Abstract
Dietary studies are essential to unravel the functioning of ecosystems and ultimately to understand biodiversity. This task, which at first may seem simple, becomes especially complex in those cases of omnivorous species with highly variable diets. In this regard, the emergence of next-generation DNA sequencing methodologies represents a powerful tool to address the problem. Here we implement a high-throughput metabarcoding strategy based on the analysis of four molecular markers aimed at sequencing both mitochondrial (animal prey) and chloroplast (diet plants) genome fragments from fecal samples of two lizard species endemic to the Balearic Archipelago (Podarcis lilfordi and P. pityusensis) obtained through non-invasive methods. The results allowed for the characterization of their diets with a high degree of taxonomic detail and have contributed a large number of new trophic records. The reported diets are based mainly on the consumption of arthropods, mollusks and plants from a diversity of taxonomic orders, as well as carrion and marine subsidies. Our analyses also reveal inter- and intra-specific differences both in terms of seasonality and geographical distribution of the sampled lizard populations. These molecular findings provide new insights into the trophic interactions of these threatened endemic lizards in their unique and isolated ecosystems.
Keywords: Balearic archipelago, COI, metabarcoding, Podarcis lilfordi, Podarcis pityusensis, psbA-trnH, rbcL, trophic ecology
Assessing trophic interactions is essential in understanding ecosystem functionality (Kartzinel and Pringle 2015). Dietary studies provide key information about both the role of species in food webs and the importance of feeding resources for their demographic viability (Alonso et al. 2014), which in turn can influence ecosystem/species management decisions (Rogers et al. 2004; de Sousa et al. 2019). This knowledge is even more relevant under the current environmental changing scenario due to the anthropogenic perturbation of ecosystems with effects on distribution, abundance, and diversity of both consumers and diet resources (Thuiller et al. 2011). However, unveiling the details of the feeding habits of the species can be complex, particularly when dealing with species with highly diverse diets (De Barba et al. 2014). In this regard, omnivorous species feeding on a variety of animal and plant species pose the greater challenge to our ecological understanding of the consumer–resource interactions (De Barba et al. 2014; da Silva et al. 2019; Tercel et al. 2021). The taxonomic identification of the diversity of food items consumed by animals has so far relied on methodologies that are not suited for revealing the full spectrum of food types from mixed diet (De Barba et al. 2014), and in most cases are based on direct observations of feeding activity or microscopic examination of feces. The former approach is time-consuming and difficult to carry out, while the latter is tedious, requires extensive training, fails to identify a variable proportion of food fragments, and does not allow the detection of soft resources owing to their high digestibility (Valentini et al. 2009; Pompanon et al. 2012; Taberlet et al. 2012). Molecular approaches such as DNA barcoding have arisen as a promising tool to investigate trophic interactions (Symondson 2002; Jurado-Rivera et al. 2009) that in combination with high-throughput sequencing result in a powerful method to decipher a broad range of associations in complex food webs (Littlefair and Clare 2016). To date, only a few DNA metabarcoding studies implementing high-throughput sequencing have investigated the diet of omnivorous animals (reviewed by Tercel et al. 2021), with a bias towards mammal species (De Barba et al. 2014; Robeson et al. 2017; Bonin et al. 2020).
Here we implement a high-throughput sequencing metabarcoding approach based on non-invasive sampling to explore the trophic interactions in two omnivorous lizard species endemic to the Balearic archipelago: Podarcis lilfordi (IUCN Endangered; Pérez-Mellado and Martínez-Solano, 2009a) and P. pityusensis (IUCN Near Threatened; Pérez-Mellado and Martínez-Solano 2009b). Both species are sister phylogenetic lineages originated ca. 5.33 Ma ago (Rodríguez et al. 2013; but see Salvi et al. (2021) for an alternative dating) with a non-overlapping distribution across the archipelago, represented by allopatric populations inhabiting coastal islands and islets of Mallorca, Menorca and Cabrera (P. lilfordi), and the main islands of Ibiza and Formentera and many of their islets (P. pityusensis). Food resources in such isolated habitats are usually scarce and unpredictable, which could have driven the evolution from their ancestral insectivorous behavior (Pérez-Mellado and Corti 1993) to their present omnivorous feeding on a broad diversity of prey animals, plants, carrion, marine resources, and even juvenile conspecifics (Pérez-Cembranos et al. 2016). Available dietary studies of Balearic lizards are based on classical approaches such as direct observations in the field (Salvador 1986a), analysis of stomach contents from dead specimens (Pérez-Mellado 1989; Pérez-Mellado and Corti 1993), or visual inspection of fecal contents (Pérez-Cembranos et al. 2016; Santamaría et al. 2020). The taxonomic resolution of the food items reported by these studies rarely reaches detailed levels such as species, genus, or family. Our study aims to overcome the limitations of traditional approaches by taking advantage of the capabilities of advanced DNA sequencing methodologies to characterize in detail the trophic spectrum of these lizard species based on a sampling that encompasses their geographic distribution. Additionally, we intend to assess the existence of regional and temporal variation in terms of trophic resources consumption both at inter- and intra-specific levels.
Materials and Methods
Sampling
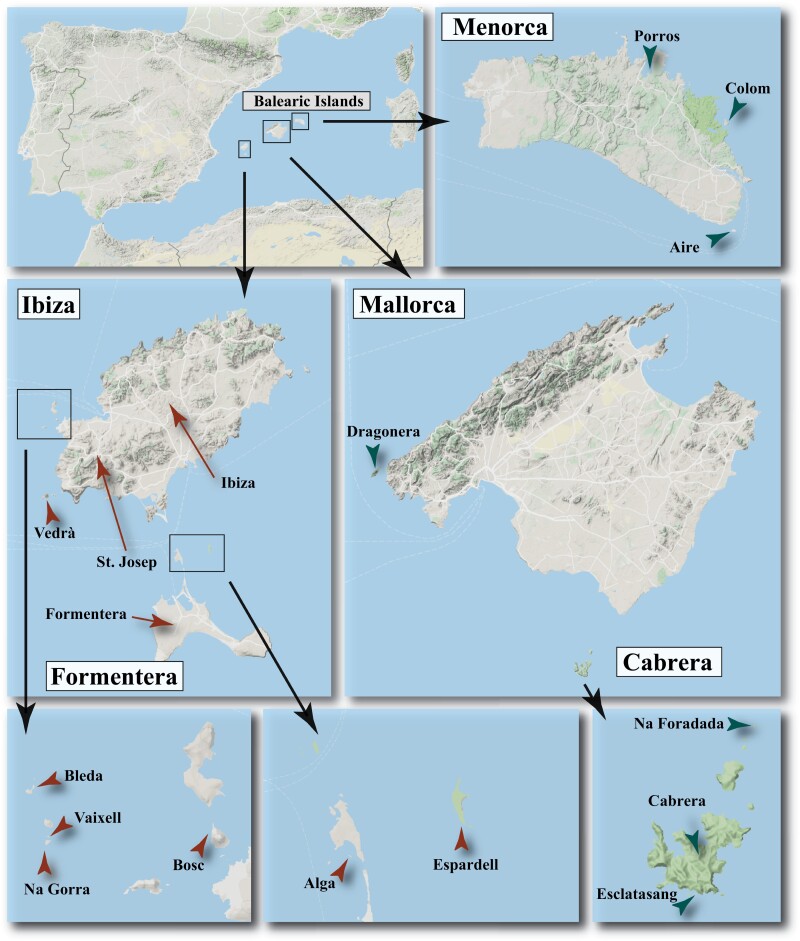
Fresh fecal samples from 242 lizard specimens (P. lilfordi: 140; P. pityusensis: 102) were collected between the spring of 2016 and the autumn of 2017 from 17 localities in the Balearic archipelago (Table 1 and Figure 1). Lizards were captured by lassoing at daytime and fresh fecal samples were directly obtained with a gentle abdominal massage and stored in absolute ethanol vials. Lizards were sexed based on their dimorphic attributes (Salvador 2009,Salvador (2009)) and immediately released at the same capture site. Samples were preserved at 4 °C in the field upon arrival to the laboratory and then stored at −20 °C until DNA extraction.
Table 1.
Sampling localities and associated metadata
| Species | Localization | Population/islet | Id | Acronym | Island area (m2) | Spring | Summer | Autumn | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Year | Female | Male | Year | Female | Male | Year | ||||||
| P. lilfordi | CABRERA | Cabrera | Cabrera | C | 1.7700.000 | – | – | – | 12 | 16 | 2017 | 6 | 7 | 2016 |
| Esclatasang | Esclatasang | E | 3.900 | – | – | – | 6 | 4 | 2017 | 4 | 4 | 2016 | ||
| Na Foradada | Na Foradada | Fo | 12.200 | – | – | – | 2 | 8 | 2017 | 2 | 5 | 2016 | ||
| MALLORCA | Dragonera | Dragonera | D | 2.880.000 | – | – | – | 9 | 8 | 2016 | 2 | 8 | 2017 | |
| MENORCA | Aire | Aire | A | 343.750 | 5 | 5 | 2017 | – | 1 | 2016 | – | – | – | |
| Colom | Colom | Co | 595.000 | 6 | 4 | 2017 | 3 | 3 | 2016 | – | – | – | ||
| Porros de Fornells | Porros | Pr | 1.600 | 4 | 6 | 2017 | – | – | – | – | – | – | ||
| P. pityusensis | FORMENTERA | Alga | Alga | Al | 11.250 | – | – | – | 2/5 | 3/5 | 2016/2017 | – | – | – |
| Espardell | Espardell | Es | 561.250 | 7 | 6 | 2017 | 5 | – | 2016 | – | – | – | ||
| Formentera | Formentera | F | 83.240.000 | 4 | 6 | 2017 | – | – | – | – | – | – | ||
| IBIZA | Bleda Plana | Bleda | Bp | 31.250 | – | – | – | – | 1 | 2016 | – | – | – | |
| Bosc de conillera | Bosc | Bc | 181.250 | 4 | 3 | 2016 | – | – | – | – | – | – | ||
| Na Gorra | Na Gorra | G | 15.625 | 5 | 5 | 2017 | 13 | 11 | 2016 | – | – | – | ||
| Sant Josep de sa Talaia | St. Josep | St | 527.600.000 | 1 | 2 | 2016 | – | – | – | – | – | – | ||
| Ses Salines | Ibiza | I | 527.600.000 | 2 | 2 | 2016 | – | – | – | – | – | – | ||
| Vaixell | Vaixell | Vx | 2.604 | – | – | – | 5 | 3 | 2017 | – | – | – | ||
| Vedrà | Vedrà | Ve | 6.250.000 | – | – | – | – | 2 | 2016 | – | – | – | ||
Figure 1.
Maps of the Balearic archipelago showing the location of the sampled Podarcis lilfordi (green arrows) and P. pityusensis populations (brown arrows). Maps were obtained with Google Maps (Map data 2020 Google) using the function ‘get_map’ in the package ‘ggmap’ version 3.0.0.902 in R version 3.6.3 (See online version for color figure).
Molecular analyses
Total DNA was extracted from individual samples using the Isolate Fecal DNA kit (Bioline, London, UK) following the manufacturer protocol and their concentrations were quantified using Qubit Fluorometric Quantitation (ThermoFisher, Foster City, CA, USA). For cost-effective reasons and given that our study seeks to describe the diet composition of each lizard population as a whole, samples from the same population/islet were pooled in equimolar concentrations by sex and collecting year. Samples were submitted to the Roy J. Carver Biotechnology Center (University of Illinois, USA) for amplification of selected markers in a microfluidic high-throughput multiplexed PCR platform (Fluidigm). For animal prey detection we selected two primer pairs: mlCOlintF/dgHCO2198 (5ʹ-GGWACWGGWTGAACWGTWTAYCCYCC-3ʹ/5ʹ-TAAACTTCAGGGTGACCAAARAAYCA-3ʹ; Meyer 2003; Leray et al. 2013) and ArtF11/ArtR17 (5ʹ-GGNKYNGGNACWGGATGAACWGTNTAYCCNCC-3ʹ/5ʹ-GGRTCAAAAAATGAWGTATTHARATTTCGRTCWGTTA-3ʹ; Mallott et al. 2015) targeting the mitochondrial cytochrome oxidase I gene (COI). For diet plant amplification we used two different primer pairs targeting chloroplast DNA regions: the RuBisCO large-subunit gene (rbcL) rbcLa_F/rbcLa_R (5ʹ-ATGTCACCACAAACAGAGACTAAAGC-3ʹ/5ʹ-GTAAAATCAAGTCCACCRCG-3ʹ; Levin et al. 2003; Kress et al. 2009) and the psbA-trnH intergenic spacer: psbA3_f/trnHf_05 (5ʹ-GTTATGCATGAACGTAATGCTC-3ʹ/5ʹ-CGCGCATGGTGGATTCACAATCC-3ʹ; Sang et al. 1997; Tate and Simpson 2003). CS1 and CS2 Fluidigm universal tags and barcode labels specific to each sample and Illumina adapters i5 and i7 were used and the resulting amplicons were validated on a Fragment Analyzer (Agilent) using the HS NGS kit (DNF-474-33). Sequencing was conducted on an Illumina MiSeq v2 platform yielding 2 × 250 paired-end reads.
Regarding animal prey analyses, a blocking oligo targeting a region within the mitochondrial COI gene was designed to minimize host amplification. We retrieved COI sequences from the most-consumed prey groups reported by previous studies on the diet of the Balearic lizards (Pérez-Cembranos et al. 2016) available at the Barcode of Life Data System (BOLD; http://www.boldsystems.org) and COI sequences from both P. lilfordi and P. pityusensis available at GenBank (Pérez-Cembranos et al. 2020). DNA sequences were aligned with MAFFT (Katoh and Standley 2013) and used for entropy analysis in BioEdit v. 7.0.5.2 (Hall 1999). Entropy plots revealed a suitable binding site encompassing the 3ʹ end of the mlCOlintF primer annealing site and a conserved sequence region in Podarcis. The blocking primer included a C3 spacer at the 3ʹ-end (5ʹ-AACTGTTTACCCCCCATTAGCCG[SpcC3]-3ʹ).
Sequence analyses and taxonomic assignment
For each marker, Micca version 1.7.2 (Albanese et al. 2015) was used for sequence merging (minimum overlapping region between R1 and R2 reads = 100 nt), trimming (removal of primer sequences and Illumina adaptors), filtering (based on the filterstats tool implemented in Micca; maximum error rate = 0.5; chimeric sequences removed) and OTU picking (cut-off value = 0.98). The taxonomic assignment of the inferred OTUs was conducted under a phylogenetic framework. As a first step the resulting DNA sequences (OTUs) were further grouped into 98% similarity clusters with Usearch version 10.0.240_i86osx32 (Edgar 2010). Usearch was also used to select a representative sequence (centroid) from each cluster that was subsequently used as a BLAST query to retrieve the 1000 most similar sequences from GenBank regardless their respective values of percentage identity. DNA matrices were built for each cluster by combining all dietary sequences in the cluster with the top 1000 hits from its centroid-based BLAST search. The resulting matrices were individually aligned with MAFFT (Katoh and Standley 2013) and used for maximum likelihood phylogenetic inference in IQTREE (Nguyen et al. 2015). Trees were explored in FigTree (Rambaut 2014) to establish the systematic position of diet sequences according to the highest taxonomic rank supported with a nodal bootstrap value ≥ 70% (Hillis and Bull 1993). Information from multiple markers targeting the same group of organisms (i.e., animals or plants) was merged following the multi-marker metabarcoding approach described in da Silva et al. (2019).
Molecular marker comparison
The performance of each of the molecular markers selected for our study was assessed based on the frequency of food items reported and identified at different taxonomic ranks (class, order, family, genus, species). Furthermore, the overlap in terms of taxonomic identifications at the above-mentioned taxonomic ranks between markers targeting the same group of dietary organisms (animals or plants) was explored using the R package VennDiagram (Chen and Boutros 2011).
Datasets
Previous research on the diet of Balearic lizards has demonstrated that food seasonality partially affects diet composition (Pérez-Mellado and Corti 1993; Pérez-Cembranos et al. 2016; Santamaría et al. 2020). In order to assess such seasonal effect and since our dataset did not include samples for all populations and for all seasons of the year due to the remote location of most of the sampled islets and the dependence on meteorological conditions to access them (Table 1), we based our analyses on two different subsets consisting of sampling locations with matching collecting seasons: Spring + Summer dataset (populations: Aire, Colom, Espardell and Na Gorra), and Summer + Autumn dataset (populations: Cabrera, Dragonera, Esclatasang and Na Foradada). In addition, we further explored geographical differences in terms of diet composition by comparing samples from the same collecting season (Spring, Summer and Autumn datasets) either combining the samples from the two lizard species or analyzing each species separately.
Diet analysis
The weighted percent of occurrence (wPOO) in our dataset was calculated following Deagle et al. (2019), a measure that considers the percentage of occurrence of each food item across the entire dataset weighted by the total count of prey items detected in each sample. This estimate has been shown to perform better than other indices when investigating taxa with a highly diverse trophic spectrum such as those of the omnivorous Balearic lizards (Andriollo et al. 2019). Niche width at both the species and populational levels was calculated using Levin’s index (Levins 1968), whereas trophic niche overlap was estimated based on Pianka’s index (Pianka 1973). We tested whether the extent of overlap was greater than expected by chance by comparing the observed values with a distribution of expected overlap values based on null model simulations using the RA3 algorithm (Lawlor 1980) implemented in the package EcoSimR (Gotelli et al. 2015). Differences were considered statistically significant when Pobs<exp < 0.05.
To assess whether there are significant differences between the surfaces of the islands and islets inhabited by the two species of lizards, we carried out an unpaired two-samples Wilcoxon test based on the surface data available on the website of the National Institute of Statistics of the Government of Spain (www.ine.es) (Table 1). In addition, we conducted Pearson correlation analyses between island area values (Table 1) and the number of different OTUs reported from their inhabiting lizard populations (Supplementary Table S1) to assess if inhabiting larger ranges imply a more diverse trophic niche by predators. Such correlation tests were performed both on the entire sampling and on each of the dataset separated by collecting season (Spring, Summer, Autumn).
Permutational Multivariate Analysis of Variance (PERMANOVA) based on Jaccard distance matrices and 999 random permutations was conducted in the vegan R package using the adonis function (Oksanen et al. 2007) to test for the effect of the sex, collecting season, and geographical location in resource consumption. The later variable was also explored at a broader scale by assigning each sampling locality (i.e., islet/population) to a main island district in the Balearic archipelago (Mallorca, Menorca, Cabrera, Eivissa or Formentera). Jaccard matrices were also projected through Principal Coordinates Analysis (PCoA) using the R package phyloseq (McMurdie and Holmes 2013) to visualize sample distances in terms of diet composition and their associate variables.
Finally, to delve into the objective of assessing the existence of variation in terms of trophic resources consumption at the inter-specific level we tested for differences in the number of diet OTUs per islet/population between lizard species for each of the orders that constitute at least 5% of the diet of either P. lilfordi or P. pityusensis (animals: Coleoptera, Diptera, Geophilomorpha, Hemiptera, Hymenoptera, Isopoda, Julida, Lepidoptera, Sarcoptiformes, Stylommatophora, Trombidiformes; plants: Asterales, Caryophyllales, Pinales, Pottiales). For each consumed order we first conducted a Fisher’s F-test homoscedasticity analysis to test for the homogeneity of the variances regarding the distribution of consumption frequencies between the populations of the two lizard species. When the variances of both samples were homogenous (Fisher’s F-test P > 0.05) we ran a Student’s two-sample t-test. By contrast, when heteroscedasticity was present (Fisher’s F-test P < 0.05) we conducted a Welch t-statistic. All analyses were carried out with the R package stats (R Core Team 2020).
Results
Sequence summary
After sequence trimming, merging, and filtering, the final dataset consisted of 989 727 (primer pair ArtF11/Art17), 544 458 (mlCIIintF/dgHCO2198), 750 494 (rbcLa_F/rbcLa_R) and 525 032 (psbA3_f/trnHf_05) high-quality paired reads. Sequence lengths averaged 236, 313, 228 and 224 bp for the arthropod-specific COI, universal COI, rbcL and psbA-trnH molecular markers, respectively.
Differential contribution of molecular markers to diet characterization
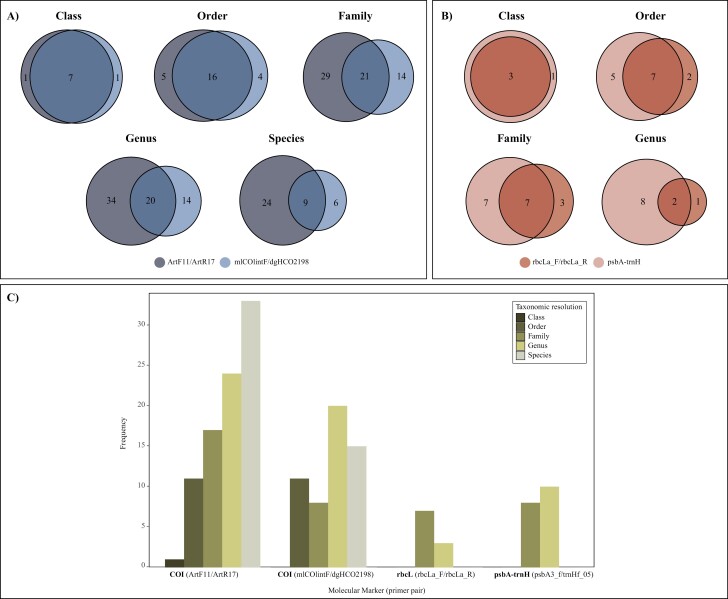
There were differences both in terms of the number of OTUs and the accuracy of the identification reported by each of the four primer pairs selected for the study. The arthropod-specific COI pair ArtF11/Art17 contributed the highest number of animal prey (86) with most of the identifications reaching the taxonomic level of species (38.4%), followed by genus (27.9%), family (19.8%), order (12.8%) and class (1.1%). Conversely, the universal COI mlCIIintF/dgHCO2198 pair yielded fewer OTUs (54) with lower taxonomic accuracy: species (27.8%), followed by genus (37%), family (14.8%), order (20.4%) and class (0%). Regarding plant-specific markers, the rbcL primer pair rbcLa_F/rbcLa_R retrieved 10 OTUs identified to genus (30%) and family (70%), whereas the oligo combination psbA3_f/trnHf_05 targeting the psbA-trnH intergenic spacer contributed more vegetal OTUs (18) with higher taxonomic resolution (genus: 55.6%, family: 44.4%).
Venn diagram analyses and frequency of food items showed the relative contribution of each primer pair in terms of OTU detection at different taxonomic ranks (Figure 2A–C). The two oligo combinations targeting animal prey produced similar results regarding low taxonomic levels (class and order), however the arthropod-specific COI primer pair ArtF11/Art17 outperformed the universal COI pair mlCIIintF/dgHCO2198 at high levels where the number of different OTUs contributed exclusively by the former was always higher than the intersection value or the number of OTUs provided by la latter. Such an effect is more noticeable as we go deeper into the taxonomic hierarchy (Figure 2A). The same pattern was observed when comparing the performance of the chloroplast-specific markers, where the psbA3_f/trnHf_05 pair targeting the psbA-trnH intergenic spacer provided the largest number of OTUs at all taxonomic levels and showed progressively more differences against rbcL marker the more detailed the taxonomic rank (Figure 2B).
Figure 2.
Venn diagrams showing the overlap in terms of taxonomic identification of prey animals (A) and diet plants (B) at different taxonomic ranks. (C) Distribution of finest OTU taxonomic identifications (i.e., taxonomic resolution) reported by each of the four molecular markers used in the study.
Diet composition
Balearic Podarcis lizards were found to exploit a wide range of food resources. Our metabarcoding approach reported 149 diet taxa of which 124 were prey animal OTUs that could be ascribed to 4 phyla, 9 classes, 25 orders, 64 families, 68 genera, and 39 species (Figure 2 and Supplementary Table S1). Phylum Arthropoda accounted for the highest number of different OTUs (113), followed by Mollusca (9), Nematoda (1) and Chordata (1). Within arthropods, insects resulted to be the most exploited class with 65 distinct OTUs mainly from orders Hemiptera (15), Lepidoptera (15), Hymenoptera (10), Coleoptera (9) and Diptera (7). Arthropod classes Arachnida (25) and Malacostraca (14) were also found to constitute an important fraction of diet of Balearic Podarcis lizards. Prey animals from phylum Mollusca were exclusively represented by gastropods from order Stylommatophora (9), while phyla Nematoda and Chordata accounted each for a single representative from orders Rhabditida and Pelecaniformes, respectively. On the other hand, our study revealed the presence in fecal samples of plant DNA from phylum Streptophyta (25 OTUs) represented by classes Bryopsida (3), Liliopsida (2), Pinopsida (3), and Magnoliopsida (17). Within the latter classes Caryophyllales (6), Apiales (2) and Asterales (3) showed the highest number of different OTUs.
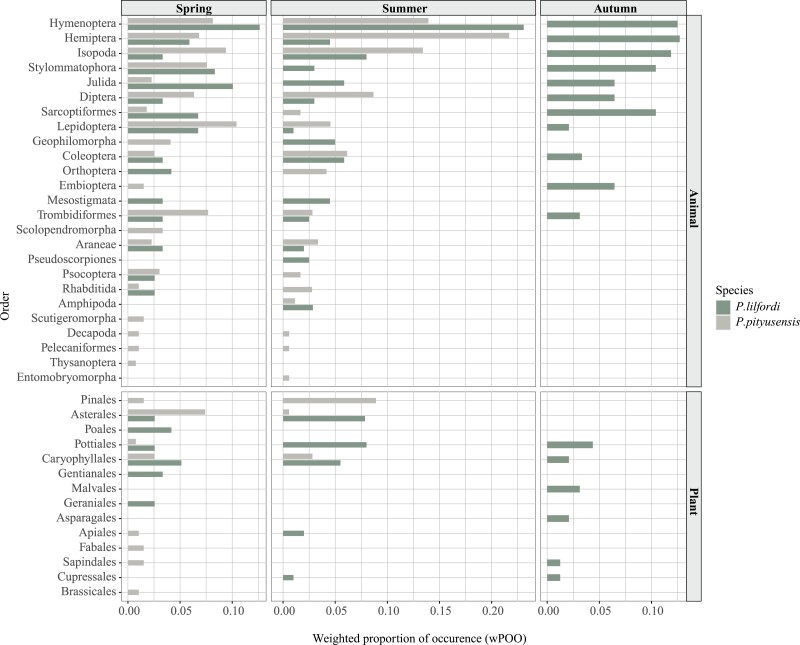
The relative use of trophic resources measured as weighted proportion of occurrence (wPOO) revealed a shared preference for certain food items regardless the lizard species or the collecting season (Figure 3). This was the case of prey animals from orders Hymenoptera, Hemiptera, Diptera, Lepidoptera, Coleoptera, Isopoda and Trombidiformes, and plants from orders Pottiales and Caryophillales. The fecal samples from P. lilfordi showed a consistent feeding throughout the seasons of the year on Hymenoptera, Hemiptera, Isopoda, Stylommatophora, Julida, Diptera, Lepidoptera, Coleoptera and Trombidiformes, as well as plants from orders Pottiales and Caryophyllales (Supplementary Table S1). On the other hand, P. pityusensis specimens were found to feed both in spring and summer on Hymenoptera, Hemiptera, Isopoda, Diptera, Sarcoptiformes, Lepidoptera, Coleoptera, Trombidiformes, Araneae, Psocoptera, Rhabditida, Decapoda and Pelecaniformes, and plant items from orders Asterales, Pinales and Caryophillales (Supplementary Table S1).
Figure 3.
Relative use of trophic resources measured as weighted proportion of occurrence (wPOO) at the order level per lizard species and season of the year.
Trophic niche analyses
Levin’s indexes for niche breadth showed similar values in P. lilfordi samples regardless the collecting season (27.53, 24.30 and 30.72 in spring, summer, and autumn, respectively). In contrast, values for P. pityusensis were comparatively high in spring (46.74) and low in summer (13.79) (autumn samples not available for P. pityusensis). Regarding Pianka’s index of niche overlap (Supplementary Table S2) all pairwise combinations analyzed provided moderate values with an average index of 0.57 (SD = 0.11, range = 0.33–0.93), although only 5 of such analyses were statistically significant when compared with a null model generated using the RA3 randomization algorithm: comparison between summer samples of P. lilfordi and P. pityusensis (Pianka’s index = 0.526; P-value = 0.036), and the pairwise analyses between summer samples of Alga and those from Bleda, Vaixell, Vedrà and Espardell (Pianka’s indexes = 0.933, 0.785, 0.714, and 0.876, respectively; P-value < 0.001 in all tests).
Island size and diet diversity
The unpaired two-samples Wilcoxon analysis to test for differences in terms of island area between the ranges inhabited by both lizard species yielded non-significant results (P-value = 0.4639). On the other hand, Pearson’s correlation analyses also rejected the hypothesis that lizard populations inhabiting larger islands show more diverse diets in terms of number of different OTUs consumed regardless of whether the analysis was based on the entire sampling (corr. coef = 0.013; P-value = 0.95) or in the datasets of each season separately (Spring: corr. coef = 0.0032, P-value = 0.99; Summer: corr. coef = 0.14, P-value = 0.68; Autumn: corr. coef = 0.84, P-value = 0.16).
Variation in the use of trophic resources
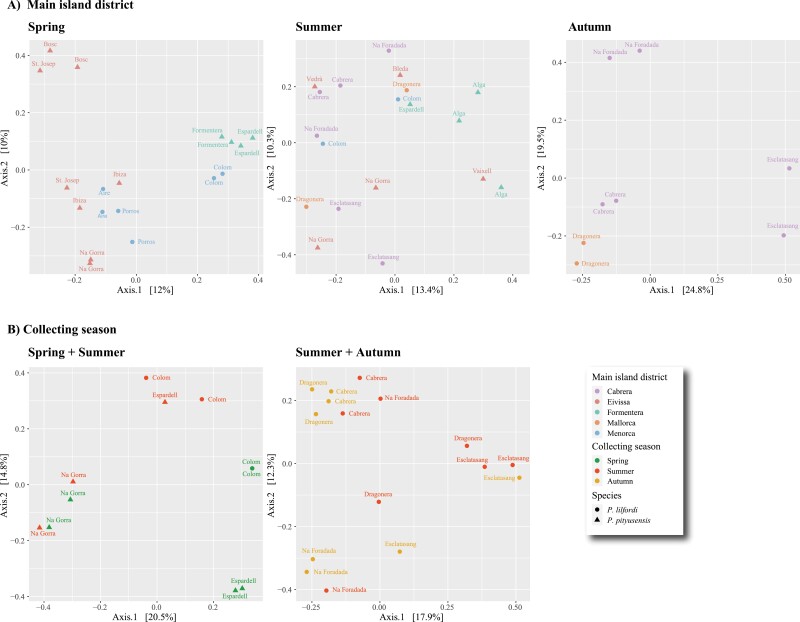
PERMANOVA analyses based on samples from the same collecting season found significant differences in terms of diet composition between P. lilfordi and P. pityusensis in both the Spring and the Summer datasets (R2 = 0.08, P-value ≤ 0.004; Table 2). The lizard species variable could not be analyzed in the Autumn dataset due to the lack of P. pityusensis samples for this season. These PERMANOVA tests also retrieved as statistically significant the geographical variables defined by both the collecting islet/population (R2 range = 0.35–0.42, P-value ≤ 0.007) and the main island district in the Balearic archipelago to which the sampling site is ascribed (R2 range = 0.11–0.19, P-value ≤ 0.035). Conversely, the sex of the individuals resulted in non-significant results in all tests. On the other hand, PERMANOVA analyses based on samples from populations with matching collecting seasons showed evidence for differences in the use of trophic resources between the two lizard species (R2 = 0.12, P-value ≤ 0.001; Table 2), among the source islets/populations (R2 range = 0.26–0.32, P-value ≤ 0.001) and in function of the collecting season variable (R2 range = 0.08–0.10, P-value ≤ 0.028) (Table 2). PCoA analyses based on Jaccard distance matrices were consistent with these findings (Figure 4).
Table 2.
Results of PERMANOVA analyses based on Jaccard distance matrices for each dataset
| Dataset | Variable | df | SumsOfSqs | MeanSqs | F.Model | R2 | Pr(>F) |
|---|---|---|---|---|---|---|---|
| Spring | Species | 1 | 0.64 | 0.64 | 1.81 | 0.08 | 0.001 |
| Sex | 1 | 0.33 | 0.33 | 0.94 | 0.04 | 0.596 | |
| Main island district | 1 | 0.86 | 0.86 | 2.42 | 0.11 | 0.001 | |
| Islet/population | 6 | 3.32 | 0.55 | 1.56 | 0.42 | 0.001 | |
| Residuals | 8 | 2.84 | 0.36 | NA | 0.36 | NA | |
| Total | 17 | 8.01 | NA | NA | 1.00 | NA | |
| Summer | Species | 1 | 0.68 | 0.68 | 1.95 | 0.08 | 0.004 |
| Sex | 1 | 0.37 | 0.37 | 1.06 | 0.04 | 0.372 | |
| Main island district | 3 | 1.63 | 0.54 | 1.56 | 0.18 | 0.001 | |
| Islet/population | 6 | 3.15 | 0.53 | 1.51 | 0.35 | 0.001 | |
| Residuals | 9 | 3.12 | 0.35 | NA | 0.35 | NA | |
| Total | 20 | 8.94 | NA | NA | 1 | NA | |
| Autumn* | Sex | 1 | 0.26 | 0.26 | 0.85 | 0.09 | 0.772 |
| Main island district | 1 | 0.55 | 0.55 | 1.76 | 0.19 | 0.035 | |
| Islet/population | 2 | 1.13 | 0.57 | 1.83 | 0.39 | 0.007 | |
| Residuals | 3 | 0.93 | 0.31 | NA | 0.32 | NA | |
| Total | 7 | 2.87 | NA | NA | 1 | NA | |
| Spring + Summer | Species | 1 | 0.64 | 0.64 | 1.90 | 0.12 | 0.001 |
| Islet/population | 2 | 1.45 | 0.73 | 2.15 | 0.26 | 0.001 | |
| Season | 1 | 0.53 | 0.53 | 1.57 | 0.10 | 0.018 | |
| Population:Season | 2 | 0.86 | 0.43 | 1.27 | 0.16 | 0.057 | |
| Residuals | 6 | 2.03 | 0.34 | NA | 0.37 | NA | |
| Total | 12 | 5.51 | NA | NA | 1 | NA | |
| Summer + Autumn | Islet/population | 3 | 2.01 | 0.67 | 1.96 | 0.32 | 0.001 |
| Season | 1 | 0.53 | 0.53 | 1.56 | 0.08 | 0.028 | |
| Population:Season | 3 | 1.09 | 0.36 | 1.06 | 0.17 | 0.318 | |
| Residuals | 8 | 2.74 | 0.34 | NA | 0.43 | NA | |
| Total | 15 | 6.37 | NA | NA | 1 | NA |
Significant P-values (< 0.05) are highlighted in bold.
*The autumn dataset contained only P. lilfordi samples and therefore was not tested for the “species” variable.
Figure 4.
Principal coordinate analyses based on the Jaccard distance matrices from the five analyzed datasets (see main text for details); (A) highlighting the main island district effect (dot colors) and lizard species (dot shapes); (B) highlighting the effect of season (dot colors) and lizard species (dot shapes) (See online version for color figure).
Fisher’s F-test, Student’s two-sample t-test and Welch t-statistic to check for statistical differences in the number of diet OTUs per islet/population between lizard species retrieved significant values regarding the consumption of Julida in the Spring dataset (Welch t-statistic P-value = 0.025; Table 3).
Table 3.
Results of the Fisher’s F-test to check for statistical differences in the number of diet OTUs per islet/population between lizard species for each of the orders that constitute at least 5% of the diet of either P. lilfordi or P. pityusensis
| Order | Spring | Summer | |||||
|---|---|---|---|---|---|---|---|
| var.test | Student’s t-test | Welch t-statistic | var.test | Student’s t-test | Welch t-statistic | ||
| Animals | Coleoptera | – | – | – | 0.111 | 0.724 | – |
| Diptera | 0.463 | 0.366 | – | 0.167 | 0.468 | – | |
| Geophilomorpha | – | – | – | 0.000 | – | 0.177 | |
| Hemiptera | 0.463 | 0.644 | – | 0.259 | 0.880 | – | |
| Hymenoptera | 0.821 | 0.351 | – | 0.259 | 0.057 | – | |
| Isopoda | 0.640 | 0.286 | – | 0.232 | 0.657 | – | |
| Julida | 0.000 | - | 0.025 | 0.000 | - | 0.070 | |
| Lepidoptera | 0.732 | 0.252 | – | – | – | – | |
| Sarcoptiformes | 0.726 | 0.407 | – | – | – | – | |
| Stylommatophora | 0.615 | 0.685 | – | – | – | – | |
| Trombidiformes | 0.821 | 0.351 | – | – | – | – | |
| Plants | Asterales | 0.821 | 0.351 | – | 0.532 | 0.166 | – |
| Caryophyllales | 0.1283 | 0.5527 | – | 0.432 | 0.824 | – | |
| Pinales | – | – | – | 0.000 | – | 0.175 | |
| Pottiales | – | – | – | 0.000 | – | 0.374 | |
Significant P-values (< 0.05) are highlighted in bold.
Discussion
The use of two different pairs of primers for each of the two main groups of organisms consumed by Balearic lizards (animals and plants) has allowed us to gather information on their trophic spectrum with a high level of taxonomic detail. Arthropods represent an important fraction of the diet of Balearic lizards (Pérez-Cembranos et al. 2016) and consistently the implementation of an arthropod-specific oligo pair has resulted in the recovery of a large number of OTUs compared to that reported by the COI universal primer pair. In addition, and despite the fact that the arthropod-specific primer pair targets a shorter COI region compared with that amplified by the universal mlCIIintF/dgHCO2198 pair (236 vs 313bp), it generated more precise taxonomic identifications that, in most cases, reached the species level. Since both primer pairs target overlapping regions of the mitochondrial COI gene, differences in terms of identification accuracy could be attributed to the greater completeness of DNA databases regarding arthropod sequences compared to other animal groups. However, and despite the greater contribution of the arthropod-specific primer pair, our results show that both COI primer pairs complement each other. This is supported by the fact that the mlCIIintF/dgHCO2198 combination detected 4 orders, 14 families, 14 genera and up to 6 species of prey animals that went unnoticed for the arthropod-specific oligos. Regarding the plant fraction of the diet, the implementation of two molecular markers widely used in the field of plant DNA Barcoding has allowed us to expand our knowledge about the herbivorous habits of the Balearic Podarcis with a level of taxonomic precision that in most cases reached the genus and/or family ranks. The highest identification efficiency of the psbA-trnH chloroplast intergenic spacer reported here is consistent with previous studies (Pang et al. 2012). In addition, psbA-trnH also reported a higher number of OTUs at all taxonomic levels compared with the rbcL marker, a fact that could be explained by the existence of priming sites within highly conserved flanking coding sequences and a highly variable non-coding region (Kress and Erickson 2007). In any case, and even though the relative performance of the rbcL marker is lower, our study has benefited from its inclusion as evidenced by the contribution of 1 plant class, 2 orders, 3 families and a genus to the final list of trophic resources consumed by the Balearic lizards.
The high-throughput sequencing metabarcoding approach implemented here supports the omnivorous and highly variable character of the diet of the Balearic Podarcis lizards. Our results show a trophic spectrum composed mainly of a diverse array of arthropod and mollusk animal prey, and vegetal tissues of terrestrial plants including mosses, monocots, and dicots. According to the weighted proportion of occurrence of diet items provided by our DNA-based approach, OTUs from orders Coleoptera, Diptera, Hemiptera, Hymenoptera, Isopoda, Lepidoptera and Trombidiformes, as well and for plant tissues from orders Caryophillales and Pottiales were consistently preyed upon by both Podarcis species across all sampled seasons. Although many of these main taxonomic groups had already been reported previously as part of the diet of the Balearic Podarcis based on macroscopic methods (Salvador 1986b; Pérez-Mellado 1989; Pérez-Mellado and Corti 1993; Traveset 1995; Pérez-Cembranos et al. 2016; Santamaría et al. 2020), the most important contribution of our study lies in the higher level of resolution of the taxonomic identifications and in the detection of new trophic records.
The resolution capacity of our DNA-based approach is clearly illustrated in the case of the ants, a trophic resource known to be particularly abundant in the diet of P. lilfordi (Pérez-Cembranos et al. 2016) and that we could ascribe to genera Crematogaster (including the species C. scutellaris), Lassius (L. lasioides), Messor (M. bouvieri), Pheidole, Tapinoma, and Tetramorium (T. semilaeve). Similarly, our DNA metabarcoding results also expands our knowledge on beetle consumption—the most important prey group in terms of ingested biomass (Pérez-Cembranos et al. 2016)—to families Anobiidae (genus Gastrallus), Scarabaeidae (Polyphylla), and Tenebrionidae including species from genera Asida and Blaps. These expansion of knowledge about the taxonomic details of the diet of Balearic lizards extends to the rest of the animal prey and plant orders detected, and it acquires special relevance in cases such as Lepidoptera, for which previous reports based on macroscopic methods have only detected poorly identified remains of larval structures (Santamaría et al. 2020) and our DNA-based analysis could ascribe to eight butterfly families including 11 genera and eight species. Such level of taxonomic resolution is highly dependent on the information available in DNA sequence databases, which currently prevent the complete taxonomic identification of many of the sequences (Wangensteen et al. 2018) (Supplementary Table S1). Thus, the identification rate of the diet DNA reads here reported will further increase as the taxonomic gaps in the databases are filled. A complementary approach in this regard would be the building of a local database of reference DNA sequences from the fauna and flora inhabiting the islets populated by the Balearic lizards. Unraveling the taxonomic details of trophic associations of P. lilfordi and P. pityusensis would not only provide valuable information about their biology but would also enable the compilation of high-quality ecological interaction networks necessary to understand ecosystem functionality (Cuff et al. 2022) and to design effective conservation strategies (Harvey et al. 2017).
In addition to a higher taxonomic resolution capacity compared to classical diet analysis, this first implementation of the metabarcoding approach in the Balearic lizards has reported new trophic records that substantially expand our knowledge about their omnivorous ecology. Our findings demonstrate the ability of the Balearic lizards to adapt to the scarce resources available in their isolated habitats as evidenced by the consumption of marine subsidies, vertebrate carrion, and mosses among other prey items from the orders Decapoda, Mesostigmata, Pelecaniformes, Psocoptera, Rhabditida, Sarcoptiformes, Scolopendromorpha, Scutigeromorpha, Geophilomorpha, Trombidiformes, and Thysanoptera. Prey decapods are represented in our results by the marbled rock crab Pachygrapsus marmoratus which is very abundant in the coastal environments of the Balearic archipelago (Crocetta et al. 2011) and the Argentine red shrimp Pleoticus muelleri which is a very common commercial shrimp in local markets whose remains could have reached the population where it has been detected (Na Gorra) through seabirds such as the Yellow-legged Gull, a very abundant seagull species in the archipelago known to feed on open-air dumps (Mas et al. 2015). The feeding on Phalacrocoracidae reported from P. pityusensis Alga and Na Gorra populations can be related to Phalacrocorax since it is the only genus of this bird family inhabiting the Balearic territory (Vicens 2012) and provides another interesting record of carrion consumption that previous studies could only report based on unidentified remains of feathers and/or hairs (Salvador 1986b). Our study also contributes the first record of feeding on psocopterans for both P. lilfordi and P. pityusensis, a prey group known to be consumed by other Podarcis species (Bombi et al. 2005). Also interesting is the case of the mites (Sarcoptiformes) detected in our analyses and represented by families and genera such as Bonomoia, Scutovertex and Tyrophagus typically associated with insects, vertebrate nests, plants, mosses, lichens, or fungi (Fan and Zhang 2007; Schäffer et al. 2010; Wirth 2016), some of them adapted to live in sun exposed rocks in salty environments (Scutovertex; Schäffer et al. 2010). This finding is consistent with previous reports of Sarcoptiformes DNA from fecal samples of lacertid lizards (Pereira et al. 2019). On the other hand, the consumption of undetermined myriapods by Balearic Podarcis demonstrated by macroscopic analyses of fecal contents (Salvador 1986b; Pérez-Mellado and Corti 1993) can be updated with a higher level of taxonomic precision based on our trophic inferences on Geophilomorpha, Scolopendromorpha (genus Cryptops), Scutigeromorpha (Scutigera coleoptrata) and Julida (Ommatoiulus). Thrip consumption (Thysanoptera) by P. pityusensis is also a new finding derived from our results that is consistent with previous reports of this insect group in the diet of other Podarcis species from the Iberian Peninsula (Escarré and Vericad 1981). New trophic records from order Trombidiformes include OTUs from families Eriophyidae (gall mites) and Tarsonemidae (thread-footed mites), both plant parasites that due to the combination of their small size and the degrading effect of the digestive processes could have gone unnoticed by classical macroscopic approaches. Our study also contributes new reports of trophic associations regarding plant groups that represent an important portion of the diversity of the diet of both P. lilfordi and P. pityusensis. They include the feeding on OTUs from two plant orders not previously reported (Brassicales and Malvales) as well as new associations with genera from orders already known to be consumed: Sonchus (Asterales), Beta and Limonium (Caryophillales), Daucus (Apiales), Asparagus (Asparagales), and Erodium (Geraniales). The feeding on mosses by the Balearic Podarcis lizards is also an unprecedented finding. These new reports represent a substantial advance in understanding the ecological role of both lizard species, whose trophic spectra are probably much broader if we consider that DNA-based inferences only provide information regarding feeding events that occurred a few hours before fecal sampling (Thuo et al. 2019). In this regard, an experimental design based on frequent long-term sampling would provide a more complete view of the highly diverse diets of these Podarcis species.
Accidental and/or secondary ingestion can be error sources when investigating omnivorous diets (Tercel et al. 2021). This could be the case of the inferred tropic associations with gymnosperms since chloroplast genomes are paternally inherited in most conifer taxa including those from orders Cupressales and Pinales reported by our study (Adams 2019). Therefore, the accumulation of pollen grains from these plant groups on the food items of the lizards could be enough for the specific PCR primers of the chloroplast genome that we have used in this study (molecular markers psbA-trnH and rbcL) to amplify them. Similarly, the finding of DNA remains from Rhabditida in the analyzed Podarcis fecal samples could not necessarily be indicative of a trophic relationship since this nematode group includes zooparasitic helminths of lizards (Moravec 2010), and therefore further research would be needed to clarify their presence in the analyzed scats.
PERMANOVA analyses based both on samples from the same collecting season (i.e., spring and summer datasets) and on samples from populations with matching collecting seasons (i.e., Spring + Summer and Summer + Autumn datasets) consistently yielded significant differences in terms of diet composition between P. lilfordi and P. pityusensis. In addition, our niche overlap analyses significantly reported a moderate resemblance in resource usage between summer samples from both lizard species (Pianka’s index 0.526), meaning that an important fraction of the diet of each species is exclusive. As evidenced by our results, such differences are not related with the size of the islands inhabited by both lizard species and could rather be explained by their evolutionary separation ca. 5.3 Ma. (Brown et al. 2008) (ca. 3.5 Ma. sensuSalvi et al. 2021) and their a non-overlapping segregated distribution in the Balearic archipelago each inhabiting well-differentiated biogeographic districts (Rivas-Martínez et al. 2017). Indeed, there exist differences in terms of flora composition between the eastern islets where P. lilfordi inhabits (Mallorca, Menorca and Cabrera district) and those located in the western part of the archipelago populated by P. pityusensis (Ibiza and Formentera district) (Rita and Bibiloni 2013). Such inter-specific dietary differences agree with previous reports based on macroscopic analyses of stomach and fecal pellet contents (Pérez-Mellado and Corti 1993). Niche breadth of P. lilfordi showed similar Levin’s index values across seasons while disparate values were observed for P. pityusensis corresponding to a hight trophic spectrum in spring and a reduced niche width in summer. These observations are in line with the analyses of seasonal effect on the diet of the Balearic Podarcis conducted by Perez-Mellado and Corti (1993) that concluded that dietary diversity is lower in summer than in spring. This could be related with the warm and dry summer conditions that characterize the Mediterranean climate in contrast with the favorable spring conditions for both flora and fauna. The seasonal dietary variation is also supported by our PERMANOVA analyses based on samples from populations with matching collecting seasons.
In addition to seasonality, the geographic distribution of lizard populations also appears to be a determining factor in explaining dietary variation. Main Balearic island district and islet/population variables were found to significantly influence the observed pattern of resources consumption, which could be related to the similarity in terms of environmental conditions and available trophic resources in nearby locations. This view is supported by our niche overlap analyses based on Pianka’s index where significantly similar resource utilization spectra were derived from pairwise diet comparisons among populations from geographically close islets in the surroundings of Ibiza and Formentera (Alga, Bleda, Espardell, Vaixell and Vedrà).
In conclusion, the metabarcoding strategy used here and based on non-invasive sampling has made it possible to characterize with a high degree of taxonomic detail the highly variable diets of the endemic Podarcis lizards of the Balearic archipelago, including new trophic records. The use of two different molecular markers for each of the main groups of organisms that make up the diet of lizards (animals and plants) has proven to be an effective strategy. The diet compositions of P. lilfordi and P. pityusensis differ inter- and intra-specifically both at the seasonal and the geographical level and are marked by the consumption of a high diversity of arthropods, mollusks, and plants, that also include feeding on carrion and on marine subsidies. Our molecular results complement those from previous studies based on macroscopic analyses of diet remains and provide a comprehensive view of the diversity of ecological interactions that characterize the populations of these threatened endemic lizards in their unique isolated ecosystems.
Supplementary Material
Acknowledgments
This study was possible thanks to the project: CGL2015-68139-C2-1-P, “Dinámica de la variación genética y respuesta adaptativa en las Podarcis insulares” financed by of the Ministerio Español de Economia y competitividad and European Regional Development Fund (ERDF). IA supported by FPI/2006/2017 and research funds from the Conselleria d’Educació, Cultura i Universitats (Govern de les Illes Balears, Spain), co-financed by the ERDF.
Contributor Information
Iris Alemany, Deptartment of Biology, Universitat de les Illes Balears, Ctra. Valldemossa km 7’5, Palma de Mallorca, 07122, Balearic Islands, Spain.
Ana Pérez-Cembranos, Department of Animal Biology, Universidad de Salamanca, Campus Miguel de Unamuno s/n, 37007, Salamanca, Spain.
Valentín Pérez-Mellado, Department of Animal Biology, Universidad de Salamanca, Campus Miguel de Unamuno s/n, 37007, Salamanca, Spain.
José Aurelio Castro, Deptartment of Biology, Universitat de les Illes Balears, Ctra. Valldemossa km 7’5, Palma de Mallorca, 07122, Balearic Islands, Spain.
Antònia Picornell, Deptartment of Biology, Universitat de les Illes Balears, Ctra. Valldemossa km 7’5, Palma de Mallorca, 07122, Balearic Islands, Spain.
Cori Ramon, Deptartment of Biology, Universitat de les Illes Balears, Ctra. Valldemossa km 7’5, Palma de Mallorca, 07122, Balearic Islands, Spain.
José A Jurado-Rivera, Deptartment of Biology, Universitat de les Illes Balears, Ctra. Valldemossa km 7’5, Palma de Mallorca, 07122, Balearic Islands, Spain.
Funding
This study was possible thanks to the project: CGL2015-68139-C2-1-P, “Dinámica de la variación genética y respuesta adaptativa en las Podarcis insulares” financed by of the Ministerio Español de Economia y competitividad and European Regional Development Fund (ERDF). IA supported by FPI CAIB 2017 and research funds from the Conselleria d’Educació, Cultura i Universitats (Govern de les Illes Balears, Spain), co-financed by the ERDF.
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
Authors’ Contributions
IA, JAJ-R, CR and VPM designed the research. IA, VPM and AP-C collected the fecal samples. IA and JAJ-R performed the molecular work, performed bioinformatics data analyses and generated the figures. CR, VPM, JAC, AP and JAJ-R obtained the funding. JAJ-R, IA and CR drafted a first version of the manuscript with contributions of other authors to the final version.
Data accessibility
Raw sequences are available in the Sequence Read Archive (SRA) database at NCBI under BioProject ID PRJNA703933.
References
- Adams RP, 2019. Inheritance of chloroplasts and mitochondria in conifers: A review of paternal, maternal, leakage and facultative inheri-tance. Phytologia 101:134–138. [Google Scholar]
- Albanese D, Fontana P, De Filippo C, Cavalieri D, Donati C, 2015. MICCA: A complete and accurate software for taxonomic profiling of metagenomic data. Sci Rep 5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso H, Granadeiro JP, Waap S, Xavier J, Symondson WOet al. , 2014. An holistic ecological analysis of the diet of Cory’s shearwaters using prey morphological characters and DNA barcoding. Mol Ecol 23:3719–3733. [DOI] [PubMed] [Google Scholar]
- Andriollo T, Landry B, Guibert B, Pastore M, Baumgart P, 2019. Nouveaux ajouts à la liste des Lépidoptères du canton de Genève. Entomo Helv 12:9–28. [Google Scholar]
- de Barba M, Miquel C, Boyer F, Mercier C, Rioux Det al. , 2014. DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: Application to omnivorous diet. Mol Ecol Resour 14:306–323. [DOI] [PubMed] [Google Scholar]
- Bombi P, Scalera R, Bologna M, Vignoli L, 2005. Food habits of Podarcis filfolensis (Reptilia, Lacertidae) on a small Mediterranean island during the dry season. Amphib-Reptilia 26:412–417. [Google Scholar]
- Bonin M, Dussault C, Taillon J, Lecomte N, Côté SD, 2020. Combining stable isotopes, morphological, and molecular analyses to reconstruct the diet of free-ranging consumers. Ecol Evol 10:6664–6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RP, Terrasa B, Pérez-Mellado V, Castro JA, Hoskisson PAet al. , 2008. Bayesian estimation of post-Messinian divergence times in Balearic Island lizards. Mol Phylogenet Evol 48:350–358. [DOI] [PubMed] [Google Scholar]
- Chen H, Boutros PC, 2011. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinf 12:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocetta F, Mifsud S, Paolini P, Piscopo J, Schembri PJ, 2011. New records of the genus Pachygrapsus (Crustacea: Decapoda) from the central Mediterranean Sea with a review of its Mediterranean zoogeography. Mediterr Mar Sci 12:75–94. [Google Scholar]
- Cuff JP, Windsor FM, Tercel MP, Kitson JJ, Evans DM, 2022. Overcoming the pitfalls of merging dietary metabarcoding into ecological networks. Methods Ecol Evol 13:545–559. [Google Scholar]
- Deagle BE, Thomas AC, McInnes JC, Clarke LJ, Vesterinen EJet al. , 2019. Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data? Mol Ecol 28:391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Escarré A, Vericad JR, 1981. Fauna alicantina. I. Saurios y ofidios. Cuadernos de la fauna alicantina. Publicaciones del Inst Estud Alicant Ser II 15:1–101. [Google Scholar]
- Fan Q, Zhang ZQ, 2007. Tyrophagus (Acari: Astigmata: Acaridae). Fauna New Zeal 56:291. Lincoln: Manaaki Whenua Press. [Google Scholar]
- Gotelli JN, Hart ME, Ellison MA, 2015. EcoSimR: Null Model Analysis for Ecological Data. R package version 0.1.0 [cited 2021 July]. Available from: http://github.com/gotellilab/EcoSimR.
- Hall TA, 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- Harvey E, Gounand I, Ward CL, Altermatt F, 2017. Bridging ecology and conservation: From ecological networks to ecosystem function. J Appl Ecol 54:371–379. [Google Scholar]
- Hillis DM, Bull JJ, 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192. [Google Scholar]
- Jurado-Rivera JA, Vogler AP, Reid CAM, Petitpierre E, Gómez-Zurita J, 2009. DNA barcoding insect-host plant associations. Proc R Soc B Biol Sci 276:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartzinel TR, Pringle RM, 2015. Molecular detection of invertebrate prey in vertebrate diets: Trophic ecology of Caribbean island lizards. Mol Ecol Resour 15:903–914. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM, 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Erickson DL, 2007. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2(6):e508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez Ret al. , 2009. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc Natl Acad Sci USA 106:18621–18626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor LR, 1980. Structure and stability in natural and randomly constructed competitive communities. Am Nat 116:394–408. [Google Scholar]
- Leray M, Yang JY, Meyer CP, Mills SC, Agudelo Net al. , 2013. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front Zool 10:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin RA, Wagner WL, Hoch PC, Nepokroeff M, Pires JCet al. , 2003. Family-level relationships of onagraceae based on chloroplast rbcL and ndhF data. Am J Bot 90:107–115. [DOI] [PubMed] [Google Scholar]
- Levins R, 1968. Evolution in Changing Environments: Some Theoretical Explorations. Princeton: Princeton University Press. [Google Scholar]
- Littlefair JE, Clare EL, 2016. Barcoding the food chain: From Sanger to high-throughput sequencing. Genome 59:946–958. [DOI] [PubMed] [Google Scholar]
- Mallott EK, Malhi RS, Garber PA, 2015. Brief communication: High-throughput sequencing of fecal DNA to identify insects consumed by wild Weddell’s saddleback tamarins (Saguinus weddelli, Cebidae, Primates) in Bolivia. Am J Phys Anthropol 156:474–481. [DOI] [PubMed] [Google Scholar]
- Mas R, Cardona E, de Pablo F, Mayol J, 2015. La població reproductora de gavina de peus grocs Larus michahellis a les Illes Balears. Anuari Ornitològic de les Illes Balears. Anu Ornitològic les Balear 30:1–16. [Google Scholar]
- McMurdie PJ, Holmes S, 2013. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer CP, 2003. Molecular systematics of cowries (Gastropoda: Cypraeidae) and diversification patterns in the tropics. Biol J Linn Soc 79:401–459. [Google Scholar]
- Moravec F, 2010. Rhabdias lacertae n. sp. (Nematoda: Rhabdiasidae), the first rhabdiasid species parasitising lizards in Europe. Syst Parasitol 77:23–27. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ, 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHHet al. , 2007. The vegan package. Community Ecol Packag 10:631–637. [Google Scholar]
- Pang X, Liu C, Shi L, Liu R, Liang Det al. , 2012. Utility of the trnH–psbA intergenic spacer region and its combinations as plant dna barcodes: A meta-analysis. PLoS ONE 7:e48833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A, Xavier R, Perera A, Salvi D, Harris DJ, 2019. DNA metabarcoding to assess diet partitioning and feeding strategies in generalist vertebrate predators: A case study on three syntopic lacertid lizards from Morocco. Biol J Linn Soc 127:800–809. [Google Scholar]
- Pérez-Cembranos A, León A, Pérez-Mellado V, 2016. Omnivory of an insular lizard: Sources of variation in the diet of Podarcis lilfordi (Squamata, Lacertidae). PLoS ONE 11:e0148947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cembranos A, Pérez-Mellado V, Alemany I, Bassitta M, Terrasa Bet al. , 2020. Morphological and genetic diversity of the Balearic lizard Podarcis lilfordi (Günther, 1874): Is it relevant to its conservation? Divers Distrib 26:1122–1141. [Google Scholar]
- Pérez-Mellado V, 1989. Estudio ecológico de la Lagartija Balear Podarcis lilfordi (Günther, 1874) en Menorca. Rev Menorca 80:455–511. [Google Scholar]
- Pérez-Mellado V, Corti C, 1993. Dietary adaptations and herbivory in lacertid lizards of the genus Podarcis from western Mediterranean islands (Reptilia: Sauria). Bonner Zool Beiträge 44:193–220. [Google Scholar]
- Pérez-Mellado V, Martínez-Solano I, 2009a. Podarcis lilfordi. IUCN Red List Threatened Species 2009:e.T17795A7481971 [cited 2022 September 11]. Available from: 10.2305/IUCN.UK.2009.RLTS.T17795A7481971.en. [DOI] [Google Scholar]
- Pérez-Mellado V, Martínez-Solano I, 2009b. Podarcis pityusensis. IUCN Red List Threatened Species 2009: e.T17800A7482971 [cited 2022 September 11]. Available from: 10.2305/IUCN.UK.2009.RLTS.T17800A7482971.en. [DOI] [Google Scholar]
- Pianka ER, 1973. The structure of lizard communities. Annu Rev Ecol Syst 4:53–74. [Google Scholar]
- Pompanon F, Deagle BE, Symondson WOC, Brown DS, Jarman SNet al. , 2012. Who is eating what: Diet assessment using next generation sequencing. Mol Ecol 21:1931–1950. [DOI] [PubMed] [Google Scholar]
- R Core Team, 2020. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; [cited 2021 July]. Available from: https://www.R-project.org/. [Google Scholar]
- Rambaut A, 2014. FigTree v. 1.4.4. Edinburgh: Institute of Evolutionary Biology, University of Edinburgh; [cited 2021 July]. http://tree.bio.ed.ac.uk/software/figtree/. [Google Scholar]
- Rita J, Bibiloni G, 2013. The flora of the islets of the Balearic Islands. In islands and plants: Preservation and understanding of flora on Mediterranean Islands. In: Cardona E, Estaún I, Comas M, Fraga P, editors. 2nd Botanical Conference in Menorca: Proceedings and Abstracts. Menorca, Spain: Institut Menorquí d’Estudis, 309–322. [Google Scholar]
- Rivas-Martínez S, Penas A, Díaz González TE, Cantó P, Río S, Costa JCet al. , 2017. Biogeographic units of the Iberian Peninsula and Baelaric Islands to district level. a concise synopsis. In: Loidi J, editor. The Vegetation of the Iberian Peninsula. Vol. 12. Plant Veg. Cham: Springer. [Google Scholar]
- Robeson MS 2nd, Khanipov K, Golovko G, Wisely SM, White MDet al. , 2017. Assessing the utility of metabarcoding for diet analyses of the omnivorous wild pig Sus scrofa. Ecol Evol 8:185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez V, Brown RP, Terrasa B, Pérez-Mellado V, Castro JAet al. , 2013. Multilocus genetic diversity and historical biogeography of the endemic wall lizard from Ibiza and Formentera Podarcis pityusensis (Squamata: Lacertidae). Mol Ecol 22:4829–4841. [DOI] [PubMed] [Google Scholar]
- Rogers ME, Abernethy K, Bermejo M, Cipolletta C, Doran Det al. , 2004. Western gorilla diet: A synthesis from six sites. Am J Primatol 64:173–192. [DOI] [PubMed] [Google Scholar]
- Salvador A, 1986a. Podarcis lilfordi (Günther, 1874). Balearen Eidechse. In: Böhme W, editor. Handbuch der Amphibien und Reptilien Europas, Echsen III (Podarcis). Wiesbaden: Aula verlag, 83–110. [Google Scholar]
- Salvador A, 1986b. Podarcis pityusensis (Boscá, 1883)—Pityusen-Eidechse. En: Böhme W, editor. Handbuch der Reptilien und Amphibien Europas. Band 2/II. Echsen (Sauria) III (Lacertidae III: Podarcis). Wiesbaden: Aula verlag, 231–253. [Google Scholar]
- Salvador A, 2009. Lagartija balear–Podarcis lilfordi (Günther, 1874). Enciclopedia Virtual de los Vertebrados Españoles. Madrid, Spain: Museo Nacional de Ciencias Naturales. http://www.vertebradosibericos.org/. [Google Scholar]
- Salvi D, Pinho C, Mendes J, Harris DJ, 2021. Fossil-calibrated time tree of Podarcis wall lizards provides limited support for biogeographic calibration models. Mol Phylogenet Evol 161:107169. [DOI] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF, 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am J Bot 84:1120–1136. [PubMed] [Google Scholar]
- Santamaría S, Enoksen CA, Olesen JM, Al E, 2020. Diet composition of the lizard Podarcis lilfordi (Lacertidae) on 2 small islands: An individual-resource network approach. Curr Zool 66:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer S, Pfingstl T, Koblmüller S, Winkler KA, Sturmbauer Cet al. , 2010. Phylogenetic analysis of European Scutovertex mites (Acari, Oribatida, Scutoverticidae) reveals paraphyly and cryptic diversity: A molecular genetic and morphological approach. Mol Phylogenet Evol 55:677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva LP, Mata VA, Lopes PB, Pereira P, Jarman SNet al. , 2019. Advancing the integration of multi-marker metabarcoding data in dietary analysis of trophic generalists. Mol Ecol Resour 19:1420–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa LL, Silva SM, Xavier R, 2019. DNA metabarcoding in diet studies: Unveiling ecological aspects in aquatic and terrestrial ecosystems. Environ DNA 1:199–214. [Google Scholar]
- Symondson WOC, 2002. Molecular identification of prey in predator diets. Mol Ecol 11:627–641. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E, 2012. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol 21:2045–2050. [DOI] [PubMed] [Google Scholar]
- Tate JA, Simpson BB, 2003. Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst Bot 28:723–737. [Google Scholar]
- Tercel MP, Symondson WO, Cuff JP, 2021. The problem of omnivory: A synthesis on omnivory and DNA metabarcoding. Mol Ecol 30:2199–2206. [DOI] [PubMed] [Google Scholar]
- Thuiller W, Lavergne S, Roquet C, Boulangeat I, Lafourcade Bet al. , 2011. Consequences of climate change on the tree of life in Europe. Nature 470:531–534. [DOI] [PubMed] [Google Scholar]
- Thuo D, Furlan E, Broekhuis F, Kamau J, Macdonald Ket al. , 2019. Food from faeces: Evaluating the efficacy of scat DNA metabarcoding in dietary analyses. PLoS ONE 14:e0225805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traveset A, 1995. Seed dispersal of Cneorum tricoccon L. (Cneoraceae) by lizards and mammals in the Balearic Islands. Acta Oecol 16:171–178. [Google Scholar]
- Valentini A, Miquel C, Nawaz MA, Bellemain E, Coissac Eet al. , 2009. New perspectives in diet analysis based on DNA barcoding and parallel pyrosequencing: The trnL approach. Mol Ecol Resour 9:51–60. [DOI] [PubMed] [Google Scholar]
- Vicens P, 2012. Primer cens hivernal de corb marí gros Phalacrocorax carbo a les zones de colgada a Balears. Anu Ornitològic les Balear 27:15–21. [Google Scholar]
- Wangensteen OS, Palacín C, Guardiola M, Turon X, 2018. DNA metabarcoding of littoral hard-bottom communities: High diversity and database gaps revealed by two molecular markers. PeerJ 6:e4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth SF, 2016. Description of developmental instars of Bonomoia sibirica sp. n.(Astigmata: Histiostomatidae) with ecological observations and phylogenetic conclusions. Acarina 24:97–111. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences are available in the Sequence Read Archive (SRA) database at NCBI under BioProject ID PRJNA703933.