupervised machine learning37. In this manner, unique gene expression profiles were first identified and then found to correlate with ACR, AMR, or no rejection by histopathology. These initial gene expression archetypes displayed AUCs of 0.78 (no rejection), 0.65 (ACR), and 0.81 (AMR). A more recent intragraft GEP study evaluated 1,320 biopsies from the 13-center INTERHEART study, defining a new gene expression archetype for ‘minor rejection’, which was specifically associated with class II DSA positivity, as well as ACR 1R (but not ACR ≥ 2R or AMR)38. These results describe the utility of intragraft transcripts to distinguish EMBs with ongoing inflammatory changes not meeting histologic CAR thresholds. As the assay requires an additional biopsy sample to be collected, the greatest clinical utility may be for those patients with prior normal biopsies who present with clinical graft dysfunction and elevated DSA or dd-cfDNA.
A similar GEP microarray methodology to measure intragraft transcripts, nCounter (nanoString, Seattle, WA) has been developed, which can be performed on formalin fixed tissue, not requiring additional tissue samples to be collected. In an AMR case-control study, a signal associated with both AMR and HLA class I and II DSA presence was identified via nCounter39. Dysregulated transcripts were implicated in natural killer (NK) cell and monocyte activation as well as donor-specific endothelial cell activation/angiogenesis. Though intragraft GEP is not widely used as a clinical tool, Alam et al. retrospectively evaluated intragraft GEP results in conjunction with dd-cfDNA and traditional EMB histopathology, finding a 61% agreement across all three modalities40. There was 84% agreement between EMB and intragraft GEP; and of the 32 EMB-negative samples that had rejection evidence on intragraft GEP, the majority had an elevation of dd-cfDNA. The evidence of ongoing molecular signals of rejection and allograft injury, despite a negative EMB, underscores EMB’s imperfection in identifying allograft injury and highlights the potential of a multi-marker approach to allograft surveillance. Finally, a study from the University of Padova evaluated miR (rather than mRNA) expression in EMB tissue samples from 33 patients with ACR, AMR, mixed rejection, or no rejection and showed the ability of 14 individual miRs to identify specific rejection types, suggesting that miR EMB profiling may be used along with GEP to identify CAR when the EMB histopathology is equivocal41.
Protein Biomarkers and Exosomal Profiling
Given the widescale availability, low-cost, and proven clinical utility of circulating cardiac biomarkers in heart failure and coronary artery disease, HT patients often undergo measurement of natriuretic peptides and cardiac troponins when there is suspected graft dysfunction or CAR. Transplanted hearts release natriuretic peptides, which peak within the first one to two months post-transplant and reach steady state within one to two years, though often this steady state is higher than that for healthy controls42. While some studies have shown associations between natriuretic peptide elevation and CAR, multiple evaluations have shown that B-type natriuretic peptide (BNP) and N-terminal pro-B-type natriuretic peptide (NT-proBNP) lack sensitivity and specificity for diagnosing CAR43–45. Evaluations of standard cardiac troponin (cTn) I and T assays have shown similar early post-transplant temporal decay patterns, but mixed results in their ability to diagnose biopsy-proven rejection46. High-sensitivity cardiac troponin (hs-cTn) has been postulated as a more sensitive diagnostic marker of CAR, but recent work showed no association between hs-cTn I and ACR ≥ 2R47. Similarly, evaluations of non-specific inflammatory biomarkers, such as C-reactive protein and interleukin-6 (IL-6), have not proven to be accurate diagnostic markers of CAR45,48. There have also been evaluations of immune and angiogenesis biomarkers within the Clinical Trials in Organ Transplantation (CTOT)-5 and CTOT-18 studies, showing an association between endothelial growth factor-C and endothelin-1 with cardiac allograft vasculopathy, as well as anti-cardiac myosin antibody and vascular endothelial growth factors-C and -A with adverse post-transplant outcomes49,50. Whether assays for circulating, targeted biomarkers can augment the accuracy of other -omics technologies for CAR remains an area of investigation.
Aside from measuring single soluble protein biomarkers, there has been growing interest in high-throughput, targeted-discovery protein expression profiling using technologies such as Olink® (Uppsala, Sweden) and Somascan® (SomaLogic, Boulder, CO). These use either antibody or nucleic acid probes that can hybridize with proteins in recipient blood samples, simultaneously evaluating hundreds to thousands of protein biomarkers with amplification of the detection signals via quantitative PCR. A recent study at Columbia University utilized protein expression profiling in 36 patients with ACR, AMR, and no rejection, identifying multiple immune modulating proteins differentially expressed in ACR and AMR51. A unique aspect of this study was the evaluation of exosomes, small intracellular vesicular bodies that hold proteins and nucleic acids that can enter the circulation as well as neighboring cells and have been implicated in cell-to-cell communication. By focusing on exosomes, we can begin to identify the cell type of origin for these deranged protein and nucleic acid signatures.
Pediatrics
Given the potentially increased risk and patient distress associated with EMB in children, the expansion of molecular rejection evaluation into pediatric HT practice will be especially important (Table 3)3. The initial dd-cfDNA work at Stanford University (described above) included both pediatric and adult HT recipients20. Additionally, investigators from the seven-center DNA-Based Transplant Rejection Test (DTRT) study group published on a combined pediatric and adult cohort, utilizing the myTAIHEART dd-cfDNA assay, which is no longer clinically available, and described a sensitivity of 65%, specificity of 93%, PPV 84.6%, NPV 81.8%, and AUC of 0.81 for pediatric patients to detect ACR ≥ 1R (a notable weakness of the study as ACR 1R is not clinically treated as rejection) or pAMR ≥121. The DTRT investigators later published an evaluation of dd-cfDNA utilizing clinically treated (rather than biopsy-proven) rejection, and the diagnostic accuracy reached a sensitivity of 95%, specificity of 56%, PPV 9%, NPV 99%, and AUC of 0.81 for pediatric patients52. Higher %dd-cfDNA at days 0 and 14 of a rejection event were associated with a composite of death, cardiac arrest, or MCS during follow-up, suggesting the clinical significance of dd-cfDNA levels in clinical rejection syndromes, regardless of EMB result. Additionally, there was an observable difference in non-rejection dd-cfDNA levels in pediatric, as compared to adult samples, suggesting more baseline immune activation post-transplant in children. Future pediatric investigations of noninvasive rejection surveillance methodologies should consider immune system development and the corresponding immunologic response.
Table 3.
Differences Between Pediatric and Adult Heart Transplantation
| Characteristic | Description | Comment |
|---|---|---|
| EMB Procedure | Most pediatric EMBs require general anesthesia; EMB in small children has potential increased risk of cardiac perforation | Minimizing EMBs by using molecular methodologies may lead to significant risk reduction |
| Fewer Transplants | Pediatric recipients comprise 10–15% of the > 6000 yearly heart transplants | Less research validating GEP, cfDNA, HLA; no studies of miR, exosomes, protein expression profiling |
| Transplant Indication | 43% of pediatric transplants worldwide are due to congenital heart disease | Previous surgeries and transfusions lead to high rates of pre- and post-transplant allosensitization |
| Immune System Considerations | Congenital heart disease patients often have congenital or acquired immunodeficiency. The immune system is underdeveloped in small children | Interpretation of GEP, dd-cfDNA, HLA profiling must be weighed with these immune considerations |
| ABO incompatible heart transplantation | Given the underdeveloped immune response, consideration of ABO incompatible transplant is standard of care in children < 2 years of age | Molecular methodologies to surveil rejection have little evidence in ABO-incompatible transplants |
| VAD-related sensitization | In the U.S., a higher proportion of children than adults with VAD go on to transplantation | Pre-transplant sensitization can result in DSA and rejection |
cfDNA, cell-free DNA; DSA, donor-specific antibody; EMB, Endomyocardial Biopsy; GEP, gene expression profiling; HLA, human leukocyte antigen; VAD, ventricular assist device.
There have been reports of the clinical use of dd-cfDNA and GEP assays from pediatric HT centers, but a pediatric-specific controlled analysis to evaluate their predictive value of EMB-proven rejection has not been performed53,54. Investigators from Children’s Hospital of Pittsburgh published initial results of dd-cfDNA evaluations used as an alternative surveillance strategy to EMB in 58 low-risk pediatric recipients53. Of the 11 patients with elevated dd-cfDNA, there were four episodes of ACR 1R with one of these patients having pAMR 2, and the remainder were ACR 0R/pAMR 0. Of the 47 patients without a dd-cfDNA elevation, there was only one episode of clinically significant rejection (ACR 2R, pAMR 0). Research in pediatric HT is limited by overall smaller numbers of HT: 10–15% of the over 6000 worldwide heart transplants per year have pediatric recipients, and most individual centers perform <10 transplants per year1,2.
Children also have unique immune considerations that must be weighed when evaluating molecular diagnostic tools. First, nearly half of the transplants performed in North America are due to congenital heart disease, and many of these patients have immune dysregulation due to protein-losing enteropathy, hepatic dysfunction, or congenital comorbidities55. As the success of congenital heart disease surgery increases, more children are living and eligible for HT, and these children are often sensitized due to the receipt of allograft material and transfusions2,56. Sensitization among pediatric recipients has increased over time (estimated at 11–57%) and is associated with increased waitlist duration and pre-transplant mortality as well as increased post-transplant rejection, CAV, and mortality17,18,56. Additionally, as multiple short- and long-term MCS devices are increasingly being used in children, it will be important to evaluate their effect on pre-transplant sensitization and post-transplant adverse outcomes. In a 2004–2014 UNOS evaluation, 19% of children received VAD prior to transplant and 42% of VAD patients became sensitized prior to transplant; however, there was no association between pre-transplant sensitization and post-transplant mortality57. Another unique immune aspect of the pediatric population which will require further study is the use of ABO-incompatible transplants as standard of care for children under two years of age.
Racial Disparities after Heart Transplant
The most striking result in stratified outcomes after HT is that Black recipients have higher post-transplant mortality rates, independent of socioeconomic status58. Among Black HT recipients, there are higher rates of pre-transplant sensitization, HLA mismatch at time of transplant, de novo DSA development, and AMR15,16,58,59. In a single-center evaluation of 137 patients (48% Black) from Emory University Hospital, Black patients were twice as likely to develop graft dysfunction, four times more likely to develop de novo DSA, and five times more likely to develop AMR than non-Black patients16. An evaluation of 63 patients (46% Black) from the GRAfT cohort showed higher dd-cfDNA levels in the first week post-transplant among Black recipients as compared to non-Black recipients, suggesting more significant early allograft injury in Black recipients28. The higher dd-cfDNA elevations in Black patients was associated with a higher risk of AMR on short-term follow-up, suggesting that elevations of dd-cfDNA in the absence of rejection may potentially trigger immune activation and lead to downstream CAR.
A post-hoc evaluation of the IMAGE trial results showed similar GEP scores among White, Black, and Other race groups (despite higher event rates in the Black and Other groups)60. Black recipients, however, had differences in individual gene expression that were significant predictors of adverse events; namely, upregulation of MARCH8 (involved in regulation of Class II HLA and inhibition of T-cell activation) and downregulation of FLT3 (involved in stimulation of NK and B cells). Given these findings, an evaluation of the OAR study results was performed, limited to Caucasian and Black recipients enrolled in the first-year post-transplant, showing that Black recipients (and not White recipients) with higher GEP scores had higher risk of mortality and those with higher tacrolimus levels had lower risk of mortality15. In White recipients, MARCH8 upregulation was associated with increased mortality, and in Black patients FLT3 upregulation was associated with increased mortality. These disparate gene expression and medication response profiles between races highlight the need for a better understanding of the genomic differences mediated by race/ethnicity, which may provide an increased precision medicine approach in HT, as has been described in other cardiovascular therapeutic applications.
HLA and non-HLA Antibody Biomarkers
HLA Typing and Antibody Determination
Histocompatibility describes the immune compatibility between donor and recipient, which is defined by the ‘immunogenetics’ of 12 highly polymorphic genetic loci (having > 35,000 HLA allele variants) on chromosome 6: three single chain class I antigen groups (HLA-A, -B, and -C) and three heterodimer class II antigen groups (HLA-DPA-DPB, -DQA-DQB, and -DRA-DRB). These genes code for the surface proteins that comprise the antigen presenting complex, which allows the body’s immune system to distinguish between self and non-self. The classical methodology to determine anti-HLA antibody (HLA-Ab) presence was the complement dependent cytotoxicity cell panel, which requires a high threshold of bound HLA antibody to elicit complement-mediated cell death. More recent HLA-Ab screening involves the use of solid-phase immunoassays, such as the Luminex- (Luminex, Austin, TX) platform, which uses multiplex bead assays that involve incubation of HLA-coated beads with recipient serum, followed by the addition of a fluorescently labeled anti-IgG detection antibody, which can then be measured as a semi-quantitative mean fluorescence intensity (MFI). Adjunct testing to aid in determining pre- or post-transplant antibody strength and potential pathogenicity include 1) serial dilutions of recipient serum, with more deleterious antibodies present at higher titer (concentration), 2) modified assays that measure activation of complement products (C3d or C1q) which, if present, likely denote more a more significant immune response and 3) confirmatory flow cytometry or complement dependent cytotoxicity crossmatch evaluations. The technology to detect HLA-Abs via solid phase immunoassays and other methodologies continues to evolve, and more consensus is needed regarding the identification and quantification of clinically relevant pre- and post-transplant HLA-Abs.
Pre-Transplant Sensitization
Prior to transplantation, recipient serum is tested to evaluate the presence and breadth of HLA sensitization. Pre-transplant HLA-Ab presence can develop from sensitizing events such as pregnancy, transfusions, or receipt of surgical allograft material (Figure 3) and is collectively reported as a calculated percentage of panel-reactive antibodies (%cPRA), which captures the HLA allele frequency in the donor population. Higher %cPRA reflects a higher proportion of HLA-incompatible donors (i.e. the likelihood of a preformed HLA Ab specific for the donor HLA antigen profile) but may not reflect the number or strength of the preformed HLA Abs. Sensitized patients wait longer for organs and have higher rates of rejection, along with pre- and post-transplant mortality61–63. Sensitization and HLA antibody-presence is dynamic, requiring multiple pre-transplant evaluations, as HLA antibody levels change with sensitizing events.
Figure 3. Determination of Human Leukocyte Antibodies Before and After Transplantation.
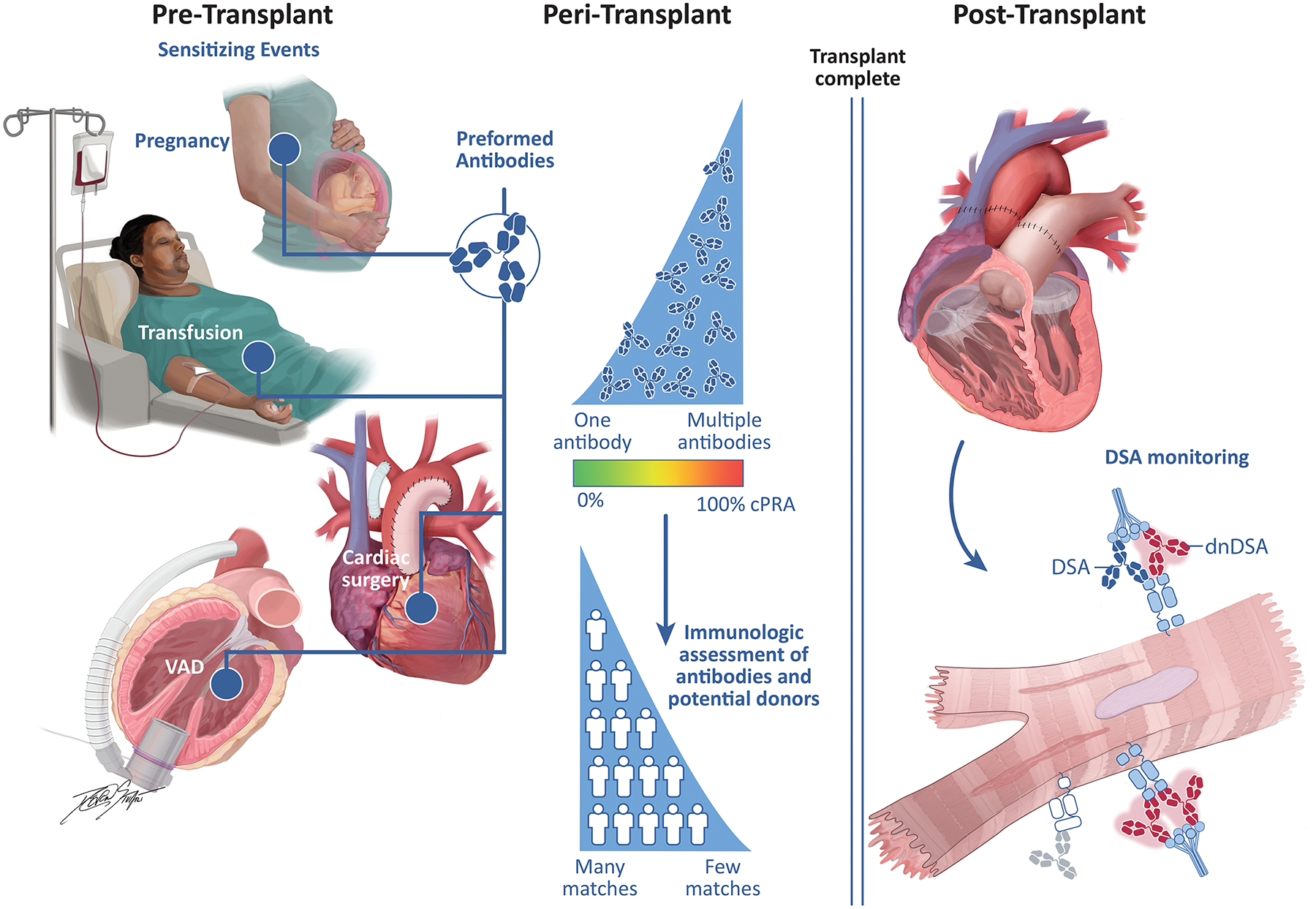
Pre-transplant sensitizing events include pregnancy, blood transfusions, ventricular assist devices, other mechanical circulatory support, as well as prosthetic materials used during cardiac surgery. As the number of pre-formed human leukocyte antigen (HLA) antibodies increases, the calculated percentage of panel-reactive antibodies (cPRA) increases, decreasing the number of potential donors. After transplantation, preformed antibodies can result in DSA (blue) or the transplant recipient can develop new antibodies, termed de novo DSA (red), both of which are associated with antibody-mediated rejection, cardiac allograft vasculopathy, allograft dysfunction and death.
cPRA, calculated percentage of panel-reactive antibodies; DSA, donor-specific antibody; dnDSA, de novo DSA; LVAD, left ventricular assist device.
Virtual Crossmatch
As travel times for donor hearts has increased, pre-transplant crossmatching has been essentially eliminated, and most transplant centers perform a virtual crossmatch at the time of donor organ consideration: evaluating the current recipient HLA-Ab profile against corresponding donor HLA antigens to determine the presence of mismatched pre-formed DSA. While mismatch at both class I and class II loci have been associated with decreased graft survival, class II mismatch is a more significant predictor of CAR, specifically AMR64,65. Multiple studies, however, have evaluated the effects of transplanting across DSA barriers and have yielded divergent results—some showing increased AMR and mortality, and others without such effects; these discrepancies may be due to different DSA strengths and characteristics, or different immunosuppression and desensitization strategies employed among these patients66. With increasing rates of sensitization, determining which DSA barriers can be safely crossed and which may require desensitization will be necessary for sensitized patients, balancing the risk of death on the waitlist with potential side effects of desensitization therapy.
Post-Transplant DSA
Post-transplant DSA detection has been associated with increased AMR, CAV, and mortality, either due to pre-formed DSA or development of de novo DSA67–70. It is estimated that 25–35% of HT recipients develop de novo DSA, which are primarily Class II, with those specific to the DQ locus being most deleterious64,71,72. These can emerge both early and late after transplantation, with their persistence (rather than the timing at which they develop) the greatest predictive factor of adverse outcomes67,72. A study of 172 HT recipients found transient de novo DSA in 15% and persistent (present in samples ≥ 30 days apart) de novo DSA in 18%. Those with persistent de novo DSA had a four-fold increased mortality risk, compared to no increased mortality risk in those with transient de novo DSA67. As more is learned about the effects of post-transplant DSA especially among high-risk patients, it will be important to evaluate how HLA-Ab detection can be used in conjunction with molecular methodologies, such as dd-cfDNA or intragraft GEP, to enhance risk stratification of DSA and patient management.
Non-HLA Antibodies
While the development of de novo DSA has been associated with AMR, many biopsy-proven AMR cases occur in patients without HLA DSA, which may be related to non-HLA antibodies73. An evaluation of 77 patients bridged to HT with VADs showed that elevated angiotensin II type 1 receptor (AT1R) antibodies (and not HLA-Abs) at time of transplant were associated with an increased composite risk of death, treated rejection, or CAV within a five-year follow-up period74. There have also been reported associations between CAR and antibodies to collagen-V, K-alpha-1 tubulin, and Major Histocompatibility Complex Class I Chain-Related Molecule A (MICA)75,76. Given the numerous potential non-HLA antibodies related to CAR, future evaluations in this area will benefit from ongoing work describing the use of protein expression profiling as well as the development of a Luminex- bead assay for non-HLA antigens. It is possible that non-HLA antibodies may synergize with de novo DSA to affect the severity of CAR.
Titration of Immunosuppression to Mitigate Risk of Infections
The combination of immunosuppressive agents (calcineurin-inhibitors, mycophenolate mofetil, corticosteroids, and various induction agents) as well as increased recipient complexity have resulted in increasing infectious complications over-time, which are now a leading cause of death after the first post-transplant month and responsible for up to 30% of overall post-transplant mortality77. Despite infectious complications, there have not been significant changes to maintenance immunosuppression over the past two decades, and the ability to safely minimize immunosuppression could improve post-transplant outcomes. This effort will require an assay quantifying a recipient’s net state of immunosuppression, the development of which has remained elusive. The most prominent example is the Immuknow® Assay (Viracor-IBT [formerly Cylex], Lenexa, Kansas) which was designed to measure T-cell activity—a primary target of calcineurin inhibition (Table 4). In this assay, peripheral CD4 T cells are stimulated with phytohemagglutinin, and bioluminescence is used to quantify ATP production, which correlates with overall T-cell activity. ATP levels ≤225 ng/mL are suggestive of a high net state of immunosuppression and have been associated with increased risk of infection78,79. Results of clinical validation studies, however, have not reliably shown high T-cell activity to be associated with CAR79,80. Despite modest data, many transplant programs continue to use this assay to guide reduction of immunosuppression, especially in the setting of infection or malignancy. Another T-cell based assay is ImmunoSpot®, which quantifies interferon response within antigen-stimulated immune cells; this assay remains limited to the research setting81. van Besouw and colleagues assayed the number of cytokine-secreting lymphocytes via ImmunoSpot® before, during, and after an episode of CAR and noted a corresponding lymphocyte rise and fall at the time of diagnosis and after treatment of rejection82. GEP has also been evaluated as a marker of immunosuppressive state and infection risk, where low GEP scores were often found at the time of significant infection (particularly with cytomegalovirus and fungal infections)83. Another biomarker that holds great promise for immunosuppression quantitation is Torque Teno Virus viral load, which has been shown to correlate with both risk of infection and allograft rejection among recipients of heart and other solid organ transplants84.
Table 4.
Description of Assays Utilized to Evaluate Immunosuppression
| Technology | Description | Commercially Available Assay | Research Assay |
|---|---|---|---|
| T Cell Assay | Bioluminescent quantification of intracellular ATP from activated CD4 T lymphocytes | Immuknow® | |
| Interferon Quantification | Solid-phase, enzyme-linked immunosorbent assay for spectrophotometric estimation of secreted antibody | ImmunoSpot® | |
| GEP of mRNA | Gene expression profiling of peripheral blood mononuclear cells | AlloMap® | |
| Torque teno virus | PCR assay for detection and quantitation of circulating torque teno virus | Biomerieux TTV R-GENE® |
ATP, adenosine triphosphate; GEP, gene expression profiling; mRNA, messengerRNA; TTV, Torque Teno Virus
Biomarkers of Primary Graft Dysfunction and Cardiac Allograft Vasculopathy
It has been hypothesized that primary graft dysfunction (PGD)—defined as acute failure of the new allograft to support the recipient’s circulation—may be a function of donor and/or recipient inflammation as well as innate immune activation85. Recently, aptamer-based protein expression profiling was performed on pre-transplant serum from 219 heart transplant recipients, analyzing a total of 354 distinct circulating protein biomarkers86. The authors found that circulating levels of C-type lectin receptor 4C were elevated in PGD as compared to controls in both a derivation and validation set, and this association remained even after adjustment for key clinical variables. C-type lectin receptor 4C is a surface marker of plasmacytoid dendritic cells, which bridge the gap between the innate and adaptive immune system and play a key role in identifying cell-free viral DNA to generate a rapid and robust interferon response. Whether patients with enrichment of plasmacytoid dendritic cells can generate a robust pro-inflammatory response to donor-derived cell free DNA is the subject of ongoing investigation. Another study which suggested a potential role for innate immunity in the development of PGD performed mass spectrometry-based proteomic profiling on microvesicles isolated from the pre-transplant serum of 88 heart transplant recipients87. The authors reported that lower levels of pre-transplant kallikrein (a serine protease involved in the inflammatory and coagulation cascades) were associated with PGD development. Lower levels of circulating kallikrein have been previously reported in other pro-inflammatory states, suggesting that these recipients may have increased innate immune activation and inflammation even prior to HT.
Unlike PGD which represents an early event in the post-transplant period, CAV is a longer-term complication of HT that is associated with significant morbidity and mortality88. Current diagnostic strategies for CAV are centered around coronary angiography, which may lack sensitivity and specificity for the diagnosis, particularly in its subclinical form. Multiple studies have demonstrated an association between DSA, specifically class II, and development of CAV69,89. There is less evidence, however, regarding other molecular biomarkers’ association with CAV. A single center study from Madrid, Spain, evaluated dd-cfDNA of 94 recipients at time of angiography and showed no association between dd-cfDNA and presence of CAV90. In another study utilizing proteomics, aptamer-based protein expression profiling identified a signature that discriminated patients with mild-moderate CAV, including proteins related to cellular injury, inflammation, and platelet activation91. As investigations continue to evaluate molecular methodologies as markers of CAR, it will be important to assess their ability to predict these other post-transplant outcomes.
Molecular Testing Implementation Barriers and Limitations
To date, the AlloMap® GEP assay is the only -omics evaluation described in this review with FDA clearance. This is due in part to the lack of standardized processes for development and approval of biomarkers, as compared to those for drug (investigational new drug [IND]) and device (investigational device exemption [IDE]) development. Within the past few years, however, genetic tests for oncologic applications have received IDE approval, and there is growing FDA interest in overseeing regulatory approval of biomarkers in solid organ transplantation92,93. While there is continued recent evidence showing the utility of dd-cfDNA and miRNAs in evaluating ACR and AMR, the PPV of these methodologies remains low when compared to EMB, leading to use primarily in the setting of a low pre-test probability for rejection (i.e., surveillance testing). For these evaluations to gain further regulatory approval and high-level evidence within professional society guidelines, they will need to be studied in appropriately powered clinical utility trials. As an example, many of these genomic assays are validated as part of the drug development process in oncology, leading to FDA approval as a companion diagnostic. Finally, although methods are being developed to reduce the costs of these assays, they are quite technically complex and cost ~$2,800 US dollars, limiting application outside of the US11.
Many of these molecular technologies have only been studied in the early post-transplant period. Future investigations need to expand beyond the first year and elucidate the implications and management of patients with persistently elevated dd-cfDNA, defining its role in assessing pathogenicity of de novo DSA and predicting long-term graft dysfunction (both systolic and diastolic) and CAV.
Comparison to other Solid Organ Transplants
Importantly, in HT as compared to other solid organ transplants, these technologies have more robust application and clinical data for their use, given that 1) the incidence of rejection in other transplanted organs is significantly lower (approximately 5–9% for renal transplantation compared to 13–24% for heart transplantation at one-year)1,2,94, 2) biomarkers of end-organ function (creatinine and AST/ALT) are reasonable markers of rejection and 3) biopsies are primarily performed for-cause rather than for surveillance as in HT. As molecular evaluation methodologies continue to develop, the lessons learned from HT will have implications for other solid organ transplant populations.
Future Directions
Current generation molecular biomarkers are used as part of a monitoring strategy post-transplant, but with further development of dd-cfDNA and miR assays, the field will likely move towards a definitive diagnosis of CAR with specific rejection subtype, allowing appropriate ACR or AMR therapy without the need for EMB (Figure 4). The GRAfT investigators demonstrated disparate guanosine:cytosine (G:C) ratio and fragment length of dd-cfDNA as a potential mechanism to distinguish between ACR or AMR, and as miR evaluation moves from bench to bedside, this methodology will likely be able to diagnose ACR or AMR7,27. Absolute quantification of dd-cfDNA, additionally, rather than thresholds of %dd-cfDNA, may enhance CAR prediction22. It is likely that the future of genomic testing will include a multi-marker approach that enhances overall specificity and PPV, while maintaining sensitivity and NPV. There remain questions regarding the frequency at which to obtain these molecular evaluations, to improve clinical utility beyond the first year after transplant.
Figure 4. Future Directions in Utilizing -Omics Technologies in Heart Transplantation.
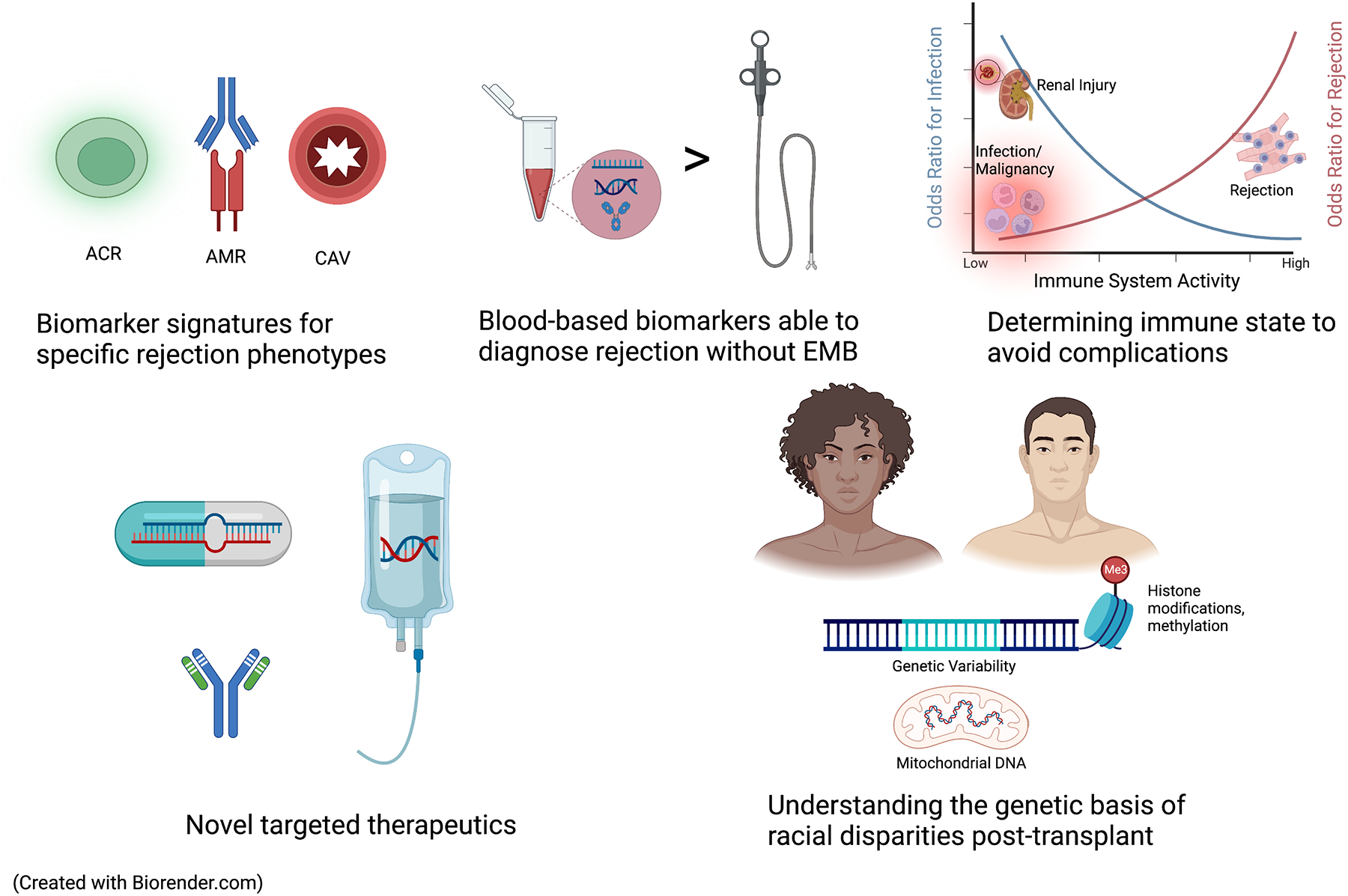
As increasing data are published regarding non-invasive modalities to detect post-transplant outcomes, they may be able to diagnose ACR, AMR, or CAV without the need for cardiac catheterization and EMB. There is growing research in novel technologies to determine the net immune state to facilitate the selection and titration of immunosuppression. Novel -omic based targets will permit the development of DNA- RNA- or monoclonal antibody-based therapeutics. Finally, an enhanced understanding of genetic polymorphisms, mitochondrial DNA patterns, and epigenetic data can shed light into mechanisms behind disparate outcomes in Black transplant patients.
ACR, acute cellular rejection; AMR, antibody-mediated rejection; CAV, cardiac allograft vasculopathy; EMB, endomyocardial biopsy
In the current era, with rejection rates below 20%, there is a growing need to provide a comprehensive evaluation of allograft health to assess overall immune state. Molecular methodologies may help improve post-transplant survival by permitting tailored immunosuppression with the goal of reducing the complications of over-immunosuppression including infection, malignancy, and chronic kidney disease. Proteomic information post-transplant can yield information regarding immune system activation and allograft injury. This can occur in concert with a more advanced evaluation of the net state of immunosuppression, potentially via quantification of Torque Teno Virus. In addition, based on work in oncology using circulating tumor DNA, there is a potential to use cell-free DNA to diagnose certain forms of cancer (e.g., post-transplant lymphoproliferative disorder) in transplant recipients prior to clinical onset.
Aside from their role as biomarkers, these genomic molecules are linked to the pathophysiology of CAR and other post-transplant complications. Much has been learned about cell-free DNA, specifically, from the COVID-19 pandemic, wherein elevated plasma cell-free DNA has been shown to be immunogenic, signaling mitochondrial production of reactive oxygen species via toll-like receptor 9; and epigenetic cell-free DNA signatures (including histone packaging and methylation) have been used to evaluate the tissue-specific immune response31,95. This knowledge will undoubtedly aid in the development of new therapeutic strategies, such as recombinant human DNase, Dornase alpha, which is used in cystic fibrosis patients to degrade cell-free DNA and improve outcomes96. Multiple specific components of the immune response can be evaluated and treated, such as the potential evaluation of IL-6 levels and complementary treatment with tocilizumab or clazakizumab97. Next-generation molecular biomarkers may also include cell-free RNA and IFNγ-induced chemokine evaluation. To reduce the disparate outcomes experienced by Black patients after transplant, there is evidence of incomplete immune suppression in these patients with current immunosuppression strategies, which may manifest with higher levels of circulating mitochondrial DNA, a potent immune system stimulant98,99.
Conclusion
Multi-omic profiling via genomics, transcriptomics, and proteomics is being increasingly used to monitor patients after HT. These technologies provide personalized risk assessments and facilitate more precise approaches to post-transplant care. While this paradigm has mostly been applied to CAR, biomarkers for additional post-transplant events, including PGD and CAV, have also been studied. Advances in dd-cfDNA have reduced patient morbidity after transplant by eliminating many surveillance EMBs. In conjunction with dd-cfDNA, miR and protein expression profiling may provide more comprehensive information about CAR, further allowing accurate blood-based diagnosis of ACR or AMR without the need for EMB. Rapidly expanding knowledge of HLA and DSA in HT will help uncover the immunologic underpinnings of AMR and allow more prompt rejection identification and treatment. The next frontier in genomic biomarkers will focus on quantifying overall immune state and mitigating consequences of immunosuppressive therapy. As molecular technologies transition from the research to clinical environment, HT clinicians are increasingly enabled to detect adverse post-transplant outcomes while simultaneously reducing post-transplant procedures and complications.
Funding Sources:
SAE is supported by the National Heart, Lung, and Blood Institute Division of Intramural Research (HHSN268201300001C); KK is supported by NIH R01CA229766 and the Gottlieb Charitable Foundation; PS is supported by an NIH K23 Career Development Award 1K23HL143179.
List of Abbreviations:
- ACR
acute cellular rejection
- AMR
antibody-mediated rejection
- CAR
cardiac allograft rejection
- dd-cfDNA
donor-derived cell-free DNA
- DSA
donor-specific antibody
- EMB
endomyocardial biopsy
- GEP
gene expression profiling
- HLA
human leukocyte antigen
- HT
heart transplantation
- miR
microRNA
Footnotes
Conflict of Interest Disclosures:
CdF = Consulting for Abbott Diagnostics, FujiRebio, Ortho/Quidel, Roche Diagnostics, Siemens Healthineers
KK = CareDx scientific advisor, speaker, grant recipient.
PS = Unrelated grant support paid to Inova from Merck, Bayer, Roche, and Abbott. Consulting for Natera, Merck and Procyrion.
All other authors report no relevant disclosures.
References
- 1.Khush KK, Cherikh WS, Chambers DC, Harhay MO, Hayes D, Hsich E, Meiser B, Potena L, Robinson A, Rossano JW, et al. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: Thirty-sixth adult heart transplantation report — 2019; focus theme: Donor and recipient size match. J Hear Lung Transplant. 2019;38:1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossano JW, Singh TP, Cherikh WS, Chambers DC, Harhay MO, Hayes D, Hsich E, Khush KK, Meiser B, Potena L, et al. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: Twenty-second pediatric heart transplantation report – 2019; Focus theme: Donor and recipient size match. J Hear Lung Transplant. 2019;38:1028–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pophal SG, Sigfusson G, Booth KL, Bacanu S-A, Webber SA, Ettedgui JA, Neches WH, Park SC. Complications of endomyocardial biopsy in children. J Am Coll Cardiol. 1999;34:2105–2110. [DOI] [PubMed] [Google Scholar]
- 4.Crespo-Leiro MG, Zuckermann A, Bara C, Mohacsi P, Schulz U, Boyle A, Ross HJ, Parameshwar J, Zakliczyski M, Fiocchi R, et al. Concordance Among Pathologists in the Second Cardiac Allograft Rejection Gene Expression Observational Study (CARGO II). Transplant J. 2012;94:1172–1177. [DOI] [PubMed] [Google Scholar]
- 5.Bermpeis K, Esposito G, Gallinoro E, Paolisso P, Bertolone DT, Fabbricatore D, Mileva N, Munhoz D, Buckley J, Wyffels E, et al. Safety of Right and Left Ventricular Endomyocardial Biopsy in Heart Transplantation and Cardiomyopathy Patients. Jacc Hear Fail. 2022;10:963–973. [DOI] [PubMed] [Google Scholar]
- 6.Khush KK, Patel J, Pinney S, Kao A, Alharethi R, DePasquale E, Ewald G, Berman P, Kanwar M, Hiller D, et al. Noninvasive detection of graft injury after heart transplant using donor‐derived cell‐free DNA: A prospective multicenter study. Am J Transplant. 2019;19:2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agbor-Enoh S, Shah P, Tunc I, Hsu S, Russell S, Feller E, Shah K, Rodrigo ME, Najjar SS, Kong H, et al. Cell-Free DNA to Detect Heart Allograft Acute Rejection. Circulation. 2021;143:1184–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng MC, Eisen HJ, Mehra MR, Billingham M, Marboe CC, Berry G, Kobashigawa J, Johnson FL, Starling RC, Murali S, et al. Noninvasive Discrimination of Rejection in Cardiac Allograft Recipients Using Gene Expression Profiling. Am J Transplant. 2006;6:150–160. [DOI] [PubMed] [Google Scholar]
- 9.Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, Cappola TP, Kao A, Anderson AS, Cotts WG, Ewald GA, et al. Gene-Expression Profiling for Rejection Surveillance after Cardiac Transplantation. New Engl J Medicine. 2010;362:1890–1900. [DOI] [PubMed] [Google Scholar]
- 10.Kobashigawa J, Hall S, Shah P, Fine B, Halloran P, Jackson AM, Khush KK, Margulies KB, Sani MM, Patel JK, et al. The Evolving Use of Biomarkers in Heart Transplantation: Consensus of an Expert Panel. Am J Transplant. 2023; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Medicare and Medicaid Services. Clinical Laboratory Fee Schedule. 2023. [Internet, accessed 2023 May 17]. Available from: https://www.cms.gov/medicare/medicare-fee-for-service-payment/clinicallabfeesched/clinical-laboratory-fee-schedule-files
- 12.Clerkin KJ, Restaino SW, Zorn E, Vasilescu ER, Marboe CC, Mancini DM. The effect of timing and graft dysfunction on survival and cardiac allograft vasculopathy in antibody-mediated rejection. J Hear Lung Transplant. 2016;35:1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kfoury AG, Snow GL, Budge D, Alharethi RA, Stehlik J, Everitt MD, Miller DV, Drakos SG, Reid BB, Revelo MP, et al. A longitudinal study of the course of asymptomatic antibody-mediated rejection in heart transplantation. J Hear Lung Transplant. 2012;31:46–51. [DOI] [PubMed] [Google Scholar]
- 14.Coutance G, Kransdorf E, Aubert O, Bonnet G, Yoo D, Rouvier P, Duong Van Huyen JP, Bruneval P, Taupin J-L, Leprince P, et al. Clinical Prediction Model for Antibody-Mediated Rejection: A Strategy to Minimize Surveillance Endomyocardial Biopsies After Heart Transplantation. Circulation Hear Fail. 2022;15:e009923. [DOI] [PubMed] [Google Scholar]
- 15.Moayedi Y, Fan C-PS, Miller RJH, Tremblay-Gravel M, Posada JGD, Manlhiot C, Hiller D, Yee J, Woodward R, McCaughan JA, et al. Gene expression profiling and racial disparities in outcomes after heart transplantation. J Hear Lung Transplant. 2019;38:820–829. [DOI] [PubMed] [Google Scholar]
- 16.Cole RT, Gandhi J, Bray RA, Gebel HM, Yin M, Shekiladze N, Young A, Grant A, Mahoney I, Laskar SR, et al. Racial differences in the development of de-novo donor-specific antibodies and treated antibody-mediated rejection after heart transplantation. J Hear Lung Transplant. 2018;37:503–512. [DOI] [PubMed] [Google Scholar]
- 17.Zuckerman WA, Zeevi A, Mason KL, Feingold B, Bentlejewski C, Addonizio LJ, Blume ED, Canter CE, Dipchand AI, Hsu DT, et al. Study rationale, design, and pretransplantation alloantibody status: A first report of Clinical Trials in Organ Transplantation in Children‐04 (CTOTC‐04) in pediatric heart transplantation. Am J Transplant. 2018;18:2135–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh TP, Cherikh WS, Hsich E, Chambers DC, Harhay MO, Hayes D, Khush KK, Perch M, Potena L, Sadavarte A, et al. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: Twenty-fourth pediatric heart transplantation report — 2021; focus on recipient characteristics. J Hear Lung Transplant. 2021;40:1050–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burnham P, Kim MS, Agbor-Enoh S, Luikart H, Valantine HA, Khush KK, DeVlaminck I. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep-uk. 2016;6:27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeVlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, Luikart H, Neff NF, Okamoto J, Bernstein D, Weisshaar D, et al. Circulating Cell-Free DNA Enables Noninvasive Diagnosis of Heart Transplant Rejection. Sci Transl Med. 2014;6:241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richmond ME, Zangwill SD, Kindel SJ, Deshpande SR, Schroder JN, Bichell DP, Knecht KR, Mahle WT, Wigger MA, Gaglianello NA, et al. Donor fraction cell-free DNA and rejection in adult and pediatric heart transplantation. J Hear Lung Transplant. 2020;39:454–463. [DOI] [PubMed] [Google Scholar]
- 22.Kim PJ, Olymbios M, Siu A, Pinzon OW, Adler E, Liang N, Swenerton R, Sternberg J, Kaur N, Ahmed E, et al. A novel donor-derived cell-free DNA assay for the detection of acute rejection in heart transplantation. J Hear Lung Transplant. 2022;41:919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moayedi Y, Foroutan F, Miller RJH, Fan CS, Posada JGD, Alhussein M, Tremblay-Gravel M, Oro G, Luikart HI, Yee J, et al. Risk evaluation using gene expression screening to monitor for acute cellular rejection in heart transplant recipients. J Hear Lung Transplant. 2019;38:51–58. [DOI] [PubMed] [Google Scholar]
- 24.Duong Van Huyen JP, Tible M, Gay A, Guillemain R, Aubert O, Varnous S, Iserin F, Rouvier P, François A, Vernerey D, et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur Heart J. 2014;35:3194–3202. [DOI] [PubMed] [Google Scholar]
- 25.Dewi IS, Celik S, Karlsson A, Hollander Z, Lam K, McManus J-W, Tebbutt S, Ng R, Keown P, McMaster R, et al. Exosomal miR-142–3p is increased during cardiac allograft rejection and augments vascular permeability through down-regulation of endothelial RAB11FIP2 expression. Cardiovasc Res. 2017;113:440–452. [DOI] [PubMed] [Google Scholar]
- 26.Constanso-Conde I, Hermida-Prieto M, Barge-Caballero E, Núñez L, Pombo-Otero J, Suárez-Fuentetaja N, Paniagua-Martín MJ, Barge-Caballero G, Couto-Mallón D, Pan-Lizcano R, et al. Circulating miR-181a-5p as a new biomarker for acute cellular rejection in heart transplantation. J Hear Lung Transplant. 2020;39:1100–1108. [DOI] [PubMed] [Google Scholar]
- 27.Shah P, Agbor-Enoh S, Bagchi P, deFilippi CR, Mercado A, Diao G, Morales DJ, Shah KB, Najjar SS, Feller E, et al. Circulating microRNAs in cellular and antibody-mediated heart transplant rejection. J Hear Lung Transplant. 2022;41:1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doshi A, Shah KB, Agbor-Enoh S, Tushak Z, Garcia V, Kong H, Jang MK, Hsu S, Feller ED, Rodrigo ME, et al. Higher levels of allograft injury in black patients early after heart transplantation. J Hear Lung Transplant. 2022;41:855–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobashigawa J, Patel J, Azarbal B, Kittleson M, Chang D, Czer L, Daun T, Luu M, Trento A, Cheng R, et al. Randomized Pilot Trial of Gene Expression Profiling Versus Heart Biopsy in the First Year After Heart Transplant. Circulation Hear Fail. 2015;8:557–564. [DOI] [PubMed] [Google Scholar]
- 30.Food and Drug Administration. FDA Device Classification, Cardiac Allograft Gene Expression Profiling Test System. [Internet, accessed 2023 Jun 27]. Available from: https://www.accessdata.fda.gov/cdrh_docs/reviews/k073482.pdf
- 31.Andargie TE, Tsuji N, Seifuddin F, Jang MK, Yuen PST, Kong H, Tunc I, Singh K, Charya A, Wilkins KJ, et al. Cell-free DNA maps COVID-19 tissue injury and risk of death, and can cause tissue injury. Jci Insight. 2021;6:e147610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gondi KT, Kao A, Linard J, Austin BA, Everley MP, Fendler TJ, Khumri T, Lawhorn SL, Magalski A, Nassif ME, et al. Single‐center utilization of donor‐derived cell‐free DNA testing in the management of heart transplant patients. Clin Transplant. 2021;35:e14258. [DOI] [PubMed] [Google Scholar]
- 33.Henricksen EJ, Moayedi Y, Purewal S, Twiggs JV, Waddell K, Luikart H, Han J, Feng K, Wayda B, Lee R, et al. Combining donor derived cell free DNA and gene expression profiling for non‐invasive surveillance after heart transplantation. Clin Transplant. 2022;e14699. [DOI] [PubMed] [Google Scholar]
- 34.Gurtan AM, Sharp PA. The Role of miRNAs in Regulating Gene Expression Networks. J Mol Biol. 2013;425:3582–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creemers EE, Tijsen AJ, Pinto YM. Circulating MicroRNAs. Circ Res. 2012;110:483–495. [DOI] [PubMed] [Google Scholar]
- 36.Duong van Huyen JP, Tible M, Gay A, Guillemain R, Aubert O, Varnous S, Iserin F, Rouvier P, François A, Vernerey D, et al. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur Heart J. 2014;35:3194–3202. [DOI] [PubMed] [Google Scholar]
- 37.Halloran PF, Potena L, Duong van Huyen JP, Bruneval P, Leone O, Kim DH, Jouven X, Reeve J, Loupy A. Building a tissue-based molecular diagnostic system in heart transplant rejection: The heart Molecular Microscope Diagnostic (MMDx) System. J Hear Lung Transplant. 2017;36:1192–1200. [DOI] [PubMed] [Google Scholar]
- 38.Halloran PF, Madill-Thomsen K, Aliabadi-Zuckermann AZ, Cadeiras M, Crespo-Leiro MG, Depasquale EC, Deng M, Gökler J, Kim DH, Kobashigawa J, et al. Many heart transplant biopsies currently diagnosed as no rejection have mild molecular antibody-mediated rejection-related changes. J Hear Lung Transplant. 2022;41:334–344. [DOI] [PubMed] [Google Scholar]
- 39.Loupy A, Duong van Huyen JP, Hidalgo L, Reeve J, Racapé M, Aubert O, Venner JM, Falmuski K, Bories MC, Beuscart T, et al. Gene Expression Profiling for the Identification and Classification of Antibody-Mediated Heart Rejection. Circulation. 2017;135:917–935. [DOI] [PubMed] [Google Scholar]
- 40.Alam A, Zyl JV, Milligan GP, McKean SM, Patel R, Hall SA. Evolving the surveillance and workup of heart transplant rejection: A real‐world analysis of the Molecular Microscope Diagnostic System. Am J Transplant. 2022;22:2443–2450. [DOI] [PubMed] [Google Scholar]
- 41.DiFrancesco A, Fedrigo M, Santovito D, Natarelli L, Castellani C, Pascale FD, Toscano G, Fraiese A, Feltrin G, Benazzi E, et al. MicroRNA signatures in cardiac biopsies and detection of allograft rejection. J Hear Lung Transplant. 2018;37:1329–1340. [DOI] [PubMed] [Google Scholar]
- 42.Kirchhoff WC, Gradaus R, Stypmann J, Deng MC, Tian TDT, Scheld HH, Breithardt G, Brisse B. Vasoactive peptides during long-term follow-up of patients after cardiac transplantation. J Hear Lung Transplant. 2004;23:284–288. [DOI] [PubMed] [Google Scholar]
- 43.Arnau-Vives MA, Almenar L, Hervas I, Osa A, Martinez-Dolz L, Rueda J, Zorio E, Urbina LM-OD, Perez JL, Mateo A, et al. Predictive value of brain natriuretic peptide in the diagnosis of heart transplant rejection. J Hear Lung Transplant. 2004;23:850–856. [DOI] [PubMed] [Google Scholar]
- 44.Hammerer-Lercher A, Mair J, Antretter H, Ruttmann E, Poelzl G, Laufer G, Puschendorf B, Hangler H. B-Type Natriuretic Peptide as a Marker of Allograft Rejection After Heart Transplantation. J Hear Lung Transplant. 2005;24:1444.e5–1444.e8. [DOI] [PubMed] [Google Scholar]
- 45.Battes LC, Caliskan K, Rizopoulos D, Constantinescu AA, Robertus JL, Akkerhuis M, Manintveld OC, Boersma E, Kardys I. Repeated Measurements of NT-pro-B-Type Natriuretic Peptide, Troponin T or C-Reactive Protein Do Not Predict Future Allograft Rejection in Heart Transplant Recipients. Transplantation. 2015;99:580–585. [DOI] [PubMed] [Google Scholar]
- 46.Hill DA, Drazner MH, deLemos JA. Do established biomarkers such as B-type natriuretic peptide and troponin predict rejection?. Curr Opin Organ Tran. 2013;18:581–588. [DOI] [PubMed] [Google Scholar]
- 47.Fitzsimons SJ, Evans JDW, Rassl DM, Lee KK, Strachan FE, Parameshwar J, Mills NL, Pettit SJ. High-sensitivity Cardiac Troponin Is Not Associated With Acute Cellular Rejection After Heart Transplantation. Transplantation. 2022;106:1024–1030. [DOI] [PubMed] [Google Scholar]
- 48.Eisenberg MS, Chen HJ, Warshofsky MK, Sciacca RR, Wasserman HS, Schwartz A, Rabbani LE. Elevated Levels of Plasma C-Reactive Protein Are Associated With Decreased Graft Survival in Cardiac Transplant Recipients. Circulation. 2000;102:2100–2104. [DOI] [PubMed] [Google Scholar]
- 49.Starling RC, Stehlik J, Baran DA, Armstrong B, Stone JR, Ikle D, Morrison Y, Bridges ND, Putheti P, et al. Multicenter Analysis of Immune Biomarkers and Heart Transplant Outcomes: Results of the Clinical Trials in Organ Transplantation‐05 Study. Am J Transplant. 2016;16:121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stehlik J, Armstrong B, Baran DA, Bridges ND, Chandraker A, Gordon R, Marco TD, Givertz MM, Heroux A, Iklé D, et al. Early immune biomarkers and intermediate‐term outcomes after heart transplantation: Results of Clinical Trials in Organ Transplantation‐18. Am J Transplant. 2019;19:1518–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kennel PJ, Saha A, Maldonado DA, Givens R, Brunjes DL, Castillero E, Zhang X, Ji R, Yahi A, George I, et al. Serum exosomal protein profiling for the non-invasive detection of cardiac allograft rejection. J Hear Lung Transplant. 2018;37:409–417. [DOI] [PubMed] [Google Scholar]
- 52.Deshpande SR, Zangwill SD, Kindel SJ, Schroder JN, Bichell DP, Wigger MA, Richmond ME, Knecht KR, Pahl E, Gaglianello NA, et al. Relationship between donor fraction cell‐free DNA and clinical rejection in heart transplantation. Pediatr Transplant. 2022;26:e14264. [DOI] [PubMed] [Google Scholar]
- 53.Feingold B, Rose‐Felker K, West SC, Zinn MD, Berman P, Moninger A, Huston A, Stinner B, Xu Q, Zeevi A, Miller SA. Early findings after integration of donor‐derived cell‐free DNA into clinical care following pediatric heart transplantation. Pediatr Transplant. 2022;26:e14124. [DOI] [PubMed] [Google Scholar]
- 54.Das BB, Chan K, Winchester RW, Zakrzewski M, Niu J. Correlation of gene expression profiling score, cardiac hemodynamics and echocardiographic parameters in asymptomatic, rejection‐free pediatric heart transplant recipients. Pediatr Transplant. 2020;24:e13673. [DOI] [PubMed] [Google Scholar]
- 55.Dykes JC, Rosenthal DN, Bernstein D, McElhinney DB, Chrisant MRK, Daly KP, Ameduri RK, Knecht K, Richmond ME, Lin KY, et al. Clinical and hemodynamic characteristics of the pediatric failing Fontan. J Hear Lung Transplant. 2021;40:1529–1539. [DOI] [PubMed] [Google Scholar]
- 56.Frandsen EL, Albers EL. Management of the sensitized pediatric heart transplant patient. Transl Pediatrics. 2019;8:302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castleberry C, Zafar F, Thomas T, Khan MS, Bryant R, Chin C, Morales DLS, Lorts A. Allosensitization does not alter post‐transplant outcomes in pediatric patients bridged to transplant with a ventricular assist device. Pediatr Transplant. 2016;20:559–564. [DOI] [PubMed] [Google Scholar]
- 58.Maredia H, Bowring MG, Massie AB, Bae S, Kernodle A, Oyetunji S, Merlo C, Higgins RSD, Segev DL, Bush EL. Better Understanding the Disparity Associated With Black Race in Heart Transplant Outcomes. Circulation Hear Fail. 2019;14:e006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris AA, Cole RT, Veledar E, Bellam N, Laskar SR, Smith AL, Gebel HM, Bray RA, Butler J. Influence of Race/Ethnic Differences in Pre-Transplantation Panel Reactive Antibody on Outcomes in Heart Transplant Recipients. J Am Coll Cardiol. 2013;62:2308–2315. [DOI] [PubMed] [Google Scholar]
- 60.Khush KK, Pham MX, Teuteberg JJ, Kfoury AG, Deng MC, Kao A, Anderson AS, Cotts WG, Ewald GA, Baran DA, et al. Gene expression profiling to study racial differences after heart transplantation. J Hear Lung Transplant. 2015;34:970–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kransdorf EP, Kittleson MM, Patel JK, Pando MJ, Steidley DE, Kobashigawa JA. Calculated panel-reactive antibody predicts outcomes on the heart transplant waiting list. J Hear Lung Transplant. 2017;36:787–796. [DOI] [PubMed] [Google Scholar]
- 62.Chiu P, Schaffer JM, Oyer PE, Pham M, Banerjee D, Woo YJ, Ha R. Influence of durable mechanical circulatory support and allosensitization on mortality after heart transplantation. J Hear Lung Transplant. 2016;35:731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clarke B, Ducharme A, Giannetti N, Kim D, McDonald M, Pflugfelder P, Rajda M, Sénéchal M, Stadnick E, Toma M, et al. Multicenter evaluation of a national organ sharing policy for highly sensitized patients listed for heart transplantation in Canada. J Hear Lung Transplant. 2017;36:491–498. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, Kransdorf E, Levine R, Patel JK, Kobashigawa JA. HLA-DQ mismatches stimulate de novo donor specific antibodies in heart transplant recipients. Hum Immunol. 2020;81:330–336. [DOI] [PubMed] [Google Scholar]
- 65.Ansari D, Bućin D, Nilsson J. Human leukocyte antigen matching in heart transplantation: systematic review and meta‐analysis. Transplant Int. 2014;27:793–804. [DOI] [PubMed] [Google Scholar]
- 66.Nikolova AP, Kobashigawa JA. Donor-specific antibodies in heart transplantation: can we afford the price or is it too steep to pay? Curr Opin Organ Tran. 2020;25:555–562. [DOI] [PubMed] [Google Scholar]
- 67.Moayedi Y, Fan CS, Tinckam KJ, Ross HJ, McCaughan JA. De novo donor‐specific HLA antibodies in heart transplantation: Do transient de novo DSA confer the same risk as persistent de novo DSA? Clin Transplant. 2018;32:e13416. [DOI] [PubMed] [Google Scholar]
- 68.Torres MF, Pando MJ, Luo C, Luikart H, Valantine H, Khush K. The role of complement‐fixing donor‐specific antibodies identified by a C1q assay after heart transplantation. Clin Transplant. 2017;31:e13121. [DOI] [PubMed] [Google Scholar]
- 69.Wang M, Patel NJ, Zhang X, Kransdorf EP, Azarbal B, Kittleson MM, Czer LSC, Kobashigawa JA, Patel JK. The effects of donor‐specific antibody characteristics on cardiac allograft vasculopathy. Clin Transplant. 2021;35:e14483. [DOI] [PubMed] [Google Scholar]
- 70.Dipchand AI, Webber S, Mason K, Feingold B, Bentlejewski C, Mahle WT, Shaddy R, Canter C, Blume ED, Lamour J, et al. Incidence, characterization, and impact of newly detected donor‐specific anti‐HLA antibody in the first year after pediatric heart transplantation: A report from the CTOTC‐04 study. Am J Transplant. 2018;18:2163–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cole RT, Gandhi J, Bray RA, Gebel HM, Morris A, McCue A, Yin M, Laskar SR, Book W, Jokhadar M, et al. De novo DQ donor‐specific antibodies are associated with worse outcomes compared to non‐DQ de novo donor‐specific antibodies following heart transplantation. Clin Transplant. 2017;31:e12924. [DOI] [PubMed] [Google Scholar]
- 72.Irving CA, Carter V, Gennery AR, Parry G, Griselli M, Hasan A, Kirk CR. Effect of persistent versus transient donor-specific HLA antibodies on graft outcomes in pediatric cardiac transplantation. J Hear Lung Transplant. 2015;34:1310–1317. [DOI] [PubMed] [Google Scholar]
- 73.Reinsmoen NL, Lai C-H, Mirocha J, Cao K, Ong G, Naim M, Wang Q, Haas M, Rafiei M, Czer L, et al. Increased Negative Impact of Donor HLA-Specific Together With Non-HLA–Specific Antibodies on Graft Outcome. Transplantation. 2014;97:595–601. [DOI] [PubMed] [Google Scholar]
- 74.Chau VQ, Flattery M, Nicholson KS, Mcdougan F, Gupta G, Uber P, Priday AG, Desai K, Kimball PM, Shah KB. Elevated AT1R Antibody and Morbidity in Patients Bridged to Heart Transplant Using Continuous Flow Left Ventricular Assist Devices. J Card Fail. 2020;26:959–967. [DOI] [PubMed] [Google Scholar]
- 75.Nath DS, Tiriveedhi V, Basha HI, Phelan D, Moazami N, Ewald GA, Mohanakumar T. A Role for Antibodies to Human Leukocyte Antigens, Collagen-V, and K-α1-Tubulin in Antibody-Mediated Rejection and Cardiac Allograft Vasculopathy. Transplantation. 2011;91:1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suárez‐Álvarez B, López‐Vázquez A, Gonzalez MZ, Fdez‐Morera JL, Díaz‐Molina B, Blanco‐Gelaz MA, Pascual D, Martínez‐Borra J, Muro M, Álvarez‐López MR, et al. The Relationship of Anti‐MICA Antibodies and MICA Expression with Heart Allograft Rejection. Am J Transplant. 2007;7:1842–1848. [DOI] [PubMed] [Google Scholar]
- 77.Khush KK, Potena L, Cherikh WS, Chambers DC, Harhay MO, Hayes D, Hsich E, Sadavarte A, Singh TP, Zuckermann A, et al. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: 37th adult heart transplantation report—2020; focus on deceased donor characteristics. J Hear Lung Transplant. 2020;39:1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kowalski RJ, Post DR, Mannon RB, Sebastian A, Wright HI, Sigle G, Burdick J, Elmagd KA, Zeevi A, Lopez-Cepero M, et al. Assessing Relative Risks of Infection and Rejection: A Meta-analysis using an Immune Function Assay. Transplantation. 2006;82:663–668. [DOI] [PubMed] [Google Scholar]
- 79.Kobashigawa JA, Kiyosaki KK, Patel JK, Kittleson MM, Kubak BM, Davis SN, Kawano MA, Ardehali AA. Benefit of immune monitoring in heart transplant patients using ATP production in activated lymphocytes. J Hear Lung Transplant. 2010;29:504–508. [DOI] [PubMed] [Google Scholar]
- 80.Rossano JW, Denfield SW, Kim JJ, Price JF, Jefferies JL, Decker JA, Smith EO, Clunie SK, Towbin JA, Dreyer WJ. Assessment of the Cylex ImmuKnow Cell Function Assay in Pediatric Heart Transplant Patients. J Hear Lung Transplant. 2009;28:26–31. [DOI] [PubMed] [Google Scholar]
- 81.Tarkowski A, Czerkinsky C, Nilsson L-Å, Nygren H, Ouchterlony Ö. Solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of IgG rheumatoid factor-secreting cells. J Immunol Methods. 1984;72:451–459. [DOI] [PubMed] [Google Scholar]
- 82.VanBesouw NM, Zuijderwijk JM, Vaessen LMB, Balk AHMM, Maat APWM, Meide PH van der, Weimar W. The direct and indirect allogeneic presentation pathway during acute rejection after human cardiac transplantation. Clin Exp Immunol. 2005;141:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moayedi Y, Gomez CA, Fan CPS, Miller RJH, Bunce PE, Tremblay-Gravel M, Foroutan F, Manlhiot C, Yee J, Shullo MA, Khush KK, et al. Infectious complications after heart transplantation in patients screened with gene expression profiling. J Hear Lung Transplant. 2019;38:611–618. [DOI] [PubMed] [Google Scholar]
- 84.DeVlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, et al. Temporal Response of the Human Virome to Immunosuppression and Antiviral Therapy. Cell. 2013;155:1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kobashigawa J, Zuckermann A, Macdonald P, Leprince P, Esmailian F, Luu M, Mancini D, Patel J, Razi R, Reichenspurner H, et al. Report from a consensus conference on primary graft dysfunction after cardiac transplantation. J Hear Lung Transplant. 2014;33:327–340. [DOI] [PubMed] [Google Scholar]
- 86.Truby LK, Kwee LC, Agarwal R, Grass E, DeVore AD, Patel CB, Chen D, Schroder JN, Bowles D, Milano CA, et al. Proteomic profiling identifies CLEC4C expression as a novel biomarker of primary graft dysfunction after heart transplantation. J Hear Lung Transplant. 2021;40:1589–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giangreco NP, Lebreton G, Restaino S, Farr MJ, Zorn E, Colombo PC, Patel J, Levine R, Truby L, Soni RK, et al. Plasma kallikrein predicts primary graft dysfunction after heart transplant. J Hear Lung Transplant. 2021;40:1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chih S, Chong AY, Mielniczuk LM, Bhatt DL, Beanlands RSB. Allograft Vasculopathy: The Achilles’ Heel of Heart Transplantation. J Am Coll Cardiol. 2016;68:80–91. [DOI] [PubMed] [Google Scholar]
- 89.Das BB, Lacelle C, Zhang S, Gao A, Fixler D. Complement (C1q) Binding De Novo Donor-Specific Antibodies and Cardiac-Allograft Vasculopathy in Pediatric Heart Transplant Recipients. Transplantation. 2018;102:502–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bravo MJ-B, Pérez-Gómez L, Hernández-Pérez FJ, Arellano-Serrano C, Torres-Sanabria M, Gómez-Bueno M, Oteo-Domínguez JF, Mingo-Santos S, Segovia-Cubero J. Lack of Usefulness of Donor-Derived Cell-Free DNA as a Biomarker for Cardiac Allograft Vasculopathy: A Prospective Study. Frontiers Cardiovasc Medicine. 2022;9:856600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Almufleh A, Zhang L, Mielniczuk LM, Stadnick E, Davies RA, Du Q, Rayner K, Liu PP, Chih S. Biomarker discovery in cardiac allograft vasculopathy using targeted aptamer proteomics. Clin Transplant. 2020;34:e13765. [DOI] [PubMed] [Google Scholar]
- 92.Food and Drug Administration. Evidence-based treatment decisions in transplantation: the right dose & regimen for the right patient/individualized treatment. 2018. [Internet, accessed 2023 June 27]. Available from: https://www.fda.gov/media/126472/download
- 93.Food and Drug Administration. Guardant 360 CDx Premarket Approval Order. 2020. [Internet, access 2023 June 27]. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200010A.pdf
- 94.Lentine KL, Smith JM, Miller JM, Bradbrook K, Larkin L, Weiss S, Handarova DK, Temple K, Israni AK, Snyder JJ. OPTN/SRTR 2021 Annual Data Report: Kidney. Am J Transplant. 2023;23:S21–S120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jackson AM, Amato-Menker C, Bettinotti M. Cell-free DNA diagnostics in transplantation utilizing next generation sequencing. Hum Immunol. 2021;82:850–858. [DOI] [PubMed] [Google Scholar]
- 96.Mogayzel PJ, Naureckas ET, Robinson KA, Mueller G, Hadjiliadis D, Hoag JB, Lubsch L, Hazle L, Sabadosa K, Marshall B. Cystic Fibrosis Pulmonary Guidelines: Chronic Medications for Maintenance of Lung Health. Am J Resp Crit Care. 2013;187:680–689. [DOI] [PubMed] [Google Scholar]
- 97.Miller CL, Madsen JC. IL-6 Directed Therapy in Transplantation. Curr Transplant Reports. 2021;8:191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kivisild T Maternal ancestry and population history from whole mitochondrial genomes. Investigative Genetics. 2015;6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boyapati RK, Tamborska A, Dorward DA, Ho G-T. Advances in the understanding of mitochondrial DNA as a pathogenic factor in inflammatory diseases. F1000research. 2017;6:169. [DOI] [PMC free article] [PubMed] [Google Scholar]


